Abstract
Thy Pol-2 intein, from Thermococcus hydrothermalis, belongs to the same allelic family as Tli Pol-2 (PI-TliI), Tfu Pol-2 (PI-TfuII) and TspTY Pol-3 mini-intein, all inserted at the pol-c site of archaeal DNA polymerase genes. This new intein was cloned, expressed in Escherichia coli and purified. The intein is a specific endonuclease (PI-ThyI) which cleaves the inteinless sequence of the Thy DNA pol gene. Moreover, PI-TliI, PI-TfuII and PI-ThyI are very similar endonucleases which cleave DNA in the same optimal conditions at 70°C yielding similar 3′-hydroxyl overhangs of 4 bp and the reaction is subject to product inhibition. The three enzymes are able to cleave the three DNA sequences spanning the pol-c site and a 24 bp consensus cleavage site was defined for the three isoschizomers. However, the exact size of the minimal cleavage site depends both on the substrate sequence and the endonuclease. The inability of the isoschizomers to cleave the inteinless DNA polymerase gene from Pyrococcus spp. KOD is due to point substitutions on the 5′ side of the pol-c site, suggesting that the absence of inteins of this allelic family in DNA polymerase genes from Pyrococcus spp. can be linked to small differences in the target site sequence.
INTRODUCTION
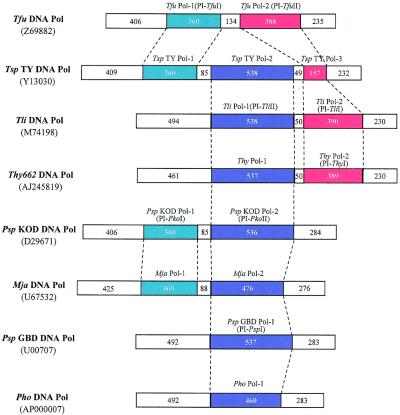
Since the first description of protein splicing in 1990 (1,2), 97 putative inteins have been identified (3,4), widely distributed in 30 different species and strains among eukarya, eubacteria and archaebacteria. Among the 32 host proteins known to date, archaeal DNA polymerases are major targets. Indeed, eight of the 23 known DNA pol α gene sequences from archaebacteria harbour one to three inteins (Fig. 1). These sequences are inserted in the DNA pol α genes at three conserved sites, pol-a, pol-b and pol-c, in motifs II, III and I of the DNA polymerases, respectively. Three allelic families have been defined based on these insertion sites. Within each allelic family, intein sequences share a high degree of homology.
Figure 1.
Inteins in DNA polymerases of archaebacteria. The three families of allelic inteins are presented. Names of inteins, lengths in amino acids of each extein and intein and GenBank accession numbers of DNA polymerases are indicated. Names for the endonuclease activities, which have been experimentally demonstrated, are indicated in parentheses after the intein names. Tfu DNA Pol, Tsp TY DNA Pol, Tli DNA Pol, Thy DNA Pol, Psp KOD DNA Pol, Mja DNA Pol, Psp GBD DNA Pol and Pho DNA Pol are the abreviations for the DNA polymerases from Thermococcus fumicolans, Thermococcus spp. TY, Thermococcus litoralis, T.hydrothermalis, Pyrococcus spp. KOD, M.jannaschii, Pyrococcus spp. GBD and Pyrococcus horikosshi OT3, respectively.
The majority of known inteins exhibit conserved motifs of the LAGLIDADG (DOD) endonucleases (5–7). This sequence conservation suggests that these inteins are homing endonucleases which, like some group I intron endonucleases, cleave the inteinless allele of their host gene, leading to the invasion of the intein coding sequence in this gene (8).
With the exception of the Tsp TY Pol-3 intein (9), which is a mini-intein missing the endonuclease core within its peptide sequence, all the archaeal DNA polymerase inteins harbour an endonuclease domain of the DOD family (7). Moreover, endonuclease activity has been demonstrated previously for seven of these inteins, i.e. at least two inteins of each allelic family: Tfu Pol-1 (PI-TfuI) and Psp KOD Pol-1 (PI-PkoI) at the pol-a insertion site, Tli Pol-1 (PI-TliII), Psp KOD Pol-2 (PI-PkoII) and Psp GBD Pol-1 (PI-PspI) at the pol-b site and Tfu Pol-2 (PI-TfuII) and Tli Pol-2 (PI-TliI) at the pol-c site (6,10–12). These highly specific endonucleases recognise and cleave a 16–30 bp sequence spanning their insertion site in the inteinless allele of the DNA polymerase gene. The DNA sequences of the DNA pol genes are highly conserved between archaebacteria. In particular, the sequences surrounding each of the intein insertion sites are ∼70% identical between species. It is known that Saccharomyces cerevisiae intein PI-SceI tolerates substitutions at several positions of its recognition site (13). Thus, given the high degree of identity between inteins of the same allelic family as well as between their insertion sites, it is reasonable to assume that endonuclease inteins of the same allelic family could be isoschizomers.
The DNA pol gene of the thermophilic archaebacteria Thermococcus hydrothermalis 662 (14) has been sequenced recently (accession number AJ245819). This polymerase harbours two inteins, Thy Pol-1 and Thy Pol-2, belonging to the allelic families of Tli Pol-1 and Tli Pol-2, inserted at the pol-b and pol-c sites, respectively (Fig. 1). Thy Pol-2 intein is highly homologous to Tli Pol-2 (PI-TliI) and Tfu Pol-2 (PI-TfuII) which are known to cleave their respective pol-c insertion site (10,12). We thus cloned and expressed in Escherichia coli the new intein Thy Pol-2 and we pointed out its endonuclease activity (PI-ThyI). Next, the endonuclease activity and the specificity of the three inteins were fully characterised in order to verify if three inteins of the same allelic family share isoschizomer properties.
MATERIALS AND METHODS
Production and purification of the inteins
The coding sequence of the Thy Pol-2 intein was amplified by PCR from the Thy genomic DNA (IFREMER) using oligonucleotides 5′-aaaatcctgcatatgagtgttactggggaaaccgaaatcat-3′ and 5′-gaagaagaattcctaattatggacgagtattccattcgc-3′. The PCR products were digested by NdeI and EcoRI and cloned into a NdeI–EcoRI digested pET26b vector (Novagen). The resulting constructs (pET26-Thy2) were sequenced by Isoprim. Escherichia coli BL21(De3)[pLysS] bacteria transformed with this expression vector were grown at 37°C in Luria Broth culture medium supplemented with 50 µg/ml kanamycin (Sigma Chemical Co.). The Thy Pol-2 intein was produced, extracted and purified as described for the recombinant Tfu Pol-2 intein (12). PI-TliI was a gift from New England Biolabs. Recombinant PI-TfuII was purified from E.coli as described (12). The homogeneous fractions of the three inteins were dialysed against 10 mM Tris–HCl pH 7.5, 50% glycerol, 0.1 mM EDTA, 1 mM DTT, 200 µg/ml BSA and 50 mM NaCl for storage.
DNA substrates for endonuclease activities
To assay Thy Pol-2 endonuclease activity, the 42 bp DNA sequence spanning its homing site was inserted in the XbaI site of plasmid pUC19 by PCR as described by Weiner (15), yielding the Thy-site construct. Three mutant sites [Thy(T9C)-site, Thy(C12T)-site and Thy(T9C+C12T)-site] were also constructed in the same way. Oligonucleotide pairs used for the different constructs are 5′-acggacggtttcttcgcgacagagtcgacctgcaggcatgc-3′ and 5′-gtcagcgtagagcactttaaagaggatccccgggtaccgag-3′ for the Thy-site, 5′-acggacggcttcttcgagagtcgacctgcaggcatgc-3′ and 5′-gtcagcgtagagaggatccccgggtaccgag-3′ for the Thy(T9C)-site, 5′-acggacggttttttcgagagtcgacctgcaggcatgc-3′ and 5′-gtcagcgtagagaggatccccgggtaccgag-3′ for the Thy(C12T)-site and 5′-acggacggctttttcgagagtcgacctgcaggcatgc-3′ and 5′-gtcagcgtagagaggatccccgggtaccgag-3′ for the Thy(T9C+C12T)-site. The sequences which hybridise to the pUC19 DNA matrix are written in bold.
The substrate for the PI-TfuII endonuclease is the plasmid Tfu-site containing a 43 bp cleavage site described before as substrate 2 (12). The substrate for PI-TliI, the plasmid pAKR7 containing a fragment of the Tli DNA pol gene in which the inteins were deleted (10,16), was a gift from New England Biolabs.
In order to assay the endonuclease activity of the inteins on the PspKOD DNA Pol gene, which harbours no intein gene at the pol-c site, three other substrates were constructed. These are either a 43 bp PspKOD pol-c site, designated Psp-site, or hybrid sites with the 5′ half of the Tfu pol-c site plus the 3′ half of the PspKOD site (Tfu.Psp-site), or the 5′ half of the PspKOD site plus the 3′ half of the Tfu site (Psp.Tfu-site). These DNA sequences were inserted in pUC19 using the oligonucleotide pairs, PspKOD-3′ (5′-gtcgctgtagattaccttaaagaggatccccgggtaccgag-3′) and PspKOD-5′ (5′-accgacggattttttgccacagagtcgacctgcaggcatgc-3′), PspKOD-3′ and S2-5′ (5′-acagacggctttttcgcaacagagtcgacctgcaggcatgc-3′) and PspKOD-5′ and S2-3′ (5′-atccgcgtacagcactttaaagaggatccccgggtaccgag-3′), respectively.
The resulting plasmids were linearised either by ScaI (New England Biolabs), or by XmnI in the case of pAKR7, and purified from a 1% agarose gel in TBE (0.09 M Tris–borate, 0.002 M EDTA) buffer. Linear substrates were diluted in water to 100 ng/µl for cleavage assays.
Endonuclease assays and minimal recognition and cleavage sites
Endonuclease activity assays were performed in a final volume of 10 µl, in various reaction buffers and temperatures ranging from 37 to 80°C. The reaction mixtures were analysed on a 1% agarose gel in TBE buffer. The amounts of undigested substrates and products were quantified with the ImageQUANT program (Molecular Dynamics Inc.).
One unit of PI-ThyI, PI-TliI or PI-TfuII endonuclease is required to digest 1 µg of linearised DNA substrate, in 1 h at 70°C, in a 50 mM Tris–acetate pH 8 buffer containing 75 mM Mg(OAc)2, 100 mM NH4OAc and 10% glycerol. Specific activities of PI-ThyI, PI-TliI and PI-TfuII were measured by incubating known amounts of linear DNA substrates with known amounts of purified endonucleases.
The endonuclease recognition sites were determined by a primer extension method as described by Wenzlau et al. (17). The sequencing and digestion procedures, using the T7 polymerase sequencing kit (Pharmacia), universal primers SeqPuc (5′-gtaacgccagggttttcc-3′) and M13Rev (5′-ggaaacagctatgaccatg-3′) and various DNA substrates as matrix, were as described previously (12).
RESULTS AND DISCUSSION
Thy Pol-2 intein is a site-specific endonuclease (PI-ThyI)
The Thy Pol-2 expression level in BL21-De3-pLysS bacteria allowed us to purify 80% homogeneous fractions of the recombinant intein. These fractions (10–20 µg/ml) were used to assay and characterise the endonuclease activity of the intein.
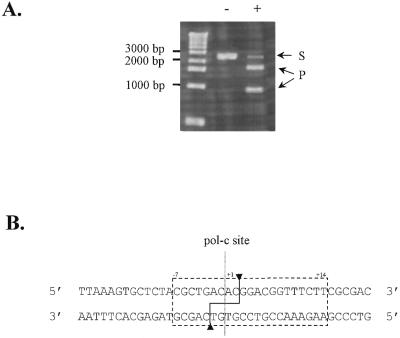
All known inteins that exhibit a specific endonuclease activity cleave a 16–40 bp sequence spanning their homing site. Thus, a 42 bp site corresponding to the sequence spanning the pol-c insertion site in the Thy DNA Pol gene was inserted in the XbaI site of pUC19, yielding the Thy-site construct, which was then ScaI linearised to serve as DNA substrate to assay Thy Pol-2 endonuclease activity. Several enzymatic assays were performed under various experimental conditions with regard to buffer pH and composition, to mono- and bivalent ions used as cofactors and to temperature. Thy Pol-2 exhibits endonuclease activity under a wide range of conditions, cleaving the linear plasmid (2730 bp) specifically into two products (940 and 1790 bp, Fig. 2A). Optimal cleavage efficiency was obtained in a 50 mM Tris–acetate pH 8 buffer containing 75 mM Mg(OAc)2, 100 mM NH4OAc and 10% glycerol, at 70°C. The specific activity of Thy Pol-2, named PI-ThyI according to the current nomenclature, on the linear substrate Thy-site, is 44 000 ± 14 000 U/mg (1.08 mol/mol.h).
Figure 2.
(A) Cleavage assay for PI-ThyI. Linearised (100 ng) Thy-site substrate were incubated either with (+) or without (–) 2 ng of PI-ThyI for 10 min at 70°C in a 50 mM Tris–acetate pH 8 buffer containing 75 mM Mg(OAc)2 and 100 mM NH4OAc. Substrate (S; 2730 bp) and products (P; 940 and 1790 bp) were separated on a 1% agarose gel in TBE buffer. (B) Minimal recognition sequence of PI-ThyI. Minimal nucleotide sequence necessary for recognition and cleavage by PI-ThyI was determined by primer extension on the plasmid Thy-site. The dotted line indicates the homing site of the intein in the DNA Pol gene. The dashed box delimitates the minimal recognition site and arrows designate the cleavage points on each DNA strand.
The minimal site for recognition and cleavage by PI-ThyI was determined using the plasmid Thy-site as DNA matrix for the primer elongation procedure. The comparison of PI-ThyI digested or undigested sequence reaction patterns yielded a 21 bp non-palindromic site corresponding to seven bases 5′ to the Thy pol-c site plus 14 bases 3′ to this site (Fig. 2B). The cleavage by PI-ThyI cleaves DNA in a fashion similar to PI-TliI (Tli Pol-2) and PI-TfuII (Tfu Pol-2) (10,12), yielding non-identical 3′ overhangs of four bases. Since the cleaved plasmid can be ligated by T4 DNA ligase, the generated ends are 5′-phosphate and 3′-hydroxyl.
Kinetic analyses of the cleavage reaction revealed that the enzyme is rapidly inactivated during the reaction (not shown). In fact, prolonged incubation of linearised Thy-site with PI-ThyI results in only partial cleavage as has been described previously for PI-TfuII (12). Since the thermococcal intein is not heat inactivated at 70°C, it is probable that PI-ThyI is inhibited by one of its digestion products as is the case for PI-TfuII.
The three inteins PI-TliI, PI-TfuII and PI-ThyI cleave the same DNA substrates
Thus, all three inteins (Tli Pol-2, Tfu Pol-2 and Thy Pol-2) from three different species of Thermococcus belong to the same allelic family and are specific endonucleases designated PI-TliI, PI-TfuII and PI-ThyI, respectively. The sequence of their cleavage sites, i.e. the sequences spanning the pol-c site in DNA pol genes, are highly conserved. Since PI-SceI is known to have stringent sequence requirements, we compared the ability of these three enzymes to recognise and cleave various substrates, in order to assess their specificity.
First, we defined the optimal cleavage conditions and minimal recognition and cleavage site for PI-TliI. We found that PI-TliI is most active in the buffer described for PI-ThyI and PI-TfuII, at 70°C, and is inhibited like the other two inteins (not shown). Under optimal conditions, PI-TliI cleaves the XmnI linearised pAKR7 substrate with a specific activity of 27 000 ± 3000 U/mg (0.66 mol/mol.h).
Hence, the three enzymes need identical conditions for catalysis. Moreover, as in the case of PI-TfuII (12), no particular DNA conformation of the substrate is preferred since supercoiled and linear DNA are cleaved with the same efficiency (not shown). Hence, the three inteins of the same allelic family have endonuclease activities which are highly similar and which likely share a common catalytic pathway. However, the specific activity of PI-TfuII is approximately 20 times higher than those of PI-ThyI and PI-TliI and the last is marginally less active than PI-ThyI (Table 1). Since the three inteins have highly related peptide sequences, PI-TfuII and PI-ThyI being 85% identical, subtle differences in intein peptide sequence must be responsible for these differences in the specific activity levels.
Table 1. Specific activities of PI-TfuII, PI-TliI and PI-ThyI endonucleases on different DNA substrates.
| Tli pol-c site | Tfu pol-c site | T9C+C12T Thy pol-c site | T9C Thy pol-c site | C12T Thy pol-c site | Thy pol-c site | |
|---|---|---|---|---|---|---|
| PI-ThyI | 5.5 ± 2.2 | 4.1 ± 0.5 | 6.5 ± 0.5 | 5 ± 1.1 | 4.9 ± 2.1 | 4.4 ± 1.4 |
| PI-TfuII | 34.5 ± 3.5 | 74.8 ± 16 | 26.5 ± 4 | 68.6 ± 5.2 | 44.4 ± 10.0 | 50.3 ± 3.8 |
| PI-TliI | 2.7 ± 0.3 | 3.3 ± 1.2 | 2.4 ± 0.4 | 4.7 ± 1.6 | 4.1 ± 1.7 | 3.8 ± 0.8 |
The specific activities are expressed in 104 U activity per mg of enzyme. One unit of enzyme is the quantity required to digest 1 µg of substrate in 1 h under optimal reaction conditions.
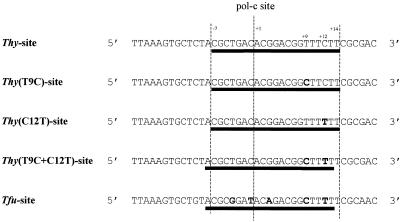
It has been reported that the endonuclease PI-TliI specifically cleaves the sequence spanning the intein insertion site (10,16). We have delineated the minimal cleavage sequence to 23 bp corresponding to 10 bp 5′ to the pol-c site plus 13 bp 3′ to the site (Fig. 3). This sequence is 2 bp longer than the PI-TfuII and PI-ThyI minimal sequence for cleavage of their own homing site.
Figure 3.
Minimal recognition sequence of PI-ThyI, PI-TfuII and PI-TliI on each pol-c site. The sequences of pol-c intein insertion sites in Thy, Tfu and Tli DNA pol genes are indicated. Minimal recognition and cleavage sequences of these three DNA sequences by PI-ThyI, PI-TfuII and PI-TliI appear in red, green and blue, respectively. Nucleotides that are not conserved in the three sequences appear in bold. The 4 bp overhangs generated by the cleavage reactions are underlined.
We then assayed the activity of each enzyme on the three substrates. It turns out that PI-ThyI, PI-TfuII and PI-TliI are able to efficiently cleave the three different target sites. Thus, these three inteins of the same allelic family are isoschizomers which tolerate point substitutions in the DNA substrate sequence. The specific activities of each endonuclease are comparable whatever the DNA substrates (Table 1). Even if these activities fluctuate on a small scale, no obvious effect of point substitutions in the substrate sequence is observable.
Finally, the primer extension method was used to determine the minimal sequence required for cleavage of each substrate by each endonuclease. The results show that the minimal recognition and cleavage sites depend on both the DNA sequence and the endonuclease (Fig. 3). All three enzymes cleave the three pol-c sites at the same position, yielding identical products with a four base 3′-hydroxyl (3′-OH) overhang, but the minimal recognition sequences vary. PI-ThyI and PI-TfuII give identical results for each target site. Both need a 21 bp sequence to cleave the Tfu and Thy substrates but this minimal recognition sequence is shifted 1 bp leftward in the case of the Tfu substrate. Both enzymes also cut the Tli substrate with the same recognition sequence of 19 bp, 2 bp shorter on the 5′ side than their recognition sequence on the Tfu site. PI-TliI recognises the same 21 bp sequence as the other two enzymes on the Tfu substrate, but it needs at least a 23 bp long sequence to cleave the Thy and Tli substrates. Its recognition site on the Thy substrate is shifted 1 bp on the 3′ side compared to the Tli site.
Hence, a 24 bp DNA sequence corresponding to 10 bases 5′ to the pol-c insertion site plus 14 bp 3′ to this site can be defined as the consensus cleavage sequence for the three isoschizomers PI-ThyI, PI-TfuII and PI-TliI (Fig. 3).
The most similar endonucleases, PI-ThyI and PI-TfuII, behave similarly on each DNA substrate in the way that they need the same minimal sequence to cleave each site. PI-TliI minimal recognition sequences have the same 3′ boundary as those of PI-TfuII and PI-ThyI but are 2 and 4 bp longer at the 5′-end on Thy and Tli pol-c sites, respectively (Fig. 3). In fact, the 3′ side of the minimal recognition sequences are the same for the three enzymes and depend on the substrate sequence while the 5′ side of the recognition sequence differs between PI-TliI and the other two enzymes.
Displacement of 3′-end of the minimal cleavage site by point nucleotide substitutions
The 3′ boundary of the recognition site is shifted 1 bp to the right on the Thy substrate for all three enzymes, compared to the other two DNA substrates. On the 3′ side of the cleavage sites, only three bases are not conserved in the three sequences. The nucleotide at position +3, 3′ to the pol-c site, obviously plays no part in the cleavage reaction as it can be indifferently G, A, T or even C in the Tfu.Psp-site hybrid substrate (see below). On the other hand, the two bases, at positions +9 and +12 from the pol-c insertion site, differ between the Thy substrate and the other two (Fig. 3). In order to see whether the displacement of the 3′-end of the recognition sequence could be attributed to these two differences in the site sequences, three mutants of the Thy-site were constructed. Either the T at position +9 or the C at position +12, or both, were changed to the bases found at these positions in the Tli and Tfu sites. We then compared the minimal recognition sequences necessary for the cleavage by PI-ThyI of these three mutated sequences and wild-type Thy and Tfu substrates.
When the T at position +9 of Thy-site is changed to a C, or when the C at position +12 is changed to a T, the recognition sequence by PI-ThyI is not modified, and cleavage occurs with the same efficiency as with the wild-type Thy substrate for all three enzymes. With the double mutant Thy substrate, the recognition sequence is shifted 1 bp to the left and becomes equivalent to the recognition sequence on the Tfu substrate (Fig. 4). Cleavage efficiency of this mutant site is optimal with PI-ThyI, but not with the other two enzymes (Table 1).
Figure 4.
Minimal cleavage sites of five different substrates by PI-ThyI. The large black lines under the five sequences indicate the minimal sequences necessary for cleavage. Nucleotides that are different from the wild-type PI-ThyI site appear in bold.
Hence, even if an adenine is tolerated at position +9 for the cleavage of Tfu.Psp-site substrate (see below), the pyrimydine pair at positions +9 and +12 appear to be directly implicated in substrate recognition. Indeed, changing the nucleotides +9 and +12 from TC to CT did not affect the cleavage efficiency by its cognate enzyme (Table 1), but only shifted the recognition sequence one nucleotide to the left. This is in line with a strong binding of endonucleases to the 3′ part of the substrate, as suggested previously by the product inhibition of the cleavage reaction (12) and as demonstrated for PI-SceI (18).
The Psp KOD Pol gene is not cleaved by the isoschizomers
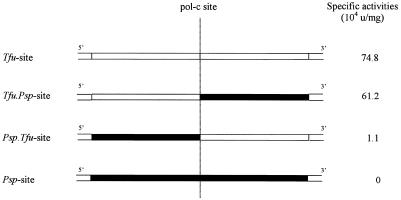
The inteinless sequences of DNA polymerase genes from the three Thermococcus species (i.e. Tli, Tfu and Thy) are specifically cleaved at the pol-c site by the three intein isoschizomers of the same allelic family. It was interesting to verify whether the DNA Pol gene of Pyrococcus KOD species (Psp KOD), which has no intein at the pol-c site, was also a substrate for these endonucleases. For this purpose, the 43 bp sequence spanning the pol-c site in Psp KOD DNA pol gene was cloned in pUC19. Two hybrid substrates composed of either the 5′ part of Tfu site plus the 3′ part of the Psp KOD site or, inversely, the 5′ part of Psp KOD site plus the 3′ part of the Tfu site were also constructed. These three substrates, called Psp-site, Tfu.Psp-site and Psp.Tfu-site, respectively, were ScaI linearised and used in standard cleavage assays by PI-TfuII endonuclease in optimal reaction conditions. The specific activities of PI-TfuII on these substrates were compared to its specific activity on its wild-type substrate Tfu-site (Fig. 5).
Figure 5.
Specific activities of PI-TfuII on four substrates, expressed in 104 U/mg. Various DNA substrates are symbolised by black or white boxes representing Psp KOD and Tfu DNA Pol gene sequences, respectively.
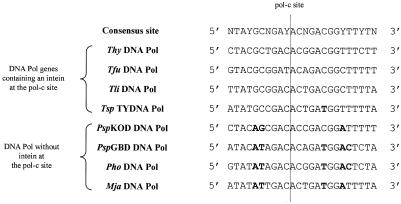
The cleavage assays show that the PspKOD sequence is not a substrate for PI-TfuII. Thus, the sequence divergences in the DNA Pol gene from PspKOD directly hinder the cleavage of this sequence. Only three nucleotides of the PspKOD sequence diverge from the 24 bp consensus cleavage site determined for the three isoschizomers (Fig. 6). The adenine at position +9, 3′ to the pol-c site, does not substantially disturb the cleavage process since PI-TfuII retains 82% of its activity on the Tfu.Psp-site hybrid substrate compared to the activity on the wild-type Tfu-site. In contrast, sequence divergences on the left half of PspKOD pol-c site result in a 98.5% loss of activity of cleavage efficiency. The GC nucleotide pair at positions –6 and –5 of the site is replaced by AG in the PspKOD sequence. This implies that this nucleotide pair, conserved in all thermococcal sequences, is directly involved in the digestion process.
Figure 6.
Sequences at the pol-c site in eight DNA Pol genes from archaebacteria. The 24 bp sequences spanning the pol-c site in DNA pol, with or without an intein at this site, are aligned with the consensus cleavage site. Nucleotides that diverge from this consensus appear in bold.
It has been shown that the endonuclease activity of PI-SceI initiates the horizontal transmission of its own coding sequence by a homing event (8). Since the three thermococcal inteins of the Tli Pol-2 allelic family and their pol-c insertion sites are highly homologous, one might expect a similar way of propagation of these inteins in the archaeal DNA polymerase genes. The pol-c site sequence from Pyrococcus species, which are phylogenetically near Thermococcus species, or from Methanococcus jannaschii only differ from Thermococcal sequences at three conserved positions –6, –5 and +9. Since we have shown that sequence divergence at positions –5 and –6 hinder the endonuclease cleavage, this could be a clue to the absence of inteins at the pol-c site in these genes which are not substrates for cleavage by inteins of Tli Pol-2 allelic family.
CONCLUSIONS
Among the eight archaeal DNA polymerases containing inteins, the four thermococcal enzymes contain an intein at the pol-c insertion site whereas enzymes from Pyrococcus species and from M.jannaschii do not. Except for the TspTY Pol-3 intein which is a mini-intein, inteins of this allelic family, i.e. Tli Pol-2, Tfu Pol-2 and Thy Pol-2, possess the endonuclease motifs characteristic of homing endonucleases and are 61.4% identical.
In the present study, we showed that the Thy Pol-2 intein is a specific endonuclease (PI-ThyI) which cleaves the inteinless form of the Thy DNA Pol gene at the pol-c site. The cleavage yields non-identical 3′-OH overhangs of four bases, which are equivalent to the one generated by PI-TliI and PI-TfuII. Furthermore, we showed that the three thermococcal inteins, PI-TliI, PI-TfuII and PI-ThyI are isoschizomers which have highly similar activities and which likely share a common catalytic pathway. However, subtle differences in the catalytic centre of PI-TfuII must be responsible for its ∼20-fold higher specific activity compared to PI-ThyI and PI-TliI.
The comparison of the minimal sequences cleaved by the three endonucleases allowed us to determine a 24 bp consensus DNA substrate for the isoschizomers family, and thus the exact substrate specificity of these enzymes. Among the 24 bp substrate sequence, eight nucleotides are variable: four positions tolerate any nucleotides whereas four require a pyrimidine. The other 16 bases of the site appear to be critical for the endonuclease activity. We showed that the substitution of nucleotides at positions –5 and –6 prevent the cleavage, but additional mutational analyses will be necessary to assess the absolute requirement of each base. Nevertheless, the specificity of the intein isoschizomers seems quite high compared to other DOD endonucleases and in particular to PI-SceI, for which only 9 bp of the substrate out of 31 bp are absolutly required (13). Since the three inteins have highly related peptide sequences, subtle differences in intein peptide sequence must be responsible for their differences in specific activity and substrate recognition.
The inteinless DNA polymerase genes from other species are not cleaved by these isoschizomers because of a limited number of point substitutions in their sequences spanning the pol-c site. Hence, the absence of inteins at the pol-c site of these genes could be linked to the fact that these genes are not substrates for these endonucleases. It has recently been shown that horizontal transmission is critical for long-term persistence of selfish genes with little or no benefit to the host organism such as homing endonucleases (19). Since persistence over long evolutionary timescales probably requires cyclical gain and loss of the homing endonuclease genes (19), finding within a small group of closely related species some with functional inteins, others with non-functional ones and species without inteins, is precisely what one would expect, reflecting the dynamic evolutionary biology of these genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Neil Jonhson for the reading of this manuscript. J.M.M. was the recipient of an incitative IFREMER grant and J.D. gratefully acknowledges support from the Conseil Régional de Bretagne.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed at Institut de Pharmacologie et Biologie Structurale, I.P.B.S./C.N.R.S., 205 Route de Narbonne, F-31077 Toulouse Cedex, France. Tel: +33 05 61 17 54 76; Fax: +33 05 61 17 59 94; Email: masson@ipbs.fr Z69882, Y13030, M74198, AJ245819, D29671, U67532, U00707, AP000007
REFERENCES
- 1.Hirata R., Ohsumi,Y., Nakano,A., Kawasaki,H., Suzuki,K. and Anraku,Y. (1990) J. Biol. Chem., 265, 6726–6733. [PubMed] [Google Scholar]
- 2.Kane P.M., Yamashiro,C.T., Wolczyk,D.F., Neff,N., Goebl,M. and Stevens,T.H. (1990) Science, 250, 651–657. [DOI] [PubMed] [Google Scholar]
- 3. Inteins-protein introns web site at http://www.blocks.fhcrc.org/∼pietro/inteins/
- 4. Inbase, the New England Biolabs intein database at http://www.neb.com/neb/inteins/ [DOI] [PMC free article] [PubMed]
- 5.Dalgaard J.Z., Klar,A.J., Moser,M.J., Holley,W.R., Chatterjee,A. and Saira Mian,I. (1997) Nucleic Acids Res., 25, 4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perler F.B., Olsen,G.J. and Adam,E. (1997) Nucleic Acids Res., 25, 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pietrokovski S. (1998) Protein Sci., 7, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gimble F.S. and Thorner,J. (1992) Nature, 357, 301–306. [DOI] [PubMed] [Google Scholar]
- 9.Niehaus F., Frey,B. and Antranikian,G. (1997) Gene, 204, 153–158. [DOI] [PubMed] [Google Scholar]
- 10.Perler F.B., Comb,D.G., Jack,W.E., Moran,L.S., Qiang,B., Kucera,R.B., Benner,J., Slatko,B.E., Nwankwo,D.O., Hempstead,S.K., Carlow,C.K.S. and Jannash,H. (1992) Proc. Natl Acad. Sci. USA, 89, 5577–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishioka M., Fujiwara,S., Takagi,M. and Imanaka,T. (1998) Nucleic Acids Res., 26, 4409–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saves I., Ozanne,V., Dietrich,J. and Masson,J.-M. (2000) J. Biol. Chem., 275, 2335–2341. [DOI] [PubMed] [Google Scholar]
- 13.Gimble F.S. and Wang,J. (1996) J. Mol. Biol., 263, 163–180. [DOI] [PubMed] [Google Scholar]
- 14.Godfroy A., Lesongeur,F., Raguénès,G., Quérellou,J., Antoine,E., Meunier,J.-R., Guezennec,J. and Barbier,G. (1997) Int. J. Syst. Bacteriol., 47, 622–626. [DOI] [PubMed] [Google Scholar]
- 15.Weiner M.P., Costa,G.L., Schoettlin,W., Cline,J., Mathur,E. and Bauer,J.C. (1994) Gene, 151, 119–123. [DOI] [PubMed] [Google Scholar]
- 16.Hodges R.A., Perler,F.B., Noren,C.J. and Jack,W.E. (1992) Nucleic Acids Res., 20, 6153–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wenzlau J.M., Saldanha,R.J., Butow,R.A. and Perlman,P.S. (1989) Cell, 56, 421–430. [DOI] [PubMed] [Google Scholar]
- 18.Wende W., Grindl,W., Christ,F., Pingoud,A. and Pingoud,V. (1996) Nucleic Acids Res., 24, 4123–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goddard M.R. and Burt,A. (1999) Proc. Natl Acad. Sci. USA, 96, 13880–13885. [DOI] [PMC free article] [PubMed] [Google Scholar]