Abstract
PAMAM dendrimers are cationic polymers that have been used for the delivery of genes and oligonucleotides to cells. However, little is known about the behavior of dendrimer–nucleic acid complexes once they reach the cell interior. To pursue this issue, we prepared dendrimers conjugated with the fluorescent dye Oregon green 488. These were used in conjunction with oligonucleotides labeled with a red (TAMRA) fluorophore in order to visualize the sub-cellular distribution of the dendrimer–oligonucleotide complex and of its components by two-color digital fluorescence microscopy. The 2′-O-methyl antisense oligonucleotide sequence used in these studies was designed to correct splicing at an aberrant intron inserted into a luciferase reporter gene; thus effective delivery of the antisense agent results in the expression of the reporter gene product. The dendrimer–oligonucleotide complex remained associated during the process of uptake into vesicular compartments and eventual entry into the nucleus. Since the pharmacological activity of the antisense compound was manifest under these conditions, it suggests that the dendrimer–oligonucleotide complex is functionally active. A surprising result of these studies was that the Oregon green 488-conjugated dendrimer was a much better delivery agent for antisense compounds than unmodified dendrimer. This suggests that coupling of relatively hydrophobic small molecules to PAMAM dendrimers may provide a useful means of enhancing their capabilities as delivery agents for nucleic acids.
INTRODUCTION
Over the past few years, a variety of non-viral methods for the delivery of genes or oligonucleotides into cells have been introduced (1–3). These include cationic lipid complexes (4,5), polypeptides (6,7), surfactants (8), liposomes (9–11) and other reagents (12–14). One promising type of delivery agent, starburst polyamidoamine (PAMAM) dendrimers, are spherical, highly ordered, multiply-branched polymers with positively charged primary amino-groups on their surface at physiological pH (15). These surface amino groups provide useful moieties for the functional modification of dendrimers since they allow covalent coupling reactions under mild reaction conditions and in a controlled fashion. Recent reports have described dendrimer–peptide (16), dendrimer–antibody (17), dendrimer–oligonucleotide (18), dendrimer–fullerence (19), dendrimer–antibody–porphyrin (20) and dendrimer–sialic acid complexes (21).
PAMAM dendrimers have been known to form stable complexes with plasmid DNAs or oligonucleotides with modest cell toxicity, and with considerable effectivness in gene transfer or oligonucleotide delivery at the cell culture level (22–25). Currently the mechanism(s) of dendrimer-mediated antisense oligonucleotide delivery is not fully understood. It has been suggested that dendrimers might act similarly to cationic lipid delivery complexes, where internalization through endocytosis is followed by oligonucleotide release from endosomes to the cytoplasm, and then rapid migration of the released oligonucleotide to the nucleus (26–28). However, the question of how oligonucleotides enter into the nucleus from endosomes and what the function of dendrimers might be in this event are not really known. It has been reported that polyamines such as PEI (29), and spermidine conjugates (14,30) enhance cell uptake of nucleic acids. Recently it was reported that endocytosed PEI, whether administered with or without DNA, undergoes translocation to the nucleus (31). The protonated amino groups of PEI were predicted to buffer acidic endosomal compartments, assisting the neutralization and dissociation of the complexes in the endosome membrane, and eventually releasing oligonucleotide to the cytoplasm (29,31).
To understand the actions of dendrimers in the delivery of oligonucleotides into cells, fluorophore-conjugated PAMAM dendrimers were prepared; these were complexed with oligonucleotides having a second, distinguishable fluorophore, and then the intracellular localization of the complexes was evaluated by fluorescence microscopy and biochemical fractionation methods. Thus, the dye Oregon green 488 was covalently conjugated to the surface amines of PAMAM dendrimer generation 5 at increasing ratios of dye to amino groups. The ability of fluorophore-conjugated dendrimers to deliver oligonucleotides in pharmacologically-active form was tested by a recently developed reporter assay that utilizes an antisense oligonucleotide to correct splicing of a mutated intron inserted into a luciferase gene stably expressed in HeLa cells (32). Surprisingly, the reporter gene assay demonstrated a dramatic increase in the luciferase expression level when antisense oligonucleotides were delivered with the Oregon green–dendrimer conjugates, as compared to unmodified PAMAM dendrimers. This suggests that simple covalent modifications of dendrimer surface moieties may allow substantial improvements in the nucleic acid delivery characteristics of these polymers.
MATERIALS AND METHODS
Materials
PAMAM dendrimers, which have ethylenediamine as an initiator core, were purchased from Dendritech (Dendritech Inc, MI) and used without further purification. Oregon Green 488 carboxylic acid succinimidyl ester 5-isomer was purchased from Molecular Probes and prepared in DMSO (1 mg/ml). Lipofectamine, as well as DMEM and Opti-MEM, were from Life Technologies (Gaithersburg, MD). Lipofectin and SuperFect were obtained from Gibco BRL (Baltimore, MD) and Qiagen (Valencia, CA), respectively. A phosphorothioate 2′-O-methyl-oligonucleotide (5′-CCUCUUACCUCAGUUACA-3′) and corresponding 3′-TAMRA-labeled derivative were purchased from the Midland Certified Reagent Company (Midland, TX). HeLa cells stably transfected with plasmid pLuc/705 were a generous gift of Dr R. Kole (University of North Carolina, Chapel Hill, NC). The ability of this particular antisense sequence to correct splicing and thus result in luciferase activity has been fully described in previous publications (22,32).
Oregon green 488 conjugation
Appropriate amounts of Oregon green 488 carboxylic acid succinimidyl ester (Molecular Probes) in DMSO (1 mg/ml) were added to the generation 5 dendrimer in 1 ml of 50 mM sodium borate buffer (pH 8.5), after which stirring continued for 3 h in the dark at room temperature. The crude reaction solution was evaporated in a Speed-Vac. This dried pellet was dissolved in a minimum volume of aqueous TFA (0.1%, v/v), and then applied to a PD-10 Sephadex G-25M column equilibrated with aqueous TFA (0.1%, v/v). Fractions recovered from the Sephadex G-25 were analyzed by TLC on a non-fluorescent silica plate using 100% MeOH as the eluent. Under long wavelength UV light, the tagged dendrimers remained almost immobile at the origin, while unconjugated Oregon green 488 migrated fairly close to the solvent front (Rf = 0.8). The conjugate products were further confirmed by measuring fluorescence intensity of each fraction at 525 nm. The purified fractions were dialyzed, the absorbance measured at the Oregron green peak, and then the material was completely dried in a Speed-Vac before measuring the weight. The approximate fluorophore to dendrimer ratios were calculated based on the extinction coefficient of Oregon green and on the reported average molecular mass of the dendrimer (provided by the supplier); it was assumed that the fluorophore made a negligible contribution to the total mass of the conjugate.
Cytotoxicity assay
A cytotoxicity assay was performed by plating cells into 24-well plates (Nunc) at 2 × 104 cells/well. Cells were incubated with complexes of oligonucleotides (0.25 µM) and dendrimers, with oligonucleotide alone (0.25 µM), or with control medium, for 24 h in serum-free or 30% serum-containing medium, rinsed twice with phosphate-buffered saline (PBS), followed by the addition of 2 ml Opti-MEM (Gibco BRL) and incubation for a further 24 h. The surviving fraction was determined by the MTT dye assay (33); 50 µl of MTT dye solution (0.5 µg/ml) was added to each well and the plates returned to the 37°C incubator for 3 h; after removing the Opti-MEM, 0.5 ml of DMSO was added to each well and incubation continued for another 30 min. Absorbance at 540 nm was quantitiated with an automated microplate reader (BioTech EL 340).
Antisense splicing correction assay
HeLa cells stably transfected with a luciferase reporter gene construct were plated in 6-well trays at a density of 3 × 105 cells/well in 3 ml of 10% FBS/DMEM and antibiotics. Cells were maintained for 24 h at 37°C in a humidified incubator (5% CO2/95% air). A 100 µl aliquot of oligonucleotide at a given concentration in Opti-MEM was combined with 100 µl of Opti-MEM containing various concentrations of PAMAM dendrimer. After being briefly mixed, the preparation was left undisturbed at room temperature for 5 min, followed by dilution to 1 ml with Opti-MEM, or Opti-MEM plus serum, before being layered on the cells. The cells were incubated for 6 h and subsequently the medium was replaced with 10% FBS/DMEM. After a further 18h, the cells were rinsed with PBS and lysed in 100 µl of lysis buffer (200 mM Tris–HCl pH 7.8, 2 mM EDTA, 0.05% Triton X-100) on ice for 15 min. Following centrifugation [13 000 r.p.m. (Eppendorf table-top centrifuge), 2 min], 5 µl of supernatant cell extract was mixed with 100 µl of luciferase assay buffer (3 mM ATP, 15 mM MgSO4, 30 mM tricine, 10 mM DTT pH 7.8) and 100 µl of luciferase substrate (1 mM d-luciferin). The light emission was quantified for 10 s using a Monolight®2010 luminometer (Analytical Luminescence Laboratory, USA). Luciferase activity was expressed as relative luminescence units (RLU) per well. Light emission was normalized to the protein concentration of each sample, determined according to the bicinchonic acid assay (Pierce Chemical Co.) for protein concentration.
Preparation of labeled oligonucleotide
A single stranded 18 base 2′-O-methyl phosphorothioate oligonucleotide was labeled with [γ-32P]ATP using T4 polynucleotide kinase (USB). Labeled oligonucleotide was purified from free [γ-32P]ATP by 16% PAGE. The band corresponding to the labeled oligonucleotide was cut from the gel and extracted with water in a microcentrifuge tube. The concentration of the labeled oligonucleotide was determined by using a UV spectrometer (BioSpec-1601, Shimadzu).
Fluorescence microscopy
Cells were seeded at a density of 5 × 104 cells/well in a 6-well plate containing glass coverslips. After 24 h, the medium in each well was replaced with the complexes of oligonucleotide–Oregon green 488-conjugated dendrimer in 1 ml Opti-MEM. After 1 h incubation at 37°C, cells were washed three times with cold PBS and then fixed by adding 3 ml of 3.5% paraformaldehyde in PBS for 1 h. Cells were washed three times with cold PBS, then additionally washed with distilled water. Cells were then mounted directly onto glass microscope slides using PermaFluor aqueous mountant (Lipshaw Immunon), sealed with clear nail polish, and processed for fluorescence microscopy to provide cellular locations of dendrimer and oligonucleotide.
Nuclei isolation by cellular fractionation
A 10-cm culture dish of 80% confluent HeLa cells stably transfected with pLuc/705 was treated with Oregon green 488-conjugated dendrimer–antisense oligonucleotide (0.25 µM) complex or oligonucleotide alone as described above. A trace amount of 32P-labeled phosphorothioate oligonucleotide was included. After incubating for 1 h, the cell monolayer was washed twice with cold PBS, scraped with a rubber policeman in 5 ml PBS, and then pelleted by centrifugation (1000 r.p.m., Eppendorf table-top centrifuge) for 5 min at 4°C. The pellets were resuspended in 0.5 ml of cold detergent-free hypotonic buffer (15 mM HEPES pH 7.6, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT and 10 mM Na2S2O5), and centrifuged for 10 min at 3000 r.p.m. (Eppendorf table-top centrifuge). The recovered cell pellets were resuspended in 0.5 ml of hypotonic buffer, allowed to swell on ice for 10 min before shearing through a 22-gauge needle. Lysed cells (>95%) were observed by addition of trypan blue to an aliquot of cells. Cells were transferred to a centrifuge tube and the nuclei were collected by centrifuging for 15 min at 4000 r.p.m. (3300 g). The recovered nuclear pellets were resuspended in 90 µl of low salt buffer (20 mM HEPES pH 7.9, 25% glycerol, 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT) and 100 µl of high salt buffer (substituting 20 mM KCl of low salt buffer with 0.8 M KCl) was added, in a dropwise fashion, while stirring slowly. The nuclei were allowed to extract for 30 min on ice with gentle mixing and then followed by centrifuging for 30 min at 14 500 r.p.m. (25 000 g). The amount of radioactive oligonucleotides and conjugated dendrimers that remained associated with the washed cells or isolated nuclei was obtained by counting 32P in a scintilation counter and measuring UV absorbance of Oregon green 488 at 502 nm, respectively. For Hoechst staining, the nuclear fraction in distilled water was mixed with Hoechst dye solution (1:1000 dilution in water) and mounted on glass slides for fluorescence microscopy.
RESULTS
Preparation of Oregon green 488-conjugated dendrimer
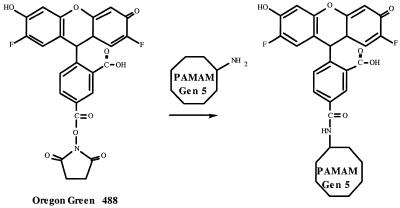
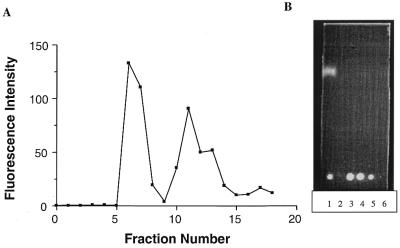
Oregon green 488, a FITC derivative, was conjugated to the surface amines of dendrimer generation 5 at various ratios (Fig. 1). The conjugated products were purified by gel filtration column chromatography and confirmed by measuring fluorescence intensities and TLC analysis of the collected fractions (Fig. 2). As indicated, column fractions 5–9 contain the conjugated dendrimer while fractions 10–15 contain unreacted fluorophore.
Figure 1.
Synthetic scheme of conjugation reaction.
Figure 2.
Purification of Oregon green 488-conjugated dendrimer generation 5 (G5-Org). The reaction solution in a minimum volume was applied to a PD-10 Sephadex G-25M column equilibrated with aquous TFA (0.1%, v/v). Fractions recovered from the PD-10 Sephadex G-25 were analyzed by fluorescence spectroscopy and TLC method. (A) An aliquot of 5 µl out of each 500 µl fraction was mixed with 1 ml of 50 mM sodium borate buffer (pH 8.5). The fluorescence intensity was measured at the emission wavelength of 525 nm after exciting at 495 nm. (B) On a fluorescent TLC plate using MeOH as the eluent, the conjugated dendrimers remained almost immobile at the origin, while unconjugated Oregon green 488 migrated fairly close to the solvent front as seen under long wave UV light. Lane 1, diluted crude reaction; lanes 2–6, fractions collected from fraction numbers 5–9, respectively, in (A).
Cytotoxicity
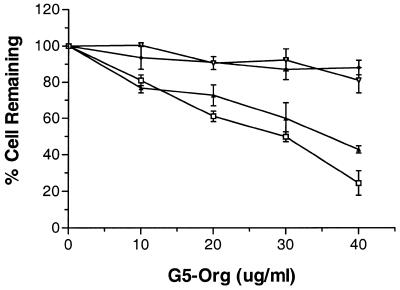
The toxic effects of Oregon green 488-conjugated dendrimer or oligonucleotide–Oregon green 488-conjugated dendrimer complexes were assessed by an MTT dye assay (33). Figure 3 shows the percent cell survival versus concentration, after treatment of cells with generation 5 dendrimer in serum-free or 30% serum-containing medium. A marked decrease in the toxicity of dendrimer or dendrimer–oligonucleotide complexes was noted when cells were treated in the presence of serum, while higher toxicity was observed for dendrimer alone or the complexes in serum-free medium.
Figure 3.
Acute toxicity assay of G5-Org. Conjugated dendrimer alone or dendrimer–oligonucleotide (OD) complexes were applied to cells with and without serum. Cell viability was expressed as percent cells remaining compared to untreated cells, based on the MTT assay. Open inverted triangles, G5-Org alone in 30% serum-containing medium; filled diamonds, G5-Org–OD complexes in 30% serum-containing medium; Filled triangles, G5-Org–OD complexes in serum-free medium; open squares, G5-Org alone in serum-free medium. Data represent the average of three experiments.
Fluoro-dendrimers enhance the cellular uptake of oligonucleotides
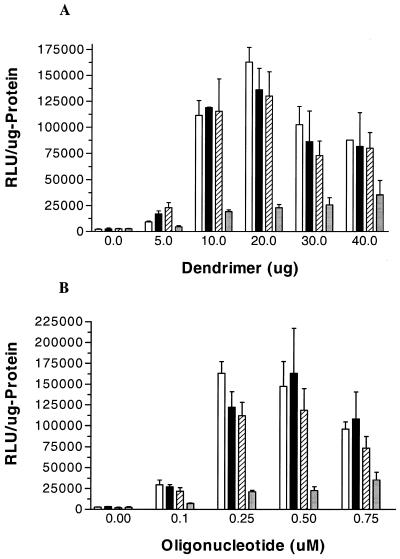
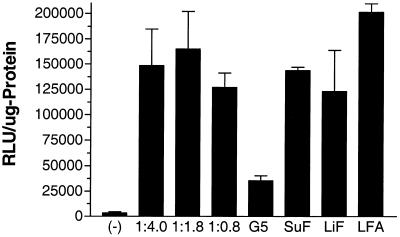
The prepared fluorophore-conjugated dendrimers were tested for their delivery efficiency by a luciferase reporter gene assay, as described in Materials and Methods. Somewhat surprisingly, after incubation with oligonucleotide–dendrimer complexes, a dramatic effect on luciferase expression was obtained for the Oregon green 488 conjugates (Fig. 4). Expression was dose-dependent for both oligonucleotide and dendrimer concentrations, with the maximum effect at 0.5 µM for oligonucleotide and 20 µg for Oregon green 488-conjugated dendrimer, respectively. Results with the conjugated dendrimers were compared to commercially available transfection agents and to unconjugated dendrimer (Fig. 5). The luciferase expression levels attained with fluorophore-conjugated dendrimer were substantially greater than those obtained with unconjugated dendrimer. Importantly, a mock conjugation reaction did not enhance the luciferase activity (data not shown). Thus, it is the addition of the relatively bulky fluorophore residues that seems to enhance the transfection ability of the dendrimer.
Figure 4.
Activation of a luciferase reporter using G5-Org–antisense complexes. (A) HeLa cells stably transfected with pLuc/705 were treated with various concentrations (µg/ml) of G5-Org at fixed concentration (0.25 µM) of a 2′-O-Me phosporothioate oligonucleotide. (B) HeLa cells stably transfected with pLuc/705 were treated with various concentrations of a 2′-O-Me phosporothioate oligonucleotide at fixed concentration (20 µg/ml) of G5-Org. The ratio of Oregon green 488 to each generation 5 dendrimer is 4:1 (open bar), 1.75:1 (filled bar), 0.8:1 (hatched bar) and unconjugated generation 5 dendrimer (shaded bar). Data represent the average of at least three experiments.
Figure 5.
Comparison of luciferase activities of fluorophore-conjugated dendrimers with commercial cytofectins at optimized concentrations of delivery agent and oligonucleotide. Ordinate: RLU/µg of protein. Control (–) followed by three preparations of dendrimer conjugated with different ratios of Oregon green 488 and with 0.25 mM oligonucleotide. G5, unconjugated dendrimer; SuF, SuperFect (with 0.5 µM oligonucleotide); LFA, Lipofectamine (with 0.25 µM oligonucleotide); LiF, Lipofectin (with 0.5 µM oligonucleotide). Vertical bars represent means and standard errors. Data represent the average of at least three experiments.
Serum effects on oligonucleotide delivery by Oregon green 488-conjugated dendrimer
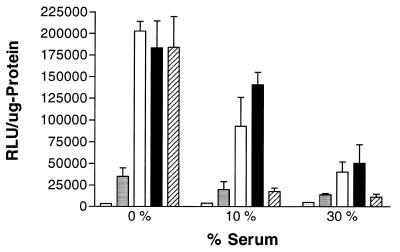
The ability of fluorophore-conjugated generation 5 dendrimer to deliver oligonucleotides to HeLa Luc/705 cells was compared with commercial cytofectins in the absence or presence of serum at a fixed concentration of reagents (0.25 µM for oligonucleotide and 20 µg for fluoro-dendrimer) (Fig. 6). The commercial dendrimer Superfect showed excellent activity in the absence of serum, and maintained activity at 30% serum concentration. Lipofectamine, a cationic lipid cytofectin, showed excellent activity in the absence of serum but displayed almost no activity in the presence of serum. The conjugated dendrimer showed similar activity to Lipofectamine or Superfect in serum-free medium, and maintained a degree of effectiveness in the presence of 30% serum that was slightly less than Superfect.
Figure 6.
Serum effects on oligonucleotide delivery by G5-Org. Serum dependence of oligonucleotide delivery by conjugated dendrimer was compared with commercial delivery agents, Lipofectamine and SuperFect. The complexes of oligonucleotide and fluorophore-conjugated dendrimer were formed at constant ratio (0.5 µM for oligonucleotide and 20 µg/ml for G5-Org). Cells treated with SuperFect–oligonucleotide or Lipofectamine–oligonucleotide complexes prepared according to the manufacturer’s instructions were used for comparison. Additional control samples underwent the same oligonucleotide treatment, but in the absence of delivery agent. The abscissa indicates the percent of fetal calf serum present during the incubation with oligonucleotides. The ordinate indicates the RLU/µg of protein. Sparsely dot-filled bar, negative control; densely dot-filled bar, unconjugated generation 5 dendrimer; open bar, 1.75:1 of Oregon green 488 to generation 5 dendrimer; filled bar, SuperFect; hatched bar, Lipofectamine. Data represent the average of at least three experiments.
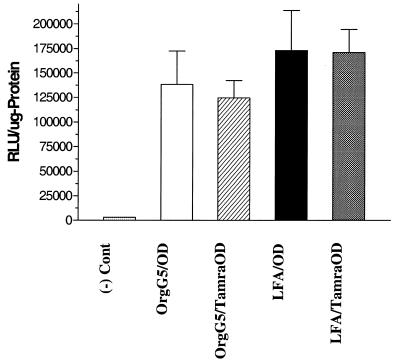
TAMRA-conjugation of the oligonucleotide does not alter pharmacological effects
Since we were interested in the relationship between sub-cellular localization and pharmacological effects, we investigated whether the labeling of the oligonucleotide with a fluorophore would affect its biological activity. The modification of the oligonucleotide at the 3′-end with TAMRA fluorophore did not alter antisense efficacy. Luciferase expression due to TAMRA-oligonucleotide was indistinguishable from that of oligonucleotide without TAMRA when delivered with either Oregon green 488-conjugated dendrimer or commercial Lipofectamine. This indicates that TAMRA-oligonucleotide, like unmodified oligonucleotide, was able to enter the nucleus and bind effectively to the complementary target sequence (Fig. 7).
Figure 7.
Comparison of luciferase activities of TAMRA-oligonucleotide with unmodified oligonucleotide. Cells were treated with conjugated dendrimer–oligonucleotide or Lipofectamine–oligonucleotide complexes as described in Materials and Methods at a concentration of conjugated dendrimer (20 µg/ml) and oligonucleotide (0.25 µM). Dot-filled bar, negative control; open bar, conjugated dendrimer–oligonucleotide complex; hatched bar, conjugated dendrimer–TAMRA-oligonucleotide; filled bar, Lipofectamine–oligonucleotide complex; shaded bar, Lipofectamine–TAMRA-oligonucleotide. Data represent the average of at least three experiments.
Cellular distribution of Oregon green 488-conjugated dendrimer and TAMRA-oligonucleotide
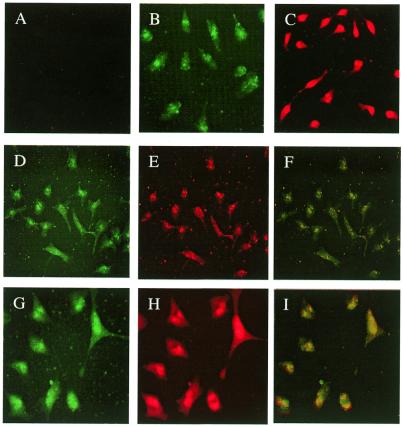
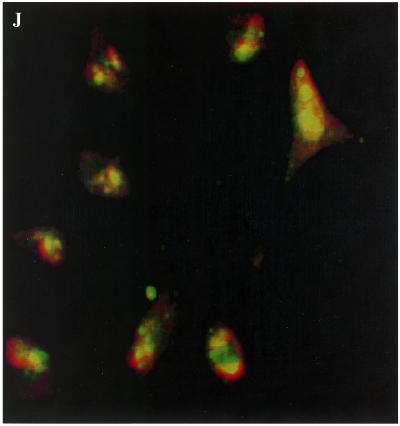
Cellular uptake of both dendrimer and oligonucleotides was investigated after incubation of Oregon green 488-conjugated dendrimer–TAMRA-oligonucleotide complexes with HeLa cells in culture. A large majority of cells showed strong fluorescence intensity in the nuclei. Digital overlay of the green (dendrimer) and red (oligonucleotide) fluorescence patterns indicated that the two types of molecule were always in close proximity, as illustrated in Figure 8. Interestingly, cells treated with Oregon green 488-conjugated dendrimer alone also showed nuclear fluoresence (Fig. 8B). At low concentrations of oligonucleotide (0.25 µM) complexed with Oregon green 488-conjugated dendrimer (20 µg in 1 ml), a homogeneous, but slightly ‘clumpy’ cellular distribution of both dendrimer and oligonucleotide was observed (Fig. 8D–F). However, at higher concentration of oligonucleotide (0.5 µM) complexed with Oregon green 488-conjugated dendrimer, strong and overlapping uniform nuclear accumulations for both dendrimer and oligonucleotides were observed (Fig. 8G–I).
Figure 8.
Cellular distribution of G5-Org and TAMRA-oligonucleotides. HeLa cells grown on glass coverslips were treated with G5-Org/3′-end TAMRA-labeled oligonucleotide (OD705) complexes at the ratio of amine to Oregon green of 50:1. Cells were incubated with 3′-TAMRA-oligonucleotide–G5-Org complexes for 1 h, fixed in 3.5% paraformaldehyde, and processed for fluoroscence microscopy. (A) TAMRA-oligonucleotide alone (0.5 µM); (B) G5-Org alone (20 µg/ml); (C) TAMRA-oligonucleotide transfected using Lipofectamine (4 µl/ml). (D–F) TAMRA-oligonucleotide (0.25 µM)–G5-Org (20 µg/ml) complexes 1 h post-incubation: (D) G5-Org through green channel; (E) TAMRA-oligonucleotides through red channel; (F) superimposition of (D) and (E). (G–I) TAMRA-oligonucleotide (0.5 µM)–G5-Org complexes 1 h post-incubation: (G) G5-Org through green channel; (H) TAMRA-oligonucleotides through red channel; (I) superimposition of G and H. (J) An expanded version of the overlap image.
Cellular fractionation
The nuclear accumulation of Oregon green 488-conjugated dendrimer–TAMRA-oligonucleotide complexes was further confirmed by subcellular fractionation (Fig. 9). Recovered nuclei contained ∼5.27% of the total dendrimers and 4.81% of oligonucleotides, respectively. The dendrimer/oligonucleotide ratio in the nucleus was similar to that of the material associated with intact cells, once again suggesting that the dendrimer and oligonucleotide traffic to the nucleus as a single entity (Table 1).
Figure 9.
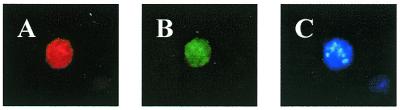
Localization of fluorophore-conjugated dendrimers and TAMRA-oligonucleotides after isolation of nuclei. (A) G5-Org in a nucleus. (B) TAMRA-oligonucleotide in a nucleus. (C) Nucleus stained with Hoechst’s reagent (Molecular Probes) as a positive control.
Table 1. Nuclear localization of oligonucleotide and conjugated dendrimer in HeLa cells.
| Whole cell (%) | Nuclei (%) | |||
|---|---|---|---|---|
| |
G5-Org |
OD |
G5-Org |
OD |
| G5-Org/OD | 14.9 | 20.7 | 3.01 | 4.81 |
| OD alone | – | 1.61 | – | 0.10 |
Nuclei were isolated as described in Materials and Methods. The percent of total radioactive oligonucleotides (OD) and conjugated dendrimers (G5-Org) in the incubation mix that remained associated with the washed cells or isolated nuclei were calculated. This was done by dividing total 32P-labeled oligonucleotide or the total amount of conjugated dendrimer (based on Oregon green absorbance at 495 nm) by 32P intensity or dendrimer amounts in the isolates. Data represent the average of two separate experiments.
DISCUSSION
There has been a substantial interest recently in the use of polycationic dendrimers to deliver both plasmid DNA and short antisense oligonucleotides (16,22–25). While significant delivery and biological effects have been reported in a number of cases, little is known about the mechanisms involved. Our initial goal in this study was to use fluorescent-tagged molecules to study the sub-cellular distribution of dendrimer–oligonucleotide complexes. Thus, both through use of fluorescence microscopy, and by cell fractionation techniques, we have found that dendrimer–oligonucleotide complexes persist as the oligonucleotide enters the cell nucleus where its pharmacological effects are manifested. This seems to be quite different from the situation with cationic lipid delivery agents, where a separation of the nucleic acid and the delivery agent seems to occur in endosomes (27,28). Although there is clearly an accumulation of dendrimer and oligonucleotide in the nucleus, a substantial amount of material remains bound at other sites in the cell, presumably the plasma membrane and endomembrane compartments. The relatively low recovery of dendrimer–oligonucleotide complexes in the nucleus in our cell fractionation studies is probably an underestimate of the amount of nuclear-associated material, since there were substantial losses in the fractionation process that were difficult to control.
A surprising result of this study is that conjugation of the dendrimer with the Oregon green fluorophore significantly enhanced the dendrimer’s abilities as a delivery agent. Thus, conjugation of one to several molecules of the fluorescent dye resulted in much stronger biological effects for the conjugated, as opposed to the unmodified, dendrimer, when tested at equal antisense oligonucleotide concentrations. The basis for this effect is unclear at present. One possibility is that the relatively hydrophobic fluor moieties enhance the ability of the dendrimer to disrupt endosomal membranes and thus traffic to the cytosol and nucleus; several agents designed to cause endosomal disruption have had positive effects on oligonucleotide delivery (5,8). Presumably conjugation of a large fraction of the surface amino groups of the dendrimer would interfere with its ability to bind nucleic acids; however, titration of a few of the amino groups seems to be compatible with retention of strong binding ability. Although this has not been directly tested, it seems unlikely that there is something unique about Oregon green 488 in terms of its ability to modify dendrimer delivery properties. Presumably conjugation with a variety of other organic molecules might have similar, or better, effects; this opens the possibility of enhancing the delivery properties of cationic dendrimers via very simple chemical modifications of the dendrimer surface.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by NIH grant CA47044 to R.L.J. We thank B. Asam for word processing and editorial assistance.
REFERENCES
- 1.Juliano R.L. and Yoo,H. (2000) Curr. Opin. Mol. Ther., 2, 297–303. [PubMed] [Google Scholar]
- 2.Luo D. and Saltzman,W.M. (2000) Nat. Biotechnol., 18, 33–37. [DOI] [PubMed] [Google Scholar]
- 3.Juliano R.L., Alahari,S., Yoo,H., Kole,R. and Cho,M. (1999) Pharm. Res., 16, 494–502. [DOI] [PubMed] [Google Scholar]
- 4.Alahari S.K., DeLong,R., Fisher,M.H., Dean,N.M., Viliet,P. and Juliano,R.L. (1998) J. Pharmacol. Exp. Ther., 286, 419–428. [PubMed] [Google Scholar]
- 5.Bennett C.F., Chiang,M.Y., Chan,H., Shoemaker,J.E. and Mirabelli,C.K. (1992) Mol. Pharmacol., 41, 1023–1033. [PubMed] [Google Scholar]
- 6.Rajur S.B., Roth,C.M., Morgan,J.R. and Yarmush,M.L. (1997) Bioconjug. Chem., 8, 935–940. [DOI] [PubMed] [Google Scholar]
- 7.Degols G., Leonetti,J.P. and Lebleu,B. (1992) Ann. NY Acad. Sci., 660, 331–333. [DOI] [PubMed] [Google Scholar]
- 8.Liang E. and Hughes,J. (1998) Biochim. Biophys. Acta, 1369, 39–50. [DOI] [PubMed] [Google Scholar]
- 9.Gao X. and Huang,L. (1995) Gene Ther., 2, 710–722. [PubMed] [Google Scholar]
- 10.De Oliveira M.C., Fattal,E., Couvreur,P., Lesieur,P., Bourgaux,C., Ollivon,M. and Dubernet,C. (1998) Biochim. Biophys. Acta, 1372, 301–310. [DOI] [PubMed] [Google Scholar]
- 11.Juliano R.L. and Akhtar,S. (1992) Antisense Drug Res. Dev., 2, 165–176. [DOI] [PubMed] [Google Scholar]
- 12.Alahari S.K., Dean,N.M., Fisher,M.H., Delong,R., Manoharan,M., Tivel,K.L. and Juliano,R.L. (1996) Mol. Pharmacol., 50, 808–819. [PubMed] [Google Scholar]
- 13.Zhao Q., Temsamani,J. and Agrawal,S. (1995) Antisense Drug Res. Dev., 5, 185–192. [DOI] [PubMed] [Google Scholar]
- 14.DeLong R.K., Yoo,H., Alahari,S.K., Fisher,M., Short,S.M., Zirbes,E., Kole,R., Janout,V., Regan,S.L. and Juliano,R.L. (1999) Nucleic Acids Res., 27, 3334–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomalia D.A., Naylor,A.M. and Goddard,W.A. (1990) Angew Chem. Int. Ed. Engl., 29, 138–175. [Google Scholar]
- 16.Haensler J. and Szoka,F.C.,Jr (1993) Bioconjug. Chem., 4, 372–379. [DOI] [PubMed] [Google Scholar]
- 17.Singh P., Moll,F.,III, Lin,S.H., Ferzli,C., Yu,K.S., Koski,R.K., Saul,R.G. and Cronin,P. (1994) Clin. Chem., 40, 1845–1849. [PubMed] [Google Scholar]
- 18.Shchepinov M.S., Udalova,I.A., Bridgman,A.J. and Southern,E.M. (1997) Nucleic Acids Res., 25, 4447–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wooley K.L., Hawker,C.J., Frechet,J.M.J., Wudl,F., Srdanov,G., Shi,S., Li,C. and Kao,M. (1993) J. Am. Chem. Soc., 115, 9836–9837. [Google Scholar]
- 20.Roberts J.C., Adams,Y.E., Tomalia,D., Mercer-Smith,J.A. and Lavallee,D.K. (1990) Bioconjug. Chem., 1, 305–308. [DOI] [PubMed] [Google Scholar]
- 21.Reuter J.D., Myc,A., Hayes,M.M., Gan,Z., Roy,R., Qin,D., Yin,R., Piehler,L.T., Esfand,R., Tomalia,D.A. and Baker,J.R.,Jr (1999) Bioconjug. Chem., 10, 271–278. [DOI] [PubMed] [Google Scholar]
- 22.Yoo H., Sazani,P. and Juliano,R.L. (1999) Pharm. Res., 16, 1799–1804. [DOI] [PubMed] [Google Scholar]
- 23.Bielinska A.U., Chen,C., Johnson,J. and Baker,J.R.,Jr (1999) Bioconjug. Chem., 10, 843–850. [DOI] [PubMed] [Google Scholar]
- 24.Helin V., Gottikh,M., Mishal,Z., Subra,F., Malvy,C. and Lavignon,M. (1999) Biochem. Pharmacol., 58, 95–107. [DOI] [PubMed] [Google Scholar]
- 25.Delong R., Stephenson,K., Loftus,T., Fisher,M., Alahari,S., Nolting,A. and Juliano,R.L. (1997) J. Pharm. Sci., 86, 762–764. [DOI] [PubMed] [Google Scholar]
- 26.Marcusson E.G., Bhat,B., Manoharan,M., Bennett,C.F. and Dean,N.M. (1998) Nucleic Acids Res., 26, 2016–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zelphati O. and Szoka,F.C.,Jr (1996) Proc. Natl Acad. Sci. USA, 93, 11493–11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zelphati O. and Szoka,F.C.,Jr (1996) Pharm. Res., 13, 1367–1372. [DOI] [PubMed] [Google Scholar]
- 29.Boussif O., Lezoualcqh,F., Zanta,M.A., Mergny,M.D., Scherman,D., Demeneix,B. and Behr,J.P. (1995) Proc. Natl Acad. Sci. USA, 92, 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guy-Caffey J.K., Bodepudi,V., Bishop,J.S., Jayaraman,K. and Chaudhary,N. (1995) J. Biol. Chem., 270, 31391–31396. [DOI] [PubMed] [Google Scholar]
- 31.Godbey W.T., Wu,K.K. and Mikos,A.G. (1999) Proc. Natl Acad. Sci. USA, 96, 5177–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang S.H., Cho,M.J. and Kole,R. (1998) Biochemistry, 37, 6235–6239. [DOI] [PubMed] [Google Scholar]
- 33.Carmichael J., DeGraff,W.G., Gazdar,A.F., Minna,J.D. and Mitchell,J.B. (1987) Cancer Res., 47, 943–946. [PubMed] [Google Scholar]