Abstract
Endothelial cells (ECs) are highly plastic, capable of differentiating into various cell types. Endothelial-to-mesenchymal transition (EndMT) is crucial during embryonic development and contributes substantially to vascular dysfunction in many cardiovascular diseases (CVDs). While targeting EndMT holds therapeutic promise, understanding its mechanisms and modulating its pathways remain challenging. Using single-cell RNA sequencing on three in vitro EndMT models, we identified conserved gene signatures. We validated original regulators in vitro and in vivo during embryonic heart development and peripheral artery disease. EndMT induction led to global expression changes in all EC subtypes rather than in mesenchymal clusters. We identified mitochondrial calcium uptake as a key driver of EndMT; inhibiting mitochondrial calcium uniporter (MCU) prevented EndMT in vitro, and conditional Mcu deletion in ECs blocked mesenchymal activation in a hind limb ischemia model. Tissues from patients with critical limb ischemia with EndMT features exhibited significantly elevated endothelial MCU. These findings highlight MCU as a regulator of EndMT and a potential therapeutic target.
Targeting mitochondrial calcium signaling preserves endothelial cell identity.
INTRODUCTION
Endothelial cells (ECs) lining the inner surfaces of blood and lymphatic vessels are exposed to a large variety of mechanical and biological stimuli that differ according to the tissue they reside in. To meet the local physiological requirements and ensure homeostasis, ECs must dynamically adapt their phenotype. Therefore, a high grade of plasticity is required.
In response to drivers (e.g., growth factors and cytokines), ECs can undergo mesenchymal activation and ultimately reprogram their identity to give rise to mesenchymal-like cells via a process named endothelial-to-mesenchymal transition (EndMT) (1). During this complex process, the basement membrane underlying ECs is disrupted, and ECs lose their cellular adhesion and delaminate from the vessel wall, switching from their usual apicobasal to a front-rear polarity (2). These complex cytoskeletal modifications and cell-to-cell junction rearrangements result in spindle-shaped mesenchymal-like cells with promigratory and proinvasive phenotype and impaired barrier function (3, 4). At the molecular level, ECs partially or totally lose the expression of canonical vascular markers [such as platelet EC adhesion molecule–1 (PECAM-1/CD31), von Willebrand factor, and vascular endothelial cadherin (VE-cadherin)] and increase the expression of typical mesenchymal proteins [such as α–smooth muscle actin (α-SMA), fibronectin, and N-cadherin] (1).
The main known EndMT inducers are members of the transforming growth factor–β (TGF-β) family and, most prominently, the TGF-β1 isoform (5, 6). However, numerous other TGF-β–independent signaling pathways are involved, resulting in either reduced or increased transcription of endothelial- and mesenchymal-specific genes by the activity of the EndMT master transcription factors SNAI1/2, ZEB1/2, and TWIST1 (7).
EndMT is a fundamental process omnipresent across the cardiovascular system beginning from the embryonic stage. During cardiac development, the formation of the endocardial cushions and the subsequent generation of the atrial septum, the interventricular septum, and the cardiac valves are dependent on ECs that undergo EndMT (8). It is therefore intuitive that aberrant EndMT may lead to developmental abnormalities that potentially contribute to valvular defects and that are the triggers of both congenital heart and adult valvular diseases (9, 10).
In adults, despite specific contexts (e.g., following ischemia) where transient EndMT is beneficial in vascular regeneration (11), EndMT is more widely recognized as a cardiovascular disease (CVD)–causal pathobiological process. The pathological vascular remodeling associated with EndMT is a common feature of a multitude of different CVDs, including cardiac fibrosis, atherosclerosis, pulmonary arterial hypertension, and a number of peripheral artery disorders (12–15). Thus, therapies designed to target EndMT hold promise to treat CVDs in which aberrant EndMT occurs. However, therapeutic approaches have faced considerable limitations due to the challenge of targeting the EndMT master-regulatory transcription factors or TGF-β signaling pathway. Identifying novel mechanisms governing this fate transition may provide more effective therapeutic interventions.
Here, we exploit single-cell RNA sequencing (scRNA-seq) to profile three experimental in vitro EndMT induction models by different inducers to mimic different degrees of remodeling leading to heterogeneous mesenchymal activation/partial EndMT. Using integrated analysis, followed by in vitro and in vivo functional validation, we identify mitochondrial calcium signaling as a previously unknown regulator of EndMT.
RESULTS
Integrated single-cell analysis identifies conserved EndMT gene signatures
To induce EndMT in vitro, we used two previously established experimental models in human umbilical vein ECs (HUVECs, abbreviated as ECs throughout the text below): first, stimulation with the potent EndMT inducer TGF-β1 (compared to untreated ECs) and, second, silencing of the endothelial transcription factor ETS-related gene (ERG) by short hairpin RNA (shRNA) (referred to as shERG compared to control pLKO-empty) (16, 17). We also set up a third model by overexpressing the essential EndMT driver SNAI1 in ECs (referred to as SNAIOE compared to control pRRL-empty). Long-term TGF-β1 treatment and ERG inhibition in ECs induced a host of mesenchymal markers, as well as a simultaneous decrease in endothelial identity markers in line with previous findings observed in HUVECs (fig. S1, A to F) (16, 17). Similar results were obtained in SNAIOE cells, suggesting that the cells underwent mesenchymal activation/partial (incomplete) EndMT as result of the concurrent expression of mesenchymal and endothelial markers (fig. S1, G to I). Concomitantly with the molecular switch in SNAIOE cells, we observed a significant increase in migration and decreased transendothelial electrical resistance (a measure of barrier function and integrity), two hallmarks of EndMT (fig. S1, J and K) (18).
For each of the three models corresponding to different grades of remodeling, thus mirroring phenotypic heterogeneity, a pooled sample of two HUVEC donors was subjected to scRNA-seq (Fig. 1A). We performed uniform manifold approximation and projection (UMAP) analysis and color-coded for canonical marker genes of previously identified EC subtypes (Fig. 1B; figs. S2, A to F, to S4, A to F; and table S1). In each model, we identified the same four EC subpopulations, namely, proliferating ECs expressing the proliferation markers MIK67 and PTTG1, tip cells displaying the previous identified signature of PGF and CXCR4 expression (19), and ECs with a mesenchymal signature (denoted as MSCs) with up-regulated mesenchymal markers SERPINE1 and FN1. We also identified cells with an intermediate signature (denoted as intermediate ECs) that formed a phenotypic continuum between the tip and MSC subtypes (figs. S2G to S4G and table S2). We then focused our analysis on ECs in a (putatively) nontransitory phenotypic state, i.e., the tip cells, proliferating cells, and MSCs, and did not further consider the ECs with the intermediate phenotype. Correlation heatmap analysis and hierarchical clustering showed that the gene signatures of tip cells, MSCs, and proliferating cells were relatively similar within each model and condition regardless of whether EndMT had occurred (under both EndMT-induced and not-induced conditions) (figs. S2H to S4H).
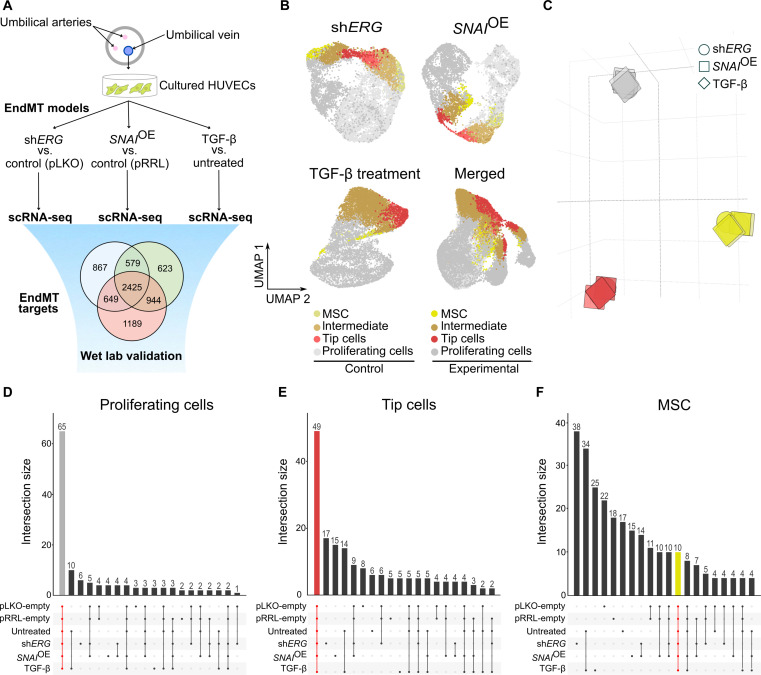
Fig. 1. scRNA-seq identifies congruent marker genes across three in vitro EndMT induction models.
(A) Graphical representation of the experimental design. (B) UMAP analysis of 32,125 cultured HUVECs from ERG silencing (shERG), SNAI1 overexpression (SNAIOE), and TGF-β experiments. (C) Principal components analysis (PCA) on the pairwise Jaccard similarity coefficients between the top 100 marker genes of cultured EC phenotypes. (D) Upset plot visualization showing the number of congruent genes (65 in gray) in the top 100 proliferating cell marker genes in cultured ECs. (E) Upset plot visualization showing the number of congruent genes (49 in red) in the top 100 tip cell marker genes in cultured ECs. (F) Upset plot visualization showing the number of congruent genes (10 in yellow) in the top 100 mesenchymal cell marker genes in cultured ECs.
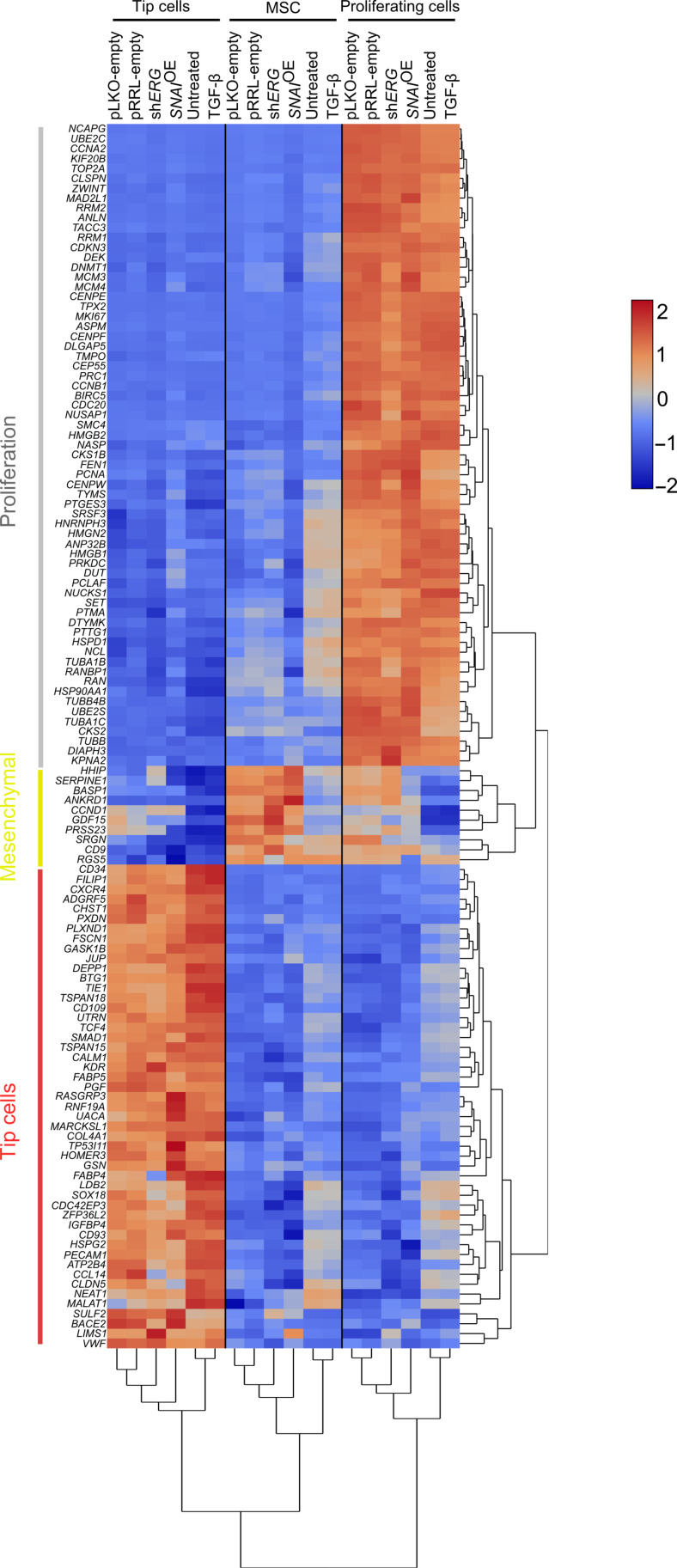
To determine whether the gene signatures were also similar across the three models (and not only within each model), we quantified marker gene set similarity. Specifically, we performed Jaccard similarity index analysis to compare marker gene sets of each subtype to all other subtypes. Our analysis indicated high congruency between the marker gene sets of proliferating, tip, and mesenchymal populations across the different models (Fig. 1C). Among the top 100 marker genes exclusively assigned to the proliferating cell cluster, 65 were shared across all conditions (e.g., induced versus not-induced EndMT and across the three models) and related to cell cycle regulation and DNA replication (e.g., CCNA2, CCNB1, DLGAP5, MCM3, TYMS, etc.) (Figs. 1D and 2 and table S3). Similarly, tip cells shared 49 common genes among the top 100 markers exclusively assigned to this cluster, including many regulators of cytoskeletal remodeling, cell movement, and angiogenesis (e.g., CDC42EP3, JUP, FSCN1, KDR, PGF, etc.) (Figs. 1E and 2 and table S3), consistent with the migratory and proangiogenic phenotype of the tip cells (20). However, the mesenchymal populations showed a more heterogeneous marker gene distribution, and only 10 marker genes of the top 100 genes exclusively assigned to this cluster were common across all three models and primarily associated with developmental and differentiation functions (Figs. 1F and 2 and table S3). Notably, none of the individual conditions altered the cell type composition, suggesting that EndMT induction does not skew phenotypic differentiation and that EndMT-like cells do not arise from the previously identified MSC population. In contrast, our results suggested that EndMT reflects global expression changes in all EC subtypes rather than affecting a specific EC subtype (figs. S2I to S4I).
Fig. 2. EndMT gene signatures are similar within each model and condition.
Heatmap and hierarchical clustering analysis of the 124 congruent marker genes of all cell states. Note: Marker genes for the intermediate cluster were not calculated since it represents a transitional phase between tip cells and MSCs.
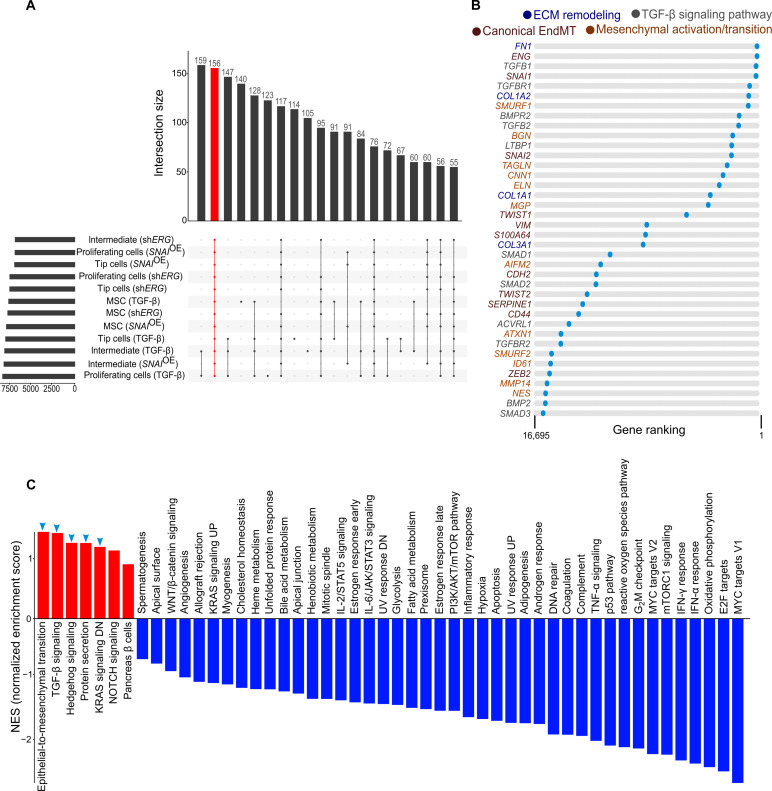
To identify genes that are specifically induced upon EndMT induction, we first performed differential gene expression analysis comparing EC phenotypes under control versus EndMT-induced conditions (fig. S5 and table S4). We then ranked each differential analysis based on fold change and used intersection analysis to identify genes that were invariably up-regulated upon EndMT induction and rank product–based meta-analysis to obtain a ranked list of genes congruently up-regulated across the conditions (e.g., induced versus not-induced EndMT) and the models irrespective of a specific EC subtype (Fig. 3A and table S5). We identified a set of 156 up-regulated genes commonly induced by EndMT induction. Among them, several were associated with transcriptional regulation (e.g., DDX3X, CTCF, GTF2I, HMGN1, PBXIP1, REST, and SETX), epigenetic networks (e.g., ANKRD11, BRD8, CHD4, CHD6, KDM3B, and SINHCAF), and developmental programs (e.g., ARL5A, EFNB3, FARP1, and FRS2) supporting a reprogramming of cell identity as the driver of EndMT in our models (table S6). Other genes were involved in protein trafficking and turnover (e.g., FBXW11, HERC1, RTN2, SEL1L, SNX27, and UBXN7), cytoskeletal remodeling (e.g., ARHGAP10, DCTN1, and TES), and metabolism (e.g., ALG8, ATP8A1, DEGS1, EPM2A, and SLC2A1), in line with the profile of (metabolically) active cells (table S6). Top-ranking genes identified by our rank product–based meta-analysis approach included all known canonical EndMT marker genes (e.g., ENG, SNAI1, SNAI2, TWIST1, VIM, S100A4, CDH2, TWIST2, SERPINE1, CD44, and ZEB2) (7), genes previously described to be broadly involved in mesenchymal activation/transition [SMURF1 and SMURF2 (21), BGN (22), TAGLN (23), CNN1 (23), ELN (24), MGP (25), AIFM2 (26), ATXN1 (27), ID1 (28), MMP14 (29), and NES (30)], and genes involved in extracellular matrix (ECM) remodeling (FN1, COL1A2, COL1A1, and COL3A1) and the TGF-β signaling pathway (TGFB1, TGFBR1, BMPR2, TGFB2, LTBP1, SMAD1, SMAD2, ACVRL1, TGFBR2, BMP2, and SMAD3) with an established role in EndMT (Fig. 3B and table S6) (7, 31). Furthermore, gene set enrichment analysis (GSEA) using 50 hallmark gene sets as input showed that the top up-regulated pathways identified in our meta-analysis were epithelial-to-mesenchymal transition; the TGF-β, Hedgehog, and NOTCH signaling pathways whose cross-talk initiates and maintains the process of mesenchymal transition (32); the KRAS signaling pathway reported to promote mesenchymal features (33); and protein secretion sustaining the increased cell’s demands during EndMT (31, 34) (Fig. 3C). On the contrary, among others, pathways involved in cell differentiation and proliferation were among the top down-regulated pathways (Fig. 3C), consistent with the inhibition of cell cycle progression and the activation of the dedifferentiation program during transition (35, 36). The GSEA also revealed changes in metabolic pathways supporting previous evidence of a metabolic switch in EndMT (37). Overall, these findings suggest that the transcriptional reprogramming upon EndMT induction occurs globally not only in all EC subtypes but also in those where EndMT was induced.
Fig. 3. EndMT induction induces expression changes independent of cellular differentiation state.
(A) Upset plot visualization showing the number of congruently up-regulated genes (red) in EndMT induction independent of cellular differentiation state. (B) String plots indicating canonical EndMT markers, genes involved in mesenchymal activation, ECM remodeling, and TGF-β signaling pathway as for color code. (C) Bar plot showing GSEA performed on the ranked list of genes of the cell state–independent EndMT rank-based meta-analysis for the hallmark gene sets. Up-regulated gene sets are indicated by blue arrowheads.
Gene ontology analysis reveals mitochondrial calcium modification as a hallmark of EndMT
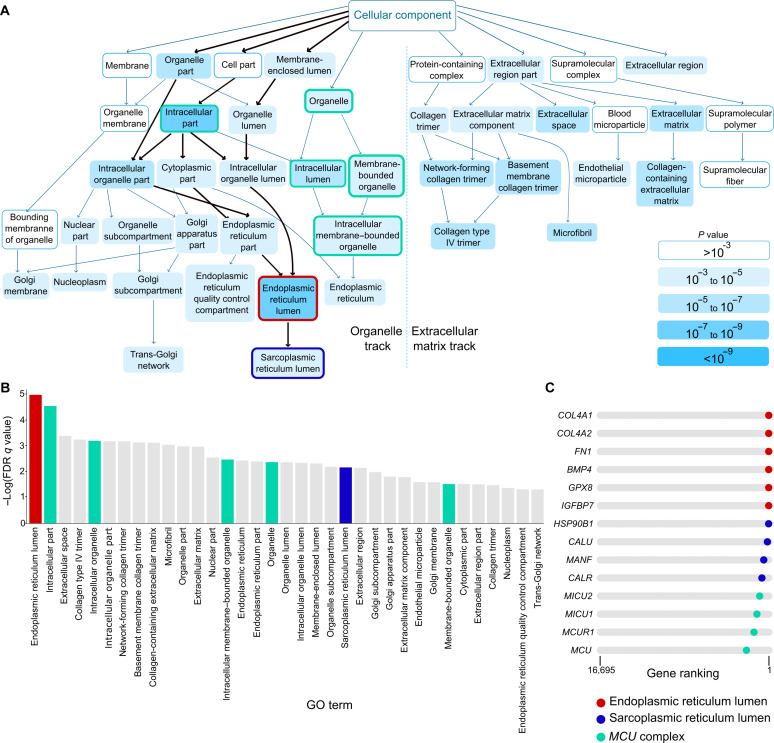
To gain further insights into the set of genes that were consistently up-regulated under the induced versus not-induced EndMT conditions and across the three models, we performed GOrilla gene ontology (GO) analysis on our meta-analysis (Fig. 4, A and B, and table S7). This revealed two major findings. First, it indicated that pathways associated with ECM formation and collagen deposition were highly up-regulated, consistent with the extensive ECM remodeling observed in EndMT in fibrotic disorders [Fig. 4, A (right) and B] (31, 34). Second, we also identified an unrecognized role for membrane-bounded organelles, including endoplasmic reticulum (ER)/sarcoplasmic reticulum (SR) and mitochondria (Fig. 4A, left). In particular, the ER and SR pathways were significantly up-regulated [Fig. 4, A (left) and B]. We focused on these findings and determined which specific genes are associated with these changes by plotting genes in the individual gene sets into our ranked list. Genes associated with the ER encoded mainly secreted proteins (COL4A1, COL4A2, FN1, BMP4, and IGFBP7) and were linked to (oxidative) protein folding (GPX8 and HSP90B1), consistent with an active secretory phenotype of mesenchymal activated cells (Fig. 4C and table S7). Genes in the SR were related to calcium (Ca2+) signaling and homeostasis (CALU, MANF, and CALR) (Fig. 4C and table S7). Ca2+ signaling is regulated by the intimate (physical) interconnection between ER/SR and mitochondria (38). When analyzing the second major intracellular membrane–bounded organelles, namely, the mitochondria, we found that genes encoding the mitochondrial Ca2+ uniporter (MCU) complex (MCU, MICU1, MICU2, and MCUR1) were highly and consistently up-regulated when EndMT was induced in each EC subtypes (Fig. 4C and table S7). Overall, these results pointed to a conserved mechanism underlying EndMT, characterized by the global activation of secretory pathways and the modulation of Ca2+ signaling across all EC subtypes. Increased secretory load has been previously linked to mesenchymal transdifferentiation (39, 40), and we therefore focused on the original observation of Ca2+ involvement in regulating EndMT.
Fig. 4. GO analysis reveals mitochondrial Ca2+ changes.
(A) Graph of enriched GO terms obtained by GOrilla GO enrichment analysis on the ranked list of genes of the cell state–independent EndMT rank-based meta-analysis. The organelle track and ECM track are separated by a vertical dotted line. The path to the ER/SR lumen is highlighted by bold arrows. The ER and SR lumen terms are highlighted by a red and a blue outlines, respectively. Terms that contain genes associated with mito-Ca2+ signaling are highlighted by a green outline. (B) Bar plot showing the order of terms according to adjusted P value of the GOrilla GO enrichment analysis performed on the ranked list of genes of the cell state–independent EndMT rank-based meta-analysis. The ER and SR lumen terms are highlighted in red and blue, respectively. Terms that contain genes associated with mito-Ca2+ signaling are highlighted in green. FDR, false discovery rate. (C) String plots indicating the rank of ER lumen, SR lumen, and MCU complex genes in the cell state–independent EndMT rank-based meta-analysis.
EndMT is accompanied by changes in mitochondrial Ca2+ homeostasis
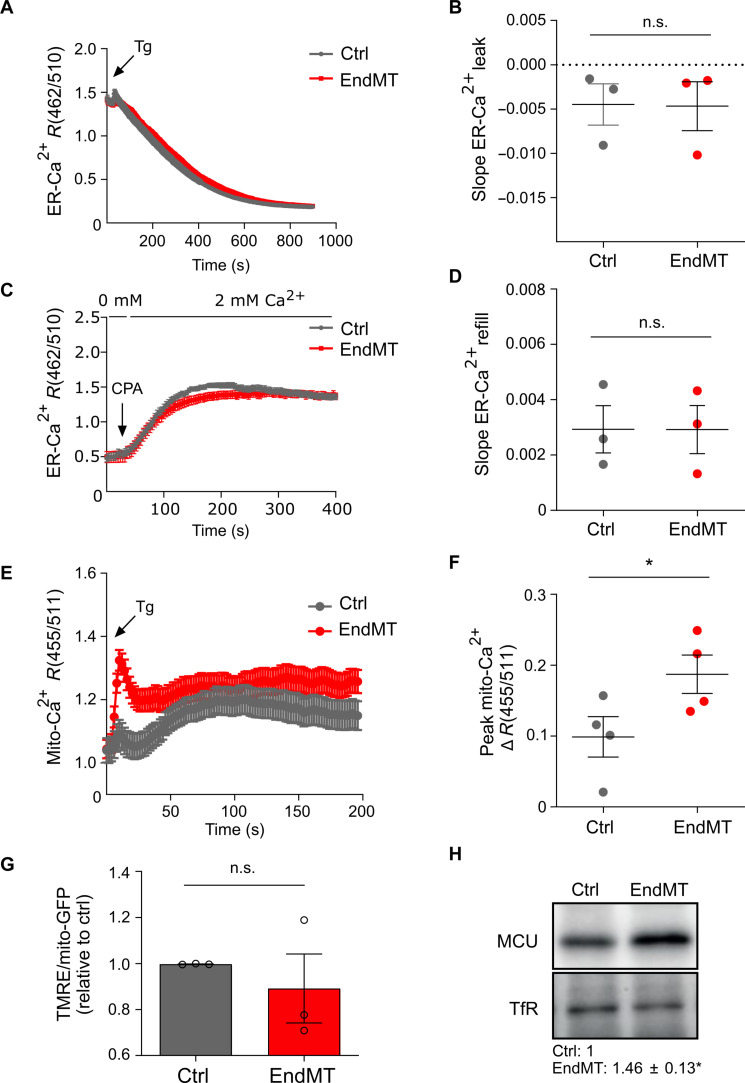
We first excluded ER stress as result of increased secretory burden by investigating the expression of ER stress sensors in our induction models (fig. S6A). We then analyzed by imaging the physical interactions between ER and mitochondria, namely, the mitochondria-associated membranes (MAMs), and detected a significant increase in the number of short-distance juxtapositions between ER and mitochondria in EndMT-derived cells as result of a closer proximity between these two organelles (fig. S6, B to D). MAMs are crucial for a correct intraorganellar communication, including the selective transmission of Ca2+ signals from the ER to mitochondria (41). We therefore investigated whether the enhancement of the ER-mitochondria interface was paralleled by changes in ER and mitochondrial Ca2+ signals during mesenchymal activation. We transduced control and EndMT-derived cells with the ER- or mitochondria-targeted ratiometric Ca2+ sensors GEM-CEPIA1 (42) or GEM-GECO1 (43), respectively, and measured Ca2+ concentrations at the resting state and upon intracellular mobilization. Confocal images and quantification of Ca2+ signals revealed similar basal Ca2+ levels between control and EndMT-derived cells in both organelles (fig. S6, E and F). We then assessed ER-mitochondria Ca2+ transfer by measuring ER-Ca2+ release and mitochondrial-Ca2+ (mito-Ca2+) uptake by thapsigargin (Tg), a well-known inducer of ER-mito Ca2+ fluxes (44). Despite no changes in the kinetics of ER leak and refilling (evaluated by measuring the slope of Tg-induced ER-Ca2+ release and cyclopiazonic acid–mediated ER-Ca2+ refill) (Fig. 5, A to D), we observed a rapid and significant increase in maximal mito-Ca2+ uptake in EndMT-derived cells compared to control ECs (Fig. 5E, F). Similar results were obtained when using histamine as a physiological stimulus of mito-Ca2+ influx (fig. S6, G and H). Mito-Ca2+ homeostasis is intrinsically modulated in EndMT irrespective of TGF-β pathway activation as comparable results were obtained in all three models of EndMT induction (fig. S6, I to L). Despite the increased mito-Ca2+ uptake, EndMT-derived cells had normal mitochondria compared to control ECs (Fig. 5G and fig. S7, A to E), suggesting that the mito-Ca2+ levels were overall maintained within a physiological range.
Fig. 5. EndMT is associated with increased mitochondrial Ca2+ uptake.
(A) ER-Ca2+ imaging traces in response to Tg (2 μM) in control cells (gray trace) and EndMT-derived HUVECs (TGF-β1 stimulation; red trace) maintained in 0 mM extracellular Ca2+ solution. Traces represent the mean CEPIA ratio (462/510 nm) ± SEM of different cells from one representative experiment. (B) Mean slope ± SEM of Tg-mediated ER Ca2+ release relative to three independent experiments. n.s., no significance. (C) ER-Ca2+ imaging traces in response to cyclopiazonic acid (CPA; 20 μM) in control cells (gray trace) and EndMT-derived HUVECs (TGF-β1 stimulation; red trace) maintained in 0 mM extracellular Ca2+ solution during stimulation, followed by the addition of 2 mM extracellular Ca2+ to measure ER refill. Traces represent the mean CEPIA ratio (462/510 nm) ± SEM of different cells from one representative experiment. (D) Mean slope ± SEM of Tg-mediated ER Ca2+ release relative to three independent experiments. (E) Mito-Ca2+ imaging traces in response to Tg (2 μM) in control (gray trace) and EndMT-derived HUVECs (TGF-β1 stimulation; red trace) maintained in 0 mM extracellular Ca2+ solution. Traces represent the mean GECO ratio (455/511 nm) ± SEM of different cells from one representative experiment. (F) Mean peak amplitude ± SEM of Tg-mediated mito-Ca2+ uptake relative to four independent experiments. *P < 0.05. (G) Quantification of the intensity of the dye tetramethylrhodamine ethyl (TMRE) that stains polarized mitochondria in control and EndMT-derived HUVECs (TGF-β1 stimulation; n = 3). Values were corrected for the mitochondrial content (mito-GFP). (H) Representative expression levels and relative quantification of immunoblot for MCU in control and EndMT-derived HUVECs (TGF-β1 stimulation; n = 4). Expression levels were normalized to transferrin receptor (TfR; loading control), and they are relative to control. *P < 0.05.
Mito-Ca2+ uptake is mediated by the MCU at the inner mitochondrial matrix (45). As expected and consistent with the scRNA-seq results, the pore-forming subunit MCU was overexpressed in EndMT-derived cells compared to control ECs (Fig. 5H). Overall, these results suggest a previously unrecognized role for mito-Ca2+ signaling in EndMT.
MCU targeting renders ECs less prone to EndMT
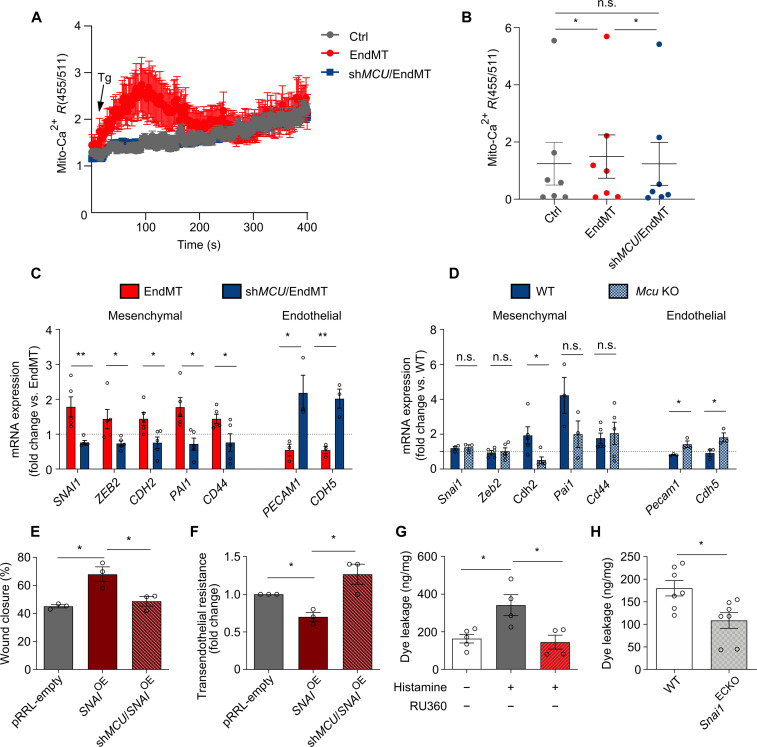
We next sought to determine whether targeting MCU-mediated Ca2+ uptake could modulate EndMT induction. We used MCU-silenced ECs (mediated by shRNA yielding >70% silencing; fig. S7F) and confirmed the functional impairment of mito-Ca2+ uptake following MCU targeting (fig. S7G). The impact of MCU silencing was subsequently tested on mito-Ca2+ uptake in EndMT-derived cells. Upon Tg stimulation, we observed an ~30% reduction in mito-Ca2+ uptake in shMCU EndMT-derived cells (Fig. 6, A and B), confirming that MCU-mediated mito-Ca2+ entry may modulate the phenotypic switch of ECs toward mesenchymal-like cells.
Fig. 6. MCU overexpression regulates EndMT.
(A) Mito-Ca2+ imaging traces in response to Tg (2 μM) in control HUVECs (gray trace) and EndMT-derived HUVECs (TGF-β1 stimulation) silenced or not for MCU (shMCU1) (blue and red traces, respectively) maintained in 0 mM extracellular Ca2+ solution. Traces represent the mean GECO ratio (455/511 nm) ± SEM of different cells from one representative experiment. (B) Mean peak amplitude ± SEM of Tg-mediated mito-Ca2+ uptake relative to seven independent experiments. *P < 0.05. (C) Reverse transcription polymerase chain reaction (RT-PCR) analysis of mRNA expression levels of mesenchymal and endothelial genes in control HUVECs and in HUVECs silenced for MCU before TGF-β1 stimulation. Expression values were normalized to 18S mRNA levels (n = 3 to 5). Dotted line indicates expression levels in EndMT. *P < 0.05 and **P < 0.01. (D) RT-PCR analysis of mRNA expression levels of mesenchymal and endothelial genes in wild-type (WT) and Mcu knockout (KO) ECs stimulated with TGF-β1 (n = 3 to 6). Dotted line indicates expression levels in EndMT in WT ECs. *P < 0.05. (E) Quantification of scratch wound assay using mitomycin C–treated control (empty), SNAIOE, and shMCU/SNAIOE HUVECs (n = 3). *P < 0.05. (F) Transendothelial electrical resistance analysis of control (empty), SNAIOE, and shMCU/SNAIOE HUVEC monolayers (n = 3). (G) Quantification of the Evans blue dye leakage in the ear of untreated and histamine-treated mice following vehicle or 100 μM RU360 topic application (n = 4 to 5). Values were normalized to milligrams of tissue. *P < 0.05. (H) Quantification of the Evans blue dye leakage in the ear of WT and Snai1ECKO histamine-treated mice (n = 7). Values were normalized to milligrams of tissue. *P < 0.05.
To further explore these findings, we analyzed the expression of mesenchymal markers upon MCU targeting. As shown in Fig. 6C and fig. S7H, genetic (by shRNA) and pharmacological inhibition of MCU (by RU360) prevented the induction of mesenchymal markers. Consistently, TGF-β1 stimulation of freshly isolated murine ECs from Mcu-deficient mice failed to induce mesenchymal activation (Fig. 6D). Functionally, these changes corresponded to decreased migration and permeability of EndMT-derived cells in vitro compared to control cells (Fig. 6, E and F, and fig. S7, I to L). Furthermore, RU360 treatment reduced vascular permeability in vivo following Ca2+ mobilization by histamine (Fig. 6G) to an extent comparable to that observed in Snai1ECKO mice in which EndMT is prevented by genetic inactivation of the master regulator Snai1 (Fig. 6H). These data confirm that MCU inhibition prevented EndMT and associated vascular dysfunctions, thus making MCU an attractive target to modulate EndMT function.
Mitochondrial Ca2+ regulates EndMT under pathophysiological conditions
EndMT is crucial for cardiac development to form specialized structures such as trabeculae and endocardial cushions (10). We therefore explored whether mito-Ca2+ signaling drives endocardial remodeling in both zebrafish and mammal embryos.
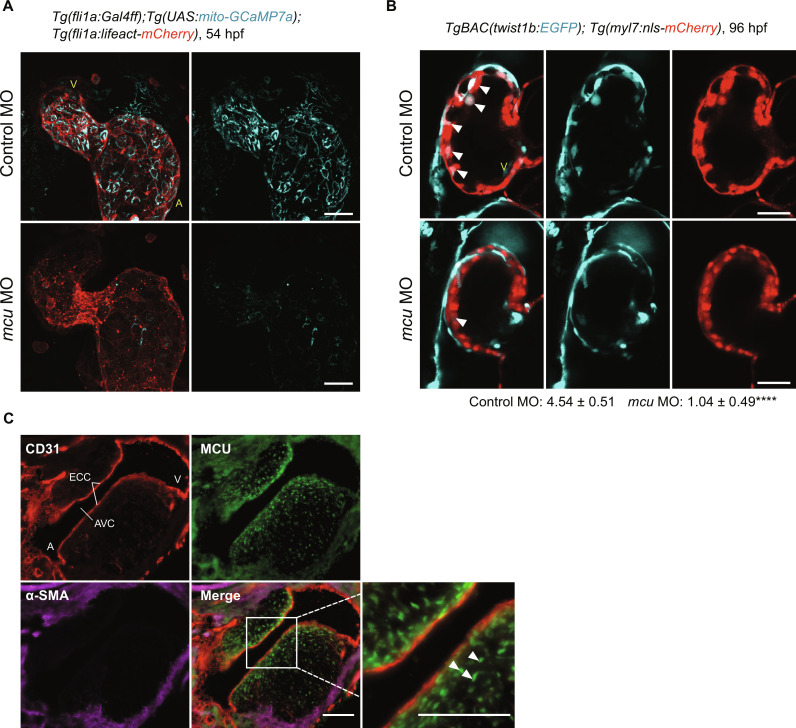
We generated a transgenic zebrafish line, Tg(fli1a:Gal4ff);Tg(uas:mito-GCaMP7a), in which mito-Ca2+ is labeled through the expression of the genetically encoded Ca2+ indicator GCaMP7a targeted to mitochondrial matrix under the control of an endocardial/EC promoter. We crossed this zebrafish line with Tg(fli1a:Lifeact-mCherry) (46) to visualize endocardial/ECs via live imaging. We observed lower GCaMP7a fluorescence intensity corresponding to decreased mito-Ca2+ influx in the endocardial cells of mcu morphants compared to control morphants (Fig. 7A), suggesting that mito-Ca2+ influx is mcu-dependent in endocardial cells during embryonic development.
Fig. 7. Endothelial MCU regulates EndMT during cardiac development.
(A) Representative three-dimensional stack confocal heart images of Tg (fli1a:Gal4ff);Tg(UAS:mito-GCamp7a);Tg(fli1a:lifeact-mCherry) embryo at 54 hours after fertilization (hpf) injected with the indicated control and mcu morpholino oligonucleotides (MOs) (n = 6). V, ventricle; A, atrium. Scale bars, 30 μm. (B) Representative single-scanned confocal ventricular heart images of TgBAC(twist1b:EGFP);Tg(myl7:nls-mCherry) embryo at 96 hpf. White arrowheads indicate twist1b+ cells quantified as indicated (n = 4). ****P < 0.0001. V, ventricle. Scale bars, 30 μm. (C) Representative heart micrographs of E11 mouse embryo stained for ECs (CD31; red), mural cells (α-SMA; purple), and MCU (green) (n = 5). Inset shows higher magnification of the boxed area. White arrowheads indicate MCU+ EndMT-derived cells migrating from the endocardium (CD31+) to the center of the cushion. ECC, endocardial cushions; AVC, atrioventricular canal; V, ventricle; A, atrium. Scale bars, 100 μm.
We next analyzed the expression of the classical epithelial/endothelial-to-mesenchymal transition marker twist1, which is important for heart development (47), using the established transgenic line TgBAC(twist1b:EGFP);Tg(myl7:nls-mCherry) (48, 49). We focused on trabeculae formation, which is initiated when the endocardial cells lining the inside of the heart tube penetrate the cardiac jelly to directly alight at the outer myocardial layer (50). Since enhanced green fluorescent protein (EGFP) is stable, we were able to observe cumulative expression patterns of twist1b. In mcu mutants, twist1b signal was significantly reduced in the trabecular region, leading to immature cardiac trabecular development (Fig. 7B).
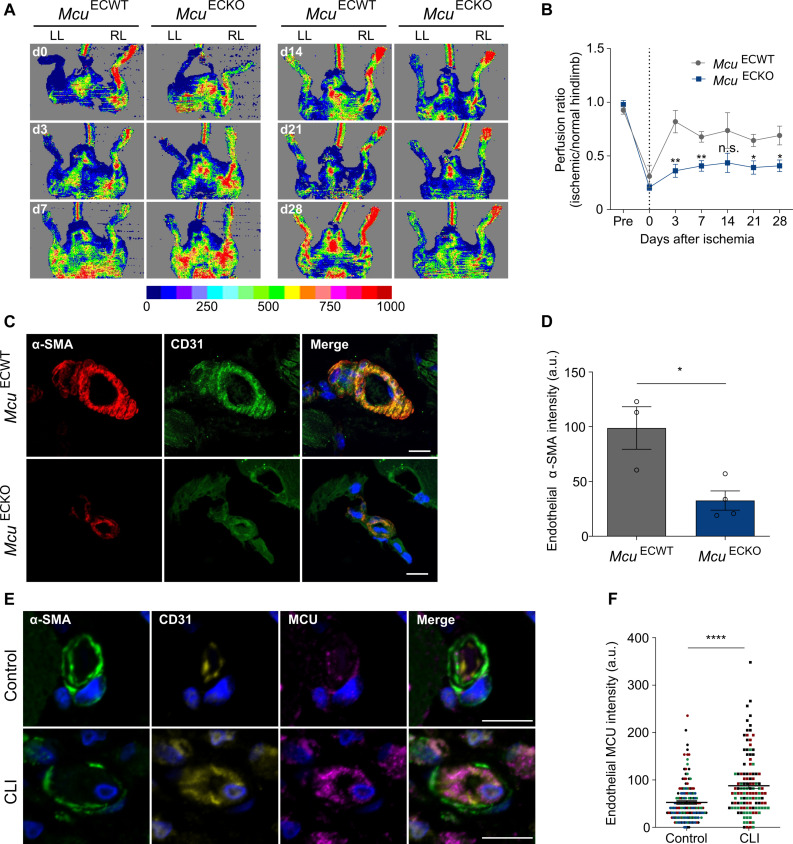
As the trabeculation process progresses, endocardial cells undergoing EndMT populate the cardiac cushions and form the primitive heart valves (51). We analyzed Mcu expression in the specialized atrioventricular structures of endocardial cushions in wild-type (WT) embryonic day 11.5 (E11.5) mouse embryos and found substantial expression along the endocardium and in EndMT-derived cells migrating from the endocardium to the center of the cushion (Fig. 7C). Together, these data indicate that MCU-mediated mito-Ca2+ supports physiological EndMT. Last, to assess whether mito-Ca2+ also regulates EndMT during adulthood under pathological conditions, we intercrossed Mculox/lox mice with the EC-specific Cre driver line Cdh5-Cre to obtain mice lacking Mcu in ECs (McuECKO) and induced hindlimb ischemia in WT and knockout (KO) mice. Neovascularization is crucial for tissue repair after ischemia. Under ischemia, transient EndMT contributes to regenerate the vascular network by supporting EC migration and clonal expansion (11). We thus investigated vessel perfusion as a readout of vessel growth and vascular regeneration. As expected, blood flow decreased precipitously upon femoral artery ligation of the left limb compared to the nonischemic control right limb (Fig. 8A). From day 3 onward, we observed a substantial and progressive improvement of ischemic limb blood flow in both WT (McuECWT) and McuECKO. However, blood flow recovery was delayed in McuECKO mice (Fig. 8B). In agreement with the blood flow measurements, immunohistological staining for CD31 and α-SMA of the ischemic limb at day 28 revealed reduced mesenchymal activation in ECs of McuECKO mice compared to the control group (Fig. 8, C and D). We also assessed MCU expression in the intramuscular arterioles of patients with lower extremity critical limb ischemia (CLI), an environment in which ECs partially transition to mesenchymal cells as evidenced by increased EC expression of N-cadherin, S100A4, and SNAIL1 (15). In line with the EndMT phenotype, the arteriolar ECs in CLI muscle showed loss of the endothelial marker VE-cadherin (fig. S8, A to C) and reduced apical-basal polarity as shown by the redistribution of the apical protein podocalyxin away from the EC apex (fig. S8, D and E). Consistent with our observations in mice, there was also increased MCU expression in the intramuscular arterioles of patients with CLI compared to control individuals (Fig. 8, E and F). Together, these data show that MCU-mediated mito-Ca2+ entry drives (arteriolar) endothelial remodeling associated with mesenchymal transdifferentiation during limb ischemia in the settings of both vessel regeneration and occlusion.
Fig. 8. Endothelial MCU regulates pathological EndMT.
(A) Representative laser Doppler images on left limb (LL) and right limb (RL) after excision of femoral artery in the left limb in McuECWT and McuECKO mice. Imaging colors from blue to red represent increased blood flow, which is defined by the color-coded heatmap. (B) Time course of mean blood flow ratios of ischemic versus normal limb measured before (pre) surgery, immediately after surgery (day 0), and days 3, 7, 14, 21, and 28 (n = 5 to 7). Dotted line indicates the day of surgery. *P < 0.05 and **P < 0.01. (C) Confocal micrographs of blood vessels from ischemic skeletal muscles of McuECWT and McuECKO mice stained for the mesenchymal marker α-SMA (red) and the endothelial marker CD31 (green). Nuclei are counterstained with DAPI (blue). Scale bars, 20 μm. (D) Quantification of α-SMA content in CD31+ vessels shown in images (n = 3 to 4). a.u., arbitrary units. *P < 0.05. (E) Confocal micrographs of small arterioles in human skeletal muscle from individuals with or without chronic limb ischemia (CLI) stained for mural cells (α-SMA; green), ECs (CD31; yellow), and MCU (purple). Nuclei are counterstained with DAPI (blue). Scale bars, 10 μm. CLI arterioles show EC thickening that markedly narrow the vessel lumen. (F) Quantification of endothelial MCU content shown in images. Data are presented as median MCU intensity and interquartile range for 160 and 120 arterioles (n = 4 control individuals and n = 3 individuals with CLI). Colors correspond to each individual. ****P < 0.0001.
DISCUSSION
Despite the growing body of evidence supporting the involvement of EndMT in the pathogenesis of several human diseases (7), our knowledge of the mechanisms regulating this process remains rudimentary. Here, by using different inducers of EC heterogeneity and plasticity, we explored the mechanism of partial transdifferentiation of ECs toward mesenchymal-like cells and revealed original insights in various aspects. First, while the single-cell analysis of three in vitro models of EndMT induction identified an MSC population, we found that EndMT induction leads to global expression changes in all EC subtypes, including the proliferating and tip clusters, rather than effects in the MSC-specific cluster.
To establish solid EndMT signatures, we extended our analysis to all induced and not-induced EndMT conditions across the three models and, thus, provided the evidence that EndMT induction does not result in changes in cell subtype composition. That is, the MSC cluster is not enriched when EndMT is induced. In the context of identifying markers that specifically denote EndMT, this observation holds high relevance.
In addition to the proliferating, tip, and MSC populations, we also identified an intermediate state that lies in between the tip and MSC phenotypes. Notably, a recent computational model predicted that tip cells undergo EndMT by previously transforming into a transitory nontip phenotype (52), consistent with the model suggesting that EndMT occurs through intermediate states (53).
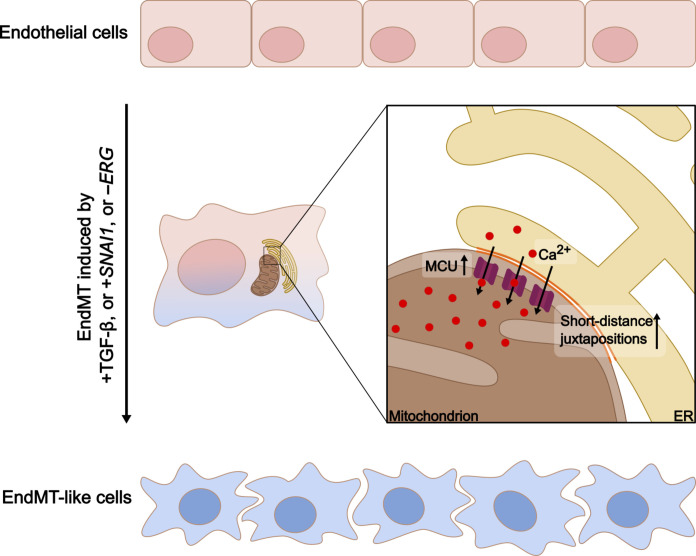
Second, we validated a previously unidentified targetable regulatory mechanism by which mito-Ca2+ controls the transdifferentiation state of ECs (Fig. 9). We observed an up-regulation of pathways associated with ECM remodeling in our meta-analysis consistent with the requested activity of ER to meet the increased cell’s demands during the vascular remodeling associated with EndMT (31, 34). Our analysis revealed an up-regulation of ER genes related to Ca2+ signaling and homeostasis. We focused on this original evidence and excluded the direct contribution of the ER (and associated Ca2+ signaling) to the EndMT-like phenotype. We showed that it is rather the physical interconnection between this organelle and the mitochondria that is important in EndMT. We demonstrated that mito-Ca2+ is a potent regulator of EndMT as shown by several forms of evidence: (i) EndMT is associated with changes in mito-Ca2+ homeostasis. We demonstrated this in the three independent models of induced EndMT, thus excluding any specific effect of the TGF-β profibrotic stimulus on MCU-mediated mito-Ca2+ uptake; (ii) mito-Ca2+ uptake by MCU overexpression regulates EndMT, and (iii) MCU inhibition prevents EndMT. While an excess of Ca2+ taken up by mitochondria leads to cell death (54), under our conditions, we did not observe any toxicity as the mitochondria displayed normal morphology and membrane potential.
Fig. 9. Schematic representation of the proposed signaling in EndMT.
ECs with a characteristic cobblestone morphology undergo mesenchymal activation following induction by TGF-β, SNAI1, or ERG silencing, resulting in EndMT-like cells with altered spindle-like morphology. This transition is associated with closer physical interactions between the mitochondria and ER (orange lines) and is characterized by MCU overexpression, which mediates increased mitochondrial Ca2+ entry. This Ca2+ signaling is crucial for the cellular transformations observed during EndMT.
Third, we demonstrated the possible translational implications of our findings to pathophysiological cardiovascular conditions in vivo. We showed that MCU-mediated mito-Ca2+ supports EndMT during cardiac development and that zebrafish mcu morphants developed abnormal cardiac trabeculae structures. Furthermore, MCU-mediated Ca2+ signaling is crucial for EndMT during vascular remodeling and regeneration of new capillaries after limb ischemia.
CLI is a severe manifestation of peripheral atherosclerotic artery disease characterized by vessel abnormalities that narrow/occlude the lumen of small arterioles leading to microvascular dysfunction (55). Unfortunately, the clinical improvement of the current therapeutic approaches is limited. Partial EndMT has recently been identified as a key remodeling process associated with luminal obstruction in patients with CLI (15). Consistent with our hypothesis that mito-Ca2+ uptake regulates EndMT, we observed higher levels of MCU in the arterioles that have undergone partial EndMT in patients with CLI. This supports the concept of MCU targeting for therapeutic purposes in these patients.
Our findings have broad implications. Mitochondrial Ca2+ homeostasis plays a major role in endothelial function and increased MCU levels have been described in a number of different CVDs, including atherosclerosis and cardiac hypertrophy (56, 57). It can therefore be envisioned that mito-Ca2+ modulation could be therapeutically beneficial under several pathological conditions. Small-molecule inhibitors of MCU have been tested therapeutically in many diseases such as cancer and neurogenerative diseases (58), yet whether MCU could be a putative pharmacological target in vascular-based disorders remains to be explored. Considering the substantial contribution of EndMT to a number of pathological conditions affecting highly vascularized organs, such as the heart, lungs, liver, kidneys, and brain (7), it is tempting to speculate that effective therapeutic intervention could be achieved by organ-specific delivery systems. On the basis of our analysis, we anticipate that other therapeutic targets can also be identified and validated. Notably, some of the congruently up-regulated genes in EndMT induction lack functional annotation.
This study also faces limitations. Our models are not strictly EndMT. Consistently with previous studies reporting an EndMT-like phenotype in cultured ECs (17, 23), we observed a down-regulation of the endothelial markers and an increase in the mesenchymal markers, suggesting that ECs show heterogeneous phenotypes and have partially undergone transition. We expect these EndMT-like cells to undergo complete differentiation to mesenchymal cells in long-term experimental settings; nevertheless, we acknowledge that a full phenotypic switch is challenging to achieve in vitro. We used TGF-β1 as major EndMT activator in HUVECs (5, 17), yet all the TGF-β family members might induce EndMT. We also anticipate that different vascular beds may show different susceptibilities to EndMT-modulating factors in line with EC transcriptional heterogeneity observed across distinct tissues (59, 60).
We assessed EndMT-like cells based on the expression of canonical markers related to different degrees of remodeling; nevertheless, they are imperfect. Readouts to uniformly assess EndMT induction are currently lacking, and our study identifies mito-Ca2+ modification as a hallmark of EndMT. Whether this is context dependent remains an open question.
Loss of MCU has been shown to have effects on endothelial function irrespective of EndMT (61–64). On the basis of our results showing that MCU targeting prevents EndMT induction, we speculate that MCU-driven EC plasticity is an early event occurring at the onset of vascular remodeling and that MCU is required for EndMT induction itself, yet the mechanisms remain be elucidated. Our GSEA showed changes in several metabolic pathways consistent with previous studies pointing out a key role of metabolism in regulating EndMT (37, 65). We speculate that mito-Ca2+ signaling may affect on the epigenome by inducing metabolic changes. This may not be unexpected given the emerging role of mito-Ca2+–dependent metabolic reprogramming in the regulation of transcriptional activation (66, 67).
MATERIALS AND METHODS
Chemicals and reagents
Recombinant human and murine TGF-β1 were purchased from Bio-Techne and R&D Systems, respectively. Tg, tunicamycin, histamine dihydrochloride, cyclopiazonic acid, Evans blue dye, mitomycin C, tamoxifen, and dimethyl sulfoxide were obtained from Sigma-Aldrich. RU360 was obtained from Merck Millipore. Tetramethylrhodamine ethyl (TMRE) was purchased from Biotium, and JC-1 was purchased from Invitrogen.
The following primary antibodies or dyes were used [dilutions for immunoblotting (IB) and immunofluorescence (IF) are given between parentheses]: anti-CD44 (IB, 1:200) (GeneTex), anti-PAI1 (IB, 1:1000) (Abcam), anti-Snai1 (IB, 1:1000) (Cell Signaling Technology), anti-glyceraldehyde-3-phosphate dehydrogenase (IB, 1:2500) (Santa Cruz Biotechnology), anti-ERG (IB, 1:1000) (Santa Cruz Biotechnology), anti–α-SMA (IB, 1:1000; IF, 1:100) (Cell Signaling Technology and Novus Biologicals), anti–α-SMA–Cy3 (IF, 1:250) (Sigma-Aldrich), anti-CD31 (89C2) (IF, 1:3200) (Cell Signaling Technology), anti-CD31 (MEC 13.3) (IF, 1:250) (BD Biosciences), anti-MCU (D2Z3B) (IF, 1:300) (Cell Signaling Technology) or anti-MCU (HPA016480) (IF, 1:100; IB, 1:1000) (Sigma-Aldrich), anti–transferrin receptor (TfR) (IB, 1:1000) (Invitrogen), anti–VE-cadherin (36-1900) (IF, 1:20) (Thermo Fisher Scientific), and anti-podocalyxin (AF1658) (IF, 1:20) (R&D Systems). Secondary horseradish peroxidase–conjugated antibodies were from Cell Signaling Technology (IB, 1:2000) or Bio-Rad (1:10,000); secondary Alexa Fluor–conjugated antibodies (IF, 1:200) were obtained from Jackson ImmunoResearch.
Cell culture
HUVECs were freshly isolated from umbilical cords obtained from multiple healthy donors with approval from the French Ministry of Higher Education, Research, and Innovation and consent of all individuals as previously described (68). HUVECs were maintained in M199 medium (Invitrogen) supplemented with 20% fetal bovine serum (PAN Biotech), 2 mM l-glutamine (Gibco), penicillin (100 IU/ml; Gibco), streptomycin (100 μg/ml; Gibco), and EC growth factor supplements (PromoCell) or in EC basal medium (EGM2) (PromoCell) supplemented with EC growth medium supplement pack (PromoCell). Cells were only used between passages 1 and 4, and all experiments were performed in HUVECs from at least three different donors.
For EndMT induction by TGF-β1, subconfluent HUVECs were cultured in 50% M199:50% EGM2 medium supplemented with TGF-β1 (10 ng/ml) or vehicle [phosphate-buffered saline (PBS)–1% bovine serum albumin] for 5 days (mRNA collection) to 7 days (for scRNA-seq and protein collection). Where indicated, cells were pretreated with RU360 (1 μM for 30 min) or silenced for MCU before EndMT induction. RU360 (2 μM) was maintained in the medium during EndMT induction.
The use of the abbreviation ECs and EndMT-derived cells throughout the experimental sections refers to control and TGF-β1–treated HUVECs, respectively.
Murine liver ECs were isolated from perfused healthy livers of WT and Mcu KO mice as described in (69) and cultured in mouse EC medium supplemented with the growth medium bullet kit (Cell Biologics). Upon adherence, cells were washed and refed with medium containing TGFβ-1 (10 ng/ml) or vehicle. RNA was extracted after 5 days of stimulation as described above.
Inactivation and overexpression strategies
Lentiviral SNAI1 expression construct was obtained by cloning the respective human cDNA into pRRLsinPPT.CMV.MCS MM WPRE vector. Ca2+ fluorescent genetic sensors (GEM-CEPIA1er and GEM-GECO1mito) (Addgene, #58217 and #32461), split GFP–based contact site sensor (SPLICS Mt-ER Short P2A) (Addgene, #164108), and mito-GFP (Addgene, #174180) were cloned into pRRLsinPPT.CMV.MCS MM WPRE vector. Validated MCU-specific TRC clones TRCN0000133861 (used in most of the experiments and indicated as shMCU1) and TRCN0000137529 (indicated as shMCU2) and ERG-specific (TRCN0000013913) shRNAs were used in the pLKO.1 vector (Addgene). Empty shRNA vectors were used as negative controls. All constructs were sequence-verified. Lentiviral particles were produced in human embryonic kidney 293T cells as previously described (70). For transductions, a multiplicity of infection of 10 was used in all the experiments. Cells were transduced overnight and refed with fresh medium the next day. Transduced cells were used in functional assays at least 3 to 4 days after transduction.
Single-cell droplet-based RNA sequencing
Single-cell suspensions of freshly isolated control and treated and transduced HUVECs from two donors were converted to separate barcoded scRNA-seq libraries using the Chromium Single Cell 3′ Library, Gel Bead & Multiplex Kit, and Chip Kit (10x Genomics, v3.1) according to the manufacturer’s instructions, aiming for 6000 cells per library. All samples were processed in parallel in the same thermal cycler. Libraries were sequenced on an MGISEQ-2000.
Mapping
Libraries were mapped to the human genome (build GRCh38) using CellRanger software (10x Genomics, version 5.0.1). Data from the raw unfiltered matrix were further analyzed using the UniApp platform (version 2.0, Unicle Biomedical Data Science, Belgium).
Quality control and data normalization
The following quality control steps were applied: Genes expressed by <3 cells were not considered; cells that had either fewer than 100 (low-quality cells), over 9000 expressed genes (possible doublets), or over 30% of unique molecular identifiers derived from the mitochondrial genome were removed. The data of the remaining cells were natural log–transformed using log1p and normalized using the Seurat package (version 3.2.2).
Data integration
To integrate the samples from different donors, the FindIntegrationAnchors function from the Seurat package was used. Canonical correlation analysis (CCA) was used to identify anchors—pairs of cells (one from each sample)—to align the samples with the following settings: number of dimensions, 30; number of neighbors to use when picking anchors, 20; number of neighbors to use when filtering anchors, 200; number of neighbors to use when scoring anchors, 30; method for nearest neighbor finding, annoy; the maximum number of features to use in nearest-neighbor finding method, 200; number of trees used by the neighbor finding method, 50; error bound on the neighbor finding algorithm, 0; and with L2 normalization on the CCA cell embeddings after dimension reduction enabled. The samples from different donors were integrated using the IntegrateData function in Seurat using the previously identified anchors as input.
In silico high-quality cell selection
After autoscaling, the normalized data were first summarized by principal components analysis (PCA) using the flashPCA package (version 2.1), and the first 30 PCs were visualized using with UMAP using the umap package (version 0.2.10.0) and the following settings: UMAP alpha, 1; number of epochs, 200; distance metric, Pearson; minimum distance, 0.3; number of neighbors, 30.
Graph-based clustering was performed to group cells according to their gene expression profiles as implemented in Seurat. Cell clusters were annotated on the basis of canonical markers. Cells that could not be unambiguously assigned to a biologically meaningful phenotype might represent low quality cells or doublets and were excluded from the analysis.
Feature selection and dimensionality reduction
After in silico selection of high-quality cells, we identified genes with high variability using the Seurat FindVariableGenes function with default parameters. The normalized data were autoscaled, and PCA was performed on variable genes, followed by UMAP to construct a two-dimensional representation of the data. This representation was only used to visualize the data.
Cluster identification
To unbiasedly group cells, we performed PCA on highly variable genes and used graph-based clustering as implemented in the FindClusters function of the Seurat package. Cluster results were visualized UMAP to verify that all visually identified clusters were captured and not underpartitioned. Overpartitioned clusters that represent the same biological phenotype were merged into a single cluster.
Pair-wise differential analysis
Differential expression analysis between two specific clusters was performed using limma as described previously (71).
Marker gene analysis
We used a two-step approach to obtaining ranked marker gene lists for each cluster. As a first criterion, marker genes for a given cluster should have the highest expression in that cluster compared to all other clusters and are therefore uniquely assigned to one cluster. Second, we ranked marker genes using a product-based meta-analysis. Briefly, we performed pair-wise differential analysis of all clusters against all other clusters separately and ranked the results of each pair-wise comparison by log2 fold change. The most up-regulated genes received the lowest rank number (top ranking marker genes), and the most down-regulated genes received the highest rank number. For each cluster, we combined the rank numbers for all genes in all pair-wise comparisons by calculating their product to obtain a final list of ranked marker genes for each cluster.
Cluster annotation
We annotated clusters based on literature-curated marker genes of canonical marker genes (table S1). In case of an entirely unknown phenotype or new sublineages of a canonical phenotype, we searched through the top 50 ranking list of markers for a coherent set of genes involved in similar biological processes.
Heatmap analysis
All heatmaps are based on cluster-averaged gene expression to account for cell-to-cell transcriptomic stochastics. Data were autoscaled for visualization.
Set intersection analysis
The intersects of congruently up-regulated genes were visualized using UpSet plots from the UpSetR package (version 1.4.0).
Jaccard similarity PCA
Jaccard similarity analysis was calculated using a custom R script.
Product-based meta-analysis
To calculate the cell state–independent EndMT signatures, we used a modification of the previously described rank-based meta-analysis, which penalizes genes that are not consistently up-regulated across all comparisons. In this case, the most up-regulated genes received the highest rank number (top ranking up-regulated genes), and the most down-regulated genes received the lowest rank number. We combined the rank numbers for all genes in all comparisons by calculating their product to obtain a final list of ranked genes in which the gene with the largest product was ranked at rank one.
Gene set enrichment analysis
We used GSEA as implemented in the clusterProfiler package to compare gene expression signatures between groups. Gene set analysis was performed using a set of vascular-related gene sets selected from the Molecular Signatures Database (MSigDB version 7.41 downloaded from http://bioinf.wehi.edu.au/software/MSigDB/), a collection of expert annotated gene sets. GSEA scores were calculated for sets with a minimum of five detected genes, and all other parameters were default.
GO analysis
We used the GOrilla web tool with default settings to perform the GO analysis (72).
RNA isolation and gene expression analysis
Total RNA was extracted with PureLink RNA mini kit (Invitrogen) according to the manufacturer’s instructions; quality and quantity were measured on a Nanodrop 1000 (Thermo Fisher Scientific). cDNA synthesis was performed with the iScript cDNA synthesis kit (Bio-Rad). Quantitative polymerase chain reaction (PCR) analyses were performed on the CFX Opus 96 (Bio-Rad) or QuantStudio 6 Flex (Thermo Fisher Scientific) Real-Time PCR System devices with human or mouse in-house–designed primer sets (Eurogentec) for which sequences are available upon request. 18S ribosomal RNA was used as housekeeping gene. For comparison of gene expression between conditions, expression (normalized to 18S ribosomal RNA endogenous control) is indicated relative to control condition.
Western blotting
Proteins were extracted in radioimmunoprecipitation assay buffer [25 mM tris-HCl (pH 7.6), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS] containing protease and phosphatase inhibitor mixes (Roche Applied Science). Protein concentration was determined using a Pierce BCA protein assay kit (Thermo Fisher Scientific).
Protein lysates were separated by SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and detected with specific primary and horseradish peroxidase–conjugated secondary antibodies in combination with Pierce enhanced chemiluminescence Western blotting substrate (Thermo Fisher Scientific). Signal was acquired with the iBright CL 1500 Imaging System (Thermo Fisher Scientific), and densitometric quantification was carried out with ImageJ (version 2.0.0) (73). Protein expression was normalized to the corresponding expression of the indicated control protein (glyceraldehyde-3-phosphate dehydrogenase or TfR was used as a loading control).
Transmission electron microscopy
We performed transmission electron microscopy on a JEOL JEM1400 (JEOL Europe BV, Zaventem, Belgium) (VIB Bio Imaging Core, Leuven Platform). For transmission electron microscopy, observations samples were fixed for 24 hours with 2.5% glutaraldehyde at pH 7.3 and buffered with 0.05 M sodium cacodylate. Before embedding in Agar 100 resin (Agar Scientific, Stansted, UK), the material was postfixed in 1% OsO4 and 1.5% potassium ferrocyanide [buffered with 0.05 M sodium cacodylate (pH 7.3)] and dehydrated in a graded ethanol series. Semithin (±1 mm) sections were cut with a Reichert Jung Ultracut E microtome and stained with 0.1% thionin–0.1% methylene blue. The ultrathin (±70 nm) sections, on copper grids, were stained with uranyl acetate and lead citrate.
In vitro assays
Transendothelial electrical resistance
Fifty thousand HUVECs were seeded on 6.5-mm 0.1% gelatin-coated polyester transwells and 0.4-μm pore size (Costar, Sigma-Aldrich). The electrical resistance was measured with an STX2-Plus electrode (World Precision Instruments) connected to an EVOM3 voltammeter (World Precision Instruments). Gelatin-coated wells without cells were used to measure the intrinsic electrical resistance of the inserts for background subtraction. Measurements were performed 4 days after transduction.
Scratch wound
One hundred and fifty thousand HUVECs were seeded in a 24-well format and were allowed to reach confluency over the next day. At time T0, the confluent monolayer was scratched with a 200-μl pipette tip and photographed. The cells were further incubated for 6 hours and photographed again at time point T6. The gap area at T0 minus the gap area at T6 was measured with ImageJ (version 2.0.0) and expressed as percentage of wound closure. Per well, two nonoverlapping regions along the scratch were analyzed.
Imaging and analysis
We performed confocal imaging with Zeiss LSM700 (Plan Apochromat 40×/1.3 oil DIC M27) or LSM880 AiryScan (Plan Neofluar 40×/1.3 oil DIC M27; Plan Apochromat 63×/1.4 oil DIC M27) confocal workstations. We analyzed mitochondrial membrane potential using TMRE (10 nM for 45 min at 37°C and 5% CO2) or JC-1 dyes (5 μM for 10 min at 37°C and 5% CO2) on living cells. TMRE imaging was done at 561-nm excitation and emission detection at 575 nm. JC-1 imaging was done at 488-nm excitation and emission detection at 520 to 550 nm (shown in green) and 579 to 633 nm (shown in red). Quantitative analyses were obtained by normalizing TMRE to the mitochondrial content of cells transduced with a mito-GFP reporter. Mitochondrial network morphology was assessed on mito-GFP–positive cells by the ImageJ tool MiNa (74) and manual counting (75).
Mitochondrial-ER contact sites were measured by the SPLICS Short-P2AMt-ER (SPLICSs) reporter detecting interactions occurring over a range of ≈8 to 10 nm (76). HUVECs expressing the reporter were imaged on a confocal system, and z-stack acquired for the whole-cell depth was processed and analyzed as previously described (76). TMRE dye was used to visualize the mitochondria.
Ca2+ imaging
ER- and mito-Ca2+ signals were monitored by using ratiometric GEM-CEPIA1er and GEM-GECO1mito sensors targeting the ER and mitochondria, respectively. Briefly, HUVECs expressing the genetic sensors were seeded on glass-bottom dishes (35 mm) and maintained at 37°C and 5% CO2 in the culture medium. Prior the experiment, cells were washed three times with a standard extracellular solution containing 140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 5 mM glucose, 0.4 mM KH2PO4−, 4 mM NaHCO3, and 10 mM Hepes (pH 7.4 adjusted with NaOH) with or without CaCl2 (referred as to 0 and 2 mM Ca2+). Solution (0 mM) also contained 0.5 mM EGTA. A blue diode laser (405 nm) was used for excitation, whereas emission was collected at the two channels: 420 nm (blue) and 530 nm (green) in line-by-line mode of the multitrack configuration of the confocal scanner. GEM-CEPIA1(462/510 nm) and GEM-GECO1(455/511 nm) ratios were recorded and analyzed to study the dynamics of the ER- and mito-Ca2+, respectively. More specifically, a decrease in the GEM-CEPIA1 ratio (462/510 nm) indicates a depletion of ER-Ca2+ store, whereas an increase in the GEM-GECO1ratio (455/511 nm) reveals mito-Ca2+ uptake. Images were acquired at 4-s intervals using a 40× objective. Tg (2 μM), cyclopiazonic acid (20 μM), or histamine (50 μM) was applied acutely during acquisition, while the cells were kept in the same standard extracellular solution described above. Image processing was carried out using ImageJ (version 2.0.0). pClamp 11.1 software (Molecular Devices Corp.) was used to further analyze fluorescence traces and to calculate peak amplitudes (Δ ratio) and slopes of Ca2+ signals upon stimulation.
Immunofluorescence
Cells/tissues
For CD31, we performed IF staining on 4% (v/v) of paraformaldehyde fixed HUVECs permeabilized for 10 min in PBS with 0.5% Triton X-100 and 0.01% Tween 20. CD31 signal was amplified by tyramide signal amplification system (Akoya Biosciences). Nuclei were visualized by 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories). We performed imaging using a 40× objective, a z-stack range between 6 and 11 μm, and a step size of 1 μm. Identical exposure and dynamic range conditions were used for all scans.
For podocalyxin and VE-cadherin
IF staining on 5-μm-thick serial paraffin sections of human skeletal muscle was performed as described with minor modifications (15, 77, 78). Briefly, sections were subjected to antigen retrieval [10 mM sodium citrate (pH 6.0), pressure-heated for 30 min] and multiplex-stained for (i) podocalyxin [detected with Alexa Fluor 488–conjugated donkey anti-goat immunoglobulin G (IgG), 1:100], (ii) VE-cadherin (detected with biotin-SP donkey anti-rabbit IgG, 1:100, and DyLight 549–conjugated streptavidin, 1:200), and (iii) mural cells (anti–α-SMA, Agilent, 1:200, detected with Alexa Fluor 647–conjugated donkey anti-mouse IgG, 1:100).
For mitochondrial calcium uniporter
paraffin sections of human skeletal muscle were subjected to antigen retrieval as for podocalyxin and VE-cadherin and multiplex stained for (i) ECs (anti-CD31), (ii) mural cells (anti–α-SMA), and (iii) MCU (anti-MCU detected with biotin-SP donkey anti-rabbit IgG, 1:100, and DyLight 549–conjugated streptavidin, 1:200). Nuclei were visualized with ProLong Glass Antifade Mountant containing NucBlue Stain (Thermo Fisher Scientific). Sections were imaged with a Nikon A1R Confocal Laser Scanning System (Nikon) using a 60× oil immersion objective and 405-, 488-, 561-, and 640-nm lasers. The z-step size was 0.150 μm and an x-y resolution of 140 nm per pixel for VE-cadherin and podocalyxin-stained sections and 0.275 μm and 70 nm per pixel for MCU-stained sections, both attained using a Galvano scanner. Maximum intensity projections were generated using NIS Elements software (Nikon). All human tissue samples and patient information were collected with written, informed consent from patients using protocols approved by the Western University Review Board for Health Sciences Research Involving Human Subjects and in accordance with the 1964 Declaration of Helsinki.
To quantify podocalyxin distribution, VE-cadherin content, and MCU content in arteriolar ECs, skeletal muscle sections were imaged and digitally scanned with a Leica Aperio VERSA Fluorescence Pathology Scanner, using a 40× objective, a z-stack range of 2 μm, and a step size of 1 μm. Identical exposure and dynamic range conditions were used for all slides with the same stains. Podocalyxin distribution was scrutinized in 20 to 37 randomly selected arterioles with distinguishable lumens per individual. An arteriole was identified as having nonpolarized ECs if fewer than half of the ECs had apically enriched podocalyxin. VE-cadherin content was quantified in 35 randomly selected arterioles per individual, assessing the integrated intensity of VE-cadherin–positive pixels using the Aperio Area Quantification FL Algorithm. Endothelial MCU content was quantified in 40 randomly selected arterioles (diameter, 10 to 20 μm) per individual based on the mean signal intensity of MCU-positive pixels within the CD31-positive area and using the Aperio Area Quantification FL Algorithm.
For CD31 and α-SMA,
IF staining was performed on 5-μm-thick serial paraffin sections of mouse skeletal muscle. We performed imaging using a 63× objective, a z-stack range between 5 to 8 μm, and a step size of 1 μm. Nuclei were visualized by DAPI (Vector Laboratories). Identical exposure and dynamic range conditions were used for all scans. α-SMA content was quantified in randomly selected vessels based on the mean signal intensity of α-SMA–positive pixels within the CD31-positive area by ImageJ (version 2.0.0).
Mouse embryos
Embryos were dated taking E0.5 as the day after the vaginal plug. Freshly dissected embryos (E11.5) were fixed in 1× PBS–4% paraformaldehyde for 1 hour. We performed immunostainings on frozen sections (10 μm). Slides were permeabilized for 15 min in 1× PBS–0.1% Triton X-100 and incubated in 1× PBS–5% horse serum for 45 min. Primary antibodies anti-CD31 (MEC 13.3) (1:250), anti-α-SMA-Cy3 (1:250), and anti-MCU (D2Z3B) (1:300) were incubated overnight at 4°C. Alexa Fluor secondary antibodies were incubated 45 min at room temperature, and slides were mounted in Dako Mounting Medium. Images were acquired at ×20 magnification of the whole section using the Axio Scan Z1 (Zeiss) and edited in ImageJ (version 2.0.0).
In vivo models
Animal housing and all experimental procedures were approved by and performed according to the National Institutional Animal Ethics Committees and institutional guidelines (APAFIS #17080-201800515357731 v13 and APAFIS #7160-2017040313471173 v4, Hauts-de-France; #21055, National Cerebral and Cardiovascular Center).
Transgenic mouse models
Mice from the EC-specific inducible Cre-driver line Pdgfb-CreERT2 (79) were crossed with Snai1flox/flox mice (80) to obtain mice with EC-specific deletion of the Snai1 gene upon tamoxifen treatment to induce Cre-mediated gene excision (Snai1ECKO mice). Correct Cre-mediated excision of the loxP flanked Snai1 segment in tamoxifen-treated Snai1ECKO mice was confirmed by PCR analysis of genomic DNA (fig. S7M). Tamoxifen treatment was done using 8-week-old mice by gavage (0.114 mg/g of body weight per day for five consecutive days and an additional day after a 2-day treatment-free interval) 1 week before the experiment. Endothelial-specific Mcu-deficient mice (McuECKO) were obtained by crossing the EC-specific Cre-driver line Cdh5-Cre (The Jackson Laboratory) with previously generated Mcuflox/flox mice (64). Correct recombination of the excised Mcu allele was confirmed as previously described (61). Mcu KO mice were previously generated from Finkel and colleagues (81).
Generation of zebrafish (Danio rerio) transgenic lines
Embryos were raised in E3 medium and incubated at 28.5°C. Developmental staging was referred to chronological and morphological criteria (82). AB strain was used as WT. The following zebrafish transgenic lines were used for experiments: Tg(fli1a:gal4ff) (83), Tg(fli1a:lifeact-mCherry) (46), Tg(myl7:nls-mCherry) (49), and TgBAC(twist1b:GFP) (48). Tg(uas:mito-GCaMP7a) was generated as described below.
To monitor mito-Ca2+ levels, we established a transgenic reporter zebrafish line expressing GCaMP7a (84) tagged to mito7, a mitochondrial matrix target sequence, under the control of Gal4-UAS system;Tg(UAS:mito-GCaMP7a). We used a DsRed2-Mito-7 vector (Addgene, #55838) and pTol2-5xUAS:GCaMP7a vector as a template.
Mito7(atgtccgtcctgacgccgctgctgctgcggggcttgacaggctcggcccggcggctcccagtgccgcgcgccaagatccattcgttg) was amplified by the following primers: Mito7, 5′-AAAAAAAAAATCAAAatgtccgtcctgacgccgct-3′ (forward) and 5′-GTGGCTGCCGCGCATcaacgaatggatcttggc-3′ (reverse). Subsequently, Mito7 oligo was inserted upstream of GCaMP7a site of pTol2-5xUAS:GCaMP7a vector by inverse PCR using the following primers: GCaMP7a-inverse, 5′-ATGCGCGGCAGCCACcaccaccacc-3′ (forward) and 5′-TTTGATTTTTTTTTTaagttggtaccctcgag-3′ (reverse) to establish the pTol2-5xUAS-mito-GCaMP7a plasmid. Tol2-mediated zebrafish transgenesis was performed by injecting 35 pg of transgene plasmid together with 50 pg of tol2 transposase mRNA, followed by subsequent screening of F1 founders and establishment of stable transgenic strains through selection in F3 generations. The following morpholino oligonucleotides (MOs; Gene Tools) were used (1.2 nl): 0.6 mM control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) and 0.6 mM mcu splice-MO (5′-CATCAAGAGTAAAGCACTGACCTGG-3′). MOs, mRNAs, and Tol2 plasmids were injected into blastomeres using IM300 Microinjector (Narishige) at one- to two-cell stage.
To obtain images of embryos, a 0.2 mM solution of 1-phenyl-2-thiourea (Sigma-Aldrich) was added in breeding E3 medium to suppress pigmentation, and MS-222 (0.1 mg/ml) was added to anesthetize the embryos during embedding. Mito-Ca2+ was visualized by the excitation of the genetically encoded Ca2+ indicator GCaMP7a-tagged to mitochondrial matrix targeting sequence (mito-GCaMP7a) using a 488-nm wavelength laser line. Excitation of EGFP and mCherry was done using 488- and 561-nm wavelength laser lines, respectively. Confocal images of 1.5-μm steps were taken with an upright FV3000 confocal microscope system (Evident) equipped with a 20× water immersion objective (Evident XLUMPlan FL_N, 1.0 numerical aperture). Dechorionated embryos were mounted in 1% low-melting agarose (Invitrogen–Thermo Fisher Scientific) dissolved in E3 medium. Images were processed with ImageJ (version 2.0.0).
Hind limb ischemia and laser Doppler perfusion imaging
We performed hind limb ischemia in 10- to 12-week-old WT and McuECKO mice as previously described (85). Briefly, the left femoral artery was ligated proximally to the deep femoral artery and distal to saphenous artery, and the femoral artery was completely removed between the two ligatures. We measured blood flow rate in each mouse’s hind limbs using a laser Doppler perfusion imager (model LDI2-IR, Moor Instruments, Wilmington, DE) at preligation and immediately afterward (day 0) and at days 3, 7, 14, 21, and 28 after ligation. An average of three measurements per animal was expressed as a ratio of perfusion in the left (ischemic) versus right (nonischemic) hind limb.
Permeability assay
Histamine-induced extravasation was obtained by intravenous injection of 300 μg of histamine in 200 μl of Evans blue (2% in PBS). Animals were euthanized 45 min later, and dye was extracted as previously described (86). Mouse ears were topically treated 24 hours before the assay with 25 μl of 100 μM RU360 in acetone/olive oil (4:1, v/v), whereas the corresponding control mice received an equivalent volume of vehicle (dimethyl sulfoxide).
Statistical analysis
Data represent means ± SEM of pooled donors or animals used in independent experiments unless otherwise stated. n values represent the number of individual donors or animals unless otherwise stated. Individual data points represent the biological replicates involving at least three technical replicates within an experiment. Statistical significance was calculated by standard two-sided Student’s t test with F testing to confirm equality of variance (Prism v10); in case of unequal variance, two-sided t test with Welch’s correction was used (Prism v10). Calcium traces were analyzed by Wilcoxon test. P < 0.05 was considered significant. No significance is indicated as n.s.
Acknowledgments
We thank W. Masselin (U1011, Institut Pasteur de Lille, France), L. Schoonjans, S. Wyns, E. Gils, P. Vanwesemael, A. Carton, and A. Manderveld (VIB-KU Leuven, Belgium); K. Kondratska, E. Desruelles, and L. Allart (U1003-PHYCEL, Université de Lille, France) for technical assistance; BioImaging Center Lille-Nord de France (BICeL) and Animal Experimentation and High Technology Platform (PLEHTA) at the Institut Pasteur de Lille for access to the equipment and facilities; BiLille platform (Université de Lille, France) for the bioinformatics support; V. Farfariello (U1003-PHYCEL, Université de Lille, France), K. De Bock (ETH Zürich, Switzerland), I. Tancevski (Medical University of Innsbruck, Austria), and F. Trottein (Center for Infection and Immunity of Lille INSERM U1019–CNRS UMR9017, France) for sharing plasmids and mouse tissues; T. Cali’ (University of Padua, Italy) for providing the SPLICS sensors; C. Vergara (UC Berkeley, CA, USA) for mouse organ perfusion and extraction; P. Lishko (UC Berkeley, CA, USA) and Y. Kirichok (UCSF, CA, USA) for providing Mcu KO mouse tissues. We also acknowledge the expert advice of P. Carmeliet (VIB-KU Leuven, Belgium), F. Soncin (CNRS IRL2820 LiMMS/IIS, France), M. Trebak (The Pennsylvania State University College of Medicine, PA, USA), and E. Meta (Unicle Biomedical Data Science, Belgium). 10x Genomics and sequencing were performed at VIB Genomics core facility (Leuven, Belgium) and BGI Genomics (Copenhagen, Denmark), respectively. The Pdgfb-CreERT2 mice were obtained from P. Carmeliet, while the Snai1flox/flox mice were a gift from A. García de Herreros (IMIM, Spain).
Funding: This work was supported by ITMO Cancer of Aviesan within the framework of the 2021-2030 Cancer Control Strategy on funds administered by Inserm, Contrat de Plan Etat-Région CPER Cancer 2015-2020, La Ligue Contre le Cancer (Pas de Calais), Agence Nationale de la Recherche Investissements d’avenir (ANR-16-IDEX-004-ULNE) to A.R.C.. M.L. and M.C. were supported by ITMO Cancer of Aviesan. E.C. is supported by the Institute Pasteur de Lille/ Région Hauts-de-France, L.B. and A.M. by the Université de Lille. G.C. is supported by the University of Torino. C.D. was a recipient of Contrat de Plan Etat-Région CPER Cancer 2015-2020 fellowship. J.G.P. was funded by Canadian Institutes of Health Research FDN-143326 and PJT-180604. M.M. received funding from the National Institutes of Health (R35GM145294, R01GM109882, R01HL142673, and R01DK135179), DOD/DHP-CDMRP PR181598P-1, and San Antonio Partnership for Precision Therapeutics (SAPPT). F.P. and H.C.D.D.l.H. were supported by the Munich Center for Systems Neurology (SyNergy EXC 2145; project ID 390857198) and the ExNet-0041-Phase2-3 (“SyNergy-HMGU”) through the Initiative and Network Fund of the Helmholtz Association. D.G. was a recipient of the Institut Universitaire de France (IUF), France Berkley Fund, Cancéropôle Nord-Ouest–AAP Projets Emergents 2023, and La Ligue Contre le Cancer, Septentrion.
Author contributions: Conceptualization: A.R.C. Methodology: A.R.C., J.G., E.Z., J.-L.D., Y.L., L.Li., L.Le., J.G.P., R.K., M.M., D.D., and F.P. Investigation: M.L., G.C., E.C., L.B., A.M., A.B., J.V.K., C.R., F.S.A., M.V., A.E., L.T., V.Ga., L.T., H.C.D.D.l.H., A.M-L., L.L’h., M.C., C.D., V.Go., V.N.S.G., D.T., H.Y., H.F., S.V., A.S., L.-C.C., F.I., and A.R.C. Visualization: A.R.C., M.L., G.C., E.C., L.B., J.G., A.B., J.V.K., C.R., L.T., H.C.D.D.l.H., A.M., M.C., C.D., V.Go., V.N.S.G., D.T., H.Y., H.F., S.V., A.S., L.-C.C., J.G.P., R.K., M.M., F.P., A.F.P., and D.G. Supervision: A.R.C., J.G., J.G.P., M.M., D.D., F.P., A.F.P., and D.G. Writing—original draft: A.R.C. Writing—review and editing: A.R.C., J.G., D.D., B.S., A.F.P., D.G.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The accession number for all raw sequencing data are available in GEO under accession number: GSE237895.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Legends for tables S1 to S7
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S7
REFERENCES AND NOTES
- 1.Dejana E., Hirschi K. K., Simons M., The molecular basis of endothelial cell plasticity. Nat. Commun. 8, 14361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez-Duffhues G., Orlova V., Ten Dijke P., In brief: Endothelial-to-mesenchymal transition. J. Pathol. 238, 378–380 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Anderberg C., Cunha S. I., Zhai Z., Cortez E., Pardali E., Johnson J. R., Franco M., Paez-Ribes M., Cordiner R., Fuxe J., Johansson B. R., Goumans M. J., Casanovas O., ten Dijke P., Arthur H. M., Pietras K., Deficiency for endoglin in tumor vsculature weakens the endothelial barrier to metastatic dissemination. J. Exp. Med. 210, 563–579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lovisa S., Fletcher-Sananikone E., Sugimoto H., Hensel J., Lahiri S., Hertig A., Taduri G., Lawson E., Dewar R., Revuelta I., Kato N., Wu C. J., Bassett R. L. Jr., Putluri N., Zeisberg M., Zeisberg E. M., LeBleu V. S., Kalluri R., Endothelial-to-mesenchymal transition compromises vascular integrity to induce Myc-mediated metabolic reprogramming in kidney fibrosis. Sci. Signal. 13, eaaz2597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arciniegas E., Sutton A. B., Allen T. D., Schor A. M., Transforming growth factor beta 1 promotes the differentiation of endothelial cells into smooth muscle-like cells in vitro. J. Cell Sci. 103, 521–529 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Pardali E., Sanchez-Duffhues G., Gomez-Puerto M. C., ten Dijke P., TGF-β-induced endothelial-mesenchymal transition in fibrotic diseases. Int. J. Mol. Sci. 18, 2157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piera-Velazquez S., Jimenez S. A., Endothelial to mesenchymal transition: Role in physiology and in the pathogenesis of human diseases. Physiol. Rev. 99, 1281–1324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberg L. M., Markwald R. R., Molecular regulation of atrioventricular valvuloseptal morphogenesis. Circ. Res. 77, 1–6 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Wirrig E. E., Yutzey K. E., Conserved transcriptional regulatory mechanisms in aortic valve development and disease. Arterioscler. Thromb. Vasc. Biol. 34, 737–741 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Gise A., Pu W. T., Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ. Res. 110, 1628–1645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manavski Y., Lucas T., Glaser S. F., Dorsheimer L., Gunther S., Braun T., Rieger M. A., Zeiher A. M., Boon R. A., Dimmeler S., Clonal expansion of endothelial cells contributes to ischemia-induced neovascularization. Circ. Res. 122, 670–677 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Zeisberg E. M., Tarnavski O., Zeisberg M., Dorfman A. L., McMullen J. R., Gustafsson E., Chandraker A., Yuan X., Pu W. T., Roberts A. B., Neilson E. G., Sayegh M. H., Izumo S., Kalluri R., Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 13, 952–961 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Evrard S. M., Lecce L., Michelis K. C., Nomura-Kitabayashi A., Pandey G., Purushothaman K. R., d’Escamard V., Li J. R., Hadri L., Fujitani K., Moreno P. R., Benard L., Rimmele P., Cohain A., Mecham B., Randolph G. J., Nabel E. G., Hajjar R., Fuster V., Boehm M., Kovacic J. C., Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. Nat. Commun. 7, 11853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Good R. B., Gilbane A. J., Trinder S. L., Denton C. P., Coghlan G., Abraham D. J., Holmes A. M., Endothelial to mesenchymal transition contributes to endothelial dysfunction in pulmonary arterial hypertension. Am. J. Pathol. 185, 1850–1858 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Chevalier J., Yin H., Arpino J. M., O'Neil C., Nong Z., Gilmore K. J., Lee J. J., Prescott E., Hewak M., Rice C. L., Dubois L., Power A. H., Hamilton D. W., Pickering J. G., Obstruction of small arterioles in patients with critical limb ischemia due to partial endothelial-to-mesenchymal transition. iScience. 23, 101251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dufton N. P., Peghaire C. R., Osuna-Almagro L., Raimondi C., Kalna V., Chauhan A., Webb G., Yang Y., Birdsey G. M., Lalor P., Mason J. C., Adams D. H., Randi A. M., Dynamic regulation of canonical TGFβ signalling by endothelial transcription factor ERG protects from liver fibrogenesis. Nat. Commun. 8, 895 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooley B. C., Nevado J., Mellad J., Yang D., St Hilaire C., Negro A., Fang F., Chen G., San H., Walts A. D., Schwartzbeck R. L., Taylor B., Lanzer J. D., Wragg A., Elagha A., Beltran L. E., Berry C., Feil R., Virmani R., Ladich E., Kovacic J. C., Boehm M., TGF-β signaling mediates endothelial-to-mesenchymal transition (EndMT) during vein graft remodeling. Sci. Transl. Med. 6, 227ra234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasparics A., Rosivall L., Krizbai I. A., Sebe A., When the endothelium scores an own goal: Endothelial cells actively augment metastatic extravasation through endothelial-mesenchymal transition. Am. J. Physiol. Heart Circ. Physiol. 310, H1055–H1063 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Goveia J., Rohlenova K., Taverna F., Treps L., Conradi L. C., Pircher A., Geldhof V., de Rooij L., Kalucka J., Sokol L., García-Caballero M., Zheng Y., Qian J., Teuwen L. A., Khan S., Boeckx B., Wauters E., Decaluwé H., de Leyn P., Vansteenkiste J., Weynand B., Sagaert X., Verbeken E., Wolthuis A., Topal B., Everaerts W., Bohnenberger H., Emmert A., Panovska D., de Smet F., Staal F. J. T., Mclaughlin R. J., Impens F., Lagani V., Vinckier S., Mazzone M., Schoonjans L., Dewerchin M., Eelen G., Karakach T. K., Yang H., Wang J., Bolund L., Lin L., Thienpont B., Li X., Lambrechts D., Luo Y., Carmeliet P., An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell 37, 21–36.e13 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Gerhardt H., Golding M., Fruttiger M., Ruhrberg C., Lundkvist A., Abramsson A., Jeltsch M., Mitchell C., Alitalo K., Shima D., Betsholtz C., VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell Biol. 161, 1163–1177 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan X., Wang Y., Fan J., Chen R., Deletion of SMURF 1 represses ovarian cancer invasion and EMT by modulating the DAB2IP/AKT/Skp2 feedback loop. J. Cell. Biochem. 120, 10643–10651 (2019). [DOI] [PubMed] [Google Scholar]
- 22.He Z., Lin J., Chen C., Chen Y., Yang S., Cai X., He Y., Liu S., Identification of BGN and THBS2 as metastasis-specific biomarkers and poor survival key regulators in human colon cancer by integrated analysis. Clin. Transl. Med. 12, e973 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tombor L. S., John D., Glaser S. F., Luxan G., Forte E., Furtado M., Rosenthal N., Baumgarten N., Schulz M. H., Wittig J., Rogg E. M., Manavski Y., Fischer A., Muhly-Reinholz M., Klee K., Looso M., Selignow C., Acker T., Bibli S. I., Fleming I., Patrick R., Harvey R. P., Abplanalp W. T., Dimmeler S., Single cell sequencing reveals endothelial plasticity with transient mesenchymal activation after myocardial infarction. Nat. Commun. 12, 681 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Xu X., Jiang Y., Hansbro N. G., Hansbro P. M., Xu J., Liu G., Elastin is a key factor of tumor development in colorectal cancer. BMC Cancer 20, 217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Y., Wang X., Zhang L., Liu W., Ning J., Gu R., Cui Y., Cai L., Xing Y., A novel epithelial-mesenchymal transition (EMT)-related gene signature of predictive value for the survival outcomes in lung adenocarcinoma. Front. Oncol. 12, 974614 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong Z., Guo M., Yu Y., Zhang F. F., Ge M. K., Chen G. Q., Shen S. M., Downregulation of AIF by HIF-1 contributes to hypoxia-induced epithelial-mesenchymal transition of colon cancer. Carcinogenesis 37, 1079–1088 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Kang A. R., An H. T., Ko J., Kang S., Ataxin-1 regulates epithelial-mesenchymal transition of cervical cancer cells. Oncotarget 8, 18248–18259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stankic M., Pavlovic S., Chin Y., Brogi E., Padua D., Norton L., Massague J., Benezra R., TGF-β-Id1 signaling opposes Twist1 and promotes metastatic colonization via a mesenchymal-to-epithelial transition. Cell Rep. 5, 1228–1242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M., Qi Y., Zhao L., Chen D., Zhou Y., Zhou H., Lv Y., Zhang L., Jin S., Li S., Zou H., Jia W., Wang C., Jiang J., Liang W., Pang L., Li F., Matrix metalloproteinase-14 induces epithelial-to-mesenchymal transition in synovial sarcoma. Hum. Pathol. 80, 201–209 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Zhao Z., Lu P., Zhang H., Xu H., Gao N., Li M., Liu C., Nestin positively regulates the Wnt/β-catenin pathway and the proliferation, survival and invasiveness of breast cancer stem cells. Breast Cancer Res. 16, 408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piera-Velazquez S., Li Z., Jimenez S. A., Role of endothelial-mesenchymal transition (EndoMT) in the pathogenesis of fibrotic disorders. Am. J. Pathol. 179, 1074–1080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J., Tian X. J., Xing J., Signal transduction pathways of EMT induced by TGF-β, SHH, and WNT and their crosstalks. J. Clin. Med. 5, 41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim R. K., Suh Y., Yoo K. C., Cui Y. H., Kim H., Kim M. J., Gyu Kim I., Lee S. J., Activation of KRAS promotes the mesenchymal features of basal-type breast cancer. Exp. Mol. Med. 47, e137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trogisch F. A., Abouissa A., Keles M., Birke A., Fuhrmann M., Dittrich G. M., Weinzierl N., Wink E., Cordero J., Elsherbiny A., Martin-Garrido A., Grein S., Hemanna S., Hofmann E., Nicin L., Bibli S. I., Airik R., Kispert A., Kist R., Quanchao S., Kurschner S. W., Winkler M., Gretz N., Mogler C., Korff T., Koch P. S., Dimmeler S., Dobreva G., Heineke J., Endothelial cells drive organ fibrosis in mice by inducing expression of the transcription factor SOX9. Sci. Transl. Med. 16, eabq4581 (2024). [DOI] [PubMed] [Google Scholar]
- 35.Lovisa S., LeBleu V. S., Tampe B., Sugimoto H., Vadnagara K., Carstens J. L., Wu C. C., Hagos Y., Burckhardt B. C., Pentcheva-Hoang T., Nischal H., Allison J. P., Zeisberg M., Kalluri R., Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat. Med. 21, 998–1009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bronson R., Lyu J., Xiong J., Transcriptome analysis reveals molecular signature and cell-type difference of Homo sapiens endothelial-to-mesenchymal transition. G3 (Bethesda) 13, jkad243 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong J., Kawagishi H., Yan Y., Liu J., Wells Q. S., Edmunds L. R., Fergusson M. M., Yu Z. X., Rovira E. L. II, Brittain M. J., Wolfgang M. J., Jurczak J. P., Fessel T. F., Finkel T., A metabolic basis for endothelial-to-mesenchymal transition. Mol. Cell 69, 689–698.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raffaello A., Mammucari C., Gherardi G., Rizzuto R., Calcium at the center of cell signaling: Interplay between endoplasmic reticulum, mitochondria, and lysosomes. Trends Biochem. Sci. 41, 1035–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reka A. K., Chen G., Jones R. C., Amunugama R., Kim S., Karnovsky A., Standiford T. J., Beer D. G., Omenn G. S., Keshamouni V. G., Epithelial-mesenchymal transition-associated secretory phenotype predicts survival in lung cancer patients. Carcinogenesis 35, 1292–1300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Y. X., Sokol E. S., del Vecchio C. A., Sanduja S., Claessen J. H., Proia T. A., Jin D. X., Reinhardt F., Ploegh H. L., Wang Q., Gupta P. B., Epithelial-to-mesenchymal transition activates Perk–Eif2α and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 4, 702–715 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Marchi S., Bittremieux M., Missiroli S., Morganti C., Patergnani S., Sbano L., Rimessi A., Kerkhofs M., Parys J. B., Bultynck G., Giorgi C., Pinton P., Endoplasmic reticulum-mitochondria communication through Ca2+ signaling: The importance of mitochondria-associated membranes (MAMs). Adv. Exp. Med. Biol. 997, 49–67 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Suzuki J., Kanemaru K., Ishii K., Ohkura M., Okubo Y., Iino M., Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat. Commun. 5, 4153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y., Araki S., Wu J., Teramoto T., Chang Y. F., Nakano M., Abdelfattah A. S., Fujiwara M., Ishihara T., Nagai T., Campbell R. E., An expanded palette of genetically encoded Ca2+ indicators. Science 333, 1888–1891 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lytton J., Westlin M., Hanley M. R., Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-Atpase family of calcium pumps. J. Biol. Chem. 266, 17067–17071 (1991). [PubMed] [Google Scholar]
- 45.Alevriadou B. R., Patel A., Noble M., Ghosh S., Gohil V. M., Stathopulos P. B., Madesh M., Molecular nature and physiological role of the mitochondrial calcium uniporter channel. Am. J. Physiol. Cell Physiol. 320, C465–C482 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakayama Y., Fukuhara S., Ando K., Matsuda M., Mochizuki N., Cdc42 mediates Bmp-induced sprouting angiogenesis through Fmnl3-driven assembly of endothelial filopodia in zebrafish. Dev. Cell 32, 109–122 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Ma L., Lu M. F., Schwartz R. J., Martin J. F., Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development 132, 5601–5611 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Fukui H., Chow R. W., Xie J., Foo Y. Y., Yap C. H., Minc N., Mochizuki N., Vermot J., Bioelectric signaling and the control of cardiac cell identity in response to mechanical forces. Science 374, 351–354 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Fukui H., Terai K., Nakajima H., Chiba A., Fukuhara S., Mochizuki N., S1P-Yap1 signaling regulates endoderm formation required for cardiac precursor cell migration in zebrafish. Dev. Cell 31, 128–136 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Manner J., Yelbuz T. M., Functional morphology of the cardiac jelly in the tubular heart of vertebrate embryos. J. Cardiovasc. Dev. Dis. 6, 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Combs M. D., Yutzey K. E., Heart valve development: Regulatory networks in development and disease. Circ. Res. 105, 408–421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinstein N., Mendoza L., Alvarez-Buylla E. R., A computational model of the endothelial to mesenchymal transition. Front. Genet. 11, 40 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao L., Dudley A. C., Fine-tuning vascular fate during endothelial-mesenchymal transition. J. Pathol. 241, 25–35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duchen M. R., Mitochondria and calcium: From cell signalling to cell death. J. Physiol. 529 (Pt. 1), 57–68 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shishehbor M. H., White C. J., Gray B. H., Menard M. T., Lookstein R., Rosenfield K., Jaff M. R., Critical limb ischemia. J. Am. Coll. Cardiol. 68, 2002–2015 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y., Yang X., Li Z., Bu K., Li T., Ma Z., Wang B., Ma L., Lu H., Zhang K., Liu L., Zhao Y., Zhu Y., Qin J., Cui J., Liu L., Liu S., Fan P., Liu X., Pyk2/MCU pathway as a new target for reversing atherosclerosis. Front. Cell Dev. Biol. 9, 651579 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kirkman D. L., Robinson A. T., Rossman M. J., Seals D. R., Edwards D. G., Mitochondrial contributions to vascular endothelial dysfunction, arterial stiffness, and cardiovascular diseases. Am. J. Physiol. Heart Circ. Physiol. 320, H2080–H2100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J., Jiang J., Hu H., Chen L., MCU complex: Exploring emerging targets and mechanisms of mitochondrial physiology and pathology. J. Adv. Res., 10.1016/j.jare.2024.02.013, (2024). [DOI] [PubMed] [Google Scholar]
- 59.Nolan D. J., Ginsberg M., Israely E., Palikuqi B., Poulos M. G., James D., Ding B. S., Schachterle W., Liu Y., Rosenwaks Z., Butler J. M., Xiang J., Rafii A., Shido K., Rabbany S. Y., Elemento O., Rafii S., Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell 26, 204–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalucka J., de Rooij L., Goveia J., Rohlenova K., Dumas S. J., Meta E., Conchinha N. V., Taverna F., Teuwen L. A., Veys K., García-Caballero M., Khan S., Geldhof V., Sokol L., Chen R., Treps L., Borri M., de Zeeuw P., Dubois C., Karakach T. K., Falkenberg K. D., Parys M., Yin X., Vinckier S., du Y., Fenton R. A., Schoonjans L., Dewerchin M., Eelen G., Thienpont B., Lin L., Bolund L., Li X., Luo Y., Carmeliet P., Single-cell transcriptome atlas of murine endothelial cells. Cell 180, 764–779.e20 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Tomar D., Dong Z., Shanmughapriya S., Koch D. A., Thomas T., Hoffman N. E., Timbalia S. A., Goldman S. J., Breves S. L., Corbally D. P., Nemani N., Fairweather J. P., Cutri A. R., Zhang X., Song J., Jana F., Huang J., Barrero C., Rabinowitz J. E., Luongo T. S., Schumacher S. M., Rockman M. E., Dietrich A., Merali S., Caplan J., Stathopulos P., Ahima R. S., Cheung J. Y., Houser S. R., Koch W. J., Patel V., Gohil V. M., Elrod J. W., Rajan S., Madesh M., MCUR1 is a scaffold factor for the MCU complex function and promotes mitochondrial bioenergetics. Cell Rep. 15, 1673–1685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mallilankaraman K., Doonan P., Cárdenas C., Chandramoorthy H. C., Müller M., Miller R., Hoffman N. E., Gandhirajan R. K., Molgó J., Birnbaum M. J., Rothberg B. S., Mak D. O., Foskett J. K., Madesh M., MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca2+ uptake that regulates cell survival. Cell 151, 630–644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoffman N. E., Chandramoorthy H. C., Shamugapriya S., Zhang X., Rajan S., Mallilankaraman K., Gandhirajan R. K., Vagnozzi R. J., Ferrer L. M., Sreekrishnanilayam K., Natarajaseenivasan K., Vallem S., Force T., Choi E. T., Cheung J. Y., Madesh M., MICU1 motifs define mitochondrial calcium uniporter binding and activity. Cell Rep. 5, 1576–1588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luongo T. S., Lambert J. P., Yuan A., Zhang X., Gross P., Song J., Shanmughapriya S., Gao E., Jain M., Houser S. R., Koch W. J., Cheung J. Y., Madesh M., Elrod J. W., The mitochondrial calcium uniporter matches energetic supply with cardiac workload during stress and modulates permeability transition. Cell Rep. 12, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu X., Wang Y., Soaita I., Lee H. W., Bae H., Boutagy N., Bostwick A., Zhang R. M., Bowman C., Xu Y., Trefely S., Chen Y., Qin L., Sessa W., Tellides G., Jang C., Snyder N. W., Yu L., Arany Z., Simons M., Acetate controls endothelial-to-mesenchymal transition. Cell Metab. 35, 1163–1178.e10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lombardi A. A., Gibb A. A., Arif E., Kolmetzky D. W., Tomar D., Luongo T. S., Jadiya P., Murray E. K., Lorkiewicz P. K., Hajnoczky G., Murphy E., Arany Z. P., Kelly D. P., Margulies K. B., Hill B. G., Elrod J. W., Mitochondrial calcium exchange links metabolism with the epigenome to control cellular differentiation. Nat. Commun. 10, 4509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.TeSlaa T., Chaikovsky A. C., Lipchina I., Escobar S. L., Hochedlinger K., Huang J., Graeber T. G., Braas D., Teitell M. A., α-Ketoglutarate accelerates the initial differentiation of primed human pluripotent stem cells. Cell Metab. 24, 485–493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaffe E. A., Nachman R. L., Becker C. G., Minick C. R., Culture of human endothelial cells derived from umbilical beins. Identification by morphologic and immunologic criteria. J. Clin. Invest. 52, 2745–2756 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sokol L., Geldhof V., García-Caballero M., Conchinha N. V., Dumas S. J., Meta E., Teuwen L. A., Veys K., Chen R., Treps L., Borri M., de Zeeuw P., Falkenberg K. D., Dubois C., Parys M., de Rooij L., Goveia J., Rohlenova K., Schoonjans L., Dewerchin M., Eelen G., Li X., Kalucka J., Carmeliet P., Protocols for endothelial cell isolation from mouse tissues: Small intestine, colon, heart, and liver. STAR Protoc. 2, 100489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carlotti F., Bazuine M., Kekarainen T., Seppen J., Pognonec P., Maassen J. A., Hoeben R. C., Lentiviral vectors efficiently transduce quiescent mature 3T3-L1 adipocytes. Mol. Ther. 9, 209–217 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Haghverdi L., Lun A. T. L., Morgan M. D., Marioni J. C., Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valente A. J., Maddalena L. A., Robb E. L., Moradi F., Stuart J. A., A simple ImageJ Macro Tool for Analyzing Mitochondrial Network Morphology in Mammalian Cell Culture. Acta Histochem. 119, 315–326 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Jin Y. N., Yu Y. V., Gundemir S., Jo C., Cui M., Tieu K., Johnson G. V., Impaired mitochondrial dynamics and Nrf2 signaling contribute to compromised responses to oxidative stress in striatal cells expressing full-length mutant huntingtin. PLOS ONE 8, e57932 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vallese F., Catoni C., Cieri D., Barazzuol L., Ramirez O., Calore V., Bonora M., Giamogante F., Pinton P., Brini M., Cali T., An expanded palette of improved SPLICS reporters detects multiple organelle contacts in vitro and in vivo. Nat. Commun. 11, 6069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balint B., Yin H., Nong Z., Arpino J. M., O'Neil C., Rogers S. R., Randhawa V. K., Fox S. A., Chevalier J., Lee J. J., Chu M. W. A., Pickering J. G., Seno-destructive smooth muscle cells in the ascending aorta of patients with bicuspid aortic valve disease. EBioMedicine 43, 54–66 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rocnik E., Saward L., Pickering J. G., HSP47 expression by smooth muscle cells is increased during arterial development and lesion formation and is inhibited by fibrillar collagen. Arterioscler. Thromb. Vasc. Biol. 21, 40–46 (2001). [DOI] [PubMed] [Google Scholar]
- 79.Claxton S., Kostourou V., Jadeja S., Chambon P., Hodivala-Dilke K., Fruttiger M., Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis 46, 74–80 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Murray S. A., Carver E. A., Gridley T., Generation of a Snail1 (Snai1) conditional null allele. Genesis 44, 7–11 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Pan X., Liu J., Nguyen T., Liu C., Sun J., Teng Y., Fergusson M. M., Rovira I. I., Allen M., Springer D. A., Aponte A. M., Gucek M., Balaban R. S., Murphy E., Finkel T., The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat. Cell Biol. 15, 1464–1472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F., Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 83.Herwig L., Blum Y., Krudewig A., Ellertsdottir E., Lenard A., Belting H. G., Affolter M., Distinct cellular mechanisms of blood vessel fusion in the zebrafish embryo. Curr. Biol. 21, 1942–1948 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Muto A., Ohkura M., Abe G., Nakai J., Kawakami K., Real-time visualization of neuronal activity during perception. Curr. Biol. 23, 307–311 (2013). [DOI] [PubMed] [Google Scholar]
- 85.Cheng Z., Garikipati V. N., Nickoloff E., Wang C., Polhemus D. J., Zhou J., Benedict C., Khan M., Verma S. K., Rabinowitz J. E., Lefer D., Kishore R., Restoration of hydrogen sulfide production in diabetic mice improves reparative function of bone marrow cells. Circulation 134, 1467–1483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schoors S., Cantelmo A. R., Georgiadou M., Stapor P., Wang X., Quaegebeur A., Cauwenberghs S., Wong B. W., Bifari F., Decimo I., Schoonjans L., De Bock K., Dewerchin M., Carmeliet P., Incomplete and transitory decrease of glycolysis: A new paradigm for anti-angiogenic therapy? Cell Cycle 13, 16–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Legends for tables S1 to S7
Tables S1 to S7