Abstract
Vocal production learning is a convergently evolved trait in vertebrates. To identify brain genomic elements associated with mammalian vocal learning, we integrated genomic, anatomical and neurophysiological data from the Egyptian fruit-bat with analyses of the genomes of 215 placental mammals. First, we identified a set of proteins evolving more slowly in vocal learners. Then, we discovered a vocal-motor cortical region in the Egyptian fruit-bat, an emergent vocal learner, and leveraged that knowledge to identify active cis-regulatory elements in the motor cortex of vocal learners. Machine learning methods applied to motor cortex open chromatin revealed 50 enhancers robustly associated with vocal learning whose activity tended to be lower in vocal learners. Our research implicates convergent losses of motor cortex regulatory elements in mammalian vocal learning evolution.
One-Sentence Summary:
A bat vocal brain region is identified and leveraged in comparative genomic analyses to reveal the evolution of mammal vocal behavior.
Vocal production learning—the ability of an organism to modify the acoustic properties of its vocalizations as a result of social experience—is an example of convergent evolution, having evolved independently within multiple lineages of birds and mammals, including humans, where it manifests as speech (Fig. 1A) (1, 2). Vocal production learning (“vocal learning”) has been extensively studied in songbirds, highlighting numerous shared behavioral features of birdsong and speech learning, such as a dependence on auditory input during a critical developmental period and a juvenile babbling phase of sensorimotor exploration prior to the maturation of the adult vocalizations (1). Convergence between song-learning birds and humans extends to neuroanatomical specializations, including direct corticobulbar projections from the vocal motor cortex analog to the hindbrain motoneurons controlling the vocal apparatus (3) and shared transcriptional specializations in analogous speech- and song-specialized brain regions (4). Thus, songbirds have become a premier model for exploring the fundamental brain anatomical, molecular, and genomic features associated with vocal learning (1, 3). An expanding literature on vocal learning behavior across mammals suggests an underappreciated diversity in the phenotypic expression of vocal learning across the taxa traditionally thought to possess it (2, 5–8). Study of the diverse forms of mammalian vocal learning behaviors could broaden our understanding of the core molecular, anatomical and physiological brain mechanisms of vocal learning and of the mechanisms underlying the convergent evolution of skilled motor behaviors more broadly.
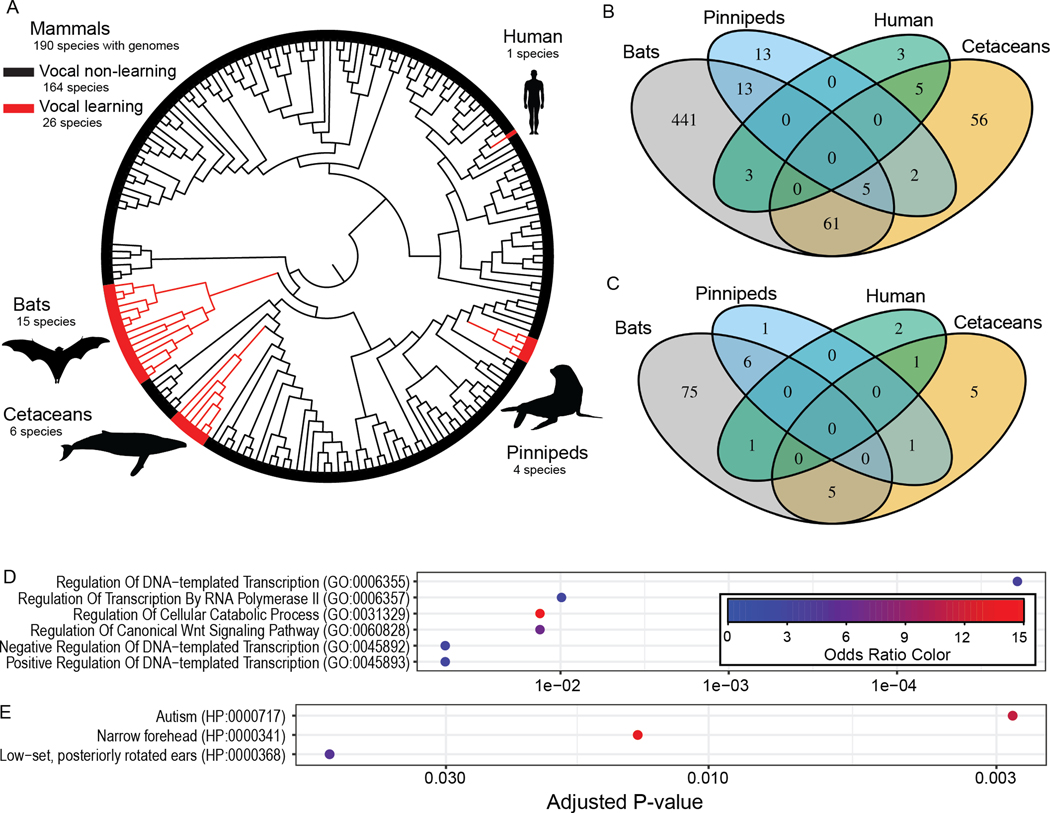
Figure 1. Convergent changes in protein sequence associated with vocal learning evolution across 215 mammalian species.
(A) A cladogram of mammalian species whose genomes were analyzed in this study highlights the convergent evolution of vocal learning species (in red) relative to non-learners (in black). The phylogenetic tree used in our analyses was derived from (98). Each of the genes implicated by RERconverge with lower (B) or higher (C) evolutionary rates in vocal learners are annotated based on whether or not they show a significant signature within the four vocal learning clades based on a Bayes factor ≥ 5 (18). All significant gene ontology categories (adjusted p < 0.10, EnrichR) are plotted for the 200 genes with conserved (D) and accelerated (E) selection in vocal learning clades, based on the combination of RERconverge and HyPhy RELAX. The points are colored by the odds ratio within the set of implicated genes relative to the genes outside of the set, which corresponds to the degree of enrichment within that set.
We evaluated evidence of convergent genomic specializations shared among four lineages of vocal learning mammals that independently evolved this trait — bats, cetaceans, pinnipeds, and humans (Fig. 1A) — using whole-genome datasets and recently developed computational approaches. Specifically, we used protein-coding sequences from genomes generated by the Zoonomia Consortium (9, 10) and models of evolutionary rate convergence (11, 12) to identify protein-coding regions repeatedly associated with the evolution of vocal learning across mammals. Although we found 200 protein-coding genes significantly associated with vocal learning, none of them showed strong evidence of selection in all four mammalian clades and only five showed strong evidence of selection in three out of the four clades. Due to individual lineages contributing disproportionately to many of the protein-coding results, we hypothesized that noncoding regulatory elements might also be under constraint for the evolution of vocal learning.. We next profiled open chromatin, a proxy for regulatory element activity (13), in multiple brain regions and somatic tissues in the Egyptian fruit bat, a mammal with robust vocal plasticity (14–16) to identify vocalization-associated regulatory genomic specializations. We accomplished this by combining anatomical tracing and electrophysiological recordings in vocalizing bats to identify a region of motor cortex associated with vocal production. The vocalization-associated epigenomic data collected from this region of this bat species — combined with hundreds of mammalian genomes (17, 18), their associated reference-free whole-genome alignments (19), and high-quality epigenomic data from the motor cortex of multiple additional mammalian species (20–22) — provided the foundation to apply a machine learning approach, the Tissue-Aware Conservation Inference Toolkit (TACIT) (23). This approach allowed us to identify putative enhancers, distal regulatory elements that tend to be highly tissue-specific, associated with the convergent evolution of vocal learning. In sum, we combined recently developed computational tools and neuroanatomical experiments in the Egyptian fruit bat and found evidence of convergent evolution in both protein-coding and noncoding DNA sequences.
Results:
Convergent Evolution in Protein Sequence Associated with Vocal Learning Behavior
To explore the possibility of shared genomic specializations associated with vocal learning, we first applied RERconverge (11) to recently released protein-coding alignments obtained for hundreds of mammals (10) to identify protein-coding genes whose relative rates of evolution differ between vocal learners and other mammals, and which may thus be under selection related to vocal learning (11, 24). We analyzed 16,209 high-quality protein-coding gene alignments across 215 boreoeutherian mammals, including 26 vocal learning species, 164 vocal non-learners, and 25 species without confident annotations (Fig. 1A, Data S1; Materials and Methods). We found evidence for lower evolutionary rates in vocal learners compared to non-learners in 804 genes and evidence for elevated evolutionary rates in 102 genes (Tau adj. p < 0.01 and permulations (24) adj. p < 0.01) (Data S2; Fig. S1). To identify which specific clades were driving the differential rates of evolution, we applied a Bayes Factor analysis that examines each clade individually for evidence of selection (18) (Data S2). Despite the large number of significant associations based on RERconverge, we found no single protein-coding gene with consistently lower or elevated evolutionary rates in all four vocal learning clades. Among the genes with reduced evolutionary rates in vocal learning species, we found only five out of 804 protein-coding genes with strong evidence of selective pressure in three out of the four vocal learning clades: CENPC, CATSPERG, MGA, TREML2, and ZCWPW1 (Bayes Factor > 5) (Fig. 1B,C). None of these proteins reached the threshold for selection in the human lineage (Materials and Methods), which could indicate different mechanisms of evolution in the laurasiatherian vocal learning clades relative to humans. Our results suggest that the vast majority of protein-coding genes that we identify are evolving much faster or much slower in one clade, but are only weakly associated with vocal learning across the other lineages.
The most strongly associated genes were CENPC (Fig. S2A; Tau = −0.30; Tau adj. p = 8.7 × 10−6; permulations adj. p <= 0.001), and GRM8 (Fig. S2B, Tau = 0.26; Tau adj. p = 3.5×10−4; permulations adj. p <= 0.001). GRM8 represents an especially promising candidate because it has previously been linked to anatomical specializations for vocal learning in songbirds (25) and is a known target of the speech-associated FOXP2 transcription factor (26). In primates and rodents, both GRM8 and FOXP2 are markers of a rare class of medium spiny neurons implicated in motor control that coexpress both DRD1 and DRD2 dopamine receptors (27–29). Overall, our results support a model where vocal learning behavior is only partially explained by differences in protein-coding gene evolutionary rates.
To further explore selection on these vocal learning-associated protein-coding genes, we applied an additional set of tools from the HyPhy package that compares non synonymous (dN) and synonymous (dS) substitution rates in the nucleotides of the amino acid sequence (30). Here, these sensitive evolutionary models of codon substitution formally compare selective regimes, modeled as dN/dS distributions, between branches annotated with the vocal learning phenotype and the rest of the phylogenetic tree (12, 30). The results were largely consistent with amino-acid level methods: the protein-coding genes with lower protein evolutionary rates in vocal learning clades also tended to be under higher constraint in vocal learning clades (Wilcoxon p = 2.5 × 10−4), and the genes with higher evolutionary rates in vocal learning species showed evidence of accelerated evolution in these same clades (Wilcoxon p = 7.9 × 10−9) (Fig. S3; Data S3). To further explore the functional trends of protein-coding genes associated with the evolution of vocal learning, we focused on the set of proteins that showed consistent behavior between RERconverge and the HyPhy RELAX model (Benjamini-Hochberg false discovery rate q < 0.05). This yielded a set of 126 proteins that were more slowly evolving in vocal learning clades and 74 with an elevated rate of evolutionary changes relative to other species. The complementary approaches of RERconverge and HyPhy RELAX identify a total of 200 vocal learning-associated genes and suggest that this behavior, vocal learning, is having a substantial impact on protein evolutionary rates.
We further interrogated evolutionary pressures across the vocal learning-associated genes by looking for evidence of diversifying position selection using the HyPhy BUSTED-PH model. Evidence of diversifying positive selection was found in 6.3% (13 transcripts, 9 genes) within the set of genes with elevated rates of evolution in vocal learning species based on RER-Converge and the HyPhy RELAX model. As expected, we identified much lower rates of diversifying positive selection in the gene with lower rates of evolution in vocal learning species (1.0%) and within the set of randomly chosen transcripts (2.0%). Among the 9 genes that showed evidence of positive selection, 8 have been associated with neurodevelopment (CCDC136, KIDINS220, LRRN1, RSG5, CYLD, GABRA5, NETO2, KIAA1109) (31–38). The gene CCDC136 has more directly been associated with multiple language-related phenotypes in humans(31, 39, 40). These results suggest that the vocal learning-associated genes across mammals may tend to play a role in human brain development and vocal behavior.
To more systematically explore the functional trends within these 126 and 74 protein-coding genes, we conducted a gene ontology analysis using EnrichR. Protein-coding genes with lower evolutionary rates in vocal learning species were associated with Regulation of DNA-templated Transcription (p=1.10×10−6, adj. p=1.9×10−5), Regulation of Canonical Wnt Signaling Pathway (p=5.1×10−5, adj. p=0.013), and the Autism human phenotype ontology (p=5.8×10−6, adj. p=0.0028) (Fig. 1F,G; Data S4). The genes with accelerated evolutionary rates were not enriched for any pathways at an adjusted p < 0.05 with at least 5 genes contributing. The enrichment of autism-associated genes among the set of genes with greater levels of conservation in vocal learners (MECP2, RAD21, DYRK1A, SIM1, FTSJ1, MEIS2, FGFR1) is particularly interesting given prevalence of speech delay and early vocal production differences in human subjects with autism (41, 42) and the previous association between autism genes and the evolution of vocal behavior in birds and bats (43, 44). Based on the association with autism, we further explored the function of the vocal learning-conserved gene set in the context of early vocal production differences. Although only four human loci have been associated with differences in early vocal production, protein-coding genes overlapping two of these loci show higher levels of conservation in vocal learning clades (INSC, DAPK3) (45).
Identification of a Vocal Production Region in Egyptian Fruit Bats
The enrichment of transcription factors in the set of vocal learning-associated proteins suggests that differences in gene regulation are likely to be a major factor in the evolution of vocal learning. Since gene regulation is often tissue-specific, we sought to identify motor regions of the brain involved in vocal production and contrast their epigenomic profiles with motor regions not involved in vocal production. We conducted this comparison in the Egyptian fruit bat, Rousettus aegyptiacus, a mammalian species with robust vocal plasticity (16, 46) and with data on its motor cortex mapping (47). To identify a candidate region, we were guided by the hypothesis that fine vocal-motor control, a key ability to vocal learning, may be associated with the anatomical specialization of the motor cortex (48–51). In particular, previous work suggested that a cortical region controlling complex vocal behavior would be characterized by a direct, monosynaptic projection onto the motoneurons controlling the vocal source (in mammals, the larynx) (48–52). Such a direct connection has been observed robustly in humans (53–56) and vocal learning birds (songbirds, parrots and hummingbirds, (57–59)), but has not been reliably found in vocal non-learning species such as chimpanzees (41) or mice (60)
We first determined whether a direct corticobulbar anatomical connection existed in R. aegyptiacus. Guided by cortical mapping experiments (47), we injected anterograde tracers into the part of the motor cortex that has been associated with orofacial motor control (ofM1) and identified fluorescently labeled descending cortical fibers in the hindbrain region where the laryngeal motoneurons reside: the nucleus ambiguus (NA) (Fig. 2A, Fig. S4A-B and Movie S1). To test the existence of a direct monosynaptic projection, we also specifically identified laryngeal motoneurons in the NA by retrogradely labeling them through bilateral muscular injection of CTB (Cholera Toxin B) into the cricothyroid muscles of the bat larynx (Fig. 2A). We validated the colocalization of descending cortical fibers and local synaptic boutons with laryngeal motoneurons using two complementary labeling approaches: one relying on immunostaining of synapses (VGLUT1) and the other one using viral labeling of synapses (SYN) (Fig. 2B-F; Fig. S5). Across five bats, 79.2% of the retrogradely labeled motoneurons (61/77) colocalized with descending cortical fibers and 26% of them (20/77) colocalized with both cortical fibers and synaptic boutons, pointing to the existence of a robust direct corticospinal projection to laryngeal motoneurons (Fig. 2G). This colocalization in the NA was consistent across the different techniques (Fig. 2G, Fig. S5) and could not be found in any other brainstem motor nuclei, including the hypoglossal nucleus, which controls the tongue and neck muscles (Fig. S4C-E). We noted that the corticobulbar fibers crossed the midline anterior to the NA at the level of the facial nucleus, offering a direct contralateral path for the innervation of the NA (Fig. S4F). These anatomical findings highlight the bat ofM1 as a possible candidate region associated with vocal production.
Figure 2. Identification of an anatomically specialized motor cortical region targeting laryngeal motoneurons in the Egyptian fruit bat.
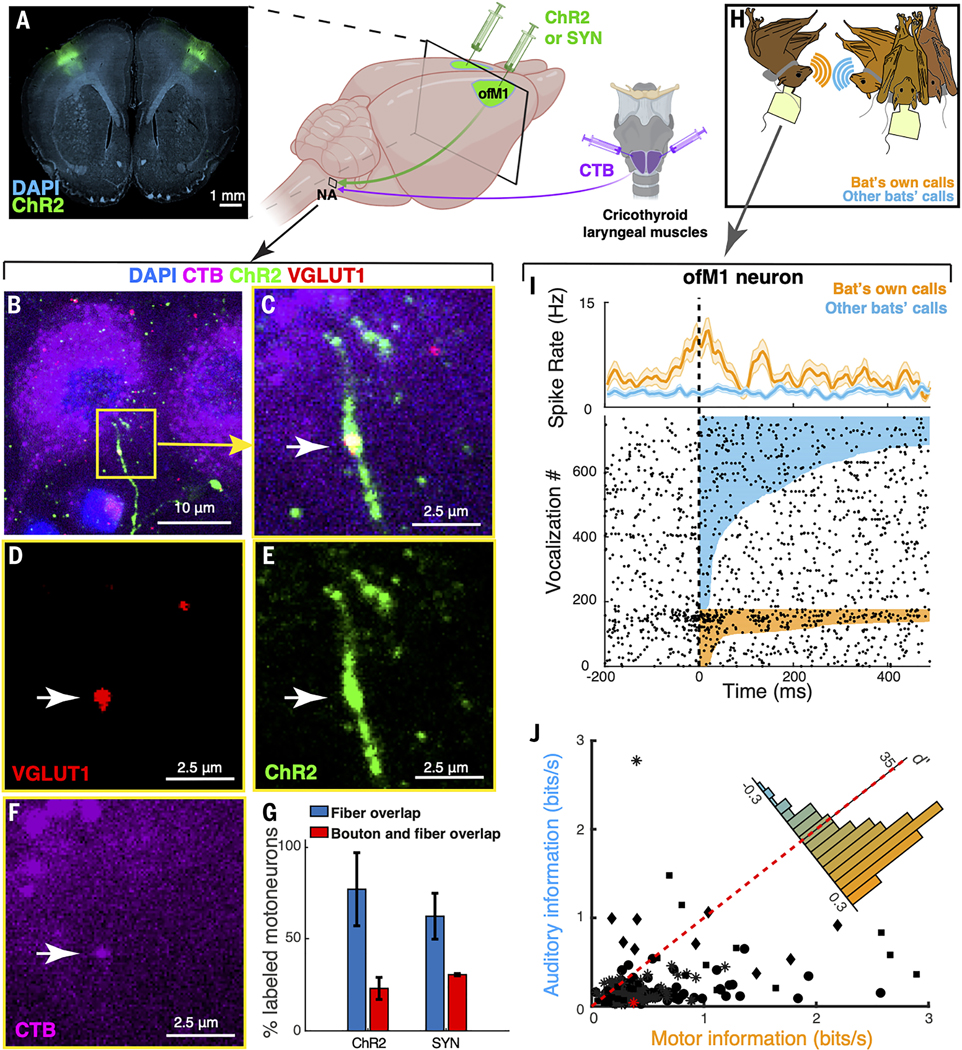
(A) Right: schematic of anatomical tracing approaches. Retrograde tracer cholera toxin B (CTB, purple) was injected bilaterally into the cricothyroid muscles to label brainstem motoneurons in nucleus ambiguus (NA). Simultaneously, an anterograde viral tracer (channelrhodopsin-2, ChR2, or Synapsin/synaptophysin dual-label, SYN; green) was injected bilaterally into the orofacial motor cortex (ofM1) to label corticobulbar projections into NA. Left: example coronal section showing cortical injection sites with anterograde tracer (ChR2, green) and DAPI labeling (cyan). (B-F) Laryngeal motoneurons in the NA identified using a retrograde tracer (CTB, purple), cortical fibers labeled with ChR2 (green), corticobulbar synapses labeled with VGLUT1 (red), and DAPI (blue). B and C are overlaid images showing colocalization of fibers with a synaptic bouton on the retrograde labeled cell (white arrow). (G) Percentage of laryngeal motoneurons labeled with CTB that are colocalized with cortical fibers (blue) or with both cortical fibers and synaptic boutons (red). Note that both tracing techniques qualitatively yielded similar results: ChR2, n = 51 cells from 3 bats; Synapsin/synaptophysin dual-label virus (SYN), n = 26 cells from 2 bats. (H) Illustration of the experimental setup during which wireless electrophysiological recordings were conducted from the identified cortical region in freely behaving and vocalizing bats. (I) Spiking activity of an example ofM1 neuron aligned to the onset of vocalizations produced (bat’s own calls, orange) or heard (other bats’ calls, blue) by the bat subject. Top row, time varying mean firing rate and corresponding raster plot below. Colored lines in the raster plot show the duration of each vocalization. Note the increased firing rate during vocal production as compared to hearing. (J) Information (see Methods) between the time varying firing rate and the amplitude of produced (x-axis) vs. heard (y-axis) vocalizations for 219 single units (marker shapes indicate bat ID, n=4 bats). The cell shown in (I) is highlighted in red. Inset shows the distribution of D-prime between motor and auditory information for the same cells. Note that the distribution is heavily skewed towards higher motor information rather than auditory information coded in the activity of the recorded neurons. Error bars are mean +/− SEM throughout the figure.
To further corroborate the role of ofM1 in vocal control, we tested whether ongoing single-cell neural activity in this area was associated with vocal production. We performed wireless electrophysiological recordings from four bats engaged in free vocal interactions with peers (Fig. 2H). Vocalizations were identified and recorded using wireless call detectors placed around the necks of the individual bats (see Materials and Methods, (46)). We found that about half of the recorded single units in ofM1 (115/237) showed a significant change in firing rates when the bats produced vocalizations as compared to staying quiet (Fig. S6A-C; ANOVA with a Poisson Generalized Linear Model per cell; p-value threshold = 0.001; Materials and Methods). In 25% of ofM1 cells that were excited during vocal production (26/104), the change of activity could not be accounted for by jaw or tongue movements, indicating that these cells were engaged in the motor control of movements specific to vocal production (Fig. S6D). Furthermore, many of the single units had a sustained increase of activity during production of vocalizations, but not during perception of vocalizations (Fig. 2I). To further assess this specific neural modulation during vocal-motor production, we quantified the information between the time-varying firing rate and the amplitude modulation of the vocalizations. This analysis confirmed that ofM1 neurons had significantly higher motor than auditory information (Fig. 2J; likelihood-ratio test on LME models, N=219, LRStat = 62.515, df = 1, p = 2.6645×10−5; average d-prime change in information gain during motor production = 0.15 ± 0.13, corresponding to an increase of 0.286 ± 0.035 bits/s). Combined, the results of the anatomical and electrophysiological study defined ofM1 as a motor cortical area associated with vocal production in R. aegyptiacus.
Epigenomic Specializations in the Vocal Production Region of the Egyptian Fruit Bat Motor Cortex
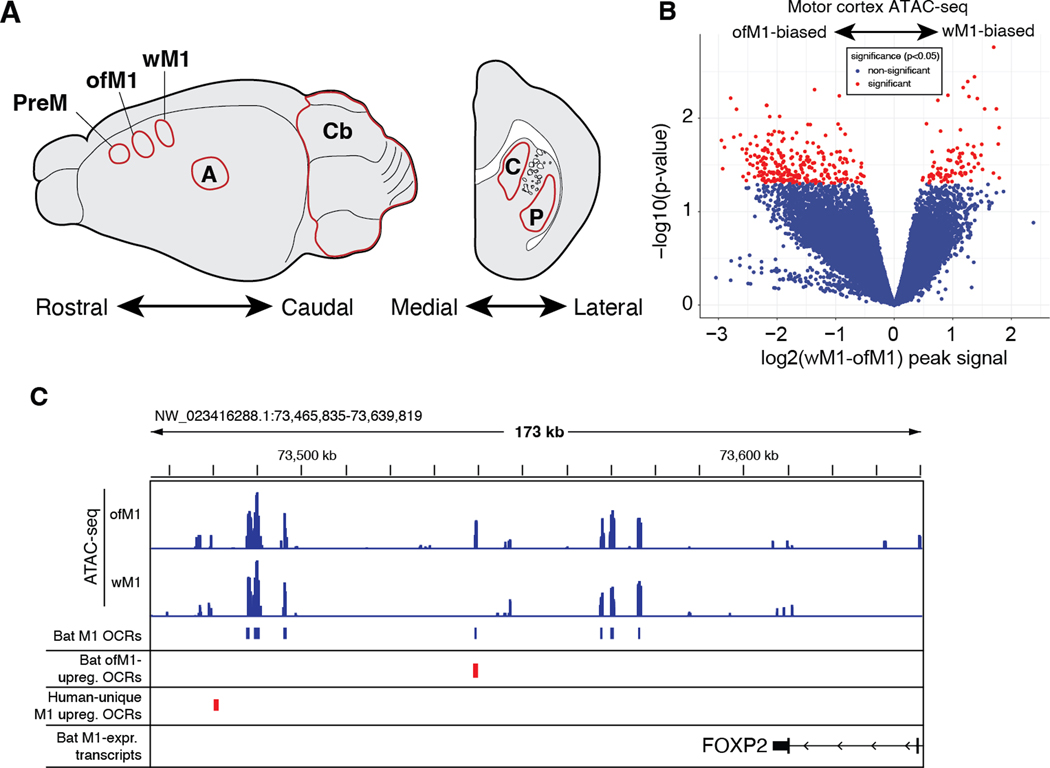
We next sought to epigenomically profile candidate regulatory elements in vocal and non-vocal brain regions in R. aegyptiacus to identify vocal learning-associated regulatory genomic specializations. We generated a multi-tissue atlas of open chromatin data — indicative of regulatory activity — by performing ATAC-seq (assay for transposase-accessible chromatin sequencing (61)) across 7 brain regions and 3 somatic tissues of R. aegyptiacus (Materials and Methods), including ofM1 (Fig. 3A). From a total set of 88,389 noncoding, non-promoter open chromatin regions (OCRs) in primary motor cortex (M1), we identified 348 candidate enhancers with differential open chromatin between orofacial motor cortex (ofM1) and wing motor cortex (wM1) (p < 0.05; Fig. 3B, Data S5, Materials and Methods). Genes proximal to OCRs with differential open chromatin between ofM1 and wM1 were significantly enriched for functional association with neuronal projections and transcriptional regulation (Data S6). These included OCRs near the genes of 51 known transcription factors (TFs), including FOXP2, a TF that has been extensively implicated in human speech and vocal learning (Fig. 3C) (62). Notably, genes near OCRs differentially open between bat ofM1 and wM1 included genes we had identified as being under convergent acceleration in vocal learners using evolutionary approaches: RERconverge analysis (n = 11) and the HyPhy RELAX analysis (n=3; GATA3, LRRN1, TNIP3) (Data S6). These specialized regions of open chromatin, coupled with an enrichment of transcription factors in the set of vocal learning-associated protein-coding genes, suggest that both cis and trans differences in gene regulation contribute to the evolution of vocal learning behaviors.
Figure 3. Differential Open Chromatin in Bat Orofacial M1 relative to Wing M1.
(A) Open chromatin was profiled from 7 dissected brain regions of Egyptian Fruit bats. (B) Volcano plot of ATAC-seq OCRs with differential activity between the orofacial and wing subregions of primary motor cortex (ofM1 and wM1, respectively) of Egyptian fruit bat. (C) Genome browser showing ofM1 and wM1 ATAC-seq traces at the 3’ end of the FOXP2 locus. Reproducible M1 open chromatin regions (OCRs) are indicated in blue, with a differentially active OCR in ofM1 relative to wM1 highlighted in red.
Convergent Evolution in Candidate Enhancer Sequences Associated with Vocal Learning Behavior
Since there is accumulating evidence that cis-regulatory differences in enhancer regions are driving the evolution of complex traits (63–65), we sought to identify OCRs whose tissue- and cell type-specificity would be shared across species of vocal learners. Detecting cis-regulatory element differences associated with trait evolution is challenging because many enhancers can preserve the same regulatory function even when the underlying genome sequence is highly divergent, and many cis-regulatory elements have tissue-specific activity (66–68). Thus, methods for convergent evolution that rely on the alignment of individual nucleotides between species (e.g. (11, 69, 70)) are likely to miss a substantial proportion of key candidate enhancers.
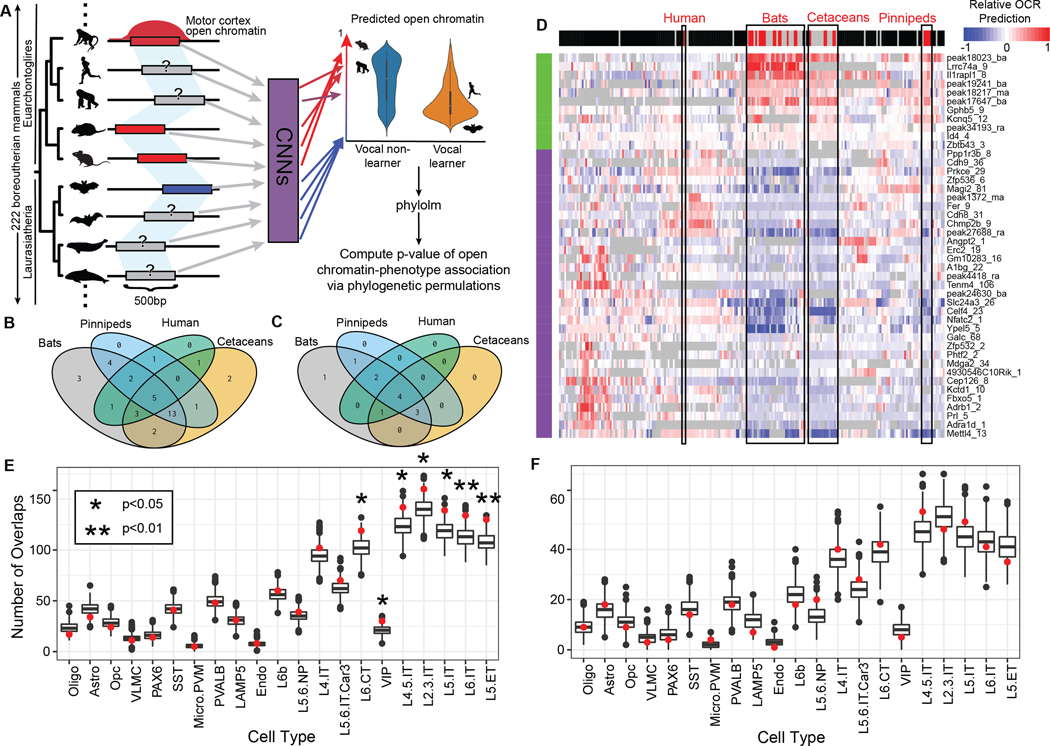
We therefore sought to extend our search for cis-regulatory elements whose evolution is associated with vocal learning behavior using a recently developed machine learning approach, TACIT (Tissue-Aware Conservation Inference Toolkit (23)). Given that it is infeasible to map the brains and collect motor cortex tissue from each vocal learning and closely related non-learning species, the TACIT approach uses machine learning models (23) to predict motor cortex open chromatin across orthologous regions of the genome (66–68). TACIT then associates predictions with vocal learning in a way that corrects for phylogenetic relationships (Fig. 4A). We used the predictions from convolutional neural networks (CNNs) that were previously trained using DNA sequence-based M1 open chromatin data obtained in this study for R. aegyptiacus with ATAC-seq and collected earlier for the mouse (21), the rat, and the macaque (20) to predict motor cortex open chromatin across 222 mammalian genomes (Materials and Methods, (23)). Given that parvalbumin has been shown to be a shared marker of brain areas critical for vocal learning in songbirds and humans (4), we also used CNNs trained to predict cell type-specific open chromatin using ATAC-seq data from mouse and human M1 parvalbumin-positive neurons (M1-PV+) (22, 23, 71). We identified regions whose predicted open chromatin was consistently lower or higher in vocal learners relative to vocal non-learners using phylogenetic logistic regression (72, 73) with phylogenetic permutations (24) (permulations adj. p < 0.1; Fig. 4A, Materials and Methods, (23)). We identified 33 open chromatin regions from our M1 CNN models that had lower predicted open chromatin in vocal learning species and 11 that had higher predicted open chromatin in vocal learning species (Fig 4D; Table S1; Data S7). From the M1-PV+ predictions, we identified five candidate enhancers that had lower predicted activity in vocal learning species and one candidate enhancer that had higher predicted regulatory activity in vocal learning species. (Table S2; Data S7). Unlike the protein-coding genes, the majority of vocal learning-associated enhancers showed evidence of higher or lower activity in at least three out of the four vocal learning clades (Fig. 4B, C, D; Data S8). Consistent with the finding that convergent vocal learning-associated gene regulation is primarily repressive (4), we found that the majority of candidate enhancers (n = 38/50 OCRs, 76%) had lower predicted open chromatin activity in vocal learning relative to vocal non-learning mammals (Fig. S7).
Figure 4. Vocal learning-associated convergent evolution in motor cortex open chromatin regions implicates specific neuron subtypes.
(A) Overview of applying the Tissue-Aware Conservation Inference Toolkit (TACIT (23)) approach to vocal learning. OCRs (left) identified in motor cortex (M1). Measured open chromatin from M1 (4 species) were used to train convolutional neural networks (CNNs) to predict M1 open chromatin from sequence alone. Red bars and corresponding arrows indicate the presence of a peak while the blue bars represent the absence. The same OCRs were then mapped across 222 mammalian genomes (left) and the identified sequences were used as input to the CNNs to predict open chromatin activity. TACIT identified OCRs whose predicted open chromatin across species was significantly associated with those species’ vocal learning status. (B-C) The 4-way Venn diagrams represent the number of OCRs implicated by TACIT (both M1 and PV+) as displaying low (B) or high (C) activity in each of the vocal learning clades based on a t-test. (D) The heatmap visualizes specific open chromatin regions along the rows (predicted higher in vocal learners in green; predicted lower in vocal learners in purple) across 222 mammals in the columns (vocal learner in red, vocal nonlearner in black, insufficient or conflicting evidence in gray). The color in each cell corresponds to the z-scored predicted open chromatin, with low open chromatin in blue, mean open chromatin in white, and high open chromatin in red. For open chromatin regions predicted to be significantly less (E) or more (F) open in vocal learning species (p<0.05), the red point shows the number of overlapping regions (y-axis) across mouse cortical cell types (x-axis). The bar-plot shows the distribution across 1,000 permutations of the peaks implicated by TACIT. The notches extend 1.58 * IQR / sqrt(n), which gives a roughly 95% confidence).
To interpret potential functions of the vocal learning-associated candidate enhancers, we annotated the nearest genes in the mouse (Data S9). In many cases, the genes closest to these putative enhancers have been associated with significant developmental delay or complete absence of speech when disrupted in humans (Tables S1-2). Four of the OCRs identified by the M1 model were proximal to genes—GALC, TCF4, TSHZ3, and ZNF536—that were also near OCRs with differential activity between bat ofM1 and wM1 (Data S6). Two of the vocal learning-associated M1 OCRs were proximal to genes—DAAM1 and VIP—previously shown to have convergent gene expression between humans and song-learning birds (4). To further explore the function of the vocal learning-associated OCRs in the motor cortex, we annotated their cell type-specificity using publicly available mouse BICCN data (74). The cell type most enriched for OCRs predicted to be lower in vocal learning species was Layer V ET (extra-telencephalic) neurons, which have previously been implicated in vocal learning (75), but strong enrichments were also found for other cortical excitatory neurons (Fig. 4E,F; Data S10).
Among genes near vocal learning-associated OCRs, the DACT1 (TACIT adj. p = 0.0014; RERconverge Tau adj. p<0.0001) and CELF4 (TACIT adj. p < 0.023; RERconverge Tau adj. p<0.0034) proteins also displayed significantly lower relative evolutionary rates in vocal learners. Despite the lack of direct evidence in the literature for its role in speech production, CELF4 has been associated with autism in the human population (76) and its function in Layer V pyramidal neurons has been linked with seizures in mice (77).
Multiple M1-PV+ interneuron OCRs associated with vocal learning are near genes previously associated with autism. For example, an OCR that is negatively associated with vocal learning evolution is in an intron of the gene CCSER1, which has nonsense mutations implicated in autism (78) and is in a locus associated with musical beat synchronization (79). An OCR that is positively associated with the evolution of vocal learning is in an intron of the gene CNTNAP4, whose deletions and copy number variation in humans and mice have been implicated in neurological disorders, including autism in humans (80, 81). To test whether these associations would have been identified by chance, we tested whether vocal learning-associated OCRs tended to be near genes associated with autism. We found the M1-PV+ OCRs with human orthologs near genes associated with autism (82) tend to be more significantly associated with vocal learning evolution than other OCRs with human orthologs (Wilcoxon p=0.0071).
Discussion:
Convergent evolution of vocal production learning has been associated with convergent evolution at the neuroanatomical level: cortical motor regions driving vocal production in humans and songbirds (human motor cortex and songbird RA) show increased connectivity with the brainstem and striatum (3). These same motor regions also show convergent evolution in patterns of gene expression, with commonly decreased gene expression found in both song-learning birds and humans (4). In this study, we investigated convergent evolution of vocal learning in mammals, both at the anatomical and the genetic level. First, we found a direct motor corticobulbar connection from a cortical region implicated in vocal production in a vocal learning bat. Second, we revealed widespread evidence of convergent evolution across vocal learning mammals in protein-coding sequences and candidate regulatory enhancers.
Our parallel study of both coding and noncoding regions linked with the vocal learning trait identifies many protein-coding genes (200) and a smaller number of noncoding regions (50), distal sites of open chromatin, that are associated with vocal learning. Although a larger number of significant protein-coding genes are identified, the vast majority of these are primarily driven by strong evidence in one of the vocal learning clades and only weak evidence in the other three. In contrast, the majority of significant noncoding regions show robust evidence of convergent selective pressure in at least three out of the four clades. The larger number of identified proteins relative to open chromatin regions could be due to better statistical power from being able to directly model nucleotide evolution in protein-coding sequence, which tend to be more stable than regulatory elements across species (11, 12). We note, however, that only 5/200 of the significantly associated protein-coding genes showed robust evidence of differential rates of evolution in at least three of four vocal learning clades. Out of these 200 proteins, many were neurodevelopmental transcription factors, which are among the most highly conserved genes in mammals (83) and thus likely to play roles in a broad range of contexts that could constrain their evolution. It is also possible that some of the identified proteins could be associated with other convergent traits that correlate with vocal learning across mammals, including echolocation (bats, cetaceans), marine adaptations (cetaceans, pinnipeds), or increased longevity (bats, cetaceans, humans).
In contrast, 33/50 vocal learning-associated OCRs had differential predicted open chromatin in at least three of four vocal learning clades; this independent convergence of gene regulatory function suggests that these OCRs may be critical for the evolution of vocal learning. Enhancers tend to have functions that are much more context- and tissue-specific (68), making them less functionally constrained than protein-coding genes, which could perhaps allow more flexibility for an individual enhancer to evolve a new role for a specific trait like vocal learning. In sum, our results suggest that the evolution of mammalian vocal learning is largely driven by changes to the noncoding regulatory elements that orchestrate gene expression rather than to the protein-coding genes themselves.
Despite the different methodologies applied to identify convergent evolution in coding and noncoding regions, both protein coding- and regulatory element-focused approaches implicated gene functional pathways associated with human autism. In our protein-coding analyses, genes with lower evolutionary rates based on both RERconverge and HyPhy RELAX were enriched for autism function. Likewise, in our analyses of regulatory evolution, multiple autism-linked genes were near human orthologs of the vocal learning-associated M1-PV OCRs. In humans, autism is often associated with speech delays and differences in social behavior, both of which could be related to the evolutionary trait of vocal learning ability(84). Broadly, this could be evidence that genomic loci associated with a complex trait across mammals may also be associated with variations in related traits within the human population.
The bulk motor cortex OCRs with lower predicted open chromatin in vocal learners show the strongest tendency to overlap with OCRs in Layer V ET neurons, which form long range projections (Fig. 4E). These results are consistent with our previous findings showing decreases in the expression of axon guidance genes in the motor cortex of vocal learning species (4). Among other functions, the Layer V ET neurons implicated by TACIT create the corticospinal projections that have been hypothesized as an anatomical landmark of vocal learners (2, 3, 48, 49, 56, 85, 86). Furthermore, the neuroethological and anatomical experiments we conducted in R. Aegyptiacus provide evidence that corticospinal projection neurons are present in the motor cortex of that bat species and that this motor cortical region participates in vocal production. Thus, consistent with previous literature, our results support a model in which the loss of regulatory element activity in the motor cortex influences axon guidance properties of long range projection neurons, which allow more robust connectivity between the cortex and the brainstem of vocal learning mammals (4, 85, 87). Alternatively, these genetic differences could relate to potential differences in the density of disynaptic connections that have been associated with skilled motor behavior, including vocalization in non-human primates (88, 89). Notably, these long range projection neurons have also been associated with predisposition to autism (90).
Methods Summary
To find vocal learning-associated convergent evolution in protein-coding sequences of the mammalian genome, we began with amino acid level multiple sequence alignments produced by the Zoonomia consortium (10). Those served as input to two classes of methods, RERconverge (11) and HyPhy (30). RERconverge with an additional permutations correction for phylogenetic structure (24) was used to find protein-coding sequences whose evolutionary rates were associated with the presence or absence of vocal learning. HyPhy RELAX was used to find protein-coding sequences that were evolving more slowly, neutrally, or faster in vocal learning species. In addition, the HyPhy BUSTED-PH method (12) was applied to find evidence of diversifying positive selection. The gene ontology analysis was performed on the intersection of the RERconverge and HyPhy results using EnrichR (91). To control for false positives across all methods, Benjamini-Hochberg false discovery rate correction (92) was applied.
To examine the existence of a direct monosynaptic projection in a vocal learning mammal, the corticobulbar projections in Egyptian fruit-bats were mapped by tracing the projection from the orofacial motor cortex and from the cricothyroid muscles of the vocal cords. Performing immunohistochemistry in the brainstem revealed that synaptic boutons of cortical projection neurons overlapped with retrogradely-labeled motoneurons - confirming the existence of a direct monosynaptic projection. The role of the orofacial motor cortex during vocal production was then validated by quantifying the information between the vocalization amplitude and single cortical neuron activity measured wirelessly in vivo while the bats produced and listened to vocalizations.
To create an atlas of open chromatin regions in the bat motor cortex (M1), several brain regions, including wing-M1 and orofacial-M1, were separately dissected. The samples were cryopreserved, then the nuclei were isolated, and subsequently ATAC-Seq was performed to measure open chromatin. The open chromatin regions from this experiment were combined with previously published experiments in macaque, rat (23), and mouse (21) to create an atlas of cross-species motor cortex open chromatin.
To find vocal learning-associated convergent evolution in noncoding regions of the Boreoeutherian mammalian genome, the TACIT machine learning approach, was applied. Orthologous regions across genomes were found by combining the CACTUS whole genome multiple sequence alignment (19), halLiftover (93) and HALPER (94). Phyloglm (72) was then used to associate predicted motor cortex and parvalbumin-positive inhibitory interneuron open chromatin with binary annotations of vocal learning behavior. Phylogenetic permutations were applied to correct for phylogenetic tree structure and Benjamini-Hochberg to correct for multiple hypothesis testing. To identify potential trends in the cell type-specificity of the implicated regions, permutations on the regions of the genome that were predicted to have significantly higher or lower open chromatin in vocal learning species were conducted.
Supplementary Material
Acknowledgments:
We would like to thank Andrew C. Halley and Leah Krubitzer for helpful discussions and critical assistance in identifying orofacial and wing subregions of the Rousettus aegyptiacus motor cortex. We would like to thank Frederic E. Theunissen for helpful discussions around the analysis of coherence of neural activity and for sharing his lab’s code. We would like to thank the other members of the Zoonomia Consortium, the Vertebrate Genomes Project Vocal Learning working group, and the Pfenning, Yartsev, and Jarvis labs for useful discussions and suggestions.
Funding:
Alfred P. Sloan Foundation Research Fellowship (ARP)
National Science Foundation grant NSF 20-525 (ARP)
National Institutes of Health grant NIDA DP1DA046585 (ARP)
National Institutes of Health grant NIDA F30DA053020 (BNP)
National Institutes of Health grant R01NS111479 (EA)
National Institutes of Health grant R01 NS121231 (BL)
National Science Foundation grant NSF IOS-2022241 (BL).
National Institutes of Health grant DP2 DP2-DC016164 (MMY).
The New York Stem Cell Foundation NYSCF-R-NI40 (MMY).
Alfred Sloan Foundation FG-2017-9646 (MMY).
Brain Research Foundation BRFSG-2017-09 (MMY).
Packard Foundation Fellowship 2017-66825 (MMY).
Klingestein-Simons Fellowship (MMY).
Human Frontiers Research Program (MMY).
Pew Charitable trust 00029645 (MMY).
McKnight Foundation 042823 (MMY).
Carnegie Mellon University Computational Biology Department Lane Fellowship (IMK).
Dana Foundation (MMY).
This work used the Extreme Science and Engineering Discovery Environment (XSEDE), through the Pittsburgh Supercomputing Center Bridges and Bridges-2 Compute Clusters, which was supported by the National Science Foundation (grant TG-BIO200055).
Footnotes
Competing interests: ARP is founder and CEO of Snail Biosciences.
Data and materials availability:
Egyptian fruit bat ATAC-seq data—including raw .fasta files, .bigWig files of genome-aligned reads, processed open chromatin .narrowpeak files, and extensive sample metadata—have been made available at GEO under accession ID GSE187366. Publicly available ATAC-Seq data was obtained from GEO GSE159815 and GSE161374. Single-nucleus open chromatin was downloaded from the NEMO archive: https://data.nemoarchive.org/biccn/grant/u19_cemba/cemba/epigenome/sncell/ATACseq/mouse/. The code for the electrophysiology and functional components of the project are available here: https://github.com/NeuroBatLab/LoggerDataProcessing/ https://github.com/NeuroBatLab/SoundAnalysisBats/ https://github.com/NeuroBatLab/LoggerDataProcessing/ https://github.com/julieelie/Kilosort2_Tetrode/configFiles/configFile16.m. The code for the convergent evolution analysis can be found in https://github.com/pfenninglab/TACIT, https://github.com/veg/hyphy, and https://github.com/veg/hyphy-analyses/tree/master/BUSTED-PH . The histological and electrophysiology source data from this study are available on FigShare under the DOI 10.6084/m9.figshare.25180430 (95). The code for conducting and visualizing the RERconverge analysis can be found here: https://zenodo.org/doi/10.5281/zenodo.10641176 (96). The code for conducting and visualizing the TACIT anlaysis can be found here: https://zenodo.org/doi/10.5281/zenodo.5952292 (97).
References and Notes:
- 1.Doupe AJ, Kuhl PK, Birdsong and human speech: common themes and mechanisms. Annu. Rev. Neurosci. 22, 567–631 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Wirthlin M, Chang EF, Knörnschild M, Krubitzer LA, Mello CV, Miller CT, Pfenning AR, Vernes SC, Tchernichovski O, Yartsev MM, A Modular Approach to Vocal Learning: Disentangling the Diversity of a Complex Behavioral Trait. Neuron 104, 87–99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarvis ED, Learned birdsong and the neurobiology of human language. Ann. N. Y. Acad. Sci. 1016, 749–777 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfenning AR, Hara E, Whitney O, Rivas MV, Wang R, Roulhac PL, Howard JT, Wirthlin M, Lovell PV, Ganapathy G, Mouncastle J, Moseley MA, Thompson JW, Soderblom EJ, Iriki A, Kato M, Gilbert MTP, Zhang G, Bakken T, Bongaarts A, Bernard A, Lein E, Mello CV, Hartemink AJ, Jarvis ED, Convergent transcriptional specializations in the brains of humans and song-learning birds. Science 346, 1256846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arriaga G, Jarvis ED, Mouse vocal communication system: are ultrasounds learned or innate? Brain Lang. 124, 96–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Searcy WA, Soha J, Peters S, Nowicki S, Variation in vocal production learning across songbirds. Philos. Trans. R. Soc. Lond. B Biol. Sci. 376, 20200257 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyack PL, A taxonomy for vocal learning. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20180406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruch H, Zürcher Y, Burkart JM, The function and mechanism of vocal accommodation in humans and other primates. Biol. Rev. Camb. Philos. Soc. 93, 996–1013 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Zoonomia Consortium, A comparative genomics multitool for scientific discovery and conservation. Nature 587, 240–245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirilenko BM, Munegowda C, Osipova E, Jebb D, Sharma V, Blumer M, Morales AE, Ahmed A-W, Kontopoulos D-G, Hilgers L, Lindblad-Toh K, Karlsson EK, Zoonomia Consortium‡, Hiller M, Integrating gene annotation with orthology inference at scale. Science 380, eabn3107 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowalczyk A, Meyer WK, Partha R, Mao W, Clark NL, Chikina M, RERconverge: an R package for associating evolutionary rates with convergent traits. Bioinformatics 35, 4815–4817 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pond S. L. Kosakovsky, Poon AFY, Velazquez R, Weaver S, Hepler NL, Murrell B, Shank SD, Magalis BR, Bouvier D, Nekrutenko A, Wisotsky S, Spielman SJ, Frost SDW, Muse SV, HyPhy 2.5-A Customizable Platform for Evolutionary Hypothesis Testing Using Phylogenies. Mol. Biol. Evol. 37, 295–299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsompana M, Buck MJ, Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7, 33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prat Y, Taub M, Yovel Y, Vocal learning in a social mammal: Demonstrated by isolation and playback experiments in bats. [Preprint] (2015). 10.1126/sciadv.1500019. [DOI] [PMC free article] [PubMed]
- 15.Prat Y, Azoulay L, Dor R, Yovel Y, Crowd vocal learning induces vocal dialects in bats: Playback of conspecifics shapes fundamental frequency usage by pups. PLoS Biol. 15, e2002556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genzel D, Desai J, Paras E, Yartsev MM, Long-term and persistent vocal plasticity in adult bats. Nat. Commun. 10, 3372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genereux DP, Seres A, Armstrong J, Johnson J, Marinescu VD, Murén E, Juan D, Bejerano G, Casewell NR, Chemnick LG, Di Palma F, Diekhans M, Fiddes IT, Garber VN, Gladyshev VN, Goodman L, Haerty W, Houck ML, Hubley R, Kivioja T, Koepfli K-P, Kuderna LFK, Lander ES, Meadows J, Murphy WJ, Nash W, Noh HJ, Nweeia M, Pfenning AR, Pollard KS, Ray D, Shapiro B, Smit A, Springer M, Steiner CC, Swofford R, Taipale J, Teeling EC, Turner-Maier J, Alfoldi J, Birren B, Ryder OA, Lewin H, Paten B, Marques-Bonet T, Lindblad-Tor K, Karlsson EK, A comparative genomics multitool for scientific discovery and conservation. Nature 579 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christmas MJ, Kaplow IM, Genereux DP, Dong MX, Hughes GM, Li X, Sullivan PF, Hindle AG, Andrews G, Armstrong JC, Bianchi M, Breit AM, Diekhans M, Fanter C, Foley NM, Goodman DB, Goodman L, Keough KC, Kirilenko B, Kowalczyk A, Lawless C, Lind AL, Meadows JRS, Moreira LR, Redlich RW, Ryan L, Swofford R, Valenzuela A, Wagner F, Wallerman O, Brown AR, Damas J, Fan K, Gatesy J, Grimshaw J, Johnson J, Kozyrev SV, Lawler AJ, Marinescu VD, Morrill KM, Osmanski A, Paulat NS, Phan BN, Reilly SK, Schäffer DE, Steiner C, Supple MA, Wilder AP, Wirthlin ME, Xue JR, Zoonomia Consortium§, Birren BW, Gazal S, Hubley RM, Koepfli K-P, Marques-Bonet T, Meyer WK, Nweeia M, Sabeti PC, Shapiro B, Smit AFA, Springer MS, Teeling EC, Weng Z, Hiller M, Levesque DL, Lewin HA, Murphy WJ, Navarro A, Paten B, Pollard KS, Ray DA, Ruf I, Ryder OA, Pfenning AR, Lindblad-Toh K, Karlsson EK, Evolutionary constraint and innovation across hundreds of placental mammals. Science 380, eabn3943 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armstrong J, Hickey G, Diekhans M, Fiddes IT, Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wirthlin ME, Kaplow IM, Lawler AJ, He J, Phan BDN, Brown AR, Stauffer WR, Pfenning AR, The Regulatory Evolution of the Primate Fine-Motor System. bioRxiv, doi: 10.1101/2020.10.27.356733 (2020). [DOI] [Google Scholar]
- 21.Srinivasan C, Phan BN, Lawler AJ, Ramamurthy E, Kleyman M, Brown AR, Kaplow IM, Wirthlin ME, Pfenning AR, Addiction-Associated Genetic Variants Implicate Brain Cell Type- and Region-Specific Cis-Regulatory Elements in Addiction Neurobiology. J. Neurosci. 41, 9008–9030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakken TE, Jorstad NL, Hu Q, Lake BB, Tian W, Kalmbach BE, Crow M, Hodge RD, Krienen FM, Sorensen SA, Eggermont J, Yao Z, Aevermann BD, Aldridge AI, Bartlett A, Bertagnolli D, Casper T, Castanon RG, Crichton K, Daigle TL, Dalley R, Dee N, Dembrow N, Diep D, Ding S-L, Dong W, Fang R, Fischer S, Goldman M, Goldy J, Graybuck LT, Herb BR, Hou X, Kancherla J, Kroll M, Lathia K, van Lew B, Li YE, Liu CS, Liu H, Lucero JD, Mahurkar A, McMillen D, Miller JA, Moussa M, Nery JR, Nicovich PR, Niu S-Y, Orvis J, Osteen JK, Owen S, Palmer CR, Pham T, Plongthongkum N, Poirion O, Reed NM, Rimorin C, Rivkin A, Romanow WJ, Sedeño-Cortés AE, Siletti K, Somasundaram S, Sulc J, Tieu M, Torkelson A, Tung H, Wang X, Xie F, Yanny AM, Zhang R, Ament SA, Behrens MM, Bravo HC, Chun J, Dobin A, Gillis J, Hertzano R, Hof PR, Höllt T, Horwitz GD, Keene CD, Kharchenko PV, Ko AL, Lelieveldt BP, Luo C, Mukamel EA, Pinto-Duarte A, Preissl S, Regev A, Ren B, Scheuermann RH, Smith K, Spain WJ, White OR, Koch C, Hawrylycz M, Tasic B, Macosko EZ, McCarroll SA, Ting JT, Zeng H, Zhang K, Feng G, Ecker JR, Linnarsson S, Lein ES, Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplow IM, Lawler AJ, Schäffer DE, Srinivasan C, Sestili HH, Wirthlin ME, Phan BN, Prasad K, Brown AR, Zhang X, Foley K, Genereux DP, Zoonomia Consortium**, Karlsson EK, Lindblad-Toh K, Meyer WK, Pfenning AR, Relating enhancer genetic variation across mammals to complex phenotypes using machine learning. Science 380, eabm7993 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saputra E, Kowalczyk A, Cusick L, Clark N, Chikina M, Phylogenetic Permulations: A Statistically Rigorous Approach to Measure Confidence in Associations in a Phylogenetic Context. Mol. Biol. Evol. 38, 3004–3021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wada K, Sakaguchi H, Jarvis ED, Hagiwara M, Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J. Comp. Neurol. 476, 44–64 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Z, Piccus Z, Zhang X, Yang H, Jarrell H, Ding Y, Teng Z, Tchernichovski O, Li X, miR-9 regulates basal ganglia-dependent developmental vocal learning and adult vocal performance in songbirds. Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savell KE, Zipperly ME, Tuscher JJ, Duke CG, Phillips RA, Bauman AJ, Thukral S, Sultan FA, Goska NA, Ianov L, Day JJ, A dopamine-induced gene expression signature regulates neuronal function and cocaine response. 10.1101/781872. [DOI] [PMC free article] [PubMed]
- 28.He J, Kleyman M, Chen J, Alikaya A, Rothenhoefer KM, Ozturk BE, Wirthlin M, Bostan AC, Fish K, Byrne LC, Pfenning AR, Stauffer WR, Transcriptional and anatomical diversity of medium spiny neurons in the primate striatum. Curr. Biol, doi: 10.1016/j.cub.2021.10.015 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnavion P, Varin C, Fakhfouri G, Olondo PM, De Groote A, Cornil A, Lopez RL, Fernandez EP, Isingrini E, Rainer Q, Xu K, Tzavara E, Vigneault E, Dumas S, de Kerchove d’Exaerde A, Giros B, Unexpected contributions of striatal projection neurons coexpressing dopamine D1 and D2 receptors in balancing motor control, bioRxiv (2023)p. 2022.04.05.487163. [Google Scholar]
- 30.Wertheim JO, Murrell B, Smith MD, Pond S. L. Kosakovsky, Scheffler K, RELAX: detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 32, 820–832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unger N, Heim S, Hilger DI, Bludau S, Pieperhoff P, Cichon S, Amunts K, Mühleisen TW, Identification of Phonology-Related Genes and Functional Characterization of Broca’s and Wernicke’s Regions in Language and Learning Disorders. Front. Neurosci. 15, 680762 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Josifova DJ, Monroe GR, Tessadori F, de Graaff E, van der Zwaag B, Mehta SG, DDD Study, Harakalova M, Duran KJ, Savelberg SMC, Nijman IJ, Jungbluth H, Hoogenraad CC, Bakkers J, Knoers NV, Firth HV, Beales PL, van Haaften G, van Haelst MM, Heterozygous KIDINS220/ARMS nonsense variants cause spastic paraplegia, intellectual disability, nystagmus, and obesity. Hum. Mol. Genet. 25, 2158–2167 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Tossell K, Andreae LC, Cudmore C, Lang E, Muthukrishnan U, Lumsden A, Gilthorpe JD, Irving C, Lrrn1 is required for formation of the midbrain-hindbrain boundary and organiser through regulation of affinity differences between midbrain and hindbrain cells in chick. Dev. Biol. 352, 341–352 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C, Hu Q, Jing J, Zhang Y, Jin J, Zhang L, Mu L, Liu Y, Sun B, Zhang T, Kong Q, Wang G, Wang D, Zhang Y, Liu X, Zhao W, Wang J, Feng T, Li H, Regulator of G protein signaling 5 (RGS5) inhibits sonic hedgehog function in mouse cortical neurons. Mol. Cell. Neurosci. 83, 65–73 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Han Y-Y, Zhou J-W, Guo Z-W, Wu Z-Q, Zhang Z-Y, Liu D-X, Long C, Multiple brain regions are involved in reaction to acute restraint stress in CYLD-knockout mice. Stress 26, 2228925 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Adak P, Banerjee N, Sinha S, Bandyopadhyay AK, Gamma-Aminobutyric Acid Type A Receptor Variants are Associated with Autism Spectrum Disorders. J. Mol. Neurosci. 73, 237–249 (2023). [DOI] [PubMed] [Google Scholar]
- 37.Mennesson M, Rydgren E, Lipina T, Sokolowska E, Kulesskaya N, Morello F, Ivakine E, Voikar V, Risbrough V, Partanen J, Hovatta I, Kainate receptor auxiliary subunit NETO2 is required for normal fear expression and extinction. Neuropsychopharmacology 44, 1855–1866 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gueneau L, Fish RJ, Shamseldin HE, Voisin N, Tran Mau-Them F, Preiksaitiene E, Monroe GR, Lai A, Putoux A, Allias F, Ambusaidi Q, Ambrozaityte L, Cimbalistienė L, Delafontaine J, Guex N, Hashem M, Kurdi W, Jamuar SS, Ying LJ, Bonnard C, Pippucci T, Pradervand S, Roechert B, van Hasselt PM, Wiederkehr M, Wright CF, DDD Study, Xenarios I, van Haaften G, Shaw-Smith C, Schindewolf EM, Neerman-Arbez M, Sanlaville D, Lesca G, Guibaud L, Reversade B, Chelly J, Kučinskas V, Alkuraya FS, Reymond A, KIAA1109 Variants Are Associated with a Severe Disorder of Brain Development and Arthrogryposis. Am. J. Hum. Genet. 102, 116–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gialluisi A, Newbury DF, Wilcutt EG, Olson RK, DeFries JC, Brandler WM, Pennington BF, Smith SD, Scerri TS, Simpson NH, SLI Consortium, Luciano M, Evans DM, Bates TC, Stein JF, Talcott JB, Monaco AP, Paracchini S, Francks C, Fisher SE, Genome-wide screening for DNA variants associated with reading and language traits. Genes Brain Behav. 13, 686–701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams AK, Smith SD, Truong DT, Willcutt EG, Olson RK, DeFries JC, Pennington BF, Gruen JR, Enrichment of putatively damaging rare variants in the DYX2 locus and the reading-related genes CCDC136 and FLNC. Hum. Genet. 136, 1395–1405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tager-Flusberg H, Paul R, Lord C, Language and communication in autism. Handbook of autism and pervasive developmental disorders 1, 335–364 (2005). [Google Scholar]
- 42.Schoen E, Paul R, Chawarska K, Phonology and vocal behavior in toddlers with autism spectrum disorders. Autism Res. 4, 177–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodenas-Cuadrado PM, Mengede J, Baas L, Devanna P, Schmid TA, Yartsev M, Firzlaff U, Vernes SC, Mapping the distribution of language related genes FoxP1 , FoxP2 , and CntnaP2 in the brains of vocal learning bat species. [Preprint] (2018). 10.1002/cne.24385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cahill JA, Armstrong J, Deran A, Khoury CJ, Paten B, Haussler D, Jarvis ED, Positive selection in noncoding genomic regions of vocal learning birds is associated with genes implicated in vocal learning and speech functions in humans. Genome Res, doi: 10.1101/gr.275989.121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.St Pourcain B, Cents RAM, Whitehouse AJO, Haworth CMA, Davis OSP, O’Reilly PF, Roulstone S, Wren Y, Ang QW, Velders FP, Evans DM, Kemp JP, Warrington NM, Miller L, Timpson NJ, Ring SM, Verhulst FC, Hofman A, Rivadeneira F, Meaburn EL, Price TS, Dale PS, Pillas D, Yliherva A, Rodriguez A, Golding J, Jaddoe VWV, Jarvelin M-R, Plomin R, Pennell CE, Tiemeier H, Davey Smith G, Common variation near ROBO2 is associated with expressive vocabulary in infancy. Nat. Commun. 5, 4831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose MC, Styr B, Schmid TA, Elie JE, Yartsev MM, Cortical representation of group social communication in bats. Science 374, eaba9584 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halley AC, Baldwin MKL, Cooke DF, Englund M, Pineda CR, Schmid T, Yartsev MM, Krubitzer L, Coevolution of motor cortex and behavioral specializations associated with flight and echolocation in bats. Curr. Biol, doi: 10.1016/j.cub.2022.04.094 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simonyan K, The laryngeal motor cortex: its organization and connectivity. Curr. Opin. Neurobiol. 28, 15–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jürgens U, Neural pathways underlying vocal control. Neurosci. Biobehav. Rev. 26, 235–258 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Simonyan K, Horwitz B, Laryngeal motor cortex and control of speech in humans. Neuroscientist 17, 197–208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martins PT, Boeckx C, Vocal learning: Beyond the continuum. PLoS Biol. 18, e3000672 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fitch WT, The Biology and Evolution of Speech: A Comparative Analysis. Annu. Rev. Linguist. 4, 255–279 (2018). [Google Scholar]
- 53.Belkhir JR, Raouf Belkhir J, Tecumseh Fitch W, Garcea FE, Chernoff BL, Sims MH, Navarrete E, Haber S, Paul DA, Smith SO, Pilcher WH, Mahon BZ, Direct electrical stimulation evidence for a dorsal motor area with control of the larynx. [Preprint] (2021). 10.1016/j.brs.2020.11.013. [DOI] [PMC free article] [PubMed]
- 54.Breshears JD, Molinaro AM, Chang EF, A probabilistic map of the human ventral sensorimotor cortex using electrical stimulation. J. Neurosurg. 123, 340–349 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Dichter BK, Breshears JD, Leonard MK, Chang EF, The Control of Vocal Pitch in Human Laryngeal Motor Cortex. Cell 174, 21–31.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuypers HGJM, Corticobulbar Connexions to the Pons and Lower Brain-Stem in Man: An Anatomical Study. Brain 81, 364–388 (1958). [DOI] [PubMed] [Google Scholar]
- 57.Wild JM, Neural pathways for the control of birdsong production. J. Neurobiol. 33, 653–670 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Paton JA, Manogue KR, Nottebohm F, Bilateral organization of the vocal control pathway in the budgerigar, Melopsittacus undulatus. J. Neurosci. 1, 1279–1288 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gahr M, Neural song control system of hummingbirds: comparison to swifts, vocal learning (Songbirds) and non-learning (Suboscines) passerines, and vocal learning (Budgerigars) and non-learning (Dove, owl, gull, quail, chicken) nonpasserines. J. Comp. Neurol. 426, 182–196 (2000). [PubMed] [Google Scholar]
- 60.Zheng D-J, Okobi DE Jr, Shu R, Agrawal R, Smith SK, Long MA, Phelps SM, Mapping the vocal circuitry of Alston’s singing mouse with pseudorabies virus. J. Comp. Neurol. 530, 2075–2099 (2022). [DOI] [PubMed] [Google Scholar]
- 61.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ, Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fisher SE, Scharff C, FOXP2 as a molecular window into speech and language. Trends Genet. 25, 166–177 (2009). [DOI] [PubMed] [Google Scholar]
- 63.King M, Wilson A, Evolution at two levels in humans and chimpanzees. [Preprint] (1975). 10.1126/science.1090005. [DOI] [PubMed]
- 64.Kvon EZ, Kamneva OK, Melo US, Barozzi I, Osterwalder M, Mannion BJ, Tissières V, Pickle CS, Plajzer-Frick I, Lee EA, Kato M, Garvin TH, Akiyama JA, Afzal V, Lopez-Rios J, Rubin EM, Dickel DE, Pennacchio LA, Visel A, Progressive Loss of Function in a Limb Enhancer during Snake Evolution. Cell 167, 633–642.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Partha R, Chauhan BK, Ferreira Z, Robinson JD, Lathrop K, Nischal KK, Chikina M, Clark NL, Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong ES, Zheng D, Tan SZ, Bower NL, Garside V, Vanwalleghem G, Gaiti F, Scott E, Hogan BM, Kikuchi K, McGlinn E, Francois M, Degnan BM, Deep conservation of the enhancer regulatory code in animals. Science 370 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Kaplow IM, Schaffer DE, Wirthlin ME, Lawler AJ, Brown AR, Kleyman M, Pfenning AR, Inferring mammalian tissue-specific regulatory conservation by predicting tissue-specific differences in open chromatin. BMC Genomics 23, 291 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai L-H, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M, Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hiller M, Schaar BT, Indjeian VB, Kingsley DM, Hagey LR, Bejerano G, A “Forward Genomics” Approach Links Genotype to Phenotype using Independent Phenotypic Losses among Related Species. Cell Rep. 2, 817–823 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A, Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li YE, Preissl S, Hou X, Zhang Z, Zhang K, Qiu Y, Poirion OB, Li B, Chiou J, Liu H, Pinto-Duarte A, Kubo N, Yang X, Fang R, Wang X, Han JY, Lucero J, Yan Y, Miller M, Kuan S, Gorkin D, Gaulton KJ, Shen Y, Nunn M, Mukamel EA, Behrens MM, Ecker JR, Ren B, An atlas of gene regulatory elements in adult mouse cerebrum. Nature 598, 129–136 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.si L Ho T, Ané C, A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Syst. Biol. 63, 397–408 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Ives AR, Garland T, Phylogenetic logistic regression for binary dependent variables. Syst. Biol. 59, 9–26 (2010). [DOI] [PubMed] [Google Scholar]
- 74.BRAIN Initiative Cell Census Network (BICCN), A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munz M, Bharioke A, Kosche G, Moreno-Juan V, Brignall A, Rodrigues TM, Graff-Meyer A, Ulmer T, Haeuselmann S, Pavlinic D, Ledergerber N, Gross-Scherf B, Rózsa B, Krol J, Picelli S, Cowan CS, Roska B, Pyramidal neurons form active, transient, multilayered circuits perturbed by autism-associated mutations at the inception of neocortex. Cell 186, 1930–1949.e31 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barone R, Fichera M, De Grandi M, Battaglia M, Lo Faro V, Mattina T, Rizzo R, Familial 18q12.2 deletion supports the role of RNA-binding protein CELF4 in autism spectrum disorders. Am. J. Med. Genet. A 173, 1649–1655 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Sun W, Wagnon JL, Mahaffey CL, Briese M, Ule J, Frankel WN, Aberrant sodium channel activity in the complex seizure disorder of Celf4 mutant mice. J. Physiol. 591, 241–255 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruzzo EK, Pérez-Cano L, Jung J-Y, Wang L-K, Kashef-Haghighi D, Hartl C, Singh C, Xu J, Hoekstra JN, Leventhal O, Leppä VM, Gandal MJ, Paskov K, Stockham N, Polioudakis D, Lowe JK, Prober DA, Geschwind DH, Wall DP, Inherited and De Novo Genetic Risk for Autism Impacts Shared Networks. Cell 178, 850–866.e26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Niarchou M, Gustavson DE, Sathirapongsasuti JF, Anglada-Tort M, Eising E, Bell E, McArthur E, Straub P, 23andMe Research Team, McAuley JD, Capra JA, Ullén F, Creanza N, Mosing MA, Hinds DA, Davis LK, Jacoby N, Gordon RL, Genome-wide association study of musical beat synchronization demonstrates high polygenicity. Nat Hum Behav 6, 1292–1309 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karayannis T, Au E, Patel JC, Kruglikov I, Markx S, Delorme R, Héron D, Salomon D, Glessner J, Restituito S, Gordon A, Rodriguez-Murillo L, Roy NC, Gogos JA, Rudy B, Rice ME, Karayiorgou M, Hakonarson H, Keren B, Huguet G, Bourgeron T, Hoeffer C, Tsien RW, Peles E, Fishell G, Cntnap4 differentially contributes to GABAergic and dopaminergic synaptic transmission. Nature 511, 236–240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Costa CIS, da Silva Montenegro EM, Zarrei M, de Sá Moreira E, Silva IMW, de Oliveira Scliar M, Wang JYT, Zachi EC, Branco EV, da Costa SS, Lourenço NCV, Vianna-Morgante AM, Rosenberg C, Krepischi ACV, Scherer SW, Passos-Bueno MR, Copy number variations in a Brazilian cohort with autism spectrum disorders highlight the contribution of cell adhesion genes. Clin. Genet, doi: 10.1111/cge.14072 (2021). [DOI] [PubMed] [Google Scholar]
- 82.Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, Packer A, SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4, 36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sullivan PF, Meadows JRS, Gazal S, Phan BN, Li X, Genereux DP, Dong MX, Bianchi M, Andrews G, Sakthikumar S, Nordin J, Roy A, Christmas MJ, Marinescu VD, Wang C, Wallerman O, Xue J, Yao S, Sun Q, Szatkiewicz J, Wen J, Huckins LM, Lawler A, Keough KC, Zheng Z, Zeng J, Wray NR, Li Y, Johnson J, Chen J, Zoonomia Consortium§, Paten B, Reilly SK, Hughes GM, Weng Z, Pollard KS, Pfenning AR, Forsberg-Nilsson K, Karlsson EK, Lindblad-Toh K, Leveraging base-pair mammalian constraint to understand genetic variation and human disease. Science 380, eabn2937 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brignell A, Morgan AT, Woolfenden S, Klopper F, May T, Sarkozy V, Williams K, A systematic review and meta-analysis of the prognosis of language outcomes for individuals with autism spectrum disorder. Autism & Developmental Language Impairments 3, 2396941518767610 (2018). [Google Scholar]
- 85.Nevue AA, Lovell PV, Wirthlin M, Mello CV, Molecular specializations of deep cortical layer analogs in songbirds. Sci. Rep. 10, 18767 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jarvis ED, Evolution of vocal learning and spoken language. Science 366, 50–54 (2019). [DOI] [PubMed] [Google Scholar]
- 87.Wang R, Chen C-C, Hara E, Rivas MV, Roulhac PL, Howard JT, Chakraborty M, Audet J-N, Jarvis ED, Convergent differential regulation of SLIT-ROBO axon guidance genes in the brains of vocal learners. J. Comp. Neurol. 523, 892–906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Strick PL, Dum RP, Rathelot J-A, The Cortical Motor Areas and the Emergence of Motor Skills: A Neuroanatomical Perspective. Annu. Rev. Neurosci. 44, 425–447 (2021). [DOI] [PubMed] [Google Scholar]
- 89.Cerkevich CM, Rathelot J-A, Strick PL, Cortical basis for skilled vocalization. Proc. Natl. Acad. Sci. U. S. A. 119, e2122345119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, Murtha MT, Bichsel C, Niu W, Cotney J, Ercan-Sencicek AG, Gockley J, Gupta AR, Han W, He X, Hoffman EJ, Klei L, Lei J, Liu W, Liu L, Lu C, Xu X, Zhu Y, Mane SM, Lein ES, Wei L, Noonan JP, Roeder K, Devlin B, Sestan N, State MW, Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, Ma’ayan A, Gene Set Knowledge Discovery with Enrichr. Curr Protoc 1, e90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Benjamini Y, Hochberg Y, Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. (1995). [Google Scholar]
- 93.Hickey G, Paten B, Earl D, Zerbino D, Haussler D, HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics 29, 1341–1342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang X, Kaplow IM, Wirthlin M, Park TY, Pfenning AR, HALPER facilitates the identification of regulatory element orthologs across species. Bioinformatics 36, 4339–4340 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Elie J, Schmid T, Yartsev M, Anatomy and neurophysiological data for “Vocal learning-associated convergent evolution in mammalian proteins and regulatory elements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Redlich R, Kowalczyk A, Pfenning AR, pfenninglab/RERconvergeVocalLearning: Vocal Learning Mammalian Protein (https://zenodo.org/doi/10.5281/zenodo.10641176).
- 97.Kaplow IM, Schaeffer D, Srinivasan C, Sestilli HH, Lawler AJ, Pfenning AR, pfenninglab/TACIT: TACIT Vocal Learning Mammal M1+PV (https://zenodo.org/doi/10.5281/zenodo.5952292).
- 98.Foley NM, Mason VC, Harris AJ, Bredemeyer KR, Damas J, Lewin HA, Eizirik E, Gates J, Zoonomia Consortium, Springer MS, Murphy WJ, A genomic timescale for placental mammal evolution. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Armstrong J, Hickey G, Diekhans M, Fiddes IT, Novak AM, Deran A, Fang Q, Xie D, Feng S, Stiller J, Genereux D, Johnson J, Marinescu VD, Alföldi J, Harris RS, Lindblad-Toh K, Haussler D, Karlsson E, Jarvis ED, Zhang G, Paten B, Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature 587, 246–251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Janik VM, Knörnschild M, Vocal production learning in mammals revisited. Philos. Trans. R. Soc. Lond. B Biol. Sci. 376, 20200244 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O’Rourke T, Martins PT, Asano R, Tachibana RO, Okanoya K, Boeckx C, Capturing the Effects of Domestication on Vocal Learning Complexity. Trends Cogn. Sci. 25, 462–474 (2021). [DOI] [PubMed] [Google Scholar]
- 102.Chikina M, Robinson J, Clark NL, Hundreds of Genes Experienced Convergent Shifts in Selective Pressure in Marine Mammals. Mol. Biol. Evol. 33, 2182–2192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kirilenko BM, Munegowda C, Osipova E, Jebb D, Sharma V, Blumer M, Morales A, Ahmed A-W, Hilgers L, Zoonomia Consortium, Hiller M, TOGA integrates gene annotation with orthology inference at scale. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Whelan S, Goldman N, A General Empirical Model of Protein Evolution Derived from Multiple Protein Families Using a Maximum-Likelihood Approach. Mol. Biol. Evol. 18, 691–699 (2001). [DOI] [PubMed] [Google Scholar]
- 105.Schliep KP, phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benjamini Y, Hochberg Y, Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995). [Google Scholar]
- 107.Smith MD, Pond S. L. Kosakovsky, RELAX: detecting relaxed selection in a phylogenetic framework. Mol. Biol. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wisotsky SR, Pond S. L. Kosakovsky, Shank SD, Muse SV, Synonymous Site-to-Site Substitution Rate Variation Dramatically Inflates False Positive Rates of Selection Analyses: Ignore at Your Own Peril. Mol. Biol. Evol. 37, 2430–2439 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yartsev MM, Witter MP, Ulanovsky N, Grid cells without theta oscillations in the entorhinal cortex of bats. Nature 479, 103–107 (2011). [DOI] [PubMed] [Google Scholar]
- 110.Genzel D, Yartsev MM, The fully automated bat (FAB) flight room: A human-free environment for studying navigation in flying bats and its initial application to the retrosplenial cortex. J. Neurosci. Methods 348, 108970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Conte WL, Kamishina H, Reep RL, Multiple neuroanatomical tract-tracing using fluorescent Alexa Fluor conjugates of cholera toxin subunit B in rats. Nat. Protoc. 4, 1157–1166 (2009). [DOI] [PubMed] [Google Scholar]
- 112.Lima SQ, Hromádka T, Znamenskiy P, Zador AM, PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS One 4, e6099 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Knowland D, Lilascharoen V, Pacia CP, Shin S, Wang EH-J, Lim BK, Distinct Ventral Pallidal Neural Populations Mediate Separate Symptoms of Depression. Cell 170, 284–297.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cui Q, Du X, Chang IYM, Pamukcu A, Lilascharoen V, Berceau BL, García D, Hong D, Chon U, Narayanan A, Kim Y, Lim BK, Chan CS, Striatal Direct Pathway Targets Npas1+ Pallidal Neurons. J. Neurosci. 41, 3966–3987 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang W, Yartsev MM, Correlated Neural Activity across the Brains of Socially Interacting Bats. Cell 178, 413–428.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shneider NA, Mentis GZ, Schustak J, O’Donovan MJ, Functionally reduced sensorimotor connections form with normal specificity despite abnormal muscle spindle development: the role of spindle-derived neurotrophin 3. J. Neurosci. 29, 4719–4735 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Elie JE, Theunissen FE, The vocal repertoire of the domesticated zebra finch: a data-driven approach to decipher the information-bearing acoustic features of communication signals. Anim. Cogn. 19, 285–315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stringer C, Pachitariu M, Steinmetz N, Reddy CB, Carandini M, Harris KD, Spontaneous behaviors drive multidimensional, brainwide activity. [Preprint] (2019). 10.1126/science.aav7893. [DOI] [PMC free article] [PubMed]
- 119.Pachitariu, Steinmetz, Kadir, Fast and accurate spike sorting of high-channel count probes with KiloSort. Adv. Neural Inf. Process. Syst. [Google Scholar]
- 120.Kilosort: Fast Spike Sorting with Drift Correction for up to a Thousand Channels (Github; https://github.com/MouseLand/Kilosort).
- 121.Hsu A, Borst A, Theunissen FE, Quantifying variability in neural responses and its application for the validation of model predictions. Network 15, 91–109 (2004). [PubMed] [Google Scholar]
- 122.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ, ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Milani P, Escalante-Chong R, Shelley BC, Patel-Murray NL, Xin X, Adam M, Mandefro B, Sareen D, Svendsen CN, Fraenkel E, Cell freezing protocol suitable for ATAC-Seq on motor neurons derived from human induced pluripotent stem cells. Sci. Rep. 6, 25474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jebb D, Huang Z, Pippel M, Hughes GM, Lavrichenko K, Devanna P, Winkler S, Jermiin LS, Skirmuntt EC, Katzourakis A, Burkitt-Gray L, Ray DA, Sullivan KAM, Roscito JG, Kirilenko BM, Dávalos LM, Corthals AP, Power ML, Jones G, Ransome RD, Dechmann DKN, Locatelli AG, Puechmaille SJ, Fedrigo O, Jarvis ED, Hiller M, Vernes SC, Myers EW, Teeling EC, Six reference-quality genomes reveal evolution of bat adaptations. Nature 583, 578–584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li Q, Brown JB, Huang H, Bickel PJ, Measuring reproducibility of high-throughput experiments. aoas 5, 1752–1779 (2011). [Google Scholar]
- 126.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 128.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang H, Song L, Wang X, Cheng H, Wang C, Meyer CA, Liu T, Tang M, Aluru S, Yue F, Liu XS, Li H, Fast alignment and preprocessing of chromatin profiles with Chromap. Nat. Commun. 12, 6566 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Granja JM, Corces MR, Pierce SE, Bagdatli ST, Choudhry H, Chang HY, Greenleaf WJ, ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yao Z, van Velthoven CTJ, Nguyen TN, Goldy J, Sedeno-Cortes AE, Baftizadeh F, Bertagnolli D, Casper T, Chiang M, Crichton K, Ding S-L, Fong O, Garren E, Glandon A, Gouwens NW, Gray J, Graybuck LT, Hawrylycz MJ, Hirschstein D, Kroll M, Lathia K, Lee C, Levi B, McMillen D, Mok S, Pham T, Ren Q, Rimorin C, Shapovalova N, Sulc J, Sunkin SM, Tieu M, Torkelson A, Tung H, Ward K, Dee N, Smith KA, Tasic B, Zeng H, A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner MM, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML, Flicek P, GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G, GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J, g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kulinskaya E, On two-sided p-values for non-symmetric distributions, arXiv [math.ST] (2008). http://arxiv.org/abs/0810.2124. [Google Scholar]
- 136.Wang Y, Zhang X, Chen F, Chen L, Wang J, Xie J, LRRK2-NFATc2 Pathway Associated with Neuroinflammation May Be a Potential Therapeutic Target for Parkinson’s Disease. J. Inflamm. Res. 14, 2583–2586 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lovell PV, Huizinga NA, Friedrich SR, Wirthlin M, Mello CV, The constitutive differential transcriptome of a brain circuit for vocal learning. BMC Genomics 19, 231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.St Pourcain B, Wang K, Glessner JT, Golding J, Steer C, Ring SM, Skuse DH, Grant SFA, Hakonarson H, Smith G. Davey, Association between a high-risk autism locus on 5p14 and social communication spectrum phenotypes in the general population. Am. J. Psychiatry 167, 1364–1372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sweetser DA, Elsharkawi I, Yonker L, Steeves M, Parkin K, Thibert R, “Pitt-Hopkins Syndrome” in GeneReviews®, Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, Amemiya A, Eds. (University of Washington, Seattle, Seattle (WA), 2012). [PubMed] [Google Scholar]
- 140.Gat-Yablonski G, Frumkin-Ben David R, Bar M, Potievsky O, Phillip M, Lazar L, Homozygous microdeletion of the POU1F1, CHMP2B, and VGLL3 genes in chromosome 3 — a novel syndrome. Am. J. Med. Genet. A 155A, 2242–2246 (2011). [DOI] [PubMed] [Google Scholar]
- 141.Bartsch O, Schindler D, Beyer V, Gesk S, van’t Slot R, Feddersen I, Buijs A, Jaspers NGJ, Siebert R, Haaf T, Poot M, A girl with an atypical form of ataxia telangiectasia and an additional de novo 3.14 Mb microduplication in region 19q12. Eur. J. Med. Genet. 55, 49–55 (2012). [DOI] [PubMed] [Google Scholar]
- 142.Chowdhury S, Bandholz AM, Parkash S, Dyack S, Rideout AL, Leppig KA, Thiese H, Wheeler PG, Tsang M, Ballif BC, Shaffer LG, Torchia BS, Ellison JW, Rosenfeld JA, Phenotypic and molecular characterization of 19q12q13.1 deletions: a report of five patients. Am. J. Med. Genet. A 164A, 62–69 (2014). [DOI] [PubMed] [Google Scholar]
- 143.Nacinovich R, Villa N, Broggi F, Tavaniello C, Bomba M, Conconi D, Redaelli S, Sala E, Lavitrano M, Neri F, 19q12q13.2 duplication syndrome: neuropsychiatric long-term follow-up of a new case and literature update. Neuropsychiatr. Dis. Treat. 13, 2545–2550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Murcia Pienkowski V, Kucharczyk M, Młynek M, Szczałuba K, Rydzanicz M, Poszewiecka B, Skórka A, Sykulski M, Biernacka A, Koppolu AA, Posmyk R, Walczak A, Kosińska J, Krajewski P, Castaneda J, Obersztyn E, Jurkiewicz E, Śmigiel R, Gambin A, Chrzanowska K, Krajewska-Walasek M, Płoski R, Mapping of breakpoints in balanced chromosomal translocations by shallow whole-genome sequencing points to EFNA5, BAHD1 and PPP2R5E as novel candidates for genes causing human Mendelian disorders. J. Med. Genet. 56, 104–112 (2019). [DOI] [PubMed] [Google Scholar]
- 145.Kraft SJ, “Genome-wide association study of persistent developmental stuttering,” thesis, University of Illinois at Urbana-Champaign (2010). [Google Scholar]
- 146.Pagnamenta AT, Khan H, Walker S, Gerrelli D, Wing K, Bonaglia MC, Giorda R, Berney T, Mani E, Molteni M, Pinto D, Le Couteur A, Hallmayer J, Sutcliffe JS, Szatmari P, Paterson AD, Scherer SW, Vieland VJ, Monaco AP, Rare familial 16q21 microdeletions under a linkage peak implicate cadherin 8 (CDH8) in susceptibility to autism and learning disability. J. Med. Genet. 48, 48–54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Karoglan A, Schanze D, Bär C, Muschke P, Zenker M, Schanze I, A 2q24.2 microdeletion containing TANK as novel candidate gene for intellectual disability. Am. J. Med. Genet. A 179, 832–836 (2019). [DOI] [PubMed] [Google Scholar]
- 148.Deriziotis P, O’Roak BJ, Graham SA, Estruch SB, Dimitropoulou D, Bernier RA, Gerdts J, Shendure J, Eichler EE, Fisher SE, De novo TBR1 mutations in sporadic autism disrupt protein functions. Nat. Commun. 5, 1–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Palumbo O, Fichera M, Palumbo P, Rizzo R, Mazzolla E, Cocuzza DM, Carella M, Mattina T, TBR1 is the candidate gene for intellectual disability in patients with a 2q24.2 interstitial deletion. Am. J. Med. Genet. A 164A, 828–833 (2014). [DOI] [PubMed] [Google Scholar]
- 150.Orsini JJ, Escolar ML, Wasserstein MP, Caggana M, “Krabbe Disease” in GeneReviews®, Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, Amemiya A, Eds. (University of Washington, Seattle, Seattle (WA), 2000). [PubMed] [Google Scholar]
- 151.Schlaubitz S, Yatsenko SA, Smith LD, Keller KL, Vissers LE, Scott DA, Cai WW, Reardon W, Abdul-Rahman OA, Lammer EJ, Lifchez CA, Magenis E, Veltman JA, Stankiewicz P, Zabel BU, Lee B, Ovotestes and XY sex reversal in a female with an interstitial 9q33.3-q34.1 deletion encompassing NR5A1 and LMX1B causing features of Genitopatellar syndrome. Am. J. Med. Genet. A 143A, 1071–1081 (2007). [DOI] [PubMed] [Google Scholar]
- 152.Hu VW, Addington A, Hyman A, Novel autism subtype-dependent genetic variants are revealed by quantitative trait and subphenotype association analyses of published GWAS data. PLoS One 6, e19067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dimassi S, Labalme A, Lesca G, Rudolf G, Bruneau N, Hirsch E, Arzimanoglou A, Motte J, de Saint Martin A, Boutry-Kryza N, Cloarec R, Benitto A, Ameil A, Edery P, Ryvlin P, De Bellescize J, Szepetowski P, Sanlaville D, A subset of genomic alterations detected in rolandic epilepsies contains candidate or known epilepsy genes including GRIN2A and PRRT2. Epilepsia 55, 370–378 (2014). [DOI] [PubMed] [Google Scholar]
- 154.Lehman A, Thouta S, Mancini GMS, Naidu S, van Slegtenhorst M, McWalter K, Person R, Mwenifumbo J, Salvarinova R, CAUSES Study, EPGEN Study, Guella I, McKenzie MB, Datta A, Connolly MB, Kalkhoran SM, Poburko D, Friedman JM, Farrer MJ, Demos M, Desai S, Claydon T, Loss-of-Function and Gain-of-Function Mutations in KCNQ5 Cause Intellectual Disability or Epileptic Encephalopathy. Am. J. Hum. Genet. 101, 65–74 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Franek KJ, Butler J, Johnson J, Simensen R, Friez MJ, Bartel F, Moss T, DuPont B, Berry K, Bauman M, Skinner C, Stevenson RE, Schwartz CE, Deletion of the immunoglobulin domain of IL1RAPL1 results in nonsyndromic X-linked intellectual disability associated with behavioral problems and mild dysmorphism. Am. J. Med. Genet. A 155A, 1109–1114 (2011). [DOI] [PubMed] [Google Scholar]
- 156.Lehalle D, Sanlaville D, Guimier A, Plouvier E, Leblanc T, Galmiche L, Radford I, Romana S, Colleaux L, de Pontual L, Lyonnet S, Amiel J, Multiple congenital anomalies-intellectual disability (MCA-ID) and neuroblastoma in a patient harboring a de novo 14q23.1q23.3 deletion. Am. J. Med. Genet. A 164A, 1310–1317 (2014). [DOI] [PubMed] [Google Scholar]
- 157.Le Guen Y, Leroy F, Philippe C, IMAGEN Consortium, Mangin J-F, Dehaene-Lambertz G, Frouin V, Enhancer Locus in ch14q23.1 Modulates Brain Asymmetric Temporal Regions Involved in Language Processing. Cereb. Cortex 30, 5322–5332 (2020). [DOI] [PubMed] [Google Scholar]
- 158.Lesca G, Rudolf G, Labalme A, Hirsch E, Arzimanoglou A, Genton P, Motte J, de Saint Martin A, Valenti M-P, Boulay C, De Bellescize J, Kéo-Kosal P, Boutry-Kryza N, Edery P, Sanlaville D, Szepetowski P, Epileptic encephalopathies of the Landau-Kleffner and continuous spike and waves during slow-wave sleep types: genomic dissection makes the link with autism. Epilepsia 53, 1526–1538 (2012). [DOI] [PubMed] [Google Scholar]
- 159.Zoghbi HY, Orr HT, Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J. Biol. Chem. 284, 7425–7429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Celestino-Soper PB, Skinner C, Schroer R, Eng P, Shenai J, Nowaczyk MM, Terespolsky D, Cushing D, Patel GS, Immken L, Willis A, Wiszniewska J, Matalon R, Rosenfeld JA, Stevenson RE, Kang S-HL, Cheung SW, Beaudet AL, Stankiewicz P, Deletions in chromosome 6p22.3-p24.3, including ATXN1, are associated with developmental delay and autism spectrum disorders. Mol. Cytogenet. 5, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Benchek P, Igo RP, Voss-Hoynes H, Wren Y, Miller G, Truitt B, Zhang W, Osterman M, Freebairn L, Tag J, Taylor HG, Chan ER, Roussos P, Lewis B, Stein CM, Iyengar SK, Association between genes regulating neural pathways for quantitative traits of speech and language disorders. npj Genomic Medicine 6, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Milani D, Sabatini C, Manzoni FMP, Ajmone PF, Rigamonti C, Malacarne M, Pierluigi M, Cavani S, Costantino MA, Microdeletion 2q23.3q24.1: exploring genotype-phenotype correlations. Congenit. Anom. 55, 107–111 (2015). [DOI] [PubMed] [Google Scholar]
- 163.Benito-Aragón C, Gonzalez-Sarmiento R, Liddell T, Diez I, d’Oleire Uquillas F, Ortiz-Terán L, Bueichekú E, Chow HM, Chang S-E, Sepulcre J, Neurofilament-lysosomal genetic intersections in the cortical network of stuttering. Prog. Neurobiol. 184, 101718 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kielb S, Kisanuki YY, Dawson E, Neuropsychological profile associated with an alpha-synuclein gene (SNCA) duplication. Clin. Neuropsychol., 1–12 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Egyptian fruit bat ATAC-seq data—including raw .fasta files, .bigWig files of genome-aligned reads, processed open chromatin .narrowpeak files, and extensive sample metadata—have been made available at GEO under accession ID GSE187366. Publicly available ATAC-Seq data was obtained from GEO GSE159815 and GSE161374. Single-nucleus open chromatin was downloaded from the NEMO archive: https://data.nemoarchive.org/biccn/grant/u19_cemba/cemba/epigenome/sncell/ATACseq/mouse/. The code for the electrophysiology and functional components of the project are available here: https://github.com/NeuroBatLab/LoggerDataProcessing/ https://github.com/NeuroBatLab/SoundAnalysisBats/ https://github.com/NeuroBatLab/LoggerDataProcessing/ https://github.com/julieelie/Kilosort2_Tetrode/configFiles/configFile16.m. The code for the convergent evolution analysis can be found in https://github.com/pfenninglab/TACIT, https://github.com/veg/hyphy, and https://github.com/veg/hyphy-analyses/tree/master/BUSTED-PH . The histological and electrophysiology source data from this study are available on FigShare under the DOI 10.6084/m9.figshare.25180430 (95). The code for conducting and visualizing the RERconverge analysis can be found here: https://zenodo.org/doi/10.5281/zenodo.10641176 (96). The code for conducting and visualizing the TACIT anlaysis can be found here: https://zenodo.org/doi/10.5281/zenodo.5952292 (97).