Abstract
The Drosophila Groucho (Gro) protein is the defining member of a family of metazoan corepressors that have roles in many aspects of development, including segmentation, dorsal/ventral pattern formation, Notch signaling, and Wnt/Wg signaling. Previous speculation has suggested that Gro may be orthologous to the yeast corepressor Tup1. In support of this idea, a detailed alignment between the C-terminal WD-repeat domains of these two proteins shows that each Gro WD repeat is most similar to the Tup1 WD repeat occupying the corresponding position in that protein. Our analysis of Gro–histone interactions provides further support for a close evolutionary relationship between Gro and Tup1. In particular, we show that, as with the N-terminal region of Tup1, the N-terminal region of Gro is necessary and sufficient for direct binding to histones. The highest affinity interaction is with histone H3 and binding is primarily observed with hypoacetylated histones. Using transient transfection assays, we show that a Gal4–Gro fusion protein containing the histone-binding domain is able to repress transcription. Deletions that weaken histone binding also weaken repression. These findings, along with our recent report that Gro interacts with the histone deacetylase Rpd3, suggest a mechanism for Gro-mediated repression.
INTRODUCTION
The Drosophila groucho (gro) gene encodes a widely used transcriptional corepressor with homologs in essentially all metazoans (1). As a corepressor, Gro lacks a DNA-binding domain, but is recruited to the DNA by specific protein–protein interactions with DNA-bound repressors. Gro interacts with a wide variety of Drosophila repressors including Hairy family bHLH factors, Runt family factors, Engrailed, Dorsal, Huckebein and Pangolin. Via these interactions, Gro plays essential roles in many developmental processes including segmentation, dorsal/ventral and terminal pattern formation, neurogenesis, sex determination and patterning of the compound eye (reviewed in 2,3).
Gro family proteins, including the human transducin-like Enhancer of split (TLE) proteins, are characterized by a WD-repeat domain that occupies the C-terminal half of most members of the family (3,4). Since WD-repeat domains generally provide interfaces for interactions with other proteins (5), it is likely that this region of Gro mediates some of the interactions required for Gro function, including interactions with DNA-bound repressors and with other corepressors. In addition to the conserved WD-repeat domain, Gro family proteins contain a highly conserved ∼130 amino acid glutamine-rich region. This domain, found at the N-terminal end of the protein, is known to mediate tetramerization, which appears to be required for Gro function (6). The WD-repeat and glutamine-rich domains are separated by a weakly conserved spacer region. Although this spacer region shows very little overall sequence conservation, it seems to be organized in a conserved manner, consisting of a glycine/proline-rich (GP) domain followed by a CcN domain, followed by a serine/proline-rich (SP) domain. The Gro GP domain is thought to contribute to repression via the recruitment of the histone deacetylase Rpd3 (7). The CcN domain is a type of nuclear localization motif characterized by a short positively charged nuclear localization signal separated by a conserved distance from putative phosphorylation sites for cdc2 kinase and casein kinase II. Finally, although the SP domain is thought to contribute to repression, little information is available about the specific biochemical functions of this domain.
The potential of these various domains to mediate repression has been explored by fusing Gro/TLE protein deletion variants to the Gal4 DNA-binding domain, thereby targeting the deletion variants to Gal4 binding site-containing reporter genes (8). These studies have revealed that the N-terminal half of the protein, excluding most of the WD-repeat domain, can repress transcription just as well as full-length Gro when artificially targeted to the template in this manner. However, since the N-terminal half of the protein contains the Q-domain, which mediates homotetramerization, it is not clear from those studies whether the N-terminus of Gro per se can repress transcription, or whether the recruitment of endogenous full-length Gro present in the host cells is responsible for the observed repression. The C-terminal WD-repeat domain is also able to weakly repress transcription in this assay suggesting the existence of multiple pathways for transcriptional repression. However, other reports indicate that the WD-repeat domain lacks repressor activity when fused to the Gal4 DNA-binding domain (7).
While it is likely that all metazoan genomes encode Gro orthologs, it is not clear if such proteins exist in single cell eukaryotes such as yeast. The best candidate for a yeast ortholog of Gro is probably Tup1. Like Gro, Tup1 functions to mediate repression by a wide variety of DNA-bound repressors (9). In addition, both Tup1 and Gro contain C-terminal WD-repeat domains of comparable length. However, the overall sequence similarity between the Gro and Tup1 WD-repeat domains is not significantly greater than the similarity between the Gro domain and WD-repeat domains found in proteins not involved in transcriptional repression (10). For example, the WD-repeat domain in β-transducin displays 23% sequence identity with Gro, while the WD-repeat domain in Tup1 displays 25% sequence identity with Gro. The N-terminal region of Tup1 (exclusive of the WD-repeat domain) does not exhibit significant homology to Gro.
As with Gro, deletion analysis of Tup1 has shown that the N-terminal region (lacking the WD repeats) can repress transcription when tethered to the DNA template via the Gal4 DNA-binding domain (11). This N-terminal region is sufficient for protein–protein interactions with hypoacetylated forms of histones H3 and H4 (12). Genetic experiments also indicate that the N-terminal histone tails are required for Tup1-dependent repression (13). The highly conserved histone tails include a series of invariant lysine residues that are the targets for acetylation (14). Hypoacetylated histones have long been associated with transcriptional repression and numerous transcriptional repressors are known to associate with histone deacetylases (14–19). It has been proposed that histone deacetylation may lead to the formation of a transcriptionally-silent form of chromatin (16,20). It is conceivable that the favorable interaction between hypoacetylated histones and Tup1 could serve to initiate and/or maintain this silenced state (12,13).
The similarities between the structure and function of Tup1 and Gro prompted us to ask whether Gro might also interact with chromatin components in a fashion similar to Tup1, despite the apparent lack of conservation between the N-terminal regions of the two proteins. Recent evidence suggests that the TLE proteins interact with the N-termini of histone H3 (21). However, the dependence of this interaction on histone acetylation was not determined; nor was it determined if the interaction was direct. Here we show that Gro and Tup1 bind directly to a similar subset of histones, and that the N-terminal region of Gro, lacking the WD repeats, is sufficient for this interaction. Furthermore, we show that Gro interacts preferentially with hypoacetylated forms of histone H3. In addition, using transient transfections in S2 cells we show that a domain of Gro that mediates the interaction with histones contributes to repression. Deletions that weaken histone binding also weaken transcriptional repression. These results, along with a detailed alignment of the Gro and Tup1 WD-repeat domains, suggest that Gro and Tup1 are closely related corepressors that may employ evolutionarily conserved mechanisms for transcriptional repression.
MATERIALS AND METHODS
Sequence analysis
Gro cDNA (accession number P16371) and Tup1cDNA (accession number NP_010007.1) were submitted to the MEME (Multiple EM for Motif Elicitation) discovery tool at the San Diego Supercomputing Center (http://www.sdsc.edu/ ) for sequence analysis.
Plasmids
Plasmids to generate the N- and C-terminal truncations of Gro for in vitro transcription/translation have been described (6). Plasmids for Escherichia coli expression of glutathione-S-transferase (GST) fusions of wild-type or mutant forms of all four core histones tails have been described previously (22). Reporter plasmids pG5DE5-37TKLuc and the internal control p–37TKLuc for S2 cell transfection have been described (6). Expression plasmids for Gal4p53Gro2–719LL and Gal4p53Gro390–719LL fusions were made by PCR amplication of the appropriate Gro-coding sequences using pET17b-Gro or the pET17b-Gro L38P, L87P double point mutant as template (6) and KpnI/BamHI linkers. PCR products were then ligated into the KpnI/BamHI sites of Gal4p53Gro121–719 (6) after excision of the Gro coding sequences. Gal4p53Gro121–719 and Gal4p53Gro390–719 were made similarly using wild-type pET17b-Gro as a template. The integrity of the fusion constructs was determined by DNA sequencing. The plasmid for expression of Gal4p53Gro-121–190 has been described (7). Plasmid pVL1392-M2Gro for expression of FLAG-Gro in Sf9 cells has been described (6).
Protein–protein interaction assays
For the GST-pulldown assays, GST–histone tail fusion proteins were expressed according to standard procedures (23). The purity and concentration of the fusion proteins was determined by SDS–PAGE followed by Coomasie Blue. Wild-type, mutant and deletion variants of in vitro transcribed/translated [35S]methionine labeled Gro were produced using the TNT7 kit (Promega) according to the manufacturer’s instructions. GST-binding assays were done at room temperature for 45 min in 0.15 M KCl HEMN buffer (40 mM HEPES, 5 mM MgCl2, 0.2 M EDTA, 1 mM DTT, 0.5 mM PMSF and 0.5% NP-40). For the Gro-histone affinity chromatography assays, FLAG-Gro was made in Sf9 cells and purified from nuclear extracts using agarose-bound anti-FLAG M2 antibodies as described (6). M2-beads (40 µl) containing purified immobilized Gro were incubated with 5 µg of purified Drosophila core histones in 300 µl of 0.15 M KCl HEMN buffer for 45 min at room temperature. After extensive washing, one-third of the bound proteins was eluted with SDS–PAGE buffer and analyzed by SDS–PAGE. The remaining two-thirds was used in a western blot assay using either α-acetylated H3 or α-acetylated H4 antibodies (Upstate Biotechnology). For GST-pulldown assays with purified FLAG-Gro, GST–H3 and GST–H4 fusion proteins were incubated with purified FLAG-Gro as described above. The amount of FLAG-Gro retained in this experiment was visualized by western blot analysis using α-FLAG M2 antibody (Sigma). For the far-western assays, 3 µg of either Drosophila histones (a kind gift of J. Kadonaga, University of California, San Diego) or calf thymus histones were resolved in 22% polyacrylamide gels and then transferred to PVDF membranes. Alternatively 10 µg of HeLa cell histones were resolved by triton acid urea (TAU) gel electrophoresis to resolve different acetylation states and then transferred to PVDF membranes. The membrane was washed twice with PBS and then for 10 min with 0.5 × 0.1 M CZ solution (20 mM HEPES pH 7.9, 17% glycerol, 0.1 M KCl, 5 mM MgCl2, 0.1 mM ZnCl2, 0.1 mM EDTA, 2 mM DTT). Crude nuclear extracts (500 µl) from Sf9 cells expressing FLAG-tagged Gro were diluted in 8 ml of 0.5 × 0.1 M CZ buffer and then incubated with the PVDF membrane for 2 h. After washing the membrane twice with TBS, bound FLAG-Gro was visualized by western blotting using α-FLAG M2 antibodies (Sigma).
Transient transfection assays
Transient transfection assays into S2 cells were done in duplicate as described by Chen et al. (6). Briefly, ∼1 × 107 S2 cells were plated in fresh media in a 100-mm dish and transfected 16–24 h later. Transcription assays were done the following day with the Dual Luciferase Reporter kit (Promega). To monitor the expression levels of transfected Gal4 fusion proteins in S2 cells, aliquots of transfected S2 cells were analyzed by western blotting with a monoclonal antibody against the N-terminal 1–147 amino acids of Gal4 (Santa Cruz Biotechnology).
RESULTS
The WD repeats in Gro and Tup1 share serial homology
We used the Multiple Expectation Maximization for Motif Elicitation (MEME) analysis tool (UCSD, supercomputing center; 24) to identify regions of similarity between Gro and Tup1. The only apparent homology between the two proteins is in the C-terminal WD-repeat region. The analysis reveals that, like Tup1 Gro has seven WD repeats, including a less conserved ‘cryptic’ fifth WD repeat in Gro (Fig. 1A), which was identified during the initial cloning of Gro (25) and which conforms loosely to the WD-repeat consensus (26) (Fig. 1A). When the repeats in Gro are classified according to which Tup1 repeat they most closely resemble, the Gro and Tup1 repeats are found to occur in roughly the same order (Fig. 1A and B), i.e. the WD-repeat domains in Tup1 and Gro share serial homology. Furthermore, the homology between the individual WD repeats in Gro and Tup1 extends to the N-terminal side of each repeat. Although the overall similarity between the WD repeats in Gro and Tup1 is low, these relationships in the WD repeats suggest that the two molecules are structurally, and therefore perhaps functionally, related. Similar analysis of the Gro and β-transducin WD repeats does not detect such serial homology (not shown).
Figure 1.
The WD repeats of Gro and Tup1 are serially related. (A) Alignment of the WD repeats of Gro and Tup1 indicating the relationship between the most similar repeats. Identical amino acids (capital letters) and conservative substitutions (+) are indicated. (B) Schematic illustration of the similarities between the WD repeats in Gro and Tup1.
Gro interacts with the N-terminal tails of core histones
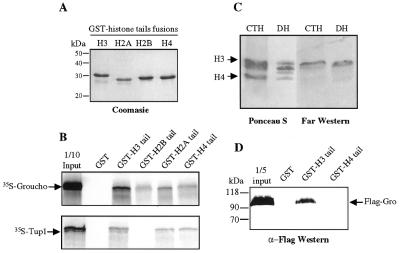
Given the previously observed interactions between Tup1 and core histones (12) and between human TLE proteins and core histones (21), we were interested in determining if Gro could interact with core histones. To investigate this possibility, we determined whether Gro could interact with the N-terminal tails of histones in a co-immobilization assay. In vitro transcribed/translated [35S]methionine labeled Gro (referred as 35S-Gro) was incubated with purified fusion proteins consisting of GST joined to the N-terminal tails of the four core histones (Fig. 2A). In these GST-pulldown assays, Gro bound strongly to the histone H3 tail, moderately to histone H2B and H4 tails, and very weakly to the histone H2A tail (Fig. 2B). In parallel experiments we observed a similar binding profile for 35S-Gro and 35S-Tup1 (12), indicating that the two proteins have similar relative affinities for the different core histone tails (Fig. 2B).
Figure 2.
Gro interacts with core histones. (A) Coomasie blue stained gel of E.coli produced and purified GST–histone tail fusion proteins. Approximately equal amounts of each of the different fusion proteins were used for the interaction assay. (B) Autoradiogram of an SDS–PAGE gel showing 35S-Gro or 35S-Tup1 retained by affinity beads containing GST or fusion proteins containing GST and various histone N-terminal tails. (C) Calf thymus histones (CTH) and Drosophila core histones (DH) were resolved in a 20% SDS–polyacrylamide gel, transferred to nitrocellulose membranes and stained with Ponceau-S or incubated with nuclear extract from Sf9 cells expressing FLAG-Gro. The Gro-interacting histones were identified by subsequent incubation with anti-FLAG antibodies. (D) GST-pulldown with bacterially made GST–H3 tail and GST–H4 tail fusion proteins and purified FLAG-Gro from baculovirus infected Sf9 cells. Bound Gro was detected by immunoblot analysis of the retained fraction.
The interactions between Gro and histones were confirmed by far western analysis using Drosophila core histones as well as calf thymus histones. Histones were resolved by SDS–PAGE, transferred to a PVDF membrane and probed with crude nuclear extracts of baculovirus-infected Sf9 cells expressing FLAG-tagged Gro (FLAG-Gro) (Fig. 2C). After extensive washing the membrane was probed with anti-FLAG M2 antibodies. In this assay, Gro interacts strongly with histone H3 and more weakly with histones H2A, H2B and H4 (Fig. 2C), confirming the observation made in the GST-pulldown assay that Gro preferentially binds to histone H3.
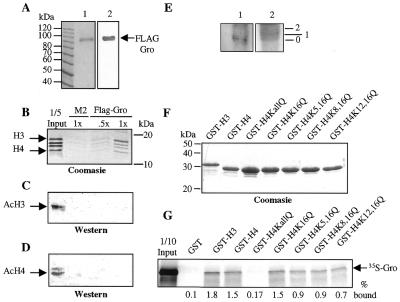
In the above-described GST-pulldown assays, the radiolabeled Gro protein was introduced in a crude reticulocyte lysate, while in the far western assays, immobilized histones were probed with crude nuclear extracts containing FLAG-tagged Gro. Similarly, the previously published experiments demonstrating interactions between TLEs and histone H3 (21) used impure proteins. The use of impure proteins opens the possibility that the observed interaction between Gro/TLE proteins and histones is mediated by a third protein present in both the nuclear extracts and the reticulocyte lysates. To address this possibility, we used highly purified components in an affinity chromatography assay. FLAG-Gro was purified to near homogeneity from baculovirus infected Sf9 cells using an anti-FLAG M2 antibody coupled to agarose beads (Fig. 3A). Immobilized FLAG-Gro was then incubated with purified Drosophila histones and after extensive washing, the retained histones were resolved by SDS–PAGE and visualized by Coomasie blue staining (Fig. 3B). In this assay, immobilized FLAG-Gro specifically retained all four histones with an enrichment of histone H3 (Fig. 3B). These results further demonstrate that Gro binds directly to core histones with a preference for histone H3.
Figure 3.
Gro interacts preferentially with hypoacetylated forms of histones. (A) SDS–PAGE gels of baculovirus produced and purified FLAG-Gro. Lane 1, Coomasie blue stained gel; lane 2, anti-FLAG western blot. The molecular masses in kDa of protein size markers are indicated. (B) Purified Drosophila histones were incubated with either 30 µl (.5x) or 60 µl (1x) of FLAG-Gro immobilized on agarose–M2 beads or with 60 µl (1x) of agarose–M2 beads alone. After incubation, one-third of the bound histones was analyzed by Coomasie blue staining. (C and D) Western blot analyses of the input and retained fractions from the affinity chromatography experiment in (B). Two-thirds of the retained histones were analyzed with anti-acetylated H3 (C) or anti-acetylated H4 (D) polyclonal antibodies. (E) Far western analysis of histone H3 resolved by TAU electrophoresis and probed with FLAG-Gro. Lane 1, far western analysis; lane 2, Coomasie blue staining. 0, 1 and 2 indicate positions of unacetylated, singly-acetylated, and doubly-acetylated species, respectively. (F) Wild-type GST–H3 and either wild-type or the indicated single or double point mutant forms of GST–H4 were produced and purified from E.coli and visualized by Coomasie blue staining. Approximately equal amounts of proteins were used for the interaction assays. (G) Autoradiogram of a GST-pulldown assay with 35S-Gro and the indicated GST–histone tail variants.
In agreement with the interactions observed in the far western assay, a very specific interaction is observed in a GST-pulldown assay using purified GST–H3 or –H4 tail fusion proteins and purified FLAG-Gro from baculovirus infected Sf9 cells. In contrast to the results seen with in vitro transcribed/translated 35S-Gro (Fig. 2B), only GST–H3 retains a detectable amount of FLAG-Gro (Fig. 2D). This apparent discrepancy suggests that a modification of Gro that does not occur in the in vitro translation system, but occurs in the Sf9 cells from which FLAG-Gro is prepared, shifts the binding in favor of histone H3, perhaps by precluding binding to the other core histones.
Gro interacts preferentially with hypoacetylated forms of histones
In vitro transcribed/translated Tup1 interacts preferentially with hypoacetylated forms of histones H3 and H4 (12). We analyzed the histone fraction retained in our FLAG-Gro affinity chromatography experiment (see above) to test whether Gro might interact with specific post-translationally modified forms of histones. A western blot using antibodies that recognize acetylated forms of histones H3 or H4 indicates that the input loaded on the FLAG-Gro column contains a readily detectable amount of acetylated histones (Fig. 3C and D). In contrast, in the fraction bound by FLAG-Gro, essentially no acetylated protein is detected with either the anti-acetylated H3 or the anti-acetylated H4 antibodies (Fig. 3C and D). Thus, Gro protein preferentially binds hypoacetylated histones. These results were further confirmed by far western analysis of triton acid urea (TAU) gels containing histone H3. These gels resolve histones based upon their acetylation state. When a TAU gel containing histone H3 was transferred to a PVDF membrane and probed with FLAG-Gro, we observed a much stronger interaction with the unmodified than with the modified forms of the histone (Fig. 3E).
Rpd3 is a histone deacetylase that potentiates Gro repression. Since Rpd3 is primarily an H4 N-terminal tail deacetylase (7,27–30), we decided to determine if specific lysines in the N-terminal tail of H4 were required for binding to Gro. The N-terminal tail of histone H4 contains four lysine residues (K5, K8, K12 and K16) that are targets for acetylation. To determine if these lysines play a role in Gro binding, we tested the binding of 35S-Gro to either a wild-type GST–H4 tail fusion protein, or to variants containing single or multiple point mutations in the histone tail that convert these highly conserved lysine residues to glutamine. Both the acetylation of lysine and the conversion of lysine to glutamine result in the conversion of a positively charged ammonium ion to a neutral amide. GST fusion proteins containing wild-type or mutant forms of the histone H4 tail were produced in E.coli, purified to near homogeneity (Fig. 3F), and tested for binding to 35S-Gro. In this assay, mutagenesis of either individual lysine residues or pairs of lysine residues to glutamine residues results in decreased binding of Gro to the H4 tail, while mutagenesis of all four lysines to glutamines almost completely abolishes the Gro interaction (Fig. 3G). Similar results were observed with 35S-Tup1 (data not shown). Thus all four lysines in the histone H4 tail are required for the detected interaction with 35S-Gro, without a particular preference.
The intact N-terminal region of Gro excluding the WD-repeat domain is required for efficient binding to core histones
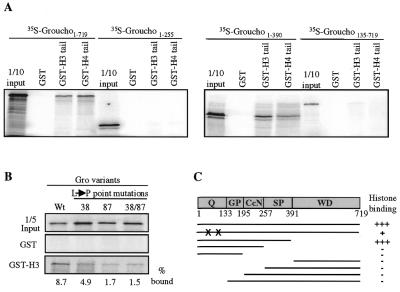
We used the GST-pulldown assay to test different in vitro transcribed/translated deletion derivatives of Gro for their ability to bind to the histone tails. We find that the WD repeats of Gro are not required for the interaction with GST–H3 or GST–H4. The deletion variant containing amino acids 1–390 binds to H3 or H4 as well as full-length Gro (Fig. 4A), while deletion of amino acids 1–390 results in a complete loss of binding to both GST fusion proteins. Deletions that remove any of the four domains within the first 390 amino acids also result in the complete loss of binding of Gro to the GST–histone fusion proteins (Fig. 4A and data not shown). These results are confirmed by far western assays (data not shown). Our finding that the entire region of Gro N-terminal to the WD repeats is necessary for histone binding is strikingly similar to findings previously obtained with Tup1 (12).
Figure 4.
The N-terminal domain of Gro is necessary and sufficient for the interaction with histones. (A) The indicated 35S-labeled Gro deletion variants were tested for binding to GST alone, the GST–H3 tail fusion protein or the GST–H4 tail fusion protein. Input and retained proteins were displayed on an SDS–PAGE gel and visualized by autoradiography. (B) In vitro transcribed/translated 35S-labeled wild-type Gro or the indicated single or double point mutants in the Q-domain were incubated with GST–H3 and the retained proteins were visualized by SDS–PAGE and autoradiography. These mutations result in loss of tetramerization and transcriptional repression by Gro. (C) Diagram summarizing the results of the GST-pulldown assays. The entire N-terminal region from 1 to 390 is necessary and sufficient for the interaction with histones. The single leucine to proline mutations in the tetramerization domain of Gro are indicated by X.
The N-terminal region of Gro includes a glutamine-rich domain at the extreme N-terminus of the protein that mediates tetramerization. To determine if tetramerization is required for histone binding, we examined the effect of point mutations in the glutamine-rich domain that were previously shown to disrupt tetramerization on the binding to GST–H3. In these point mutants the conversion of specific leucines (Leu38 and Leu87) to prolines is believed to disrupt putative amphipathic α-helices required for tetramerization. Mutation of one or both leucines resulted in a significant reduction in binding to GST–H3 (Fig. 4B). These findings suggest that tetramerization is a prerequisite for binding. Perhaps formation of the tetramer is necessary for the assembly of a high-affinity platform for histone H3 recruitment. The fact that the mutant tetramerization domain is still sufficient for at least low-affinity binding suggests that this region may have some role in H3 binding apart from its role in tetramerization.
Deletions that eliminate histone binding also compromise repression
To determine if there is a correlation between the ability to bind histones and the ability to repress transcription, we decided to employ a transient transfection assay in which Gro deletion variants are targeted to the template by fusion to the Gal4 DNA-binding domain. For these experiments, we replaced the N-terminal tetramerization domain of Gro with the heterologous tetramerization domain from p53. This heterologous tetramerization domain was previously shown to substitute partially for the Gro tetramerization domain in transcriptional repression (6). By using the heterologous tetramerization domain we avoid the complication caused by the ability of the Gro tetramerization domain to recruit endogenous Gro to the template.
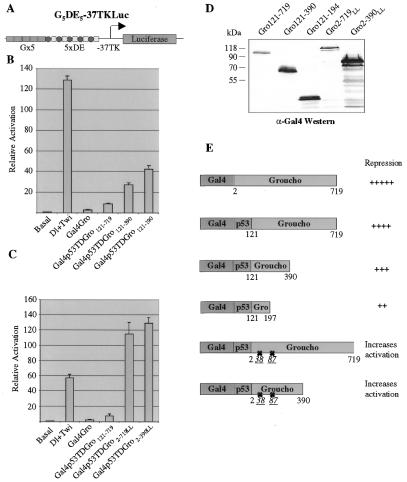
As previously shown, full-length Gro (Gal4-Gro) is a slightly better repressor than the p53 tetramerization domain/Gro chimera (Gal4p53Gro121–719) (Fig. 5B), implying that the N-terminal 121 amino acids of Gro may carry out some biochemical function in addition to repression (6). Further deletion analysis reveals that Gal4p53Gro121–390 represses transcription to nearly the same extent as Gal4p53Gro121–719 and more efficiently than Galp53Gro121–194 (Fig. 5B). Since amino acids 121–194 in Gro interact with Rpd3 just as efficiently as amino acids 121–390 (7), we believe that the stronger repression by amino acids 121–390 is not simply due to the more efficient recruitment of Rpd3. Thus, these results show that sequences outside the tetramerization and Rpd3-recruitment domains are required for efficient transcriptional repression just as they are required for histone binding. Immunoblots from transfected cells reveals that all the Gal4Gro derivatives were expressed to at least the same level as Gal4p53Gro121–719 (Fig. 5D).
Figure 5.
Functional dissection of the repression domains in Gro. (A) Diagram of the reporter construct used in this analysis. (B and C) S2 cells were transfected with either the reporter vector alone, or in combination with Dorsal and Twist (60 and 20 ng, respectively) expression vectors and the indicated Gal4Gro derivatives (5 µg). In addition an internal control reporter (–37tkRLuc; 0.1 µg) was included in all the transfections for normalization. The bars represent the average and standard deviation of two independent duplicate experiments. (D) Aliquots of transfected S2 cells were analyzed by western blot with a monoclonal α-Gal4 antibody. (E) Summary of the various Gal4Gro derivatives used in the co-transfection assays and their relative repression levels.
As mentioned above, chimeras in which the Gro glutamine-rich domain is replaced by the p53 tetramerization domain do not repress transcription quite as well as Gro proteins containing the glutamine-rich domain, suggesting that the glutamine-rich domain may have some function in repression apart from its ability to mediate tetramerization. In an effort to explore this possibility, we decided to create p53/Gro chimeras containing the mutant form of the glutamine-rich domain that is unable to mediate tetramerization. In such chimeras, the tetramerization function would be supplied by the p53 domain, while the mutant glutamine-rich domain would still be able to perform other functions required for repression (such as histone H3 binding). However, when we examined these chimeras in the transfection assay we got a very surprising result. Unexpectedly, co-transfection of these Gro derivatives resulted in a slight increase in transcriptional activity, rather than repression (Fig. 5C). The expression of these Gro derivatives was also confirmed by western blot analysis of transfected cells. These findings imply that the context of the tetramerization domain is critical for its ability to mediate transcriptional repression.
DISCUSSION
We have studied the interaction of Gro with core histones and found that Gro binds to all four core histones with a preference for histone H3. The N-terminal region of Gro lacking the WD repeats is sufficient for the interaction with all histones. We also show that the interaction of Gro with histones is direct and that the acetylation state of histones is important in modulating the interaction with Gro. Deletions of Gro that abolish histone binding, also weaken transcriptional repression in transient transfection assays.
Similarities between Gro and the Tup1 corepressor
Sequence analysis reveals that, like Tup1, Gro contains a total of seven C-terminal WD repeats. Alignment of the WD repeats in Gro and Tup1 reveals serial homology. In other words, each WD repeat of Gro is most closely related to the Tup1 WD repeat that occupies the corresponding position in that protein. These findings suggest that the evolutionary and therefore functional relationship between the two proteins may be closer than is suggested by the overall level of sequence homology.
The analysis presented here of the region of Gro N-terminal to the WD repeats reinforces the notion that there is a close functional relationship between Gro and Tup1. Like Tup1 (12), Gro interacts directly with core histones and preferentially with histone H3. We also found that, as with Tup1, the interaction of Gro with histones is modulated by acetylation. Both proteins bind preferentially to hypoacetylated histones. Furthermore, mutagenesis of the lysines in histone H4 known to be targets of acetylation abolishes the interaction between the histone H4 tail and Gro. Deletion analysis indicates that, as with Tup1, the entire N-terminal region of Gro excluding the WD repeats is necessary and sufficient for the interaction with histones. These findings suggest that, despite the lack of sequence homology, the N-terminal domains of the two proteins may assume similar folds designed to interact with histones in similar manners.
A further similarity between Gro and Tup1 comes from the finding that both appear to function as tetramers (6,31). In each case, tetramerization is mediated by putative coiled-coil motifs in the N-terminal domain of the protein. In the case of Tup1, the tetramer is probably associated with a single molecule of Ssn6. While not absolutely essential for Tup1 function, Ssn6 does enhance repression, perhaps by helping to direct Tup1 to target promoters (32,33). While it is not yet clear if Drosophila Gro interacts with a Drosophila Ssn6 homolog, the TLE proteins have been shown to bind UTY/X proteins, which could be mammalian homologs of Ssn6 (34). A search of the Drosophila genome reveals a gene encoding a protein with extensive similarity to the UTY/X proteins.
In conclusion, Gro and Tup1 show strikingly similar histone binding and oligomerization properties. Taken together with the serial homology in the WD-repeat domains, this functional similarity suggests that the two proteins should be considered orthologs of one another.
Critical positioning of the Gro tetramerization domain
Our attempt to determine if the Gro Q domain might mediate functions other than tetramerization led to an unexpected finding. In particular, we found that we could not restore function to full-length Gro containing a mutant tetramerization domain by adding the p53 tetramerization to the N-terminus of the protein. In contrast, addition of the p53 domain to a deletion variant of Gro completely lacking the Q domain largely restores repression. This finding suggests that the positioning of the tetramerization domain with respect to the remainder of the protein is critical for repression. One possible interpretation of this finding is that the proper juxtaposition of the four subunits of the repression domain, as determined by the position of the tetramerization domain, is critical for function. It remains to be determined whether or not Tup1 function exhibits similar structural requirements.
A model for Gro-mediated repression
We have recently shown that Gro interacts directly with the histone deacetylase Rpd3 via the GP domain in Gro, and that the activity of Rpd3 enhances Gro repression in S2 cells (7). These results suggest that Rpd3 might be a component of the Gro repressor complex. However, the phenotype of Gro-deficient embryos is more severe than the phenotype of Rpd3-deficient embryos (7,35). Thus, it is possible that Rpd3 only functions in a subset of Gro repression activities, or that other histone deacetylases compensate for the lack of Rpd3 function. It is interesting to note that disruption of yeast Rpd3 function alone does not impair Tup1-mediated repression and conversely, Rpd3 repression occurs in the absence of Tup1 (36). However, disruption of three major deacetylase complexes in yeast at the same time severely abrogates Tup1-mediated repression (S.Roth, personal communication). Thus, in Drosophila as well as in yeast, Rpd3, which is primarily a histone H4 deacetylase (27–30), may partially mediate Gro/Tup-1 repression. In agreement with this conclusion, the interaction between Gro and histone H4 depends upon the lysines in H4 that are targeted by Rpd3. While Rpd3 is a histone H4 deacetylase, it may be a part of a complex with other deacetylases that target other histones (14,37).
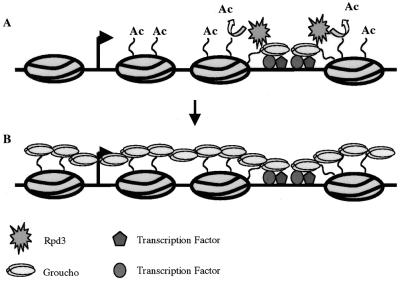
Recruitment of histone deacetylase by DNA-bound repressors in yeast causes deacetylation of histones H3 and H4 only within one or two nucleosomes of the repressor binding site (38). A model that reconciles this short-range deacetylation with the known ability of Gro to function as a long-range repressor (3) is shown in Figure 6. In particular, the initial recruitment of Gro, along with histone deacetylases could result in the local deacetylation of the template, which could, in turn, result in the further recruitment of Gro thereby enlarging the Gro-bound domain. The reiteration of this process could then generate a large transcriptionally-silent domain. An intriguing possibility is that the binding of Gro to histone tails might inhibit the activity of histone acetyl transferases, thus maintaining the hypoacetylated state that is correlated with repression.
Figure 6.
A speculative model for Gro-mediated repression. (A) Multiple transcription factors can individually or synergistically recruit Gro to the DNA template. Gro in turn recruits a histone deacetylase activity. (B) Polymerization of Gro along the chromatin template facilitated by interactions with deacetylated histones allows the establishment of a large transcriptionally-silent domain.
The mechanism for long-range repression by Gro proposed here is, in many ways, similar to the mechanism that has been proposed for silencing by the yeast Sir complex (22). This complex contains components (Sir3 and Sir4) that, like Gro, interact with the hypoacetylated tails of core histones, an interaction that has been proposed to mediate the spreading of heterochromatin to generate a transcriptionally-silent domain. In addition, recent experiments demonstrate that another component of the Sir complex (Sir2) functions as a histone deacetylase (39). It may thus be playing a role similar to that which we have proposed for Rpd3, i.e., by deacetylating histones it may strengthen the interaction between the corepressor complex and the chromatin template thus helping to maintain the silenced state. These observations suggest that there may be common features to all long range transcriptional silencing.
Acknowledgments
ACKNOWLEDGEMENTS
We thank James Kadonaga for providing purified and recombinant Drosophila histones, Andrew Carmen and Michael Grunstein for GST–histone tail expression plasmids and extensive technical advice, and Sharon Roth for the Tup1 plasmid for in vitro transcription. We thank James Posakony for pointing out sequence similarities between Gro and Tup1. This research was supported by NIH grant GM44522 and by a grant from the March of Dimes. R.D.F.-S. was partially supported by a CONACyT fellowship.
REFERENCES
- 1.Fisher A.L. and Caudy,M. (1998) Genes Dev., 12, 1931–1940. [DOI] [PubMed] [Google Scholar]
- 2.Parkhurst S.M. (1998) Trends Genet., 14, 130–132. [DOI] [PubMed] [Google Scholar]
- 3.Chen G. and Courey,A.J. (2000) Gene, 249, 1–16. [DOI] [PubMed] [Google Scholar]
- 4.Fisher A.L. and Caudy,M. (1998) Genes Dev., 12, 1931–1940. [DOI] [PubMed] [Google Scholar]
- 5.Smith T.F., Gaitatzes,C., Saxena,K. and Neer,E.J. (1999) Trends Biochem. Sci., 24, 181–185. [DOI] [PubMed] [Google Scholar]
- 6.Chen G., Nguyen,P.H. and Courey,A.J. (1998) Mol. Cell. Biol., 18, 7259–7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher A.L., Ohsako,S. and Caudy,M. (1996) Mol. Cell. Biol., 16, 2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahi M., Komachi,K. and Johnson,A.D. (1998) Cold Spring Harbor Symp. Quant. Biol., 63, 447–457. [DOI] [PubMed] [Google Scholar]
- 10.Treitel M.A. and Carlson,M. (1995) Proc. Natl Acad. Sci. USA, 92, 3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzamarias D. and Struhl,K. (1994) Nature, 369, 758–761. [DOI] [PubMed] [Google Scholar]
- 12.Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- 13.Huang L., Zhang,W. and Roth,S.Y. (1997) Mol. Cell. Biol., 17, 6555–6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davie J.R. (1998) Curr. Opin. Genet. Dev., 8, 173–178. [DOI] [PubMed] [Google Scholar]
- 15.Steger D.J. and Workman,J.L. (1996) Bioessays, 18, 875–884. [DOI] [PubMed] [Google Scholar]
- 16.Wolffe A.P. and Pruss,D. (1996) Cell, 84, 817–819. [DOI] [PubMed] [Google Scholar]
- 17.Edmondson D.G. and Roth,S.Y. (1998) Methods, 15, 355–364. [DOI] [PubMed] [Google Scholar]
- 18.Grant P.A., Duggan,L., Côté,J., Roberts,S.M., Brownell,J.E., Candau,R., Ohba,R., Owen-Hughes,T., Allis,C.D., Winston,F., Berger,S.L. and Workman,J.L. (1997) Genes Dev., 11, 1640–1650. [DOI] [PubMed] [Google Scholar]
- 19.Kuo M.H., Brownell,J.E., Sobel,R.E., Ranalli,T.A., Cook,R.G., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Nature, 383, 269–272. [DOI] [PubMed] [Google Scholar]
- 20.Luger K., Mäder,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 21.Palaparti A., Baratz,A. and Stifani,S. (1997) J. Biol. Chem., 272, 26604–26610. [DOI] [PubMed] [Google Scholar]
- 22.Hecht A., Laroche,T., Strahl-Bolsinger,S., Gasser,S.M. and Grunstein,M. (1995) Cell, 80, 583–592. [DOI] [PubMed] [Google Scholar]
- 23.Dubnicoff T., Valentine,S.A., Chen,G., Shi,T., Lengyel,J.A., Paroush,Z. and Courey,A.J. (1997) Genes Dev., 11, 2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baily T.L. and Elkan,C. (1994) Ismb, 2, 28–36. [PubMed] [Google Scholar]
- 25.Delidakis C. and Artavanis-Tsakonas,S. (1992) Proc. Natl Acad. Sci. USA, 89, 8731–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Voorn L. and Ploegh,H.L. (1992) FEBS Lett., 307, 131–134. [DOI] [PubMed] [Google Scholar]
- 27.Burgess S.M., Ajimura,M. and Kleckner,N. (1999) Proc. Natl Acad. Sci. USA, 96, 6835–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rundlett S.E., Carmen,A.A., Suka,N., Turner,B.M. and Grunstein,M. (1998) Nature, 392, 831–835. [DOI] [PubMed] [Google Scholar]
- 29.Vermaak D., Wade,P.A., Jones,P.L., Shi,Y.B. and Wolffe,A.P. (1999) Mol. Cell. Biol., 19, 5847–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rundlett S.E., Carmen,A.A., Kobayashi,R., Bavykin,S., Turner,B.M. and Grunstein,M. (1996) Proc. Natl Acad. Sci. USA, 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jabet C., Sprague,E.R., VanDemark,A.P. and Wolberger,C. (2000) J. Biol. Chem., 275, 9011–9018. [DOI] [PubMed] [Google Scholar]
- 32.Cooper J.P., Roth,S.Y. and Simpson,R.T. (1994) Genes Dev., 8, 1400–1410. [DOI] [PubMed] [Google Scholar]
- 33.Smith R.L., Redd,M.J. and Johnson,A.D. (1995) Genes Dev., 9, 2903–2910. [DOI] [PubMed] [Google Scholar]
- 34.Grbavec D., Lo,R., Liu,Y., Greenfield,A. and Stifani,S. (1999) Biochem. J., 337, 13–17. [PMC free article] [PubMed] [Google Scholar]
- 35.Mannervik M. and Levine,M. (1999) Proc. Natl Acad. Sci. USA, 96, 6797–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadosh D. and Struhl,K. (1997) Cell, 89, 365–371. [DOI] [PubMed] [Google Scholar]
- 37.Pazin M.J. and Kadonaga,J.T. (1997) Cell, 89, 325–328. [DOI] [PubMed] [Google Scholar]
- 38.Kadosh D. and Struhl,K. (1998) Mol. Cell. Biol., 18, 5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imai S., Armstrong,C.M., Kaeberlein,M. and Guarente,L. (2000) Nature, 403, 795–800. [DOI] [PubMed] [Google Scholar]