Abstract
The DNA methyltransferase of bacteriophage T4 (T4 Dam MTase) recognizes the palindromic sequence GATC, and catalyzes transfer of the methyl group from S-adenosyl-l-methionine (AdoMet) to the N6-position of adenine [generating N6-methyladenine and S-adenosyl-l-homocysteine (AdoHcy)]. Pre-steady state kinetic analysis revealed that the methylation rate constant kmeth for unmethylated and hemimethylated substrates (0.56 and 0.47 s–1, respectively) was at least 20-fold larger than the overall reaction rate constant kcat (0.023 s–1). This indicates that the release of products is the rate-limiting step in the reaction. Destabilization of the target-base pair did not alter the methylation rate, indicating that the rate of target nucleoside flipping does not limit kmeth. Preformed T4 Dam MTase–DNA complexes are less efficient than preformed T4 Dam MTase–AdoMet complexes in the first round of catalysis. Thus, this data is consistent with a preferred route of reaction for T4 Dam MTase in which AdoMet is bound first; this preferred reaction route is not observed with the DNA-[C5-cytosine]-MTases.
INTRODUCTION
Biological methylation of DNA is an important process in living cells and also represents an interesting example of sequence-specific DNA–protein interactions. These reactions are catalyzed by DNA methyltransferases (MTases), which recognize short palindromic sequences and transfer a methyl group from S-adenosyl-l-methionine (AdoMet) to a target adenine or cytosine base (1). Bacteriophage T4 encodes a DNA-[N6-adenine] MTase (T4 Dam MTase) that methylates adenine in the sequence GATC, as well as in some non-canonical GAY sequences, in cytosine, 5-methylcytosine and 5-hydroxymethylcytosine-containing DNAs (2–4).
Biochemical studies of DNA MTases generally use naturally occurring highly polymeric DNA as the substrate to determine the kinetic parameters of the reaction. However, methylation of highly polymeric DNA, even if it contains only a single recognition site, is complicated by the facilitated diffusion of the bound enzyme in a 1-D search for its specific recognition site (5). The use of relatively short oligonucleotide duplexes (usually in the range 12–30 bp) containing the specific recognition site simplifies the experimental conditions and allows one to obtain more precise data to calculate the reaction parameters. In addition, one can introduce substitutions or other modifications into the duplex structure in order to elucidate their effects on enzyme–DNA interactions.
We previously investigated the ability of the T4 Dam MTase to bind and methylate substrates in which different structural components of the recognition site were altered or eliminated. We also examined the effect of various defects in the synthetic oligonucleotide duplexes on the steady state kinetic parameters of methylation (6–8). We showed that addition of T4 Dam MTase to a complete reaction mixture containing a duplex with a native recognition site resulted in a ‘burst’ of product formation, followed by a slower constant rate of product formation. This indicated that release of product methylated DNA from the enzyme is the rate-limiting step in the reaction. Certain defects in duplex structure significantly reduced the kcat, but resulted in only small changes in the Km value and in the ability of the enzyme to bind the DNA duplex. Other altered duplexes were poorly bound by the enzyme, although the kcat remained largely unchanged. Thus, these studies allowed us to establish which modified structural elements interact with the T4 Dam MTase and, to some extent, how they interact.
Pre-steady state analysis is a powerful tool in studying complex enzyme-catalyzed reactions (9). This is particularly true for bisubstrate reactions in which numerous microscopic rate constants can contribute to kcat and Km. However, only a few DNA MTases have been studied using this technique (10). In this study, we used pre-steady state analysis to obtain further insight into the kinetic behavior of T4 Dam, and applied this method to characterize the pre-steady state steps of the interaction between the enzyme and various duplex substrates with canonical or defective recognition sites.
MATERIALS AND METHODS
Enzymes and chemicals
[3H-CH3]-AdoMet was purchased from Amersham. Unlabeled AdoMet (Sigma) was purified further by chromatography on a C18-reversed-phase column as described previously (11). Oligonucleotides were synthesized and purchased from Midland Certified Reagent Company (Midland, TX). They were additionally purified as described (11). Oligonucleotide concentrations were determined spectrophotometrically from the molar extinction coefficients of individual nucleotides and the known sequences. T4 Dam MTase was purified to homogeneity as previously described (11). Protein concentrations were determined by the Bradford method (12).
Synthetic duplexes were obtained by heating and annealing complementary oligonucleotide chains from 90–20°C over a 7–12 h period (13). Variant duplexes were obtained by hybridizing oligonucleotides complementary to a standard 20mer oligonucleotide I (Table 1), designated as the upper strand. Oligonucleotides I and II, which contain the canonical T4 Dam MTase recognition site, GATC, in the center of the sequence, are completely complementary and capable of forming the stable ‘specific’ 20mer duplex 1 (Table 2). Oligonucleotide III has base substitutions (relative to II) that alter the canonical recognition site to GTAC; it was combined to form the ‘double-mismatch’ duplex 2 (Table 2). Additional duplexes were prepared containing methylated A in the GATC of the top strand and unmethylated A in the GATC of the bottom strand (duplex 1m), as well as a substitution of 2-aminopurine (N) for A (duplex 3).
Table 1. Oligodeoxyribonucleotides used in this study.
| Oligonucleotide | 5′-Sequence-3′ |
|---|---|
| I | CAGTTTAGGATCCATTTCAC |
| Im | CAGTTTAGGMTCCATTTCAC |
| II | GTGAAATGGATCCTAAACTG |
| IIn | GTGAAATGGNTCCTAAACTG |
| III | GTGAAATGGTACCTAAACTG |
Nucleotide residues included in recognition site, GATC, are underlined. In IIm, M is N6-methyladenine; in IIn, N is 2-aminopurine.
Table 2. Pre-steady state kinetic parameters of T4 Dam MTase interaction with duplexes containing a native or modified recognition site at limiting enzyme concentrationa.
| Oligonucleotide substrate | Combination of duplexes | Recognition sequenceb | Burstc | kmeth (s–1)c | kcat (s–1)c |
|---|---|---|---|---|---|
| Canonical sites | |||||
| -G-A-T-C- | 0.92 | 0.56 | 0.023 | ||
| 1 | I + II | -C-T-A-G- | (0.05) | (0.10) | (0.002) |
| -G-M-T-C- | 0.85 | 0.47 | 0.021 | ||
| 1m | Im + II | -C-T-A-G- | (0.05) | (0.09) | (0.002) |
| -G-M-T-C- | 0.84 | 0.14 | 0.021 | ||
| 1md | Im + II | -C-T-A-G- | (0.07) | (0.04) | (0.002) |
| Substituted sites | |||||
| -G-A-T-C- | 0.86 | 0.49 | 0.0051 | ||
| 2 | I + III | -C-A-T-G- | (0.05) | (0.09) | (0.0005) |
| -G-A-T-C- | 0.90 | 0.67 | 0.0038 | ||
| 3 | I + IIn | -C-T-N-G- | (0.03) | (0.12) | (0.0005) |
aEnzyme was preincubated with [3H-CH3]-AdoMet prior to the addition of duplex.
bN is 2-aminopurine; M is N6-methyladenine.
cValues in parentheses correspond to standard deviations.
dEnzyme was preincubated with duplex 1m before the addition of the [3H-CH3]-AdoMet.
Steady state and pre-steady state assays
Methyl transfer assays were carried out at 25°C as described (11). Reaction mixtures contained 100 mM Tris–HCl, pH 8.0, 10 mM EDTA, 10 mM DTT and 0.2 mg/ml BSA. In order to determine the kinetic parameters of the reaction, the concentration of duplexes and enzyme were varied, as well as the order of the pre-incubation of various components of the reaction. In the steady state assay, 20 µl aliquots were withdrawn at time intervals and spotted on DE81 anion-exchange filter paper (Whatman, 2.0 cm). The molar concentration of [3H-CH3]-groups incorporated into DNA was quantitated as described earlier (14).
The micro-volume rapid quench instrument ‘KinTek Corp. RQF-3’ was used for the pre-steady state assay of T4 Dam MTase. Syringes, mixers and age-loops were equilibrated to 25°C. The feeding syringe containing the enzyme preparation was kept at 0°C to avoid inactivation of the T4 Dam MTase during the experiment. Sodium dodecyl sulfate (SDS) [0.05% (w/v)] in 25 mM Tris–HCl (pH 8.3) was used as a quench solution. The quenched samples were collected in Eppendorf tubes and evaporated to a volume of 100 µl using an Eppendorf vacuum concentrator. Duplicate 50 µl aliquots were spotted onto DE81 filters for 3H-counting. Kinetic parameters were obtained by using the computer program ‘Scientist’ (MicroMath Inc.) for non-linear regression analysis to fit the experimental data.
RESULTS
Methylation of unmodified substrates
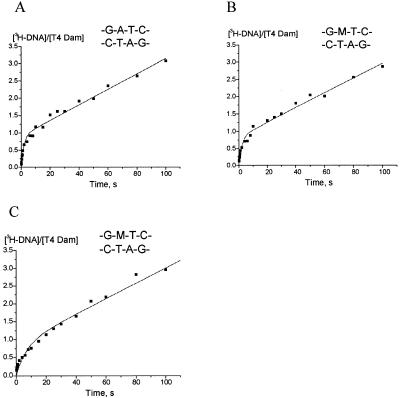
T4 Dam product formation over time for native unmethylated and hemimethylated substrate under various reaction conditions is shown in Figure 1. Assays were performed under initial velocity conditions of saturating substrate (Fig. 1A, B and C). Reactions were started by the addition of DNA substrates to preformed T4 Dam-AdoMet complexes (Fig. 1A and B) or by addition of AdoMet to preformed T4 Dam-DNA complexes (Fig. 1C).
Figure 1.
Pre-steady state kinetics of the MTase using the pre-formed MTase–AdoMet complex (A and B) or the pre-formed MTase–DNA complex (C). The concentrations of T4 Dam and duplexes were 0.158 and 1.0 µM, respectively; [3H-CH3] AdoMet was at 8 µM. The amount of 3H-labeled CH3 groups incorporated into 20mer duplexes was quantitated as indicated in the text. The data were fit to equation 1.
The assay conditions used, presence of saturating concentrations of AdoMet (8 µM, Km = 0.49 µM) and DNA (1 µM, Km = 5.3 – 12.9 nM) (8,16), assured absence of any free form of the enzyme. This allowed the whole methylation reaction,
E + D + AdoMet ![]() E·AdoMet·D
E·AdoMet·D ![]() E·AdoHcy·mD
E·AdoHcy·mD
![]() E + AdoHcy + mD to be shortened and described by the kinetic scheme of Gutfreund (15):
E + AdoHcy + mD to be shortened and described by the kinetic scheme of Gutfreund (15):
where E is T4 Dam with bound substrate AdoMet (or bound product AdoHcy), D is the substrate duplex, mD is the methylated product duplex, k2 is the methylation rate constant and k3 is the rate constant for release of products mD and AdoHcy. According to this scheme, product mD accumulation is described by the equation:
P = k22 (1 – e(–(k2 + k3)*t))/(k2 + k3)2 + k2k3t/(k2 + k3) 2
where P = [mD]/[Eo] and Eo is the total enzyme concentration. While this equation fits our experimental data satisfactorily, a better fit was obtained with the equation in its more general form:
P = B (1 – e–kmeth*t) + kcat*t 3
where B is the burst of product normalized to [Eo], kmeth is the rate of methyl transfer and kcat is the steady-state reaction rate constant.
Figure 1A shows T4 Dam product formation over time with unmethylated DNA (duplex 1, Table 1). Kinetic parameters derived from the data according to equation (3) are shown in Table 2. The steady-state rate of product formation kcat of 0.023 s–1 (Table 2) is in agreement with the value of 0.015 s–1 previously determined under initial velocity conditions (16). The value of kmeth (0.56 s–1) was 24-fold higher than kcat and a burst of product formation was observed. The burst value B of 0.92 methyl groups transferred per enzyme molecule was close to the theoretical value of 1.0. The non-zero time course intercept indicates that catalysis was limited by an event after methyl transfer. This is similar to results found for M.EcoRI (10), M.PvuII (17), M.HhaI (18) and other DNA MTases.
Figure 1B shows the time course of product formation for hemimethylated DNA (duplex 1m, Table 1). The derived kinetic parameters (Table 2) were similar to those for the unmethylated substrate, with a kcat of 0.021 s–1 and kmeth of 0.47 s–1. If monomeric T4 Dam MTase were to bind in a random orientation to hemimethylated duplex 1m, then a non-productive complex should be formed in half of the binding events. However, the burst value of 0.85 is not consistent with this assumption.
Under saturating substrate conditions, the order of pre-incubation of T4 Dam MTase with AdoMet or hemimethylated 20mer duplex 1m had no influence on the burst value or kcat. However, kmeth was ∼3.5-fold lower when the enzyme was pre-incubated with duplex 1m than when it was pre-incubated with AdoMet (Fig. 1C; Table 2). This suggests that in the first round of catalysis, the preformed T4 Dam MTase–DNA complex is less efficient than the preformed enzyme–AdoMet complex. An alternative explanation would be that the preformed enzyme–DNA complex is not catalytically competent, must dissociate, bind AdoMet and then rebind DNA, which could slow down productive complex formation.
Rapid kinetic analysis of methylation of duplexes with modified sites
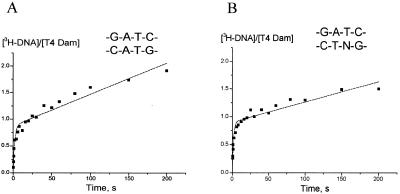
We previously investigated T4 Dam MTase binding and steady state methylation of oligonucleotides containing modified recognition sites (6,8,16). Here we extend those studies by means of rapid kinetic analysis. These experiments were also performed under initial velocity conditions of saturating substrate, since they permit determination of burst values and a comparison of kmeth versus kcat.
Having a central double mismatch in the GATC sequence (duplex 2) had relatively minor effects on the parameters B and kmeth but reduced kcat by 4.5-fold (Fig.2A; Table 2). The double mismatch is expected to facilitate target adenine flipping out of the DNA helix. However, since this defect only mildly decreased kmeth, we can conclude that adenine flipping is not the rate-limiting step in the methyl transfer reaction catalyzed by T4 Dam.
Figure 2.
Methyl group incorporation into duplexes containing base substitutions using the pre-formed T4 MTase–AdoMet complex. The concentrations of T4 Dam MTase and oligonucleotide duplexes were 0.158 and 1.0 µM, respectively, except for (B) in which the enzyme concentration was 0.27 µM; [3H-CH3] AdoMet was at 8 µM. The data from (A) and (B) were fit to equation 1.
A→N substitution of one of the target adenine residues (duplex 3) eliminates the N6-amino group and shifts one Watson–Crick H-bond. This alteration had no effect on the burst value, while increasing kmeth slightly (from 0.56 to 0.67 s–1; Fig. 2B; Table 2); however, kcat was decreased 6-fold, in agreement with previous steady state experiments (8). Since release of product from the central complex (enzyme–methylated DNA–AdoHcy) is the rate limiting step of the reaction for the native duplex, this suggests that A→N substitution results in a 6-fold slower release of products from the central complex.
DISCUSSION
Burst analysis under conditions of limiting enzyme concentration
Only a small number of experiments have been done to determine the rate-limiting step of the reaction catalyzed by DNA MTases. The primary source of this data comes from characterizing the rate of accumulation of methylated DNA product using steady-state reaction conditions. If a clear burst of product formation is observed, then the chemical step is not rate limiting and is faster than any subsequent step. Such bursts have been observed for several DNA MTases, including EcoRI (19), PvuII (17) and T4 Dam (16), indicating that release of product methylated DNA is the rate-limiting step in this reaction. Pre-steady state experiments (10) showed that for EcoRI MTase, the kmeth is >300-fold faster than kcat. In contrast, TaqI MTase transfers the methyl group with a kmeth of 0.05 s–1, whereas the release of the methylated DNA product from the enzyme complex has a rate constant of 5 s–1 (E.Weinhold, personal communication). It should be noted here that the TaqI MTase results were for reactions carried out at 25°C; consequently, the above kcat value differs from the previously published one of 0.73 s–1 obtained at 60°C (20). For T4 Dam, kmeth is at least 20-fold higher than kcat with duplex 1 or duplex 1m (Table 2). Therefore, release of methylated product from the complex (and not methyl transfer) is the rate-limiting step; thus, quantitatively, it is intermediate between the EcoRI DNA and TaqI MTases.
A natural substrate for DNA MTase is asymmetric hemimethylated DNA; thus, it is not surprising that these enzymes show asymmetry in DNA recognition. Most of the substrate duplexes that we used in our experiments have only one adenine residue available for methylation. Therefore, for these substrates a monomeric MTase should be correctly orientated for methylation in only half of binding events, assuming non-preferential orientation in binding a target site; this should result in a burst value of 0.5. However, we observed burst magnitudes varying between 0.84 and 0.90 for duplexes 1m, 2 and 3, which have only one methylatable adenine residue (Table 2). One explanation for these results is that under the conditions of these experiments, T4 Dam has a preference for binding with hemimethylated DNA such that the majority of the enzyme reaches a productive orientation. Alternatively, if no such initial binding discrimination occurs, those molecules initially bound in the incorrect orientation could rapidly reorient on the DNA without formal dissociation from the complex. This would result in subsequent methylation and, thus, yield a higher than expected burst magnitude. Such a model suggests that the T4 Dam–DNA complex is not a static structure, but is in rapid dynamic equilibrium between two states of the complex, E*·D↔D*·E, which have different enzyme–DNA orientations.
Studies with a duplex having a structural defect in the recognition site
We made a duplex having a structural defect in the recognition site in order to determine how this might affect enzyme activity; viz., a central, double base-mismatch within GATC (duplex 2). This had only a slight effect on kmeth, but it decreased kcat by ∼4.5-fold (Fig. 2A; Table 2). Since the target adenine residue is unpaired in this duplex, extra-helical base flipping should occur more readily. Therefore, if target base flipping were the rate-limiting step in determining the methylation rate constant, we would expect to see an increase in the kmeth. However, this was not the case, so we must conclude that flipping of the target adenine is not rate limiting, and that unpairing of the target adenine has little effect on methyl group transfer. The same conclusions were made for the EcoRI MTase because it has a kmeth of 41 s–1 (10) and a base-flipping rate of at least 195 s–1 (21). Furthermore, it was shown that the methylation rate is limited by the preceding step of the enzyme–substrates complex isomerization (21,22). Although the absence of a base complementary to the target adenine had little effect on kmeth, the kcat was reduced 4.5-fold. Thus, this defect reduces the speed of a step or steps involved in product release, but leaves the essential features of catalysis itself largely unaltered.
A proposed kinetic scheme for the T4 Dam MTase
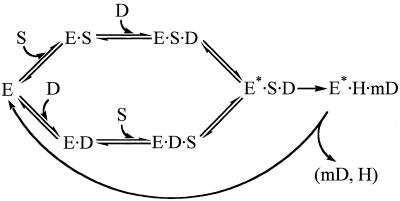
At limiting enzyme concentration, we observed 3.5-fold slower kmeth when T4 Dam was pre-incubated with substrate duplex 1m, compared to pre-incubation with AdoMet (Table 2). From this we can conclude that although the enzyme may have a random order of substrate binding, the route via the enzyme–AdoMet complex is kinetically more efficient than via the enzyme–DNA complex. That is, preformed T4 Dam MTase–DNA complexes are less efficient than preformed T4 Dam MTase–AdoMet complexes in the first round of catalysis, suggesting a preferred route of reaction for T4 Dam MTase in which AdoMet is bound first. In this regard, Adams and Blumenthal (17) failed to observe a burst with M·PvuII when the enzyme was pre-incubated with buffer only, but obtained a burst when it was pre-incubated with [3H-CH3]-AdoMet. Although these results suggest that M·PvuII had a preferred route analogous to T4 Dam, an alternative explanation could account for their results; e.g., the majority of the PvuII MTase was isolated containing bound (cold)-AdoMet. Based on our preliminary kinetic analyses, we propose the reaction scheme presented in Figure 3. The conversion of the preliminary E·D·S and E·S·D complexes to the productive complex E*·S·D could theoretically proceed via isomerization steps (23). The isomerization step, E·D·S↔E*·S·D, is proposed to be several-fold slower than the isomerization step of the more adaptable complex, E·S·D↔E*·S·D. We must now verify this scheme by standard techniques, such as the use of reaction inhibitors.
Figure 3.
Proposed kinetic scheme for the T4 Dam MTase. E, T4 Dam Mtase; S, AdoMet; D, DNA duplex; H, AdoHcy; mD, methylated DNA duplex. The isomerization step, E·D·S↔E*·S·D, is proposed to be several-fold slower than the isomerization step of the more adaptable complex, E·S·D↔E*·S·D.
A detailed understanding of both the specificity and the catalysis of an enzyme requires the assignment of rate constants for the individual reaction steps. Of extreme interest are the first steps, involving extrahelical nucleoside flipping and stabilization of the target base within the enzyme’s active site, and the restacking of the modified base inside the double-stranded helix. Fortunately, data on base flipping can be obtained by performing stopped flow fluorescence measurements using DNA containing 2-aminopurine (22). This can be correlated with the data obtained by pre-steady state chemical analysis of the methylation process. Using these techniques, we should achieve a better understanding of the DNA methylation process.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a US Public Health Service grant from the Fogarty International Center (No. TW00529), a grant from the Russian Foundation for Fundamental Researches (No. 95-04-12671), a US Public Health Service grant GM29227 from the National Institutes of Health (to S.H.) and an NSF grant MCB-9603567 (to N.R.).
REFERENCES
- 1.Wilson G.G. (1991) Nucleic Acids Res., 19, 2539–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlagman S.L. and Hattman,S. (1983) Gene, 22, 139–156. [DOI] [PubMed] [Google Scholar]
- 3.Schlagman S.L., Miner,Z., Fehér,Z. and Hattman,S. (1988) Gene, 73, 517–530. [DOI] [PubMed] [Google Scholar]
- 4.Schlagman S.L. and Hattman,S. (1989) Nucleic Acids Res., 17, 9101–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surby M.A. and Reich,N.O. (1996) Biochemistry, 35, 2209–2217. [DOI] [PubMed] [Google Scholar]
- 6.Malygin E.G., Petrov,N.A., Gorbunov,Y.A., Kossykh,V.G. and Hattman,S.M. (1997) Nucleic Acids Res., 25, 4393–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zinoviev V.V., Ovechkina,L.G. and Malygin,E.G. (1996) Mol. Biol., 30, 724–726. [Google Scholar]
- 8.Malygin E.G., Zinoviev,V.V., Petrov,N.A., Evdokimov,A.A., Kossykh,V.G., Jen-Jacobson,L. and Hattman,S. (1999) Nucleic Acids Res., 27, 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson K.A. (1992) Enzyme, 20, 1–60. [Google Scholar]
- 10.Reich N.O. and Mashhoon,N. (1993) J. Biol. Chem., 268, 9191–9193. [PubMed] [Google Scholar]
- 11.Kossykh V.G., Schlagman,S.L. and Hattman,S. (1995) J. Biol. Chem., 270, 14389–14393. [DOI] [PubMed] [Google Scholar]
- 12.Bradford M. (1976) Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 13.Szczelkun M.D. and Connolly,B.A. (1995) Biochemistry, 34, 10724–10733. [DOI] [PubMed] [Google Scholar]
- 14.Thielking V., Du Bois,S., Eritja,R. and Guschlbauer,W. (1997) Biol. Chem., 378, 407–415 [DOI] [PubMed] [Google Scholar]
- 15.Gutfreund H. (1955) Discuss Faraday Soc., 20, 167–173 [Google Scholar]
- 16.Zinoviev V.V., Evdokimov,A.A., Gorbunov,Y.A., Malygin,E.G., Kossykh,V.G. and Hattman,S. (1998) Biol. Chem., 379, 481–488. [DOI] [PubMed] [Google Scholar]
- 17.Adams G.M. and Blumenthal,R.M. (1997) Biochemistry, 36, 8284–8292. [DOI] [PubMed] [Google Scholar]
- 18.Lindstrom W.M. Jr, Flynn,J. and Reich,N.O. (2000) J. Biol. Chem., 275, 4912–4919. [DOI] [PubMed] [Google Scholar]
- 19.Reich N.O. and Mashhoon,N. (1991) Biochemistry, 30, 2933–2939. [DOI] [PubMed] [Google Scholar]
- 20.Pues H., Bleimling,N., Holz,B., Wölcke,J. and Weinhold,E. (1999) Biochemistry, 38, 1426–1434. [DOI] [PubMed] [Google Scholar]
- 21.Allan B.W., Reich N.O. and Beechem,J.M. (1999) Biochemistry, 38, 5308–5314. [DOI] [PubMed] [Google Scholar]
- 22.Allan B.W., Beechem,J.M., Lindstrom,W.M. and Reich,N.O. (1998) J. Biol. Chem., 273, 2368–2373. [DOI] [PubMed] [Google Scholar]
- 23.Fersht A. (1977) Enzyme Structure and Mechanism. W.H. Freeman and Company, Reading and San Francisco, Chapter 7.