Abstract
The 5′ cap and 3′ poly(A) tail of eukaryotic mRNAs cooperate to synergistically stimulate translation initiation in vivo. We recently described mammalian cytoplasmic extracts which, following ultracentrifugation to partially deplete them of ribosomes and associated initiation factors, reproduce cap–poly(A) synergy in vitro. Using these systems, we demonstrate that synergy requires interaction between the poly(A)-binding protein (PABP) and the eukaryotic initiation factor (eIF) 4F holoenzyme complex, which recognises the 5′ cap. Here we further characterise the requirements and constraints of cap–poly(A) synergy in reticulocyte lysates by evaluating the effects of different parameters on synergy. The extent of extract depletion and the amounts of different initiation factors in depleted extracts were examined, as well as the effects of varying the concentrations of KCl, MgCl2 and programming mRNA and of adding a cap analogue. The results presented demonstrate that maximal cap–poly(A) synergy requires: (i) limiting concentrations of ribosome-associated initiation factors; (ii) precise ratios of mRNA to translation machinery (low concentrations of ribosome-associated initiation factors and low, non-saturating mRNA concentrations); (iii) physiological concentrations of added KCl and MgCl2. Additionally, we show that the eIF4G–PABP interaction on mRNAs which are capped and polyadenylated significantly increases the affinity of eIF4E for the 5′ cap.
INTRODUCTION
The majority of eukaryotic mRNAs carry an m7GpppN cap at their 5′-end (1) and a poly(A) tail at their 3′-end (for reviews see 2,3), both of which regulate mRNA stability. In addition, either the cap or poly(A) tail alone enhances translation initiation and the two elements together cooperate to synergistically stimulate translation initiation in vivo (4–8), at the stage of 40S ribosomal subunit recruitment (8). The 5′ cap and 3′ poly(A) tail are recognised, respectively, by the eukaryotic initiation factor (eIF) 4F holoenzyme complex [consisting of the cap-binding protein (eIF4E) and an ATP-dependent RNA helicase (eIF4A) bound to a central scaffold molecule (eIF4G)] (for reviews see 3,9) and by the poly(A)-binding protein (PABP) (8). The C-terminal domain of eIF4G also interacts with eIF3, a complex which can directly associate with 40S ribosomal subunits. The eIF4F complex thus plays a pivotal role in recruiting the 40S ribosomal subunit to the capped mRNA 5′-end. Recently, evidence for a direct interaction between eIF4G and PABP in yeast and mammalian extracts was obtained (10–12) and circularisation of capped and polyadenylated transcripts by purified yeast eIF4E, eIF4G and PABP was visualised by high resolution microscopy (13). Based on these observations, non-exclusive hypotheses were proposed that cap–poly(A) synergy results from mRNA circularisation, due to either enhanced formation of initiation factor–mRNA complexes or facilitated ribosome recycling (see for example 14,15). Evidence in support of the former hypothesis came from the demonstration that the interaction of wheatgerm PABP with eIF4F increases the affinity of eIF4E for a cap analogue by some 40-fold and, similarly, that the affinity of eIF4F-complexed plant PABP for poly(A) is greater than that of free PABP (15,16).
Cap–poly(A) synergy can be reproduced in a variety of in vitro cell-free extracts derived from eukaryotic cells (7,17,18). The majority of such systems exhibit synergy only in the presence of endogenous competitor mRNAs. In the absence of competition, the positive effects of capping and polyadenylation on translation are at best only additive (7,17,19). We recently described the development of in vitro mammalian cell-free translation systems which exhibit cap–poly(A) synergy in the absence of competitor mRNAs (20). Using these systems we demonstrated that integrity of the eIF4G–PABP interaction was required for cap–poly(A) cooperativity and for poly(A)-mediated stimulation of uncapped mRNA translation. In addition, we demonstrated that ribosome arrival at the mRNA 3′-end was not a prerequisite for cap–poly(A) cooperativity, excluding a role of direct, continuous ribosome recycling from the mRNA 3′-end back to the cap in synergy.
Here we report a detailed characterisation of the requirements for poly(A) dependency in rabbit reticulocyte lysate (RRL) cell-free translation extracts. The data presented strongly suggest that synergy arises, at least in part, from increased affinities of capped and polyadenylated mRNAs for certain initiation factors. Furthermore, we present direct evidence that the functional affinity of the eIF4E–cap interaction is considerably increased upon interaction of eIF4G with PABP on capped and polyadenylated mRNA.
MATERIALS AND METHODS
Plasmid construction and in vitro transcription
The construction of the monocistronic plasmids pB2 and p0p24 has been described elsewhere (20). The pB2 plasmids contain the cDNA for Xenopus laevis cyclin B2 [from the beginning of the 5′-untranslated region (5′-UTR) up to the stop codon] followed by cDNA corresponding to the 3′-UTR of the influenza virus NS mRNA, under control of the bacteriophage T7 φ10 promoter. The p0p24 plasmids contain (also under control of the T7 φ10 promoter) a short oligonucleotide-derived 5′-UTR, followed by the region coding for the human immunodeficiency virus (HIV-1Lai) p24 protein and the influenza virus NS 3′-UTR (Fig. 1A). Two versions of each of these plasmids differ only in the presence or absence of an A50 tract inserted at the unique EcoRI site, 24 nt downstream of the authentic polyadenylation signal. In vitro transcription and quantification and purification of capped and uncapped in vitro transcripts were performed as described (20) using pB2 and p0p24 plasmids which had been linearized with EcoRI.
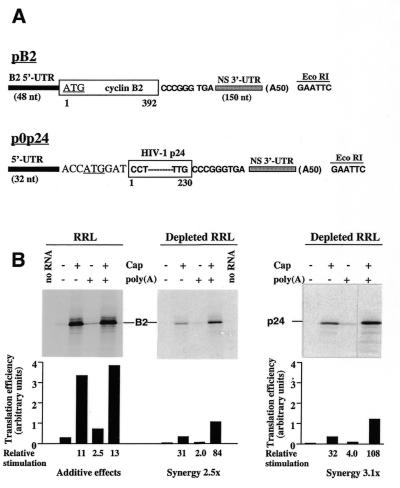
Figure 1.
Cap–poly(A) synergy in ribosome-depleted RRL. (A) Schematic representation of the plasmids used in this work. The Xenopus laevis cyclin B2 and HIV-1 p24 coding regions are shown as open boxes. Numbers below coding regions refer to the first and last amino acids of each reporter gene product. The 5′- and 3′-UTRs are depicted as black and speckled boxes, respectively; the translation initiation codon is underlined. Clones were constructed in duplicate, differing by the presence or absence of an A50 insertion (in parentheses) at the EcoRI site used for linearisation prior to transcription. (B) Standard RRL (left) or ribosome-depleted RRL (middle and right) was programmed with in vitro transcribed RNAs derived from the pB2 or p0p24 plasmids in the form indicated above each lane [final RNA concentrations 6.1 µg/ml for B2 mRNAs and 4 µg/ml (the molar equivalent) for 0p24 mRNAs]. The final concentrations of added KCl and MgCl2 were 115 and 0.6 mM, respectively. Control reactions were programmed with water (no RNA lane). Translations were processed as described in Materials and Methods. An autoradiograph of the dried 20% polyacrylamide gel is shown. The position of the cyclin B2 or HIV-1p24 translation product is marked. Translation efficiency derived from densitometric quantification is plotted below each lane. Relative stimulation of translation was calculated by comparing the translation efficiency of capped and/or polyadenylated RNA to that of the –/– RNA. Cap–poly(A) synergy was calculated by the formula: (relative stimulation of +/+) ÷ [(relative stimulation of +/–) + (relative stimulation of –/+)].
Preparation of translation extracts and in vitro translation
Nuclease-treated RRL (Promega) was partially depleted of ribosomes by ultracentrifugation in a Beckman TL-100 benchtop ultracentrifuge essentially as described previously (20). In experiments aimed to evaluate the effects of different levels of ribosome depletion on cap–poly(A) synergy, the speed and duration of ultracentrifugation were varied (85 000 or 90 000 r.p.m., 15–45 min). In all cases the post-centrifugation supernatant was carefully removed without disturbing the pellet of ribosomes/associated initiation factors and was aliquoted and stored at –80°C.
HeLa cell S10 and S100 extracts were prepared essentially as described previously (21), except that cells were not starved of methionine before extract preparation. HeLa cell high salt S100 extracts (HS100) were prepared by making S10 extracts 0.5 M in KCl by the slow addition of 4 M KCl on ice. After 30 min incubation at 4°C, HS100 was prepared by ultracentrifugation following the procedure used for standard S100 extracts. All extracts were dialysed overnight against H100 buffer (10 mM HEPES–KOH pH 7.5, 100 mM KCl, 1 mM MgCl2, 0.1 mM EDTA and 7 mM β-mercaptoethanol) prior to treatment with micrococcal nuclease as described (22).
In vitro translation reactions were performed in the presence of [35S]methionine. Reactions contained 50% by volume flexi-reticulocyte lysate (Promega) or ribosome-depleted RRL and 20 or 30% by volume H100 buffer or HeLa cell extracts in H100 buffer. Reactions were programmed with the indicated concentrations of in vitro transcribed mRNAs. Final concentrations of added KCl and MgCl2 were 125 and 0.6 mM, respectively, unless otherwise stated. In certain experiments various concentrations of a cap analogue (Ambion Inc), diluted in H100 buffer, or a fragment of recombinant NSP3 encompassing amino acids 163–313 [which had been overexpressed in Escherichia coli and purified exactly as described previously (12) and was a gift of Dr D. Poncet] were added to reactions simultaneously with the programming RNAs.
Translations were performed for 90 min at 30°C and the translation products were analysed by SDS–PAGE as described previously (23), using gels containing 20% (w/v) acrylamide. Dried gels were exposed to Biomax MR film (Kodak) for 1–4 days depending on the particular experiment. Densitometric quantification of translation products was as described previously (24) using multiple exposures of each gel to ensure that the linear response range of the film was covered and that low levels of translation could be accurately quantified.
Antibodies, western blotting and co-immunoprecipitation
Rabbit anti-eIF4G peptide 7 (raised against residues 327–342) and rabbit anti-eIF4E antisera were a gift of Dr R. Rhoads and have been described previously (25,26). Monoclonal antibody 10E10 against human PABP was a gift of Dr M. Görlach and has been described previously (27). Mouse monoclonal anti-eIF4A antibody was kindly donated by Dr H. Trachsel. Western blot analysis of the components of the eIF4F complex in RRL or ribosome-depleted RRL was performed as described previously (24). Blots were developed using the commercial DAB peroxidase substrate kit (Vector Laboratories). Co-immunoprecipitation of eIF4G and PABP from reticulocyte lysates was performed exactly as described previously (20) by immunoprecipitating the eIF4F complex with antibodies directed against eIF4G followed by western blot analysis of immunoprecipitates using antibodies directed against PABP.
RESULTS
We recently reported the development of a nuclease-treated, ribosome-depleted RRL translation system in which the mRNA 5′ cap and 3′ poly(A) tail cooperate to synergistically stimulate translation in vitro (20). The aim of the present study was a detailed characterisation of the requirements and constraints of cap–poly(A) synergy in ribosome-depleted RRL.
Since the effects of the poly(A) tail on translation can be measured in two ways, by comparing the translation efficiency of uncapped mRNAs with and without a poly(A) tail or by examining the synergy obtained upon addition of both poly(A) and a cap to an mRNA (28), we compared the translation efficiency of four versions of each mRNA: neither capped nor polyadenylated (–/–), capped and non-polyadenylated (+/–), uncapped and polyadenylated (–/+), both capped and polyadenylated (+/+). These were synthesized in vitro from cDNA transcription templates which only differed by an oligonucleotide-derived homopolymer A50 insertion preceeding a unique EcoRI site at the end of the 3′-UTR (see Materials and Methods and Fig. 1A). Thus, the polyadenylated versions of the transcripts terminate with an A50GAAUU tail. It has previously been shown that 50 A residues suffice to demonstrate the roles of the poly(A) tail in translation initiation (5,7).
Evaluation of cap–poly(A) synergy in ribosome-depleted RRL as a function of the extract depletion and programming mRNA concentration
Several laboratories have previously demonstrated that the positive effects of capping and polyadenylation on translation initiation in standard nuclease-treated RRL are at best additive (17,19). Translation of in vitro transcribed mRNAs in such systems is highly efficient and is significantly stimulated by capping and to a lesser extent by polyadenylation (Fig. 1B, left). When RRL is partially depleted of its ribosomes and associated initiation factors by ultracentrifugation, the resulting post-centrifugation supernatant exhibits significantly reduced translation capacities irrespective of the cap and poly(A) status of the mRNA (Fig. 1B, middle). However, the cap and polyadenylation status of the mRNAs determines the extent to which translation is reduced in depleted RRL: to <5% for –/– and –/+ B2 mRNAs, to <10% for +/– B2 mRNA and to 35% for +/+ B2 mRNA (Fig. 1B). As a consequence of these differential reductions in translation efficiencies, cap–poly(A) cooperative stimulation of translation is observed (calculated as the relative stimulation of +/+ RNA divided by the sum of the relative stimulations of –/+ and +/– RNAs; a synergy of 2.5-fold for the data presented in Fig. 1B). Quantitatively similar results in terms of cap–poly(A) synergy were obtained upon translation of a second series of mRNAs derived from the p0p24 cDNAs in depleted RRL (Fig. 1B, right).
The concentrations of 40S and 60S ribosomal subunits which remain in the ribosome-depleted RRL following standard ultracentrifugation (90 000 r.p.m. for 20 min; see Materials and Methods) are below the detection limit of conventional assays (data not shown; 20). However, an indirect measure of the degree of extract depletion can be obtained by comparing the translation efficiency of a control mRNA translated in depleted as opposed to standard RRL. To assess cap–poly(A) cooperativity as a function of the residual translation capacity of the extract, a total of 10 aliquots of RRL were depleted of ribosomes by different conditions of ultracentrifugation (see Materials and Methods). The four different versions of the pB2-derived mRNAs were translated at 6.1 µg/ml (final RNA concentration) in the various resulting post-centrifugation supernatants. Extract depletion was evaluated by comparing the translation efficiency of the +/– B2 RNA in standard RRL and each of the 10 ribosome-depleted extracts. Figure 2A depicts the cap–poly(A) synergy observed in each extract plotted as a function of this residual translation capacity. Significant cap–poly(A) synergy was observed in all extracts which retained between 4 and 8% of the translation activity of non-depleted RRL and was much diminished in those extracts which were more, or less, depleted (as measured by their capacity to translate +/– mRNA).
Figure 2.
Cap–poly(A) synergy as a function of mRNA concentration and extract depletion. (A) Ten aliquots of ribosome-depleted RRL prepared under various centrifugation conditions were programmed with the four different versions of pB2-derived mRNAs as described in the legend to Figure 1. Translation products were analysed as described in the legend to Figure 1 and cap–poly(A) synergy in each extract was calculated. Cap–poly(A) synergy is plotted against the translation efficiency of +/– mRNA translation in the particular batch of depleted RRL relative to that observed in standard RRL (residual translation activity; a value of 1.0 reflects retention of 100% translation activity). Differences in preparation of batches of depleted RRL were as follows: centrifugation at 90 000 r.p.m. for 15, 20, 25, 40 or 45 min (aliquots 9, 6, 3, 2 and 1, respectively, counting from the left) or 85 000 r.p.m. for 15 or 35 min (aliquots 10 and 7). Aliquot 8 was prepared by supplementing aliquot 1 with 0.075× ribosome pellet (with respect to starting extract) recovered after centrifugation. Aliquots 4 and 5 were derived by mixing aliquots 3 and 9 in the ratios 95:5 and 90:10, respectively. (B) Ribosome-depleted extract number 4 [marked with an asterisk in (A)] was programmed with the indicated final RNA concentrations of the four different versions of B2 mRNA under the conditions described in the legend to Figure 1. Translation products were analysed and quantified as described in the legend to Figure 1 and cap–poly(A) synergy in each extract was calculated. RNA concentration is plotted against translation efficiency (open triangles, –/– mRNAs; diamonds, +/–; filled triangles, –/+; circles, +/+; left-hand y-axis) and calculated cap–poly(A) synergy (thick line; right-hand y-axis).
We previously demonstrated that cap–poly(A) cooperativity could be significantly amplified in depleted RRL by programming translation reactions with low, limiting concentrations of mRNA (20). Figure 2B recapitulates the results of an experiment in which different final concentrations of mRNA were translated in ribosome-depleted RRL. The four versions of B2 mRNAs were translated in ribosome-depleted extract 4 (marked with an asterisk in Fig. 2A) at four different final mRNA concentrations and cap–poly(A) synergy was calculated at each mRNA concentration. Cap–poly(A) synergy was highest at the lowest RNA concentration tested (1.25 µg/ml) and was abolished at the highest concentration (10 µg/ml; Fig. 2B). This reduction in observed cap–poly(A) synergy with increasing RNA concentration resulted mainly from the fact that the extracts were more rapidly saturated by the +/+ mRNAs than by the +/– or –/+ mRNAs and that the –/–, –/+ and +/– mRNAs showed considerable non-linearity of the RNA dose–response effect over the lower range of RNA concentrations (20). Taken together, the results shown in Figure 2 demonstrate that cap–poly(A) synergy requires limiting concentrations of available ribosomes/associated factors and non-saturating, low concentrations of programming mRNA.
Cap–poly(A) synergy in the depleted RRL system does not result from differences in mRNA stability
Many laboratories have previously demonstrated the relatively long physical half-lives of mRNAs translated in standard RRL (see for example 19). However, it cannot be formally excluded that cap–poly(A) synergy as detected here results from differences in mRNA stability which are exaggerated in ribosome-depleted RRL. To address this possibilty, we chose to evaluate the functional half-life of the different mRNAs translated in depleted RRL, rather than to examine the integrity of the pool of programming mRNA after translation. Effectively, minor physical modifications of mRNA structure, such as cap removal or poly(A) tail shortening, would have a profound effect on translation capacity, but would go undetected in experiments designed to examine mRNA length after re-extraction of RNA from translation extracts. Thus, we consider that translation efficiency is dependent upon the stability of initiation-competent mRNAs, rather than that of the whole mRNA pool.
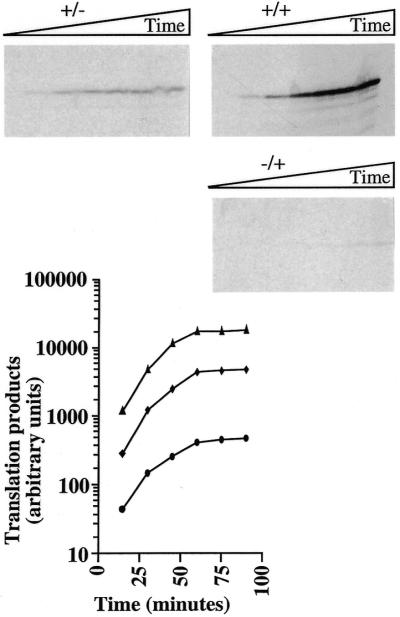
Depleted RRL was programmed with equal concentrations of either +/–, –/+ or +/+ mRNAs derived from p0p24 and aliquots of the translation reactions were analysed at various times for the quantity of p24 synthesised (Fig. 3). No significant differences in functional stability could be detected between the three different 0p24 mRNAs. In effect, the yield of translation products from all three mRNAs increased with similar rates for the first 60 min of translation and then reached a plateau which was maintained between 60 and 90 min (most probably because of exhaustion of the translation extracts). Thus, while precise functional half-lives cannot be calculated for each mRNA from such data, the results demonstrate that cap–poly(A) synergy derives from dramatic differences in translation efficiency rather than from differences in functional mRNA stability between the capped and/or polyadenylated versions of these mRNAs.
Figure 3.
Time course of protein synthesis in ribosome-depleted RRL. Ribosome-depleted RRL reactions were programmed with +/–, –/+ or +/+ mRNAs derived from the p0p24 plasmids (final RNA concentration 4 µg/ml) as described in the legend to Figure 1. Aliquots were removed at 15 min intervals and the reactions stopped prior to analysis of translation products as described in the legend to Figure 1. Control reactions programmed with water gave no detectable protein synthesis (data not shown). The yield of translation products for each mRNA (triangles, +/+; diamonds, +/–; circles, –/+) is plotted against time of incubation.
The effects of KCl and MgCl2 concentration on cap–poly(A) synergy in vitro
Protein synthesis in vitro has a strong requirement for K+ ions (29). However, the absolute potassium concentration which is optimal for translation varies according to the mRNA species tested, with uncapped mRNAs in general exhibiting lower KCl optima than their capped counterparts (30). In the light of such data, we evaluated the effects of altering KCl or MgCl2 concentration on translation efficiency of the four different versions of B2 mRNA. Figure 4 shows the results of such an experiment.
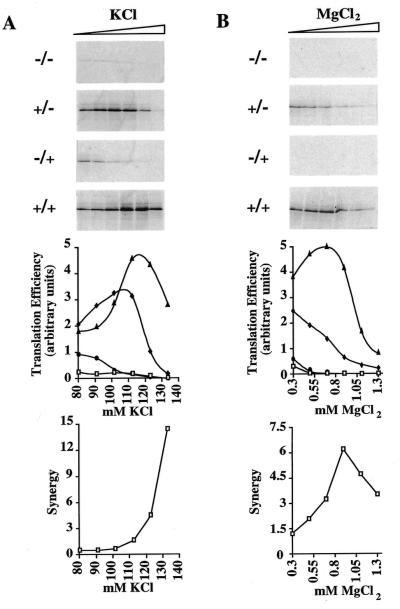
Figure 4.
Influence of KCl and MgCl2 concentrations on cap–poly(A) synergy in ribosome-depleted RRL. Ribosome-depleted RRL was programmed with mRNAs derived from pB2 (final RNA concentration 6.1 µg/ml) transcribed in the form indicated alongside each panel. Translation reactions contained 0.65 mM added MgCl2 and varying concentrations of added KCl (80–133 mM; left) or 125 mM added KCl and varying concentrations of added MgCl2 (0.3–1.3 mM; right). Translation products were analysed as described in the legend to Figure 1. The translation efficiencies of the various RNAs (squares, –/–; diamonds, +/–; circles, –/+; triangles, +/+) plotted as a function of salt concentration and the cap–poly(A) synergy calculated at each salt concentration are shown below the two series of panels.
With respect to KCl concentration, the +/+, +/–, –/+ and –/– mRNAs had very different KCl optima for translation (optima of 112–120 mM for +/+, 100–110 mM for +/– and <80 mM for the –/+ and –/– mRNAs). Thus, while the effects of capping and polyadenylation were at best additive below 100 mM added KCl, between 100 and 130 mM added KCl cap–poly(A) synergy was observed and increased exponentially with increasing KCl concentrations to exceed 10-fold at 130 mM added KCl (Fig. 4A). It should be noted that cap–poly(A) synergy was highest at added KCl concentrations in excess of the optimum for translation of the +/+ mRNA. This apparent paradox results from the fact that translation of +/– mRNA at such KCl concentrations was exceptionally inefficient and underscores the fact that synergy reflects the stimulatory effects of polyadenylating a capped mRNA and is thus highest when the difference in translation efficiency between capped, non-polyadenylated versus capped, polyadenylated mRNA translation is greatest. Overall, these results demonstrate clearly that cap–poly(A) cooperativity is greatest at physiological KCl concentrations. In this respect it should be noted that the particular batch of RRL used for all of the experiments described in this report had an unusually high KCl requirement (as measured with standard mRNAs) and exhibited optimal translation at concentrations 10–20 mM higher than observed with previous batches.
A similar pattern of effects on translation was observed with increasing MgCl2 concentrations (Fig. 4B). With the exception of the +/+ mRNA (optimum of 0.6–0.7 mM added MgCl2), the MgCl2 optima for the different B2 mRNAs were below the range tested (as judged by the absence of a peak of translation for the +/–, –/+ and –/– mRNAs). Thus, once again, cap–poly(A) synergy increased with increasing salt concentration (in the range 0.3–0.9 mM MgCl2) and only began to diminish at MgCl2 concentrations exceeding 1.0 mM. Globally, the results presented in this section clearly demonstrate that cap–poly(A) cooperativity requires near physiological concentrations of both KCl and MgCl2. Furthermore, since the four versions of the B2 mRNAs tested here are structurally identical except for the presence or absence of a 5′ cap and 3′ poly(A) tail, the significantly elevated optima of the +/+ mRNA with respect to salt concentrations (as compared to the other three RNAs) is highly indicative of increased affinities of RNA–protein or protein–protein interactions on capped and polyadenylated mRNA.
Cap–poly(A) synergy requires limiting concentrations of ribosome-associated translation factors
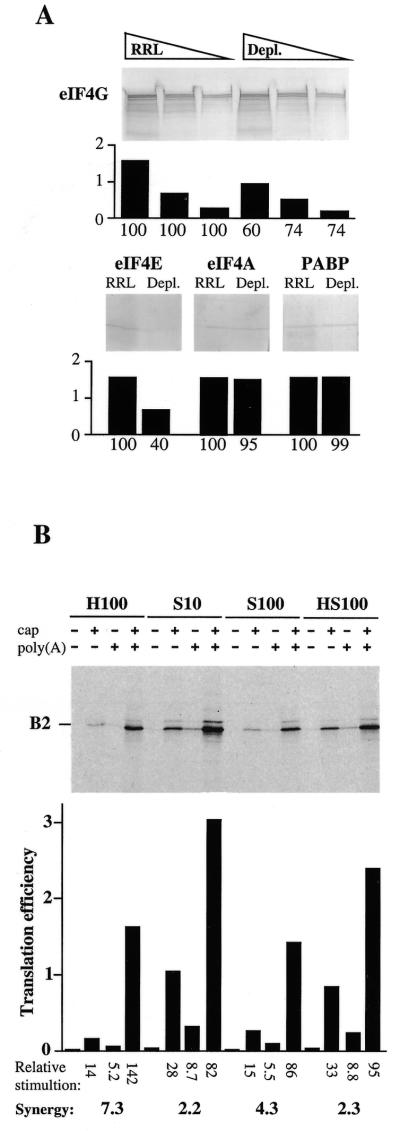
Depletion of ribosomal subunits by ultracentrifugation without prior high salt treatment of translation extracts results in depletion of any protein which associates tightly with ribosomes (see for example 31). Thus, we compared the concentrations of various translation factors in standard RRL and ribosome-depleted RRL by western blotting. Since cap–poly(A) cooperativity requires integrity of the eIF4G–PABP complex (20) and presumably mRNA circularisation via the resulting cap–eIF4E–eIF4G–PABP–poly(A) interaction, we confined this comparison to components of the eIF4F complex (eIF4A, eIF4E, eIF4G and PABP). With respect to standard RRL, ribosome-depleted RRL contained significantly reduced concentrations of eIF4E and eIF4G (Fig. 5A). This observation is in agreement with previous reports suggesting that the concentration of eIF4E is limiting in reticulocyte lysates and that the major fraction of eIF4E is associated with eIF4G, which itself can associate with ribosomes (32,33). Conversely, ribosome depletion of extracts had very little effect on the concentrations of PABP and eIF4A. Both of these proteins are abundant in cells (27,34,35) and a major proportion of eIF4A is not complexed with eIF4G (34).
Figure 5.
(A) Western blot analysis of equal volumes of RRL or ribosome-depleted RRL was performed as described in Materials and Methods with antibodies against the indicated translation factors. Volumes of 5, 2.5 and 1.25 µl of RRL and ribosome-depleted RRL were analysed to ensure linearity of the immunological responses. For membranes developed with antibodies against eIF4E, eIF4A and PABP, only the lanes loaded with 1.25 µl of each extract are shown. Developed membranes were analysed by densitometry and the intensity of the immunological signal is plotted (in arbitrary units) below each panel. The percentage of each protein remaining in depleted RRL is indicated below each plot (100% is the signal observed with each antibody against the same volume of standard RRL). (B) Ribosome-depleted RRL was programmed with the indicated forms of pB2-derived mRNAs (final RNA concentration 6.1 µg/ml) and supplemented with 20% (v/v) H100 buffer, HeLa cell S10 extract, S100 extract or HS100 extract as indicated (see Materials and Methods; final salt concentrations 125 and 0.8 mM added KCl and MgCl2, respectively). Translation products were analysed as described in the legend to Figure 1. The translation efficiency, relative stimulation (as compared to the –/– mRNA in each set of conditions) and calculated cap–poly(A) synergy in each extract are indicated.
Next, we evaluated whether the poly(A) dependency of ribosome-depleted extracts resulted from reduced concentrations of free ribosomes or rather from limiting concentrations of available ribosome-associated initiation factors. To this end, ribosome-depleted RRL was supplemented either with H100 buffer or with nuclease-treated HeLa cell S10 extract (containing ribosomes and both ribosome-associated and free initiation factors), S100 extract (containing free initiation factors but neither ribosomes nor exclusively ribosome-associated translation factors) or HS100 (containing all translation factors but no ribosomes) (see Materials and Methods; Fig. 5B). To ensure significant cap–poly(A) synergy in the depleted RRL, translation reactions were performed in near optimal concentrations of KCl and MgCl2. Under these conditions cap–poly(A) synergy in extracts supplemented with control H100 buffer exceeded 7-fold. While synergy was only modestly reduced upon addition of HeLa cell S100 extract (synergy 4.3-fold), it was significantly reduced upon addition of either HeLa cell HS100 or S10 (synergy 2-fold in both cases; Fig. 5B). Thus, synergy was significantly reduced when ribosome-associated factors (with or without ribosomes) were added back to the depleted extracts. These results support the conclusion that maximal cap–poly(A) cooperativity in ribosome-depleted RRL requires depletion of ribosome-associated initiation factors and not merely ribosomal subunits.
Capped, polyadenylated mRNAs have an elevated functional affinity for eIF4E
As mentioned above, the high salt optima of +/+ mRNA (as compared to +/–, –/+ and –/– mRNAs) strongly suggest that the protein–RNA or protein–protein interactions which form on capped, polyadenylated mRNAs are of a particularly high affinity. In addition, western blot analysis demonstrated that, of the known proteins which interact with mRNA 5′- or 3′-ends, the concentration of eIF4E was the most reduced in ribosome-depleted RRL, as compared to standard RRL. Thus, we examined the relative sensitivities of +/– and +/+ mRNAs with respect to cap analogue inhibition of translation in ribosome-depleted RRL.
Depleted RRL reactions were programmed with equal, low concentrations of either +/– or +/+ mRNAS derived from p0p24 and supplemented with increasing concentrations of the cap analogue m7G(5′)ppp(5′)G. The experiment was performed in parallel in the absence or presence of 10 µg/ml of a recombinant fragment of rotavirus NSP3 protein. NSP3 has previously been shown to interact with eIF4G and displace PABP from the functional eIF4F complex (12; Fig. 6A) and in doing so to abrogate cap–poly(A) synergy in the depleted RRL system (20). It was verified that the concentration of NSP3 used was sufficient to evict PABP from eIF4G, by immunoprecipitation of eIF4G and western blot analysis of the immunocomplex for the presence of PABP (Fig. 6B). NSP3 at 10 µg/ml displaces >80% of PABP from eIF4G. As reported previously, the addition of higher concentrations of NSP3 does not eliminate the residual PABP from the eIF4F complex (20; data not shown).
Figure 6.
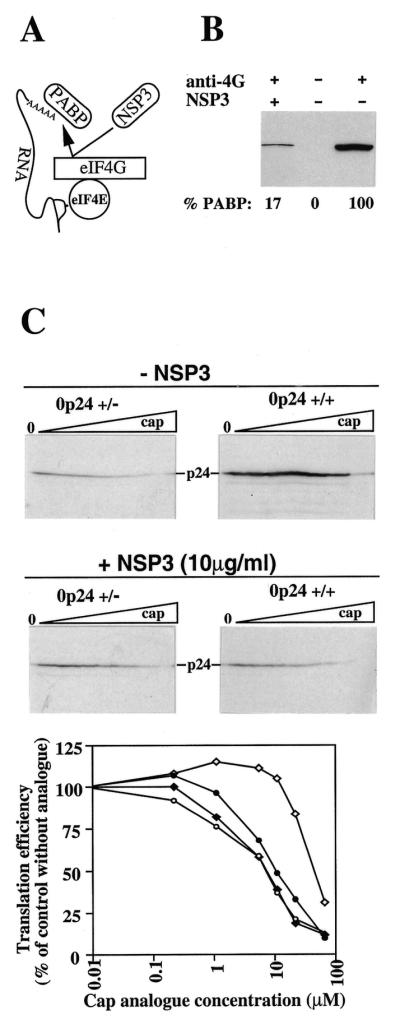
Sensitivity of polyadenylated and non-polyadenylated capped mRNA translation to cap analogue inhibition in depleted RRL. (A) Schematic representation of the action of rotavirus NSP3 protein, which abolishes the possibility of mRNA circularisation via the cap–eIF4E–eIF4G–PABP–poly(A) tail interaction. (B) RRL translation reactions, supplemented with NSP3 protein in H100 buffer (NSP3 concentration 10 µg/ml in the reaction) or H100 buffer were immunoprecipitated with antibody against eIF4G or preimmune sera (as indicated, + and –). The immunoprecipitates were analysed for the presence of PABP by western blotting. A photograph of the developed membrane is shown. The percentage of PABP in the immunoprecipitate is given below each lane. (C) Ribosome-depleted RRL was programmed with the indicated forms of p0p24-derived mRNAs (final RNA concentration 3.15 µg/ml) and supplemented with H100 buffer or NSP3 protein in H100 buffer (as indicated; final salt concentrations 125 and 0.8 mM added KCl and MgCl2, respectively). Reactions then received increasing concentrations of cap analogue in H100 buffer (final concentrations from left to right: 0, 0.22, 1.1, 5.5, 11, 22 and 66 µM). Translation products were analysed as described in the legend to Figure 1. The translation efficiencies of the +/+ (diamonds) and +/– (circles) mRNAs, relative to the efficiencies observed in the absence of cap analogue, are plotted against cap analogue concentration. Open and filled symbols represent, respectively, the translation efficiencies in the absence and presence of NSP3.
In control depleted RRL, +/+ mRNA was translated some 5- to 6-fold more efficiently than +/– mRNA (0 lanes, Fig. 6C, upper). In the absence of NSP3 a significant difference in the relative sensitivities of the two forms of RNA to inhibition by the cap analogue was evident. In effect, 8- to 10-fold higher concentrations of analogue were required to inhibit +/+ mRNA translation by 50%, as compared to inhibition of +/– mRNA translation. The addition of NSP3 to translation reactions reduced +/+ mRNA translation efficiency to approach that of its +/– counterpart, as reported previously (20). More importantly, +/+ mRNA translation was then as sensitive to cap analogue inhibition as was the +/– mRNA translated either with or without NSP3. This formally demonstrates that the increased capacity of +/+ mRNA translation to withstand cap analogue inhibition requires integrity of the eIF4G–PABP interaction.
DISCUSSION
The 5′ cap and 3′ poly(A) tail cooperate to synergistically stimulate mRNA translation in vivo. We recently described nuclease-treated RRL translation extracts which reproduce cap–poly(A) synergy after partial depletion of ribosomes and associated translation initiation factors by ultracentrifugation (20). In this study we present a detailed characterisation of the requirements and constraints for cap–poly(A) synergy in such extracts. Cap–poly(A) synergy is a common feature of mRNAs translated in ribosome-depleted RRLs which have been sufficiently depleted of ribosomes and associated factors (Fig. 2). However, it should be noted that minor variations exist between batches of RRL with respect to their ability to translate the capped and polyadenylated forms of mRNA after ribosome depletion (data not shown). Given the extreme sensitivity of mRNA translation in depleted RRL to the concentrations of added KCl and MgCl2 (Fig. 4), it is likely that such minor variations reflect differences in endogenous salt concentrations of commercial RRL preparations.
A kinetic assay of mRNA translation in depleted RRL confirmed that synergy is the product of dramatically enhanced translation efficiencies on capped, polyadenylated mRNAs and does not reflect differences in mRNA functional stability, at least over the duration of a standard in vitro translation. In addition, it was shown that optimal synergy in a given extract requires the combination of limiting concentrations of components of the translation machinery coupled with low, non-saturating concentrations of programming mRNA. This finding is in agreement with the results from in vivo studies which demonstrated that the positive effects of polyadenylation are greatest when ribosomes and initiation factors are limiting (36). The nature of the limiting components was further examined by supplementing depleted RRL with different HeLa cell fractions containing combinations of ribosomes and ribosome-associated or free translation initiation factors. It was found that depletion of extracts of ribosome-associated translation factors is a prerequisite for maximal cap–poly(A) synergy. The initiation factor eIF4E, as compared to the other components of the eIF4F complex, was most reduced in concentration in depleted as opposed to standard RRL.
Two different mechanistic explanations for cap–poly(A) synergy have previously been advanced (14,15): facilitation of ribosome recycling from the 3′-end back to the 5′ cap on mRNAs which are circularised by the cap–eIF4E–eIF4G–PABP–poly(A) tail interaction or increased affinity of the eIF4F–PABP complex for mRNA 5′- or 3′-ends leading to enhanced de novo recruitment of 40S ribosomal subunits. Here we have presented data which indicate that capped, polyadenylated mRNAs have particularly high affinities for certain translation components. Firstly, the concentration of +/+ mRNA required to saturate ribosome-depleted RRL reactions is at least 2- to 3-fold lower than that of the equivalent +/– mRNA (see for example Fig. 2B; and data not shown). Secondly, the elevated salt optima of the capped and polyadenylated mRNA are highly suggestive of enhanced protein–protein and/or protein–RNA binding affinities. Indeed, we found that the affinity of +/+ mRNA for eIF4E is several-fold higher than that of +/– mRNA, since capped, polyadenylated mRNA translation was 8- to 10-fold less sensitive to inhibition by a cap analogue than that of its non-polyadenylated counterpart (Fig. 6). Interruption of the eIF4G–PABP interaction by rotaviral NSP3 protein rendered +/+ mRNA translation sensitive to cap analogue inhibition. A logical interpretation of these data is that the interaction between eIF4G and PABP and the resulting mRNA circularisation via a cap–eIF4E–eIF4G–PABP–poly(A) tail interaction increases the affinity of the eIF4E–cap interaction. The experiments described here measured the functional capacity of eIF4E/eIF4G to direct translation initiation rather than the direct biochemical binding affinity between two purified components. This increase in functional affinity of eIF4E for the cap could stem from two different effects. It is conceivable that the PABP–eIF4G interaction alters the conformation of eIF4E so that the latter has an intrinsically higher affinity for 5′ caps. Alternatively, it is possible that the eIF4G–PABP interaction serves to tether the whole eIF4F complex to the mRNA and increase the local concentration of eIF4E near the cap, thus increasing the apparent affinity of the latter for the 5′ cap. Unfortunately, it is impossible to discriminate between these two non-exclusive hypotheses from the data presented here. However, it is worth noting that biochemical binding assays using a fluorescent cap analogue and purified initiation factors demonstrated that wheatgerm PABP–eIF4F, as opposed to eIF4F alone, has a 40-fold enhanced affinity for the cap analogue (15). Thus, one might reasonably predict that this advantage of mRNA 5′–3′ end cross-talk is conserved between plants and mammals. It remains to be determined whether the functional or direct affinities of other components of the eIF4F complex for mRNA are also increased by the eIF4G–PABP interaction in the RRL system. Nevertheless, the data presented here strongly suggest that cap–poly(A) synergy results, at least in part, from an increased capacity of eIF4E to recognise capped, polyadenylated mRNAs, which presumably leads to enhanced recruitment of 40S ribosomal subunits.
Acknowledgments
ACKNOWLEDGEMENTS
We thank D. Poncet for the gift of the recombinant fragment of rotavirus NSP3 protein and R. Rhoads, M. Görlach and H. Trachsel for the gifts of antibodies. We are grateful to Sylvie van der Werf and Cécile Malnou for their interest in this work. Work in our laboratory is supported by the Programme de Recherche Clinique de l’Institut Pasteur, by a Contrat d’Incitation à la Recherche en vue d’Applications (CCV no. 8) from the Pasteur Institute and by grant no. 6495 from the Association Française contre les Myopathies.
REFERENCES
- 1.Banerjee A.K. (1980) Microbiol. Rev., 44, 175–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson R.J. (1993) Cell, 74, 9–14. [DOI] [PubMed] [Google Scholar]
- 3.Rhoads R.E. (1988) Trends Biochem. Sci. 13, 52–56. [DOI] [PubMed] [Google Scholar]
- 4.Doel M.T. and Carey,N.H. (1976) Cell, 8, 51–58. [DOI] [PubMed] [Google Scholar]
- 5.Gallie D.R. (1991) Genes Dev., 5, 2108–2116. [DOI] [PubMed] [Google Scholar]
- 6.Gallie D.R. (1998) Gene, 216, 1–11. [DOI] [PubMed] [Google Scholar]
- 7.Preiss T. and Hentze,M.W. (1998) Nature, 392, 516–520. [DOI] [PubMed] [Google Scholar]
- 8.Tarun S.Z. Jr and Sachs,A.B. (1995) Genes Dev., 9, 2997–3007. [DOI] [PubMed] [Google Scholar]
- 9.Morley S.J., Curtis,P.S. and Pain,V.M. (1997) RNA, 3, 1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 10.Imataka H., Gradi,A. and Sonenberg,N. (1998) EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarun S.Z. Jr and Sachs,A.B. (1996) EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 12.Piron M., Vende,P., Cohen,J. and Poncet,D. (1998) EMBO J., 17, 5811–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wells S.E., Hillner,P.E., Vale,R.D. and Sachs,A.B. (1998) Mol. Cell., 2, 135–140. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson A. (1996) In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–480.
- 15.Wei C.-C., Balasta,M.L., Ren,J. and Goss,D.J. (1998) Biochemistry, 37, 1910–1916. [DOI] [PubMed] [Google Scholar]
- 16.Le H., Tanguay,R.L., Balasta,M.L., Wei,C.C., Browning,K.S., Metz,A.M., Goss,D.J. and Gallie,D.R. (1997) J. Biol. Chem., 272, 16247–16255. [DOI] [PubMed] [Google Scholar]
- 17.Iizuka N., Najita,L., Franzusoff,A. and Sarnow,P. (1994) Mol. Cell. Biol., 14, 7322–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebauer F., Corona,D.F.V., Preiss,T., Becker,P.B. and Hentze,M.W. (1999) EMBO J., 18, 6146–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munroe D. and Jacobson,A. (1990) Mol. Cell. Biol., 10, 3441–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel Y.M., Poncet,D., Piron,M., Kean,K.M. and Borman,A.M. (2000) J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 21.Borman A., and Jackson,R.J. (1992) Virology, 188, 685–696. [DOI] [PubMed] [Google Scholar]
- 22.Jackson R.J. and Hunt,T. (1983) Methods Enzymol., 96, 50–74. [DOI] [PubMed] [Google Scholar]
- 23.Dasso M.C. and Jackson,R.J. (1989) Nucleic Acids Res., 17, 6485–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borman A.M., Kirchweger,R., Zeigler,E., Rhoads,R.E., Skern,T. and Kean,K.M. (1997) RNA, 3, 186–196. [PMC free article] [PubMed] [Google Scholar]
- 25.Kerekatte V., Smiley,K., Hu,B., Gelder,F. and DeBenedetti,A. (1995) Int. J. Cancer, 64, 27–31. [DOI] [PubMed] [Google Scholar]
- 26.Yan R., Rychlik,W., Etchison,D. and Rhoads,R.E. (1992) J. Biol. Chem., 267, 23226–23231. [PubMed] [Google Scholar]
- 27.Görlach M., Burd,C.G. and Dreyfuss,G. (1994) Exp. Cell Res., 211, 400–407. [DOI] [PubMed] [Google Scholar]
- 28.Tarun S.Z. Jr, Wells,S.E., Deardorff,J.A. and Sachs,A.B. (1997) Proc. Natl Acad. Sci. USA, 94, 9046–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelham H. and Jackson,R.J. (1976) Eur. J. Biochem., 67, 247–256. [DOI] [PubMed] [Google Scholar]
- 30.Jackson R.J. (1991) Biochim. Biophys. Acta, 1088, 345–358. [DOI] [PubMed] [Google Scholar]
- 31.Grifo J.A., Tahara,S.M., Morgan,M.A., Shatkin,A.J. and Merrick,W.C. (1983) J. Biol. Chem., 258, 5804–5810. [PubMed] [Google Scholar]
- 32.Hiremath L.S., Webb,N.R. and Rhoads,R.E. (1985) J. Biol. Chem., 260, 7843–7849. [PubMed] [Google Scholar]
- 33.Goss D.J., Carberry,S.E., Dever,T.E., Merrick,W.C. and Rhoads,R.E. (1990) Biochemistry, 29, 5008–5012. [DOI] [PubMed] [Google Scholar]
- 34.Duncan R., Milburn,S.C. and Hershey,J.W.B. (1987) J. Biol. Chem., 262, 380–388. [PubMed] [Google Scholar]
- 35.Duncan R. and Hershey,J.W.B. (1983) J. Biol. Chem., 258, 7228–7235. [PubMed] [Google Scholar]
- 36.Proweller A. and Butler,J.S. (1997) J. Biol. Chem., 272, 6004–6010. [DOI] [PubMed] [Google Scholar]