Abstract
Background
Colorectal cancer (CRC) is a prevalent cause of death from malignant tumors. This study aimed to develop a nicotinamide adenine dinucleotide (NAD+) metabolism and immune-related prognostic signature, providing a theoretical foundation for prognosis and therapy in CRC patients.
Methods
NAD + metabolism-related and immune-related subtypes of CRC patients were identified by consistent clustering. Differentially expressed genes (DEGs) between the two subtypes of CRC were identified by overlapping. A risk signature was constructed using univariate Cox and least absolute shrinkage and selection operator (LASSO) regression analyses. Independent prognostic predictors were authenticated by Cox analysis. Gene set variation analysis (GSVA) and single-sample gene set enrichment analysis (ssGSEA) were applied to investigate the connection between the prognostic signature and the immune microenvironment. Chemotherapy drug sensitivity and immunotherapy responsiveness were projected using the ‘pRRophetic’ package and Tumor Immune Dysfunction and Exclusion (TIDE) website. The Human Protein Atlas (HPA) database was used to assess the protein expression of prognostic genes in CRC and normal tissues.
Results
Using bioinformatics methods, three prognostic genes related to immune-related NAD + metabolism were identified, and the results were used to establish and verify a prognostic signature related to immune-related NAD + metabolism in CRC patients. Cox regression analysis confirmed that the risk score was a reliable independent prognostic predictor. GSVA and ssGSEA indicated that the prognostic signature was associated with the immune microenvironment. TIDE analysis suggested that the signature might act as an immunotherapy predictor. Chemotherapy sensitivity analysis revealed that COMP was correlated with chemotherapy sensitivity in CRC patients and might be a potential therapeutic target.
Conclusion
This study identified NAD + metabolism-immune-related prognostic genes (MOGAT2, COMP, and DNASE1L3) and developed a prognostic signature for CRC prognosis, which is significant for clinical prognosis prediction and treatment strategy decisions for CRC patients.
Keywords: Colorectal cancer, NAD+ metabolism-related genes, Immune-related genes, Prognostic signature
1. Introduction
Colorectal cancer (CRC) is a prevalent malignant tumor globally and ranks as the second leading cause of cancer-related deaths. Its incidence and mortality rates continue to rise each year [1]. The most commonly observed type is adenocarcinoma, with squamous cell carcinoma, adenosquamous carcinoma, spindle cell carcinoma, and undifferentiated carcinoma occurring less frequently [2]. The primary treatment approaches for CRC include preoperative chemoradiotherapy or combined radiotherapy and chemotherapy following radical surgery in the early stages, chemotherapy in tandem with targeted therapy or immunotherapy in the middle and late stages, or neoadjuvant therapy [3]. Nonetheless, the overall survival rate for CRC patients remains considerably low, primarily due to tumor progression involving invasion and metastasis. With advancements in biotechnology, biomarkers can be employed to guide prognosis and treatment decisions, facilitating tailored treatment options to enhance patients' quality of life [4]. Consequently, early diagnosis and intervention play a pivotal role in reducing the incidence rate and improving the cure rate. Therefore, the identification of novel prognostic markers and models is of utmost importance. This is not only critical for improving patient prognosis but also crucial for providing patients with accurate treatment strategies.
Nicotinamide adenine dinucleotide [5] is an important metabolite and coenzyme that facilitates redox reactions in various metabolic pathways and cellular processes. It plays a central role in energy metabolism [6]. Studies have indicated that increased levels of NAD in CRC tissue can reduce reactive oxygen species levels, maintain cell stemness, and decrease the sensitivity of CRC cells to chemotherapy, thus impacting tumor growth [7]. Furthermore, NAD anabolism can enhance aging-associated secretory phenotypes, which in turn promote breast tumorigenesis [8]. In terms of tumor immunotherapy, nicotinamide adenine dinucleotide (NAD+) metabolism helps maintain inducible PD-L1 expression, drives tumor immune evasion, and sustains the activity and expression of the methylcytosine dioxygenase Tet1, mainly through α-ketoglutarate (α-KG). This ultimately reduces resistance to anti-PD-L1 antibody immunosuppressants [9]. Consequently, further exploration of the role of NAD + -related genes in the occurrence and progression of CRC is needed.
The immune system plays a crucial role in recognizing and eliminating tumors. The development, recurrence, and metastasis of tumors are closely associated with immune dysfunction, particularly abnormalities in cellular immune function, in which T cells are key players. Colorectal carcinogenesis is influenced by genetic and epigenetic alterations in tumor cells, as well as tumor-host interactions. Notably, robust lymphocyte responses observed in CRC tissues, characterized by high densities of CD3+ T cells and T-cell subsets, are frequently associated with positive clinical results. This finding underscores the central role of T-cell-mediated immunity in suppressing tumor progression [10,11]. Additionally, a study [12] demonstrated that the abundance of tumor-infiltrating T cells correlated with specific molecular features of CRC. However, the association between NAD + -related genes and tumor immunity in the occurrence and progression of CRC requires further investigation.
To address these gaps in knowledge, this study used bioinformatics methods to identify NAD + metabolism- and immune-related subtypes in CRC patients. Subsequently, prognostic features related to NAD + metabolism and immunity were developed for CRC patients, with the aim of predicting patient prognosis and informing clinical treatment decisions. These findings offer insights into the associations among NAD + metabolism, immunity, and the mechanisms influencing the occurrence and progression of CRC.
2. Materials and methods
2.1. Dataset and gene source
We integrated transcriptomic data and clinical information from The Cancer Genome Atlas-Colorectal Cancer (TCGA-CRC) dataset, which consisted of 380 CRC tissue samples and 51 normal tissue samples. This integration was carried out using the UCSC database (http://xena.ucsc.edu/). Among these samples, 372 CRC samples were included in consensus clustering and survival analysis because they contained complete survival time records. In addition, we used two additional CRC datasets, GSE17538 and GSE115261, which were extracted from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The GSE17538 dataset, which included 232 CRC samples with survival information, was used to validate our prognostic signature. The GSE115261 dataset, consisting of transcriptomic data from 10 CRC samples and 10 normal samples, was utilized for the validation of the expression of prognostic genes.
To identify NAD + metabolism-related genes (NMRGs), we mined data from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (pathway: hsa00760, Nicotinate and nicotinamide metabolism) (https://www.genome.jp/kegg/pathway.html) and the Reactome database (pathway: R-HSA-196807, Nicotinate metabolism) (https://reactome.org/). Immune-related genes (IRGs) were identified from the ImmPort database (https://immport.niaid.nih.gov).
2.2. Consensus clustering
For consensus clustering, we used the 'ConsensusClusterPlus' package [13] to classify the 372 CRC patients in the TCGA-CRC cohort based on the expression of NMRGs or IRGs. The parameters were set as follows: maxK = 6, reps = 1000, pItem = 0.8, clusterAlg = ‘pam’, distance = ‘canberra’, and innerLinkage = ‘complete'.
2.3. Identification of differentially expressed genes (DEGs)
We identified the differentially expressed genes (DEGs) through the ‘limma’ package (version 3.48.3 [14] based on the following threshold values: p value < 0.05 and |log2FoldChange| > 1.
2.4. Functional annotation analysis
We used the R package ‘clusterProfiler’ (version 4.0.5) [15] for Gene Ontology (GO) and KEGG enrichment analysis. GO terms were categorized into cellular component [3], molecular function (MF), and biological process (BP) terms. The significance criterion was an adjusted p value ≤ 0.05.
2.5. Creation of a prognostic signature related to immune-related NAD + metabolism in CRC
The TCGA-CRC dataset included a total of 372 patients whose survival information was available. Through randomization (ratio 7:3), the data were divided into a training set (n = 261) and a testing set (n = 111). In the training set, we performed univariate Cox analysis using the ‘survminer’ package (version 0.4.9) and least absolute shrinkage and selection operator (LASSO) regression analysis using the ‘glmnet’ package (version 4.0–2) to identify NAD + metabolism-immune-relevant prognostic genes. The RiskScore = formula was used to separate patients into high-risk and low-risk subgroups based on the coefficient obtained by LASSO and the optimal threshold calculated using the surv_cutpoint function. We evaluated the predictive efficiency of the prognostic signature by analyzing Kaplan‒Meier (K‒M) curves, Receiver Operating Characteristic (ROC) analysis, and risk curves.
2.6. Relevance analysis of the gene signature and clinical parameters
Risk scores for the different clinical feature subgroups were compared with the Wilcoxon test (two groups) or Kruskal‒Wallis test (more than two groups).
2.7. Independent prognostic analysis and nomogram construction
Risk score and clinical characteristic factors were included in the Cox analysis (univariate Cox and multivariate Cox) to determine independent prognostic predictors. A nomogram integrating the independent prognostic predictors was constructed using the R language ‘rms’ to predict survival at 1, 3 and 5 years in CRC patients. The calibration curves were employed to assess the precision of the prediction.
2.8. Correlation analysis of the prognostic signature and tumor immunity
The scores of the 13 immune-related pathways in the two risk subgroups were calculated by the gene set variation analysis (GSVA) algorithm [16]. The scores of 24 types of infiltrating immune cells for each sample in the two risk subgroups were assessed by the single-sample gene set enrichment analysis (ssGSEA) algorithm [17]. The correlation between prognostic genes and immune cells was measured through the TIMER database (https://cistrome.shinyapps.io/timer/).
2.9. Therapy analysis based on the prognostic signature
We inferred and assessed the sensitivity of the two risk subgroups to immune checkpoint inhibitor [18] therapy using the TIDE algorithm [19]. Furthermore, using the ‘pRRophetic’ R package [20], we computed the half maximal inhibitory concentration (IC50) values for each patient in the two risk subgroups to analyze the correlation between prognostic genes and chemotherapy drug sensitivity.
2.10. Analysis and verification of the expression of prognostic genes
We initially observed a discrepancy in the expression of prognostic genes in CRC and normal samples in the external dataset GSE115261. Box-line plots illustrating this discrepancy were created using the ‘ggpubr’ package (version 0.4.0). To further determine the protein expression levels of prognostic genes in normal and CRC tissues, we utilized immunohistochemistry images from the Human Protein Atlas (HPA) database. We obtained human normal colonic epithelial cells (CCD814) and three human CRC cell lines (HCT-116, LOVO, and SW480) from iCell Bioscience, Inc. (Shanghai, China). The cells were incubated at 37 °C in an atmosphere of 5 % CO2. Total RNA was extracted from the four cell lines in the logarithmic growth phase using TRIzol Reagent following the instructions from Ambion (USA). Total RNA was then reverse transcribed into cDNA using the SweScript First-Strand cDNA Synthesis Kit from Servicebio (China). qPCR was subsequently performed using 2 × Universal Blue SYBR Green qPCR Master Mix according to the manufacturer's directions (Servicebio, China). The sequences of primers used for qPCR are displayed in Table 1. The expression levels were normalized to those of the internal reference gene GAPDH and calculated using the 2−ΔΔCt method [21].
Table 1.
Primer sequences for qPCR.
| Primer | Sequences |
|---|---|
| MOGAT2 For | CTGTTACTGCGGAACCGAAAG |
| MOGAT2 Rev | CCATGAAAGAGTGGGAGGGAG |
| DNASE1L3 For | CGTGAAACACCGCTGGAAGG |
| DNASE1L3 Rev | TTGGGAACAACAGAACTGACGATT |
| COMP For | CCGAGTCCGCTGTATCAACA |
| COMP Rev | TATGTTGCCCGGTCTCACAC |
| GAPDH For | CCCATCACCATCTTCCAGG |
| GAPDH Rev | CATCACGCCACAGTTTCCC |
2.11. Statistical analysis
A Venn diagram was generated using the Jvenn website (http://jvenn.toulouse.inra.fr/app/example.html) [22]. All bioinformatics analyses were performed using R language. The Wilcoxon test (for two groups) or Kruskal‒Wallis test (for more than two groups) was used to compare the data from different groups.
3. Results
3.1. Recognition of NAD + metabolism-related and immune-related subtypes of CRC
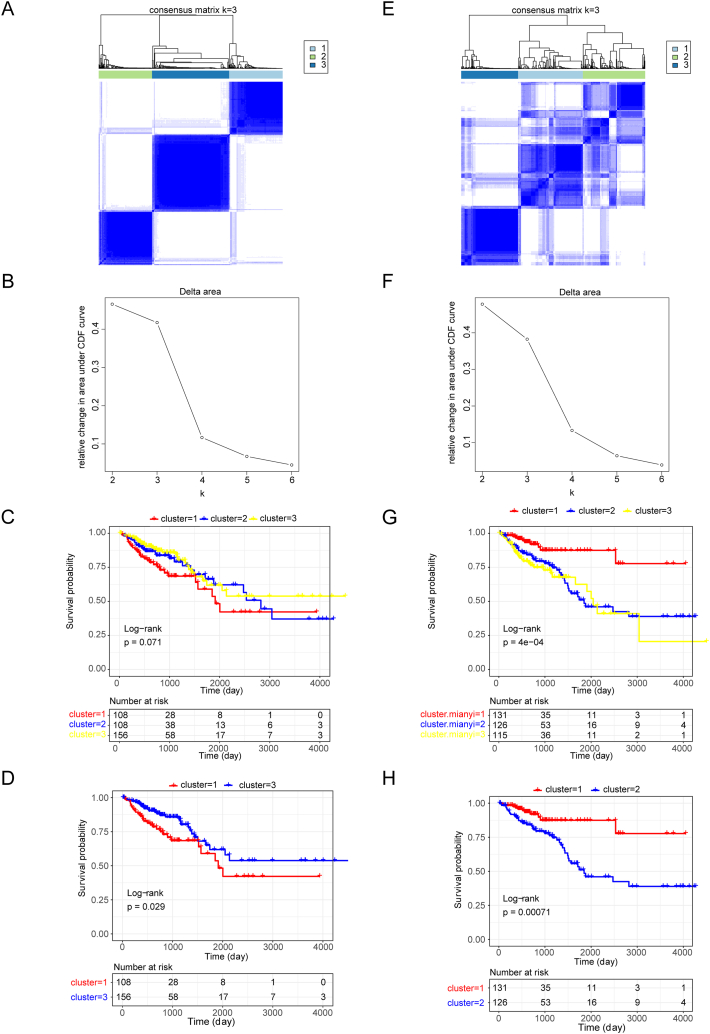
Based on the expression of 35 NMRGs (Supplementary Table 1) detected in the TCGA-CRC dataset, we categorized the 372 CRC patients in the TCGA-CRC dataset into three NAD + metabolism-related subtypes through consensus clustering Fig. 1A and B). Survival analysis revealed significant differences in survival between cluster 1 and cluster 3 (p value = 0.029) (Fig. 1C and D). Furthermore, we classified these 372 CRC patients into three immune-related subtypes based on the expression of 1057 IRGs detected in the TCGA-CRC dataset (Fig. 1E and F). K‒M curves revealed significant differences in survival between the three immune-related subtypes, with more pronounced survival disparities between cluster 1 and cluster 2 (p value = 0.00071) (Fig. 1G and H).
Fig. 1.
Recognition of nicotinamide adenine dinucleotide (NAD+) metabolism-related and immune-related subtypes of colorectal cancer (CRC). (A) Consensus matrix heatmap depicting consensus values for each cluster (k) for NAD + metabolism-related subtypes. Clustering according to a consensus at K = 3. (B) Delta area plot reflecting the relative changes in the area under the cumulative distribution function (CDF) curve for the NAD + metabolism-related subtypes. (C) Survival curves of CRC patients with three clusters of NAD + metabolism-related subtypes. (D) Survival curves of patients with CRC stratified according to the NAD + metabolism-related subtypes in cluster 1 and cluster 3. (E) Consensus matrix heatmap depicting consensus values for each cluster (k) for immune-related subtypes. Clustering according to a consensus at K = 3. (F) Delta area plot reflecting the relative changes in the area under the CDF curve for immune-related subtypes. (G) Survival curves of CRC patients with three clusters of immune-related subtypes. (H) Survival curves of CRC patients with immune-related subtypes in cluster 1 and cluster 2.
3.2. Identification of NAD + metabolism- and immune-related DEGs
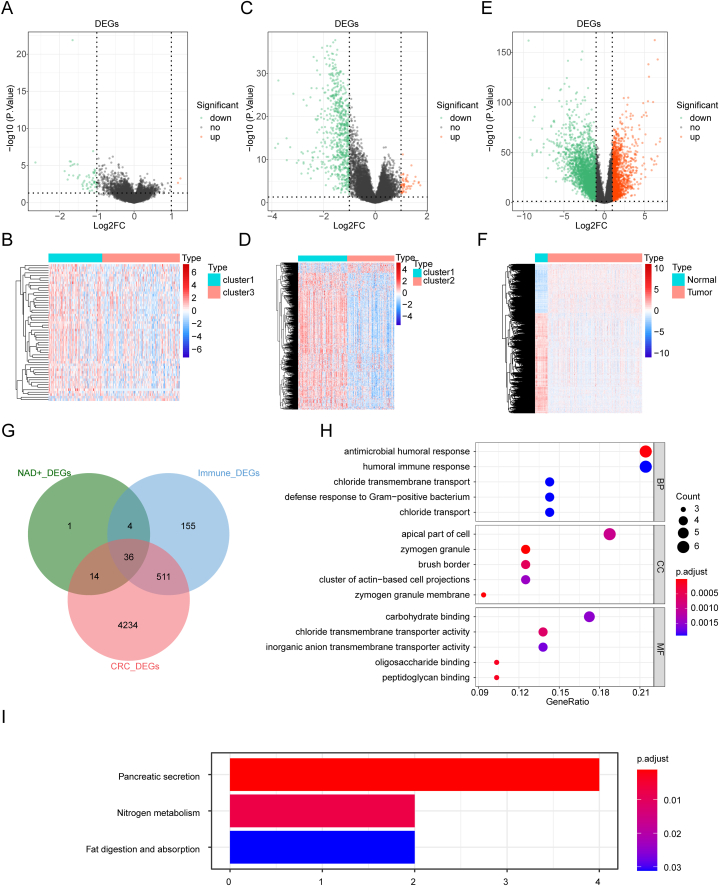
Hence, we identified 55 DEGs between NAD + metabolism-related cluster 3 and cluster 1 (cluster 3 vs cluster 1). Among these DEGs, 2 genes were upregulated and 53 genes were downregulated in cluster 3 at the expression level Fig. 2A and B). Moreover, we discovered 706 DEGs between immune-related cluster 2 and cluster 1 (cluster 2 vs cluster 1). These DEGs consisted of 49 upregulated genes and 657 downregulated genes in cluster 2 Fig. 2C and D). Furthermore, we identified 4795 DEGs between CRC and normal samples in the TCGA-CRC dataset (tumor vs normal). Among these DEGs, 1577 genes were upregulated, and 3218 genes were downregulated (Fig. 2E and F). By intersecting these three sets of DEGs, we obtained 36 common genes related to NAD + metabolism and immune function (Fig. 2G–Table 2).
Fig. 2.
Identification of differentially expressed genes (DEGs) related to NAD + metabolism and immune responses (A) Volcano plot of DEGs between NAD + metabolism-related cluster 3 and cluster 1. (B) Heatmap of DEGs between NAD + metabolism-related cluster 3 and cluster 1. (C) Volcano plot of DEGs between immune-related cluster 2 and cluster 1. (D) Heatmap of DEGs between immune-related cluster 2 and cluster 1. (E) Volcano plot of DEGs between CRC and normal samples in The Cancer Genome Atlas-Colorectal Cancer (TCGA-CRC) dataset. (F) Heatmap of DEGs between CRC and normal samples in the TCGA-CRC dataset. (G) Candidate genes were obtained from the Venn diagram of NAD + metabolic DEGs, immune-related DEGs and CRC DEGs. (H) Gene Ontology (GO) enrichment pathway of candidate genes. (I) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment pathway of candidate genes.
Table 2.
The list of NAD + metabolism and immune-related differentially expressed genes (DEGs) in colorectal cancer (CRC).
| Symbol | logFC | AveExpr | t | P.Value | adj.P.Val | B |
|---|---|---|---|---|---|---|
| IGJ | −3.75E+00 | 9.94E+00 | −1.27E+01 | 4.30E-29 | 1.63E-26 | 5.54E+01 |
| DNASE1L3 | −2.65E+00 | 3.80E+00 | −1.10E+01 | 2.77E-23 | 3.79E-21 | 4.22E+01 |
| DMBT1 | −3.43E+00 | 9.94E+00 | −8.55E+00 | 1.10E-15 | 5.90E-14 | 2.50E+01 |
| ITLN1 | −3.76E+00 | 6.26E+00 | −8.11E+00 | 2.06E-14 | 9.72E-13 | 2.21E+01 |
| CLCA4 | −3.29E+00 | 5.07E+00 | −7.83E+00 | 1.30E-13 | 5.69E-12 | 2.03E+01 |
| PLAC8 | −1.88E+00 | 8.49E+00 | −7.83E+00 | 1.30E-13 | 5.70E-12 | 2.03E+01 |
| CLCA1 | −3.87E+00 | 7.65E+00 | −7.78E+00 | 1.83E-13 | 7.88E-12 | 2.00E+01 |
| FCGBP | −2.61E+00 | 1.18E+01 | −7.50E+00 | 1.08E-12 | 4.29E-11 | 1.82E+01 |
| MUC4 | −2.36E+00 | 9.27E+00 | −7.43E+00 | 1.59E-12 | 6.18E-11 | 1.79E+01 |
| PLA2G2A | −2.85E+00 | 9.29E+00 | −7.41E+00 | 1.85E-12 | 7.13E-11 | 1.77E+01 |
| B3GNT6 | −2.79E+00 | 4.87E+00 | −7.29E+00 | 3.88E-12 | 1.44E-10 | 1.70E+01 |
| HEPACAM2 | −2.40E+00 | 5.81E+00 | −6.93E+00 | 3.45E-11 | 1.09E-09 | 1.48E+01 |
| DUOXA2 | −2.42E+00 | 7.54E+00 | −6.66E+00 | 1.62E-10 | 4.75E-09 | 1.33E+01 |
| DHRS9 | −2.07E+00 | 6.38E+00 | −6.56E+00 | 3.02E-10 | 8.46E-09 | 1.27E+01 |
| SPINK4 | −2.77E+00 | 7.41E+00 | −6.51E+00 | 4.00E-10 | 1.10E-08 | 1.25E+01 |
| MUC2 | −3.03E+00 | 1.11E+01 | −6.45E+00 | 5.55E-10 | 1.50E-08 | 1.21E+01 |
| ZG16 | −2.77E+00 | 5.45E+00 | −6.38E+00 | 8.23E-10 | 2.17E-08 | 1.18E+01 |
| SLC6A14 | −2.05E+00 | 6.83E+00 | −6.34E+00 | 1.04E-09 | 2.71E-08 | 1.15E+01 |
| BEST2 | −1.96E+00 | 3.01E+00 | −6.31E+00 | 1.22E-09 | 3.14E-08 | 1.14E+01 |
| CA4 | −2.51E+00 | 4.44E+00 | −6.29E+00 | 1.35E-09 | 3.45E-08 | 1.13E+01 |
| CA1 | −2.48E+00 | 3.34E+00 | −6.07E+00 | 4.49E-09 | 1.05E-07 | 1.01E+01 |
| FAM55A | −2.07E+00 | 6.46E+00 | −6.03E+00 | 5.69E-09 | 1.32E-07 | 9.88E+00 |
| C6orf105 | −1.72E+00 | 6.49E+00 | −6.00E+00 | 6.64E-09 | 1.52E-07 | 9.73E+00 |
| SI | −2.25E+00 | 5.09E+00 | −5.92E+00 | 1.03E-08 | 2.25E-07 | 9.31E+00 |
| MS4A12 | −2.22E+00 | 4.04E+00 | −5.86E+00 | 1.43E-08 | 3.04E-07 | 8.99E+00 |
| PIGR | −2.58E+00 | 1.38E+01 | −5.86E+00 | 1.44E-08 | 3.06E-07 | 8.98E+00 |
| CEACAM7 | −1.91E+00 | 9.83E+00 | −5.47E+00 | 1.06E-07 | 1.85E-06 | 7.05E+00 |
| REG1B | −2.61E+00 | 4.79E+00 | −5.41E+00 | 1.43E-07 | 2.44E-06 | 6.76E+00 |
| REG1A | −2.85E+00 | 8.18E+00 | −5.38E+00 | 1.71E-07 | 2.89E-06 | 6.59E+00 |
| B4GALNT2 | −1.72E+00 | 2.99E+00 | −5.11E+00 | 6.37E-07 | 9.42E-06 | 5.32E+00 |
| CHGA | −1.81E+00 | 4.13E+00 | −4.78E+00 | 2.90E-06 | 3.58E-05 | 3.87E+00 |
| MOGAT2 | −1.19E+00 | 5.53E+00 | −4.30E+00 | 2.39E-05 | 2.26E-04 | 1.86E+00 |
| VSIG2 | −1.43E+00 | 6.73E+00 | −3.61E+00 | 3.67E-04 | 2.27E-03 | −7.18E-01 |
| SLC26A3 | −1.49E+00 | 9.28E+00 | −3.51E+00 | 5.34E-04 | 3.09E-03 | −1.07E+00 |
| FAM55D | −1.30E+00 | 7.02E+00 | −3.24E+00 | 1.36E-03 | 6.69E-03 | −1.93E+00 |
| COMP | 1.19E+00 | 5.63E+00 | 3.40E+00 | 7.84E-04 | 4.24E-03 | −1.43E+00 |
To investigate the functions of these 36 genes, we performed functional enrichment analysis. Table 3, Table 4 revealed 48 GO terms (21 BP terms, 11 CC terms, and 16 MF terms) and 3 KEGG pathways. We visualized the top 5 items for each GO category in a bubble diagram (Fig. 2H). GO analysis indicated that these genes are involved in immune-related processes, ion transport, digestion, and glycosylation (Fig. 2H–Table 3). KEGG pathway analysis revealed the involvement of these genes in “pancreatic secretion,” “nitrogen metabolism,” and “fat digestion and absorption” Fig. 2I–Table 4).
Table 3.
The Gene Ontology (GO) enrichment results of NAD + metabolism- and immune-related DEGs.
| ONTOLOGY | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID |
|---|---|---|---|---|---|---|---|---|
| BP | GO:0019730 | antimicrobial humoral response | 6/28 | 142/18862 | 5.38E-08 | 2.64E-05 | 2.05E-05 | CHGA/ITLN1/REG1A/PLA2G2A/REG1B/DMBT1 |
| BP | GO:1902476 | chloride transmembrane transport | 4/28 | 96/18862 | 1.17E-05 | 1.90E-03 | 1.48E-03 | CLCA4/BEST2/SLC26A3/CLCA1 |
| BP | GO:0050830 | defense response to Gram-positive bacterium | 4/28 | 98/18862 | 1.27E-05 | 1.90E-03 | 1.48E-03 | CHGA/ZG16/PLA2G2A/DMBT1 |
| BP | GO:0006959 | humoral immune response | 6/28 | 380/18862 | 1.66E-05 | 1.90E-03 | 1.48E-03 | CHGA/ITLN1/REG1A/PLA2G2A/REG1B/DMBT1 |
| BP | GO:0006821 | chloride transport | 4/28 | 109/18862 | 1.94E-05 | 1.90E-03 | 1.48E-03 | CLCA4/BEST2/SLC26A3/CLCA1 |
| BP | GO:0098661 | inorganic anion transmembrane transport | 4/28 | 120/18862 | 2.83E-05 | 2.31E-03 | 1.80E-03 | CLCA4/BEST2/SLC26A3/CLCA1 |
| BP | GO:0015701 | bicarbonate transport | 3/28 | 43/18862 | 3.47E-05 | 2.43E-03 | 1.89E-03 | CA1/CA4/SLC26A3 |
| BP | GO:0007586 | digestion | 4/28 | 138/18862 | 4.90E-05 | 3.00E-03 | 2.33E-03 | MOGAT2/SI/MUC4/MUC2 |
| BP | GO:0016266 | O-glycan processing | 3/28 | 61/18862 | 9.95E-05 | 5.42E-03 | 4.20E-03 | B3GNT6/MUC4/MUC2 |
| BP | GO:0015698 | inorganic anion transport | 4/28 | 171/18862 | 1.13E-04 | 5.52E-03 | 4.28E-03 | CLCA4/BEST2/SLC26A3/CLCA1 |
| BP | GO:0042742 | defense response to bacterium | 5/28 | 344/18862 | 1.36E-04 | 6.08E-03 | 4.71E-03 | CHGA/ZG16/PLAC8/PLA2G2A/DMBT1 |
| BP | GO:0015711 | organic anion transport | 5/28 | 376/18862 | 2.06E-04 | 8.43E-03 | 6.54E-03 | CA1/CA4/SLC26A3/SLC6A14/PLA2G2A |
| BP | GO:0022600 | digestive system process | 3/28 | 99/18862 | 4.18E-04 | 1.43E-02 | 1.11E-02 | MOGAT2/MUC4/MUC2 |
| BP | GO:0006486 | protein glycosylation | 4/28 | 250/18862 | 4.80E-04 | 1.43E-02 | 1.11E-02 | B4GALNT2/B3GNT6/MUC4/MUC2 |
| BP | GO:0043413 | macromolecule glycosylation | 4/28 | 250/18862 | 4.80E-04 | 1.43E-02 | 1.11E-02 | B4GALNT2/B3GNT6/MUC4/MUC2 |
| BP | GO:0030277 | maintenance of gastrointestinal epithelium | 2/28 | 22/18862 | 4.82E-04 | 1.43E-02 | 1.11E-02 | MUC4/MUC2 |
| BP | GO:0006493 | protein O-linked glycosylation | 3/28 | 105/18862 | 4.96E-04 | 1.43E-02 | 1.11E-02 | B3GNT6/MUC4/MUC2 |
| BP | GO:0070085 | glycosylation | 4/28 | 263/18862 | 5.81E-04 | 1.58E-02 | 1.23E-02 | B4GALNT2/B3GNT6/MUC4/MUC2 |
| BP | GO:0010669 | epithelial structure maintenance | 2/28 | 29/18862 | 8.42E-04 | 2.17E-02 | 1.68E-02 | MUC4/MUC2 |
| BP | GO:0006730 | one-carbon metabolic process | 2/28 | 40/18862 | 1.60E-03 | 3.79E-02 | 2.94E-02 | CA1/CA4 |
| BP | GO:0009101 | glycoprotein biosynthetic process | 4/28 | 347/18862 | 1.63E-03 | 3.79E-02 | 2.94E-02 | B4GALNT2/B3GNT6/MUC4/MUC2 |
| CC | GO:0042588 | zymogen granule | 4/32 | 13/19520 | 4.21E-09 | 3.28E-07 | 2.44E-07 | ZG16/CLCA1/REG1A/DMBT1 |
| CC | GO:0042589 | zymogen granule membrane | 3/32 | 10/19520 | 4.76E-07 | 1.86E-05 | 1.38E-05 | ZG16/CLCA1/DMBT1 |
| CC | GO:0005903 | brush border | 4/32 | 101/19520 | 2.17E-05 | 5.65E-04 | 4.19E-04 | CA4/SLC26A3/SI/ITLN1 |
| CC | GO:0045177 | apical part of cell | 6/32 | 414/19520 | 4.98E-05 | 9.72E-04 | 7.21E-04 | CA4/CLCA4/SLC26A3/CEACAM7/SI/DUOXA2 |
| CC | GO:0098862 | cluster of actin-based cell projections | 4/32 | 156/19520 | 1.19E-04 | 1.43E-03 | 1.06E-03 | CA4/SLC26A3/SI/ITLN1 |
| CC | GO:0031526 | brush border membrane | 3/32 | 59/19520 | 1.22E-04 | 1.43E-03 | 1.06E-03 | CA4/SLC26A3/ITLN1 |
| CC | GO:0030667 | secretory granule membrane | 5/32 | 305/19520 | 1.28E-04 | 1.43E-03 | 1.06E-03 | CA4/ZG16/CLCA1/PIGR/DMBT1 |
| CC | GO:0016324 | apical plasma membrane | 5/32 | 351/19520 | 2.47E-04 | 2.41E-03 | 1.79E-03 | CA4/CLCA4/SLC26A3/CEACAM7/SI |
| CC | GO:0005796 | Golgi lumen | 3/32 | 102/19520 | 6.15E-04 | 5.33E-03 | 3.96E-03 | ZG16/MUC4/MUC2 |
| CC | GO:0031253 | cell projection membrane | 4/32 | 337/19520 | 2.14E-03 | 1.67E-02 | 1.24E-02 | CA4/SLC26A3/ITLN1/REG1A |
| CC | GO:0031225 | anchored component of membrane | 3/32 | 170/19520 | 2.67E-03 | 1.90E-02 | 1.41E-02 | CA4/CEACAM7/ITLN1 |
| MF | GO:0070492 | oligosaccharide binding | 3/29 | 16/18337 | 1.96E-06 | 1.53E-04 | 8.12E-05 | ITLN1/REG1A/REG1B |
| MF | GO:0042834 | peptidoglycan binding | 3/29 | 18/18337 | 2.86E-06 | 1.53E-04 | 8.12E-05 | ZG16/REG1A/REG1B |
| MF | GO:0015108 | chloride transmembrane transporter activity | 4/29 | 101/18337 | 1.85E-05 | 6.61E-04 | 3.51E-04 | CLCA4/BEST2/SLC26A3/CLCA1 |
| MF | GO:0030246 | carbohydrate binding | 5/29 | 267/18337 | 5.62E-05 | 1.50E-03 | 7.99E-04 | ZG16/SI/ITLN1/REG1A/REG1B |
| MF | GO:0015103 | inorganic anion transmembrane transporter activity | 4/29 | 145/18337 | 7.64E-05 | 1.63E-03 | 8.68E-04 | CLCA4/BEST2/SLC26A3/CLCA1 |
| MF | GO:0005254 | chloride channel activity | 3/29 | 74/18337 | 2.14E-04 | 3.32E-03 | 1.76E-03 | CLCA4/BEST2/CLCA1 |
| MF | GO:0004089 | carbonate dehydratase activity | 2/29 | 14/18337 | 2.17E-04 | 3.32E-03 | 1.76E-03 | CA1/CA4 |
| MF | GO:0005229 | intracellular calcium activated chloride channel activity | 2/29 | 16/18337 | 2.86E-04 | 3.40E-03 | 1.81E-03 | CLCA4/CLCA1 |
| MF | GO:0061778 | intracellular chloride channel activity | 2/29 | 16/18337 | 2.86E-04 | 3.40E-03 | 1.81E-03 | CLCA4/CLCA1 |
| MF | GO:0005253 | anion channel activity | 3/29 | 86/18337 | 3.33E-04 | 3.57E-03 | 1.89E-03 | CLCA4/BEST2/CLCA1 |
| MF | GO:0005539 | glycosaminoglycan binding | 4/29 | 228/18337 | 4.33E-04 | 4.21E-03 | 2.24E-03 | ZG16/COMP/REG1A/REG1B |
| MF | GO:0008509 | anion transmembrane transporter activity | 5/29 | 459/18337 | 6.95E-04 | 6.20E-03 | 3.29E-03 | CLCA4/BEST2/SLC26A3/CLCA1/SLC6A14 |
| MF | GO:0022839 | ion gated channel activity | 2/29 | 42/18337 | 2.00E-03 | 1.65E-02 | 8.74E-03 | CLCA4/CLCA1 |
| MF | GO:0008376 | acetylgalactosaminyltransferase activity | 2/29 | 47/18337 | 2.50E-03 | 1.91E-02 | 1.01E-02 | B4GALNT2/B3GNT6 |
| MF | GO:0016836 | hydro-lyase activity | 2/29 | 64/18337 | 4.58E-03 | 3.27E-02 | 1.74E-02 | CA1/CA4 |
| MF | GO:0016835 | carbon-oxygen lyase activity | 2/29 | 79/18337 | 6.90E-03 | 4.61E-02 | 2.45E-02 | CA1/CA4 |
Table 4.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment results of NAD + metabolism- and immune-related DEGs.
| KEGG | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID |
|---|---|---|---|---|---|---|---|---|
| hsa04972 | Pancreatic secretion | 4.54E+04 | 102/8149 | 3.75E-05 | 1.13E-03 | 9.08E-04 | 22802/1811/1179/5320 | |
| hsa00910 | Nitrogen metabolism | 4.53E+04 | 17/8149 | 4.83E-04 | 7.25E-03 | 5.85E-03 | 759/762 | |
| hsa04975 | Fat digestion and absorption | 4.53E+04 | 43/8149 | 3.11E-03 | 3.11E-02 | 2.51E-02 | 80168/5320 |
3.3. The NAD + metabolism-immune-related prognostic signature for CRC
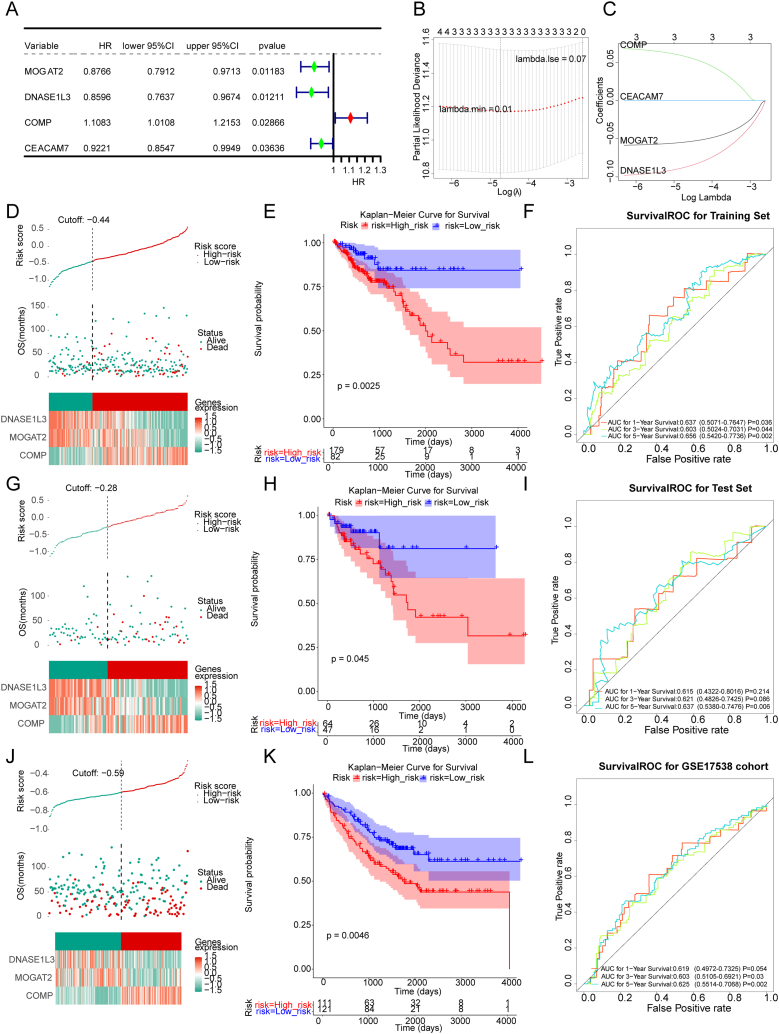
To identify NAD + metabolism-immune-related genes associated with the overall survival (OS) of CRC patients, we conducted univariate Cox analysis using the 36 genes in the training set. Four genes were found to be significantly associated with OS in CRC patients (p value < 0.05). Among these genes, COMP was identified as a risk factor for CRC prognosis (hazard ratio (HR) > 1), whereas MOGAT2, DNASE1L3, and CEACAM7 were protective factors (HR < 1) (Fig. 3A). The four genes were further analyzed using LASSO analysis. Fig. 3B and C demonstrates that when the lambda min was 0.01, the optimal number of genes determined was three. Consequently, MOGAT2, DNASE1L3, and COMP were selected as the three optimal NAD + metabolism-immune-relevant prognostic genes for establishing a prognostic signature. Subsequently, we derived a prognostic signature using the following formula: .
Fig. 3.
The NAD + metabolism-immune-relevant prognostic signature for CRC. (A) Univariate forest plot of the correlation between prognostic gene expression and overall survival (OS) in CRC patients. (B) Deviance plot of partial likelihood determined by least absolute shrinkage and selection operator (LASSO) Cox regression analysis. (C) Graph of the gene coefficients. The abscissa is log(Lambda), and the ordinate is the coefficient corresponding to the gene. Risk curve, scatter plot, model gene expression heatmap, Kaplan‒Meier (K–M) survival analysis and receiver operating characteristic (ROC) curve of patients with high or low risk in different sets. (D–F) Training set. (G–I) Test set. (J–L) External verification set.
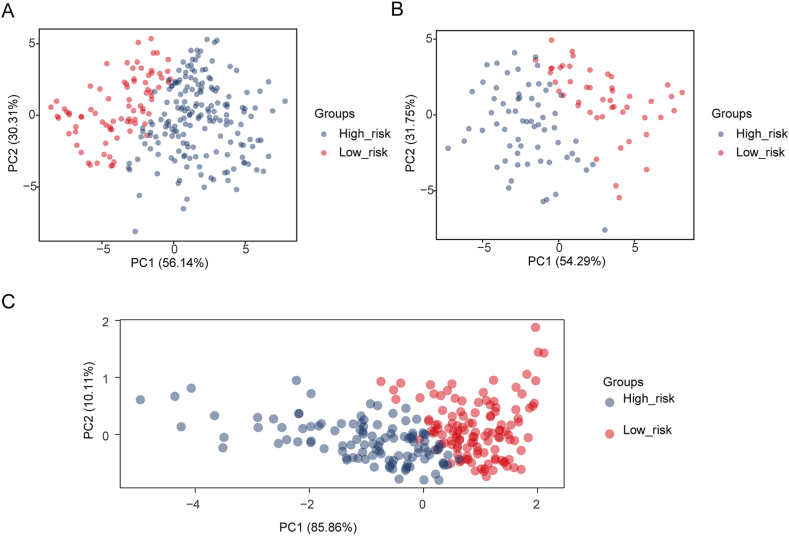
. Using this formula, we computed the risk score for each CRC patient in the training set and classified them into two risk subgroups (high-risk and low-risk) using the optimal cutoff value (Fig. 3D). Principal component analysis (PCA) revealed a clear distinction between the high- and low-risk groups based on the expression of the three prognostic genes (Supplementary Fig. 1A). The Kaplan‒Meier curve demonstrated that patients at greater risk had significantly poorer survival than those at lower risk (Fig. 3E). ROC curves were plotted to assess the predictive efficiency of the signature. The area under the curve (AUC) values for OS in the training set were all greater than 0.6 (1-, 3-, and 5-year), indicating decent accuracy (Fig. 3F). Fig. 3D displays the distribution of the ranked risk score and survival status for each patient in the training set. Survival status revealed that as the risk score increased, patients had a relatively greater risk of death. The expression heatmap showed that COMP was highly expressed in patients with higher risk scores, whereas DNASE1L3 and MOGAT2 were highly expressed in patients with lower risk scores (Fig. 3D). To further validate the applicability and reliability of the risk signature, the above analysis was also conducted in the testing set and external validation set (GSE17538). A consistent trend was observed in the testing set and external validation set (Fig. 3G-L, Supplementary Fig. 1B-C. These results indicate that the NAD + metabolism-immune-related prognostic signature serves as a valid survival predictor for CRC patients.
3.4. Relativity analysis of clinical parameters and the NAD + metabolism-immune-relevant prognostic signature
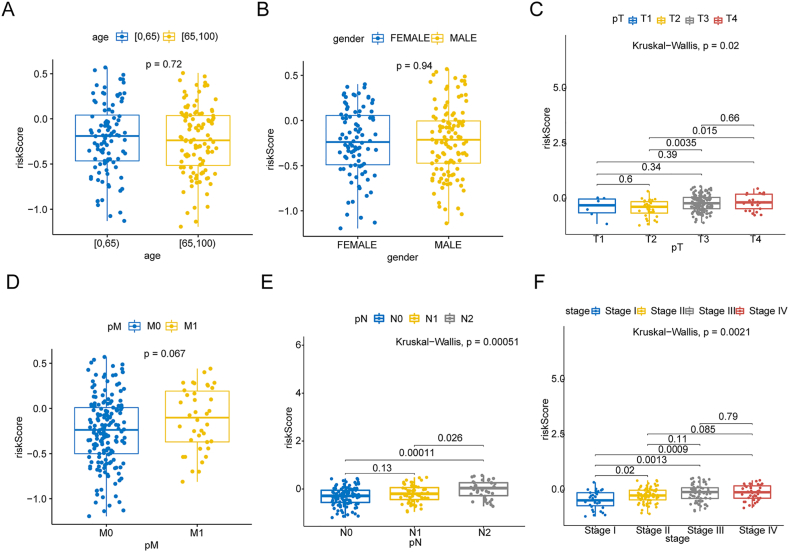
To explore the associations between the NAD + metabolism-immune-related prognostic signature and clinical factors, we compared the risk scores of patients in different clinical subgroups. As shown in Fig. 4A-F, the NAD + metabolism-immune-related risk score was correlated with pathologic T stage, pathologic N stage, and stage (p value < 0.05) but was not correlated with age, sex, or pathologic M stage.
Fig. 4.
Risk score analysis of different clinicopathological features. The distribution of risk scores between high- and low-risk patients was stratified according to (A) age, (B) sex, (C) pathological T stage, (D) pathological M stage, (E) pathological N stage and (F) stage.
3.5. The nomogram containing the NAD + metabolism-immune-relevant risk score as an independent prognostic predictor
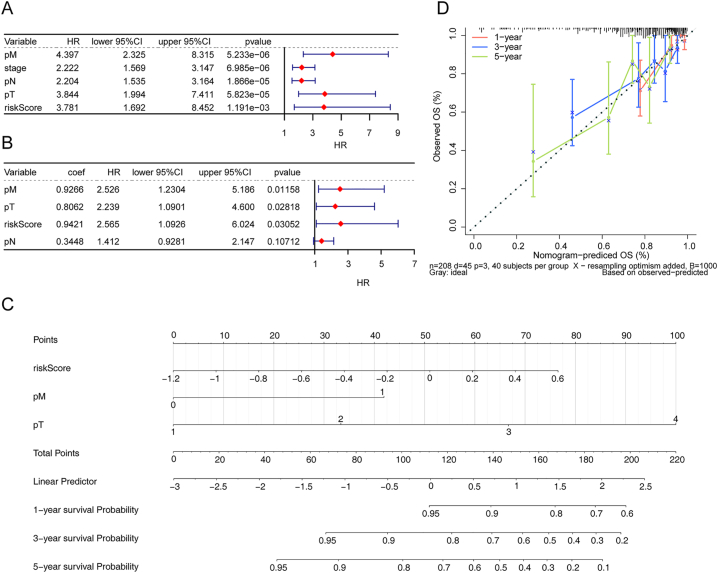
Cox analysis (univariate Cox and multivariate Cox) revealed that the risk score, pathologic T stage, and pathologic M stage were independent predictors of prognosis for CRC patients Fig. 5A-B, p value < 0.05). A nomogram incorporating these independent prognostic predictors was generated Fig. 5C). The C-index of the nomogram was 0.7582962, and calibration curves demonstrated the satisfactory performance of the nomogram in predicting survival at 1, 3, and 5 years for CRC patients (Fig. 5D).
Fig. 5.
TCGA-CRC independent prognosis forest map. (A) Univariate Cox analyses. The red diamond squares on the transverse lines indicate the hazard ratio (HR), and the blue transverse lines indicate the 95 % confidence interval (CI). (B) Multivariate Cox analyses. (C) Independent prognosis model nomogram. Nomograms that integrate the prediction of the probability of patient survival for 1-, 3- or 5-year OS. (D) Calibration curves of the nomogram for predicting survival outcomes at 1, 3, and 5 years. The 45-degree line represents the ideal prediction.
3.6. Association of the NAD + metabolism-immune-related prognostic signature with the immune microenvironment
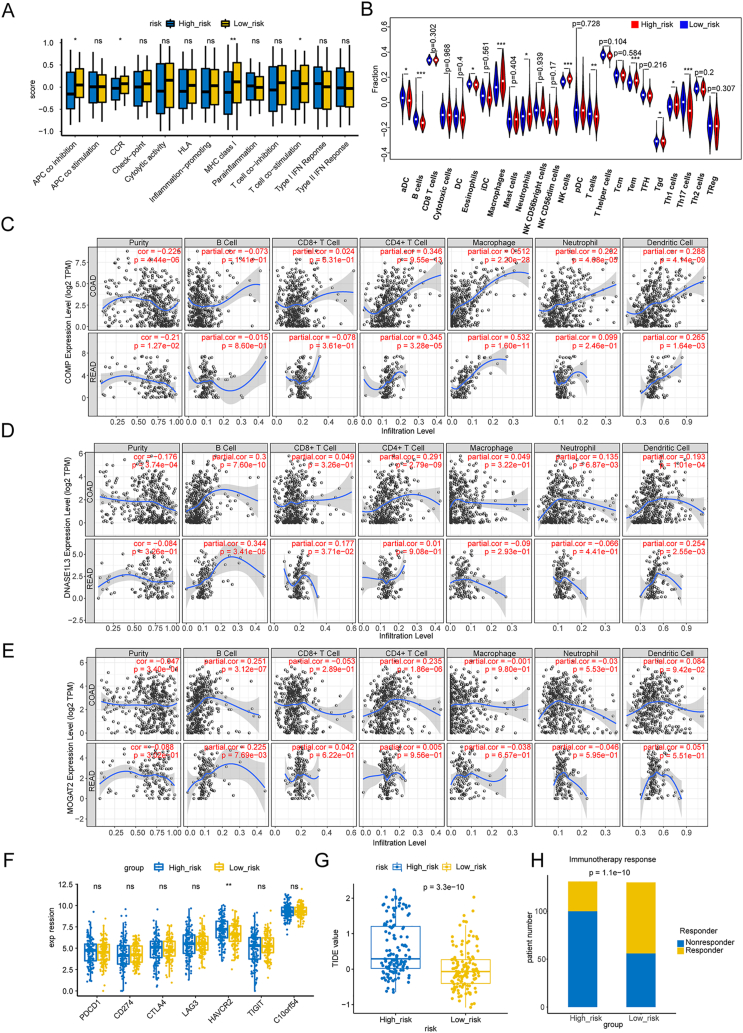
The GSVA algorithm demonstrated that patients in the low-risk subgroup had higher scores for immune-related pathways, such as APC coinhibition, CCR, MHC class I, and T-cell costimulation Fig. 6A). Using the ssGSEA algorithm, we calculated the fraction of each immune infiltration cell for samples in both risk subgroups. As shown in Fig. 6B, the fractions of macrophages, neutrophils, NK cells, Tems, Tgds, and Th1 cells were increased in patients with higher risk scores, whereas the fractions of aDCs, B cells, eosinophils, T cells, and Th17 cells were increased in patients with lower risk scores. To further investigate the associations between prognostic genes and immune cells in CRC, we utilized the TIMER database Fig. 6C–E). Based on thresholds of |cor| > 0.3 and p < 0.05, we found that COMP was notably positively correlated with CD4+ T cells and macrophages Fig. 6C), whereas DNASE1L3 was significantly positively correlated with B cells (Fig. 6D).
Fig. 6.
Association of the NAD + metabolism-immune-related prognostic signature with the immune microenvironment. (A) Differences in 13 immune-related pathways in the high-to low-risk groups. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant. (B) Differences in the levels of 24 kinds of tumor-infiltrating immune cells (TIICs). (C–E) Correlations between gene expression and immune cells. The expression of COMP (C), DNASE1L3 (D), and MOGAT2 (E) correlated with that of six immune cell types. (F) Comparison of immune checkpoint expression between the high- and low-risk groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, not significant. (G–H) Analysis of tumor immune dysfunction and exclusion (TIDE) scores in the high- and low-risk groups. (G) Comparison of the results of the rank sum test of the TIDE score. (H) Is the number of people in the high-low-expression group who responded to immunotherapy and those who did not respond. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.7. Association of the NAD + metabolism-immune-related prognostic signature with therapy
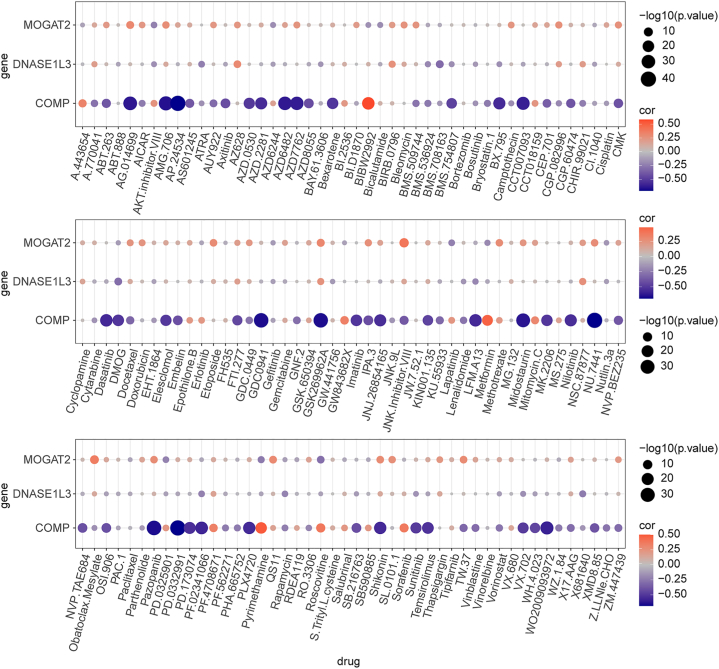
The immune checkpoint molecule HAVCR2 was found to be expressed at higher levels in patients with higher risk scores Fig. 6F). According to the TIDE results, patients in the low-risk group might respond better to immunotherapy than patients in the high-risk group Fig. 6G and H). We also investigated the correlation between prognostic genes and sensitivity to chemotherapy drugs (IC50 values) using the ‘pRRophetic’ package. We observed that COMP was associated with multiple drugs, including bryostatin.1, camptothecin, cytarabine, dasatinib, pazopanib, and shikonin (Fig. 7).
Fig. 7.
Association of the NAD + metabolism-immune-related prognostic signature with therapy The red dots are positively correlated, and the blue dots are negatively correlated.
3.8. The expression of NAD + metabolism-immune-related prognostic genes
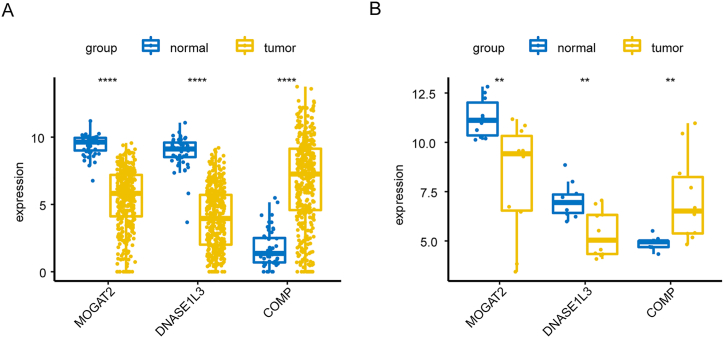
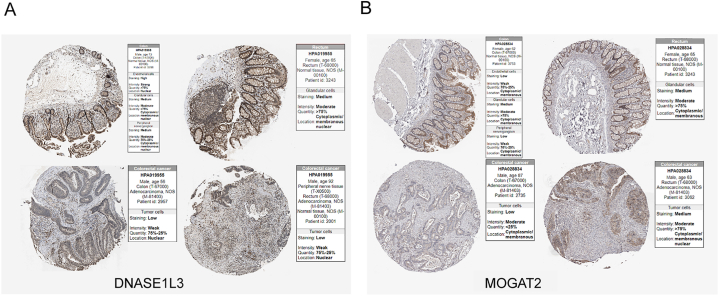
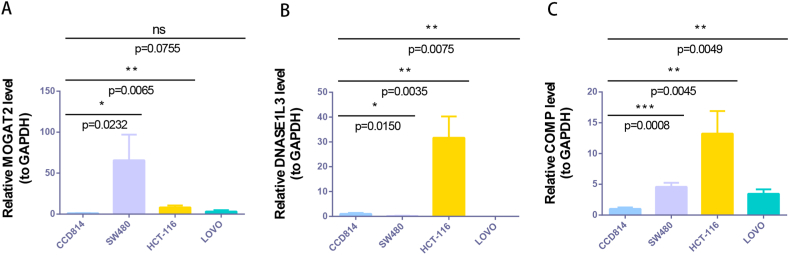
In the TCGA-CRC dataset, MOGAT2 and DNASE1L3 were downregulated, whereas COMP showed elevated expression in CRC tissues compared to normal tissues Fig. 8A). We further confirmed these expression trends in the external validation set GSE115261 Fig. 8B). To determine the changes in the expression of prognostic genes at the protein level, we obtained immunohistochemistry images from the HPA database. Unfortunately, we did not detect immunohistochemical results for COMP in CRC. However, we found that the protein expression of DNASE1L3 and MOGAT2 was lower in CRC tissues than in normal tissues Fig. 9A and B). We also analyzed the mRNA expression of prognostic genes in human normal colonic epithelial cells (CCD814) and three human CRC cell lines (HCT-116, LOVO, and SW480). Consistent with the results from public databases, COMP was upregulated in HCT-116, LOVO, and SW480 cells, whereas DNASE1L3 was downregulated in LOVO and SW480 cells (Fig. 10 A-C). However, the expression of MOGAT2 in the cell lines was inconsistent with that in the tissue samples, possibly due to the complexity of the tumor tissue.
Fig. 8.
Expression levels of prognostic genes in the TCGA and validation sets GSE115261. (A) TCGA-CRC. (B) GSE115261. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 9.
Protein expression levels of prognostic characteristic genes in the Human Protein Atlas (HPA) database. (A) DNASE1L3. (B) MOGAT2.
Fig. 10.
mRNA expression levels of NAD + metabolic immune-related prognostic genes in CCD814, HCT-116, LOVO and SW480 cells. (A) MOGAT2. (B) DNASE1L3. (C) COMP. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, not significant.
4. Discussion
CRC is the third most prevalent malignant tumor globally, accounting for approximately one-tenth of all cancer patients, with an estimated one million new cases reported annually worldwide. It is also a prominent contributor to cancer-related deaths, accounting for 9.2 % of deaths [23]. The primary cause of death in CRC patients is the occurrence of distant metastasis. Patients without distant metastasis exhibit a 5-year survival rate of 80–90 %, whereas those with distant metastasis have a significantly lower rate of 10–20 %. During tumor cell proliferation, a substantial energy supply is needed, which mainly involves the production of mitochondrial acetyl-CoA from glucose. This raw material is then subjected to oxidation reactions via the citric acid cycle under the influence of NAD+/NADH, resulting in ATP production. These metabolic processes provide energy for tumor cell proliferation [24]. NAD + metabolism is also involved in the regulation of diverse biological functions, including immune regulation and aging [18,25]. Immune T cells have been found to be closely associated with the occurrence, development, metastasis, and invasion of tumor cells as immune recognition improves. NAD+ is capable of regulating the expression of the tumor immune checkpoint PD-L1 through epigenetic modification. This, in turn, promotes the secretion of IFNγ by T cells and facilitates tumor immune evasion. Sirt1, an NAD + metabolic enzyme, can activate cytokine secretion in antigen-presenting cells by regulating the acetylation of IRF1. Consequently, PD-L1 expression is promoted, and tumor immunity is mediated [10,26]. The immune microenvironment and immune components significantly impact the prognosis of cancer patients during the adaptive response to antitumor therapy [27]. However, the precise mechanisms through which NAD + metabolism regulates immune function in CRC remain unclear. Hence, we established a robust prognostic model for CRC based on NAD + metabolism and immune-related genes. This was accomplished through thorough bioinformatics analysis, immune checkpoint analysis, immune therapy response prediction, immune infiltration analysis, chemotherapy drug sensitivity analysis, and qRT‒PCR validation. Our objective was to investigate the mechanisms underlying the association between NAD + metabolism and immune function in CRC.
First, a total of 36 DEGs related to NAD + metabolism and immunity were identified in CRC through differential expression analysis. These DEGs included CA1, MS4A12, CA4, CHGA, and CLCA4, among others. In CRC, the downregulation of CA1 and upregulation of ANXA4 promote the accumulation of ANXA4 on the cell membrane. Furthermore, CA1 expression is associated with the malignant proliferation, development, differentiation, and metastasis of cancer cells [28,29]. MS4A12 is a specific storage-operated calcium channel (SOC) in the colon that facilitates Ca2+ influx through epidermal growth factor (EGF). This promotes chloride nitrogen metabolism, cell proliferation, and cell motility but significantly impairs chemotactic invasion [30,31]. The CLCA4 protein is expressed in secretory epithelial cells of the small intestine and affects chloride ion transmission, cell apoptosis, cell cycle control, and tumor metastasis [32].
Three genes related to NAD + metabolism and immunity were subsequently identified as prognostic genes. This was determined through univariate Cox and LASSO regression analyses. The identified genes were MOGAT2, DNASE1L3, and COMP. MOGAT2 is a gene involved in fatty acid synthesis and is predominantly expressed in the human small intestine, liver, and white fat. It serves as a key rate-limiting enzyme for triglyceride synthesis in the small intestine. A study by Valerie [5] demonstrated that increased expression of adipogenic genes, including MOGAT2, promotes adipogenesis, proliferation, and the inflammatory response in hepatocellular carcinoma (HCC) mice, leading to liver proliferation and steatosis. Additionally, MOGAT2 has been identified as a gene with double methylation in inflammatory breast cancer [33]. This study provides the first evidence that MOGAT2 is a relevant prognostic gene in CRC.
Recombinant deoxyribonuclease I-like protein 3 (DNASE1L3) is a highly expressed secreted enzyme that plays a crucial role in regulating DNA and chromatin autoimmunity. It has been observed that DNASE1L3 can degrade the extracellular chromatin of apoptotic bodies. Studies have shown that mutations in DNASE1L3 are closely associated with the phenotype of lupus [34]. Moreover, there is evidence linking DNASE1L3 deficiency to inflammatory bowel disease [34].
Recent research has suggested that DNASE1L3 is involved in cell apoptosis, proliferation, invasion, and metastasis. It has emerged as a potential prognostic biomarker in lung adenocarcinoma and colon cancer studies, although its specific mechanism of action remains unclear [35,36]. Notably, Han's study [37] highlighted the regulatory effect of DNASE1L3 in HCC. DNASE1L3 is expressed in the endoplasmic reticulum, and it is secreted from the cell. Following apoptosis induction, its endoplasmic reticulum-targeted gene sequence is cleaved and translocated to the nucleus. DNASE1L3 also acts as a plasma nuclease in the digestive cycle, aiding in the evasion of phagocytosis after apoptosis.
In the context of HCC, Li [38] discovered that DNASE1L3 negatively regulates disease. By binding to β-chain proteins, DNASE1L3 inhibits their nuclear translocation, thereby restraining the proliferation and metastasis of HCC cells. Mechanistically, DNASE1L3 promotes β-ubiquitination-related degradation of catenin and disrupts the recruitment of the β-catenin disruption complex. Consequently, downstream targets such as c-Myc, P21, and P27 are inhibited, effectively controlling the cell cycle and EMT signaling. Research by SUN [39] further supports these findings by demonstrating that DNASE1L3 significantly reduces HCC cell proliferation, colony formation, migration, and invasion in vitro. It also inhibited the formation of subcutaneous tumors in nude mice in vivo. In a mouse model, overexpression of DNASE1L3 inhibited AKT/NRASV12-induced liver cancer, whereas DNASE1L3 deficiency exacerbated DEN/CCl-induced liver cancer in DNASE1L3 mice. Systemic analysis revealed that DNASE1L3 impairs HCC cell cycle progression by interacting with CDK2 and inhibiting CDK2-stimulated E2F1 activity. C-terminal deletion of DNASE1L3 reduces its interaction with CDK2 and eliminates its inhibitory effect on HCC.
Furthermore, Li [40] established a mouse model of colon cancer and demonstrated that the absence of DNASE1L3 in the tumor microenvironment is associated with tumor occurrence and development. Additionally, it enhances antitumor immunity.
Cartilage oligomeric matrix protein (COMP) is an extracellular matrix protein that plays a crucial role in cellular phenotype regulation during histogenesis and remodeling. Recent research has focused on investigating the involvement of COMP in various diseases, such as liver fibrosis and pulmonary fibrosis [41,42]. In the context of colon cancer-related diseases, experimental studies have confirmed that the overexpression of COMP is associated with the carcinogenic effects of colon cancer, as well as with the infiltration of CAFs and M2 macrophages [43]. In liver malignancies, Li [44] discovered that the COMP/CD36 signaling pathway leads to the phosphorylation of ERK and AKT, resulting in the upregulation of markers associated with EMT, including MMP-2/9, Slug, and Twist, in HCC cells. This, in turn, influences tumor development. Furthermore, Blom's research [45] revealed a positive correlation between COMP expression and TNM stage and tumor differentiation. Additionally, a negative correlation has been identified between COMP expression and the presence of PD-L1 on tumor cells and immune cells. Tumor fibrosis is associated with high levels of COMP expression in the extracellular matrix, and tumors with dense fibrosis and elevated COMP expression exhibit less infiltration of immune cells. Moreover, Dakhova [46] demonstrated that knockdown of the COMP gene affects the expression of genes associated with the reactive matrix in prostate stromal cell lines, leading to a decrease in tumor cell development. Our study indicated that CRC patients with elevated COMP expression have significantly shorter OS than those with low COMP expression. Furthermore, we observed reduced T-cell infiltration in tumors with high COMP expression. Notably, we also conducted qRT‒PCR experiments to validate the expression of MOGAT2, DNASE1L3, and COMP in CRC cells, confirming their significant difference in expression between human colon cancer cells (HCT116 and SW480) and normal colon epithelial cells (CCD814). However, no difference was observed in LOVO cells. These findings suggest that MOGAT2, DNASE1L3, and COMP may play a role in cell apoptosis, proliferation, invasion, and metastasis in CRC, providing potential clinical prognostic markers for this disease.
In addition, we observed significant differences in 11 types of immune cells (aDCs, B cells, eosinophils, macrophages, neutrophils, NK cells, T cells, Tem cells, Tgd cells, Th1 cells, and Th17 cells) between the CRC and control groups. We also identified four immune pathways (APC coinhibition, CCR, MHC class I, and T-cell costimulation pathways) that exhibited significant differences. Hence, it is reasonable to speculate that MOGAT2, DNASE1L3, and COMP may exert their effects on these 11 immune cells through the four immune signaling pathways mentioned above, ultimately regulating cell apoptosis, proliferation, invasion, and metastasis in CRC.
By comparing the rank sum test results of TIDE scores, we obtained correlations between MOGAT2, DNASE1L3, and COMP model genes and IC50. Among them, we found that the correlations between COMP and AG.014699, AMG.706, AP.24534, AZD6482, AZD7762, CCT007093, GDC0941, GSK26962A, midostaurin, NU.7441, pazopanib, and PD.0332991 were less than −0.6, indicating a significant strong negative correlation. Based on the results of the immunotherapy and chemotherapy sensitivity analyses, MOGAT2, DNASE1L3, and COMP inhibited the occurrence and development of CRC. Therefore, these three prognostic genes play a crucial role in the clinical treatment of patients. Interfering with MOGAT2, DNASE1L3, and COMP can regulate the development of tumors and improve patient quality of life. Additionally, this study provides new insights for the development of COMP-related chemotherapy drugs for the clinical treatment of CRC.
This study has several notable advantages. First, we employed various analytical methods, including differential gene expression analysis, enrichment analysis, Cox analysis, and the LASSO algorithm, and validated the results using multiple datasets to ensure accuracy and reliability. Second, we established and clinically validated a prognostic model, highlighting the significance and effectiveness of the model genes in determining CRC prognosis. Finally, by analyzing immune cells and pathways, we revealed immune mechanisms relevant to CRC prognosis, offering new perspectives for personalized treatment. Overall, this study excelled in methodology, results, and clinical applications, providing valuable implications for the treatment and prognosis improvement of CRC patients. However, there are several limitations to consider. First, the number of clinical samples collected in our study was limited, and future validation will require a larger sample size. Second, additional in vitro experiments were not conducted to validate the potential mechanisms of CRC identified in this study, necessitating further experimental validation of the study's findings.
5. Conclusion
In summary, this study revealed that NAD + metabolism and immune-related genes (MOGAT2, DNASE1L3, and COMP) are associated with CRC prognosis. Additionally, a prognostic signature based on NAD + metabolism and immune-related factors was developed for stratifying the survival risk of CRC patients. These findings provide valuable insights for future investigations on NAD + metabolism, immune-related prognostic characteristics, and the potential to minimize unnecessary steps and economic losses in experimental research. Moving forward, further exploration of the underlying mechanisms involving the three identified genes (MOGAT2, DNASE1L3, and COMP) in the progression of CRC is warranted. However, it is important to note that this study is retrospective and relies on public database data. To effectively apply the analysis results and prognostic models, more clinical samples and extensive data support and validation are necessary. Moreover, additional experimental research is required to fully understand the mechanisms of action of the identified genes related to NAD + metabolism and immune-related prognosis.
Ethics approval
This study was conducted in accordance with the World Medical Association Declaration of Helsinki. The data from TCGA and GEO are all accessible to the public; the current research followed the data access policies and publishing guidelines of TCGA and GEO. This study was reviewed and deemed exempt by our local ethics committee.
Consent to participate
The data was obtained through public database, which has been widely agreed, all personal information had already been anonymized to protect the individuals' privacy. For these reasons, the study protocol was exempt from the requirement to obtain informed consent from the participants.
Funding
This work was supported by (1) Guiyang Science and Technology Bureau, the First Affiliated Hospital of Guizhou University of Traditional Chinese Medicine, Great Health Science and Technology Cooperation Project (Zhuke Contract [2019] 9-2-8); (2) Research Project of Traditional Chinese Medicine and Ethnic Medicine Science and Technology of Guizhou Provincial Administration of Traditional Chinese Medicine (No. QZYY-2019-18); (3) National Natural Science Foundation of China Post-subsidy Funds for Scientific Research and Innovation Exploration Special Project (No. 2019YFC171250403); (4) Scientific Research Start-up Fund Project of Guizhou University of Traditional Chinese Medicine (GuiZhongYiKeYuanNei No. [2019]19); (5) Doctoral Startup Fund of the First Affiliated Hospital of Guizhou University of Traditional Chinese Medicine (No. GYZYYFY-BS-2018(10) and No. GYZYYFY-BS-2019(03)); (6) National Innovation and Entrepreneurship Training Program for College Students (GuiZhongYiDaChuangHeZi (2022) No.1).
Data availability statement
The datasets used in this study were available in online repositories. Here were the URLs for the databases used in this research. The datasets used and analyzed in the current study are available from the UCSC database (https://xenabrowser.net), GEO database (https://www.ncbi.nlm.nih.gov/gds), KEGG database (https://www.genome.jp/kegg/pathway.html), Reactome database (https://reactome.org/) and ImmPort database (https://immport.niaid.nih.gov), TIMER database (https://cistrome.shinyapps.io/timer/), Jvenn website (http://jvenn.toulouse.inra.fr/app/example.html).
CRediT authorship contribution statement
Tao Ye: Writing – review & editing, Writing – original draft, Resources, Funding acquisition, Data curation, Conceptualization. Hong Huang: Writing – review & editing, Writing – original draft, Data curation, Conceptualization. Kangli Chen: Software, Methodology. Yuanao Yu: Software, Methodology. Dongqin Yue: Validation. Li Jiang: Validation. Huixian Wu: Validation. Ning Zhang: Visualization, Supervision, Resources, Project administration, Methodology, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
All authors would like to thank the reviewers for their helpful comments on this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e34403.
Abbreviations
- CRC
colorectal cancer
- NAD+
nicotinamide adenine dinucleotide
- LASSO
least absolute shrinkage and selection operator
- GSVA
gene set variation analysis
- ssGSEA
single sample gene set enrichment analysis
- TIDE
tumor immune dysfunction and exclusion
- HPA
Human Protein Atlas
- TCGA-CRC
The Cancer Genome Atlas-Colorectal Cancer
- GEO
Gene Expression Omnibus
- NMRGs
NAD + metabolism-related genes
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- IRGs
immune-related genes
- DEGs
differentially expressed genes
- GO
Gene Ontology
- CC
cellular component
- MF
molecular function
- BP
biological process
- ROC
receiver operating characteristic
- K‒M
Kaplan‒Meier
- ICI
immune checkpoint inhibitor
- IC50
half maximal inhibitory concentration
- OS
overall survival
- HR
hazard ratio
- AUC
area under curve
- PCA
principal component analysis
- SOCs
storage-operated calcium channels
- EGF
epidermal growth factor
- MOGAT2
monoacylglycerol O-acyltransferase 2
- HCC
hepatocellular carcinoma
- DNASE1L3
deoxyribonuclease I-like protein 3
- COMP
cartilage oligomeric matrix protein
- CI
confidence interval
- CDF
cumulative distribution function
- SE
standard.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
Fig. S1.
PCA results for the training set, test set, and external validation set. (A) The training set focused on PCA results from two groups of patients in the high-to low-risk group. (B) The test set focused on PCA results from two groups of patients in the high-to low-risk group. (C) The external validation set focused on PCA results from two groups of patients in the high-to low-risk group.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Fleming M., Ravula S., Tatishchev S.F., Wang H.L. Colorectal carcinoma: pathologic aspects. J. Gastrointest. Oncol. 2012;3(3):153–173. doi: 10.3978/j.issn.2078-6891.2012.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buccafusca G., Proserpio I., Tralongo A.C., Rametta Giuliano S., Tralongo P. Early colorectal cancer: diagnosis, treatment and survivorship care. Crit. Rev. Oncol. Hematol. 2019;136:20–30. doi: 10.1016/j.critrevonc.2019.01.023. [DOI] [PubMed] [Google Scholar]
- 4.Sveen A., Kopetz S., Lothe R.A. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat. Rev. Clin. Oncol. 2020;17(1):11–32. doi: 10.1038/s41571-019-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanc V., Riordan J.D., Soleymanjahi S., Nadeau J.H., Nalbantoglu I., Xie Y., Molitor E.A., Madison B.B., Brunt E.M., Mills J.C., Rubin D.C., Ng I.O., Ha Y., Roberts L.R., Davidson N.O. Apobec1 complementation factor overexpression promotes hepatic steatosis, fibrosis, and hepatocellular cancer. J. Clin. Invest. 2021;131(1) doi: 10.1172/jci138699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covarrubias A.J., Perrone R., Grozio A., Verdin E. NAD(+) metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021;22(2):119–141. doi: 10.1038/s41580-020-00313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X., Liu Y., Liu Z., Lin C., Meng F., Xu L., Zhang X., Zhang C., Zhang P., Gong S., Wu N., Ren Z., Song J., Zhang Y. CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner. Mol. Cancer. 2021;20(1):114. doi: 10.1186/s12943-021-01412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy J.P., Giacomantonio M.A., Paulo J.A., Everley R.A., Kennedy B.E., Pathak G.P., Clements D.R., Kim Y., Dai C., Sharif T., Gygi S.P., Gujar S. The NAD(+) salvage pathway supports PHGDH-driven serine biosynthesis. Cell Rep. 2018;24(9) doi: 10.1016/j.celrep.2018.07.086. 2381-91.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lv H., Lv G., Chen C., Zong Q., Jiang G., Ye D., Cui X., He Y., Xiang W., Han Q., Tang L., Yang W., Wang H. NAD(+) metabolism maintains inducible PD-L1 expression to drive tumor immune evasion. Cell Metabol. 2021;33(1) doi: 10.1016/j.cmet.2020.10.021. 110-27.e5. [DOI] [PubMed] [Google Scholar]
- 10.Bruni D., Angell H.K., Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer. 2020;20(11):662–680. doi: 10.1038/s41568-020-0285-7. [DOI] [PubMed] [Google Scholar]
- 11.Jung G., Hernández-Illán E., Moreira L., Balaguer F., Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020;17(2):111–130. doi: 10.1038/s41575-019-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llosa N.J., Cruise M., Tam A., Wicks E.C., Hechenbleikner E.M., Taube J.M., Blosser R.L., Fan H., Wang H., Luber B.S., Zhang M., Papadopoulos N., Kinzler K.W., Vogelstein B., Sears C.L., Anders R.A., Pardoll D.M., Housseau F. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43–51. doi: 10.1158/2159-8290.Cd-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2(3) doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., Angell H., Fredriksen T., Lafontaine L., Berger A., Bruneval P., Fridman W.H., Becker C., Pagès F., Speicher M.R., Trajanoski Z., Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Chiarugi A., Dölle C., Felici R., Ziegler M. The NAD metabolome--a key determinant of cancer cell biology. Nat. Rev. Cancer. 2012;12(11):741–752. doi: 10.1038/nrc3340. [DOI] [PubMed] [Google Scholar]
- 19.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., Liu J., Freeman G.J., Brown M.A., Wucherpfennig K.W., Liu X.S. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Bardou P., Mariette J., Escudié F., Djemiel C., Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinf. 2014;15(1):293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trosko J.E. On the potential origin and characteristics of cancer stem cells. Carcinogenesis. 2021;42(7):905–912. doi: 10.1093/carcin/bgab042. [DOI] [PubMed] [Google Scholar]
- 25.Garten A., Schuster S., Penke M., Gorski T., de Giorgis T., Kiess W. Physiological and pathophysiological roles of NAMPT and NAD metabolism. Nat. Rev. Endocrinol. 2015;11(9):535–546. doi: 10.1038/nrendo.2015.117. [DOI] [PubMed] [Google Scholar]
- 26.Yang H., Lee S.M., Gao B., Zhang J., Fang D. Histone deacetylase sirtuin 1 deacetylates IRF1 protein and programs dendritic cells to control Th17 protein differentiation during autoimmune inflammation. J. Biol. Chem. 2013;288(52):37256–37266. doi: 10.1074/jbc.M113.527531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pagès C., Tosolini M., Camus M., Berger A., Wind P., Zinzindohoué F., Bruneval P., Cugnenc P.H., Trajanoski Z., Fridman W.H., Pagès F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 28.Yuan S., Wang P., Zhou X., Xu J., Lu S., Chen Y., Zhang Y. Differential proteomics mass spectrometry of melanosis coli. Am J Transl Res. 2020;12(7):3133–3148. [PMC free article] [PubMed] [Google Scholar]
- 29.Peng Y., Zhang Z., Zhang A., Liu C., Sun Y., Peng Z., Liu Y. Membrane-cytoplasm translocation of annexin A4 is involved in the metastasis of colorectal carcinoma. Aging (Albany NY) 2021;13(7):10312–10325. doi: 10.18632/aging.202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koslowski M., Sahin U., Dhaene K., Huber C., Türeci O. MS4A12 is a colon-selective store-operated calcium channel promoting malignant cell processes. Cancer Res. 2008;68(9):3458–3466. doi: 10.1158/0008-5472.Can-07-5768. [DOI] [PubMed] [Google Scholar]
- 31.Koslowski M., Türeci O., Huber C., Sahin U. Selective activation of tumor growth-promoting Ca2+ channel MS4A12 in colon cancer by caudal type homeobox transcription factor CDX2. Mol. Cancer. 2009;8:77. doi: 10.1186/1476-4598-8-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loewen M.E., Forsyth G.W. Structure and function of CLCA proteins. Physiol. Rev. 2005;85(3):1061–1092. doi: 10.1152/physrev.00016.2004. [DOI] [PubMed] [Google Scholar]
- 33.Van der Auwera I., Yu W., Suo L., Van Neste L., van Dam P., Van Marck E.A., Pauwels P., Vermeulen P.B., Dirix L.Y., Van Laere S.J. Array-based DNA methylation profiling for breast cancer subtype discrimination. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tusseau M., Lovšin E., Samaille C., Pescarmona R., Mathieu A.L., Maggio M.C., Selmanović V., Debeljak M., Dachy A., Novljan G., Janin A., Januel L., Gibier J.B., Chopin E., Rouvet I., Goncalves D., Fabien N., Rice G.I., Lesca G., Labalme A., Romagnani P., Walzer T., Viel S., Perret M., Crow Y.J., Avčin T., Cimaz R., Belot A. DNASE1L3 deficiency, new phenotypes, and evidence for a transient type I IFN signaling. J. Clin. Immunol. 2022;42(6):1310–1320. doi: 10.1007/s10875-022-01287-5. [DOI] [PubMed] [Google Scholar]
- 35.Chen J., Ding J., Huang W., Sun L., Chen J., Liu Y., Zhan Q., Gao G., He X., Qiu G., Long P., Wei L., Lu Z., Sun Y. DNASE1L3 as a novel diagnostic and prognostic biomarker for lung adenocarcinoma based on data mining. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.699242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J., Yi J., Zhang Z., Cao D., Li L., Yao Y. Deoxyribonuclease 1-like 3 may be a potential prognostic biomarker associated with immune infiltration in colon cancer. Aging (Albany NY) 2021;13(12):16513–16526. doi: 10.18632/aging.203173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grassi G.G. Clinical aspects of the relationship between antibiotic usage and resistance. J. Antimicrob. Chemother. 1977;3(Suppl C):77–84. doi: 10.1093/jac/3.suppl_c.77. [DOI] [PubMed] [Google Scholar]
- 38.Li B., Ge Y.Z., Yan W.W., Gong B., Cao K., Zhao R., Li C., Zhang Y.W., Jiang Y.H., Zuo S. DNASE1L3 inhibits proliferation, invasion and metastasis of hepatocellular carcinoma by interacting with β-catenin to promote its ubiquitin degradation pathway. Cell Prolif. 2022;55(9) doi: 10.1111/cpr.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun J., Wang X., Shen Q., Wang M., Chen S., Zhang X., Huang Y., Zhang Z., Li W., Yuan Y., Huang Z. DNASE1L3 inhibits hepatocellular carcinoma by delaying cell cycle progression through CDK2. Cell. Oncol. 2022;45(6):1187–1202. doi: 10.1007/s13402-022-00709-1. [DOI] [PubMed] [Google Scholar]
- 40.Li W., Nakano H., Fan W., Li Y., Sil P., Nakano K., Zhao F., Karmaus P.W., Grimm S.A., Shi M., Xu X., Mizuta R., Kitamura D., Wan Y., Fessler M.B., Cook D.N., Shats I., Li X., Li L. DNASE1L3 enhances antitumor immunity and suppresses tumor progression in colon cancer. JCI Insight. 2023;8(17) doi: 10.1172/jci.insight.168161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vuga L.J., Milosevic J., Pandit K., Ben-Yehudah A., Chu Y., Richards T., Sciurba J., Myerburg M., Zhang Y., Parwani A.V., Gibson K.F., Kaminski N. Cartilage oligomeric matrix protein in idiopathic pulmonary fibrosis. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0083120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magdaleno F., Arriazu E., Ruiz de Galarreta M., Chen Y., Ge X., Conde de la Rosa L., Nieto N. Cartilage oligomeric matrix protein participates in the pathogenesis of liver fibrosis. J. Hepatol. 2016;65(5):963–971. doi: 10.1016/j.jhep.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma H., Qiu Q., Tan D., Chen Q., Liu Y., Chen B., Wang M. The cancer-associated fibroblasts-related gene COMP is a novel predictor for prognosis and immunotherapy efficacy and is correlated with M2 macrophage infiltration in colon cancer. Biomolecules. 2022;13(1) doi: 10.3390/biom13010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q., Wang C., Wang Y., Sun L., Liu Z., Wang L., Song T., Yao Y., Liu Q., Tu K. HSCs-derived COMP drives hepatocellular carcinoma progression by activating MEK/ERK and PI3K/AKT signaling pathways. J. Exp. Clin. Cancer Res. 2018;37(1):231. doi: 10.1186/s13046-018-0908-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blom A.M., Gialeli C., Hagerling C., Berntsson J., Jirström K., Papadakos K.S. Expression of Cartilage Oligomeric Matrix Protein in colorectal cancer is an adverse prognostic factor and correlates negatively with infiltrating immune cells and PD-L1 expression. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1167659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dakhova O., Rowley D., Ittmann M. Genes upregulated in prostate cancer reactive stroma promote prostate cancer progression in vivo. Clin. Cancer Res. 2014;20(1):100–109. doi: 10.1158/1078-0432.Ccr-13-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in this study were available in online repositories. Here were the URLs for the databases used in this research. The datasets used and analyzed in the current study are available from the UCSC database (https://xenabrowser.net), GEO database (https://www.ncbi.nlm.nih.gov/gds), KEGG database (https://www.genome.jp/kegg/pathway.html), Reactome database (https://reactome.org/) and ImmPort database (https://immport.niaid.nih.gov), TIMER database (https://cistrome.shinyapps.io/timer/), Jvenn website (http://jvenn.toulouse.inra.fr/app/example.html).