Abstract
The crystal structure of the chimerical decamer d(CCACTAGTG)r(G), bearing a 3′-terminal ribo-guanidine, has been solved and refined at 1.8 Å resolution (R-factor 16.6%; free R-factor 22.8%). The decamer crystallizes in the orthorhombic space group P212121 with unit cell constants a = 23.90 Å, b = 45.76 Å and c = 49.27 Å. The structure was solved by molecular replacement using the coordinates of the isomorphous chimera r(GCG)d(TATACGC). The final model contains one duplex and 77 water molecules per asymmetric unit. Surprisingly, all residues adopt a conformation typical for A-form nucleic acids (C3′-endo type sugar pucker) although the all-DNA analog, d(CCACTAGTGG), has been crystallized in the B-form. Comparing circular dichroism spectra of the chimera and the corresponding all-DNA sequence reveals a similar trend of the former molecule to adopt an A-like conformation in solution. The results suggest that the preference of ribonucleotides for the A-form is communicated into the 5′-direction of an oligonucleotide strand, although direct interactions of the 2′-hydroxyl group can only be discerned with nucleotides in the 3′-direction of a C3′-endo puckered ribose. These observations imply that forces like water-mediated contacts, the concerted motions of backbone torsion angles, and stacking preferences, are responsible for such long-range influences. This bi-directional structural communication originating from a ribonucleotide can be expected to contribute to the stability of the A-form within all-RNA duplexes.
INTRODUCTION
Nucleic acids can adopt a variety of helical conformations, which are classified as A, B and Z. While a number of environmental factors, such as the water activity and counter-ion identity and concentration, are known to influence the relative stabilities of these forms, less is known about the influence of the base sequence, base modifications, or the chemical nature of the sugar units on the helices. Our group has been investigating the latter factors for some time. In the course of these studies it was shown that a single substitution of a dG•dC by a dA•T (where dA•T/T•dA or A•T/A•T means dA•dT/dT•dA) base pair (1,2) and the methylation of two cytosines (3) in a G-C-alternating DNA decamer can convert the structure from the left-handed Z-form to the right-handed A-form. We have also analyzed a number of mixed DNA/RNA molecules in order to elucidate the influence of the 2′-hydroxyl groups on the oligomeric structures (4–6). Studies based on the C/G-only sequence CCGGCGCCGG have shown that while the all-DNA analog crystallizes as a B-form duplex (7) introduction of ribose residues in the center (4) or at both termini (5) lead to an adoption of the A-form. Conversely, a structural conversion from the A-form to the B-form could be evoked by the binding of minor-groove directed drugs to a chimerical oligomer demonstrating for the first time ribonucleotides in a B-form background (8). Various other single crystal structures of hybrid duplexes (9) and of several chimeras (10–12) have been determined and all have been found to adopt the A-form. However, in none of these latter cases has the structure of the all-DNA analog been determined for comparison.
All the previously investigated chimeras contain RNA nucleotides followed on the 3′-side by DNA nucleotides, suggesting that the main incentive for the adoption of the A-form may stem from the directional interactions of the ribose O2′ with the nucleotide unit on its 3′-side. Most natural chimeras, e.g. Okazaki fragments, have a similar arrangement of 5′RNA–3′DNA residues. Recently, however, stretches of RNA have been found covalently embedded within the DNA background of the origin of replication of the human cytomegalovirus genome (13). These origins of replication therefore exhibit RNA–DNA hybrid regions bordering DNA duplexes and consequently display both 5′RNA–3′DNA and 5′DNA–3′RNA steps. Similar chimerical structures are observed in reverse splicing processes (14) and in synthetic therapeutic oligomers (15). Besides for the natural occurrence of such chimeras, it would also be interesting to determine what structural influence a ribose at the 3′-end of a helix may exert, because the findings could be consequential for the inherent stability of RNA helices. Furthermore, in instances like the Okazaki fragments, the RNA primer is preceded by a DNA strand which could be induced to adapt to the A-form helix following on the 3′-side. Finally, we are interested in oligonucleotides which may exhibit a B→A transition, which so far has eluded crystallographic investigations. Here we report the crystal structure of the decamer d(CCACTAGTG)r(G) (‘d’ denotes deoxyribonucleotides and ‘r’ stands for ribonucleotides) which has been refined at 1.8 Å resolution.
MATERIALS AND METHODS
Nucleic acid synthesis, purification, crystallization and data collection
The chimera, d(CCACTAGTG)r(G), and its all-DNA analog, d(CCACTAGTGG), were synthesized on Applied Biosystems nucleic acid synthesizers (Models 381 and 391; Foster City, CA) and purified as described (16). Screening for crystallization conditions was performed at room temperature with a 24-trial set-up (16). Crystals with round edges, lacking a well-defined morphology, appeared after 3 days when 2 mM oligomer (double-stranded concentration), 50 mM spermine tetrachloride, 5 mM MgCl2, 20 mM Na cacodylate (pH 7.0) and 10% 2-methyl-2,4-pentanediol (MPD) were vapor-equilibrated in a hanging drop with a reservoir containing 60% MPD. The same recipe yielded large, sharp-edged orthorhombic crystals after 1 week, when the amounts of all ingredients were increased 10-fold and crystallization was performed in a sitting drop. A single specimen, measuring 0.3 × 0.3 × 1.0 mm3, was mounted in a thin-walled special glass capillary with a drop of mother liquor at one end. Diffraction data were collected up to 1.8 Å resolution at room temperature on a Siemens four-circle goniostat, equipped with a multiwire proportional counter and a MacScience rotating anode, producing graphite-monochromated CuKα radiation (λ = 1.5418 Å) at 40 kV/100 mA. The data were processed with the program package XENGEN 2.0 (17) and the space group was determined as orthorhombic P212121 (a = 23.9 Å, b = 45.8 Å and c = 49.3 Å), suggesting one duplex per asymmetric unit (volume per base pair: 1347 Å3). Data were reduced with an Rsym of 3.1%. Reflections above 2σ in F (4624) were used in the structure determination and refinement.
Structure solution and refinement
The cell constants suggested a similar crystal packing to other A-DNA decamers in the orthorhombic system. The chimera r(GCG)d(TATACGC) (10) showed the closest match with the cell constants of the present decamer, and despite its quite different sequence it turned out to be a suitable model for the rotation/translation search and the subsequent refinement in X-PLOR (18), omitting the 2′-hydroxyl groups from the coordinates. Rigid body refinement, using data between 8.0 and 4.0 Å resolution and treating the whole duplex as a single unit, was used to globally readjust the molecule in the cell. The refinement cycle started with the 10 strongest reflections and gradually included all data in the resolution range. Treating all 20 nt as individual rigid bodies, the resolution was extended to 3.0 Å with an R-factor of 40.8%. Because excellent geometry and crystal packing were maintained, positional refinement was performed with all data to 2.0 Å resolution, after which the R-factor assumed a value of 38.4%. A single round of positional and temperature factor refinement reduced the R-factor to 34.1%. The sequence was now mutated to that of the present decamer, and a 2′-hydroxyl group was attached to the 3′-terminal residues. Positional and temperature factor refinement yielded an R-value of 26.5%. The 2Fo–Fc and Fo–Fc maps at this stage indicated that the terminal rG10•dC11 should have a much reduced twist angle with respect to the preceding base pair. Model building, guided by Fo–Fc ‘omit’ maps, on a Silicon Graphics Indigo 2 workstation with the molecular graphics program CHAIN (19) achieved the required adjustments, and further positional and temperature factor refinement proceeded smoothly to an R-factor of 26.3% for all reflections between 8.0 and 1.8 Å. To reduce a possible model bias, a round of simulated annealing (20) (4000–300 K, 0.0005 fs sampling intervals) was carried out, bringing the R-factor down to 25.3%. At this stage water molecules were placed into spherical peaks of the Fo–Fc difference densities with peak heights above 3σ. Waters were incorporated in four groups of 27, 22, 18 and 10 with intermittent positional and B-value refinement, yielding successive R-factors of 22.5, 20.1, 17.6 and 16.6%. All waters maintained hydrogen-bonded contacts to nucleic acid hetero atoms or other water molecules during the refinement. The solvent molecules were afterwards confirmed in Fo–Fc ‘omit’ maps, with 10 waters at a time removed from the model. All final water temperature factors were below three times the average phosphate group temperature factors with many approaching those of the nucleic acid atoms. The final model consists of one decamer duplex and 77 water molecules per asymmetric unit (Table 1). Data collection and refinement statistics are summarized in Table 1. The geometrical parameters of the duplex were extracted with the program CURVES (21) (Table 2). The final coordinates have been deposited with the Nucleic Acid Data Bank under ID code AH0011 (22).
Table 1. Crystallographic data.
| Space group | P212121 (orthorhombic) |
| Unit cell dimensions | a = 23.9 Å, b = 45.8 Å, c = 49.3 Å |
| Resolution range | 8.0–1.8 Å |
| Number of reflections (F ≥ 2σ(F)) | |
| Observed | 4634 |
| % Completeness (all data) | 87.4 |
| % Completeness (1.9–1.8 Å) | 67.2 |
| Rsym | 3.1% |
| Asymmetric unit | 1 Duplex |
| R-factors | |
| Rcryst | 16.6% |
| Rfree | 22.8% |
| Parameter/topology files | Param_nd.dna |
| Top_ndbx.dna | |
| Deviations from ideal geometry | |
| Bonds | 0.01 Å |
| Angles | 2.0° |
| Dihedral angles | 31.9° |
| Improper angles | 1.9° |
| Final model | |
| Nucleic acid atoms | 405 |
| Water oxygens | 77 |
Table 2. Helical parameters.
| Base Pair | Twist (°) | Roll (°) | Tilt (°) | Rise (Å) | Slide (Å) | Inclination (°) | x-displacement (Å) | Propeller twist (°) |
|---|---|---|---|---|---|---|---|---|
| dC1•rG20 (1) |
|
|
|
|
|
10 |
–5.1 |
–7 |
| |
31 |
1 |
–4 |
3.2 |
–2.1 |
|
|
|
| dC2•dG19 (2) |
|
|
|
|
|
16 |
–4.7 |
–8 |
| |
30 |
3 |
–1 |
3.3 |
–1.8 |
|
|
|
| dA3•T18 (3) |
|
|
|
|
|
20 |
–4.3 |
–6 |
| |
28 |
0 |
0 |
3.4 |
–2.0 |
|
|
|
| dC4•dG17 (4) |
|
|
|
|
|
21 |
–4.1 |
–11 |
| |
33 |
3 |
–3 |
3.2 |
–2.2 |
|
|
|
| T5•dA16 (5) |
|
|
|
|
|
14 |
–5.8 |
–13 |
| |
29 |
18 |
–3 |
2.9 |
–1.7 |
|
|
|
| dA6•T15 (6) |
|
|
|
|
|
9 |
–7.2 |
–15 |
| |
29 |
8 |
4 |
3.2 |
–1.9 |
|
|
|
| dG7•dC14 (7) |
|
|
|
|
|
14 |
–5.6 |
–11 |
| |
28 |
1 |
1 |
3.4 |
–2.2 |
|
|
|
| T8•dA13 (8) |
|
|
|
|
|
15 |
–5.5 |
–9 |
| |
30 |
5 |
1 |
3.4 |
–2.0 |
|
|
|
| dG9•dC12 (9) |
|
|
|
|
|
15 |
–5.9 |
–3 |
| |
23 |
7 |
–2 |
3.8 |
–1.9 |
|
|
|
| rG10•dC11 (10) |
|
|
|
|
|
10 |
–7.4 |
–2 |
| |
|
|
|
|
|
|
|
|
| Average | 29 | 5 | –1 | 3.3 | –2.0 | 14 | –5.6 | –9 |
Circular dichroism (CD) spectroscopy
The nucleic acids were dissolved in 50 mM Na cacodylate (pH 7.0), 25 mM NaCl, 2 mM MgCl2, to yield 100 µM solutions. CD spectra of the present chimera and its all-DNA analog were recorded at 10°C with a J-715 spectropolarimeter (JASCO Corp., Tokyo, Japan). For each sequence 10 spectra between 220 and 300 nm (0.2 nm stepsize) were averaged and smoothed by the manufacturer’s software. Care was taken not to introduce artifacts by the smoothing procedure.
RESULTS
Molecular structure
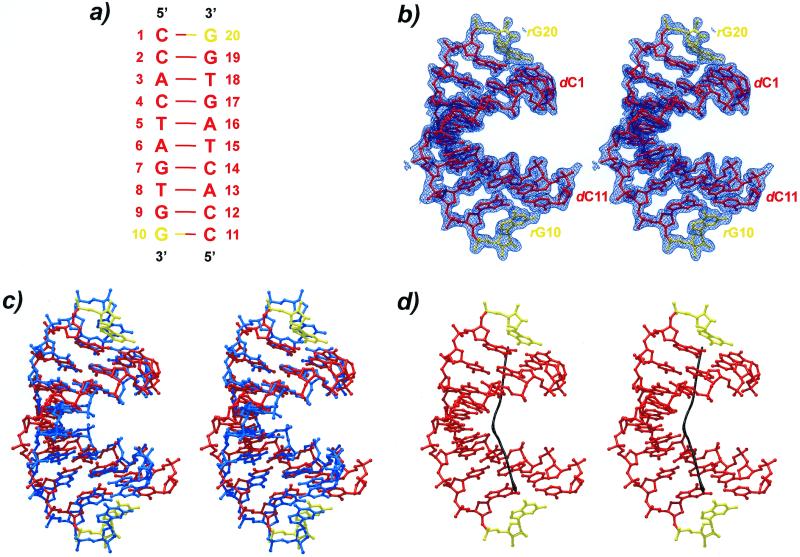
Composition and numbering of the chimerical decamer duplex are indicated in Figure 1 (RNA residues in yellow). The self-complementary molecule, d(CCACTAGTG)r(G), crystallized as a right-handed duplex with an overall A-form geometry. The structure superimposes on fiber A-DNA with an r.m.s. deviation of 1.9 Å and a mean distance of 1.7 Å between common atoms (Fig. 1), exhibiting considerable local flexibility in helical and backbone parameters. A wide and shallow minor groove results from the unusually large x-displacement of the base pairs (average –5.6 Å), leaving the major groove deep and narrow (Table 2). Unlike in many of the A-DNA single crystal structures, the base pairs are strongly inclined by an average 14.4°, approaching the fiber A-DNA value of 19–20°. The rise is high for an A-form duplex (average 3.3 Å), while the average tilt adopts a more common value (average 29°). The roll, measuring the opening of the base pair steps into one of the grooves, is positive everywhere (average 5°). The propeller twist increases monotonously from either end towards the center, averaging –9°.
Figure 1.
(a) Schematics of the sequence and numbering. DNA residues are shown in red and RNA nucleotides in yellow. (b) Stereo view of the final 2Fo–Fc electron density map at 1.8 Å resolution contoured at the 1σ-level. (c) Stereo superposition of the present structure with a fiber A-DNA model of the same sequence (blue). (d) Stereo picture of the helix axis calculated with the program CURVES (21). A strong kink is clearly visible in the center of the duplex.
The average helical parameters obscure the variability of the structure on the local level. The helix is quite underwound at the terminal (dG9-rG10)•(dC11-dC12) step, with a twist angle of 23°. At the same time, the terminal rG10•dC11 base pair exhibits a very large displacement into the shallow groove (–7.4 Å). A similarly large displacement (–7.2 Å) is found at the central dA6•T15 base pair. These distortions are correlated with the sites involved in crystal packing interactions (see below).
Bending of the duplex
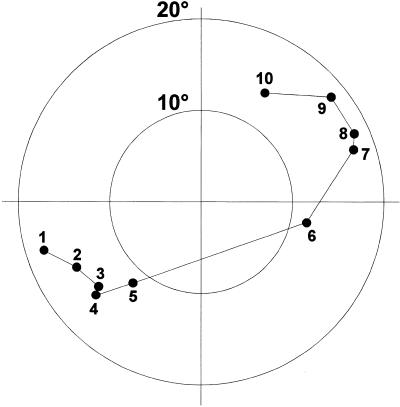
One of the major sources of difference compared to the fiber A-DNA conformation stems from a severe bending of the duplex. When comparing angles between the partial helical axes of successive portions of the chimera, a 27° kink is seen at the (T5-dA6)•(T15-dA16) step (Fig. 1). This bend becomes particularly obvious in a plot of the base pair normal vectors (Fig. 2). The position of the curve correlates with an all-trans backbone conformation about torsion angles α and γ of residue dA16, which is unmatched in the opposite strand of the duplex. As a consequence of the extended backbone conformation the distance between the pertaining consecutive phosphate groups lengthens to 6.8 Å while it is only 5.5 Å at this position in the opposite strand. At the same time we observe a large roll angle (18°) at precisely this (T5-p-dA6)•(T15-p-dA16) step (Table 2), which indicates a wide opening of the base pairs towards the minor groove. Both the uncompensated extended backbone conformation and the roll angle deviations have previously been described as sources of DNA and RNA bending (23–25). They are likely a conformational response of the oligomer to external stress in the form of packing interactions, which ultimately results in the bending of the duplex. It is conceivable that ligand interactions, e.g. in a protein–nucleic acid complex, could substitute for packing interactions and induce the bending, a basis for the indirect readout of nucleic acid sequences.
Figure 2.
Normal vector plot for the decamer duplex viewed down the helix axis. The tips of unit vectors perpendicular to the best planes through the base pairs are indicated after shifting to a common origin (for base pair numbering see Table 2). The concentric circles indicate deviations of 10° and 20° from the helical axis. The large step between base pairs 5 and 6 across the plot corresponds to the bend at the (T5-dA6)•(T15-dA16) step.
While it may be surmised that the above kinking is mostly due to packing forces acting on the duplex (25–28), the precise location probably marks a conformationally malleable spot of the duplex. Interestingly, it appears at a T-dA step, reminiscent of the bending of B-DNA duplexes at A•T-tracts (29). Bending or kinking may be facilitated at the T-dA step, because there is little stacking overlap at this position. Such ‘weak’ pyrimidine-purine steps can explain DNA roll-bending both in the A-form (30–32) as well as in the B-form (33). It has been shown that bending at the T-dA step is not an intrinsic feature of the double helix but rather induced by the environment (29,34–36).
Hydration
A total of 77 water molecules have been modeled in the present structure. The typical A-DNA packing contacts (Fig. 3) shield much of the shallow groove of the duplexes from the aqueous environment (see below). Where accessible for the solvent, N2 and N3 of the purines and O2 of the pyrimidines in the shallow groove attract water molecules as is typically observed (37–39). The preferred hydration sites at O6/N6 and N7 of the purines and O4/N4 of the pyrimidines in the deep groove are often occupied by waters. Furthermore, the anionic phosphate oxygens, in particular O1P, are heavily hydrated.
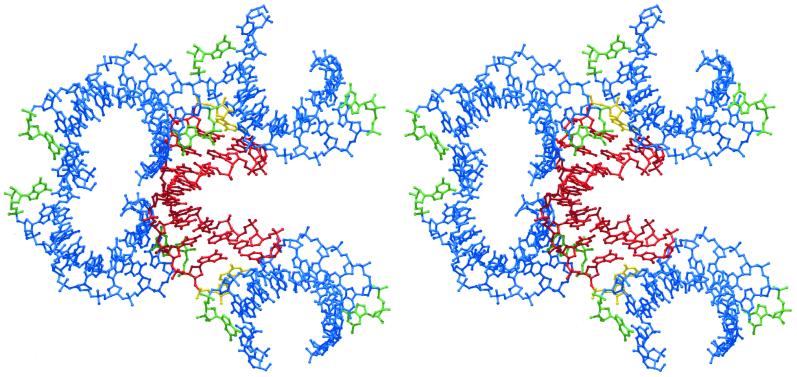
Figure 3.
Stereo view of the molecular packing arrangement in the present orthorhombic crystal structure. A reference duplex (red) receives contacts from the termini of two neighboring duplexes in its shallow groove (left) and donates via its own ends the analogous interactions to other symmetry related molecules (top and bottom; symmetry related molecules in blue). The ribonucleotides, corresponding to the 3′-ends of the strands, are shown in yellow for the reference duplex and in green for the symmetry related molecules.
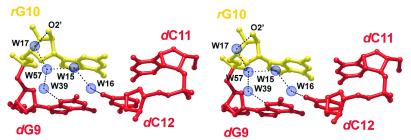
An interesting hydration feature involves one terminus of the duplex including the 2′-hydroxyl group of the pertaining rG10 residue (Fig. 4). A network of water molecules links this hydroxyl group to both the preceding nucleotide, dG9, as well as a residue in the opposite strand, dG12. Viewed from the terminal rG10 residue, the water-mediated interactions extend into the 5′-direction and may constitute one means by which the 3′-terminal ribose communicates its structural preferences to the remainder of the molecule. The equivalent sites of the opposite terminus of the duplex are occupied by packing contacts (see below) and therefore do not show a similar hydration pattern.
Figure 4.
Hydration pattern involving the terminal base pair step dG9-dG10 (and dC11-dC12). Water molecules link the pertaining 2′-hydroxyl group to residues in the same and the opposite strand. The ribonucleotide rG10 is shown in yellow and the DNA parts in red.
Packing interactions
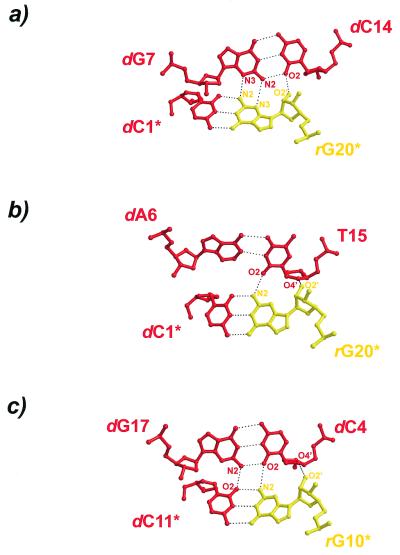
The chimerical duplexes pack in a typical A-form manner with the terminal base pairs abutting the shallow grooves of neighboring 21-screw related duplexes (Fig. 3). The main interactions occur between base pairs dG7•dC14 and (dC1•rG20)* [dG7-(dC1•rG20)* base triple, Fig. 5a], between dA6•T15 and (dC1•rG20)* [T15-(dC1•rG20)* base triple, Fig. 5b], and between dG17•dC4 and (rG10•dC11)* (base quadruple, Fig. 5a; asterisks denote symmetry related molecules). Interestingly, while one terminus, (dC1•rG20)*, engages in two base triple interactions, there is a quadruple formed at the opposite end, (rG10•dC11)*, similar to the situation in the structure of d(CCGGC)r(G)d(CCGG) (4). Although the same type of nucleotides are involved in the base multiples formed between the duplex termini (rG•dC base pairs) and base pairs dG7•dC14 (Fig. 5a) and dG17•dC4 (Fig. 5c), respectively, a triple is formed in the first case while the latter association leads to a quadruple. Therefore, small positional adjustments due to the local environment can affect the details of the nucleic acid self-interactions and packing modes.
Figure 5.
Detailed crystal packing interactions of the present chimerical decamer duplex. Both termini are involved in the formation of base multiples through the interaction with the shallow grooves of neighboring duplexes. (a and b) Base triple interactions. (c) Base quadruple. Deoxyribonucleotides are depicted in red and ribonucleotides are shown in yellow.
The indicated contacts of the 2′-hydroxyl groups (Fig. 5) are only strong in the first triple (2.5 Å distance to the dC14 O2 atom). In the latter two base multiples, contacts of the 2′-hydroxyls of rG20* and rG10* to O4′ of T15 and dC4, respectively, are borderline hydrogen bonds with distances ~3.5 Å. The involvement of the 2′-hydroxyl groups may therefore be fortuitous in the present case, indicating that their structural influence (see below) is likely not exerted through these packing contacts.
It should be noted that in the T15-(dC1•rG20)* triple only a single base–base hydrogen bond connects the two base pairs. In this case the adenine base, dA6, lacking an appropriate 2-substituent, may oppose the formation of an additional hydrogen bond to yield a quadruple and lead to a shifting of the contact to the pyrimidine, T15. However, the sugar units are possible additional latching points and in the present case a weak hydrogen bonding contact is seen to stabilize the triple interaction (O2′ of rG20* to O4′ of T15; Fig. 5b).
Conformation in solution
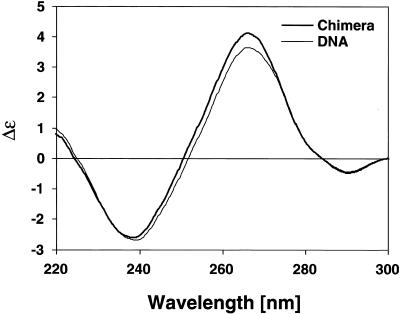
Because the crystallization conditions for the present chimera with a high spermine content and the crystal packing may strongly influence the observed conformation (see Discussion), we tried to assess the structural role of the 3′-ribonucleotide in solution and in the absence of polyamines. Figure 6 shows CD spectra between 220 and 300 nm of the present chimera and the corresponding all-DNA sequence. Although the detected differences are small, they are clearly attributable to a change in conformation rather than a concentration difference because the heights of both maximum (∼266 nm) and the minimum (∼239 nm) in the scans are shifted upward. Furthermore, the minimum of the chimera spectrum is slightly displaced towards lower wavelengths (by ~1–2 nm) and the zero line crossover is at 251 rather than 253 nm as seen for the all-DNA analog. All the trends in the amplitudes and positions and location of the crossover points are more or less indicative of a more A-like conformation for the chimera compared to the B-DNA molecule in solution. The magnitude of the effects is comparable to those seen with the incorporation of 2′-fluoro substituents into a DNA or DNA–RNA hybrid background of comparable length, which similarly favor an A-like conformation (40).
Figure 6.
CD spectra of d(CCACTAGTGG) (thin line) and the present chimera, d(CCACTAGTG)r(G) (thick line). The increase in the maximum, the shift of the zero crossover point to a lower wavelength and the reduced minimum, in combination with a slight shift of this minimum to a lower wavelength, are all suggestive of a more A-like conformation in the chimera than B-DNA.
DISCUSSION
Nucleic acid duplexes containing both DNA and RNA components are frequently encountered in vivo, for example in Okazaki fragments or during RNA synthesis and reverse transcription. Because DNA seems to prefer the B-form in solution while RNA usually adopts an A-conformation, such chimeras and hybrids face the structural dilemma of which conformation to maintain. These considerations prompted the single crystal structure determinations of chimerical and hybrid molecules. All the investigated mixed DNA/RNA molecules have been found in the A-form except for the minor groove drug-complexed chimeras (8). However, only for the sequence CCGGCGCCGG both chimeras (4,5) and the all-DNA analog (7) have been investigated. Although the DNA was found in the B-conformation, the chimeras crystallized in the A-form. These studies therefore suggested that chimerical molecules generally adopt the A-form because of the structural influence of the ribose sugars. It can be objected, however, that the tested decamers may have an inherent inclination towards the A-form, because of their G/C-rich sequence. Indeed, it came as a surprise to find d(CCGGCGCCGG) in the B-form, since it has been suggested that dG-dG steps in DNA fragments favor the A-conformation (7,41–44).
All so far investigated chimerical sequences contain ribose residues either on the 5′-end, on both termini, or in the center of the oligomer. It is known that the 2′-hydroxyl group of the ribose interacts through direct or water-mediated hydrogen bonds with residues on the 3′-side and/or with other atoms of its own nucleotide unit (45–47). Such contacts may be the prime incentive for the molecules to maintain the A-conformation. Here we observe that, upon introduction of a 3′-terminal ribose, the structure of the sequence CCACTAGTGG also switches from B (35) to A. This finding is particularly significant because the oligomer is rather rich in A/T (40%), indicating that its sequence may not be particularly prone to adopt the A-form. Assuming an active role for the ribose in the conversion process, its influence would have to be communicated into the 5′-direction of the sugar unit. Such a control by the 2′-hydroxyl groups cannot be exerted through direct interactions but may occur through the correlated torsion angles of the backbone or the base stacking as a consequence of the C3′-endo sugar pucker. One other critical mediator we observe is the water structure, which links the 2′-OH to preceding residues of both strands.
It is known that the nucleic acid conformation is dependent on the ionic environment. Therefore, the composition of the crystallization solutions may have influenced the outcome of our experiments. The crystallization set-ups for the present A-form chimera and the analogous B-form DNA contained the same ingredients at the same pH (35). However, the magnesium and spermine concentrations were approximately reversed and polyethylene glycol 200 was replaced by MPD (35). The high spermine concentrations in our mother liquor may be another reason why the A-form is observed for the chimera. However, so far it has not been possible to crystallize the same all-DNA oligonucleotide sequences both in the A-form and the B-form. It is unlikely that the crystallization solution alone is responsible for the observed effects.
For oligomers crystallized in the A-conformation the question of the influence of packing forces is particularly prevalent (25–28,32,44). In general, the observation that many sequences, including the all-DNA analog of the present structure, crystallize in the B-form while others prefer the A-form shows that certain sequences have a predisposition for the A-form. CD spectra of some chimeras suggest that the A-form can also be adopted in solution (4,5). In corroboration of these findings, comparing the CD spectra of the present chimera to those of an all-DNA molecule of the same sequence we also detected an inclination of the 3′-r(G)-molecule towards the A-conformation in solution, independent from intermolecular interactions.
Taken together, there seem to be both inherent chemical driving forces, e.g. the base sequence and the type of sugar unit, as well as environmental factors, like the ionic composition of the medium and crystal contacts, whose combined effects determine the molecular conformation of a nucleic acid duplex. We cannot quantitatively estimate the relative contributions from each individual of these factors to the stabilization of the A-form. However, we observe that the introduction of a single atom, the 2′-hydroxyl oxygen, can shift the balance in the present case toward the A-form despite an A/T content of 40%. This observation clearly suggests a structural influence for the 2′-hydroxyl group. The results of the present study therefore indicate that the A-conformation within an all-RNA helix should be particularly stable. It seems that each ribonucleotide reinforces this conformation by communicating it into both directions of the strands.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Elisabeth Weyher-Stingl for her help in acquiring the CD spectra. We gratefully acknowledge support through NIH grant GM-17378 and an Ohio Eminent Scholar Endowment to M.S. by the Board of Regents of Ohio. We also thank the Ohio Regents Hayes Investment Fund for the partial support to purchase an R-axis IIc imaging plate. M.C.W. has been sponsored by an OSU Presidential Fellowship and postdoctoral stipends from the Deutsche Forschungsgemeinschaft and the Engelhorn-Stiftung.
NDB no. AH0011
REFERENCES
- 1.Ban C., Ramakrishnan,B. and Sundaralingam,M. (1996) Biophys. J., 71, 1215–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ban C., Ramakrishnan,B. and Sundaralingam,M. (1996) Biophys. J., 72, 1222–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tippin D.B., Ramakrishnan,B. and Sundaralingam,M. (1997) J. Mol. Biol., 270, 247–258. [DOI] [PubMed] [Google Scholar]
- 4.Ban C., Ramakrishnan,B. and Sundaralingam,M. (1994) J. Mol. Biol., 236, 275–285. [DOI] [PubMed] [Google Scholar]
- 5.Ban C., Ramakrishnan,B. and Sundaralingam,M. (1994) Nucleic Acids Res., 22, 5466–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong Y. and Sundaralingam,M. (1998) Structure, 6, 1493–1501. [DOI] [PubMed] [Google Scholar]
- 7.Heinemann U., Alings,C. and Bansal,M. (1992) EMBO J., 11, 1931–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Ramakrishnan,B. and Sundaralingam,M. (1995) Nature Struct. Biol., 2, 2–4. [Google Scholar]
- 9.Horton N.C. and Finzel,B.C. (1996) J. Mol. Biol., 264, 521–533. [DOI] [PubMed] [Google Scholar]
- 10.Wang A.H.J., Fujii,S., van Boom,J.H., van der Marel,G.A., van Boeckel,S.A.A. and Rich,A. (1982) Nature, 299, 601–604. [DOI] [PubMed] [Google Scholar]
- 11.Egli M., Usman,N., Zhang,S. and Rich,A. (1992) Proc. Natl Acad. Sci. USA, 89, 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egli M., Usman,N. and Rich,A. (1993) Biochemistry, 32, 3221–3237. [PubMed] [Google Scholar]
- 13.Prichard M.N., Jairath,S., Penfold,M.E.T., Jeor,S.S., Bohlman,M.C. and Pari,G.S. (1998) J. Virol., 72, 6997–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J., Zimmerly,S., Perlman,P.S. and Lambowitz,A.M. (1996) Nature, 381, 332–335. [DOI] [PubMed] [Google Scholar]
- 15.Cole-Strauss A., Yoon,K., Xiang,Y., Byrne,B.C., Rice,M.C., Gryn,J., Holloman,W.K. and Kmiec,E.B. (1996) Science, 273, 1386–1388. [DOI] [PubMed] [Google Scholar]
- 16.Wahl M.C., Ramakrishnan,B., Ban,C.G., Chen,X. and Sundaralingam,M. (1996) Acta Crystallogr., D52, 668–675. [DOI] [PubMed] [Google Scholar]
- 17.Howard A.J., Nielsen,C. and Xuang,N.H. (1985) Methods Enzymol., 144, 211–237. [Google Scholar]
- 18.Brünger A.T. (1992) X-PLOR – A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- 19.Sack J. and Quiocho,F.A. (1992) CHAIN – Crystallographic Modeling Program. Baylor College of Medicine, Houston, TX.
- 20.Brünger A.T. (1988) In Isaacs,N.W. and Taylor,M.R. (eds), Crystallographic Computing 4: Techniques and New Technologies. Clarendon Press, Oxford, UK, pp. 126–140.
- 21.Lavery R. and Sklenar,H. (1988) J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- 22.Berman H.M., Olson,W.K., Westbrook,J., Gelbin,A., Demeny,T. and Beveridge,D. (1994) The Nucleic Acid Data Base: A Guide to the Use of a Relational Database of Nucleic Acid Crystal Structures. Center for Computational Chemistry, Rutgers University, New Brunswick, NJ and Wesleyan University, Middletown, CN.
- 23.Haran T.E., Shakked,Z., Wang,A.H.-J. and Rich,A. (1987) J. Biomol. Struct. Dyn., 5, 199–217. [DOI] [PubMed] [Google Scholar]
- 24.Dock-Bregeon A.C., Chevrier,B., Podjarny,A., Johnson,J., deBear,J.S., Gough,G.R., Gilham,P.T. and Moras,D. (1989) J. Mol. Biol., 209, 459–474. [DOI] [PubMed] [Google Scholar]
- 25.Ramakrishnan B. and Sundaralingam,M. (1993) Biochemistry, 32, 11458–11468. [DOI] [PubMed] [Google Scholar]
- 26.Shakked Z., Guerstein-Guzikevich,G., Eisenstein,M., Frolow,F. and Rabinovich,D. (1989) Nature, 342, 456–460. [DOI] [PubMed] [Google Scholar]
- 27.Heinemann U. (1991) J. Biomol. Struct. Dyn., 8, 801–811. [DOI] [PubMed] [Google Scholar]
- 28.Ramakrishnan B. and Sundaralingam,M. (1993) J. Biomol. Struct. Dyn., 11, 11–26. [DOI] [PubMed] [Google Scholar]
- 29.Goodsell D.S., KaczorGrzeskowiak,M. and Dickerson,R.E. (1994) J. Mol. Biol., 239, 79–97. [DOI] [PubMed] [Google Scholar]
- 30.Shakked Z., Rabinovich,D., Cruse,W.B.T., Egert,E., Kennard,O., Sala,G., Salisbury,S.A. and Viswamitra,M.A. (1981) Proc. R. Soc. Lond. B, 213, 479–487. [DOI] [PubMed] [Google Scholar]
- 31.Shakked Z., Rabinovich,D., Kennard,O., Cruse,W.B.T., Salsibury,S.A. and Viswamitra,M.A. (1983) J. Mol. Biol., 166, 183–201. [DOI] [PubMed] [Google Scholar]
- 32.Jain S. and Sundaralingam,M. (1989) J. Biol. Chem., 264, 12720–12784. [Google Scholar]
- 33.Goodsell D.S., Kopka,M.L., Cascio,D. and Dickerson,R.E. (1993) Proc. Natl Acad. Sci. USA, 90, 2930–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain S., Zon,G. and Sundaralingam,M. (1991) Biochemistry, 30, 3567–3576. [DOI] [PubMed] [Google Scholar]
- 35.Shakked Z., Guzikevich-Guerstein,G., Frolow,F., Rabinovich,D., Joachimiak,A. and Sigler,P.B. (1994) Nature, 368, 469–473. [DOI] [PubMed] [Google Scholar]
- 36.Guzikevich-Guerstein G. and Shakked,Z. (1996) Nature Struct. Biol., 3, 32–37. [DOI] [PubMed] [Google Scholar]
- 37.Westhof E. (1987) Annu. Rev. Biophys. Biophys. Chem., 17, 125–144. [DOI] [PubMed] [Google Scholar]
- 38.Westhof E. (1988) Int. J. Biol. Macromol., 9, 185–192. [Google Scholar]
- 39.Westhof E., Dumas,P. and Moras,D. (1988) Biochimie, 70, 145–165. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt S., Niemann,A., Krynetskaya,N.F., Oretskaya,T.S., Metelev,V.G., Suchomlinov,V.V., Shabarova,Z.A. and Cech,D. (1992) Biochim. Biophys. Acta, 1130, 41–46. [DOI] [PubMed] [Google Scholar]
- 41.McCall M., Brown,T. and Kennard,O. (1985) J. Mol. Biol., 183, 385–396. [DOI] [PubMed] [Google Scholar]
- 42.Lauble H., Frank,R., Blöcker,H. and Heinemann,U. (1988) Nucleic Acids Res., 16, 7799–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frederick C.A., Quigley,G.J., Teng,M.-K., Coll,M., van der Marel,G.A., van Boom,J.H., Rich,A. and Wang,A.H.-J. (1989) Eur. J. Biochem., 181, 295–307. [DOI] [PubMed] [Google Scholar]
- 44.Wahl M.C. and Sundaralingam,M. (1997) Biopolymers, 44, 45–63. [DOI] [PubMed] [Google Scholar]
- 45.Portmann S., Usman,N. and Egli,M. (1995) Biochemistry, 34, 7569–7575. [DOI] [PubMed] [Google Scholar]
- 46.Wahl M.C., Ban,C., Sekharudu,C., Ramakrishnan,B. and Sundaralingam,M. (1996) Acta Crystallogr., D52, 655–667. [DOI] [PubMed] [Google Scholar]
- 47.Biswas R., Wahl,M.C., Ban,C. and Sundaralingam,M. (1997) J. Mol. Biol., 267, 1149–1156. [DOI] [PubMed] [Google Scholar]