Abstract
Vascular Ehlers-Danlos Syndrome (vEDS) is an inherited connective tissue disorder caused by COL3A1 gene, mutations that encodes type III collagen, a crucial component of blood vessels. vEDS can be life-threatening as these patients can have severe internal bleeding due to arterial rupture. Here, we generated induced pluripotent stem cell (iPSC) lines from two vEDS patients carrying a missense mutation in the COL3A1 (c.226A > G, p. Asn76Asp) gene. These lines exhibited typical iPSC characteristics including morphology, expression of pluripotency markers, and could differentiate to all three germ layer. These iPSC lines can serve as valuable tools for elucidating the pathophysiology underlying vEDS.
Keywords: vascular Ehlers-Danlos Syndrome, Induced pluripotent stem cells, COL3A1
1. Resource utility
Patients carrying a missense variation in the COL3A1 gene (c.226A > G, p.Asn76Asp) showed vascular Ehlers-Danlos Syndrome (vEDS) disease phenotype. iPSCs generated from these patients would serve as a valuable tool to model vEDS disease in a dish for prospective drug screening.
2. Resource details
Ehlers-Danlos Syndrome (EDS) is a group of inherited disorders that primarily affect the connective tissues in the body such as the skin, joints (ligaments, cartilage), and blood vessels. Patient with EDS usually exhibit joint flexibility and stretchy, fragile skin. Among EDS, vascular Ehlers-Danlos Syndrome (vEDS) is the most severe form of the disorder that is characterized by a defect in the synthesis of type III collagen, a critical component of the blood vessel walls that maintains the structural integrity of arteries and internal organs. These defects are due to mutations in the COL3A1 gene that encodes the pro-alpha1 chains of type III collagen. As a result, there is disruption in the assembly of type III collagen fibrils leading to weakened blood vessels and tissues that are prone to rupture. Indeed, patients with a single nucleotide missense variation in the COL3A1 gene have exhibited severe vascular complications such as hypertension and varicose veins and carry an increased risk of arterial rupture that can lead to hemorrhage and subcutaneous bleeding. Currently, there are no specific medications that can correct the underlying collagen defect in vEDS, however, for patients with symptomatic or high-risk arterial aneurysms, vascular grafting is considered a necessary surgical intervention to repair the weakend blood vessels (Frank et al., 2019; Frank et al., 2015).
By utilizing patient-specific induced pluripotent stem cells (iPSCs), we can establish a robust in vitro screening platform to model the vEDS phenotype in a dish. This approach involves generating iPSCs from patients carrying the missense mutations in the COL3A1 (c.226A > G, p. Asn76Asp) gene associated with vEDS. These iPSCs can then be differentiated into endothelial cells (iPSC-ECs) and vascular smooth muscle cells (iPSC-VSMCs) that can recapitulate the cellular components of blood vessels affected by vEDS and model the associated disease phenotype in-a-dish. Moreover, this iPSC platform can enable us to conduct high-throughput drug screening assays, which can provide insights into potential therapeutic interventions for vEDS-associated vascular complications (Alqahtani et al., 2022).
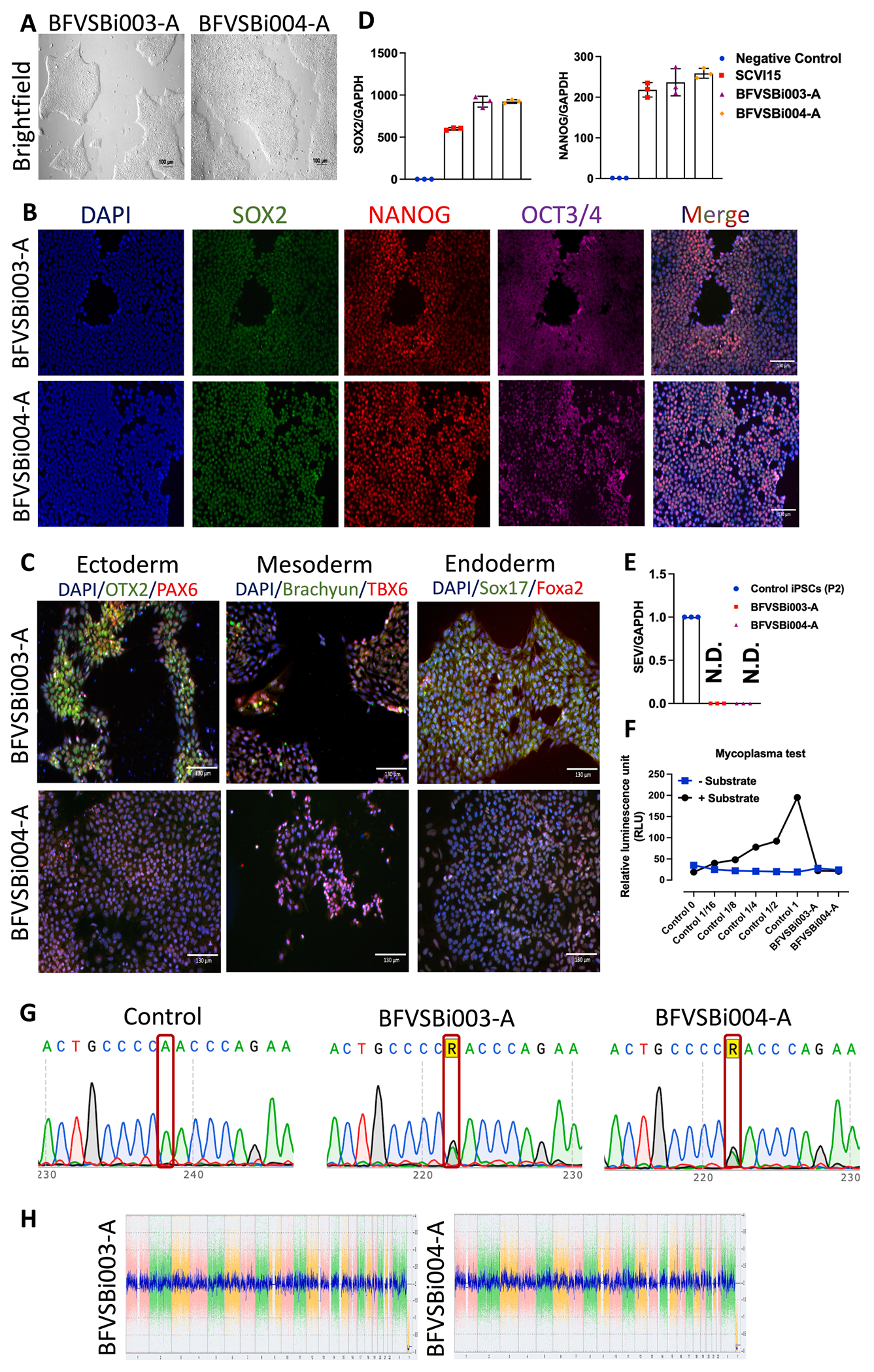
Here, we generated iPSC lines from two female patients, a 49-year-old (BFVSBi003-A) and a 53-year-old (BFVSBi004-A), both carrying the COL3A1 (c.226A > G, p.Asn76Asp) gene missense mutation (Table 1). Patient’s peripheral blood mononuclear cells (PBMCs) were reprogrammed into iPSCs using a Sendai virus vector containing Oct3/4, Sox2, Klf4, and c-Myc (Yamanaka factors). These iPSC clones exhibited typical iPSC morphology (Fig. 1A) and expressed pluripotency markers OCT3/4, NANOG, and SOX2, as demonstrated by immunostaining (Fig. 1B). Subsequent differentiation assays confirmed the ability of these iPSC lines to differentiate into endoderm, mesoderm, and ectoderm lineages (Fig. 1C). Furthermore, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis revealed expression of NANOG and SOX2 at the mRNA level in both lines (Fig. 1D). Importantly, the iPSC lines were free of the Sendai virus vector (Fig. 1E). The presence of the COL3A1 gene (c.226A > G, p.Asn76Asp) single nucleotide missense variation was confirmed by Sanger sequencing (Fig. 1G). Karyotype analysis using KaryoStat assays demonstrated normal karyotypes for both lines (Fig. 1H), and mycoplasma contamination was absent (Fig. 1F). Lastly, short tandem repeat (STR) analysis confirmed the genetic origin of these iPSC lines, matching that of their respective donor PBMCs (submitted in archive with journal).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography Bright field | Normal | Fig. 1A |
| Phenotype | Qualitative analysis (Immunocytochemistry) | Positive expression of pluripotency markers: Oct3/4, NANOG, SOX2 | Fig. 1B |
| Quantitative analysis (RT-qPCR) | mRNA expression of SOX2 and NANOG | Fig. 1D | |
| Genotype | Karyotype: Whole genome array (KaryoStat™ Assay) Resolution 1–2 Mb | Normal karyotype: 46 XX for BFVSBi003-A XX for BFVSBi004-A |
Fig. 1H |
| Identity | Microsatellite PCR (mPCR) or STR analysis | N/A | N/A |
| 16 loci tested, 100 % matching indentity | Submitted in archive with journal | ||
| Mutation analysis | Sequencing | COL3A1 gene (c.226A > G, p.Asn76Asp) | Fig. 1G |
| Southern Blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Luminescence: Negative | Fig. 1F |
| Differentiation potential | Directed differentiation, Immunofluorescence staining for 2 markers per germ layer | Positive Immunofluorescence staining of three germ layer markers Ectoderm: PAX6, OTX2 Endoderm: SOX17, FOXA2, Mesoderm: BRACHYURY, TBX6 |
Fig. 1C |
| Donor screening | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
Fig. 1.
3. Materials and methods
3.1. Isolation, culture, and reprogramming of PBMCs to iPSCs
PBMCs were isolated from whole blood using Percoll density gradient medium (GE Healthcare, #17089109) as previously described (Tripathi et al., 2024). The isolated PBMCs were purified with Dulbecco’s Phosphate Buffered Saline (DPBS) and plated in 24-well plate. The culture medium consisted of StemPro®-34 Serum-Free Medium (SFM) (ThermoFisher Scientific, #10639011) supplemented with StemPro®-34 Nutrient Supplement (ThermoFisher Scientific). Specific growth factors and cytokines, including Stem Cell Factor (SCF) (Peprotech, 100 ng/mL), FLT3 ligand (FLT3) (ThermoFisher Scientific, 100 ng/mL), Interleukin-3 (IL-3) (Peprotech, 20 ng/mL), Interleukin-6 (IL-6) (ThermoFisher Scientific, 20 ng/mL), and Erythropoietin (EPO) (ThermoFisher Scientific, 20 ng/mL), were added to the culture medium to support cell proliferation. Briefly, 2.5 × 105 PBMCs were plated and reprogrammed using the CytoTune-iPSC 2.0 Sendai Reprogramming Kit (ThermoFisher Scientific, #A16517). Transduced PBMCs were plated onto Matrigel-coated plates and cultured in StemPro™-34 medium (Thermo Fisher Scientific). After seven days, the medium was switched to StemMACS™ iPS-Brew medium (Miltenyi Biotec, #130 104-368), and cells were maintained for an additional ten to fifteen days. Colonies were picked, and clones were expanded as previously described (Sayed et al., 2020).
3.2. Maintenance of Induced Pluripotent Stem Cells (iPSCs)
iPSCs were cultured in StemMACS™ iPS-Brew XF medium supplemented as specified (#130-104-368, Miltenyi Biotec) at 37 °C in a humidified atmosphere until they were 95 % confluent. The cells were detached using 0.5 mM EDTA, resuspended in a ROCK inhibitor medium (Selleck Chemicals, #Y27632), and replated onto Matrigel-coated plates. The medium was refreshed after 24 h and every other day until the cells reached confluence.
3.3. Trilineage differentiation assay
To assess the pluripotency of iPSCs, cells were differentiated into endoderm, mesoderm, and ectoderm using a trilineage differentiation kit (STEMCELL Technologies, #05110).
3.4. Immunofluorescence staining
iPSCs or iPSC-differentiated germ layers were fixed with 4 % paraformaldehyde, permeabilized with digitonin, and blocked with Bovine Serum Albumin (BSA) and serum (Donkey Serum or Goat Serum). After overnight incubation with primary antibodies (Table 2), cells were incubated with respective secondary antibodies. Nuclei were counter-stained with NucBlue (ThermoFisher Scientific, #R37606) before imaging.
Table 2.
Reagents details.
| Antibodies used for Immunocytochemistry | ||||
|---|---|---|---|---|
| Antibody | Dilution | Company Cat # | RRID | |
|
| ||||
| Pluripotency Markers | Rabbit Anti-NANOG | 1:200 | Proteintech Cat# 142951-1-AP, | AB_1607719 |
| Mouse IgG2bκ Anti-OCT-3/4 | 1:200 | Santa Cruz Biotechnology Cat# sc-5279 | AB_628051 | |
| Mouse IgG1κ Anti-SOX2 | 1:200 | Santa Cruz Biotechnology Cat# sc-365823 | AB_10842165 | |
| Ectoderm Markers | Goat Anti-OTX2 | 1:200 | R&D Systems Cat# 963273 | AB_2157172 |
| Rabbit Anti-Pax6 | 1:100 | Thermo Fisher Scientific Cat# 42-6600 | AB_2533534 | |
| Endoderm Markers | Goat Anti-SOX17 | 1:200 | R&D Systems Cat# 963121 | AB_355060 |
| Rabbit Anti-Foxa2 | 1:250 | Thermo Fischer Scientific Cat# 701698 | AB_2576439 | |
| Mesoderm Markers | Goat Anti-Brachyury | 1:200 | R&D Systems Cat# 963427 | AB_2200235 |
| Rabbit Anti-Tbx6 | 1:200 | Thermo Fischer Scientific cat # PA5-35102 | AB_2552412 | |
| Secondary Antibodies | Alexa Fluor 488 Goat Anti-Mouse IgG1 | 1:1000 | Thermo Fisher Scientific #A-21121 | AB_2535764 |
| Alexa Fluor 488 Donkey Anti-Goat IgG (H + L) | 1:1000 | Thermo Fisher Scientific #A-11055 | AB_2534102 | |
| Alexa Fluor 555 Goat Anti-Rabbit IgG (H + L) | 1:500 | Thermo Fisher Scientific #A-21428 | AB_141784 | |
| Alexa Fluor 647 Goat Anti-Mouse IgG2b | 1:250 | Thermo Fisher Scientific #A-21242 | AB_2535811 | |
| Primers | ||||
| Target | Forward/Reverse primer (5′-3′) | |||
|
| ||||
| Sendai Virus | Sendai Virus genome | Mr04269880_mr | ||
| Genotyping | COL3A1 gene (c.226A > G, p.Asn76Asp) | Fwd: TTTCAAACCTTTTCAACTTTGGC Rev: CTTACTGGATCTCCCTTGGGG |
||
| House-Keeping Gene | GAPDH | HS02758991_g1 | ||
| SOX2 | HS01053049_s1 | |||
| NANOG | HS02387400_g1 | |||
3.5. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted from iPSCs using TRIzol® and Direct-zol™ RNA Miniprep Kit (Zymo Research, #R2052) at passage 14. cDNA was synthesized using the iScript™ cDNA Synthesis Kit (BioRad, #1708891), followed by RT-PCR analysis of NANOG, SOX2, and SEV using TaqMan™ Gene Expression Assay (Applied Biosystems, #4444556).
3.6. Karyotyping
At passage 12, iPSCs were collected and analyzed for chromosomal abnormalities using the KaryoStat™ assay (ThermoFisher).
3.7. Short Tandem Repeat (STR) analysis
Genomic DNA was isolated from PBMCs and iPSCs using the DNeasy Blood & Tissue Kit (Qiagen, #56304) at passage 15. DNA amplification was performed using the CLA IdentiFiler™ Direct PCR Amplification Kit (Thermo Fisher, #A44660), and the products were analyzed by capillary electrophoresis.
3.8. Sequencing
Genomic DNA extracted from iPSC lines was subjected to an amplification with the help of NEB High-Fidelity PCR kit (New England Biolabs, #M0541S) using customized primers (Table 2) at passage 13. The PCR products were sequenced using the ABI3130xl platform (Stanford PAN facility) after purification with the QIAquick Purification Kit (Qiagen, #28706).
3.9. Mycoplasma detection
Mycoplasma contamination was assessed using the MycoAlert Detection Kit (Lonza, #LT07-705).
Resource Table
| Unique stem cell lines identifier | 1. BFVSBi003-A 2. BFVSBi004-A |
| Alternative name(s) of stem cell lines | 1. VS-18 2. VS-19 |
| Institution | Baszucki Family Vascular Surgery Biobank |
| Contact information of distributor | Dr. Nazish Sayed sayedns@stanford.edu |
| Type of cell lines | iPSC |
| Origin | Human |
| Additional origin info (Applicable for human ESC or iPSC) | 1. BFVSBi003-A; Age:49, Sex: Female; Ethnicity: Caucasian 2. BFVSBi004-A; Age:53, Sex: Female; Ethnicity: Caucasian |
| Cell Source | PBMCs |
| Clonality | Clonal |
| Method of reprogramming | Integration-free Sendai virus expressing human OCT4, SOX2, KLF4, and c-MYC |
| Genetic Modification | Yes |
| Type of Genetic Modification | Spontaneous mutation |
| Evidence of the reprogramming transgene loss | RT/q-PCR |
| Associated disease | vascular Ehlers-Danlos Syndrome (vEDS) |
| Gene/locus | 1. COL3A1 (c.226A > G, p.Asn76Asp) |
| Date archived/stock date | 1. BFVSBi003-A: 12-17-2023 2. BFVSBi004-A: 11-23-2023 |
| Cell line repository/bank | 1. https://hpscreg.eu/cell-line/BFVSBi003-A 2. https://hpscreg.eu/cell-line/BFVSBi004-A |
| Ethical approvals | The Administrative Panel approved the generation of the lines on Human Subjects Research (IRB) under IRB #62122, “Human Induced Pluripotent Stem Cells for Studying Cardiac and Vascular Diseases.” |
Acknowledgments
This study was supported by research grants from the National Institutes of Health (NIH) R01 HL158641, R01 HL161002, and American Heart Association (AHA) SFRN grant 869015 (NS); and AHA Postdoctoral fellowship 23POST1020812 (AM).
Footnotes
CRediT authorship contribution statement
Amit Manhas: Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Dipti Tripathi: Writing – review & editing, Validation, Resources, Project administration, Methodology, Formal analysis, Data curation. Chikage Noishiki: Methodology. David Wu: Methodology. Lu Liu: Methodology. Karim Sallam: Validation, Investigation. Jason T. Lee: Project administration, Funding acquisition. Eri Fukaya: Resources. Nazish Sayed: Writing – review & editing, Visualization, Validation, Supervision, Resources, Project administration, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Alqahtani M, Claudinot A, Gaudry M, Bartoli A, Barral PA, Vidal V, Boyer L, Busa T, Cadour F, Jacquier A, De Masi M, Bal L, 2022. Endovascular management of vascular complications in ehlers-danlos syndrome type IV. J. Clin. Med 11 (21) 10.3390/jcm11216344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Albuisson J, Ranque B, Golmard L, Mazzella JM, Bal-Theoleyre L, Fauret AL, Mirault T, Denarie N, Mousseaux E, Boutouyrie P, Fiessinger JN, Emmerich J, Messas E, Jeunemaitre X, 2015. The type of variants at the COL3A1 gene associates with the phenotype and severity of vascular Ehlers-Danlos syndrome. Eur. J. Hum. Genet 23 (12), 1657–1664. 10.1038/ejhg.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Adham S, Seigle S, Legrand A, Mirault T, Henneton P, Albuisson J, Denarie N, Mazzella JM, Mousseaux E, Messas E, Boutouyrie P, Jeunemaitre X, 2019. Vascular Ehlers-Danlos syndrome: long-term observational study. J. Am. Coll. Cardiol 73 (15), 1948–1957. 10.1016/j.jacc.2019.01.058. [DOI] [PubMed] [Google Scholar]
- Sayed N, Liu C, Ameen M, Himmati F, Zhang JZ, Khanamiri S, Moonen JR, Wnorowski A, Cheng L, Rhee JW, Gaddam S, Wang KC, Sallam K, Boyd JH, Woo YJ, Rabinovitch M, Wu JC, 2020. Clinical trial in a dish using iPSCs shows lovastatin improves endothelial dysfunction and cellular cross-talk in LMNA cardiomyopathy. Sci. Transl. Med 12 (554) 10.1126/scitranslmed.aax9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi D, Manhas A, Noishiki C, Wu D, Adkar S, Sallam K, Fukaya E, Leeper NJ, Sayed N, 2024. Generation of induced pluripotent stem cell line from a patient suffering from arterial calcification due to deficiency of CD73 (ACDC). Stem Cell Res. 75, 103285 10.1016/j.scr.2023.103285. [DOI] [PMC free article] [PubMed] [Google Scholar]


