Abstract
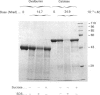
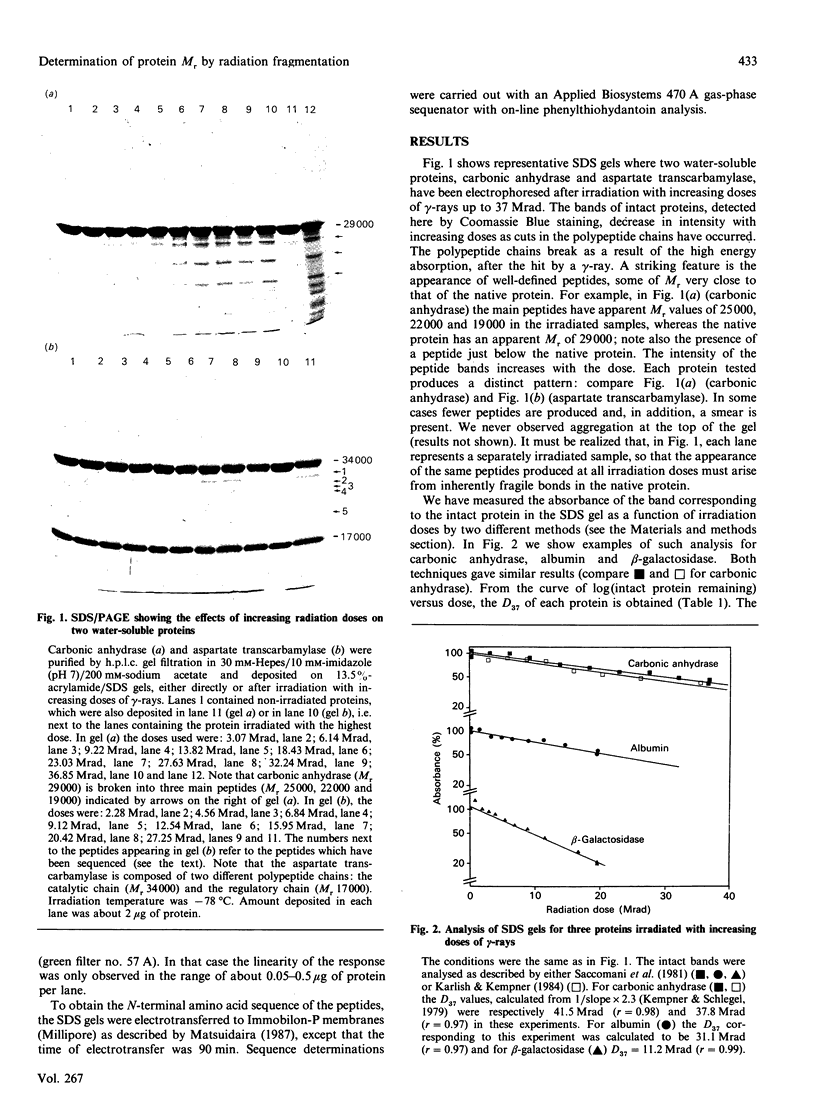
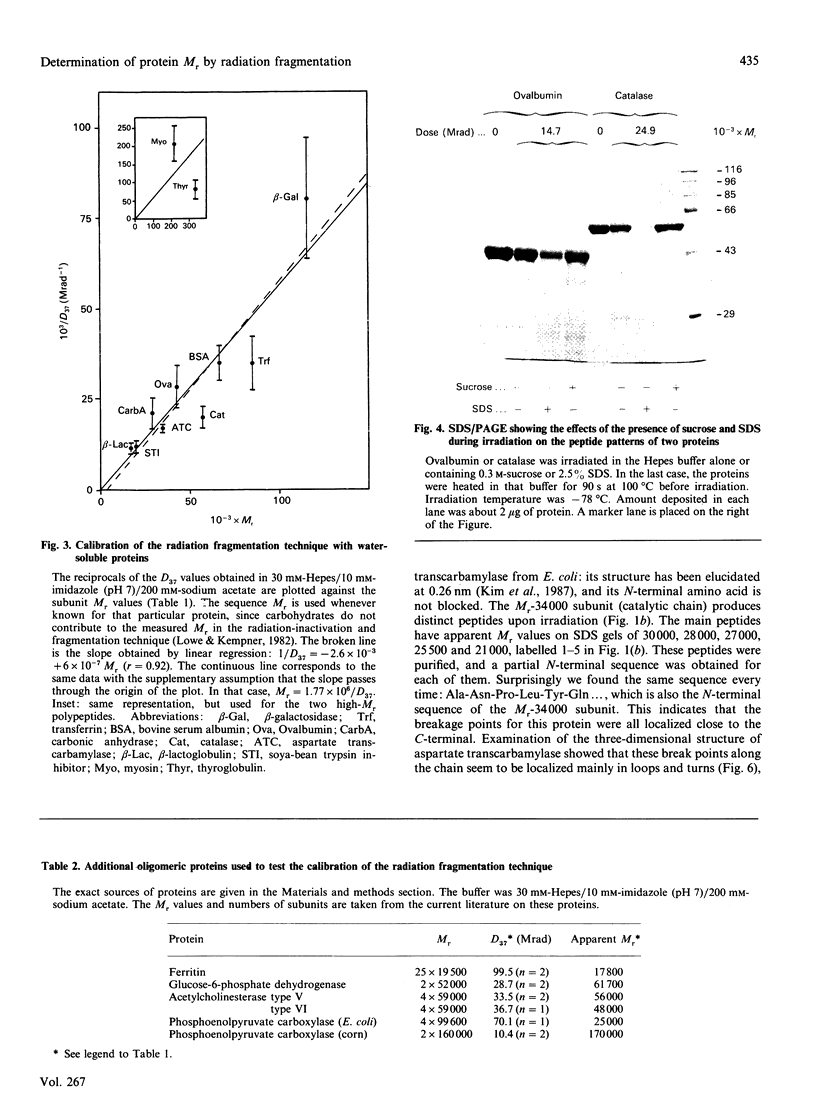
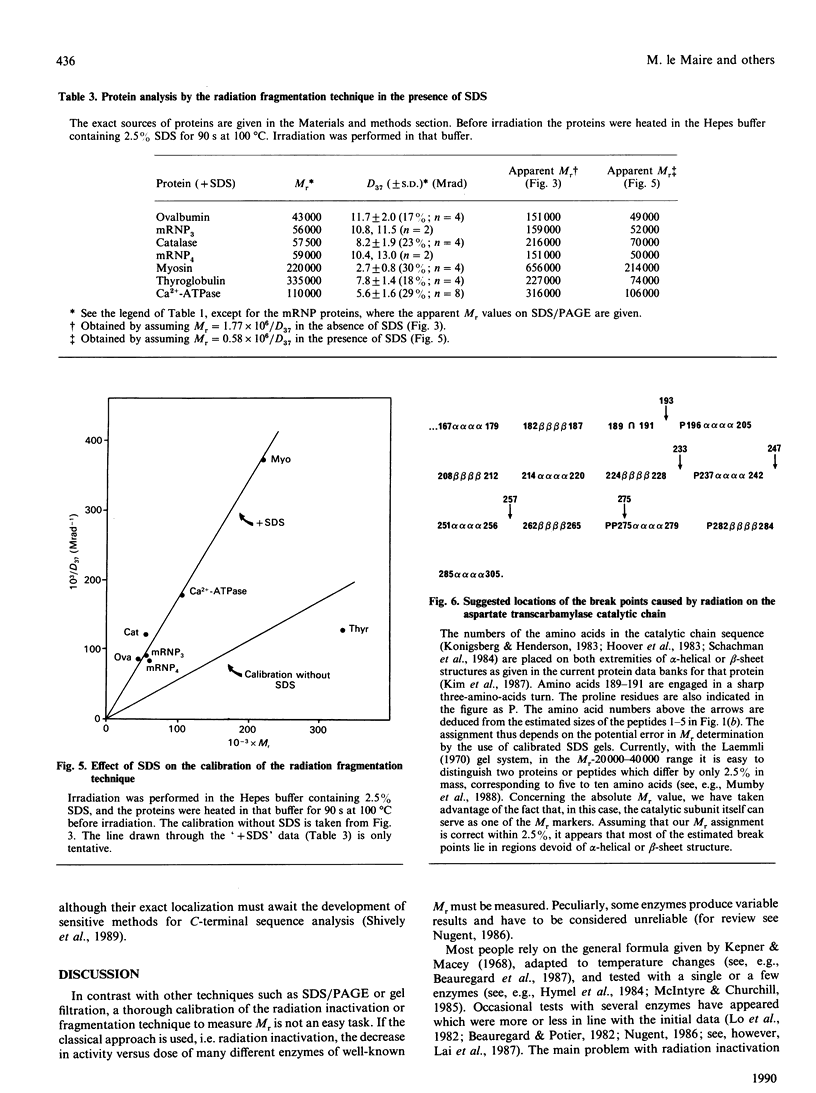
We have reinvestigated the use of ionizing radiations to measure the molecular mass of water-soluble or membrane proteins. The test was performed by using the most straightforward aspect of the technique, which consists of SDS/PAGE analysis of the protein-fragmentation process. We found that exposure of purified standard proteins to increasing doses of ionizing radiation causes progressive fragmentation of the native protein into defined peptide patterns. The coloured band corresponding to the intact protein was measured on the SDS gel as a function of dose to determine the dose (D37.t) corresponding to 37% of the initial amount of unfragmented protein deposited on the gel. This led to a calibration curve between 1/D37.t and the known molecular mass of the standard proteins whose best fit gave Mr = 1.77 x 10(6)/D37.t at -78 degrees C, i.e. 35% higher than the generally accepted value at that temperature obtained from inactivation studies. However, we have to conclude that this method is useless to determine the state of aggregation of a protein, since, for all the oligomers tested, the best fit was obtained by using the protomeric molecular mass, suggesting that there is no energy transfer between promoters. Furthermore, SDS greatly increases the fragmentation rate of proteins, which suggests additional calibration problems for membrane proteins in detergent or in the lipid bilayer. But the main drawback of the technique arises from our observation that some proteins behaved anomalously, leading to very large errors in the apparent target size as compared with true molecular mass (up to 100%). It is thus unreliable to apply the radiation method for absolute molecular-mass determination. We then focused on the novel finding that discrete fragmentation of proteins occurs at preferential sites, and this was studied in more detail with aspartate transcarbamylase. N-Terminal sequencing of several radiolysis fragments of the catalytic chain of the enzyme revealed that breaks along the polypeptide chains are localized close to the C-terminal end. Examination of the three-dimensional structure of aspartate transcarbamylase suggests that radiolysis sites (fragile bonds) might be localized in connecting loops.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen J. P., Vilsen B. Radiation inactivation analysis of sarcoplasmic reticulum Ca-ATPase in membrane-bound form and in detergent-solubilized monomeric states. FEBS Lett. 1988 Jul 4;234(1):120–126. doi: 10.1016/0014-5793(88)81316-4. [DOI] [PubMed] [Google Scholar]
- Beauregard G., Giroux S., Potier M. Target size analysis by radiation inactivation: a large capacity tube rack for irradiation in a Gammacell 220. Anal Biochem. 1983 Jul 15;132(2):362–364. doi: 10.1016/0003-2697(83)90021-0. [DOI] [PubMed] [Google Scholar]
- Beauregard G., Maret A., Salvayre R., Potier M. The radiation inactivation method as a tool to study structure-function relationships in proteins. Methods Biochem Anal. 1987;32:313–343. doi: 10.1002/9780470110539.ch6. [DOI] [PubMed] [Google Scholar]
- Beauregard G., Potier M. Radiation inactivation of enzymes at low dose rates: identical molecular weights of rat liver cytosolic and lysosomal neuraminidases. Anal Biochem. 1982 May 15;122(2):379–384. doi: 10.1016/0003-2697(82)90299-8. [DOI] [PubMed] [Google Scholar]
- Beauregard G., Potier M. Radiation inactivation of membrane proteins: molecular weight estimates in situ and after Triton X-100 solubilization. Anal Biochem. 1984 Aug 1;140(2):403–408. doi: 10.1016/0003-2697(84)90185-4. [DOI] [PubMed] [Google Scholar]
- Beauregard G., Potier M. Temperature dependence of the radiation inactivation of proteins. Anal Biochem. 1985 Oct;150(1):117–120. doi: 10.1016/0003-2697(85)90448-8. [DOI] [PubMed] [Google Scholar]
- Bowman B. J., Berenski C. J., Jung C. Y. Size of the plasma membrane H+-ATPase from Neurospora crassa determined by radiation inactivation and comparison with the sarcoplasmic reticulum Ca2+-ATPase from skeletal muscle. J Biol Chem. 1985 Jul 25;260(15):8726–8730. [PubMed] [Google Scholar]
- Chamberlain B. K., Berenski C. J., Jung C. Y., Fleischer S. Determination of the oligomeric structure of the Ca2+ pump protein in canine cardiac sarcoplasmic reticulum membranes using radiation inactivation analysis. J Biol Chem. 1983 Oct 10;258(19):11997–12001. [PubMed] [Google Scholar]
- Darnbrough C. H., Ford P. J. Identification in Xenopus laevis of a class of oocyte-specific proteins bound to messenger RNA. Eur J Biochem. 1981 Jan;113(3):415–424. doi: 10.1111/j.1432-1033.1981.tb05081.x. [DOI] [PubMed] [Google Scholar]
- Edwards P. A., Kempner E. S., Lan S. F., Erickson S. K. Functional size of rat hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase as determined by radiation inactivation. J Biol Chem. 1985 Aug 25;260(18):10278–10282. [PubMed] [Google Scholar]
- Fluke D. J. Temperature dependence of ionizing radiation effect on dry lysozyme and ribonuclease. Radiat Res. 1966 Jul;28(3):677–693. [PubMed] [Google Scholar]
- Gerhart J. C., Holoubek H. The purification of aspartate transcarbamylase of Escherichia coli and separation of its protein subunits. J Biol Chem. 1967 Jun 25;242(12):2886–2892. [PubMed] [Google Scholar]
- Hoover T. A., Roof W. D., Foltermann K. F., O'Donovan G. A., Bencini D. A., Wild J. R. Nucleotide sequence of the structural gene (pyrB) that encodes the catalytic polypeptide of aspartate transcarbamoylase of Escherichia coli. Proc Natl Acad Sci U S A. 1983 May;80(9):2462–2466. doi: 10.1073/pnas.80.9.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymel L., Maurer A., Berenski C., Jung C. Y., Fleischer S. Target size of calcium pump protein from skeletal muscle sarcoplasmic reticulum. J Biol Chem. 1984 Apr 25;259(8):4890–4895. [PubMed] [Google Scholar]
- Jensen J., Nørby J. G. Membrane-bound Na,K-ATPase: target size and radiation inactivation size of some of its enzymatic reactions. J Biol Chem. 1988 Dec 5;263(34):18063–18070. [PubMed] [Google Scholar]
- Kempner E. S., Miller J. H. Radiation inactivation of glutamate dehydrogenase hexamer: lack of energy transfer between subunits. Science. 1983 Nov 11;222(4624):586–589. doi: 10.1126/science.6635656. [DOI] [PubMed] [Google Scholar]
- Kempner E. S., Miller J. H. Radiation-damaged tyrosinase molecules are inactive. Biophys J. 1989 Jan;55(1):159–162. doi: 10.1016/S0006-3495(89)82787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempner E. S. Molecular size determination of enzymes by radiation inactivation. Adv Enzymol Relat Areas Mol Biol. 1988;61:107–147. doi: 10.1002/9780470123072.ch3. [DOI] [PubMed] [Google Scholar]
- Kempner E. S., Schlegel W. Size determination of enzymes by radiation inactivation. Anal Biochem. 1979 Jan 1;92(1):2–10. doi: 10.1016/0003-2697(79)90617-1. [DOI] [PubMed] [Google Scholar]
- Kepner G. R., Macey R. I. Membrane enzyme systems. Molecular size determinations by radiation inactivation. Biochim Biophys Acta. 1968 Sep 17;163(2):188–203. doi: 10.1016/0005-2736(68)90097-7. [DOI] [PubMed] [Google Scholar]
- Kim K. H., Pan Z. X., Honzatko R. B., Ke H. M., Lipscomb W. N. Structural asymmetry in the CTP-liganded form of aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1987 Aug 20;196(4):853–875. doi: 10.1016/0022-2836(87)90410-4. [DOI] [PubMed] [Google Scholar]
- Konigsberg W. H., Henderson L. Amino acid sequence of the catalytic subunit of aspartate transcarbamoylase from Escherichia coli. Proc Natl Acad Sci U S A. 1983 May;80(9):2467–2471. doi: 10.1073/pnas.80.9.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Maire M., Aggerbeck L. P., Monteilhet C., Andersen J. P., Møller J. V. The use of high-performance liquid chromatography for the determination of size and molecular weight of proteins: a caution and a list of membrane proteins suitable as standards. Anal Biochem. 1986 May 1;154(2):525–535. doi: 10.1016/0003-2697(86)90025-4. [DOI] [PubMed] [Google Scholar]
- Le Maire M., Moller J. V., Tanford C. Retention of enzyme activity by detergent-solubilized sarcoplasmic Ca2+ -ATPase. Biochemistry. 1976 Jun 1;15(11):2336–2342. doi: 10.1021/bi00656a014. [DOI] [PubMed] [Google Scholar]
- Lo M. M., Barnard E. A., Dolly J. O. Size of acetylcholine receptors in the membrane. An improved version of the radiation inactivation method. Biochemistry. 1982 Apr 27;21(9):2210–2217. doi: 10.1021/bi00538a033. [DOI] [PubMed] [Google Scholar]
- Lowe M. E., Kempner E. S. Radiation inactivation of the glycoprotein, invertase. J Biol Chem. 1982 Nov 10;257(21):12478–12480. [PubMed] [Google Scholar]
- Makino S. Interaction of proteins with amphiphilic substances. Adv Biophys. 1979;12:131–184. [PubMed] [Google Scholar]
- Margossian S. S., Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 1982;85(Pt B):55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- McGrew S. G., Inui M., Chadwick C. C., Boucek R. J., Jr, Jung C. Y., Fleischer S. Comparison of the calcium release channel of cardiac and skeletal muscle sarcoplasmic reticulum by target inactivation analysis. Biochemistry. 1989 Feb 7;28(3):1319–1323. doi: 10.1021/bi00429a056. [DOI] [PubMed] [Google Scholar]
- McIntyre J. O., Churchill P. Glucose-6-phosphate dehydrogenase from Leuconostoc mesenteroides is a reliable internal standard for radiation-inactivation studies of membranes in the frozen state. Anal Biochem. 1985 Jun;147(2):468–477. doi: 10.1016/0003-2697(85)90300-8. [DOI] [PubMed] [Google Scholar]
- McIntyre J. O., Churchill P., Maurer A., Berenski C. J., Jung C. Y., Fleischer S. Target size of D-beta-hydroxybutyrate dehydrogenase. Functional and structural molecular weight based on radiation inactivation. J Biol Chem. 1983 Jan 25;258(2):953–959. [PubMed] [Google Scholar]
- Mumby S., Pang I. H., Gilman A. G., Sternweis P. C. Chromatographic resolution and immunologic identification of the alpha 40 and alpha 41 subunits of guanine nucleotide-binding regulatory proteins from bovine brain. J Biol Chem. 1988 Feb 5;263(4):2020–2026. [PubMed] [Google Scholar]
- Nozaki Y., Schechter N. M., Reynolds J. A., Tanford C. Use of gel chromatography for the determination of the Stokes radii of proteins in the presence and absence of detergents. A reexamination. Biochemistry. 1976 Aug 24;15(17):3884–3890. doi: 10.1021/bi00662a036. [DOI] [PubMed] [Google Scholar]
- Nugent J. H. Molecular-size standards for use in radiation-inactivation studies on proteins. Biochem J. 1986 Oct 15;239(2):459–462. doi: 10.1042/bj2390459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten F., Gordy W. TEMPERATURE EFFECTS ON FREE RADICAL FORMATION AND ELECTRON MIGRATION IN IRRADIATED PROTEINS. Proc Natl Acad Sci U S A. 1960 Aug;46(8):1137–1144. doi: 10.1073/pnas.46.8.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J. A., Tanford C. The gross conformation of protein-sodium dodecyl sulfate complexes. J Biol Chem. 1970 Oct 10;245(19):5161–5165. [PubMed] [Google Scholar]
- Saccomani G., Sachs G., Cuppoletti J., Jung C. Y. Target molecular weight of the gastric (H+ + K+)-ATPase functional and structural molecular size. J Biol Chem. 1981 Aug 10;256(15):7727–7729. [PubMed] [Google Scholar]
- Schachman H. K., Pauza C. D., Navre M., Karels M. J., Wu L., Yang Y. R. Location of amino acid alterations in mutants of aspartate transcarbamoylase: Structural aspects of interallelic complementation. Proc Natl Acad Sci U S A. 1984 Jan;81(1):115–119. doi: 10.1073/pnas.81.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively J. E., Paxton R. J., Lee T. D. Highlights of protein structural analysis. Trends Biochem Sci. 1989 Jul;14(7):246–252. doi: 10.1016/0968-0004(89)90056-x. [DOI] [PubMed] [Google Scholar]
- Swillens S. Inactivation of macromolecules by ionizing radiation. Deterministic single-hit or stochastic multievent process? Biochem J. 1986 Feb 1;233(3):655–659. doi: 10.1042/bj2330655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Malathi P., Preiser H., Jung C. Y. Radiation inactivation studies on the rabbit kidney sodium-dependent glucose transporter. J Biol Chem. 1985 Sep 5;260(19):10551–10556. [PubMed] [Google Scholar]
- Tanford C., Nozaki Y., Reynolds J. A., Makino S. Molecular characterization of proteins in detergent solutions. Biochemistry. 1974 May 21;13(11):2369–2376. doi: 10.1021/bi00708a021. [DOI] [PubMed] [Google Scholar]
- le Maire M., Ghazi A., Møller J. V., Aggerbeck L. P. The use of gel chromatography for the determination of sizes and relative molecular masses of proteins. Interpretation of calibration curves in terms of gel-pore-size distribution. Biochem J. 1987 Apr 15;243(2):399–404. doi: 10.1042/bj2430399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Maire M., Rivas E., Møller J. V. Use of gel chromatography for determination of size and molecular weight of proteins: further caution. Anal Biochem. 1980 Jul 15;106(1):12–21. doi: 10.1016/0003-2697(80)90112-8. [DOI] [PubMed] [Google Scholar]
- le Maire M., Viel A., Møller J. V. Size exclusion chromatography and universal calibration of gel columns. Anal Biochem. 1989 Feb 15;177(1):50–56. doi: 10.1016/0003-2697(89)90012-2. [DOI] [PubMed] [Google Scholar]