Abstract
One-fifth of meningiomas classified as benign by World Health Organization (WHO) histopathological grading will behave malignantly. To better diagnose these tumors, several groups turned to DNA methylation, whereas we combined RNA-sequencing (RNA-seq) and cytogenetics. Both approaches were more accurate than histopathology in identifying aggressive tumors, but whether they revealed similar tumor types was unclear. We therefore performed unbiased DNA methylation, RNA-seq, and cytogenetic profiling on 110 primary meningiomas WHO grade I and II). Each technique distinguished the same three groups (two benign and one malignant) as our previous molecular classification; integrating these methods into one classifier further improved accuracy. Computational modeling revealed strong correlations between transcription and cytogenetic changes, particularly loss of chromosome 1p, in malignant tumors. Applying our classifier to data from previous studies also resolved certain anomalies entailed by grouping tumors by WHO grade. Accurate classification will therefore elucidate meningioma biology as well as improve diagnosis and prognosis.
Molecular classification identifies aggressive WHO grade I and II meningiomas and explains anomalies in prior studies.
INTRODUCTION
Over the past decade, next-generation sequencing (NGS) has transformed our understanding of many tumors of the central nervous system (CNS) (1), leading to better diagnosis and targeted therapies. For example, medulloblastomas are now classified by molecular characteristics such as sonic hedgehog pathway activation (2), which has led to the development of rational therapies that are currently in clinical trials (3, 4). Similarly, diffuse gliomas are now classified on the basis of isocitrate dehydrogenase mutation (5) and chromosomal losses (1p/19q codeletion) (6), while the discovery of a subset of tumors with H3K27M mutations shed light on their dismal prognosis and, again, led to promising therapies (7).
Unfortunately, the field of meningioma has been slow to benefit from parallel efforts. NGS studies have associated certain mutations with specific histologic grades (8–12), and a number of epigenetic studies have linked methylation patterns to clinical outcomes (13–16). In the largest such study to date, Sahm et al. (14) performed unsupervised clustering of nearly 500 tumors across World Health Organization (WHO) grades and found two major epigenetic groups composed of six subclasses with distinct clinical and genomic features. In a similar vein, we performed whole-exome sequencing (WES), RNA sequencing (RNA-seq), and cytogenetic profiling in 160 tumors across all three WHO grades to develop a molecular classification system that identified three types of meningioma (17). Type A tumors, which rarely recur, harbor mutations in TRAF7 (tumor necrosis factor receptor-associated factor 7), AKT1, or KLF4 but show no chromosomal losses. Type B meningiomas also exhibit a benign clinical course but are deficient in NF2/Merlin (the protein encoded by NF2, the neurofibromatosis 2 gene) and are characterized by loss of the chromatin-modifying enzyme PRC2 (polycomb repressive complex 2) (8, 17). Type C tumors recur frequently despite gross total resection, and they are both NF2-deficient and characterized by chromosomal instability (CIN), particularly loss of chromosome (Chr) 1p. These tumors lose the repressive form of the cell-cycle regulating DREAM complex and instead feature its MuvB core bound to FOXM1 and MYBL2, which leads to activation of the cell cycle (17, 18).
Both the methylation and our transcriptomic-based classification systems predicted tumor recurrence more accurately than the WHO histopathological grading system (19), which remains the clinical standard despite the fact that 20% of histologically benign (WHO grade I) tumors will behave aggressively (20). The difference classification can make was underscored by our finding that whereas in the WHO grading system, malignant tumors (grade III) nearly always evolve from grade I and II tumors, our system classified malignant tumors as molecular type C from their very first appearance. This suggests a very different model of meningioma biology.
If we are to understand meningioma behavior, then we must start with accurate classification. We noticed that both our transcriptional profiling and the methylation classification of Sahm et al. (14) segregate many of the common genomic alterations seen in meningioma, such as recurrent somatic mutations (NF2, TRAF7, and AKT1) and large-scale chromosomal deletions (1p and 22q), in similar ways (14, 17). This raises a few obvious questions: First, are these two classification systems identifying similar subsets? If so, could DNA methylation changes underlie the transcriptional differences we identified? If not, what does?
In this study, we set out to answer these questions. We performed a comprehensive analysis of 110 primary meningiomas using DNA methylation profiling, RNA-seq, and WES. We first used unsupervised approaches to analyze DNA methylation profiles and identified three clusters of meningioma (see Materials and Methods). These clusters were highly concordant with our prior transcriptional types (17), which supports the notion that there are three biologically distinct meningioma groups. Next, we used a multipronged approach to identifying “true” meningioma groups. We then used computational modeling to distinguish the relative contributions of DNA methylation and CIN to the transcriptional characteristics of each tumor type. Last, we reexamined several publicly available meningioma datasets to determine whether more accurate classification might alter the interpretation of results based on histopathological grading.
RESULTS
DNA methylation profiling identifies three major meningioma clusters
To determine how DNA methylation might relate to our prior transcriptional types, we performed DNA methylation profiling on 110 primary meningiomas that had undergone our RNA-seq–based transcriptional classification (17). We elected not to include recurrences for two reasons: First, we wanted to avoid overweighting the classifier with multiple samples from the same tumor; second, we need to improve prognostication of primary tumors, rather than those that have already declared themselves malignant by recurring. As a result, we had no WHO grade III tumors in this cohort. (Of the 450 primary meningiomas resected at our institution over the past 10 years, only one was grade III; primary grade III meningiomas are exceedingly rare.) In addition, we selected only tumors that had received no prior treatment, to avoid the likely confounding effects of radiation or radiosurgery. Using an unsupervised approach, we identified three methylation clusters of meningioma that were stable across multiple algorithms and sets of CpG sites (see Materials and Methods, fig. S1A, and table S1). These clusters (hereafter labeled Meth 1, Meth 2, and Meth 3) were each defined by a methylation signature, i.e., those sites that most strongly identified them (Fig. 1A). Comparing the methylation patterns of each cluster demonstrated some noteworthy differences. Across all signature probes, Meth 1 tumors showed a balanced methylation pattern (table S2A). Meth 2 was notably hypomethylated, mainly because of nonpromoter sites (table S2B). Conversely, Meth 3 was hypermethylated, most prominently at promoter sites, which were enriched within the Meth 3 signature. Since CpG island promoter hypermethylation has been observed in numerous cancers and typically acts to repress gene transcription (21), we looked specifically at the subset of probes located on CpG island promoters (table S2C). Not only are Meth 3 tumors significantly hypermethylated across the CpG island promoters contained within our methylation signatures (86%), but they are also hypermethylated across all CpG island promoters throughout the genome (60%).
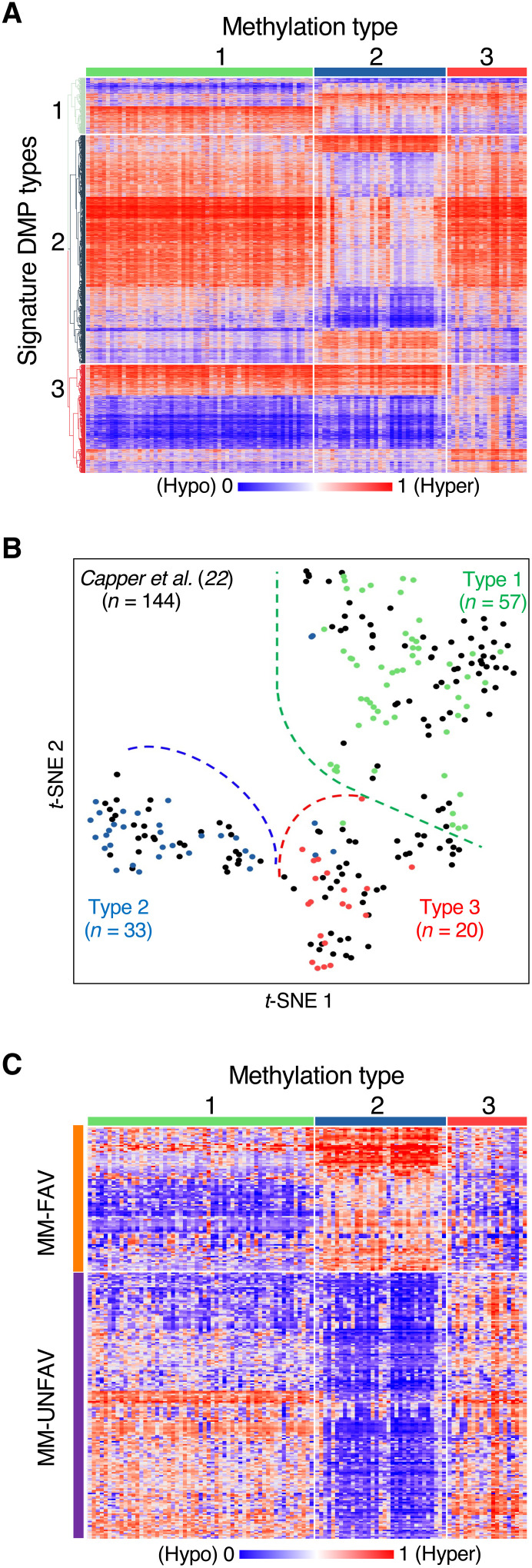
Fig. 1. Meningiomas comprise three methylation clusters.
(A) Heatmap of methylation data for the signature probes of each methylation cluster. (B) t-distributed stochastic neighbor embedding (t-SNE) of our cohort and the meningiomas from the Heidelberg CNS tumor classifier (22) across our signature probes. (C) Heatmap of methylation data for the MM-FAV (favorable) and MM-UNFAV (unfavorable) probes from Olar et al. (13) across our three methylation clusters.
To ensure that our data were consistent with previously published cohorts, we processed the publicly available raw data for all meningiomas from the Heidelberg CNS tumor classifier (22). The Heidelberg samples (n = 144) mirrored our data across our signature probes (Fig. 1B). We performed unsupervised non-negative matrix factorization (NMF) on the Heidelberg samples alone across our signature probes, and again three clusters emerged as optimal (fig. S1B). Furthermore, comparing these clusters to assignments made by a random forest classifier trained on our samples showed that 137 (95%) of the Heidelberg meningiomas were similarly assigned (fig. S1C and table S3). Thus, across our signature probes for this external dataset, our methylation clusters were stable, whether they were derived from our own data or the Heidelberg samples alone.
Several groups have reported that DNA methylation can be used for meningioma classification and prognosis (13–15). We wanted to compare our methylation clusters to previously published classifiers, but only Olar et al. (13) provided probe-level data, which defined two methylation subgroups classified by a set of 283 CpG sites. These sites were associated with either a favorable (MM-FAV, n = 98) or unfavorable (MM-UNFAV, n = 185) prognosis when hypermethylated. With regard to methylation status in aggregate, our Meth 2 tumors were conferred a uniformly favorable prognosis by the probes from Olar et al. (13), as all favorable probes were hypermethylated and all unfavorable probes were hypomethylated relative to all other tumors (Fig. 1C). On the other hand, Meth 3 tumors were hypermethylated in 93% of unfavorable probes and only 32% of favorable probes, thereby earning an unfavorable prognosis in the Olar et al. (13) system. These prognoses were corroborated by the outcomes for our methylation types: Meth 2 tumors rarely recurred, but the median progression-free survival for Meth 3 tumors was only 37 months (see below). Given the relatively homogenous methylation pattern of Meth 2 across these probes compared to the heterogeneous patterns seen across Meth 1 and Meth 3, it seems that Meth 2 meningiomas in the Olar et al. (13) cohort were the dominant factor in their identification of favorable and unfavorable probes.
Identification of three biologically distinct meningioma groups (MenG)
The aforementioned methylation analysis was performed blinded to the transcriptional types from our molecular profiling (17), so our next step was to compare different classification schemes of meningiomas. We first classified all tumors with our transcriptional classifier. Given that large-scale deletions of Chr 1p and Chr 22q are the most common cytogenetic changes seen in meningioma (8, 12) and bore a strong relationship with our transcriptional types (17), we also classified tumors cytogenetically as having “no loss,” “22q loss,” or “1p/22q loss” (no tumors had Chr 1p loss without Chr 22q loss), defining “loss” as involving at least one-third of a chromosomal arm. Next, we compared these two classifications to our methylation clusters and found strong concordance, as 83 (75%) tumors were assigned similarly by all three classifications. To improve the precision of our classification for the 27 discordant cases, we created an additional classifier to stratify meningiomas into groups according to NF2 status and level of CIN (8). Tumors with normal levels of NF2, based on a control group of publicly available arachnoid granulation cell data (23), were classified as NF2 intact; tumors with low levels of NF2 but only one or two large-scale chromosomal deletions were considered NF2-deficient, low CIN; those with low levels of NF2 and more than two large-scale deletions were classified as NF2-deficient, high CIN. This system also agreed with the prior three, as 77 (70%) tumors were categorized consistently across all four classifications (Fig. 2 and table S1).
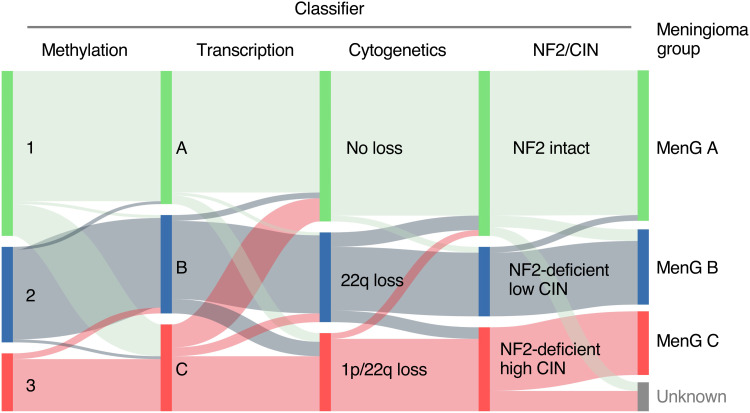
Fig. 2. Epigenetic, transcriptional, and chromosomal meningioma classifications converge on three biologically distinct MenGs.
Sankey plot demonstrating concordance between each subclassification leading to MenG assignment.
This concordance, along with the fact that three categories are sufficient in the different systems to capture differences in tumor behavior, suggests that there are three biologically distinct types of meningioma that display consistent differences across the genome, epigenome, and transcriptome. We assigned each of the 100 tumors (91%) for which there was agreement among at least three of the four classifications (methylation, transcription, cytogenetics, and NF2/CIN) to one of three Meningioma Groups (MenG): MenG A, MenG B, or MenG C (Fig. 3).
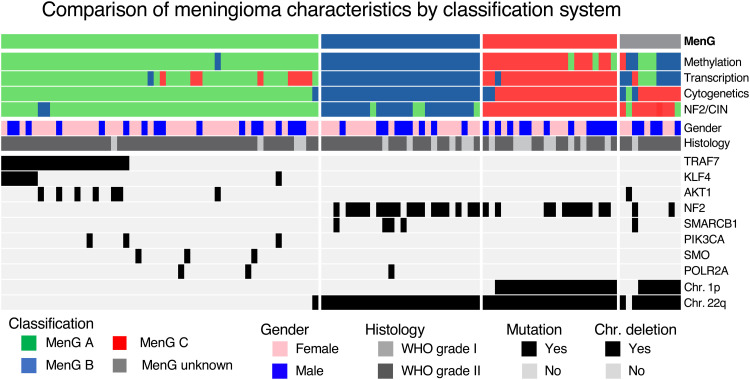
Fig. 3. Oncoprint of the cohort.
Subclassifications and differences among gender, histologic grade, common somatic mutations, and deletions of Chr 1p and Chr 22q according to MenGs. KLF4, Kruppel-like factor 4; SMARCB1, SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily B, Member 1; POLR2A, RNA Polymerase II Subunit A; SMO, Smoothened, Frizzled Class Receptor; PIK3CA, Phosphatidylinositol-4, 5-Bisphosphate 3-Kinase Catalytic Subunit Aalpha.
If we assume MenG as the “ground truth” classification and exclude the 10 tumors that eluded MenG assignment (table S4), then transcriptional profiling correctly classified 90 (90%) of tumors (tables S1 and S5). The greatest challenge for transcriptional profiling was correctly distinguishing MenG A and MenG C, as eight MenG A tumors were misclassified as type C. Given the malignant behavior of MenG C, this is an important distinction to make. Because of this low specificity, the transcriptional classifier’s positive predictive value (PPV) for MenG C was only 72%, although its negative predictive value (NPV) was 99% (Table 1). We suspect that, because of their CIN, MenG C tumors are the most genetically heterogeneous, and it will take much larger cohorts to distinguish transcriptional signatures that are consistent across this class. By comparison, DNA methylation, cytogenetic profiling, and NF2/CIN were each remarkably accurate on their own (Table 1). With regard to aggressive meningiomas, NPV was excellent across all classification modalities (>96%), while the methylation, cytogenetic, and NF2/CIN classifications each achieved 100% PPV. Although all four classifications can effectively identify the molecular group of a meningioma, the integrated approach reduces outliers.
Table 1. Diagnostic accuracy of individual classifiers.
| Classifier |
Diagnostic accuracy
overall |
Diagnostic accuracies for MenG C specifically | ||||
| Accuracy | Sensitivity | Specificity | PPV | NPV | ||
| Transcription | 90% | 91% | 95% | 90% | 72% | 99% |
| Cytogenetics | 97% | 98% | 91% | 100% | 100% | 98% |
| Methylation | 96% | 97% | 86% | 100% | 100% | 96% |
| NF2/CIN | 94% | 100% | 100% | 100% | 100% | 100% |
Clinical characteristics of the three meningioma groups
These MenGs display significant differences in clinical variables (table S5). MenG A and MenG B showed the typical female preponderance (67 and 65%, respectively), while MenG C featured more males (55%). This recapitulated our previous findings (17), as 71% of patients with either type A or B tumors were female, but 56% of patients with type C tumors were male. There was much greater CIN among MenG C, while NF2 expression was retained in MenG A and decreased in the other groups (Figs. 3 and 4).
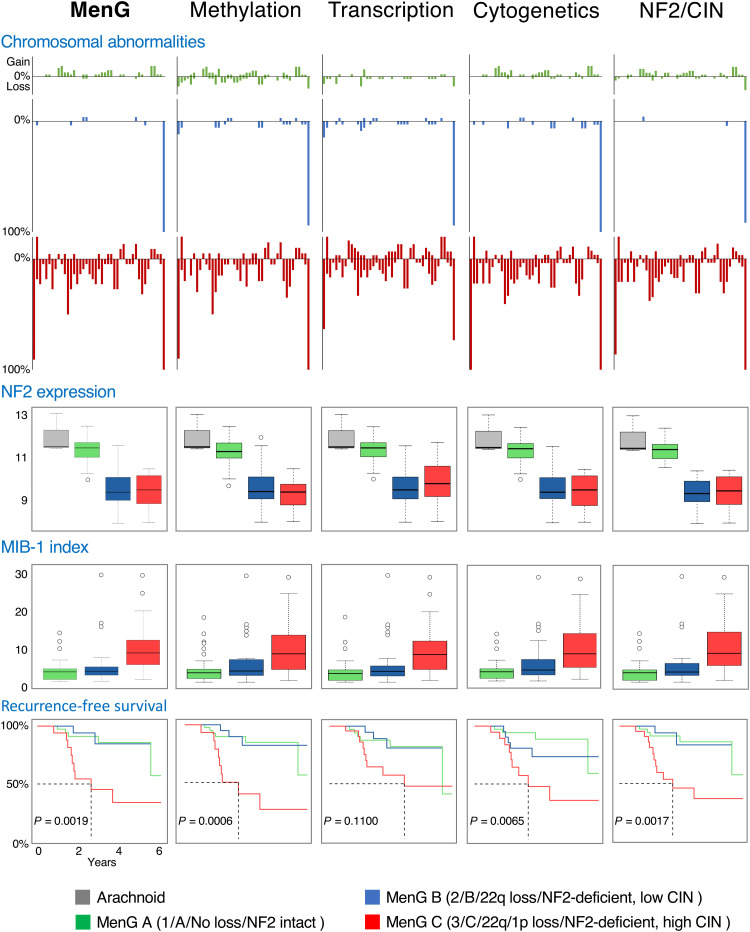
Fig. 4. Genomic and clinical outcomes differ substantially between MenGs.
Large-scale chromosomal abnormalities (A), NF2 expression (B), mitotic index (MIB-1) (C), and RFS (D) all differ substantially between MenGs but are maintained across methodologies (methylation, RNA-seq, cytogenetics, and NF2/CIN).
Similarly, mitotic activity differed among the groups. The median MIB-1 index, a measure of mitotic activity that is associated with WHO grade and has prognostic value (24, 25), ranged from 3% in MenG A to 8.4% in MenG C (Fig. 4 and table S5). Given the association between WHO grade and MIB-1, we performed subgroup analyses stratified by WHO grades I and II. The same trend of increasing median MIB-1 was seen among both WHO grade I tumors (2.9% in MenG A, 3.2% in MenG B, and 5.7% in MenG C) and WHO grade II tumors (8.8, 10.3, and 15.2%, respectively). In parallel, necrosis was modest in MenG A and MenG B tumors (only 4 and 15%, respectively) but present in nearly half (41%) of MenG C tumors.
Since our cohort was limited to primary tumors and lacked WHO grade III tumors, we added samples from the Heidelberg CNS tumor classifier (22) to determine the relationship between WHO grades and our groups. We excluded meningiomas that featured discordant methylation and chromosomal classifications, which are the two classifications we can derive from DNA methylation data alone. Expectedly, MenG A featured WHO grade I tumors almost exclusively, but 44% of MenG C tumors were also WHO grade I, in accord with our previous findings (fig. S2A and table S5) (17). Conversely, 17% of WHO grade III meningiomas were classified as MenG A or MenG B.
Comparing recurrence-free survival (RFS) rates within our cohort, patients with WHO grade I or II tumors showed similar rates of recurrence (fig. S2B). MenG A and MenG B were indistinguishable in this regard, but MenG C was associated with significantly lower RFS (log-rank P value of 0.0019) (fig. S2B). Because of higher rates of subtotal resection in MenG A and MenG C (table S5), we reanalyzed RFS for only those tumors that underwent gross total resection (Simpson grades I to III). Within this subset, the difference in RFS was even more pronounced: 44% of MenG C tumors recurred, whereas very few MenG A or MenG B tumors recurred (2.6 and 4.8%, respectively; fig. S2C). To account for the influence of WHO grade on tumor recurrence, we compared RFS within each histologic grade. Among WHO grade I tumors, those we classified as MenG C demonstrated significantly lower RFS (3.1 years, log-rank P value of 0.019), and a similar trend was seen among WHO grade II tumors, although it did not reach significance because of the small size of this subset (fig. S2D).
DNA methylation and cytogenetic changes correlate closely with gene expression in meningioma
DNA methylation changes and CIN affect gene expression in cancer (26, 27). Given the strong concordance of our classifiers, we asked how differential promoter methylation or copy number variability (CNV) correlated with the differential gene expression seen in our cohort using a partial least squares (PLS) regression model. PLS is often used to integrate high-dimensional genomic data (28, 29) and is similar to principal component analysis (PCA) in that it finds the factors that explain the maximal variation in the data, but it does so for both the dependent and independent data. It then projects the data onto a simplified space defined by those components and performs linear regression. This technique is particularly suitable when there is collinearity between model components, which is often the case for promoter CpG sites, as they are adjacent to one another and can be similarly methylated.
Each gene is modeled individually using the methylation status of every promoter site, the relative amplification/deletion of its locus, and the log-transformed expression value. We limited this analysis to the 67 samples that had gene-level CNV data (derived from WES) and were categorized consistently by all four of our classifications (Fig. 3 and table S5) to select those samples most representative of each underlying MenG. This also reduced the possibility of including any low-quality data from one of the profiling methods, which could lead to erroneous classification. Last, each model was evaluated by both the R-squared value (R2), as an overall measure of goodness of fit, and the variable importance in projection (ViP) scores, which measure the relative importance of each covariate.
To establish a baseline, we modeled every gene for which we had promoter methylation, relative CNV, and expression data (n = 15,373; Table 2). We defined models as being “highly accurate” if the R2 was more than 2 SDs above the median (R2 = 0.29; fig. S3A). A highly accurate model signifies a close correlation between a gene’s expression and its promoter methylation and/or CNV status, which implies that those factors regulate the transcription of that gene. We used the relative ViP scores (see Materials and Methods) and the correlation coefficient for each covariate to determine whether each highly accurate model was more likely to be “driven” by methylation, CNV, or both (fig. S3B).
Table 2. Relative contributions of promoter methylation and copy number variation to changes in gene expression in meningioma, by MenG.
Boldface text indicates the dominant association for highly accurate models in each subset of genes. The only exception (marked by asterisk) is the differentially methylated genes for MenG C for which there is a notably lower proportion of highly accurate models compared to the genes for MenG A/B and which is only marginally greater than the baseline of all genes. For convenience, we refer here to the different classes of gene expression changes as driven by methylation, CNV, or both, but this is in the context of computational modeling; we have not experimentally demonstrated causality.
| Highly accurate models | Methylation-driven | CNV-driven |
Driven by both
methylation and CNV |
|
| All genes | 7.5% | 59.8% (690) | 38.7% (446) | 1.5% (17) |
| Most variably methylated genes | 14.6% | 87.7% (128) | 11.6% (17) | 0.7% (1) |
| Chr 1p/22q genes | 33.0% | 14.7% (61) | 82.9% (343) | 2.4% (10) |
| Transcriptional classifier genes | 15.4% | 91.7% (77) | 7.1% (6) | 1.2% (1) |
| DEGs | 16.4% | 60.9% (277) | 38% (173) | 1.1% (5) |
| MenG A | 15.0% | 64.3% (36) | 32.1% (18) | 3.6% (2) |
| MenG B | 14.7% | 76.9% (40) | 23.1% (12) | 0% (0) |
| MenG C | 33.3% | 14.1% (9) | 84.4% (54) | 1.6% (1) |
| Differentially methylated genes | 23.7% | 85.1% (189) | 10.8% (24) | 4.1% (9) |
| MenG A | 22.6% | 84.3% (43) | 13.7% (7) | 2% (1) |
| MenG B | 22.5% | 87.6% (85) | 8.2% (8) | 4.1% (4) |
| MenG C | *12.3% | 88.2% (15) | 5.9% (1) | 5.9% (1) |
We used two scenarios to establish the validity of our approach. First, we identified the most variably methylated CpG promoter site for each gene and sorted all genes in order of decreasing promoter site variance. As expected, as promoter variance decreased, so did the prevalence of highly accurate models and the relative proportion of models that strongly correlate with methylation (fig. S3C). To validate our model, we first compared the set of 1000 genes with the highest promoter site variation to the baseline of all genes. As expected, this set was significantly enriched for highly accurate models (14.6% versus 7.5%; P = 1.4 × 10−15) that were strongly associated with methylation (88% versus 60%; P = 9.8 × 10−11) (Table 2). Next, we reasoned that genes located on Chr 1p and Chr 22q, the two most common large-scale deletions in meningioma (25 and 53% in our cohort, respectively), should be more strongly associated with CNV. As expected, these genes had a substantial proportion of highly accurate models (33% versus 7.5%; P = 2.1 × 10−193), and most were associated with CNV (83% versus 39%; P = 3.0 × 10−53) (Table 2).
Having verified the accuracy of our model, we investigated how our transcriptional types and differential expression in general might be affected by DNA methylation and/or underlying copy number variations. We analyzed the 611 genes used by the random forest classifier to define our transcriptional types, of which 546 were able to be modeled. This set was enriched for highly accurate models (15.4%; P = 2.4 × 10−11), most of which were associated with methylation (92%; P = 1.3 × 10−8) (Table 2), demonstrating that DNA methylation correlated closely to the transcriptional classification. Despite consistent differences in chromosomal gains and losses between our transcriptional types, hardly any genes in the classifier correlated most strongly with CNV (7%; P = 1.4 × 10−8). That said, three of the top 10 genes—specifically, those ranked first (MPPED1, Chr 22q), second (LEPR, Chr 1p), and eighth (ALPL, Chr 1p)—were subject to CNV. Therefore, Chr 1p and Chr 22q losses still played a significant role in our transcriptional classification despite the low prevalence of CNV-driven genes. Together, these findings explain the substantial concordance among our methylation, transcription, and cytogenetic classifications.
To investigate how DNA methylation and CNV contribute to differential expression within our cohort, we identified the differentially expressed genes (DEGs) using the RNA-seq data from three publicly available arachnoid samples as a control group (23). To identify DEGs, we performed pairwise comparisons between all tumors and the arachnoid controls and then between each MenG and the arachnoid controls, with a false discovery rate of 1 × 10−6. Across all DEGs (n = 2777), there was enrichment of highly accurate models (16%; P = 1.1 × 10−51). Of these, a greater proportion was associated with DNA methylation (61%) than with CNV (31%), similar to the distribution among all genes (Table 2).
Aggressive tumors are more influenced by CIN, particularly Chr1p loss
To evaluate the influence of DNA methylation and CNV within each subgroup, we selected those DEGs that were unique to each group. Compared to all DEGs, those unique to MenG A and MenG B showed similar rates of highly accurate models (~15.0%), most of which correlated with methylation (64 and 77%, respectively) rather than CNV (32 and 23%, respectively). DEGs unique to MenG C showed a markedly higher rate of highly accurate models (33%) (Table 2), and these were more influenced by CNV (84%) than by methylation (14%). Chr 1p contained a large proportion of MenG C DEGs (48%) but only 6 and 7% of DEGs unique to MenG A and MenG B, respectively. Moreover, of these MenG C DEGs on Chr 1p, 61% were highly accurate, and nearly all (93%) were correlated with CNV, demonstrating a marked influence of Chr 1p loss on the gene expression changes seen in MenG C tumors.
While this analysis suggests that CNV plays an outsized role in the differential expression of MenG C tumors, the low rate of methylation-driven DEGs raises the question of how methylation correlates with gene expression within MenG C. (Recall that these tumors showed substantial promoter hypermethylation.) To answer this question, we performed a reciprocal analysis from the alternative perspective of unique differentially methylated promoters. We first identified all CpG promoter sites that were differentially methylated between each MenG and all other tumors (false discovery rate of 1 × 10−6 and minimum difference in mean β of 0.1) and then aggregated their corresponding genes (n = 936). As expected, these genes were enriched for highly accurate models (23.7%; P = 7.0 × 10−67), with a large proportion influenced by methylation (85.1%; P = 1.2 × 10−12) (Table 2). We then analyzed genes that were differentially methylated in only one MenG. The models for genes unique to MenG A and MenG B yielded similar results in terms of highly accurate models (22.6 and 22.5%, respectively) and the proportion associated with methylation (84.3 and 87.6%, respectively). Unexpectedly, the proportion of highly accurate models for genes unique to MenG C (12.3%) was significantly less than those for MenG A and MenG B (P = 0.022 and P = 0.014, respectively), although it was greater than the baseline of all genes (P = 0.049) (Table 2). Therefore, methylation appears to play less of a regulatory role in gene expression for MenG C than it does in MenG A and MenG B, despite the substantial promoter hypermethylation seen in these aggressive tumors.
MenG classification can resolve anomalies in published findings that relied on WHO grade
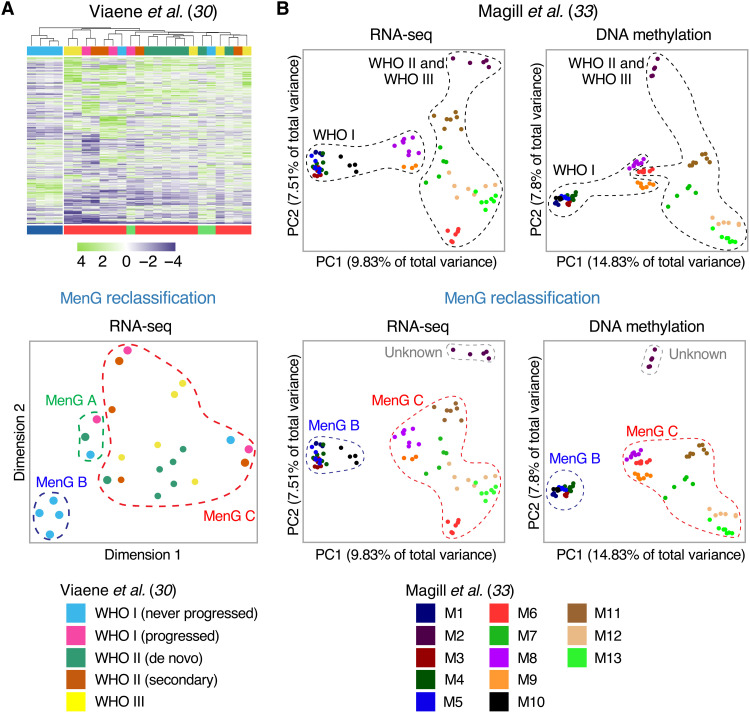
Given that most meningioma studies rely on WHO grading for tumor classification, we asked how viewing these data through the lens of MenG classification might alter our understanding of meningioma biology. One of the central questions in meningioma research is whether malignant tumors are malignant from their inception or whether they evolve from more benign tumors. In our previous study (17), we found that malignant tumors were molecularly distinct from the start. That is, the first occurrence was classified as group C and so was each subsequent recurrence. There were relatively few such group C tumors, however, so we were particularly interested in whether a slightly larger dataset could shed light on this question. Viaene et al. (30) had performed RNA-seq on a diverse set of 25 meningiomas, featuring low-grade tumors that progressed, along with their recurrences; low-grade tumors that did not progress; and de novo high-grade tumors (table S6). There were six WHO grade I tumors that did not progress. The authors concluded that these were molecularly distinct from the tumors that did progress, based on a cluster of four meningiomas that was composed solely of “WHO I not progressed” tumors (Fig. 5A). The remaining two WHO I not progressed tumors clustered, rather inconveniently, with “WHO I progressed” tumors and their recurrences, along with de novo high-grade tumors.
Fig. 5. Accurate MenG classification of publicly available datasets sheds light on previous studies based on WHO grading.
(A) Heatmap (top) and t-SNE (bottom) of the top 1000 most DEGs from a cohort of meningiomas studied by Viaene et al. (30). The WHO grade I tumors that did not progress were all MenG B and separate clearly from the other tumors. (B) PCA of RNA-seq and DNA methylation data of 86 spatially distinct samples from 13 meningiomas studied by Magill et al. (33), segregated by WHO grade, as in the original study (top) and by MenG (bottom). MenG demonstrates markedly better cluster separation.
Integrated MenG classification neatly resolves this puzzle and proves the authors’ conclusion fundamentally correct, because it turns out that the distinct cluster of four WHO I not progressed tumors is composed of the only MenG B tumors in this entire cohort. Of the four tumors that recurred, three were MenG C, along with all of their recurrences (which were of higher WHO grades). The fourth was classified as MenG A, while its recurrence was MenG C. Because transcriptional profiling on its own was the least accurate of our four methodologies, particularly with respect to differentiating types A and C [this study and (17)], we believe that this tumor was misclassified as MenG A and is actually MenG C. This underscores the benefits of integrating DNA methylation and cytogenetic analysis with RNA-seq to achieve more accurate classification. Regardless, this dataset supports our finding that it is predominantly MenG C tumors that recur and that these tumors maintain their molecular identity even as they progress through WHO grades over time.
Integrated classification sheds light on intra- and intertumoral heterogeneity
Although a number of studies have found substantial intratumoral heterogeneity in higher-grade and recurrent meningiomas (31, 32), few have used RNA-seq or DNA methylation, and even fewer have made their raw data freely available. One that did explored intratumoral heterogeneity both molecularly and radiographically (33), profiling 86 spatially distinct samples from 13 meningiomas (M1 to M13) across all three WHO grades. Slightly more than half their cohort were recurrences (54% of tumors and 56% of samples), of which four had undergone radiation; all tumors were located supratentorially along the midline to allow for multiple stereotactic sampling (only 37% of our tumors were similarly located) (table S7). Given the high proportion of malignant tumors, we were keen to learn what integrated classification would reveal.
We applied our methodology to their raw data and found that 12 of the 13 tumors were either MenG B or MenG C (as would be expected, since MenG A is primarily located along the skull base) (table S7) (17). One tumor, M2, was unclassifiable because its cytogenetic, transcriptional, and methylation profiles differed completely from those of any other tumor we have analyzed (fig. S4A). When we applied the Heidelberg classifier to this tumor (see Materials and Methods), none of its samples matched to meningioma, and only two samples could even be matched to the family containing meningioma (table S7). We therefore suspect that this tumor is either a rare variant of meningioma not captured in any of the datasets we analyzed or not a meningioma at all. The only tumor that was not clearly assigned a molecular group was M11, with five of seven samples classified as “unknown” due to a split between the Chr- and gene-level methodologies (MenG B and MenG C, respectively). The remaining two samples exhibited loss of Chr 1p and were thereby uniformly designated as MenG C by all four modalities. As M11 was a recurrent WHO grade II tumor, we suspect MenG C to be the correct classification. Otherwise, there was remarkable consistency in MenG classification among a given tumor’s samples.
Magill et al. (33) judged intratumoral heterogeneity by how closely the data points for samples of a given tumor cluster on a PCA plot (Fig. 5B). The awkwardness of grouping the clusters by WHO grade is apparent: Two of the low-grade samples (M8 and M9) lie closer to the high-grade group than to the rest of the WHO grade I tumors on the RNA-seq PCA plot (Fig. 5B, top left). Even more puzzlingly, these two tumors appear within the high-grade group on the DNA methylation PCA plot, while the remaining low-grade tumors again form a separate cluster (Fig. 5B, top right). MenG classification reorganizes and clarifies both plots (Fig. 5B, bottom): The tight cluster of WHO grade I tumors turns out to be the five MenG B tumors within this cohort. In addition, having found M2 to be unclassifiable, the MenG C cluster is more cohesive and coherent than was the high-grade cluster, and there is no longer any ambiguity with tumors M8 and M9. We also note that the degree of sample dispersion is very similar between low- and high-grade tumors (Fig. 5B, top) but differs starkly between MenG C and MenG B (Fig. 5B, bottom)—there is hardly any dispersal at all in the MenG B tumors. Furthermore, the MenG B group as a whole is tightly clustered and covers a very narrow area, signifying a high degree of transcriptional and epigenetic homogeneity not only within tumors but also among tumors within this group. MenG C tumors cover a considerably larger area, which is consonant with their marked genomic instability. Cytogenetically, MenG classification also more sharply differentiates tumors with minimal instability (MenG B tumors, mean CIN = 1.13 large-scale deletions; WHO grade I tumors, mean CIN = 2.04 large-scale deletions).
Integrated molecular classification therefore reveals substantial differences between biological types that were obscured by histopathological grade. Even discrepancies in subclassifications can reveal nuances of meningioma biology and generate new hypotheses. Consider M9: Its methylation, transcriptional, and NF2/instability profiles all indicate MenG C, but the chromosomal classification depends on how one defines a chromosomal loss. By our criterion for a large-scale deletion (at least one-third of a chromosomal arm), M9 does not qualify as showing Chr 1p loss, making its chromosomal classification align with MenG B. However, all seven samples feature a small deletion at the telomeric end of Chr 1p (fig. S4B). Therefore, if Chr 1p loss is indeed a crucial step in MenG C tumorigenesis, the segment deleted in this tumor may contain the biologically relevant portion of Chr 1p.
DISCUSSION
A welter of meningioma classification systems have appeared in just the past few years. Some have attempted to combine molecular techniques with histology (34, 35); others have sought to replace histopathological grades with groups based on methylation (14) or combined modalities (36); systems have been devised with anything from two to six distinct types of tumor, in contradistinction to the 15 histopathological subtypes. To our knowledge, there has been no effort to discern whether different modalities or classification systems were hinting at the same underlying biology. Our comprehensive profiling of 110 primary meningiomas, with analysis of an additional 255 samples from published datasets, strongly suggests that there are three biologically distinct groups of meningiomas that can be identified best by integrating DNA methylation, RNA-seq, and CIN/cytogenetics. Given that we need better prognostication when a tumor first appears (and before it declares itself malignant by recurring), we focused on primary tumors, but when we supplemented our cohort with an outside dataset enriched for recurrent tumors (33), k-means clustering subdivided our MenG C tumors into two subclasses (fig. S4A). Both still quite clearly fall under the aegis of high-CIN tumors with NF2 and Chr 1p/22q loss, however, and we consider this broad category more important from a clinical point of view. Moreover, thanks to the work of Magill et al. (33), we can see that MenG C tumors display considerable intra- and intertumoral heterogeneity, but their biologic identity remains stable despite molecular and chromosomal changes throughout the tumor. MenG B tumors, however, exhibit little such heterogeneity. This is reassuring from a clinical standpoint, as it indicates that a patient can be given a more accurate prognosis with molecular classification, even if the entire tumor is not sampled. For practical purposes, cytogenetic analysis (losses of Chr 1p and Chr 22q), with or without DNA methylation profiling, would provide very accurate, clinically useful classification with 100% PPV for aggressive meningiomas.
There are notable parallels between our multimodal classification and the largest, most comprehensive methylation-based classification to date (14). While the data from this study is not readily available, their “MC ben-2” class appears largely synonymous with our MenG A: These tumors feature the fewest CNVs and nearly all have TRAF7, AKT1, or KLF4 mutations. Our MenG B category most closely aligns with their “MC ben-1” tumors, nearly all of which bear Chr 22q deletions but retain Chr 1p. Last, MenG C resembles all of their high-risk subgroups (MC int-A, MC int-B, and MC mal), the vast majority of which demonstrate Chr 1p and Chr 22q loss. We likely had few, if any, “MC mal” tumors, which are predominantly WHO grade III, because we deliberately limited our cohort to primary tumors. In addition, while “MC int-A” and “MC int-B” feature similar names and clinical behavior, they belong to different major epigenetic groups (groups A and B, respectively) and presumably feature substantially different methylation profiles. We speculate that our MenG C may represent only one of these classes.
Subsequent work by the same group (37) explored molecular features of these epigenetic classes. In their aggressive tumors, they noted elevated expression of FOXM1—as we did in our MenG C tumors (log2 fold change = 1.15, adjusted P = 1.9 × 10−6) and as we previously reported (17)—but they also found a molecular signature (AC3) that indicates defective homologous recombination repair. This signature was distributed throughout the genome and closely correlated with the degree of genomic instability, thereby implicating defective homologous repair as a possible cause. Given our finding that CIN, particularly Chr 1p loss, drives many transcriptional changes in MenG C, DNA misrepair could be a primary event in the biology of these aggressive tumors.
Vasudevan et al. (18) performed multiplatform profiling on a meningioma cohort enriched for high-grade tumors, including RNA-seq (n = 42), DNA methylation profiling (n = 26), and WES (n = 24). Their transcriptional profiling identified two clusters with differences in gender and WHO grade, although there was no difference in RFS between groups. Their methylation-based clustering, on the other hand, identified three clusters with increasing methylation levels; their “high methylation” cluster was associated with significantly lower RFS, consistent with our MenG C group. They, too, identified FOXM1 as a key transcription factor associated with meningioma recurrence and propose that genomic, epigenomic, and transcriptomic mechanisms converge on FOXM1/Wnt signaling to drive aggressive tumors. Specifically, they found hypermethylation of a promoter of the Wnt antagonist SFRP1 and low SFRP1 expression in tumors with elevated FOXM1 expression. We also find decreased expression of SFRP1 in MenG C (log2 fold change = −2.93, adjusted P = 8.4 × 10−9), but our PLS model suggests that this is likely not due to promoter hypermethylation. All promoter sites judged to be of high importance by our PLS model featured positive correlations between methylation and expression. Individual inspection of all SFRP1 promoter sites (n = 19) among our PLS cohort identified 10 probes for which MenG C tumors are hypermethylated, of which six show statistically significant (P < 0.05) negative correlations with expression upon linear regression. This was primarily due to a single MenG C tumor with a unique, marked cytogenetic loss at the gene locus; exclusion of this sample left only one SFRP1 promoter with a statistically significant negative correlation, as opposed to 10 promoters with a significant positive correlation. It is possible that hypermethylation of this specific promoter site could be silencing this gene, but the overall picture of promoter hypermethylation for SFRP1 seems to be the opposite within our data. Interactions between the epigenome and transcriptome therefore cannot be assumed but must be rigorously explored, one gene at a time.
As we were finalizing this manuscript, Nassiri et al. (36) published comprehensive profiling of 124 meningiomas and defined four molecular groups with cluster-of-cluster assignments using CNV derived from WES, DNA methylation profiling, and mRNA sequencing. As far as we can tell (the data are not accessible), these groups are identical to ours [this study and (17)]; their molecular group 1 (immunogenic) corresponds to our MenG B, their molecular group 2 (benign NF2 wild type) to our MenG A, and their molecular group 3 (hypermetabolic) and group 4 (proliferative) to our MenG C. Half their cohort were either WHO grade II (35%) or grade III (16%) tumors, most likely recurrences, which allowed them to distinguish two subtypes of our MenG C as we did and much like three high-risk groups of Sahm et al. (14). We suspect that the “proliferative” tumors identified by Nassiri et al. (36) arise from the “hypermetabolic” group, with the accumulation of more CNVs due to their inherent genomic instability. Last but not least, Nassiri et al. (36) find their proliferative tumors respond to vorinostat, which is consistent with our previous work showing that histone deacetylase inhibitors prolong survival in a patient-derived orthotopic xenograft (PDOX) model of type C meningioma (38).
The identification of these remarkably similar molecular groups of meningiomas by multiple teams, using completely independent datasets and parallel but different approaches, strongly supports our contention that there are three primary biological classes of meningioma. Greater public availability of data and collaboration between groups would substantially increase the sample sizes for validation studies. This would likely best be accomplished by a multicenter study using a consistent genomics pipeline. Larger cohorts numbering in the thousands would be necessary to capture rarities such as primary grade III tumors, but validation cohorts of completely resected tumors with long-term outcome data are likely within reach and would more accurately define the prognoses of MenG A and MenG B, as there are few recurrences among these groups. Comparisons of multiple types of data should allow the field to determine the most efficient means to identify these groups in a clinical setting. Cytogenetic analysis of Chr 1p and Chr 22q would be low cost and readily translatable to clinical practice. As we were preparing this work, two new studies demonstrated that cytogenetic data, particularly loss of Chr 1p, significantly improves prognostication when integrated with WHO histopathologic grade (34, 35). Even this simple step, as we previously proposed (17), would increase the likelihood of detecting supposedly benign meningiomas with a high rate of recurrence despite complete resection. Equally important, our molecular groups could help avoid unnecessary treatment in patients with a low risk of recurrence (MenG A or MenG B).
MATERIALS AND METHODS
Study design
The objective of this study was to compare our previously published molecular classification of meningiomas (17) to clusters obtained from DNA methylation profiling. We had hypothesized significant overlap between these groupings but had no estimate of how closely they would align; therefore, statistical power and sample sizes were not computed. We did not have a comprehensively profiled cohort to validate our complete methodology, but we used publicly available DNA methylation data (22) to validate our DNA methylation classification.
Sample selection and preparation
Tumor samples consisted of fresh-frozen tissue from 110 primary meningiomas in 108 patients treated at Baylor College of Medicine (BCM). Of those samples, 101 were included in the previously published cohort for our molecular classification (17). All patients provided written informed consent, and tumor tissues were collected under an institutional review board (IRB)–approved protocol at BCM by the Human Tissue Acquisition and Pathology Core (protocol H-14435). As previously described (17), all meningiomas were reviewed by a board-certified neuropathologist before sample preparation and graded according the 2016 WHO guidelines. Using representative fresh-frozen blocks with an estimated purity of ≥95%, DNA and RNA extraction was performed on 20 to 30 mg of tumor tissue using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Patient data review
In accordance with our IRB-approved protocol, we reviewed the following data: patient age at surgery, sex, race, tumor size, tumor location, preoperative embolization, extent of resection, histologic grade, MIB-1 index, and presence of brain invasion. Diagnostic imaging was rereviewed to define tumor location, extent of resection, and presence/date of local recurrence. Local recurrence after gross total resection was defined as local development of any contrast enhancement on subsequent brain imaging, while recurrence following subtotal resection was defined as any measurable growth of residual tumor. We included only primary meningiomas in our sample, reasoning that having several recurrences of the same tumor would give that tumor too much weight in the classifier. Histologically, 90 tumors were WHO grade I, 20 were WHO grade II, and none were WHO grade III (the vast majority of grade III tumors are recurrences). One patient had a history of childhood radiation, and she featured two distinct meningiomas (sphenoid wing and petroclival), both of which were included in this analysis. A detailed summary of clinical information is available in table S5.
DNA methylation extraction and processing
We used the Illumina 850K EPIC BeadChip to conduct DNA methylation analysis on extracted tumor DNA, as per the manufacturer’s instructions at the Génome Québec Innovation Centre (Montreal, Canada). We quantified DNA with Picogreen and used 500 ng for DNA bisulfite treatment. DNA was amplified for 20 to 24 hours at 37°C and then hybridized for 16 to 20 hours at 48°C. Arrays were scanned using Illumina iScan with the Methylation NXT setting. We analyzed methylation data with GenomeStudio v2011.1 and Methylation module 1.9.0.
We imported, processed, and normalized the methylation raw data (.idat files) using the Chip Analysis Methylation Pipeline (ChAMP) R package (39), which uses the minfi package (40, 41) to load the data. Probes were filtered out if they failed to hybridize (detection P > 0.01), had <3 beads in 5% of samples, were not at CpG sites, were defined as multihit probes, were located on sex Chrs, or were associated with single-nucleotide polymorphisms (42). We assessed sample quality and determined that none warranted exclusion. The β values were calculated and normalized using the BMIQ (Beta MIxture Quantile dilation) method (43). We performed singular value decomposition (SVD) analysis to look for batch effects but found none that warranted correction (44). In a separate analysis to evaluate the robustness of our epigenetic types to a different processing method, we processed the raw methylation data with the SeSAMe package (45) using default settings.
Whole-exome RNA-seq extraction and processing
WES had been performed as previously described (17) and was available for 90 samples. In 101 samples, RNA-seq had been performed as previously described using the Illumina platform. We obtained RNA-seq data for the remaining nine tumors from Tempus, which entailed sending tumor samples along with saliva for processing according to their protocol (46).
Bioinformatic analyses for methylation data
To ensure that our methylation clusters were not dependent on either the algorithm or the set of CpG probes used to derive them, we used two different algorithms and two different sets of probes. For algorithms, we used NMF using the NMF package (47) and also k-means clustering using the ConsensusClusterPlus package (48), both with default settings. Regarding our different probe sets, we sorted all probes in order of decreasing overall variance to select both a “small” and a “large” set of most variably methylated probes. For the small set, rather than choose an arbitrary number of probes, we used the condition number of the normalized β matrix to inform our decision. The condition number is a measure of the sensitivity of a function’s output to small changes in the input, and a well-conditioned matrix (low condition number) is more stable to small perturbations in its input. The kappa function from base R (version 4.0.3) was applied to the β matrix with an increasing number of probes, demonstrating exponential decay of the condition number as more probes were added. As the curve approached its asymptote around 4000, we selected the local minimum from 3500 to 5000 as our initial number of most variably methylated probes, which occurred at 4230 probes. For the large set of probes, we selected the top 10% of most variably methylated probes (n = 73,544).
We used both NMF and k-means clustering as above onto both sets of probes, finding three to be the optimal number of clusters in all cases (fig. S1B). Moreover, there was high concordance between these clusters, as 91% of samples were assigned identically across all four analyses (table S1).
We then identified the “methylation signature” of each cluster. Samples that did not cluster similarly across our four analyses (n = 10) were excluded, and we performed pairwise comparisons between each cluster and all other tumors to identify differentially methylated probes (DMPs). We identified these DMPs using tumors using the champ.DMP function [which uses the limma package (49, 50)] with a false discovery rate of 1 × 10−6 and a minimum difference in mean β of 0.25. DMPs that appear for multiple clusters were excluded, leaving three signatures that were unique to each cluster. These signatures comprised 7552 DMPs: 1072 for cluster 1, 4396 DMPs for cluster 2, and 2084 DMPs for cluster 3. Only 903 signature probes (12%) were in the small set, while 6985 (93%) were in the large set. Therefore, limiting an analysis to only the first few thousand most variably methylated probes (as is often done when identifying epigenetic groups) seems to exclude a substantial number of potentially important sites, while the top 10% captures the vast majority.
Having identified these three methylation signatures, we then defined our final epigenetic clusters, specifically for those samples that had discordant preliminary clusters. We used NMF on all 110 samples across the methylation signature probes to define our final methylation clusters/types.
Given the high rate of large-scale chromosomal losses in our cohort, we also processed the raw data using the SeSAMe pipeline (45), which accounts for artifacts caused by germline or somatic chromosomal deletions. The raw data were loaded using the openSesame function with default settings to generate a β matrix. All analyses previously described were performed again to generate Sesame methylation clusters, which were 95% concordant (table S1).
To define probes as being hypermethylated or hypomethylated for a specific methylation type, we identified the type that featured the most divergent mean β value. We did this rather than compare each type mean to the cohort mean so that each probe could have only one designation (hypermethylated or hypomethylated for a single type). For the analysis of all CpG promoter islands, we removed any probes for which the mean β values for all of the methylation types were within 0.01 of one another, i.e., probes with highly similar methylation levels across all three types. Last, promoter sites were defined as those located at “exon start,” “transcription start site 200,” and “transcription start site 1500” sites.
We identified DMPs a second time to identify genes featuring promoters that were differentially methylated among MenG A, MenG B, and MenG C. We again used the champ.DMP function with a false discovery rate of 1 × 10−6; this time, however, we used a 0.1 minimum difference in mean β. We aggregated all genes with a promoter identified as being differentially methylated for a given MenG and removed any genes belonging to multiple groups, thereby leaving genes with promoter site(s) differentially methylated in only a single MenG. All methylation heatmaps were generated using the ComplexHeatmap package (51).
Bioinformatic analyses for RNA-seq data
We processed raw reads from the RNA-seq data with an in-house pipeline that uses TopHat2 (RRID:SCR_013035) for read alignment; FastQC (RRID:SCR_014583) and RSeQC (RRID:SCR_005275) for read and alignment quality assessment; HTSeq (RRID:SCR_005514) for expression count; and GATK (RRID:SCR_001876) for variant calling. The reads were aligned to GRCh38 Human reference genome and mapping to the human transcriptome according to UCSC gene annotations. We then normalized the RNA-seq read counts for genes, applied a variance stabilizing transformation, and identified DEGs using the DESeq2 package (52).
CNV identification
WES had been used to identify CNVs in 90 samples, as previously described (17). We also calculated CNVs from DNA methylation data using the conumee package (53). For samples with WES data, we compared the two sets of CNVs and found strong agreement, validating both methods against one another. To identify large-scale CNV events, which we defined as involving at least one-third of a chromosomal arm, we used the WES-based CNV data if available; otherwise, we used the data derived from methylation. These data were reviewed independently and separately by two co-authors (J.C.B. and A.S.H.).
Analysis of publicly available data arachnoid cell data (23)
We downloaded RNA-seq raw data (fastq files) from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus, accession number GSE139651, for the three arachnoid granulation samples (AG01, AG02, and AG03) and processed it according to the same method as detailed above. Regarding the NF2 cutoff used to distinguish types in our NF2/CIN classification, we defined “NF2 deficient” as being 1 SD below the median of the log-transformed expression levels of these three arachnoid granulation cells (median = 11.96, SD = 0.90, cutoff = 11.06).
PLS model creation and classification
We implemented PLS models with the caret package (54) using the function train and method “pls.” The independent data consisted of the normalized β values of all promoter CpG sites (defined as exon start, transcription start site 200, and transcription start site 1500) and the locus-specific relative CNV inferred from WES data, while the dependent data were the variance-transformed (log-scale) RNA-seq counts. The relevant characteristics of each model were the R2 and the ViP scores. The ViP is based on the regression coefficients weighted by the reduction in the residual sum of squares, signifying the relative importance of each covariate to the model, and is scaled so that the highest ranked covariate has a score of 100.
To establish a baseline distribution of R2 values, we modeled all genes for which the necessary data were available. We then classified models as being highly accurate if the R2 value was more than two SDs greater than the median (>0.29). These highly accurate models were further classified as being associated primarily with methylation, CNV, or both, depending on the relative ViP scores (fig. S3B).
Any covariates with a ViP score greater than 90 were considered a possible “driver” of the model; however, a PLS model evaluates only the presence of a correlation and not the direction of that correlation. Therefore, we checked for the appropriate direction of correlation between a covariate with a ViP of >90 and gene expression—positive in the case of CNV (since copy number loss/gain is expected to lead to down-/up-regulation, respectively) and negative in the case of promoter methylation (since hypermethylation is expected to lead to down-regulation)—by performing linear regression between that covariate and the transformed counts data. Of the 464 genes for which CNV featured a R2 above the cutoff, only 1 (0.22%) featured a correlation opposite to what we expected. Of the 786 promoter sites featuring a R2 above the cutoff, 69 (8.8%) featured the correlation opposite to what we expected.
Last, models were classified as being driven by CNV, methylation, or both based on the classifications of all identified drivers of that model (Table 2). A model was deemed “CNV-driven” (n = 446) if CNV was the only driver of the model and the correlation was in the expected direction. A model was “methylation-driven” (n = 690) if promoter sites were the only drivers of the model and all featured the expected direction of correlation. Models were “both-driven” (n = 17) if both CNV and promoter sites formed strong correlations, all in the expected directions. Only models classified as one of these three types were considered highly accurate. Models with a correlation opposite from expectation were classified as “CNV-opposite” (n = 1) or “methylation-opposite” (n = 60). Last, there were 19 models that featured promoter sites with both the expected and opposite directions of correlation, and these were classified as “mixed drivers.” Details for the model results, classifications, β values, mean group expression values, and correlations are available in table S8.
As this is a novel approach, we had no previous guidance for the selection of the R2 or ViP cutoffs. We therefore performed multiple analyses, varying the values of both parameters. We found that the relative proportions of each type of model (highly accurate, methylation-driven, and CNV-driven) remained similar and that the conclusions of our analyses were robust to changes in these values.
Analysis of publicly available data from the Heidelberg CNS tumor classifier (22)
DNA methylation raw data (.idat files) were downloaded from the NCBI Gene Expression Omnibus, accession number GSE109381. We identified all tumors diagnosed as meningiomas from this cohort (n = 144 of 3905) and processed their raw data with the ChAMP pipeline as described above. As these samples were analyzed using the Illumina 450K array (as opposed to the EPIC 850K array for our samples), the two datasets had only 3573 of the 7552 methylation signature probes in common. We used this shared subset as the methylation signature for this analysis. We performed t-distributed stochastic neighbor embedding (t-SNE) across the signature probes for the Heidelberg samples combined with our own using the Rtsne package (55). We then performed NMF using default settings among the Heidelberg samples across the signature probes, finding three groups to be optimal. Last, we trained a random forest model (base R function randomForest) across the signature probes using our cohort and their methylation types. We applied this model to the Heidelberg samples, finding that the assignments were highly concordant with the clusters generated by NMF of the Heidelberg samples alone (table S3).
Utilization of DNA methylation probe sets from Olar et al. (13)
We downloaded the list of probes (n = 283) from the online supplementary materials and found that 277 of them were included in our processed data. To determine what relative prognosis a probe conferred for each methylation type, we compared the mean β value of a given type to the mean of all other tumors. If the mean β value of a favorable probe (MM-FAV) was greater for that group (hypermethylated), it conferred a favorable prognosis; if the mean β value was less for that group (hypomethylated), it conferred an unfavorable prognosis. For unfavorable probes (MM-UNFAV), on the other hand, a greater mean β value conferred an unfavorable prognosis, and a lower mean β value conferred a favorable prognosis.
Analysis of data from Viaene et al. (30)
After receiving permission and access from the authors, we downloaded the raw data (FASTQ files) from CAVATICA of the Center for Data Driven Discovery in Biomedicine at the Children’s Hospital of Philadelphia Research Institute. We combined these files with the raw data from our samples and processed this cohort as stated above for our cohort. Because there was only RNA-seq data, we applied the transcriptional classifier from our prior study to these outside samples to assign their transcriptional type and their presumed MenG. We generated the heatmap using the ComplexHeatmap package (51) over the 1000 genes with the greatest variance in expression; we used the default settings of the built-in unsupervised hierarchical clustering algorithm to cluster the samples.
Analysis of publicly available data from Magill et al. (33)
We downloaded DNA methylation raw data (.idat files) from the NCBI Gene Expression Omnibus, accession number GSE151067. We combined these files with our own raw data and processed them with the ChAMP pipeline as described above. No samples were of low quality to warrant exclusion, and no significant batch effects were seen to warrant correction. We also downloaded the RNA-seq raw data in the form of raw counts from the NCBI Gene Expression Omnibus, accession number GSE151921. We combined these data with the raw counts from our own samples and then normalized and variance-transformed the data with the DESeq2 package (52).
When we reviewed the DNA methylation, RNA-seq, and cytogenetic profile of samples from their M2 tumor, we found it to be sui generis in all modalities. We therefore uploaded the data to Heidelberg CNS tumor methylation classifier (22) at https://molecularneuropathology.org/mnp. While the samples scored higher for meningioma than other tumor types, none of the samples were matched to meningioma, and only two could even be matched to the tumor family containing meningioma (table S7). One of the samples, M2-5, was matched by the classifier to “control tissue, hemispheric cortex,” suggesting that stereotactic biopsy sampled adjacent brain tissue. Given these abnormal findings for M2, we uploaded all other samples to the classifier as well. Of the 82 such samples, 76 (93%) were matched to meningioma. One tumor, M6, featured four of the nonmatched samples; these all featured relatively high scores for meningioma that did not meet the matching criteria. The remaining two nonmatched samples, both from M7, did not appear to be good meningioma samples. One sample, M7-1, matched to “control tissue, inflammatory tumor microenvironment,” while the other, M7-3, featured equal scores between that diagnosis and meningioma. We therefore removed these two samples from our analysis. Notably, these two samples were most divergent among M7, and their exclusion significantly reduces the dispersion (and therefore heterogeneity) of M7 on the DNA methylation PCA. In the analysis by Magill et al. (33), M7 featured the second-highest degree of heterogeneity for DNA methylation, and this finding appears to have been exaggerated by these two samples, although their conclusion of increased heterogeneity still stands when these samples are excluded.
To classify the epigenetic types of these samples, we first trained a random forest model (base R function randomForest) across the methylation signature probes using our samples and their methylation types and then applied this model to the samples from Magill et al. (33), to define their methylation types. To classify these samples transcriptionally, we applied the classifier (also a random forest model) from our prior study (17) to the processed RNA-seq data. CNV data were derived from DNA methylation using the conumee package (53) to classify the samples by chromosomal and NF2/instability (in conjunction with NF2 expression data). For fig. S4A, we used k-means clustering with k = 5 using the ConsensusClusterPlus package (48) to define clusters and organize the dendrogram.
Statistical analyses
All statistical analyses were performed in R (version 4.0.3); RFS analysis and subsequent survival data visualization used the survival and survminer packages (56, 57). Analysis of variance (ANOVA) and the chi-square test were used to compare clinical variables between groups, while the chi-square test was used to compare the proportions of PLS model types. For all analyses, we considered a P < 0.05 to be significant.
Acknowledgments
We thank the patients who entrusted us with care and participated in this study. We thank Magill et al. and Capper et al. for making NGS raw data freely available. We thank A. Resnick at the University of Pennsylvania for enabling us to access the data from Viaene et al. We thank K. A. Heck and J. C. Goodman for expert neuropathology. We also thank V. L. Brandt for discussions and commitment to clarity in editing the manuscript.
Funding: This work was supported by the NINDS (K08NS102474 to A.J.P. and R25NS070694 to J.C.B.), the Roderick D. MacDonald Fund (to A.J.P.), the Hamill Foundation (to A.J.P.), and P30 Cancer Center Support Grant NCI-CA125123. The Human Tissue Acquisition & Pathology Core at BCM is funded through P30 Cancer Center Support Grant NCI-CA125123.
Author contributions: Conceptualization: J.C.B., T.J.K., and A.J.P. Methodology: J.C.B., A.S.H., and A.J.P. Investigation: J.C.B., C.C.H., A.S.H., and A.J.P. Software: J.C.B., A.O.H., and A.S.H. Formal analysis: J.C.B., A.O.H., A.S.H., and A.J.P. Data curation: J.C.B., C.C.H., A.O.H., A.S.H., and A.J.P. Visualization: J.C.B., T.J.K., and A.J.P. Resources: A.O.H., A.S.H., T.J.K., and A.J.P. Writing—original draft: J.C.B. Writing—review and editing: A.S.H., T.J.K., and A.J.P. Funding acquisition: J.C.B. and A.J.P. Supervision: A.S.H., T.J.K., and A.J.P.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All raw and processed methylation and RNA-seq data have been deposited in the Gene Expression Omnibus database, https://ncbi.nlm.nih.gov/geo, under the following accession numbers: GSE189673 (SuperSeries), GSE189521 (DNA methylation), and GSE189672 (RNA-seq). All code for the analyses in this study has been deposited at Zenodo, https://doi.org/10.5281/zenodo.5809773. In addition, the code, along with subsequent updates and additions, is available at https://github.com/jbayley/meningiomas.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Table S2
Legends for tables S1, S3 to S8
Other Supplementary Material for this manuscript includes the following:
Tables S1, S3 to S8
REFERENCES AND NOTES
- 1.Ballester L. Y., Olar A., Roy-Chowdhuri S., Next-generation sequencing of central nervous systems tumors: The future of personalized patient management. Neuro Oncol. 18, 308–310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Northcott P. A., Korshunov A., Witt H., Hielscher T., Eberhart C. G., Mack S., Bouffet E., Clifford S. C., Hawkins C. E., French P., Rutka J. T., Pfister S., Taylor M. D., Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 29, 1408–1414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ClinicalTrials.gov, “Reduced craniospinal radiation therapy and chemotherapy in treating younger patients with newly diagnosed WNT-driven medulloblastoma,” Identifier NCT02724579, 3 March 2016; http://clinicaltrials.gov/ct2/show/NCT02724579.
- 4.Robinson G. W., Orr B. A., Wu G., Gururangan S., Lin T., Qaddoumi I., Packer R. J., Goldman S., Prados M. D., Desjardins A., Chintagumpala M., Takebe N., Kaste S. C., Rusch M., Allen S. J., Onar-Thomas A., Stewart C. F., Fouladi M., Boyett J. M., Gilbertson R. J., Curran T., Ellison D. W., Gajjar A., Vismodegib exerts targeted efficacy against recurrent sonic hedgehog-subgroup medulloblastoma: Results from phase II pediatric brain tumor consortium studies PBTC-025B and PBTC-032. J. Clin. Oncol. 33, 2646–2654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons D. W., Jones S., Zhang X., Lin J. C.-H., Leary R. J., Angenendt P., Mankoo P., Carter H., Siu I.-M., Gallia G. L., Olivi A., McLendon R., Rasheed B. A., Keir S., Nikolskaya T., Nikolsky Y., Busam D. A., Tekleab H., Diaz L. A., Hartigan J., Smith D. R., Strausberg R. L., Marie S. K. N., Shinjo S. M. O., Yan H., Riggins G. J., Bigner D. D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W., An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahm F., Reuss D., Koelsche C., Capper D., Schittenhelm J., Heim S., Jones D. T. W., Pfister S. M., Herold-Mende C., Wick W., Mueller W., Hartmann C., Paulus W., von Deimling A., Farewell to oligoastrocytoma: In situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol. 128, 551–559 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Wierzbicki K., Ravi K., Franson A., Bruzek A., Cantor E., Harris M., Homan M. J., Marini B. L., Kawakibi A. R., Ravindran R., Teodoro R., Yadav V. N., Koschmann C., Targeting and therapeutic monitoring of H3K27M-mutant glioma. Curr. Oncol. Rep. 22, 19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harmancı A. S., Youngblood M. W., Clark V. E., Coşkun S., Henegariu O., Duran D., Erson-Omay E. Z., Kaulen L. D., Lee T. I., Abraham B. J., Simon M., Krischek B., Timmer M., Goldbrunner R., Omay S. B., Baranoski J., Baran B., Carrión-Grant G., Bai H., Mishra-Gorur K., Schramm J., Moliterno J., Vortmeyer A. O., Bilgüvar K., Yasuno K., Young R. A., Günel M., Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat. Commun. 8, 14433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark V. E., Erson-Omay E. Z., Serin A., Yin J., Cotney J., Ozduman K., Avşar T., Li J., Murray P. B., Henegariu O., Yilmaz S., Günel J. M., Carrión-Grant G., Yilmaz B., Grady C., Tanrikulu B., Bakircioğlu M., Kaymakçalan H., Caglayan A. O., Sencar L., Ceyhun E., Atik A. F., Bayri Y., Bai H., Kolb L. E., Hebert R. M., Omay S. B., Mishra-Gorur K., Choi M., Overton J. D., Holland E. C., Mane S., State M. W., Bilgüvar K., Baehring J. M., Gutin P. H., Piepmeier J. M., Vortmeyer A., Brennan C. W., Pamir M. N., Kiliç T., Lifton R. P., Noonan J. P., Yasuno K., Günel M., Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 339, 1077–1080 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark V. E., Harmancı A. S., Bai H., Youngblood M. W., Lee T. I., Baranoski J. F., Ercan-Sencicek A. G., Abraham B. J., Weintraub A. S., Hnisz D., Simon M., Krischek B., Erson-Omay E. Z., Henegariu O., Carrión-Grant G., Mishra-Gorur K., Durán D., Goldmann J. E., Schramm J., Goldbrunner R., Piepmeier J. M., Vortmeyer A. O., Günel J. M., Bilgüvar K., Yasuno K., Young R. A., Günel M., Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat. Genet. 48, 1253–1259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brastianos P. K., Horowitz P. M., Santagata S., Jones R. T., McKenna A., Getz G., Ligon K. L., Palescandolo E., Van Hummelen P., Ducar M. D., Raza A., Sunkavalli A., Macconaill L. E., Stemmer-Rachamimov A. O., Louis D. N., Hahn W. C., Dunn I. F., Beroukhim R., Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat. Genet. 45, 285–289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bi W. L., Greenwald N. F., Abedalthagafi M., Wala J., Gibson W. J., Agarwalla P. K., Horowitz P., Schumacher S. E., Esaulova E., Mei Y., Chevalier A., Ducar M., Thorner A. R., van Hummelen P., Stemmer-Rachamimov A., Artyomov M., Al-Mefty O., Dunn G. P., Santagata S., Dunn I. F., Beroukhim R., Genomic landscape of high-grade meningiomas. NPJ Genom. Med. 2, 15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olar A., Wani K. M., Wilson C. D., Zadeh G., DeMonte F., Jones D. T. W., Pfister S. M., Sulman E. P., Aldape K. D., Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma. Acta Neuropathol. 133, 431–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahm F., Schrimpf D., Stichel D., Jones D. T. W., Hielscher T., Schefzyk S., Okonechnikov K., Koelsche C., Reuss D. E., Capper D., Sturm D., Wirsching H.-G., Berghoff A. S., Baumgarten P., Kratz A., Huang K., Wefers A. K., Hovestadt V., Sill M., Ellis H. P., Kurian K. M., Okuducu A. F., Jungk C., Drueschler K., Schick M., Bewerunge-Hudler M., Mawrin C., Seiz-Rosenhagen M., Ketter R., Simon M., Westphal M., Lamszus K., Becker A., Koch A., Schittenhelm J., Rushing E. J., Collins V. P., Brehmer S., Chavez L., Platten M., Hänggi D., Unterberg A., Paulus W., Wick W., Pfister S. M., Mittelbronn M., Preusser M., Herold-Mende C., Weller M., von Deimling A., DNA methylation-based classification and grading system for meningioma: A multicentre, retrospective analysis. Lancet Oncol. 18, 682–694 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Nassiri F., Mamatjan Y., Suppiah S., Badhiwala J. H., Mansouri S., Karimi S., Saarela O., Poisson L., Gepfner-Tuma I., Schittenhelm J., Ng H.-K., Noushmehr H., Harter P., Baumgarten P., Weller M., Preusser M., Herold-Mende C., Tatagiba M., Tabatabai G., Sahm F., von Deimling A.; International Consortium on Meningiomas, Zadeh G., Aldape K. D., DNA methylation profiling to predict recurrence risk in meningioma: Development and validation of a nomogram to optimize clinical management. Neuro Oncol. 21, 901–910 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Choudhury, S. T. Magill, C. D. Eaton, B. C. Prager, W. C. Chen, K. Seo, C.-H. G. Lucas, J. E. Villanueva-Meyer, T.-C. Lam, J. K.-S. Pu, L.-F. Li, G. K.-K. Leung, H. N. Vasudevan, S. J. Liu, J. W. Chan, Z. Qiu, M. Y. Zhang, M. V. Martin, M. S. Susko, S. E. Braunstein, N. A. Oberheim Bush, J. Schulte, N. Butowski, P. K. Sneed, M. S. Berger, A. Perry, J. J. Phillips, D. A. Solomon, J. F. Costello, M. W. McDermott, J. N. Rich, D. R. Raleigh, Meningioma epigenetic grouping reveals biologic drivers and therapeutic vulnerabilities. medRxiv 2020.11.23.20237495 [Preprint]. 27 November 2020; 10.1101/2020.11.23.20237495. [DOI]
- 17.Patel A. J., Wan Y.-W., Al-Ouran R., Revelli J.-P., Cardenas M. F., Oneissi M., Xi L., Jalali A., Magnotti J. F., Muzny D. M., Doddapaneni H., Sebastian S., Heck K. A., Goodman J. C., Gopinath S. P., Liu Z., Rao G., Plon S. E., Yoshor D., Wheeler D. A., Zoghbi H. Y., Klisch T. J., Molecular profiling predicts meningioma recurrence and reveals loss of DREAM complex repression in aggressive tumors. Proc. Natl. Acad. Sci. U.S.A. 116, 21715–21726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasudevan H. N., Braunstein S. E., Phillips J. J., Pekmezci M., Tomlin B. A., Wu A., Reis G. F., Magill S. T., Zhang J., Feng F. Y., Nicholaides T., Chang S. M., Sneed P. K., McDermott M. W., Berger M. S., Perry A., Raleigh D. R., Comprehensive molecular profiling identifies FOXM1 as a key transcription factor for meningioma proliferation. Cell Rep. 22, 3672–3683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis D. N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W. K., Ohgaki H., Wiestler O. D., Kleihues P., Ellison D. W., The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 131, 803–820 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Rogers L., Barani I., Chamberlain M., Kaley T. J., McDermott M., Raizer J., Schiff D., Weber D. C., Wen P. Y., Vogelbaum M. A., Meningiomas: Knowledge base, treatment outcomes, and uncertainties. A RANO review. J. Neurosurg. 122, 4–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes L. A. E., Melotte V., de Schrijver J., de Maat M., Smit V. T. H. B. M., Bovée J. V. M. G., French P. J., van den Brandt P. A., Schouten L. J., de Meyer T., van Criekinge W., Ahuja N., Herman J. G., Weijenberg M. P., van Engeland M., The CpG island methylator phenotype: What’s in a name? Cancer Res. 73, 5858–5868 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Capper D., Jones D. T. W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D. E., Kratz A., Wefers A. K., Huang K., Pajtler K. W., Schweizer L., Stichel D., Olar A., Engel N. W., Lindenberg K., Harter P. N., Braczynski A. K., Plate K. H., Dohmen H., Garvalov B. K., Coras R., Hölsken A., Hewer E., Bewerunge-Hudler M., Schick M., Fischer R., Beschorner R., Schittenhelm J., Staszewski O., Wani K., Varlet P., Pages M., Temming P., Lohmann D., Selt F., Witt H., Milde T., Witt O., Aronica E., Giangaspero F., Rushing E., Scheurlen W., Geisenberger C., Rodriguez F. J., Becker A., Preusser M., Haberler C., Bjerkvig R., Cryan J., Farrell M., Deckert M., Hench J., Frank S., Serrano J., Kannan K., Tsirigos A., Brück W., Hofer S., Brehmer S., Seiz-Rosenhagen M., Hänggi D., Hans V., Rozsnoki S., Hansford J. R., Kohlhof P., Kristensen B. W., Lechner M., Lopes B., Mawrin C., Ketter R., Kulozik A., Khatib Z., Heppner F., Koch A., Jouvet A., Keohane C., Mühleisen H., Mueller W., Pohl U., Prinz M., Benner A., Zapatka M., Gottardo N. G., Driever P. H., Kramm C. M., Müller H. L., Rutkowski S., von Hoff K., Frühwald M. C., Gnekow A., Fleischhack G., Tippelt S., Calaminus G., Monoranu C.-M., Perry A., Jones C., Jacques T. S., Radlwimmer B., Gessi M., Pietsch T., Schramm J., Schackert G., Westphal M., Reifenberger G., Wesseling P., Weller M., Collins V. P., Blümcke I., Bendszus M., Debus J., Huang A., Jabado N., Northcott P. A., Paulus W., Gajjar A., Robinson G. W., Taylor M. D., Jaunmuktane Z., Ryzhova M., Platten M., Unterberg A., Wick W., Karajannis M. A., Mittelbronn M., Acker T., Hartmann C., Aldape K., Schüller U., Buslei R., Lichter P., Kool M., Herold-Mende C., Ellison D. W., Hasselblatt M., Snuderl M., Brandner S., Korshunov A., von Deimling A., Pfister S. M., DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prager B. C., Vasudevan H. N., Dixit D., Bernatchez J. A., Wu Q., Wallace L. C., Bhargava S., Lee D., King B. H., Morton A. R., Gimple R. C., Pekmezci M., Zhu Z., Siqueira-Neto J. L., Wang X., Xie Q., Chen C., Barnett G. H., Vogelbaum M. A., Mack S. C., Chavez L., Perry A., Raleigh D. R., Rich J. N., The meningioma enhancer landscape delineates novel subgroups and drives druggable dependencies. Cancer Discov. 10, 1722–1741 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozen O., Demirhan B., Altinörs N., Correlation between histological grade and MIB-1 and p53 immunoreactivity in Meningiomas. Clin. Neuropathol. 24, 219–224 (2005). [PubMed] [Google Scholar]
- 25.Olar A., Wani K. M., Sulman E. P., Mansouri A., Zadeh G., Wilson C. D., DeMonte F., Fuller G. N., Aldape K. D., Mitotic index is an independent predictor of recurrence-free survival in Meningioma. Brain Pathol. 25, 266–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen K. D., Timp W., Bravo H. C., Sabunciyan S., Langmead B., McDonald O. G., Wen B., Wu H., Liu Y., Diep D., Briem E., Zhang K., Irizarry R. A., Feinberg A. P., Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 43, 768–775 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao X., Lv N., Liao J., Long J., Xue R., Ai N., Xu D., Fan X., Copy number variation is highly correlated with differential gene expression: A pan-cancer study. BMC Med. Genet. 20, 175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boulesteix A.-L., Strimmer K., Partial least squares: A versatile tool for the analysis of high-dimensional genomic data. Brief. Bioinform. 8, 32–44 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Li W., Zhang S., Liu C.-C., Zhou X. J., Identifying multi-layer gene regulatory modules from multi-dimensional genomic data. Bioinformatics 28, 2458–2466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viaene A. N., Zhang B., Martinez-Lage M., Xiang C., Tosi U., Thawani J. P., Gungor B., Zhu Y., Roccograndi L., Zhang L., Bailey R. L., Storm P. B., O’Rourke D. M., Resnick A. C., Grady M. S., Dahmane N., Transcriptome signatures associated with meningioma progression. Acta Neuropathol. Commun. 7, 67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfisterer W. K., Hank N. C., Preul M. C., Hendricks W. P., Pueschel J., Coons S. W., Scheck A. C., Diagnostic and prognostic significance of genetic regional heterogeneity in Meningiomas. Neuro Oncol. 6, 290–299 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juratli T. A., Thiede C., Koerner M. V. A., Tummala S. S., Daubner D., Shankar G. M., Williams E. A., Martinez-Lage M., Soucek S., Robel K., Penson T., Krause M., Appold S., Meinhardt M., Pinzer T., Miller J. J., Krex D., Ely H. A., Silverman I. M., Christiansen J., Schackert G., Wakimoto H., Kirsch M., Brastianos P. K., Cahill D. P., Intratumoral heterogeneity and TERT promoter mutations in progressive/higher-grade meningiomas. Oncotarget 8, 109228–109237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magill S. T., Vasudevan H. N., Seo K., Villanueva-Meyer J. E., Choudhury A., John Liu S., Pekmezci M., Findakly S., Hilz S., Lastella S., Demaree B., Braunstein S. E., Bush N. A. O., Aghi M. K., Theodosopoulos P. V., Sneed P. K., Abate A. R., Berger M. S., McDermott M. W., Lim D. A., Ullian E. M., Costello J. F., Raleigh D. R., Multiplatform genomic profiling and magnetic resonance imaging identify mechanisms underlying intratumor heterogeneity in meningioma. Nat. Commun. 11, 4803 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Driver J., Hoffman S. E., Tavakol S., Woodward E., Maury E. A., Bhave V., Greenwald N. F., Nassiri F., Aldape K., Zadeh G., Choudhury A., Vasudevan H. N., Magill S. T., Raleigh D. R., Abedalthagafi M., Aizer A. A., Alexander B. M., Ligon K. L., Reardon D. A., Wen P. Y., Al-Mefty O., Ligon A. H., Dubuc A. M., Beroukhim R., Claus E. B., Dunn I. F., Santagata S., Bi W. L., A molecularly integrated grade for meningioma. Neuro Oncol. 2021, noab213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maas S. L. N., Stichel D., Hielscher T., Sievers P., Berghoff A. S., Schrimpf D., Sill M., Euskirchen P., Blume C., Patel A., Dogan H., Reuss D., Dohmen H., Stein M., Reinhardt A., Suwala A. K., Wefers A. K., Baumgarten P., Ricklefs F., Rushing E. J., Bewerunge-Hudler M., Ketter R., Schittenhelm J., Jaunmuktane Z., Leu S., Greenway F. E. A., Bridges L. R., Jones T., Grady C., Serrano J., Golfinos J., Sen C., Mawrin C., Jungk C., Hänggi D., Westphal M., Lamszus K., Etminan N., Jungwirth G., Herold-Mende C., Unterberg A., Harter P. N., Wirsching H.-G., Neidert M. C., Ratliff M., Platten M., Snuderl M., Aldape K. D., Brandner S., Hench J., Frank S., Pfister S. M., Jones D. T. W., Reifenberger G., Acker T., Wick W., Weller M., Preusser M., von Deimling A., Sahm F.; German Consortium on Aggressive Meningiomas (KAM) , Integrated molecular-morphologic meningioma classification: A multicenter retrospective analysis, retrospectively and prospectively validated. J. Clin. Oncol. 39, 3839–3852 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nassiri F., Liu J., Patil V., Mamatjan Y., Wang J. Z., Hugh-White R., Macklin A. M., Khan S., Singh O., Karimi S., Corona R. I., Liu L. Y., Chen C. Y., Chakravarthy A., Wei Q., Mehani B., Suppiah S., Gao A., Workewych A. M., Tabatabai G., Boutros P. C., Bader G. D., de Carvalho D. D., Kislinger T., Aldape K., Zadeh G., A clinically applicable integrative molecular classification of meningiomas. Nature 597, 119–125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paramasivam N., Hübschmann D., Toprak U. H., Ishaque N., Neidert M., Schrimpf D., Stichel D., Reuss D., Sievers P., Reinhardt A., Wefers A. K., Jones D. T. W., Gu Z., Werner J., Uhrig S., Wirsching H.-G., Schick M., Bewerunge-Hudler M., Beck K., Brehmer S., Urbschat S., Seiz-Rosenhagen M., Hänggi D., Herold-Mende C., Ketter R., Eils R., Ram Z., Pfister S. M., Wick W., Weller M., Grossmann R., von Deimling A., Schlesner M., Sahm F., Mutational patterns and regulatory networks in epigenetic subgroups of meningioma. Acta Neuropathol. 138, 295–308 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Zhang H., Qi L., Du Y., Huang L. F., Braun F. K., Kogiso M., Zhao Y., Li C., Lindsay H., Zhao S., Injac S. G., Baxter P. A., Su J. M., Stephan C., Keller C., Heck K. A., Harmanci A., Harmanci A. O., Yang J., Klisch T. J., Li X.-N., Patel A. J., Patient-derived orthotopic xenograft (PDOX) mouse models of primary and recurrent meningioma. Cancers 12, 1478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris T. J., Butcher L. M., Feber A., Teschendorff A. E., Chakravarthy A. R., Wojdacz T. K., Beck S., ChAMP: 450k chip analysis methylation pipeline. Bioinformatics 30, 428–430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aryee M. J., Jaffe A. E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A. P., Hansen K. D., Irizarry R. A., Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fortin J.-P., Triche T. J., Hansen K. D., Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 33, 558–560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W., Laird P. W., Shen H., Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 45, e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teschendorff A. E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S., A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29, 189–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teschendorff A. E., Menon U., Gentry-Maharaj A., Ramus S. J., Gayther S. A., Apostolidou S., Jones A., Lechner M., Beck S., Jacobs I. J., Widschwendter M., An epigenetic signature in peripheral blood predicts active ovarian cancer. PLOS ONE 4, e8274 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou W., Triche T. J., Laird P. W., Shen H., SeSAMe: Reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 46, e123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beaubier N., Tell R., Lau D., Parsons J. R., Bush S., Perera J., Sorrells S., Baker T., Chang A., Michuda J., Iguartua C., MacNeil S., Shah K., Ellis P., Yeatts K., Mahon B., Taxter T., Bontrager M., Khan A., Huether R., Lefkofsky E., White K. P., Clinical validation of the tempus xT next-generation targeted oncology sequencing assay. Oncotarget 10, 2384–2396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaujoux R., Seoighe C., A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11, 367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkerson M. D., Hayes D. N., ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth G. K., Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article 3 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Wettenhall J. M., Smyth G. K., limmaGUI: A graphical user interface for linear modeling of microarray data. Bioinformatics 20, 3705–3706 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.V. Hovestadt, M. Zapatka, conumee: Enhanced copy-number variation analysis using Illumina DNA methylation arrays (2017); http://bioconductor.org/packages/conumee/. [DOI] [PMC free article] [PubMed]
- 54.M. Kuhn, J. Wing, S. Weston, A. Williams, C. Keefer, A. Engelhardt, T. Cooper, Z. Mayer, B. Kenkel, R Core Team, M. Benesty, R. Lescarbeau, A. Ziem, L. Scrucca, Y. Tang, C. Candan, T. Hunt, caret: Classification and regression training (2021); https://CRAN.R-project.org/package=caret.
- 55.J. H. Krijthe, Rtsne: T-distributed stochastic neighbor embedding using a Barnes-Hut Implementation (2015); https://github.com/jkrijthe/Rtsne.
- 56.T. Therneau, T. Lumley, A. Elizabeth, C. Cynthia, survival: Survival analysis (2021); https://cran.r-project.org/web/packages/survival/index.html.
- 57.A. Kassambara, M. Kosinski, P. Biecek, S. Fabian, survminer: Drawing survival curves using ‘ggplot2’ (2021); https://CRAN.R-project.org/package=survminer.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Table S2
Legends for tables S1, S3 to S8
Tables S1, S3 to S8