Abstract
Glucagon-like peptide 1 (GLP1), which is mainly processed and cleaved from proglucagon in enteroendocrine cells (EECs) of the intestinal tract, acts on the GLP1 receptor in pancreatic cells to stimulate insulin secretion and to inhibit glucagon secretion. However, GLP1 processing is not fully understood. Here, we show that reticulon 4B (Nogo-B), an endoplasmic reticulum (ER)-resident protein, interacts with the major proglucagon fragment of proglucagon to retain proglucagon on the ER, thereby inhibiting PCSK1-mediated cleavage of proglucagon in the Golgi. Intestinal Nogo-B knockout in male type 2 diabetes mellitus (T2DM) mice increases GLP1 and insulin levels and decreases glucagon levels, thereby alleviating pancreatic injury and insulin resistance. Finally, we identify aberrantly elevated Nogo-B expression and inhibited proglucagon cleavage in EECs from diabetic patients. Our study reveals the subcellular regulatory processes involving Nogo-B during GLP1 production and suggests intestinal Nogo-B as a potential therapeutic target for T2DM.
Subject terms: Mechanisms of disease, Endocrine system and metabolic diseases
Glucagon-like peptide 1 (GLP1) is processed and cleaved from proglucagon to stimulate insulin secretion. Here, authors show that Nogo-B interacts with proglucagon to retain it on the ER, thereby inhibiting PCSK1-mediated cleavage of proglucagon and secretion of GLP1.
Introduction
Type 2 diabetes (T2DM) is characterized by progressive defects in insulin secretion in a context of relative insulin resistance1. Insulin resistance results in disorders of glucolipid metabolism and an elevated blood glucose level, inducing systemic inflammation as well as oxidative damage to pancreatic β cells, which further reduces insulin secretion (creating a vicious pathogenic cycle)2,3. Glucagon is a peptide hormone secreted by pancreatic α cells that activates gluconeogenesis to raise blood glucose upon hypoglycemia4. Elevated glucagon levels in T2DM lead to sustained glycogenolysis and gluconeogenesis, leading to liver damage and exacerbating hyperglycemia and insulin resistance4.
Glucagon-like peptide 1 (GLP1) is produced through the differential and tissue-specific processing of proglucagon (proGCG) in different tissues, which released in response to nutrient ingestion that potentiates the glucose-induced insulin response5. Interestingly, GLP1 and glucagon share the same precursor molecule, proGCG, which is cleaved by proprotein convertase subtilisin/kexin type 2 (PCSK2) to synthesize glucagon in pancreatic α cells or is cleaved by PCSK1 to synthesize GLP1 in enteroendocrine cells (EECs)6–8. Some studies have indicated that the gut is the primary source of circulating GLP1, but GLP1 derived from islet α cells also significantly contribute to the regulation of glucose homeostasis9,10. In addition to stimulating insulin secretion and inhibiting glucagon secretion, GLP1 also has extra pancreatic functions that ultimately induce a feeling of satiety and thereby reducing food intake6,7. In T2DM patients, dietary stimulation of GLP1 levels is significantly reduced, but its effect is relatively preserved11. Consequently, the majority of pharmaceutical endeavors targeting the treatment of T2DM by augmenting the effect of GLP1 have concentrated on GLP1 receptor (GLP1R) agonists11. Given the side effects associated with GLP1R agonists and the high rate of drug discontinuation, it becomes imperative to devise pharmaceutical interventions capable of enhancing endogenous GLP1 levels12. However, the regulatory mechanism of the GLP1 production process is not fully understood. Further understanding of the processing of GLP1 production contributes to the discovery of drug targets.
Reticulon 4 (RTN4), also called Nogo, has three isoforms (Nogo-A, Nogo-B, and Nogo-C) with each generated as an alternatively spliced transcript variant13 (derived both from differential splicing and differential promoter usage). Nogo-A and Nogo-C are predominantly expressed in the central nervous system, while Nogo-B is expressed in a variety of tissues, including liver, heart, and vessel14–17. Studies during the past years have revealed the function of Nogo-B in metabolic diseases and processes such as alcoholic liver disease18, cholestasis19, obesity20, white adipogenesis21, and ChREBP-mediated lipogenesis19. Nogo-B whole-body genetic deletion improves glucose intolerance and insulin resistance in mice fed a high-fat or high-carbohydrate diet16,20. Nogo-B knockout in normal chow-fed mice significantly activates the hepatic INSR-IRS-AKT pathway with no clear mechanism16. Overexpression of Nogo-B receptor (NgBR) activates the insulin signaling pathway and attenuates T2DM progression, but NgBR acts independently of Nogo-B22. While dysregulation of Nogo-B has been implicated in multiple metabolic diseases, its physical interactions and impacts on insulin signaling have remained unknown.
In this study, we found that the Nogo-B levels are aberrantly increased in the liver, pancreas, and small intestines of T2DM mice. Knockdown of Nogo-B in db/db mice increased both insulin and GLP1 levels and alleviated pancreatic injury and insulin resistance. Biochemically, intestinal Nogo-B binds to the major proglucagon fragment (MPGF) of proGCG to retain proGCG on the endoplasmic reticulum (ER), thereby inhibiting PCSK1-mediated cleavage of proGCG in the Golgi. Tissue-specific deletion of Nogo-B in the intestine increased GLP1 and insulin levels, leading to decreased glucagon levels in T2DM mice, resulting in the expected alleviation of both pancreatic injury and insulin resistance. In humans, we also identified increased expression of Nogo-B and inhibited cleavage of proGCG in EECs from diabetic patients. Collectively, our data established an unrecognized role for Nogo-B in the regulation of GLP1 production and suggested that Nogo-B has potential as a target for the treatment of T2DM.
Results
siRNA-mediated knockdown of Nogo-B ameliorates spontaneous T2DM in db/db mice
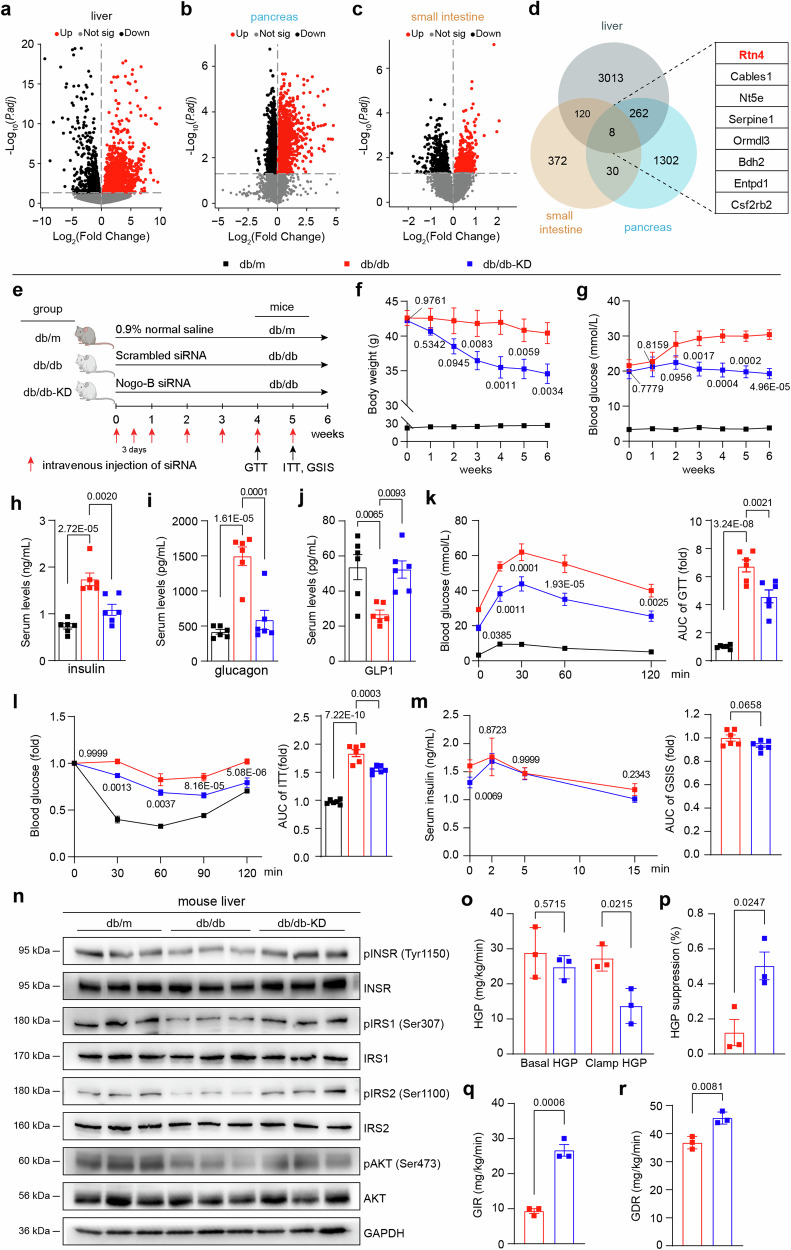
Defective insulin secretion by pancreatic β cells and the inability of insulin-sensitive tissues to respond appropriately to insulin are the two main factors that combine to cause T2DM23. The pancreas, small intestine, and liver all exert functions in regulating insulin signaling. To test the potential role of Nogo-B in the insulin signaling pathway and T2DM, we identified differentially expressed genes in the liver24 (GSE171269), pancreas25 (GSE31953), and small intestine26 (GSE145052) of T2DM model mice (Fig. 1a–d). A total of 8 genes were commonly increased in the three organs of T2DM mice (Fig. 1d), of which Rtn4 attracted our attention. In these three tissues as well as in the brain, Nogo-A is highly expressed mainly in the brain, whereas Nogo-B is widely expressed (Supplementary Fig. 1a), indicating that the upregulated protein in the three tissues of T2DM mice is the Nogo-B isoform.
Fig. 1. Nogo-B knockdown ameliorates spontaneous T2DM in db/db mice.
Volcano plot depicting differential transcriptome in the liver (a), pancreas (b), and small intestine (c) of db/db mice. Red dots represent up-regulated genes, while black dots represent down-regulated genes. d Identification of candidate genes upregulated in the liver, pancreas, and small intestine of T2DM mice. e Experimental design for siRNA injection in vivo. db/db mice were injected with scrambled siRNA or Nogo-B siRNA, and db/m mice were injected with 0.9% normal saline as the healthy control. f Body weight of mice (n = 6 mice per group). g Fasted glucose levels of mice (n = 6 mice per group). h–j Serum levels of insulin, glucagon and GLP1 of mice (n = 6 mice per group). k Oral glucose tolerance test (OGGT) and area under the curve (AUC) analysis of mice (n = 6). l Insulin tolerance test (ITT) and AUC analysis of mice (n = 6 mice per group). m Glucose stimulated insulin secretion (GSIS) test and AUC analysis of mice (n = 6 mice per group). n pINSR, INSR, pIRS1, IRS1, pIRS2, IRS2, pAKT and AKT protein levels in mouse liver (n = 6 mice per group). o Hepatic glucose production (HGP) under basal and clamp conditions of mice (n = 3 mice per group). HGP suppression (p), glucose infusion rate (GIR, q),and glucose disposal rate (GDR, r) of mice (n = 3 mice per group). Data are expressed as the mean ± SEM. The p values of (a–c) were calculated by two-sided test and adjusted for p values by the Benjamini-Hochberg method to control for false positives due to multiple comparisons. The p values of (f–l) were calculated by one-way ANOVAs. The p values of (m, o–r) were calculated by two-tailed Student’s t test. The p values of (f, g, k, l) are indicated as db/db-KD group vs. db/db group. Source data are provided as a Source Data file.
To determine whether knockdown of Nogo-B confers benefit in alleviating T2DM, we intravenously injected scrambled siRNA and siRNA against Nogo-B into db/db mice, with db/m mice as healthy controls (Fig. 1e). We found that Nogo-B levels in db/db mice were significantly higher in the liver, pancreas, and small intestine compared to db/m mice, and siRNA treatment reduced Nogo-B levels in these tissues (Supplementary Fig. 1b, c). Notably, Nogo-B knockdown significantly reduced body weight (Fig. 1f) and attenuated hyperglycemic symptoms in db/db mice (Fig. 1g). We also assessed insulin, glucagon and GLP1 levels and found that Nogo-B knockdown attenuated hyperinsulinemia (Fig. 1h), hyperglucagonemia (Fig. 1i) and increased GLP1 levels (Fig. 1j). To further evaluate glucose homeostasis, we performed a glucose tolerance test (GTT) and an insulin tolerance test (ITT), which showed that Nogo-B knockdown improved glucose tolerance (Fig. 1k) and increased insulin sensitivity (Fig. 1l).
In addition, the glucose-stimulated insulin secretion (GSIS) assay showed that Nogo-B knockdown resulted in increased insulin secretion two minutes after glucose injection (Fig. 1m, left), but did not affect total secretion due to hyperinsulinemia in db/db mice as indicated by AUC (Fig. 1m, right). Nogo-B knockdown significantly increased the phosphorylation levels of insulin receptor (INSR), Insulin Receptor Substrate 1 (IRS1), IRS2, and AKT, which were reduced in liver and muscle of db/db mice (Fig. 1n, Supplementary Fig. 2a–c). To measure the effect of Nogo-B knockdown on insulin sensitivity more accurately, we performed hyperinsulinemic-euglycemic clamp experiments. Nogo-B knockdown significantly enhanced insulin-stimulated suppression of hepatic glucose production (HGP), while increasing glucose infusion rate (GIR) and glucose disposal rate (GDR). GIR and GDR can reflect whole-body insulin sensitivity. Reduction in islet volume, apoptosis of β cells and loss of β-cell granulation are also features of T2DM27. Immunofluorescence analysis showed that db/db mice had distorted islet structure, less insulin staining, smaller β-cell fractions (Supplementary Fig. 2d, e) and significant apoptosis (Supplementary Fig. 2f, g) compared to db/m mice, and that Nogo-B knockdown alleviated these defects.
The liver coordinates systemic metabolic homeostasis and adaptation to nutrient supply and deprivation28. Reversal of liver injury is also associated with remission of T2DM. We found that Nogo-B knockdown reduced liver injury and lipid accumulation in db/db mice (Supplementary Fig. 3a–e). The recovery of serum cholesterol, triglyceride, free fatty acid, LDL-C and transaminase levels also supported the conclusion of reduced hepatic lipid accumulation (Supplementary Information: Table S1). Nogo-B knockdown also suppressed appetite in db/db mice (Supplementary Fig. 3i) and increased their respiratory exchange rate (RER) and energy expenditure, suggesting that energy expenditure is more exuberant in Nogo-B knockdown db/db mice (Supplementary Fig. 3f–l). These results suggest that Nogo-B knockdown attenuates hyperglycemia and insulin resistance, reduces pancreatic and hepatic injury in db/db mice, and influences levels of insulin, glucagon, and GLP1.
Nogo-B deficiency enhances glucose metabolism and triggers the activation of the insulin signaling pathway
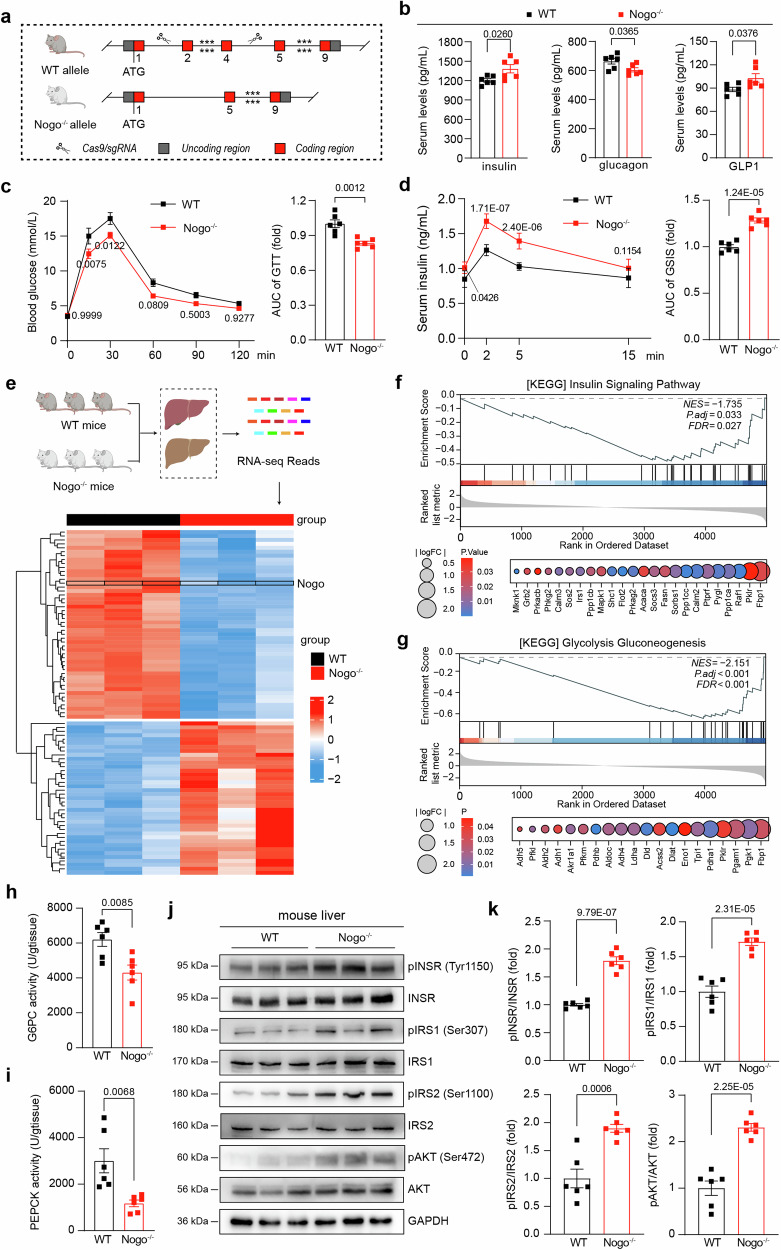
The increased expression of Nogo-B and subsequent alleviation of diabetic symptoms following Nogo-B knockdown in db/db mice prompted us to further investigate Nogo-B’s role in the insulin signaling pathway and its impact on relevant hormone levels. The three isoforms of Nogo are generated through RNA splicing events13, thus deletion of Nogo-B exons 2-4 leads to deficiency of other Nogo members, resulting in Nogo knockout (Nogo−/−) mice (Fig. 2a, Supplementary Fig. 4a). We compared physiological parameters of Nogo-/- mice to wild-type (WT) littermates at 5 to 8 weeks of age and found no significant differences in weekly body weight and blood glucose levels (Supplementary Fig. 4b, e). However, Nogo−/− mice showed a significant reduction in body weight gain and food consumption (Supplementary Fig. 4c, d). Furthermore, Nogo−/− mice showed increased insulin and GLP1 levels, decreased glucagon levels (Fig. 2b), and no changes in inflammatory factors (Supplementary Fig. 4f, g). GTT and GSIS assays revealed that Nogo−/− mice exhibited both enhanced glucose clearance (Fig. 2c) and insulin secretion (Fig. 2d) compared to WT littermates.
Fig. 2. Nogo-B deficiency promotes glucose metabolism and activates the insulin signaling pathway.
a A strategy for generating Nogo−/− mice. b Serum levels of insulin, glucagon and GLP1 of normal chow-fed 8-week-old Nogo−/− mice and their wild-type (WT) littermates (n = 6 mice per group). c OGTT and AUC analysis of normal chow-fed 8-week-old Nogo−/− mice and their WT littermates (n = 6 mice per group). d GSIS and AUC analysis of normal chow-fed 8-week-old Nogo−/− mice and their WT littermates (n = 6 mice per group). e Heatmap analysis of bulk RNA-seq data from livers of normal chow-fed 8-week-old Nogo−/− mice and their WT littermates (n = 3 mice per group). f, g GSEA in insulin signaling pathway and the glycolysis-gluconeogenesis pathway. The enzyme activity of G6PC (h) and PEPCK (i) in the liver of normal chow-fed 8-week-old Nogo−/− mice and their WT littermates (n = 6 mice per group). j, k pINSR, INSR, pIRS1, IRS1, pIRS2, IRS2, pAKT, and AKT protein levels in mouse liver and analysis of band density, the experiment was repeated at least three times (n = 6 mice per group). Data are expressed as the mean ± SEM. The p values were calculated by two-tailed Student’s t test. Source data are provided as a Source Data file.
Since the liver plays a critical role in the systemic response to insulin and regulates glucose and lipid metabolism29, we conducted RNA-seq of livers isolated from 8-week-old Nogo−/− mice and WT littermates (Fig. 2e). To enrich our RNA-Seq data, we obtained the KEGG pathway dataset from the Molecular Signatures Database (MSigDB) Collections database and performed gene enrichment analysis (GSEA) (Supplementary Fig. 5a). Analysis of the Hallmark gene matrix revealed significant enrichment of the insulin signaling pathway (Fig. 2f) and the glycolysis-gluconeogenesis pathway (Fig. 2g). To validate the RNA-seq results, we assessed the activities of the gluconeogenesis-related enzymes glucose-6-phosphatase catalytic subunit (G6PC) and phosphoenolpyruvate carboxykinase (PEPCK), revealing that Nogo-B deficiency significantly inhibited gluconeogenesis (Fig. 2h, i). Nogo-B deficiency also significantly increased the phosphorylation levels of INSR, IRS1, IRS2, and AKT in liver and muscle (Fig. 2j, k, Supplementary Fig. 5b, c). These results suggest that Nogo-B deficiency affects the levels of insulin, glucagon and GLP1, thereby promoting glucose metabolism and activating insulin signaling pathways.
Nogo-B interacts with proGCG but not insulin
Nogo-B knockdown/deficiency significantly alters insulin, glucagon, and GLP1 levels (Fig. 1h–j, Fig. 2b). Considering Nogo-B located in the endoplasmic reticulum (ER) and plasma membrane, and the ER is responsible for processing secreted proteins, the receptors for the hormones are located on the plasma membrane. Therefore, we hypothesize that Nogo-B may influences the production of these hormones, or competitively binds to the receptors of these hormones, thereby affecting their levels.
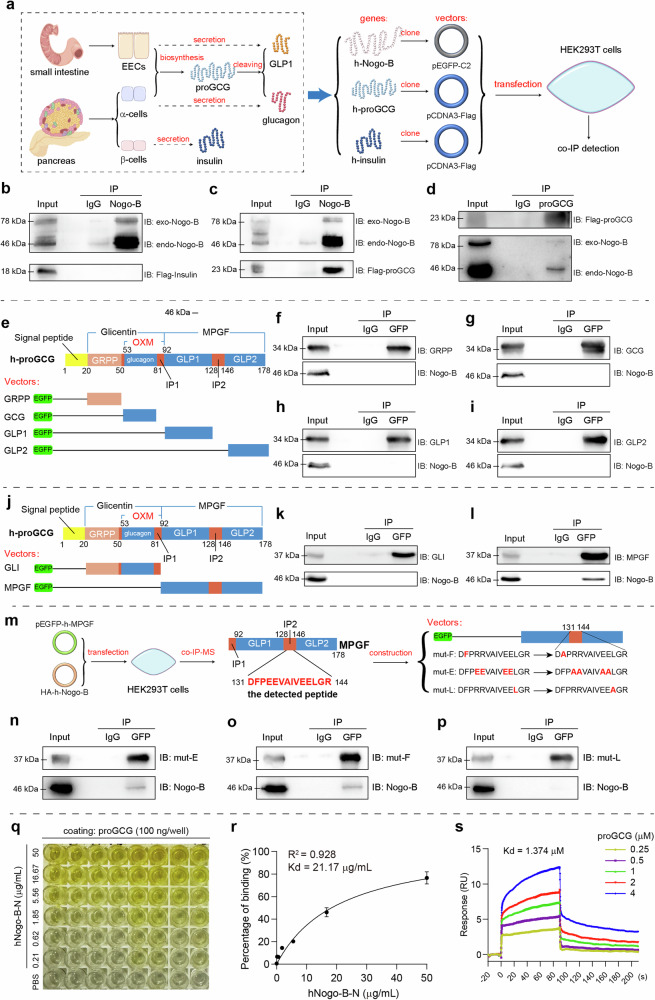
Since proteins with similar sequences often have similar three-dimensional structures and functions, we assessed whether Nogo-B interacts with the receptors of these hormones by comparing sequence similarity and using immunoprecipitation-mass spectrometry (IP-MS) assays. Notably, both glucagon and GLP1 originate from the same peptide, proGCG, which undergoes tissue-specific post-translational modifications (Fig. 3a)6. We compared the amino acid sequences of Nogo-B with those of proGCG and insulin. The comparisons revealed significant sequence similarity between Nogo-B and various species of proGCG and insulin (Supplementary Fig. 6a, b), indicating a potential similarity in structure between Nogo-B and these hormones. Based on the abundant expression of insulin, proGCG, and their respective receptors in the liver and pancreas, we performed IP-MS assays using a Nogo-B antibody and extracts from excised liver and pancreas samples of C57BL/6J mice (Supplementary Fig. 6c, d). Our results showed that Nogo-B interacted with insulin (score: 4.6, reflecting the degree of matching of the secondary peptide profiles) and proGCG (score: 34.5) in the pancreas (Supplementary Fig. 6e), but not with any of the receptors.
Fig. 3. Nogo-B interacts with proGCG.
a Tissue-specific post-translational modifications of glucagon and GLP1, and the strategy for the co-IP experiments. h in “h-Nogo-B, h-proGCG and h-insulin” means human-derived. b Immunoprecipitation assay with Nogo-B and exogenous insulin in HEK293T cells (n = 3 biological replicates). c, d Immunoprecipitation assay with Nogo-B and exogenous proGCG in HEK293T cells (n = 3 biological replicates). e Construction of EGFP-tagged expression vectors for proGCG-sheared basic short peptides (GRPP, glucagon, GLP1, and GLP2). f–i Immunoprecipitation assay with Nogo-B and proGCG-sheared basic short peptides (GRPP, glucagon, GLP1, and GLP2) (n = 3 biological replicates). j Construction of EGFP-tagged expression vectors for glicentin (GLI) and major proglucagon fragment (MPGF). k, l Immunoprecipitation assay with Nogo-B and GLI and MPGF (n = 3 biological replicates). m Mass spectrometric detection of the peptide bound to MPGF by Nogo-B, where the peptide with the highest secondary structure similarity to the profile was in the IP2 region. Construction of EGFP-tagged expression vectors for the MPFG with mutations in the hydrophobic amino acid site. n–p Immunoprecipitation assay with Nogo-B and MPFG with mutations in the hydrophobic amino acid site (n = 3 biological replicates). q ELISA for the binding of Nogo-B to proGCG. r Non-linear fit analysis of ELISA results (n = 6 biological replicates). s Affinity constants of Nogo-B and proGCG detected by SPR. All experiments of (b–d, f–I, k, l, n–p) were repeated at least three times Data are expressed as the mean ± SEM. Source data are provided as a Source Data file.
To validate our findings, we generated expression vectors containing tagged human-derived Nogo-B, insulin, or proGCG, and performed co-IP experiments in HEK293T cells (Fig. 3a). The co-IP assays confirmed the interaction between Nogo-B and proGCG in HEK293T cells (Fig. 3c, d), whereas no interaction was detected between Nogo-B and insulin (Fig. 3b). These findings suggest that Nogo-B specifically interacts with proGCG, which could affect the processing of proGCG into glucagon and GLP1, and merits further investigation.
Nogo-B interacts with proGCG through the major proglucagon fragment (MPGF)
Besides glucagon and GLP1, the proGCG peptide serves as a precursor for other biologically relevant peptides such as GLP2, oxyntomodulin, glicentin, glicentin-related pancreatic polypeptide (GRPP), major proglucagon fragment (MPGF), and intervening peptides (IP1 and IP2)6 (Fig. 3e). To identify the specific region(s) of proGCG that interact with Nogo-B, we constructed expression vectors for EGFP-tagged short peptides derived from human proGCG (GRPP, glucagon, GLP1, and GLP2) and overexpressed them in HEK293T cells (Fig. 3e). The co-IP assays revealed no interaction between Nogo-B and any of these peptides (Fig. 3f–i), indicating that Nogo-B does not bind directly to these peptides but instead interacts with their associated intervening peptides, IP1 or IP2. We then constructed expression vectors for glicentin (GLI) and MPGF, which are peptides that give rise to glucagon and GLP1, respectively (Fig. 3j). The co-IP results showed that although Nogo-B did not bind directly to GLP1 or GLP2, it did bind to MPGF, suggesting that Nogo-B interacts with the IP2 region (Fig. 3k, l).
proGCG is cleaved by various polyprotein convertases (PCSKs) in different tissues (Fig. 3m)6. PCSK2, which is highly expressed in α cells, primarily produces glucagon. In EECs, high expression of PCSK1 leads to the production of GLP1, oxyntomodulin (OXM), and GLP26. Considering that Nogo-B interacts with MPGF but not with GLP1 or GLP2, we hypothesized that Nogo-B might bind to the IP2 region to compete with PCSK1 for the cleavage site30. To investigate this, we generated expression vectors with cleavage site mutations based on each of the two cleavage sites within the IP2 region of MPGF (Supplementary Fig. 7a) and conducted separate co-IP experiments for each mutant. However, Nogo-B bound to MPGF regardless of the mutation in the binding site (Supplementary Fig. 7b–d). These results suggest that Nogo-B is binding to the IP2 region of MPGF, but does not compete for the binding site of PCSK1.
Nogo-B interact with the leucine residue at position 142 of proGCG
To determine the specific binding sites of Nogo-B in the IP2 region, we co-transfected MPGF and Nogo-B into HEK293T cells, and performed co-IP-MS assays using either GFP antibody or Nogo-B antibody (Fig. 3m, Supplementary Fig. 7e). Analysis of the results of co-IP-MS assay with the Nogo-B antibody revealed that peptide DFPEEVAIVEELGR had the highest score among the Nogo-B binding peptides, which is located at positions 131–141 and is found exclusively within the IP2 region (Supplementary Fig. 7f). Since protein–protein interactions are typically mediated by hydrophobic amino acids, we mutated phenylalanine (F, position 132) and leucine (L, position 142) to alanine (A) within the proGCG131-144 segment. Given the high frequency of glutamate (E) in this segment, we mutated the residues at positions 134, 135, 140, and 141 to alanine. We conducted co-IP assays to evaluate the binding of these mutated forms of MPGF to Nogo-B. Notably, mutations of phenylalanine and glutamate to alanine did not prevent Nogo-B binding to MPGF. However, mutation of leucine at position 142 to alanine abolished this interaction. These findings suggest that the leucine at position 142 in proGCG is crucial for its interaction with Nogo-B and may play a role in the cleavage and maturation of GLP1.
We then analyzed the co-IP-MS results with the GFP antibody and found that the Nogo-B fragment that binds to proGCG may be at position 25-58 (Supplementary Fig. 7g), which is located in the N-terminal structural domain (amino acids 1-185)13. Therefore, to further validate the interaction between Nogo-B and proGCG, we purified the N-terminal structural domain of human Nogo-B protein (hNogo-B-N) and the proGCG protein (hproGCG), and assessed their binding kinetics by enzyme-linked immunosorbent assay (ELISA) and surface plasmon resonance (SPR) experiments. In ELISA experiments, a significant increase in absorbance was observed with increasing concentration of hNogo-B-N (Fig. 3q). Non-linear regression analysis was used to characterize the affinity between hNogo-B-N and hproGCG, yielding a dissociation constant (Kd) of 21.17 μg/mL with a high goodness of fit (R2 = 0.928) (Fig. 3r). In addition, SPR analysis confirmed the specificity of the protein–protein interaction with a measured affinity constant of 1.374 μM, consistent with the expected affinity range of protein–protein interactions under physiological conditions (Fig. 3s). We also found that there is no interaction between Nogo-B and PCSK1 (Supplementary Fig. 7h). Taken together, these findings underline the interaction between Nogo-B and proGCG.
Nogo-B inhibits PCSK1-mediated cleavage of proGCG by retaining it on the ER
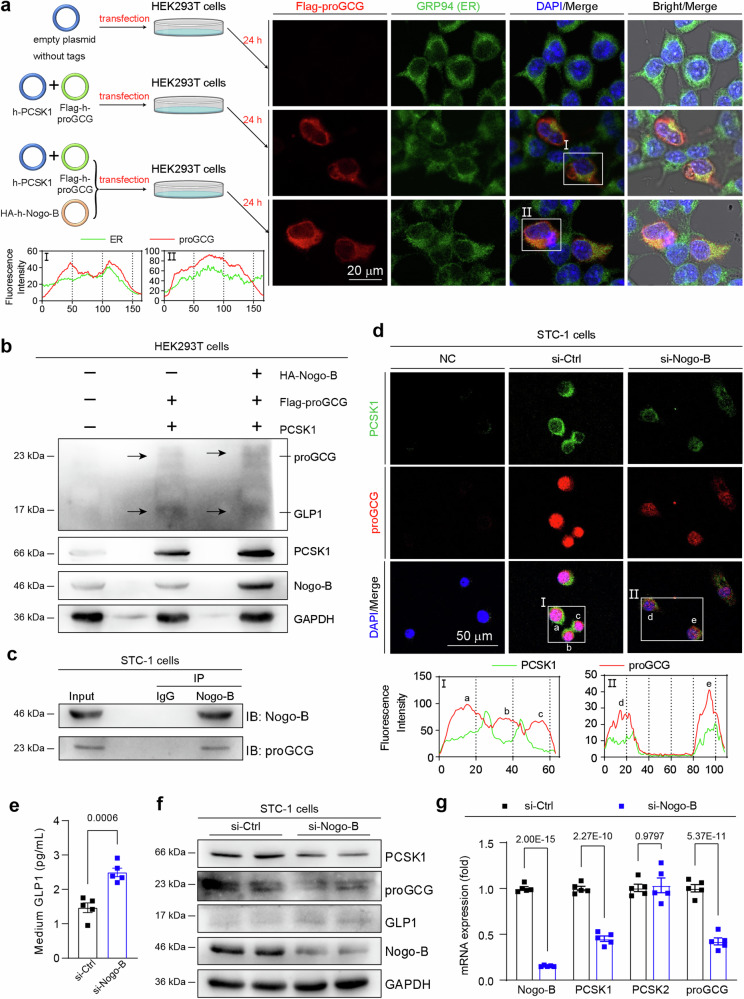
Considering that Nogo-B is localized within the ER, while GLP1 is a secreted protein, it is necessary for the precursor proGCG to undergo transportation from the ER to the Golgi apparatus after processing and folding within the ER31. This transport is followed by cleavage by PCSK1 in the Golgi32. To investigate the effect of Nogo-B on the transport of proGCG between the ER and Golgi, we co-transfected HEK293T cells with PCSK1 and proGCG expression vectors (Fig. 4a, left panel). Overexpression of Nogo-B was found to result in proGCG retention at the ER, in contrast to normal HEK293T cells in which proGCG is transported outside the ER (Fig. 4a, right panel). This indicates that excessive Nogo-B retains proGCG on the ER, impeding its transit to the Golgi. Furthermore, the levels of proGCG and GLP1 were evaluated after overexpression of Nogo-B to examine the effrct of its PCSK1 cleavage efficiency. It was observed that Nogo-B overexpression increased proGCG levels while decreasing GLP1 levels, suggesting that Nogo-B inhibits the cleavage of proGCG by PCSK1, consequently reducing the production of GLP1 (Fig. 4b, Supplementary Fig. 8a).
Fig. 4. Nogo-B inhibits PCSK1-mediated cleavage of proGCG in STC-1 cells.
a Representative images of immunofluorescence staining (ER, green; proGCG, red; DAPI, blue) of HEK293T cells transfected with human-derived proGCG and PCSK1. Overexpression of Nogo-B detects the localization of proGCG on the ER. I-II: The intensity distribution of the marker scans in the boxed area shows the spatial distribution and fluorescence intensity of the different markers. Enhanced co-localization is indicated by the resemblance in shape of the intensity distribution curves and the increased number of overlaps. The experiment was conducted independently on three occasions. b Detection of Nogo-B overexpression on exogenous PCSK1 cleavage of exogenous proGCG by Western blot in HEK293T cells (n = 3 biological replicates). c Endogenous co-IP assay with Nogo-B and proGCG in STC-1 cells (n = 3 biological replicates). d Representative images of immunofluorescence staining (PCSK1, green; proGCG, red; DAPI, blue) of STC-1 cells transfected with scrambled siRNA (si-Ctrl) or Nogo-B siRNA (si-Nogo-B) to detect co-localization of proGCG and PCSK1. I-II: The intensity distribution of the marker scans in the boxed area shows the spatial distribution and fluorescence intensity of the different markers. Enhanced co-localization is indicated by the resemblance in shape of the intensity distribution curves and the increased number of overlaps. The experiment was conducted independently on three occasions. e GLP1 levels in the culture medium of STC-1 cells transfected with si-Ctrl or si-Nogo-B (n = 5 biological replicates). f PCSK1, proGCG, GLP1 and Nogo-B protein levels in STC-1 cells transfected with si-Ctrl or si-Nogo-B (n = 6 biological replicates). g Nogo-B, PCSK1, PCSK2 and proGCG mRNA levels in STC-1 cells transfected with si-Ctrl or si-Nogo-B (n = 5 biological replicates). All experiments were repeated at least three times. Data are expressed as the mean ± SEM. The p values were calculated by two-tailed Student’s t test. Source data are provided as a Source Data file.
To confirm the binding of Nogo-B to proGCG in enterocytes, STC-1 cells (an EEC cell line) were utilized (Fig. 4c). In STC-1 cells, co-IP Western blot assays were performed to further confirm the interaction between Nogo-B and proGCG. Co-staining of PCSK1 (green) and proGCG (red) in STC-1 cells revealed that Nogo-B knockdown resulted in decreased protein levels of proGCG and PCSK1, but increased co-localization between them (Fig. 4d). This suggests that reduction of Nogo-B promotes the interaction between proGCG and PCSK1 in STC-1 cells. The retention of proGCG in the ER by Nogo-B may induce the unfolded protein response (UPR) or ER stress. We tested the expression of UPR and ER stress-related proteins in STC-1 cells and showed that overexpression of Nogo-B activated the UPR and ER stress (Supplementary Fig. 8b, c), whereas inhibition of Nogo-B had no significant effect (Supplementary Fig. 8d, e). Meanwhile, Nogo-B knockdown in STC-1 cells resulted in an increase in the level of GLP1 in the medium (Fig. 4e). This knockdown also led to decreased protein and mRNA levels of proGCG and PCSK1, and increased GLP1 protein levels (Fig. 4f, g, Supplementary Fig. 9d). Conversely, Nogo-B overexpression led to a decrease in GLP1 levels in the medium, increased protein and mRNA levels of proGCG and PCSK1, and decreased GLP1 protein levels (Supplementary Fig. 9a–c). These findings suggest that the abundance of Nogo-B in STC-1 cells modulates the cleavage of proGCG by PCSK1, consequently affects the production of GLP1. Excessive Nogo-B inhibits the cleavage of proGCG by PCSK1, leading to a reduction in GLP1 levels.
Although GLP1 is mainly secreted by intestinal EEC cells, the contribution of islet α cell-derived GLP1 in regulating blood glucose homeostasis is noteworthy5. Given the similarity in the cleavage and processing of GLP1 in both islet α cells and intestinal EEC cells5, we examined the expression of Nogo-B in the pancreas of db/db mice. Immunofluorescence analysis revealed that in db/m mice, Nogo-B expression was relatively low in islet α cells, whereas a significant positive signal was observed in islet β cells (Supplementary Fig. 10, middle panel). Conversely, db/db mice exhibited an increase in Nogo-B expression within islet α cells, accompanied by enhanced co-localization with proGCG (Supplementary Fig. 10, down panel). These findings suggest that Nogo-B expression is upregulated in islet α cells in mice with T2DM, potentially impeding GLP1 production in a manner akin to that observed in EEC cells.
Intestinal Nogo-B knockout increases GLP1 levels in mice
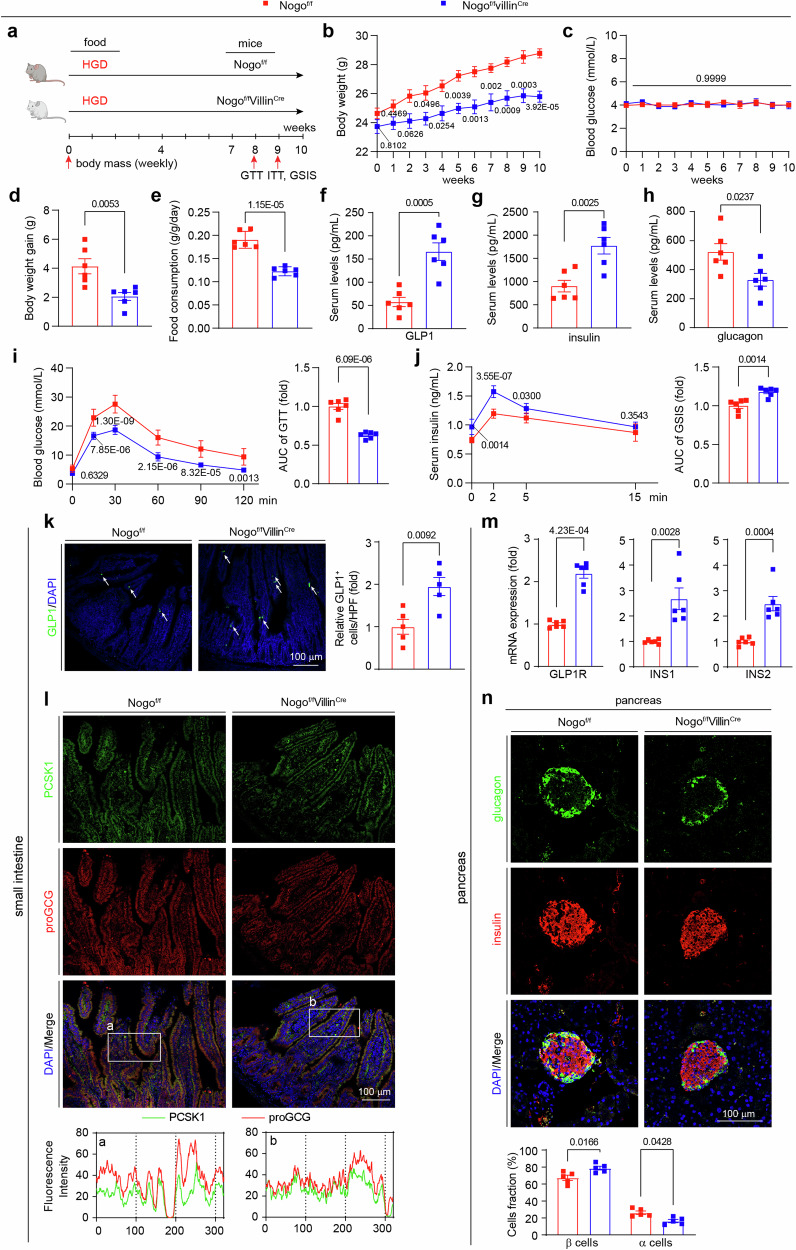
To investigate the effect of intestinal Nogo-B reduction on GLP1 production, we generated and validated mice with selective knockout of intestinal Nogo-B (Nogof/fVillinCre) and their littermate controls (Nogof/f) (Supplementary Fig. 11a, b). GLP1 is secreted in response to glucose levels, stimulating insulin secretion and suppressing appetite. We subjected the mice to a high glucose diet (HGD) for 10 weeks and examined GLP1 levels and insulin-related pathways (Fig. 5a). In addition to regulating the insulin signaling pathway, GLP1 inhibits appetite, reduces food intake and body weight through its receptor GLP1R33. Compared to Nogof/f mice, the Nogof/fVillinCre mice exhibited lower body weight (Fig. 5b), slower weight gain (Fig. 5d), and reduced food consumption (Fig. 5e), but no change in blood glucose levels (Fig. 5c). Intestinal Nogo-B knockout increased GLP1 and insulin levels and decreased glucagon levels. GTT and ITT assays indicated improved glucose tolerance without affecting insulin resistance in Nogof/fVillinCre mice (Fig. 5i, Supplementary Fig. 11c). GSIS assays also demonstrated that intestinal Nogo-B knockout enhanced insulin secretion (Fig. 5j). In addition, gluconeogenesis-related enzyme activities were inhibited (Supplementary Fig. 11d, e) and hepatic and muscle insulin sensitivity was activated (Supplementary Fig. 11f–i) in Nogof/fVillinCre mice.
Fig. 5. Intestinal Nogo-B knockout increases GLP1 levels in mice.
a Experimental design in vivo. Nogof/fVillinCre and their littermate controls (Nogof/f) received high glucose diet (HGD) for 10 weeks and were conducted the assays as scheduled. b Body weight of Nogof/fVillinCre and Nogof/f mice (n = 6 mice per group). c Fasted glucose levels of mice (n = 6). d Body weight gain of Nogof/fVillinCre and Nogof/f mice after 10 weeks on HGD (n = 6 mice per group). e Food consumption of Nogof/fVillinCre and Nogof/f mice (n = 6 mice per group). f–h Levels of GLP1, insulin and glucagon of Nogof/fVillinCre and Nogof/f mice (n = 6 mice per group). i OGTT and AUC analysis of Nogof/fVillinCre and Nogof/f mice (n = 6 mice per group). j GSIS and AUC analysis of Nogof/fVillinCre and Nogof/f mice (n = 6 mice per group). k Representative images of immunofluorescence staining for GLP1 (GLP1, green; DAPI, blue) of small intestine from HGD-fed Nogof/fVillinCre and Nogof/f mice. White arrowheads indicate GLP1-positive signals. Quantification of results were expressed as the mean numbers of GLP1-positive cells in 5 high-power fields (HPFs) (n = 5 mice per group, right panel). l Representative images of immunofluorescence staining PCSK1 and proGCG (PCSK1, green; proGCG, red; DAPI, blue) of small intestine from HGD-fed Nogof/fVillinCre and Nogof/f mice. a, b The intensity distribution of the marker scans in the boxed area shows the spatial distribution and fluorescence intensity of the different markers. Enhanced co-localization is indicated by the resemblance in shape of the intensity distribution curves and the increased number of overlaps. The experiment was conducted independently on three occasions. m The mRNA levels of GLP1R, INS1, and INS2 in the pancreas of Nogof/fVillinCre and Nogof/f mice after 10 weeks on HGD (n = 5 mice per group). n Islet images of Nogof/fVillinCre and Nogof/f mice after 10 weeks on HGD (glucagon, green; insulin, red; DAPI, blue). Analysis of β-cell or α-cell as the percentage of pancreatic cells (n = 5 mice per group, bottom panel). Data are expressed as the mean ± SEM. The p values were calculated by two-tailed Student’s t test. Source data are provided as a Source Data file.
Immunofluorescence staining of the small intestine revealed that intestinal Nogo-B knockout increased the number of GLP1-positive cells (Fig. 5k) and promoted co-localization of proGCG and PCSK1 (Fig. 5i). GLP1 acts on the pancreas to activate GLP1R and insulin transcription, and we observed increased mRNA levels of pancreatic GLP1R, INS1, and INS2 in the Nogof/fVillinCre mice (Fig. 5m). Changes in insulin and glucagon levels coincided with increased β-cell fractions and decreased α-cell fractions in the pancreas (Fig. 5n). Meanwhile, 10 weeks of HGD did not cause significant liver injury (Supplementary Fig. 12a–c), and biochemical analysis revealed no significant differences in blood lipid levels and liver transaminase levels (Supplementary Information: Table S2). Metabolic cage analysis demonstrated similar oxygen consumption, energy expenditure and heat production between the two groups of mice (Supplementary Fig. 12d–i). These findings indicate that intestinal Nogo-B knockout indeed promotes GLP1 levels in mice, subsequently stimulating insulin secretion.
Intestinal Nogo-B knockout alleviates the characteristic manifestations of T2DM induced by STZ
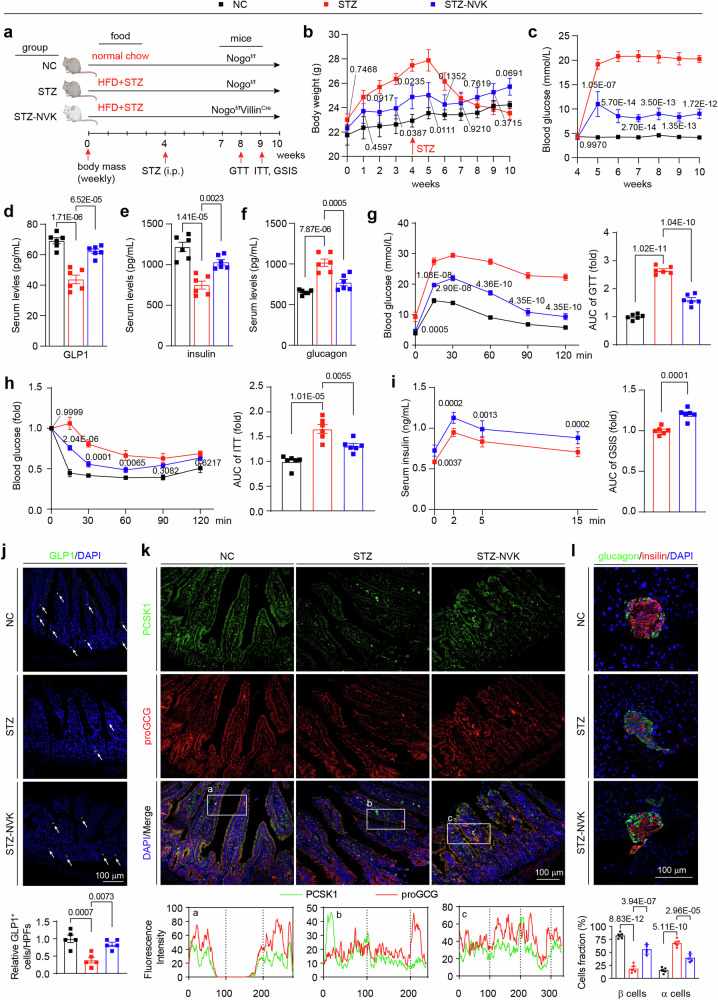
Based on the fact that intestinal Nogo-B knockout increases GLP1 and insulin levels, to further explore the therapeutic effects of intestinal Nogo-B knockout in T2DM, we used single high-dose intraperitoneal injections of streptozotocin (STZ) in conjunction with high-fat diet induction to recapitulate the characteristic manifestations of T2DM, including impaired glucose tolerance, insufficient insulin secretion and insulin resistance34,35. T2DM was induced in Nogof/f mice (STZ group) and Nogof/fVillinCre mice (STZ-NVK group), with Nogof/f mice serving as healthy controls (NC group) (Fig. 6a). Prior to the STZ injection, the body weight of the STZ-NVK group was already lower compared to the STZ group (Fig. 6b). Intestinal Nogo-B knockout significantly improved STZ-induced hyperglycemia (Fig. 6c), increased GLP1 and insulin levels and reduced glucagon levels (Fig. 6d–f). Glucose tolerance, insulin sensitivity, and insulin secretion were also significantly recovered in the STZ-NVK group (Fig. 6g–i). Intestinal Nogo-B knockout restored the phosphorylation levels of INSR, IRS1, IRS2 and AKT (Supplementary Fig. 13a–d). Hyperinsulinemic-euglycemic clamp experiments (Supplementary Fig. 13e–h) and gluconeogenesis-related enzyme activity assays (Supplementary Fig. 14a, b) further demonstrated that intestinal Nogo-B knockout inhibited hepatic glucose production and increased insulin sensitivity.
Fig. 6. Intestinal Nogo-B knockout alleviates STZ-induced T2DM.
a Experimental design in vivo. Nogof/fVillinCre (STZ-NVK group) and Nogof/f (STZ group) mice received STZ and fed on HFD to induce T2DM, using the Nogof/f mice as healthy control (NC group). b Body weight of mice in NC, STZ, and STZ-NVK groups (n = 6 mice per group). c Fasted glucose levels of mice in NC, STZ, and STZ-NVK groups (n = 6 mice per group). d–f Levels of insulin, glucagon and GLP1 of mice in NC, STZ, and STZ-NVK groups (n = 6 mice per group). g OGTT and AUC analysis of mice in NC, STZ, and STZ-NVK groups (n = 6 mice per group). h ITT and AUC analysis of mice in NC, STZ, and STZ-NVK groups (n = 6 mice per group). i GSIS and AUC analysis of mice in NC, STZ, and STZ-NVK groups (n = 6 mice per group). j Representative images of immunofluorescence staining (GLP1, green; DAPI, blue) of small intestine from mice in NC, STZ, and STZ-NVK groups. White arrowheads indicate GLP1-positive signals. Quantification of results were expressed as the mean numbers of GLP1-positive cells in 5 HPFs (n = 5 mice per group, right panel). k Representative images of immunofluorescence staining (PCSK1, green; proGCG, red; DAPI, blue) of small intestine from mice in NC, STZ, and STZ-NVK groups. a–c The intensity distribution of the marker scans in the boxed area shows the spatial distribution and fluorescence intensity of the different markers. Enhanced co-localization is indicated by the resemblance in shape of the intensity distribution curves and the increased number of overlaps. The experiment was conducted independently on three occasions. l Islet images of mice in NC, STZ, and STZ-NVK groups (glucagon, green; insulin, red; DAPI, blue). Analysis of β-cell or α-cell as the percentage of pancreatic cells (n = 5 mice per group, bottom panel). Data are expressed as the mean ± SEM. The p values were calculated by one-way ANOVAs. The p values of (b, c, g, h) were indicated STZ-KD group vs. STZ group. Source data are provided as a Source Data file.
Immunofluorescence analysis of the small intestine revealed that STZ-NVK mice exhibited an increase in the number of GLP1-positive cells (Fig. 6j) and higher levels and co-localization of proGCG and PCSK1 (Fig. 6k). In the pancreas, intestinal Nogo-B knockout restored the decreased transcript levels of GLP1R, INS1, and INS2 induced by STZ (Supplementary Fig. 14c–e), increased fraction of islet β-cells (Fig. 6l), and reduced islet apoptosis (Supplementary Fig. 14f). STZ injection led to liver whitening, enlargement, damage, and severe lipid accumulation, but these effects were ameliorated in the STZ-NVK group (Supplementary Fig. 15a). Quantification of liver-to-body ratio, liver TG levels, and inflammatory factor levels further confirmed these findings (Supplementary Fig. 15b–e). The restoration of blood lipids and transaminases indicated that intestinal Nogo-B knockout restored the balance of glycolipid conversion and improved liver injury (Supplementary Information: Table S3). Metabolic cage analysis revealed that the STZ-NVK group had higher RER, energy expenditure, and heat production, suggesting an overall improvement in their metabolism (Supplementary Fig. 15f–k). Additionally, the reduction in food consumption contributed to the relief of symptoms (Supplementary Fig. 15l). Collectively, these findings further establish the involvement of intestinal Nogo-B in the regulation of GLP1 levels, and demonstrate that intestinal Nogo-B knockout ameliorates the STZ-induced characteristic manifestations of T2DM.
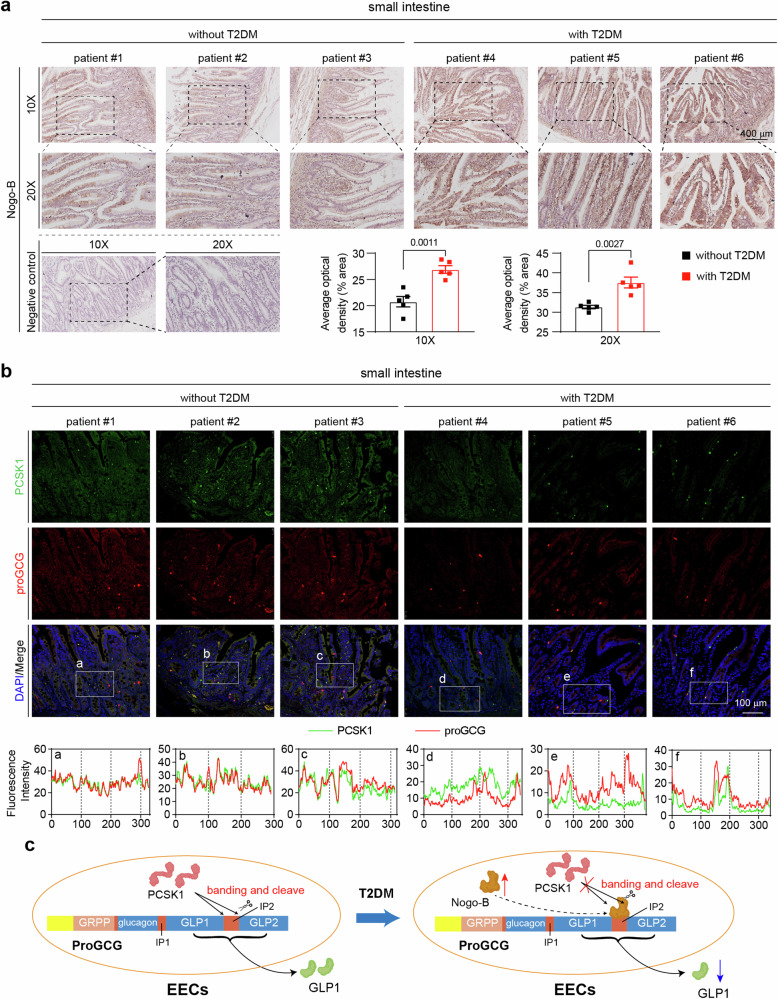
Nogo-B expression is increased in the small intestine of T2DM patients
To investigate whether Nogo-B expression is upregulated in the small intestine of diabetic patients, we collected and prepared paraffin sections of the duodenum from patients undergoing pancreatic resection for pancreatic occupancy, both with and without T2DM (specific inclusion criteria in the Methods section). Immunohistochemical staining was performed, revealing a notable increase in Nogo-B signal intensity in the small intestine of patients with T2DM (Fig. 7a). These results indicate that Nogo-B expression is significantly increased in the small intestine of T2DM patients, mirroring the findings observed in T2DM mice. Furthermore, to explore the impact of elevated Nogo-B on proGCG cleavage by PCSK1 in T2DM patients, we examined the levels and co-localization of PCSK1 and proGCG in the small intestine. Consistent with previous findings, both PCSK1 and proGCG levels were reduced in the T2DM patients, and the co-localization was also diminished (Fig. 7b).
Fig. 7. Nogo-B expression is elevated in the small intestine of T2DM patients.
a Representative images of immunohistochemical staining of Nogo-B in small intestine from patients with or without T2DM. Quantification of Nogo-B density from 5 individual images (n = 5 patients per group). b Representative images of immunofluorescence staining (PCSK1, green; proGCG, red; DAPI, blue) of small intestine from patients with or without T2DM. a–f The intensity distribution of the marker scans in the boxed area shows the spatial distribution and fluorescence intensity of the different markers. Enhanced co-localization is indicated by the resemblance in shape of the intensity distribution curves and the increased number of overlaps. The experiment was conducted independently on three occasions. c working model of Nogo-B affecting GLP1 levels. Data are expressed as the mean ± SEM. The p values were calculated by two-tailed Student’s t test. Source data are provided as a Source Data file.
Meanwhile, MPGF generates GLP2 in addition to GLP1 cleaved by PCSK1. Dipeptidyl peptidase-4 (DPP4) degrades GLP1, thereby affecting its levels12. We examined the levels of GLP2 and DPP4 and showed that Nogo-B expression also affected serum GLP2 levels (Supplementary Fig. 16a–d) but not DPP4 levels (Supplementary Fig. 16e–h). These observations suggest that Nogo-B is elevated in the EECs of T2DM patients and interacts with the IP2 region of proGCG, thereby interfering with the cleavage of MPGF by PCSK1. Consequently, this disruption leads to reduced secretion of GLP1 (Fig. 7c).
Discussion
Inadequate insulin secretion and reduced insulin sensitivity are the two main features of T2DM23. GLP1 secreted from the intestine acts on the pancreas to stimulate insulin secretion and inhibit glucagon secretion, while insulin and glucagon act (primarily) on the liver to regulate blood glucose levels36. Drugs that enhance insulin sensitivity, stimulate insulin secretion or the incretin axis, or inhibit hepatic gluconeogenesis are active in more than 7000 trials worldwide using mechanisms of action37. The development of mechanisms and targets is currently the focus and hotspot of T2DM research37,38. In this study, we found the Nogo-B levels were aberrantly increased in the liver, pancreas and small intestine of T2DM mice, and Nogo-B knockdown alleviated hyperglycemia in db/db mice (Fig. 1). Serum insulin and GLP1 levels were increased and glucagon levels were decreased in Nogo-B knockout mice, and Nogo-B deficiency promotes glucose metabolism and activates the insulin signaling pathway (Fig. 2), indicating Nogo-B is involved in maintaining the homeostasis of blood glucose. We also demonstrated that intestinal Nogo-B regulates GLP1 production, which binds to the MPGF of proGCG to retain proGCG on the ER, thereby inhibiting PCSK1-mediated cleavage of proGCG at the Golgi. Intestinal Nogo-B knockout increased GLP1 levels to stimulate insulin secretion and alleviate symptoms in T2DM mice.
Nogo-B is an ER-resident membrane protein involved in maintaining ER structure: deficiency of Nogo-B has been shown to reduce the number of ER tubules and to restrict the expansion of peripheral lamellae39,40. The ER is known to be the major site of protein synthesis, folding, and secretion41,42. The proGCG gene is transcribed, translated, and then cleaved by various PCSKs to produce a variety of biologically active short peptides, including glucagon and GLP136. In HEK293T cells, we found that Nogo-B binds to the IP2 region of the MPGF fragment, notably without competitively binding to the known PCSK1 cleavage site in the IP2 region. We also found that increased Nogo-B retained proGCG on the ER, significantly inhibited PCSK1 cleavage of proGCG and reduced GLP1 secretion. Given that Nogo-B is localized to the ER and PCSK1 is enriched in the Golgi apparatus43,44, it is possible that Nogo-B retains uncleaved proGCG in the ER, preventing it from reaching the Golgi apparatus for cleavage by PCSK1. This may explain the significant increase in the co-localization of proGGC with PCSK1 after Nogo-B knockdown/knockout in STC-1 cells and intestine.
Our results also show that in normal STC-1 cells, Nogo-B knockdown increased GLP1 levels but resulted in decreased protein and mRNA levels for PCSK1 and proGCG (Fig. 4). However, serum GLP1 levels, intestinal proGCG and intestinal PCSK1 expression were decreased in diabetic mice (Fig. 6). Similarly, intestinal proGCG and PCSK1 expression was decreased in T2DM patients (Fig. 7). In normal STC-1 cells, when Nogo-B knockdown promoted GLP1 secretion, the cells reduced PCSK1 and proGCG levels through negative feedback regulation to maintain homeostasis of GLP16. In the intestine of T2DM patients and mice, EECs are damaged by systemic inflammation and oxidative stress caused by metabolic dysfunction, which in turn reduces the expression of PCSK1 and proGCG45,46. Our results highlight the involvement of Nogo-B in the regulation of GLP1 production under both physiological and pathological conditions.
Nogo-B knockdown by siRNA decreased food intake in db/db mice. It is commonly believed that siRNAs cannot cross the blood-brain barrier47, and Nogo-B siRNA did not affect Nogo-B expression in the brain of db/db mice (Supplementary Fig. 1). In the central nervous system, GLP1 acts as a neurotransmitter signaling satiety along the brainstem-hypothalamus pathway48. We discovered that intestinal Nogo-B deficiency increases serum GLP1 levels and suppresses food intake in mice, indicating that intestinal Nogo-B, rather than brain Nogo-B, directly regulates intestinal GLP1 production, thereby affecting appetite and food intake (Supplementary Figs. 12, 15). Systematic monitoring of weekly body weight and serum GLP1 levels in mice has provided intriguing insights into the regulatory role of Nogo-B on body weight and its potential dependence on GLP1 (Supplementary Fig. 16i–l). We discovered that Nogo-B deficiency upregulates GLP1 levels and suppresses appetite, leading to lower body weight in mice. This effect was observed regardless of the pathological state of the mice.
Non-alcoholic fatty liver disease (NAFLD) is strongly associated with insulin resistance and T2DM, and there is a bidirectional facilitative relationship between NAFLD and T2DM prevalence49,50. Previous studies have shown that Nogo-B is associated with a variety of pathophysiological processes in the liver, with demonstrated impacts of Nogo-B on liver regeneration51, alcoholic liver18, liver fibrosis52, glucolipid metabolism16, obesity20, and cholestasis19. Nogo-B global knockout significantly activates hepatic INSR-INS-AKT pathway and alleviates the symptoms of NAFLD induced by high-fat or high-sugar diets, but the exact mechanism remains unknown16. Our current study indicates that intestinal Nogo-B knockdown increases GLP1 levels (thus stimulating insulin secretion), which explains the activation of the hepatic INSR-INS-AKT pathway in Nogo−/− mice. However, our results show that knockdown of intestinal Nogo-B in normal mice does not affect blood glucose levels, suggesting that the body is able to maintain blood glucose homeostasis through the regulation of other hormone levels. Most of the drugs used clinically to treat T2DM control blood glucose homeostasis by increasing insulin levels. The lipogenic effect of insulin exacerbates lipid accumulation and insulin resistance in the liver, which makes the treatment of T2DM a symptomatic rather than a curative approach53. Study have shown that restoring functional β cells is a therapeutic goal in the treatment of diabetes54. Intestinal Nogo-B knockout stimulates pancreatic insulin secretion and repairs damaged islets by promoting GLP1 production, while hepatic Nogo-B knockout inhibits de novo lipid synthesis in the liver and ameliorates liver damage thereby alleviating insulin resistance. Thus, Nogo-B could be a multi-organ target for the treatment of T2DM that alleviates T2DM by stimulating insulin secretion, repairing damaged islets, and reducing insulin resistance.
Some limitations need to be mentioned. Due to the limitations of the Nogo antibody, we were only able to detect Nogo-A and Nogo-B. Although the peptide fragment bound to proGCG was not present in Nogo-C, we were unable to definitively rule out an effect of Nogo-C on GLP1 without further investigation, which is worthy exploring in future studies. Although we found that the increased expression of Nogo-B in islet α cells of T2DM mice may also inhibit the production of islet α cells-derived GLP1, we were unable to further verify the function of pancreatic Nogo-B due to the lack of suitable cell lines and the almost undetectable activity of pancreatic GLP155. Furthermore, as the protein structure of Nogo-B is still unknown, we cannot develop a screen to identify agonists/antagonists/stability-affecting molecules of Nogo-B to block its function in maturation of GLP1 and T2DM.
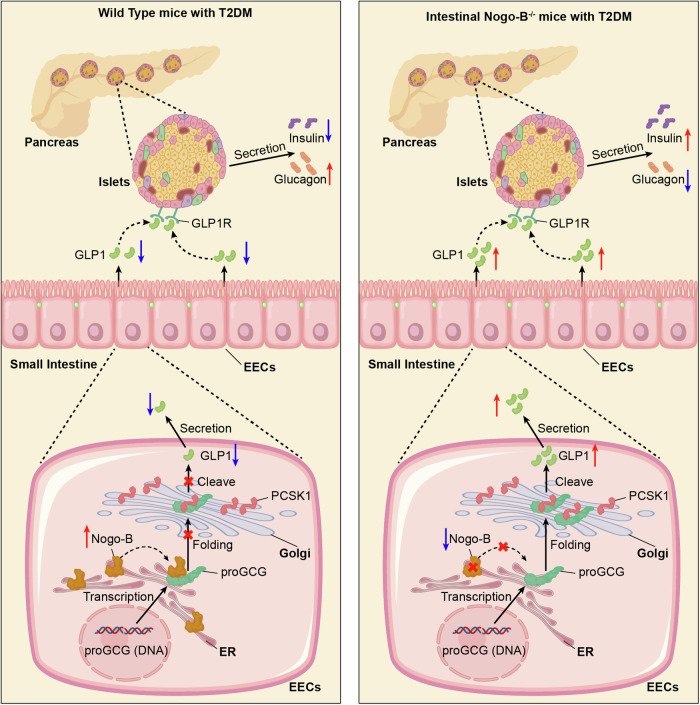
In conclusion, we have identified the role of Nogo-B in the regulation of glucose homeostasis, which may be one of the etiological processes in T2DM. Furthermore, the results that Nogo-B binds to the IP2 region of MPGF on the ER to inhibit PCSK1 shearing of proGCG to produce GLP1 further refine the physiology of GLP1 synthesis (Fig. 8).
Fig. 8. Proposed model.
This study demonstrates that the ER-resident protein, Nogo-B, interacts with the MPGF of proGCG to retain proglucagon within the ER, thereby inhibiting the PCSK1-mediated cleavage of proGCG in the Golgi. The knockout of intestinal Nogo-B in mice with T2DM results in elevated levels of GLP1 and insulin, while reducing glucagon levels. Consequently, this alleviates pancreatic injury and improves insulin resistance.
Methods
Human duodenum
This study included 10 patients aged 40–60 years with pancreatic occupations from June 2022 to December 2022 at the China-Japan Friendship Hospital (Beijing, China). All patients underwent their first surgery without prior radiotherapy or chemotherapy. The 10 patients with pancreatic occupations were divided into two groups, one without T2DM and the other with T2DM. Patients with a history of other malignancies or other abnormal metabolic diseases (except T2DM) were excluded at the time of inclusion. Paraffin sections were prepared from the normal duodenum, which was removed at the same time as the dominant pancreatic resection. Each patient provided signed informed consent, and the study was approved by the Ethics Committee of China-Japan Friendship Hospital (No 2018-72-K52). The samples used in this study were collected at different times during clinical procedures and then embedded for long-term storage. Paraffin sections of small bowel samples from each patient were prepared and stained simultaneously before the experiment. As there was only one small bowel sample per patient, only one section per patient was stained and analyzed. Basic patient information is provided in the Supplementary Information: Table S6.
Animals
All animal experiments conducted in this study were performed in compliance with the ARRIVE guidelines and followed the “Guide for the Care and Use of Laboratory Animals (the Guide)” published by the National Institute of Health (NIH Publications No. 8023, revised 1978). Ethical approval for these experiments was obtained from the Ethics Committee of Hefei University of Technology (HFUT20210901001). The db/db mice, db/m mice, Nogo global deficient (Nogo−/−) mice, and Nogo conditional knockout (Nogof/f) mice were purchased from GemPharmatech Co., Ltd. (Nanjing, China). The Villin-Cre (VillinCre) mice were purchased from Shanghai Model Organisms Center, Inc. (Shanghai, China). Nogo intestinal-specific deficient (Nogof/fVillinCre) mice were generated by crossing Nogof/f with VillinCre mice.
Mice were housed in a controlled environment with a temperature of 23 ± 1 °C, a relative humidity of 60–70%, and a 12-h light/12-h dark cycle. Only male mice were used for the experiments, and they were approximately 8 weeks old at the beginning of the study. Prior to the experiments, the mice were allowed to acclimate for at least 7 days. Specific details regarding food, feeding times, and experimental arrangements for glucose and insulin tolerance tests, as well as metabolic cage assays (PhenoMasterNG, Bad Homburg, Germany), can be found in the corresponding Figure legends. At the conclusion of the experiments, all mice were euthanized by CO2 asphyxiation. Relevant samples were collected for further analysis. The activities of alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), and serum triglycerides, cholesterol, high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C) were measured using an automated biochemical analyzer (3100, Hitachi High-Technologies Corporation, Tokyo, Japan).
In vivo siRNA interference
The male db/m mice, aged 8 weeks, served as a healthy control group and were injected intravenously with 0.9% normal saline (db/m group). The male db/db mice, also aged 8 weeks, were randomly divided into two groups: one group received an intravenous injection of scrambled siRNA (db/db group), and the other group received an intravenous injection of siRNA against Nogo-B (db/db-KD group). The Entranster™-in vivo transfection reagent, obtained from Engreen Biosystem Co., Ltd. (Beijing, China), was used for injections according to the manufacturer’s instructions. Specific details regarding the injection strategies can be found in the respective Figure legends. Both scrambled siRNA and Nogo-B siRNA were synthesized by GENEWIZ (Suzhou, China). The Nogo-B siRNA was a mixture of three different target pairs, named si-Nogo-B #1, #2, and #3. The sequences for scrambled siRNA and si-Nogo-B #1, #2, and #3 are provided in the Supplementary Information: Table S4.
Cell lines
HEK293T cells (Cat# CRL-3216, ATCC) and STC-1 cells (Cat# CRL-3254, ATCC) were all maintained in DMEM/high glucose medium containing 10% fetal bovine serum (FBS) and 50 g/mL penicillin/streptomycin. Cells were cultured at 37 °C and 5% CO2 in a humidified cell incubator.
Reagents
Anti-Flag (Cat# M20008), anti-GFP (Cat# M20004), anti-GLP1 (Cat# T56696), and anti-HA (Cat# M20003), anti-INSR (Cat# T55803), anti-IRS1 (Cat# T55830), anti-pIRS1 (Cat# TA3272), anti-IRS2 (Cat# PA5455), anti-pIRS2 (Cat# TA8383), anti-AKT (Cat# T55561), anti-pAKT (Cat# T40067), anti-PERK (Cat# TP52759), anti-pPERK (Cat# TA4499), anti-IRE1 (Cat# TA7651), anti-pIRE1 (Cat# T55605), anti-ATF4 (Cat# T55873), anti-XBP1 (Cat# T56725) antibodies were purchased from Abmart. Anti-cleaved caspase3 (Cat# 9664) antibody was purchased from Cell Signaling Technology. Anti-Nogo (Cat# IMG-5346A) antibody was purchased from Novus. Anti-GAPDH (Cat# HRP-60004), anti-pINSR (Cat# 31133-1-AP), anti-GCG (Cat# 15954-1-AP), anti-GRP94 (Cat# 14700-1-AP), anti-HSP90 (Cat# 13171-1-AP), anti-Rabbit IgG (Cat# 30000-0-AP), fluorescein (FITC)–conjugated Affinipure Goat anti-Mouse IgG(H + L) (Cat# SA00003-1), fluorescein (FITC)–conjugated Affinipure Goat anti-Rabbit IgG(H + L) (Cat# SA00003-2), Rhodamine (TRITC)–conjugated Goat anti-Mouse IgG(H + L), (Cat# SA00007-1), and Rhodamine (TRITC)–conjugated Goat anti-Rabbit IgG(H + L) (Cat# SA00007-2) antibodies were purchased from Proteintech. Anti-insulin (Cat# sc-377071) and anti-PCSK1 (Cat# sc-100578) antibodies were purchased from Santa Cruz Biotechnology.
DMEM (Cat# 01-052-1 A) and Fetal bovine serum (Cat# 04-010-1 A) were purchased from Biological Industries. 4% formaldehyde (Cat# BL539A), Penicillin-Streptomycin Solution (Cat# BL505A), Total RNA Isolation Reagent(Cat# BS258A), Cell lysis buffer (Cat# BL509A) and Commassie Brilliant Blue Staining Solution (Cat# BL605A) were purchased from Biosharp. EntransterTM-in vivo (Cat# 18668-11-1) was purchased from Engreen Biosystem. DAPI (Cat# HY-D0814) was purchased from MCE. Streptozotocin (STZ, Cat# 572201) and Oil red O (Cat# O9755) were purchased from Sigma-Aldrich. Insulin (Cat# I8830) and D-glucose (Cat# G8150) were purchased from Solarbio. Hieff TransTM Liposomal Transfection Reagent (Cat# 40802ES03) and GoldBand Plus 3-color Regular Range Protein Marker (Cat# 20350ES) were purchased from Yeasen Biotechnology. rProtein A/G MagPolyBeads (Cat# A10002) was purchased from Abmart. Mouse GLP1 Elisa kit (Cat# RX203401M), Mouse glucagon Elisa kit (Cat# RXJ202477M), and Mouse insulin Elisa kit (Cat# RX202485M) were purchased from Ruixin Biological Technology. Triglyceride labassay (Cat# 290-63701) was purchased from Wako Chemical. High Fat Diet 60% (Cat# MD12015A) was purchased from Medicience. Urea (Cat# U5128), dithiothreitol (DTT) (Cat# D9779) and iodoacetamide (IAA) (Cat# I1149) were purchased from Sigma. Sequencing grade modified trypsin (Cat# V5111) was purchased from Promega. Sep-Pak C18 cartridges (Cat# WAT023590) were obtained from Waters.
Bioinformatics analysis
We obtained three microarray datasets (GSE171269, GSE31953, and GSE145052) from the Gene Expression Omnibus database. To analyze the differential expression of genes, we utilized the limma package (version 3.26.8) in the R Bioconductor project. Genes showing significant differential expression were identified based on criteria of |fold change | > 1 and an adjusted p-value (FDR < 0.05). Subsequently, we visualized the genes that were significantly up-regulated across all three datasets. Venn diagrams were generated using VennDiagram software (version 1.7.3).
RNA sequencing analysis
Livers from 8-week-old Nogo−/− mice and their wild-type (WT) littermates fed a normal chow diet, were isolated and rinsed with phosphate-buffered saline (PBS). Approximately 100 mg of liver tissue was collected and flash-frozen in liquid nitrogen for subsequent RNA-seq analysis. RNA-seq analysis was performed by Novogene Bioinformatics Technology Co., Ltd (Beijing, China). The raw RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE236979. To conduct gene set enrichment analysis (GSEA), we utilized the ClusterProfiler package for clustering analysis, the MSigDB package to acquire reference gene sets, and the MSigDB Collections database for gene set analysis56,57. These bioinformatics tools allowed for a more comprehensive and detailed understanding of the differences in gene expression58.
Glucose tolerance tests (GTT)
For the GTT, mice were transferred to clean cages the day before the experiment and fasted for 16 h from 6 pm to 10 am. Throughout the fasting period, the mice had access to regular drinking water. At 10 am on the following day, basal blood glucose levels were measured from the tail tip using a glucometer. The mice were then weighed, and each mouse received an oral gavage of D-glucose solution (1 g/kg) dissolved in sterile saline. Blood glucose levels were measured at 15, 30, 60, 90, and 120 min after administration of the D-glucose solution.
Insulin tolerance tests (ITT)
For the ITT, mice were transferred to clean cages before the experiment and fasted for 6 h from 8 am to 2 pm. Throughout the fasting period, the mice had access to regular drinking water. At 2 pm, basal blood glucose levels were measured at the tail end using a glucometer. The mice were then weighed, and each mouse injected intraperitoneally with insulin (0.5 UI/kg) diluted in sterile saline. Blood glucose levels were measured at 15, 30, 60, 90, and 120 min after administration of the insulin solution.
Glucose-stimulated insulin secretion (GSIS)
For the GSIS assays, mice were transferred to clean cages and fasted for 6 h, starting at 8 am. During the fasting period, mice had access to normal drinking water. At 2 pm, mice were administered D-glucose (3 g/kg) intraperitoneally to start the timer. Blood samples were taken from the facial venous plexus at 0, 2, 5, and 15 min after injection, coinciding with the first phase of insulin secretion. These samples were then centrifuged at 5000 g for 15 min at 4 °C to separate the plasma, which was stored at −80 °C for later analysis. Plasma insulin levels were determined using an ultra-sensitive insulin assay kit, following the manufacturer’s instructions.
Hyperinsulinemic/euglycemic clamp tests
Hyperinsulinemic/euglycemic clamp tests were conducted by Shanghai Andubio Co, Ltd. The experimental procedure was divided into two main stages: intubation surgery and the clamp test itself.
Intubation surgery
After 4 h of fasting, mice were anaesthetized by intraperitoneal injection of tribromoethanol (200 μL/10 g), and the right internal jugular vein was cannulated under aseptic conditions, with the catheter filled with 50 U/mL heparin and secured with an external vest after wrapping around the neck. After surgery, each mouse was kept in separate cages and washed every 8 h with sterile saline containing 50 U/mL heparin to prevent tube clogging. The clamp tests were started after approximately 1–2 days of recovery.
Clamp test
Mice were fasted for 5 h, kept awake, and on the day of the clamp tests, a high dose of insulin (at a rate of 300 mU/kg/min) was initially administered via jugular vein cannulation within 1 min and then maintained at a rate of 2.5 mU/kg/min. After insulin infusion, 20% glucose infusion was started at an initial rate of 50 mg/kg/min, with subsequent adjustment of the glucose infusion rate (GIR) according to blood glucose levels. [13 C]glucose was infused at 0.03 mg/kg/min (after calculation based on the body weight of the mice). In general, the steady-state period was reached at about 80 min and the final plateau period was ensured to be stable for more than 30 min so that the blood glucose level was maintained at 16–18 mmol/L. Blood samples were taken from the end of the mice’s tails in heparinized capillary tubes at 0, 90, 100, 110, and 120 min to obtain 20–30 μL of blood samples, and then the serum was collected after standing for 2–3 h at 4 °C. The serum was analyzed by LC-MS for the amount of labeled glucose and the final data were calculated.
Amino acid sequence comparison
Amino acid sequence comparisons are made using the Multiple Sequence Comparison by Log-Expectation tool (https://www.novopro.cn/tools/muscle.html)59 to compare protein sequences. Identical amino acids are highlighted in red and marked with an *.
Immunohistochemistry and immunofluorescence staining
At the end of the experiment, a portion of the liver, pancreas, and small intestine were carefully excised and fixed in 4% paraformaldehyde. The fixed tissues were then embedded in paraffin. For the assessment of target protein expression by immunohistochemistry, 5-μm paraffin sections were prepared and processed following established protocols60. Immunofluorescent staining was performed on frozen tissue sections of tissue or cell slides, following established protocols60. In each experiment, a negative control (NC) was included, where rabbit normal IgG was used instead of the primary antibody, while keeping the remaining steps unchanged. The resulting images were visualized and captured using either a ZEISS Scope A1 fluorescence microscope (Oberkochen, Germany) or a Leica confocal microscope (Leica, Wetzlar, Germany).
After image capture, various experiments were analyzed using different methodologies with the assistance of Image J software. The average percentage of β-cell and α-cell fractions per pancreas was determined by calculating the ratio of the area occupied by insulin+ and glucagon+ cells to the total area of pancreatic tissue. The average percentage of CC3+ cells was calculated by dividing the number of CC3+ cells by the total number of cells in the islet area. The quantification of GLP1+ cells in the small intestine was expressed as the mean number of GLP1+ cells observed in five high-power fields (HPFs). Co-localization analysis involved examining the intensity distribution of markers within a selected area to visualize the spatial distribution and fluorescence intensity of defined markers. A higher degree of co-localization was indicated by the presence of similar shapes of the intensity distribution curves and a greater proportion of overlap.
Construction of expression vector and transfection
The nucleotide sequences of all proteins or peptides were cloned into the corresponding plasmids by GENEWIZ (Suzhou, China). The nucleotide sequences of these proteins or peptides were obtained from NCBI, and the specific sequences can be found in the Supplementary Information: Table S5.
Cells were seeded into plates, with or without cell slides, for transfection. Once the cell density reached 70–80%, the cells were transferred to fresh medium without antibiotics but supplemented with 10% fetal bovine serum. The expression vectors were transfected into cells using Hieff TransTM Liposomal Transfection Reagent (YEASEN, Shanghai, China). After 24–48 h, the cells were ready for the subsequent steps of the experiment.
Co-immunoprecipitation-mass spectrometry (co-IP-MS)
To perform co-IP-MS on mouse tissues, proteins were extracted from the liver and pancreas of wild-type mice. Two individual samples from each tissue were incubated with either rabbit normal IgG (3 μg) or anti-Nogo-B antibody (3 μg). The mixtures were incubated overnight at 4 °C. Subsequently, rProtein A/G MagPolyBeads (Abmart, Shanghai, China) were added to the mixtures, followed by an addition incubation at 4 °C for 2 h. After a brief centrifugation, the beads were collected and washed with PBS. Subsequently, 50 μL loading buffer was added into one tenth of the bead pellets, and the protein samples were eluted into the buffer by boiling at 100 °C for 5 min. The resulting eluates (10 μL) were applied to SDS-PAGE, and the presence of proteins was confirmed through silver staining. The remaining bead samples were denatured in 6 M urea buffer, reduced by 20 mM DTT at 56 °C for 30 min and alkylated by 40 mM IAA at room temperature for 30 min in the dark. The samples were diluted to 1 M urea final concentration and digested by trypsin (1/50, w/w) at 37 °C for 20 h. Tryptic peptides were desalted by C18 cartridges. Eluted peptides were concentrated by SpeedVac and stored at −20 °C until MS detection.
The MS detection was performed on an UltiMate 3000 RSLC nano-LC system coupled with an Orbitrap Fusion Lumos Mass Spectrometer (Thermo Fisher Scientific, Waltham, Mass, USA). Prior to LC-MS/MS analysis, tryptic peptides were dissolved in 0.1% (v/v) formic acid in 2% ACN. Solvent A was 0.1% FA in water and solvent B was 0.1% FA in 80% ACN. Samples were loaded with a Reversed Phase C18 trapping column (Thermo Scientific Acclaim PepMap, 75 μm × 20 mm, 3 μm, 100 Å) and then separated by an analytical RP C18 column (Thermo Scientific Acclaim PepMap RSLC, 75 μm × 25 cm, 2 μm, 100 Å). Gradient increased from 2% solvent B to 6% in 4 min, to 8% in 8 min, to 30% in 58 min and finally to 90% in 62 min at a flow rate of 0.3 μL/min. For MS1 scanning, a resolution of 60,000 FWHM was used, the maximum injection time was 50 ms, an AGC target of 4 × 105, the m/z scan range was set from 350 to 1500. For MS2 scanning, the collision energy of higher-energy collisional dissociation (HCD) was set to 30%, maximum injection time was 35 ms at an AGC target of 1 × 104 and acquired by ion trap with an m/z scan range from 200 to 1800. The acquired proteomics raw data were processed by MaxQuant software (v1.6.17.0) with Mus musculus protein database (17063 entries) downloaded from Uniprot.
To perform co-IP-MS on HEK293T cells, the cells were transfected with the pEGFP-h-MPGF and HA-h-Nogo-B expression vector, respectively. After 36 h, total proteins were extracted from the cells. The extracted protein samples were then incubated with either rabbit normal IgG (3 μg) or anti-Nogo-B antibody (3 μg) and incubated overnight at 4 °C. Following the incubation, rProtein A/G MagPolyBeads (Abmart, Shanghai, China) were added to the mixtures and incubated for an additional 2 h at 4 °C. After a brief centrifugation, the beads were collected and washed with PBS. Subsequently, 50 μL of loading buffer was added, and the protein samples were eluted into the buffer by boiling at 100 °C for 5 min. The entire eluted protein samples were subjected to SDS-PAGE. Following electrophoresis, the gel was stained with Coomassie brilliant blue solution. A portion of the gel containing the protein samples of interest was carefully excised and subjected to protein extraction and subsequent MS detection (Shanghai Applied Protein Technology Co. Ltd).
Gel pieces were cut from SDS PAGE, destained with 30% ACN/100 mM NH4HCO3 until the gels were destained. The gels were dried in a vacuum centrifuge. The in-gel proteins were reduced with dithiothreitol (10 mM DTT/100 mM NH4HCO3) for 30 min at 56 °C, then alkylated with iodoacetamide (200 mM IAA/100 mM NH4HCO3) in the dark at room temperature for 30 min. Gel pieces were briefly rinsed with 100 mM NH4HCO3 and ACN, respectively. Gel pieces were digested overnight in 12.5 ng/μL trypsin in 25 mM NH4HCO3. The peptides were extracted three times with 60% ACN/0.1% TFA. The extracts were pooled and dried completely by a vacuum centrifuge.
Each fraction was subjected to nano LC-MS/MS analysis using a Thermo Scientific Q Exactive mass spectrometer coupled to an Easy nLC system. Peptides were loaded onto a reverse phase trap column (Thermo Scientific Acclaim PepMap 100, 100 μm × 2 cm, nano Viper C18) and separated on a C18 analytical column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin). The separation employed a linear gradient of buffer B (84% acetonitrile, 0.1% formic acid) in buffer A (0.1% formic acid) at a flow rate of 300 nL/min, controlled by IntelliFlow technology.
The mass spectrometer operated in positive ion mode, acquiring MS data with a data-dependent top20 method. The survey scan ranged from 300 to 1800 m/z, and the most abundant ions were selected for HCD fragmentation. The AGC target was set to 1 × 106, with a maximum inject time of 50 ms and one scan range. Dynamic exclusion was enabled for 30 s. For the HCD spectra, the resolution was set to 17,500 at m/z 100, with an AGC target of 1 × 105. The isolation width was 1.5 m/z, micro scans set to 1, and the maximum injection time was 50 ms. The normalized collision energy was 27 eV. The underfill ratio was set to 0.1% to ensure adequate precursor ion selection at maximum fill time. Peptide recognition mode was enabled throughout the analysis.
MS/MS spectra were searched using MASCOT engine (Matrix Science, London, UK; version 2.2) against a nonredundant International Protein Index Arabidopsis sequence database v3.85 (released at September 2011, 39679 sequences) from the European Bioinformatics Institute (http://www.ebi.ac.uk/). For protein identification, the following options were used. Peptide mass tolerance = 20 ppm, MS/MS tolerance = 0.1 Da, Enzyme = Trypsin, Missed cleavage = 2, Fixed modification: Carbamidomethyl (C), Variable modification: Oxidation (M).
Co-IP-Western blot assays
For the co-IP-Western blot assays, cellular protein was extracted from either cells or cells transfected with expression vectors, following the method described earlier. A portion of the protein extract was saved as the input sample. The rProtein G MagPoly Beads were inverted several times to ensure thorough mixing. An appropriate volume of the magnetic bead suspension was pipetted into a centrifuge tube, placed on a magnetic separator, and left to stand for about 1 min until the solution clarified, after which the supernatant was aspirated. This washing step was repeated twice. Rabbit normal IgG (3 μg) or the specific antibodies of interest (3 μg) were added to the pre-treated magnetic beads, followed by vertexing to mix and incubating on a rotator at room temperature for approximately 30 min to ensure full contact and adsorption. After incubation, the tubes were placed back on the magnetic separator until the solution clarified, and the supernatant was aspirated.
The remaining sample solution was added to the magnetic beads and gently inverted to evenly disperse the antigen and bead-antibody complexes. The tube was then placed on a rotating mixer and incubated overnight at 4 °C. Once incubation was complete, the tubes were placed back on the magnetic separator and the supernatant was aspirated. Five volumes of wash buffer were added to the centrifuge tube containing the magnetic beads, vortexed to resuspend, and placed on the magnetic separator for about 1 min before the supernatant was discarded. This washing procedure was repeated five times. After washing, an equal volume of 1x SDS-PAGE Sampling Buffer was added to the magnetic beads, and the mixture was thoroughly combined. The mixture was heated at 95 °C for 5 min, followed by magnetic separation to collect the supernatant for Western blot analysis.
Expression and purification of hproGCG and hNogo-B-N protein
The hproGCG protein, which has a C-terminal fusion of the Strep-tag II tag, was cloned into the pHLsec expression vector. The expression vector containing hproGCG was transfected into 293 F cells. After transfection, the 293 F cells were cultured in suspension at 37 °C for four days. Following the culture period, the cell culture supernatant was collected. Affinity chromatography was performed using StrepTactin sepharose resin, which exploits the specific binding between the Strep-tag II and StrepTactin to purify the hproGCG protein from the supernatant, a step that was carried out overnight at 4 °C. Further purification of hproGCG protein was achieved by gravity column chromatography, with the flow-through fraction collected. The resin was washed with PBS to remove impurities, followed by elution of hproGCG protein using a 2.5 mM desthiobiotin solution. The eluted hproGCG protein was concentrated and dissolved in PBS solution to yield a high purity target protein.
The gene for the hNogo-B-N protein (1–185), which has an N-terminal fusion of the mouse heavy chain antibody signal peptide, was cloned into an expression vector containing a human Fc fragment. The expression vector containing the hNogo-B-N gene was transfected into 293 F cells. Post-transfection, the 293 F cells were cultured in suspension at 37 °C for four days. Upon completion of the culture, the cell culture supernatant was collected. Affinity chromatography using Protein A magnetic beads was employed to purify the hNogo-B-N protein from the supernatant based on the specific interaction between the Fc fragment and Protein A, with this step also conducted overnight at 4 °C. The supernatant and Protein A magnetic beads were separated using a magnetic stand to facilitate subsequent washing and elution steps. The magnetic beads were washed with PBS to remove unbound proteins and other impurities. The hNogo-B-N protein was eluted from the Protein A magnetic beads using a 0.1 M glycine solution (pH 3.0). The eluted protein was rapidly neutralized using a 1 M Tris-HCl solution (pH 8.5) to stabilize its structure and activity. The neutralized hNogo-B-N protein was then concentrated and buffer-exchanged into PBS solution to obtain a high-purity target protein.
Enzyme-linked immunosorbent assay (ELISA)
For the ELISA detection, the protein proGCG was diluted to a working concentration of 100 ng/well and coated onto an ELISA plate, followed by overnight incubation at 4 °C to ensure uniform distribution. The plate was subjected to a blocking step at 37 °C for 1 h to prevent non-specific interactions. After thorough washing with PBST, a series of dilutions of hNogo-B were prepared and added to the corresponding wells, including a control well without hNogo-B, and incubated at room temperature for 2 h. After another round of washing, anti-Nogo-B primary antibodies were introduced and incubated at room temperature for 1 h. The plate was then washed again to remove unbound primary antibodies, after which HRP-conjugated secondary antibodies were added and incubated under the same conditions. A final wash step removed any unbound secondary antibodies, and the chromogenic substrate was added, initiating a color development reaction at 37 °C for 15 min. The reaction was halted by the addition of a stop solution, and the optical density (OD) of each well was measured at 450 nm using a microplate reader. The OD values were converted into binding percentage by analyzing the OD values corresponding to different concentrations of hNogo-B: (ODmax-ODmin)/(OD sample-ODmin) × 100%. Concentration-response curves were plotted to assess the strength of the interaction between the two and their affinity.
Surface plasmon resonance (SPR) affinity assay
For the SPR affinity assay, a 1× HBS-EP+ buffer is formulated as the running buffer for the experiment. The hNogo-B-N protein is diluted to a concentration of 5 μg/mL using the running buffer and immobilized on the Fc2 channel of the SPR instrument via ligand capture for a duration of 40 s, until a signal of approximately 290 resonance units (RU) is achieved. The analyte, proGCG protein, is diluted to a starting concentration of 4000 nM in the running buffer and subjected to a continuous two-fold dilution series. The analyte is then injected over both the experimental and reference channels, with an association phase lasting 90 s at a flow rate of 30 μL/min, followed by a dissociation phase of 120 s. The system is regenerated using a 10 mM glycine-HCl (pH 1.5) regeneration buffer, applied for 30 s at the same flow rate to remove bound analytes and restore the sensor surface activity. Data analysis involves the kinetic evaluation of at least five consecutive concentration points, employing either a 1:1 binding model or a steady-state analysis approach to determine the affinity constants between the ligand and the analyte.
Western blot
Proteins were extracted from cell or tissue samples using a protein lysis buffer supplemented with protease inhibitors (PMSF and cocktail). Western blot analysis was performed to detect the protein expression levels of Nogo-B, Nogo-A, Flag, GFP, proGCG, GLP1, PCSK1, GAPDH, and HSP90, following established protocols61. The blots were normalized using either GAPDH or HSP90 as the loading control. For quantitative analysis of band density, all Western blot results were scanned using Photoshop software and normalized to the density of the loading control molecule in the corresponding samples.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from cell or tissue samples using Trizol reagent, following the established protocol60. Subsequently, cDNA was synthesized using 1 μg of total RNA and a reverse transcription kit. The mRNA expression levels of the relevant genes were assessed using qPCR, employing SYBR green PCR master mix. The primers used for qPCR amplification are listed in Supplementary Information: Table S4.
Statistics and reproducibility
The data analysis was conducted by a technician who was blind to the experimental design. The results were presented as the mean ± standard error of the mean (SEM) for each group or normalized to the mean of the experimental control group. Biological experimental replicates in each group are shown in the figure legends. Normality and equal variance tests were performed to determine whether parametric or nonparametric analysis should be used with the Prism software. Two-tailed Student’s t tests were employed for comparisons between two groups, while one-way ANOVAs with post-hoc tests were used for comparisons among more than two groups. In the analysis, a significant difference is considered when p < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This study was supported by China NSFC grants U22A20272 and 82173807 to Y.D., 82304503 to S.Z., 82300516 to Z.Y., and the Excellent Scientific and Technological Innovation Training Program of Shenzhen (RCYX20210706092040048) to L.D.
Author contributions
K.G., C.X, and Y.D. designed the study. K.G., C.X., and Y.D. drafted and edited the manuscript; K.G. and C.X. performed most of the experiments; Z.F. performed the bioinformatics analysis. Y.G. and L.D. conducted the IP-MS assay and data analysis. H.C., Y.C., S.Z., J.T., and C.L. provided suggestions for the experimental procedure. RR.P contributed to the in vitro and in vivo experiments. M.W., S.C. assisted in the experimental operation or data collection. L.J., L.L. performed protein purification experiments. Z.Y., L.M., and Y.I. performed the statistical analysis. B.W. provided the clinical samples required for the experiments.
Peer review
Peer review information
Nature Communications thanks Furkan Burak, Ming-Wei Wang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
There are no restrictions on data availability. All data supporting the findings of this study are available within the main text, supplementary information, and Source data. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact. Plasmids generated in this study are available from the lead contact upon request. RNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE236979. The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium via the PRlDE partner repository with the dataset identifier PXD046104 and PXD046118. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ke Gong, Chao Xue.
Change history
9/10/2024
A Correction to this paper has been published: 10.1038/s41467-024-52287-5
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51352-3.
References
- 1.Cantley, J. & Ashcroft, F. M. Q&A: insulin secretion and type 2 diabetes: why do beta-cells fail? BMC Biol.13, 33 (2015). 10.1186/s12915-015-0140-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demir, S., Nawroth, P. P., Herzig, S. & Ekim Ustunel, B. Emerging targets in type 2 diabetes and diabetic complications. Adv. Sci.8, e2100275 (2021). 10.1002/advs.202100275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weir, G. C., Gaglia, J. & Bonner-Weir, S. Inadequate beta-cell mass is essential for the pathogenesis of type 2 diabetes. Lancet Diabetes Endocrinol.8, 249–256 (2020). 10.1016/S2213-8587(20)30022-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haedersdal, S., Andersen, A., Knop, F. K. & Vilsboll, T. Revisiting the role of glucagon in health, diabetes mellitus and other metabolic diseases. Nat. Rev. Endocrinol.19, 321–335 (2023). 10.1038/s41574-023-00817-4 [DOI] [PubMed] [Google Scholar]
- 5.Holst, J. J. Glucagon-like peptide-1: are its roles as endogenous hormone and therapeutic wizard congruent? J. Intern. Med.291, 557–573 (2022). 10.1111/joim.13433 [DOI] [PubMed] [Google Scholar]
- 6.Sandoval, D. A. & D’Alessio, D. A. Physiology of proglucagon peptides: role of glucagon and GLP-1 in health and disease. Physiol. Rev.95, 513–548 (2015). 10.1152/physrev.00013.2014 [DOI] [PubMed] [Google Scholar]
- 7.Holst, J. J. The physiology of glucagon-like peptide 1. Physiol. Rev.87, 1409–1439 (2007). 10.1152/physrev.00034.2006 [DOI] [PubMed] [Google Scholar]
- 8.Holst, J. J., Albrechtsen, N. J. W., Rosenkilde, M. M. & Deacon, C. F. Physiology of the incretin hormones, GIP and GLP-1-regulation of release and posttranslational modifications. Compr. Physiol.9, 1339–1381 (2019). 10.1002/cphy.c180013 [DOI] [PubMed] [Google Scholar]
- 9.Chambers, A. P. et al. The role of pancreatic preproglucagon in glucose homeostasis in mice. Cell Metab.25, 927–934.e923 (2017). 10.1016/j.cmet.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song, Y. et al. Gut-proglucagon-derived peptides are essential for regulating glucose homeostasis in mice. Cell Metab.30, 976–986.e973 (2019). 10.1016/j.cmet.2019.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drucker, D. J. & Nauck, M. A. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet368, 1696–1705 (2006). 10.1016/S0140-6736(06)69705-5 [DOI] [PubMed] [Google Scholar]
- 12.Smits, M. M. & Holst, J. J. Endogenous glucagon-like peptide (GLP)-1 as alternative for GLP-1 receptor agonists: Could this work and how? Diab. Metab. Res. Rev. 39, e3699 (2023). [DOI] [PubMed]
- 13.GrandPré, T., Nakamura, F., Vartanian, T. & Strittmatter, S. M. Identification of the Nogo inhibitor of axon regeneration as a reticulon protein. Nature403, 439–444 (2000). 10.1038/35000226 [DOI] [PubMed] [Google Scholar]
- 14.Acevedo, L. et al. A new role for Nogo as a regulator of vascular remodeling. Nat. Med.10, 382–388 (2004). 10.1038/nm1020 [DOI] [PubMed] [Google Scholar]
- 15.Manzo, O. L. et al. Rewiring endothelial sphingolipid metabolism to favor S1P over ceramide protects from coronary atherosclerosis. Circ. Res.134, 990–1005 (2024). [DOI] [PMC free article] [PubMed]
- 16.Zhang, S. et al. Reduced Nogo expression inhibits diet-induced metabolic disorders by regulating ChREBP and insulin activity. J. Hepatol.73, 1482–1495 (2020). 10.1016/j.jhep.2020.07.034 [DOI] [PubMed] [Google Scholar]
- 17.Li, J., Wu, W., Xin, Y., Zhao, M. & Liu, X. Inhibition of Nogo-B promotes cardiac hypertrophy via endoplasmic reticulum stress. Biomed. Pharmacother.104, 193–203 (2018). 10.1016/j.biopha.2018.05.039 [DOI] [PubMed] [Google Scholar]
- 18.Park, J. K. et al. An endoplasmic reticulum protein, Nogo-B, facilitates alcoholic liver disease through regulation of kupffer cell polarization. Hepatology65, 1720–1734 (2017). 10.1002/hep.29051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, S. et al. Rosiglitazone alleviates intrahepatic cholestasis induced by alpha-naphthylisothiocyanate in mice: The role of circulating 15-deoxy-Delta(12,14) -PGJ(2) and Nogo. Br. J. Pharmacol.177, 1041–1060 (2020). 10.1111/bph.14886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, X. et al. Inhibition of high-fat diet-induced obesity via reduction of ER-resident protein Nogo occurs through multiple mechanisms. J. Biol. Chem.298, 101561 (2022). 10.1016/j.jbc.2022.101561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, J. et al. Nogo-B deficiency suppresses white adipogenesis by regulating beta-catenin signaling. Life Sci.321, 121571 (2023). 10.1016/j.lfs.2023.121571 [DOI] [PubMed] [Google Scholar]
- 22.Chen, Y. et al. NGBR is required to ameliorate type 2 diabetes in mice by enhancing insulin sensitivity. J. Biol. Chem.296, 100624 (2021). [DOI] [PMC free article] [PubMed]
- 23.Galicia-Garcia, U. et al. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci.21, 6275 (2020). 10.3390/ijms21176275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo, K. et al. Dendrocalamus latiflorus and its component rutin exhibit glucose-lowering activities by inhibiting hepatic glucose production via AKT activation. Acta Pharm. Sin. B12, 2239–2251 (2022). 10.1016/j.apsb.2021.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, I. M. et al. Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol. Syst. Biol.8, 594 (2012). 10.1038/msb.2012.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Z. et al. Microbiota and adipocyte mitochondrial damage in type 2 diabetes are linked by Mmp12+ macrophages. J. Exp. Med.219, e20220017 (2022). 10.1084/jem.20220017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao, R. H. Diabetes in the undernourished: coincidence or consequence? Endocr. Rev.9, 67–87 (1988). 10.1210/edrv-9-1-67 [DOI] [PubMed] [Google Scholar]
- 28.Titchenell, P. M., Lazar, M. A. & Birnbaum, M. J. Unraveling the regulation of hepatic metabolism by insulin. Trends Endocrinol. Metab.28, 497–505 (2017). 10.1016/j.tem.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yecies, J. L. et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab.14, 21–32 (2011). 10.1016/j.cmet.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dey, A. et al. Significance of prohormone convertase 2, PC2, mediated initial cleavage at the proglucagon interdomain site, Lys70-Arg71, to generate glucagon. Endocrinology146, 713–727 (2005). 10.1210/en.2004-1118 [DOI] [PubMed] [Google Scholar]
- 31.McGirr, R., Guizzetti, L. & Dhanvantari, S. The sorting of proglucagon to secretory granules is mediated by carboxypeptidase E and intrinsic sorting signals. J. Endocrinol.217, 229–240 (2013). 10.1530/JOE-12-0468 [DOI] [PubMed] [Google Scholar]
- 32.Hayashi, H. et al. Glucagon-like peptide-1 production in the GLUTag cell line is impaired by free fatty acids via endoplasmic reticulum stress. Metabolism63, 800–811 (2014). 10.1016/j.metabol.2014.02.012 [DOI] [PubMed] [Google Scholar]
- 33.Drucker, D. J. GLP-1 physiology informs the pharmacotherapy of obesity. Mol. Metab.57, 101351 (2022). 10.1016/j.molmet.2021.101351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, Q. et al. Impaired lipophagy induced-microglial lipid droplets accumulation contributes to the buildup of TREM1 in diabetes-associated cognitive impairment. Autophagy19, 2639–2656 (2023). 10.1080/15548627.2023.2213984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu, T. et al. PRDM16 exerts critical role in myocardial metabolism and energetics in type 2 diabetes induced cardiomyopathy. Metabolism146, 155658 (2023). 10.1016/j.metabol.2023.155658 [DOI] [PubMed] [Google Scholar]
- 36.Drucker, D. J., Habener, J. F. & Holst, J. J. Discovery, characterization, and clinical development of the glucagon-like peptides. J. Clin. Investig.127, 4217–4227 (2017). 10.1172/JCI97233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perreault, L., Skyler, J. S. & Rosenstock, J. Novel therapies with precision mechanisms for type 2 diabetes mellitus. Nat. Rev. Endocrinol.17, 364–377 (2021). 10.1038/s41574-021-00489-y [DOI] [PubMed] [Google Scholar]
- 38.Artasensi, A., Pedretti, A., Vistoli, G. & Fumagalli, L. Type 2 diabetes mellitus: a review of multi-target drugs. Molecules25, 1987 (2020). 10.3390/molecules25081987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voeltz, G. K., Prinz, W. A., Shibata, Y., Rist, J. M. & Rapoport, T. A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell124, 573–586 (2006). 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- 40.Jozsef, L. et al. Reticulon 4 is necessary for endoplasmic reticulum tubulation, STIM1-Orai1 coupling, and store-operated calcium entry. J. Biol. Chem.289, 9380–9395 (2014). 10.1074/jbc.M114.548602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellgaard, L. & Helenius, A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol.4, 181–191 (2003). 10.1038/nrm1052 [DOI] [PubMed] [Google Scholar]
- 42.Benham, A. M. Protein secretion and the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol.4, a012872 (2012). 10.1101/cshperspect.a012872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seidah, N. G. & Chretien, M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res.848, 45–62 (1999). 10.1016/S0006-8993(99)01909-5 [DOI] [PubMed] [Google Scholar]
- 44.Seidah, N. G. et al. The activation and physiological functions of the proprotein convertases. Int. J. Biochem. Cell Biol.40, 1111–1125 (2008). 10.1016/j.biocel.2008.01.030 [DOI] [PubMed] [Google Scholar]
- 45.Tian, F. et al. Ginsenoside compound K increases glucagon-like peptide-1 release and L-cell abundance in db/db mice through TGR5/YAP signaling. Int. Immunopharmacol.113, 109405 (2022). 10.1016/j.intimp.2022.109405 [DOI] [PubMed] [Google Scholar]
- 46.Daryabor, G., Atashzar, M. R., Kabelitz, D., Meri, S. & Kalantar, K. The effects of type 2 diabetes mellitus on organ metabolism and the immune system. Front. Immunol.11, 1582 (2020). 10.3389/fimmu.2020.01582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong, Y. Z., Siegwart, D. J. & Anderson, D. G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliver Rev.144, 133–147 (2019). 10.1016/j.addr.2019.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vrang, N. & Larsen, P. J. Preproglucagon derived peptides GLP-1, GLP-2 and oxyntomodulin in the CNS: Role of peripherally secreted and centrally produced peptides. Prog. Neurobiol.92, 442–462 (2010). 10.1016/j.pneurobio.2010.07.003 [DOI] [PubMed] [Google Scholar]
- 49.Ferguson, D. & Finck, B. N. Emerging therapeutic approaches for the treatment of NAFLD and type 2 diabetes mellitus. Nat. Rev. Endocrinol.17, 484–495 (2021). 10.1038/s41574-021-00507-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Younossi, Z. M. et al. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: a systematic review and meta-analysis. J. Hepatol.71, 793–801 (2019). 10.1016/j.jhep.2019.06.021 [DOI] [PubMed] [Google Scholar]
- 51.Gao, L. et al. Reticulon 4B (Nogo-B) facilitates hepatocyte proliferation and liver regeneration in mice. Hepatology57, 1992–2003 (2013). 10.1002/hep.26235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumashiro, N. et al. Cellular mechanism of insulin resistance in nonalcoholic fatty liver disease. Proc. Natl Acad. Sci. USA108, 16381–16385 (2011). 10.1073/pnas.1113359108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujikawa, T. et al. Leptin engages a hypothalamic neurocircuitry to permit survival in the absence of insulin. Cell Metab.18, 431–444 (2013). 10.1016/j.cmet.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dirice, E. et al. Inhibition of DYRK1A stimulates human beta-cell proliferation. Diabetes65, 1660–1671 (2016). 10.2337/db15-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galvin, S. G. et al. The human and mouse islet peptidome: effects of obesity and type 2 diabetes, and assessment of intraislet production of glucagon-like Peptide-1. J. Proteome Res.20, 4507–4517 (2021). 10.1021/acs.jproteome.1c00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics27, 1739–1740 (2011). 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst.1, 417–425 (2015). 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA102, 15545–15550 (2005). 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res.32, 1792–1797 (2004). 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duan, Y. et al. Peroxisome Proliferator-activated receptor gamma activation by ligands and dephosphorylation induces proprotein convertase subtilisin kexin type 9 and low density lipoprotein receptor expression. J. Biol. Chem.287, 23667–23677 (2012). 10.1074/jbc.M112.350181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu, Y. et al. 25-Hydroxycholesterol activates the expression of cholesterol 25-hydroxylase in an LXR-dependent mechanism. J. Lipid Res.59, 439–451 (2018). 10.1194/jlr.M080440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are no restrictions on data availability. All data supporting the findings of this study are available within the main text, supplementary information, and Source data. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact. Plasmids generated in this study are available from the lead contact upon request. RNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE236979. The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium via the PRlDE partner repository with the dataset identifier PXD046104 and PXD046118. Source data are provided with this paper.