Abstract
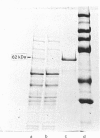
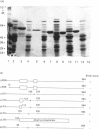
Xylanase A (XYLA) from Pseudomonas fluorescens subspecies cellulosa shows sequence conservation with two endoglucanases from the same organism. The conserved sequence in XYLA, consisting of the N-terminal 234 residues, is not essential for catalytic activity. Full-length XYLA and a fusion enzyme, consisting of the N-terminal 100 residues of XYLA linked to mature alkaline phosphatase, bound tightly to crystalline cellulose (Avicel), but not to xylan. The capacity of truncated derivatives of the xylanase to bind polysaccharides was investigated. XYLA lacking the first 13 N-terminal amino acids did not bind to cellulose. However, a catalytically active XYLA derivative (XYLA'), in which residues 100-234 were deleted, bound tightly to Avicel. Substrate specificity, cellulose-binding capacity, specific activity and Km for xylan hydrolysis were evaluated for each of the xylanases. No differences in any of these parameters were detected for the two enzymes. It is concluded that XYLA contains a cellulose-binding domain consisting of the N-terminal 100 residues which is distinct from the active site. Spatial separation of the catalytic and cellulose-binding domains is not essential for the enzyme to function normally.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biely P., Mislovicová D., Toman R. Soluble chromogenic substrates for the assay of endo-1,4-beta-xylanases and endo-1,4-beta-glucanases. Anal Biochem. 1985 Jan;144(1):142–146. doi: 10.1016/0003-2697(85)90095-8. [DOI] [PubMed] [Google Scholar]
- Ghangas G. S., Wilson D. B. Cloning of the Thermomonospora fusca Endoglucanase E2 Gene in Streptomyces lividans: Affinity Purification and Functional Domains of the Cloned Gene Product. Appl Environ Microbiol. 1988 Oct;54(10):2521–2526. doi: 10.1128/aem.54.10.2521-2526.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert H. J., Jenkins G., Sullivan D. A., Hall J. Evidence for multiple carboxymethylcellulase genes in Pseudomonas fluorescens subsp. cellulosa. Mol Gen Genet. 1987 Dec;210(3):551–556. doi: 10.1007/BF00327211. [DOI] [PubMed] [Google Scholar]
- Gilbert H. J., Sullivan D. A., Jenkins G., Kellett L. E., Minton N. P., Hall J. Molecular cloning of multiple xylanase genes from Pseudomonas fluorescens subsp. cellulosa. J Gen Microbiol. 1988 Dec;134(12):3239–3247. doi: 10.1099/00221287-134-12-3239. [DOI] [PubMed] [Google Scholar]
- Hall J., Gilbert H. J. The nucleotide sequence of a carboxymethylcellulase gene from Pseudomonas fluorescens subsp. cellulosa. Mol Gen Genet. 1988 Jul;213(1):112–117. doi: 10.1007/BF00333406. [DOI] [PubMed] [Google Scholar]
- Hall J., Hazlewood G. P., Huskisson N. S., Durrant A. J., Gilbert H. J. Conserved serine-rich sequences in xylanase and cellulase from Pseudomonas fluorescens subspecies cellulosa: internal signal sequence and unusual protein processing. Mol Microbiol. 1989 Sep;3(9):1211–1219. doi: 10.1111/j.1365-2958.1989.tb00271.x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langsford M. L., Gilkes N. R., Singh B., Moser B., Miller R. C., Jr, Warren R. A., Kilburn D. G. Glycosylation of bacterial cellulases prevents proteolytic cleavage between functional domains. FEBS Lett. 1987 Dec 10;225(1-2):163–167. doi: 10.1016/0014-5793(87)81150-x. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavin M., Forsberg C. W. Catalytic and substrate-binding domains of endoglucanase 2 from Bacteroides succinogenes. J Bacteriol. 1989 Jun;171(6):3310–3315. doi: 10.1128/jb.171.6.3310-3315.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Teeri T. T., Lehtovaara P., Kauppinen S., Salovuori I., Knowles J. Homologous domains in Trichoderma reesei cellulolytic enzymes: gene sequence and expression of cellobiohydrolase II. Gene. 1987;51(1):43–52. doi: 10.1016/0378-1119(87)90472-0. [DOI] [PubMed] [Google Scholar]
- Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem. 1988 Jan 4;170(3):575–581. doi: 10.1111/j.1432-1033.1988.tb13736.x. [DOI] [PubMed] [Google Scholar]
- Warren R. A., Beck C. F., Gilkes N. R., Kilburn D. G., Langsford M. L., Miller R. C., Jr, O'Neill G. P., Scheufens M., Wong W. K. Sequence conservation and region shuffling in an endoglucanase and an exoglucanase from Cellulomonas fimi. Proteins. 1986 Dec;1(4):335–341. doi: 10.1002/prot.340010407. [DOI] [PubMed] [Google Scholar]
- West C. A., Elzanowski A., Yeh L. S., Barker W. C. Homologues of catalytic domains of Cellulomonas glucanases found in fungal and Bacillus glycosidases. FEMS Microbiol Lett. 1989 May;50(1-2):167–172. doi: 10.1016/0378-1097(89)90479-5. [DOI] [PubMed] [Google Scholar]