Abstract
The neurodevelopmental disorder known as Rett syndrome has recently been linked to the methyl-CpG-binding transcriptional repressor, MeCP2. In this report we examine the consequences of these mutations on the function of MeCP2. The ability to bind specifically to methylated DNA and the transcription repression capabilities are tested, as well as the stability of proteins in vivo. We find that all missense mutations (R106W, R133C, F155S, T158M) within the methyl-binding domain impair selectivity for methylated DNA, and that all nonsense mutations (L138X, R168X, E235X, R255X, R270X, V288X, R294X) that truncate all or some of the transcriptional repression domain (TRD) affect the ability to repress transcription and have decreased levels of stability in vivo. Two missense mutations, one in the TRD (R306C) and one in the C-terminus (E397K), had no noticeable effects on MeCP2 function. Together, these results provide evidence of how Rett syndrome mutations can affect distinct functions of MeCP2 and give insight into these mutations that may contribute to the disease.
INTRODUCTION
Mutations in the transcriptional repressor and methyl-CpG-binding protein, MeCP2, have recently been proposed to contribute to the neurodevelopmental disorder known as Rett syndrome, one of the most common causes of mental retardation in young females (1–5). This disease typically involves a period of apparently normal growth and development (until the age of 6–18 months), followed by regression with loss of speech and purposeful hand use (6). Other characteristics of Rett syndrome include autism, ataxia, and stereotypic hand movements, such as hand-washing and wringing. MeCP2 has been reported to be required for early embryonic development in mice, and the abundance of MeCP2 protein in the brain, as compared to other somatic tissues, gives support to the idea that mutations in MeCP2 may affect human development, resulting in a neurological phenotype (7).
MeCP2 belongs to a family of methyl-CpG-binding proteins all of which contain a conserved methyl-CpG-binding domain (MBD) (8). The MBD can bind methyl-CpG dinucleotides and MeCP2 is capable of binding selectively to a single methylated CpG dinucleotide. The relevance of this interaction is becoming increasingly more apparent. Methylation of CpGs is the major modification in vertebrate genomes that leads to stable alterations in gene expression during development (9–11). A tight correlation has already been established between repression of transcription and CpG methylation. Methylated DNA templates, when introduced into Xenopus oocytes and mammalian cells, become strongly repressed while unmethylated ones do not (12,13).
MeCP2 confers repression through its transcriptional repression domain (TRD), which may repress transcription by several mechanisms. MeCP2 has been shown to cofractionate with Sin3 and histone deacetylases and is likely to be involved in recruiting them to methylated promoters (14,15). MeCP2 may also make direct contacts with the basal transcription machinery, such as TFIIB, leading to repression in a deacetylase-independent manner (16,17). Indirect pathways of repression by MeCP2 could also exist, including inhibition of the binding of other transcription factors to methylated promoters already occupied by MeCP2.
Mutations in MeCP2 corresponding to Rett syndrome have been identified in 76% of the sporadic cases and in 45% of familial cases of the disease (6). Most of the first mutations identified were missense mutations located within the MBD (1). It was later discovered, however, that Rett patients commonly carried mutations located within the TRD and C-terminus as well, including nonsense and deletion mutations (2–5). Correlation between mutation type and severity of phenotype has been unclear, however, there is some evidence to suggest that nonsense mutations resulting in early truncation of the protein and larger deletions are more likely to be lethal earlier than missense mutations (4). Other evidence, such as that from studies on milder variants of Rett syndrome, shows that similar mutations may contribute to Rett syndrome with differing severity (18). Additionally, it is unclear what is contributing to Rett syndrome in the 25–50% of patients who have apparently normal MeCP2, although there is some speculation that modifier loci and the untranslated regions of MeCP2 may be involved (6,18).
In this work we investigate the consequences of various mutations associated with Rett syndrome on the function of the MeCP2 protein, including its ability to bind DNA and repress transcription. Previously we have investigated four point mutations within the MBD of Xenopus MeCP2 (xMeCP2) and discovered that three of the mutations completely inhibited the selective binding to methylated DNA, while the fourth only moderately inhibited binding (19). Here we examine the effects of the same missense mutations within the MBD of human MeCP2 (hMeCP2), as well as missense mutations outside the MBD and a series of nonsense mutations. The binding properties and transcriptional repression capabilities of hMeCP2 with Rett syndrome mutations are examined and may provide insight into how these mutations lead to the development of this disorder.
MATERIALS AND METHODS
Construction of mutants
The cDNA of hMeCP2 was cloned from human leukemia CCRF cells by RT–PCR and sequenced. All constructs used in this study were generated from this hMeCP2 construct. Point mutations and truncations were introduced by PCR, and the mutant and wild-type (WT) MeCP2 DNAs were subcloned into the NdeI/EcoRI sites of pTYB1 (New England Biolabs) as chitin-binding protein fusions and sequenced. The Gal4 DNA-binding domain (DBD) and cloning sites were isolated by PCR from pMSIIGal4 (14) and inserted into the PmeI sites of pcDNA3.1 (Invitrogen). WT and mutant MeCP2s were digested from pTYB1 and inserted downstream of the Gal4 DBD.
Recombinant protein purification
Recombinant WT and mutant MeCP2s were expressed in Escherichia coli BL21(DE3) and induced with 0.5 mM IPTG at 16°C overnight. Extracts were prepared by resuspending the bacteria in column buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl) supplemented with 0.1% Triton X-100 and Complete protease inhibitors (Roche Molecular Biochemicals), followed by sonication and centrifugation at 12 000 g for 30 min. Extracts were bound to chitin beads (New England Biolabs) and were washed three times with column buffer. Fusions were cleaved on the column overnight with column buffer supplemented with 50 mM DTT. Eluted fraction were pooled and dialyzed with column buffer plus 10% glycerol.
Southwestern assay
The southwestern procedure is based on a protocol previously described (20). Equal molar amounts of WT and mutant proteins were resolved by 4–20% SDS–PAGE and transferred at 20 V overnight to PVDF membranes. After transfer, immobilized proteins were denatured for 5 min in binding buffer (20 mM HEPES pH 7.5, 3 mM MgCl2, 40 mM KCl and 10 mM 2-mercaptoethanol) containing 6 M guanidine hydrochloride, followed by four successive 2-fold dilutions with binding buffer only. After two additional washes with binding buffer alone, the filters were pre-blocked for 10 min with 2% non-fat dried milk in binding buffer and washed again with binding buffer alone. The filters were incubated for 1 h at room temperature in the presence of 32P-labeled oligonucleotides [GAC12 5′-GATCC(GAC)11GATC-3′ or GAM12 5′-GATCM(GAM)11GATC-3′] (21) in binding buffer (~2 × 106 c.p.m. ml–1), together with 0.1% Triton X-100 and non-specific competitor DNA (20 µg/ml native E.coli DNA, 2 µg/ml denatured E.coli DNA). Finally, the filters were washed four times in binding buffer supplemented with 0.01% Triton X-100. After air-drying for 5 min, the filters were exposed to X-OMAT film (Kodak). Following the southwestern assays, the filters were probed with MeCP2 antibodies raised against the MBD of Xenopus MeCP2 (a kind gift from P. L. Jones, NIH, Bethesda, MD).
Transcription assays in Xenopus oocytes
Xenopus stage VI oocytes were prepared and maintained in MBSH as previously described (22). Constructs containing the Gal4 fusions were linearized with StuI at ∼1 kb downstream of the cloning site and 5′-capped mRNAs were transcribed in vitro from the T7 promoter using the mMessageMachine kit (Ambion). RNAs were then polyadenylated in vitro using E.coli poly-A polymerase (Life Technologies) and subsequently used for oocyte injections. WT and mutant RNAs were injected into the cytoplasm of 30–40 oocytes each, and 4 h later a mix containing 500 ng of G5-HSVtk-CAT and 50 ng of CMV-CAT DNA was injected into the nuclei (14). After 16 h, the oocytes were collected and homogenized in 20 mM Tris–HCl pH 8.0 using 10 µl per oocyte. A portion of the homogenate was removed, centrifuged at 14 000 g for 15 min, and the clarified extract used in western blotting. RNA was isolated from the remaining homogenate using RNAzol (Cinna Scientific).
Extension reactions were made as previously described (14). A 30mer oligonucleotide (5′-CCACCATATAGGTCACTAAAAAAAGAGGTA-3′) complementary to CAT mRNA was used for the CMV-CAT and G5-HSVtk-CAT reporters. An H4 primer (5′-GAGGCCGGAGATGCGCTTGAC-3′) that anneals to the endogenous H4 transcript was used as an internal control for RNA recovery and loading. RNA corresponding to three oocytes were mixed with 1 pmol each of 32P-labeled primer and annealed at 65°C (10 min), 55°C (30 min) and 37°C (20 min). Following annealing, extension products were transcribed at 42°C for 1 h using SuperScript II-RT (Life Technologies) and were separated on 6% polyacrylamide sequencing gels, visualized by autoradiography and quantitated using ImageQuant (Molecular Dynamics).
Cell culture and transfection
The human kidney cell line, Bosc 23, was obtained from ATCC and maintained in DMEM supplemented with 10% FCS at 37°C with 5% CO2. Transient transfections were made using FUGENE (Roche Molecular Biochemicals) transfection reagent following manufacturer’s protocol. WT and mutant Gal4–hMeCP2 fusions were cotransfected with G5-HSVtk-CAT, and a CMV-βGAL construct was transfected in parallel to determine transfection efficiency. At 48 h after transfection, cells were washed twice in 1× PBS and harvested. A portion of the cells was removed and placed in 2× SDS loading buffer for western blotting. RNA was isolated from the remaining cells using RNAzol. Total RNA was quantitated by UV spectrophotometry and equal amounts were used for primer extension reactions, as described above.
Protein degradation assay
Gal4–hMeCP2 fusion RNAs were injected into the cytoplasm of 40 oocytes and translation was allowed to proceed for 16 h. Oocytes were then placed in MBSH containing 50 µg/ml of cycloheximide as previously reported to arrest translation (23). Six oocytes were removed at the indicated time points and extracts were made, resolved on 4–20% SDS–PAGE and western blotting performed using anti-Gal4 DBD antibodies (RK5C1-Santa Cruz). This experiment was repeated three times, and results were scanned, quantitated using ImageQuant and averaged.
RESULTS
Rett syndrome mutations in hMeCP2
MeCP2 is the archetypical methyl-CpG binding protein in vertebrates and previous work has identified two distinct functional domains, MBD and TRD (8). Although it is not known if there exists a third functional domain, the C-terminus has been implicated in contributing to enhanced DNA binding (24). Mutations that affect MeCP2 function, therefore, are likely to occur in one of these three regions.
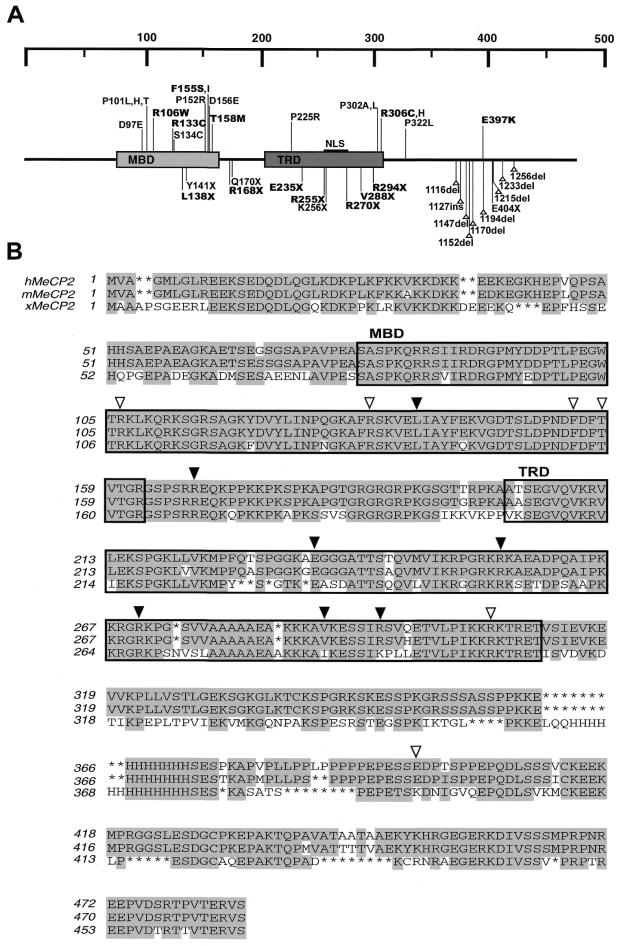
Combined data from different groups investigating Rett syndrome show that there is in fact a wide range of mutations occurring in MeCP2 located in these distinct regions (Fig. 1A) (1–5). Missense mutations predominantly cluster around the MBD, which is highly conserved among methyl-CpG binding proteins. Nonsense mutations center around the TRD, whereas deletion mutations are confined to the C-terminus. Eight of the nonsense mutations delete some or all of the nuclear localization signal (NLS) and are not expected to be actively transported to the nucleus (4–5).
Figure 1.
Schematic representation and sequence alignment of MeCP2 with Rett syndrome mutations identified. (A) MeCP2 with its two functional domains MBD and TRD indicated. Rett syndrome associated mutations found to date are marked, with missense mutations located on the top, and nonsense and deletion/insertion mutations on the bottom. Mutations used in this study are in bold type. The NLS is marked. (B) Sequence alignment of MeCP2 from human (h), mouse (m) and Xenopus (x). Conserved residues are shaded and mutations used in this study are marked by open triangles for missense mutations and closed triangles for nonsense mutations.
Sequence alignment of MeCP2 of human, mouse and Xenopus MeCP2 proteins show a high degree of conservation throughout most of the MeCP2 molecule (Fig. 1B). Most of the mutations are found at conserved residues, however, in some Rett syndrome cases, the E397 residue is mutated to a lysine, the residue found in Xenopus MeCP2 (2). Because of this, and the fact that this mutation is not within the MBD or TRD suggests that it may not contribute to loss of MeCP2 function and to Rett syndrome. To investigate the functional significance of Rett syndrome mutations in MeCP2, we focused on exploring the effects of these mutations on methyl-CpG binding, the ability to repress transcription and protein stability in vivo. Dissection of the MBD of MeCP2 in earlier studies has shown that the MBD alone is sufficient for methylated DNA binding, and minor deletions of the MBD prevented its binding to methylated sequences (25). Random point mutations within the MBD of MBD1 have also been found to inhibit methylation specific binding (26).
Rett mutations in the MBD of hMeCP2 abolish its selectivity for methylated DNA
Previously we have shown that three Rett mutations in the MBD of Xenopus MeCP2 (R106W, R133C, F155S) completely disrupt its ability to bind specifically to methylated DNA while a fourth (T158M) only moderately affects binding (19). To determine if the Rett mutations in the MBD of human MeCP2 affect binding to methylated DNA, Rett mutations were introduced into hMeCP2 and recombinant proteins were expressed and purified. Four missense mutations within the MBD (R106W, R133C, F155S, T158M), one in the TRD (R306C), and another just outside the TRD (E397K) were generated (Fig. 1B, open triangles), as well as seven nonsense mutations which truncate the protein at various sites, one within the MBD, one between the MBD and TRD, and five within the TRD (Fig. 1B, closed triangles).
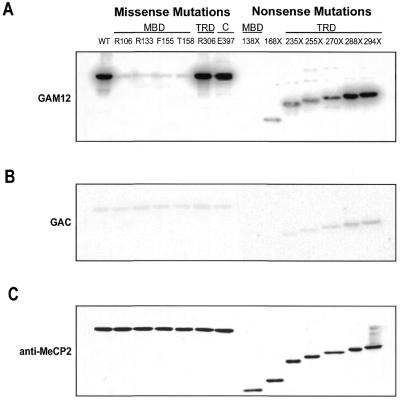
Binding to methylated DNA was determined by southwestern analysis using unmethylated (GAC12) and methylated (GAM12) oligonucleotides. In contrast to the earlier studies using Xenopus MeCP2, all of the four missense mutations within the MBD of hMeCP2 completely abolished selectivity for methylated DNA, while missense mutations outside of the MBD did not (Fig. 2A). Binding of these mutant MeCP2 proteins to the methylated GAM12 probe was similar to that observed on the unmethylated GAC12, suggesting that the binding to the methylated probe was due to non-specific interactions, which occur on unmethylated DNA as well. This suggests that selectivity for methylated DNA is lost in the proteins, not DNA binding altogether. Furthermore, only the truncation that occurred in the MBD (L138X) was unable to bind DNA, while all of the other nonsense mutations retained the ability to bind. Binding was enhanced for the longer proteins, particularly V288X and R294X, both of which showed binding comparable to WT, even though they are missing the entire C-terminal portion of the protein. This suggests that the C-terminus may not be involved in facilitating DNA binding, but possibly in protein stability. Furthermore, it is likely that the region between the MBD and the end of the TRD (near residues V288 and R284) is necessary for enhancing DNA binding. Most mutant proteins showed slight binding to unmethylated GAC12 probes, however, unmethylated binding was almost undetectable for the smaller truncations (R168X, E235X and partially R255X) (Fig. 2B). This suggests that non-specific binding may require the region between the MBD and residue R255, and confirms previous work demonstrating that the regions flanking the MBD are responsible for non-specific DNA interactions (21). Western analysis of the blots following southwestern showed equal amounts of protein (Fig. 2C). The solution structure of the MBD of MeCP2 and MBD1 predicts that these mutations affect residues that are critical for making direct contacts with the major groove of the DNA. Whether or not these residues are in direct contact with methyl groups on methylated DNA is not known. It is not surprising, therefore, that in our experiments mutations at these sites completely abolish methylated DNA binding specificity. It is unclear, however, why partial binding occurs for the T158M mutation for Xenopus MeCP2.
Figure 2.
Analysis of Rett syndrome mutations of MeCP2 on its affinity for methylated DNA. MeCP2 proteins with Rett syndrome mutations were analyzed for their binding to methylated or unmethylated oligonucleotide probes by southwestern analysis. (A) Binding to methylated GAM12 probes was lost when missense or nonsense mutations were in the MBD. Missense mutations outside the MBD did not affect binding, and binding for nonsense mutations outside the MBD occurred and increased with increase in protein size. (B) Non-specific binding to unmethylated GAC12 probes was apparent for all proteins except for two nonsense mutations that resulted in the smallest MeCP2 proteins (138X and 168X). (C) Western blotting of membranes following southwestern with anti-MeCP2 antibodies shows equal amounts of proteins.
Rett deletions of the TRD of hMeCP2 abolish its ability to repress transcription
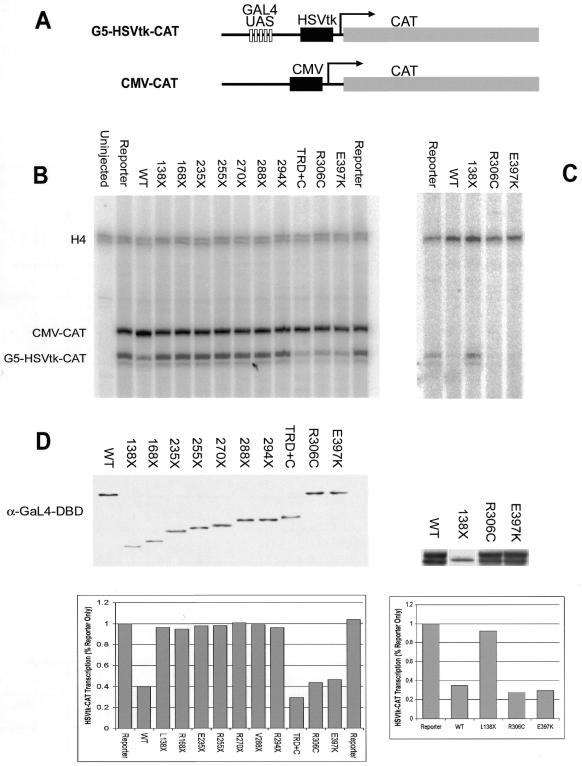
To investigate the effects of Rett syndrome mutations in MeCP2 on its ability to repress transcription we used Xenopus oocytes. This system has previously been shown to be useful in investigating transcriptional properties of MeCP2 (14). To test the ability to repress transcription independent of DNA binding, Rett hMeCP2 DNAs were fused to the Gal4 DBD and analyzed for their ability to repress transcription from a promoter containing Gal4 binding sites. RNA of the Gal4–hMeCP2 fusions were injected into oocytes, followed by a second nuclear injection of a reporter containing five Gal4 binding sites upstream of an HSVtk promoter (G5-HSVtk-CAT) (Fig. 3A). A second reporter that does not contain Gal4 binding sites (CMV-CAT) was also injected to monitor non-specific repression as well as transcription efficiency of the oocytes. At 16 h after injection of the reporter constructs, RNA was extracted. Transcription of the reporters was measured by primer extension using a CAT probe that yields two distinct size products for each reporter. A histone H4 primer was also used to control for RNA loading.
Figure 3.
Analysis of Rett syndrome mutations of MeCP2 and its ability to repress transcription. MeCP2 constructs containing Rett syndrome mutations were fused to the Gal4 DBD and analyzed for their ability to repress transcription in Xenopus oocytes. (A) Schematic representation of reporter constructs used in these analyses, one containing five Gal4 upstream activation sequences (G5-HSVtk-CAT) and another without (CMV-CAT). Oocytes were injected with RNA of the Gal4–hMeCP2 constructs, followed by a mix of both reporters. RNA from the oocytes was extracted and expression of the reporters analyzed by primer extension. (B) Primer extension using primers against CAT show changes in expression from the G5-HSVtk-CAT reporter. Oocytes containing the reporters only (‘Reporter’) show basal levels of transcription. In the presence of WT MeCP2 or the TRD+C of MeCP2, expression of G5-HSVtk-CAT is repressed. All nonsense mutations in MeCP2 failed to repress transcription. Missense mutations in the TRD (R306C) or C-terminus (E397K) repressed transcription at levels comparable to WT. Activity from the CMV-CAT promoter remained unchanged in the presence of MeCP2 and was used as a control for oocyte transcription ability and injection efficiency. RNA loading and integrity was monitored using a primer against endogenous H4. (C) The ability of R306C and E397K to repress transcription was confirmed by transient transfections into human Bosc 23 cells. Gal4–MeCP2 constructs were co-transfected with G5-HSVtk-CAT and RNA was analyzed by primer extension. (D) Western analyses of oocyte or cell extracts using anti-Gal4 DBD antibodies show relative levels of Gal4–MeCP2 proteins. Results of the primer extensions were counted and represented in histograms.
Analysis of the primer extension products reveals that the basal transcription seen in the oocytes without any MeCP2 (reporter only) is repressed in the presence of WT Gal4-MeCP2, and not with the truncation constructs (Fig. 3B). All of the Gal4 nonsense mutations, including the R294X mutation, which has only six amino acids missing from its TRD, fail to cause repression of the reporter containing the Gal4 binding sites. The TRD and C-terminus (TRD+C) caused the greatest amount of repression, while R306C and E397K caused repression at levels comparable to WT. Additionally, one MBD missense mutation (R106W) was also tested and, as expected, showed repression comparable to WT (data not shown). Although repression of the reporter only reaches ~3-fold, we believe this is significant based on previous experiments with this system (14). These results suggest that the missense mutations that contribute to Rett syndrome do not affect the ability of these MeCP2 proteins to cause repression.
Because of differences between the human and Xenopus systems, and to confirm that these missense mutations can repress in a human system, transient transfections were made using the same Gal4 constructs and reporter. The WT, L138X, R306C and E397K constructs were cotransfected into the human kidney Bosc 23 cell lines. At 48 h post-transfection, total RNA was extracted and analyzed by primer extension. As seen in Xenopus oocytes, the WT and missense mutations repressed transcription of the reporter, while the L138X mutation did not (Fig. 3C). Western blotting of the oocytes and cell extracts show relative protein levels and quantitation of the above results are provided (Fig. 3D).
These results show that the nonsense mutations within the MBD and more importantly those within the TRD fail to repress transcription. The two missense mutations (R306C and E397K), however, can repress transcription and have thus far behaved similarly to the wild-type hMeCP2. The corepressor Sin3 has been previously shown to bind to various sites within the MeCP2 molecule and can interact with MeCP2 in the absence of the TRD (15). It would therefore be surprising for a single missense mutation to affect corepressor interactions and its ability to repress transcription.
The C-terminus of hMeCP2 is required for protein stability
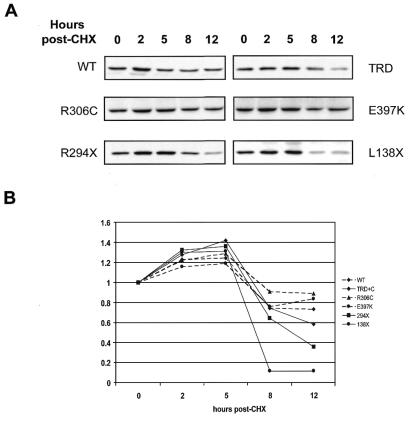
During the transcription assays in oocytes, it became apparent that more RNA for the nonsense mutations had to be injected than the full-length constructs to obtain the same protein levels, raising the possibility of a role for the C-terminus of hMeCP2 in protein stability. It is likely that the truncated hMeCP2 proteins may be degraded faster in vivo, contributing to their loss of function. To investigate this, the Rett hMeCP2 constructs that were used in the transcription assays were also used to analyze the rates of protein degradation in oocytes. Three times more RNA was injected into the oocytes than for the transcription assays, and translation was allowed to proceed for 16 h. The oocytes were then placed in buffer containing cyclohexamide, which has been previously shown to arrest translation (23), and oocytes were collected at various time points after. Protein levels were monitored by western analysis. The shortest truncation (L138X) and the longest truncation (R294X) were tested, as well as the two missense mutations outside of the MBD (R306C, E397K) and the WT.
Western analyses indicated that there is a decrease in stability for the truncated proteins (Fig. 4). The smallest deletion showed the largest decrease in protein levels, falling to almost 10% of the initial amount by 8 h, with rapid degradation after 5 h. The longest truncation, R294X, was also less stable than full-length protein, falling below 40% of the initial amount by 12 h. The TRD+C showed some degradation, falling to 60% of starting levels by 12 h. The full-length proteins (WT, R306C, E397K), however, maintained levels between 70 and 80% of the initial amount, with little degradation by 12 h. This indicates that truncated MeCP2 proteins are being degraded faster than the full-length constructs and that the TRD+C also experience some degradation. No decrease in stability was observed for proteins with missense mutations (R306C and E397K).
Figure 4.
Effects of mutations in MeCP2 on its stability in vivo. RNA from MeCP2 constructs were injected into Xenopus oocytes and allowed to translate. Oocytes were then placed in buffer containing cyclohexamide, in order to arrest protein translation, and collected at various time points after. Extracts were prepared and protein levels monitored by western blotting. (A) Western blots at indicated time points show full-length proteins (WT, R306C and E397K) have the greatest stability at 12 h, while L138X becomes largely degraded between 5 and 8 h. R294X shows moderate degradation as does the TRD+C. (B) The experiment was repeated twice and results were counted, averaged and graphed.
DISCUSSION
The discovery that Rett syndrome may be caused by mutations in the methyl-CpG-binding protein, MeCP2, has prompted a search for a better understanding of how methylation contributes to normal developmental process (1–5). Understanding how mutations in MeCP2 alter its ability to act as a transcriptional repressor helps in determining how MeCP2 functions normally, and may provide strategies for therapeutic intervention. In this work, we investigate the functional consequences of mutations in MeCP2 that are associated with Rett syndrome by examining methyl-CpG-binding, transcriptional repression capabilities and protein stability in vivo. It was expected that the mutations tested would affect at least one of these functions, however, it is equally important to see if any of the mutations do not.
Binding to methylated CpG dinucleotides is a critical function for MeCP2, as for other MBDs (27). All missense mutations found within the MBD of MeCP2 (R106W, R133C, F155S, T158M), as well as one nonsense mutation (L138X), abrogated methylation specific binding to the DNA template. Loss of this specificity can have several consequences. Target genes may not get repressed by the MeCP2 mutants due to lack of DNA binding, and MeCP2 may have a greater tendency to bind unmethylated sequences in a non-specific manner, thereby altering normal gene expression. It has been previously shown that MeCP2 does not require DNA binding to interact with corepressors (14–15). MeCP2 that is unable to bind DNA could interact with corepressor complexes causing indirect effects. Further binding studies of the Rett syndrome mutants to nucleosomal DNA and a more detailed analysis of corepressor interactions would be necessary to determine if these effects occur.
A second major function for MeCP2 is in repression of transcription mediated by its TRD. We analyzed a series of nonsense mutations (L138X, R186X, E235X, R255X, R270X, V288X, R294X) and two missense mutations, one inside the TRD (R306C) and one in the C-terminus (E397K) for their ability to repress transcription. All of the nonsense mutations that resulted in premature truncations were unable to repress transcription. In contrast, both missense mutations retained their ability to repress transcription comparable to WT. Because patients with Rett syndrome have been found without carrying mutations in MeCP2, it is possible that these missense mutations are not true mutations, but rather polymorphisms. It is difficult to conceive of a way that the E397K mutation in particular, which changes a non-conserved residue found in the human sequence to one in the Xenopus sequence, could contribute to loss of function. In fact others have speculated that this mutation is not a disease causing mutation (2). Further work needs to be done to identify what other factors could lead to Rett syndrome, as well as what MeCP2 complexes exist, particularly in different cell types in the brain.
The nonsense mutations, which are able to bind methylated DNA yet are unable to repress transcription, may also have indirect effects. The decrease in stability of the nonsense mutations, questions, however, if these truncated proteins survive long enough to cause any secondary changes.
It has been suggested that a correlation between mutation and disease severity exists, and that mutations that truncate most of the protein are lethal at an early age (4). If true, it is likely that complete loss of MeCP2 function contributes to lethality, and partial loss yields a range of phenotypic severity. Until more Rett patients are screened and a true correlation between the mutation type and phenotype is proven, it is difficult to know the effects of each mutation or if they contribute differently to the clinical outcome of the disease. Mutations outside the coding region of MeCP2, such as within the 3′-UTR, could have an effect on MeCP2 expression and localization, and may also contribute to Rett syndrome (6). MeCP2 contains an evolutionarily conserved 3′-UTR which exhibits alternative polyadenylation dependent on tissue type and developmental stage (28). Knowing how methyl-CpG-binding proteins, like MeCP2, function in various regions of the brain, as well as the identification of target genes, is needed to better understand how MeCP2 contributes to Rett syndrome.
REFERENCES
- 1.Amir R.E., Van den Veyver,I.B., Wan,M., Tran,C.Q., Francke,U. and Zoghbi,H.Y. (1999) Nature Genet., 23, 185–188. [DOI] [PubMed] [Google Scholar]
- 2.Wan M., Lee,S.S., Zhang,X., Houwink-Manville,I., Song,H.R., Amir,R.E., Budden,S., Naidu,S., Pereira,J.L., Lo,I.F., Zoghbi,H.Y., Schanen,N.C. and Francke,U. (1999) Am. J. Hum. Genet., 65, 1520–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang F., Buervenich,S., Nicolao,P., Bailey,M.E., Zhang,Z. and Anvret,M. (2000) J. Med. Genet., 37, 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheadle J.P., Gill,H., Fleming,N., Maynard,J., Kerr,A., Leonard,H., Krawczak,M., Cooper,D.N., Lynch,S., Thomas,N., Hughes,H., Hulten,M., Ravine,D., Sampson,J.R. and Clarke,A. (2000) Hum. Mol. Genet., 9, 1119–1129. [DOI] [PubMed] [Google Scholar]
- 5.Huppke P., Laccone,F., Kramer,N., Engel,W. and Hanefeld,F. (2000) Hum. Mol. Genet., 9, 1369–1375. [DOI] [PubMed] [Google Scholar]
- 6.Van den Veyver I.B. and Zoghbi,H.Y. (2000) Curr. Opin. Genet. Dev., 10, 275–279. [DOI] [PubMed] [Google Scholar]
- 7.Tate P., Skarnes,W. and Bird,A. (1996) Nature Genet., 12, 205–208. [DOI] [PubMed] [Google Scholar]
- 8.Meehan R.R., Lewis,J.D. and Bird,A.P. (1992) Nucleic Acids Res., 20, 5085–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird A. and Tweedie,S. (1995) Phil. Trans. R. Soc. London Ser. B, 349, 249–253. [DOI] [PubMed] [Google Scholar]
- 10.Yoder J.A., Walsh,C.P. and Bestor,T.H. (1997) Trends Genet., 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 11.Razin A. (1998) EMBO J., 17, 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buschhausen G., Wittig,B., Graessmann,M. and Graessman,A. (1987) Proc. Natl Acad. Sci. USA, 84, 1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kass S.U., Landsberger,N. and Wolffe,A.P. (1997) Curr. Biol., 7, 157–165. [DOI] [PubMed] [Google Scholar]
- 14.Jones P.L., Veenstra,G.J., Wade,P.A., Vermaak,D., Kass,S.U., Landsberger,N., Strouboulis,J. and Wolffe,A.P. (1998) Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- 15.Nan X., Ng,H.H., Johnson,C.A., Laherty,C.D., Turner,B.M., Einsenmann,R.N. and Bird,A. (1998) Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 16.Yu F., Thiesen,J. and Stratling,W.H. (2000) Nucleic Acids Res., 28, 2201–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaludov N.K. and Wolffe,A.P. (2000) Nucleic Acids Res., 28, 1921–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Bona C., Zappella,M., Hayek,G., Meloni,I., Vitelli,F., Bruttini,M., Cusano,R., Loffredo,P., Longo,I. and Renieri,A. (2000) Eur. J. Hum. Genet., 8, 325–330. [DOI] [PubMed] [Google Scholar]
- 19.Ballestar E., Yusufzai,T.M. and Wolffe,A.P. (2000) Biochemistry, 39, 7100–7016. [DOI] [PubMed] [Google Scholar]
- 20.Lewis J.D., Meehan,R.R., Henzel,W.J., Maurer-Fogy,I., Jeppensen,P., Klein,G. and Bird,A. (1992) Cell, 69, 905–914. [DOI] [PubMed] [Google Scholar]
- 21.Nan X., Meehan,R.R. and Bird,A. (1993) Nucleic Acids Res., 21, 4886–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almouzni G. and Wolffe,A.P. (1993) Genes Dev., 7, 2033–2047. [DOI] [PubMed] [Google Scholar]
- 23.Bellini M. and Gall,J.G. (1999) Mol. Biol. Cell., 10, 3425–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandler S.P., Guschin,D., Landsberger,N. and Wolffe,A.P. (1999) Biochemistry, 38, 7008–7018. [DOI] [PubMed] [Google Scholar]
- 25.Wakefield R.I., Smith,B.O., Nan,X., Free,A., Soteriou,A., Uhrin,D., Bird,A.P. and Barlow,P.N. (1999) J. Mol. Biol., 291, 1055–1065. [DOI] [PubMed] [Google Scholar]
- 26.Ohki I., Shimotake,N., Fujita,N., Nakao,M. and Shirakawa,M. (1999) EMBO J., 18, 6653–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendrich B. and Bird,A. (1998) Mol. Cell. Biol., 18, 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coy J.F., Sedlacek,Z., Bachner,D., Delius,H. and Poustka,A. (1999) Hum. Mol. Genet., 8, 1253–1262. [DOI] [PubMed] [Google Scholar]