Abstract
We have investigated the relative merits of two commonly used methods for target site selection for ribozymes: secondary structure prediction (MFold program) and in vitro accessibility assays. A total of eight methylated ribozymes with DNA arms were synthesized and analyzed in a transient co-transfection assay in HeLa cells. Residual expression levels ranging from 23 to 72% were obtained with anti-PSKH1 ribozymes compared to cells transfected with an irrelevant control ribozyme. Ribozyme efficacy depended on both ribozyme concentration and the steady state expression levels of the target mRNA. Allylated ribozymes against a subset of the target sites generally displayed poorer efficacy than their methylated counterparts. This effect appeared to be influenced by in vivo accessibility of the target site. Ribozymes designed on the basis of either selection method displayed a wide range of efficacies with no significant differences in the average activities of the two groups of ribozymes. While in vitro accessibility assays had limited predictive power, there was a significant correlation between certain features of the predicted secondary structure of the target sequence and the efficacy of the corresponding ribozyme. Specifically, ribozyme efficacy appeared to be positively correlated with the presence of short stem regions and helices of low stability within their target sequences. There were no correlations with predicted free energy or loop length.
INTRODUCTION
Hammerhead ribozymes are potentially powerful tools for sequence-specific inhibition of target gene expression (1). Their intrinsic cleavage activity makes them theoretically superior to traditional antisense oligodeoxynucleotides (ODNs) in terms of inhibitory capacity. Recent advances (1–3) have extended the range of targets so far that virtually any limited stretch of RNA is now likely to contain a useful target. However, other problems, including methods of delivery and target site selection, remain to be solved. The latter, in particular, appears to be a critical step in the design of antisense or ribozyme molecules for suppression of target gene expression, as there may exist only a few sites within any mRNA that are accessible to hybridization (4,5). We have compared two commonly employed methods for the rational selection of target sites, secondary structure prediction and in vitro accessibility assays. As target we have used the mRNA for a novel human protein kinase, PSKH1 (6).
RNA secondary structure prediction algorithms based mainly on energy minimization have intrinsic limitations, although subject to continuous improvement (7,8). An established theory for selection of target sites based on predicted structure is also required. The possible modulation of RNA structures by protein binding in vivo (9,10) cannot yet be modeled and has so far limited their use in designing antisense ODNs or ribozymes. A positive correlation between the inhibitory effect of an antisense RNA and low local folding potential has been noted (high ΔG) (11). Together with more recent data (12–14), this suggests that extended single-stranded or unstructured regions may be the best targets for antisense ODNs. However, although ribozymes targeting predicted loop regions have proved efficacious in some cases (15,16), there are also examples of failures (17). A recent systematic analysis of the hybridization of tRNA-Phe to a set of complementary ODNs determined that all high-yield heteroduplexes involved RNA sequences forming both double-stranded stems and single-stranded regions, and that bases of the latter regions were often stacked onto the stems (18). This suggests the requirement for at least some degree of helical conformation in the secondary structures of favorable targets.
The accessibility of different stretches of the mRNA for hybridization with short antisense ODNs may be determined in vitro by RNase H-mediated cleavage of the RNA at regions where the ODNs hybridize to the target transcript (14,19–22). Screening of a large set of ODNs targeting potential ribozyme cleavage sites has been employed to select the most promising sites for ribozyme targeting (19). Accessibility assays with specific ODNs have also been performed in cell extracts on endogenous transcripts in order to better approximate the in vivo situation (14,22). The most accessible sites within any target RNA may be selected by performing the RNase H-assay with a randomized set of ODNs (20,21) or with a target-specific set of cDNA fragments prepared by partial DNase I-digestion (23). ODN libraries in previous studies (20,21) have been more or less completely randomized. We wanted to identify specific sites amenable to cleavage with standard hammerhead ribozymes, and to restrict the analysis to those triplets that are cleaved most efficiently. Since the hammerhead ribozyme seems to prefer a purine in the first position of the cleavage triplet and a C or an A in the third position (24), we decided to concentrate on the triplets GUC, GUA, AUC and AUA in our analysis. Four ODN libraries, each specific for one of these cleavage triplets, were synthesized and used to screen in vitro transcribed PSKH1 RNA for accessible sites.
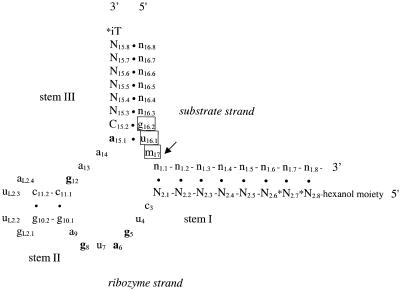
Based on a study of the most active ribozymes expressed in vivo from a randomized library (25), the ribozymes used in the present study were designed to have symmetric 8+8 nt arms (Fig. 1) (26). Chemically synthesized ribozymes of similar arm lengths have subsequently been successfully employed (19,27–29). The stem II structure (Fig. 1) was chosen to be 2bp long (30). The activity of such truncated ribozymes in cell culture has been demonstrated (27,29). For increased nuclease stability of the ribozymes, we have retained unmodified ribonucleotides in only five catalytically important positions (31,32). Deoxyribonucleotides were used in the flanking arms (33,34) and 2′-O-alkylated ribonucleotides in the core and stem–loop II (31,32,35) (Fig. 1). Nuclease stability of ribozymes was increased further by an inverted thymidine at the 3′-end (28,29,32) and by a hexanol moiety at the extreme 5′-end (36). Ribozymes used in vivo frequently include short stretches of phosphorothioate linkages at the 5′-end (28,29), 3′-end (34) or both (27) for stabilization against exonucleases. We included two phosphorothioates at the 5′-end and one at the 3′-end. DNA nucleotides in the arms have been reported to increase catalytic efficiencies of ribozymes in vitro, most likely due to increased product dissociation rates (37–39). DNA-armed ribozymes may recruit RNase H activity upon hybridization with the target RNA and thus enhance their apparent activity (40). We are here primarily interested in the accessibility of cleavage sites. Anything that reduces the importance of intrinsic ribozyme cleavage activity and increases the importance of target-specific inhibition of expression is desirable.
Figure 1.
Schematic of the hammerhead ribozyme hybridizing to its target mRNA. The arrow indicates the position of cleavage in the mRNA. Numbering is according to the nomenclature of Hertel et al. (26). Unmodified ribonucleotides are in bold lower case, 2′-O-alkylated ribonucleotides in plain lower case, and deoxynucleotides in upper case. Phosphorothioates are indicated by asterisk, while iT denotes an inverted 3′–3′ thymidine. Bases of the GUM target triplet, where M is C or A, are boxed.
Cellular delivery of ribozymes is commonly accomplished with various cationic liposome formulations (16,19,27–29). We have used the cationic liposome reagent lipofectamine to co-transfect HeLa cells with a mixture of ribozyme, a firefly luciferase reporter gene construct containing the complete coding cDNA of the target PSKH1, and a Renilla luciferase-encoding plasmid serving as an internal transfection control. We have succeeded in constructing a ribozyme against PSKH1 mRNA which reduced the activity of the corresponding reporter gene to 20–25% in a concentration dependent manner. A correlation is observed with certain features of the predicted secondary structure of the target mRNA.
MATERIALS AND METHODS
All restriction enzymes were from New England Biolabs.
Synthesis and purification of hammerhead ribozymes
Automated RNA and DNA synthesis was carried out on an Applied Biosystems model 394 DNA/RNA synthesizer. 2′-O-Alkyl ribozymes containing five unmodified purine ribonucleotides were synthesized by solid phase β-cyanoethyl phosphoramidite chemistry (41), using the 2′-O-tert-butyldimethylsilyl protection strategy for the ribonucleotides (42,43). Syntheses were performed on controlled pore glass bearing an inverted thymidine linkage (Glen Research). A lipophilic capture tag, 1-[Diisopropyl(DL-α-tocopheryloxy)silyl]oxy-6-(2-cyanoethyl N,N-diisopropylamino-phosphinoxy)hexane, was added at the 5′-end of the oligomer as described (36). Cleavage from the support and release of all base labile protecting groups (44,45), reverse phase HPLC purification (Pharmacia Source 5RPC 10/10 column, using a flow-rate of 1 ml/min, or a µBondapac C-18 column), lyophilization, desilylation (46), butanol-precipitation and counter-ion exchange with NaClO4 were performed essentially as described (36). Ribozymes, retaining a 5′ hexanol-linker, were desalted (NAP-10 columns, Pharmacia) and quantified by UV spectroscopy. Molar extinction coefficients were calculated based on the nearest-neighbor method (Biopolymer Calculator at http://paris.chem.yale.edu ). Exact molecular weights were calculated. Ribozymes were controlled by denaturing 15% polyacrylamide gel electrophoresis prior to their application in cell culture experiments.
Sequence and modification of hammerhead ribozymes
PSKH1-specific 2′-O-methylated ribozymes (MRz) were designed targeting a total of eight sites. Allylated ribozymes (ARz) targeting a subset of these sites were also synthesized in order to evaluate the relative effects of the two types of modification on the efficacy of ribozymes. For both types of ribozymes, controls having the same chemical composition and length of hybridizing arms were designed targeting an irrelevant mRNA (human tissue factor). Ribozymes are named after their type of modification and cleavage position. Thus MRz-519 is a methylated ribozyme cleaving after guc519, while ARz-519 is the corresponding allylated ribozyme. The respective control ribozymes are MRz-TF and ARz-TF. The unique sequences (flanking arms) of the ribozymes were as follows (the conserved sequence of the core is indicated by <core> for all but the first ribozyme sequence): MRz-TF/ARz-TF, A*A*T-C-T-C-C-T-c-u-g-a-u-g-a-g-g-g-u-u-a-c-c-g-a-a-a-C-T-T-A-G-T*G-iT; MRz-118, T*C*G-G-G-A-A-G- <core> -a-C-C-T-T-G-C*T-iT; MRz-238, C*C*G-G-C-T-T-T <core> -a-C-A-G-G-G-C*C-iT; MRz-287, A*G*T-C-G-G-G-G- <core> -a-C-C-G-G-G-G*C-iT; MRz-465/ARz-465, T*G*A-T-G-G-C-A- <core> -a-C-G-G-C-T-G*C-iT; MRz-519/ARz-519, G*C*A-G-C-T-C-C- <core> -a-C-T-C-A-C-A*C-iT; MRz-539/ARz-539, A*C*G-C-A-C-C-C- <core> -a-C-G-C-A-G-C*A-iT; MRz-548, G*T*T-G-G-C-A-T- <core> -a-C-G-C-A-C-C*C-iT; MRz-712, A*G*A-T-A-C-C-G- <core> -a-C-G-C-C-A-T*C-iT. Modified and unmodified ribonucleotides are in plain and bold lower case, respectively, while deoxyribonucleotides are in upper case. An asterisk denotes a phosphorothioate linkage, while iT indicates an inverted 3′–3′ thymidine (Fig. 1).
Specific and semi-randomized DNA ODNs
Semi-randomized 13mer antisense ODNs specific for each of the target triplets GUC, GUA, AUC and AUA were synthesized and HPLC-purified. The ODNs were designed to mimic the hybridizing arms of symmetrically armed (6+6 nt) hammerhead ribozymes and had the following sequences (N denotes a randomized position): GUC-specific library, (N)6GAC(N)4; GUA-specific library, (N)6TAC(N)4; AUC-specific library, (N)6GAT(N)4; AUA-specific library, (N)6TAT(N)4. Specific 13mer antisense ODNs targeting the selected ribozyme cleavage sites were also synthesized. These ODNs are identified by the cleavage position of their corresponding ribozymes, and have the following sequences: O-118, GGGAAGGACCTTG; O-238, GGCTTTGACAGGG; O-287, TCGGGGGACCGGG; O-465, ATGGCATACGGCT; O-519, AGCTCCGACTCAC; O-539, GCACCCGACGCAG; O-548, TGGCATGACGCAC; O-712, ATACCGGACGCCA.
Plasmids
The cDNA of the coding sequences of PSKH1 was previously cloned in our lab and inserted into the expression vector pcDNA3 (Invitrogen) downstream of a T7 promoter, producing the plasmid pcDNA3-PSK. A Renilla luciferase expression plasmid, pEF1-Rluc, constructed by inserting the EF1α promoter into the multiple cloning site of the pRL-null vector (Promega), was used as an internal control in co-transfection experiments. This plasmid was a kind gift from Professor A.-B. Kolstø.
Preparation of PSKH1-luciferase fusion constructs
The full-length coding sequence of PSKH1 cDNA was cloned in-frame with firefly luciferase cDNA into the BglII/NcoI sites of the pGL3 Enhancer expression vector (Promega) as a BamHI–NcoI PCR fragment, producing the plasmid pPSK-Luc. The PSK-Luc fusion was cloned into the tetracycline response plasmid pTRE (Clontech) in two steps. A KpnI–BamHI fragment of pPSK-Luc was first transferred to the EGFP-1 vector (Clontech). Subsequently, an EcoRI–BamHI fragment from this clone was excised and cloned into the same sites of the pTRE vector, giving pTRE-PSK-Luc. Finally, PSK-Luc cDNA was also cloned into the pcDNA3 expression vector as an EcoRI–XbaI fragment for higher-level expression. This expression plasmid was termed pcDNA3-PSK-Luc.
In vitro transcription of PSKH1 RNA
For in vitro transcription of PSKH1 RNA with T7 RNA polymerase, the pcDNA3-PSK plasmid was linearized internally with EcoNI, producing a 1.03 kb transcript containing 47 nt of 5′ vector-derived sequence, or downstream with XbaI (producing a 1.37 kb transcript). Protein and salt were removed from restriction reactions using JetQuick columns (Genomed). Purified DNA was eluted in DEPC-treated water (DEPC-H2O), precipitated with ethanol, and resuspended in DEPC-H2O. Run-off transcription of uniformly 32P-labeled RNA was performed in 50 µl reactions containing 2 µg template DNA, 0.5 mM each of GTP, ATP, CTP and UTP (Boehringer Mannheim), 50 U RNasin (Promega), 1–2 µl 10 µCi/µl [α-32P]rATP or rUTP (Amersham) and 50 U T7 RNA polymerase (New England Biolabs) in RNA polymerase buffer (40 mM Tris–HCl, 6 mM MgCl2, 2 mM spermidine, 10 mM DTT, pH 7.9). Reactions were incubated for 2 h at 37°C. RNA was then desalted by centrifugation through RNase-free G50 Sephadex QuickSpin Columns (Boehringer Mannheim) and deproteinated by phenol extraction. RNA was then precipitated with isopropanol and resuspended in RNase-free water. Alternatively, RNA was purified by LiCl-precipitation (2.5 M final concentration) following transcription. Yield and concentration of RNA were calculated from the percentage of incorporated radioactivity.
Antisense ODN and RNase H-mediated in vitro cleavage of PSKH1 RNA
RNase H-mediated cleavage assays with antisense ODN libraries were performed in 10 µl reactions containing 40 µM ODN library, 50 nM PSKH1 mRNA and 0.25 U RNase H (Promega) in a buffer containing 40 mM Tris–HCl pH 7.5, 10 mM MgCl2 and 10 mM DTT. A stock dilution of RNA in buffer was heated to 95°C for 60 s and then preincubated at 37°C for 15 min before adding enzyme. An 8.0 µl mixture of RNA, buffer and enzyme was then mixed with 2.0 µl 200 µM ODN library, yielding the indicated final concentrations of components. Reactions were incubated for 30 min at 37°C, and quenched on ice with 10 µl of denaturing loading buffer (8 M urea, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). Samples were heated to 95°C for 2 min prior to loading onto a 4% denaturing polyacrylamide gel. RNA size markers were prepared by run-off in vitro transcription of differently linearized (NcoI, XmnI, BsaA1, NaeI, BstUI, FokI and DdeI) pcDNA3 templates. Samples and standards were analyzed by electrophoresis for 3–4 h at 50 W at room temperature on 40 cm sequencing gels. The gels were then transferred to Whatman 3MM paper, wrapped in plastic foil and exposed overnight in a Phosphor Screen (Molecular Dynamics) prior to analysis on a radioanalytical scanner (Storm 860, Molecular Dynamics). Fragment sizes were calculated from their measured mobilities and those of the size markers. Reactions with specific antisense ODNs (200 nM, 4-fold molar excess) were performed essentially as for the ODN libraries.
Cultured cells
HeLa cells (from ATCC) were maintained in Dulbecco’s Minimal Essential Medium supplemented with 2 mM glutamine and 10% fetal calf serum (all reagents from Gibco BRL). Upon thawing, the cells were grown to near confluency for 2–3 days and passaged at least once before they were used for experiments. Cells were routinely passaged every 3–4 days.
Transient co-transfections
Cells were plated in 12-well (Costar) or 24-well (Sarstedt) plates at 30–40% confluency and transfected at an estimated 60–80% confluency the following day. Transfections were performed with 100 nM ribozyme (1.23–1.30 µg/ml), 0.40 µg/ml reporter construct (pTRE-PSK-Luc or pcDNA3-PSK-Luc) and 8 ng/ml internal control plasmid (pEF1-Rluc). Nucleic acids were complexed with lipofectamine (Gibco BRL) (a final concentration of 8.2–8.5 µg/ml lipid) at a constant 1:5 w/w ratio, following optimization with the transfection agent. Complexes were formed by mixing equal volumes of medium-diluted nucleic acids (ribozyme + DNA) and lipofectamine and incubating at room temperature for 30 min. The complexes were subsequently diluted to the final volume with serum-free medium and added to pre-washed cells (250 and 500 µl for 16 and 22 mm wells, respectively) for 5 h. The complexes were subsequently replaced with full medium.
Uptake of FITC-labeled ribozymes
Uptake of FITC-labeled ribozymes under the standard transfection conditions was determined by fluorescence spectroscopy. Lipofectamine complexes were diluted with medium and chilled on ice prior to addition to cells (in 12-well plates) preincubated (for at least 30 min) either at 37°C or on ice. Following transfection for 5 h, cells were harvested by washing three times with ice cold phosphate-buffered saline (PBS) and lysed with 1.5 ml PBS–TDS (1% Triton, 0.5% deoxycholate and 1% SDS in PBS) for at least 15 min at room temperature under vigorous shaking to achieve complete lysis. Parallels were combined and fluorescence recorded on a spectrometer (LS-5, Perkin Elmer) at excitation and emission wavelengths of 492 and 522 nm, respectively.
Luciferase activity assays
Cells were harvested 24 h after initiation of transfection and washed twice in cold PBS prior to lysis for 30 min on ice with a passive lysis buffer supplied with the Dual-Luciferase® Reporter Assay System kit (Promega). Following brief centrifugation to pellet cell debris, luciferase assays were performed in white non-transparent 96-well plates (Nunc) using a plate-reading luminometer (MicroLumat Plus, EG&G Berthold) equipped with two injectors. Dual-luciferase assays were performed on 25 µl lysate supernatant. Firefly and Renilla luciferase activities were recorded following the respective injections of 100 µl LAR II and 100 µl Stop&Glow reagents of the assay system according to the manufacturer’s instructions. The instrumental background levels of luminescence were recorded for empty wells.
RESULTS
Selection of ribozyme target sites by MFold secondary structure prediction
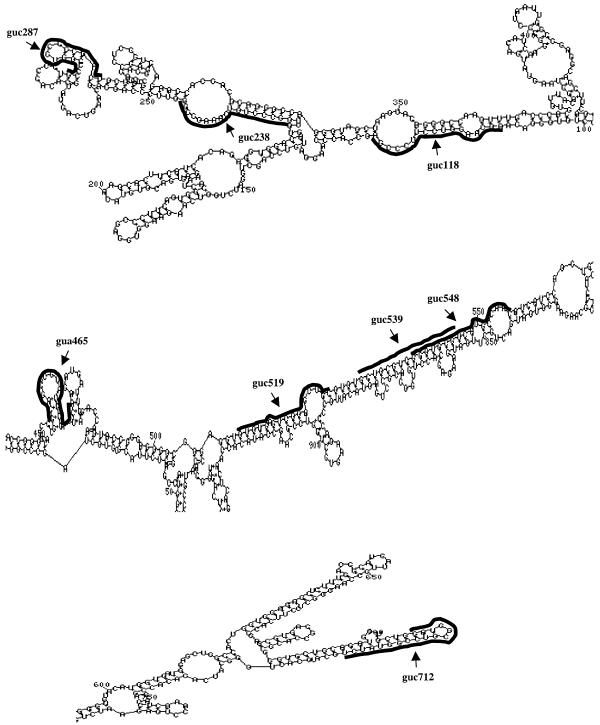
We have used the MFold program (version 3.0 at http://mfold2.wustl.edu ) (7,8) to predict the secondary structure of PSKH1 mRNA as a means of selecting GUC sites for ribozyme targeting. The MFold program generated many suboptimal secondary structures that differed minimally in energy from the optimal folding. Although these secondary structures displayed varying degrees of folding differences, certain substructures could be identified that demonstrated a considerable degree of conservation. High conservation of these substructures among alternate suboptimal structures may increase their likelihood of being ‘true’ structures (i.e. contained in the actual secondary structure of the mRNA in vivo). Therefore, in selecting GUC sites for hammerhead ribozyme targeting, we have screened 10 optimal and suboptimal secondary structures of the first 1.0 kb of the PSKH1 transcript, as transcribed from pTRE-PSK-Luc, for recurring substructures. All selected target sites were located within identical substructures in at least six out of the 10 most energetically stable structures, including the optimal structure (Table 1). For some target sites, several of the suboptimal structures diverged only slightly from the consensus in ways that did not significantly change the basic characteristics of the substructure. Due to the lack of a consensus on what kind of structures make the best targets, we have chosen GUC targets (guc238, guc287, guc519 and guc712) that are predicted to be located within very different substructures (Fig. 2). The guc238 cleavage triplet is located within a large internally looped stem in which the 5′ arm (stem I) of the corresponding ribozyme is complementary to the nucleotides of the loop. The guc287 target sequence, on the other hand, is predicted to fold into a short stem–loop structure. The guc519 target site is located at the end of what is essentially a very long basepaired region that is interrupted by occasional short bulges. A ribozyme was also designed targeting guc712, located at a particularly stable hairpin structure at the end of a very long, and presumably very stable, stem. This target site was included due to the high conservation of its secondary structure among suboptimal structures of the target RNA (identical in 10 out of 10 and 19 out of the 20 energetically most favorable structures). In addition to the above target sites, the first GUC site in the RNA (guc118) was selected due to its close proximity to the translation initiation region (only ~20 nt downstream of the initiation codon). This region has often been targeted (12,33,47,48) because the RNA in this region might be relatively open due to the need for binding of the components of the translation machinery. The predicted secondary structure for the guc118 target consists of a stem with two internal loops (Fig. 2).
Table 1. Features of the predicted secondary structure of selected ribozyme target sites.
| guc118 | guc238 | guc287 | gua465 | guc519 | guc539 | guc548 | guc712 | |
|---|---|---|---|---|---|---|---|---|
| Structural frequency | 6/10 | 6/10 | 6/10 | 6/10 | 6/10 | 7/10 | 7/10 | 10/10 |
| Major loop | 5 | 8 | 4 | 6 | 3 | 0 | 3 | 3 |
| Major stem | 6 | 8 | 3 | 4 | 6 | 17 | 10 | 11 |
| ΔG (helix)a | –12.7 | –20.1 | –5.7 | –8.8 | –8.1 | –11.4 | –9.7 | –13.1 |
| ΔG (target)b | –13.2 | –19.4 | –6.7 | –9.4 | –14.4 | –18.1 | –15.4 | –8.3 |
| ΔG (duplex)c | –32.9 | –36.0 | –40.2 | –35.2 | –34.1 | –36.2 | –34.0 | –31.7 |
Structural frequency denotes the number of times the target site substructure occurred unchanged in the 10 energetically most stabilized foldings of the mRNA. Major loop and stem are the longest stretches of predicted unpaired and paired target sequence bases, respectively. Free energies (ΔG) were calculated for the most stable helical region within the target sequencea, for the target sequence as a wholeb, and for the hammerhead–RNA duplexc, as detailed in the text. Energies are in kcal/mol.
Figure 2.
Excerpts of the energetically optimal secondary structure of the first 1000 bases of the pTRE-PSK-Luc transcript, as predicted by MFold version 3.0 (7,8). Target sequences of the ribozymes are highlighted. Arrows indicate position of cleavage.
In vitro accessibility assays with semi-randomized ODN libraries and specific ODNs
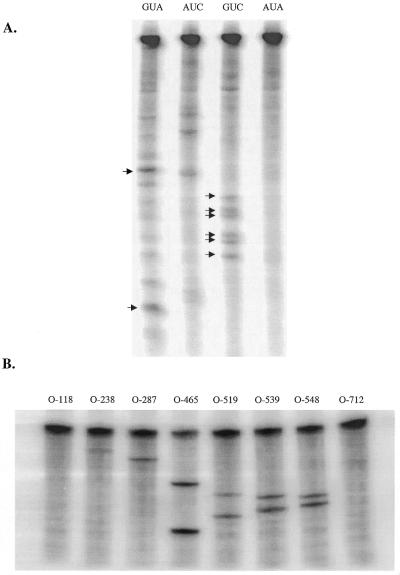
Target triplet-specific, semi-randomized ODN libraries were used to screen in vitro transcribed PSKH1 RNA for hybridization-accessible GUC, GUA, AUC and AUA sites. Accessible sites were identified by RNase H-mediated cleavage of the RNA at the RNA–DNA hybrids generated by hybridization of antisense ODNs to target RNA (19–21). Screening of a 1.03 kb PSKH1 transcript from the pcDNA3-PSK plasmid was performed separately with each of the four ODN libraries (Fig. 3A). Since the transcript was uniformly labeled, cleavage at any position produced two cleavage products. This complicated the interpretation of the data since each pair of cleavage fragments suggested two possible sites of cleavage. Screening reactions were subsequently performed on a longer RNA transcript (1.37 kb) that differed from the first transcript in its 3′-end. This facilitated the interpretation of cleavage data, as 5′ cleavage fragments were identical for the two RNAs. Screening of two differently sized transcripts increased confidence in the results. Any accessible sites that are not identified in the longer transcript are likely to be due to structural features of the specific transcript that may not be represented in the full-length mRNA. Such sites are more likely to be encountered at the ends of the transcripts since the local folding of these regions will be most strongly affected by the lack or presence of additional sequences. Consequently, any cleavage sites originating within the first or last 200 nt of the transcript were ignored in the selection of accessible sites by library screening, as were cleavage sites that could not be detected in the longer transcript.
Figure 3.
In vitro accessibility assays. (A) Separate screening of a 1.03 kb PSKH1 in vitro transcript with triplet-specific ODN libraries. Reactions were performed with 50 nM RNA and 40 µM ODN library as detailed in Materials and Methods. Arrows indicate pairs of fragments resulting from cleavage at subsequently selected ribozyme target sites. (B) Cleavage of PSKH1 RNA (50 nM) with molar excess (200 nM) of specific ODNs against ribozyme target sites selected by oligo library screening (O-465, O-519, O-539, O-548) and structural considerations.
The screening assays demonstrated several reproducible cleavage sites that obeyed the above restrictions. Most cleavage sites were identified with the GUA and GUC libraries, while no useful sites were identified with the AUA library (Fig. 3A). Screening of PSKH1 RNA with the GUA library produced two very prominent cleavage fragments that were not produced by any of the other libraries, demonstrating their GUA triplet specificity. The sizes of these two fragments were estimated at ~615 and 405 nt. Their combined estimated size of 1020 nt was in good agreement with the transcript length (which was 1030 nt). The shorter fragment was shown to be the 5′ cleavage fragment, as it was also present when screening the longer RNA transcript (data not shown). The size of this fragment was consistent with cleavage near a site corresponding to position 460 in the PSKH1 cDNA. Examination of the sequence indicated a suitable GUA triplet at position 463–465 as the only probable site of cleavage. Screening with the GUC library produced six closely spaced fragments with estimated sizes ranging from 470 to 560 nt which fitted nicely into three pairs with combined sizes of the expected length (1020–1030 nt). By a similar analysis as for the GUA library cleavage fragments, the putative cleavage sites were identified as guc519, guc539 and guc548, respectively. One of these sites, guc519, had previously also been selected on the basis of MFold secondary structure predictions (see above) prior to the antisense library screening. The AUC library screening also produced several fragment pairs, but these were either weaker than their GUA and GUC counterparts, or could not be unambiguously assigned to a specific triplet. The screening assays were performed with a ratio of ODN (40 µM) to target RNA (50 nM) of 800:1, which given the complexity (410 = 1 × 106) of the libraries corresponds to a ratio of specific ODN (40 pmol) to RNA of roughly 1:1250. This might seem too low a ratio to reasonably account for the degree of cleavage observed. However, the observed cleavage would correspond to a turnover of only ~100 molecules/h. Furthermore, the concentration of ODNs that can productively hybridize to a given target site may be significantly higher as mismatches corresponding to the ends of the ODNs are not expected to severely impair hybridization efficiency. Non-triplet-specific cleavage events (involving ODNs with mismatched or wobble-paired triplets, as well as partial hybridization of only the random-armed portions) may also conceivably occur, although they are less likely to account for the major cleavage events. Non-specific cleavage events would also be expected to result from screening with more than just one library. Examples of such cleavage events were observed, but did not include the selected cleavage sites. Nonetheless, to ensure that putative ribozyme target sites were properly identified, the provisional identifications were subsequently confirmed by cleavage of the RNA with the corresponding specific antisense ODNs. In all cases, the cleavage fragments produced by the specific ODNs co-migrated with the corresponding cleavage fragments produced by the library screenings (data not shown). These results demonstrate the utility of triplet-specific semi-randomized ODN libraries in identifying potential target sites.
In vitro cleavage assays were performed with specific ODNs against all sites targeted by ribozymes to investigate whether the sites that were selected by library screening were more accessible than target sites selected by alternative means. The specific ODNs, like those of the semi-randomized libraries, were designed to have the same recognition sequences as a corresponding hypothetical 6+6 armed hammerhead ribozyme. The in vitro cleavage assays demonstrated that all the ODNs targeting sites that were selected by library screening resulted in stronger cleavage than the best among the ODNs targeting sites selected by theoretical/structural considerations (Fig. 3B). Furthermore, the strongest cleavage was achieved with the ODN (O-465) corresponding to the GUA site that was cleaved most strongly in the library screenings (Fig. 3A). Thus accessibility data obtained from library screening assays were representative for the hybridization efficiencies of the corresponding unique ODNs. Consequently, major cleavage events do not appear to be substantially influenced by the presence of non-specific ODNs in the library screening. This suggests that neither positive nor negative cooperativity of binding (49) occurs to a significant degree under our assay conditions.
Cellular uptake of ribozyme
FITC-labeled control ribozymes were used to investigate the cellular uptake of ribozyme under the optimized co-transfection conditions by spectroscopy. In order to distinguish between intracellular uptake and membrane association by spectroscopic analysis, uptake experiments were performed in parallel by incubating cells on ice as well as at 37°C. Similar amounts of MRz-FITC and ARz-FITC ribozymes were found to be associated with cells following incubation both on ice (26–28% of total added ribozyme) and at 37°C (58–59% of added ribozyme) (data not shown). The increase in cell-associated fluorescence at the higher temperature, suggests that in excess of 30% of ribozyme has been internalized. The cellular association of ribozyme following incubation on ice was shown to have a linear dependency on ribozyme concentration in the range 0–100 nM (data not shown). This is consistent with a state of equilibrium between membrane-bound and free (in medium) ribozyme. Assuming that this equilibrium is maintained upon internalization of ribozyme at the permissive temperature, the amount of internalized ribozyme is determined by the formula I = T(β–α)/(T–α), where T is the total amount of ribozyme added to cells, and α and β are the measured cell-associations upon incubation on ice and at 37°C, respectively. Net uptake of ribozyme under the given transfection conditions was estimated at 42–45% (data not shown). Co-transfection optimization experiments performed at various w/w ratios (1:1 to 6:1) of lipofectamine to total nucleic acids demonstrated that uptake was only slightly influenced by lipofectamine concentration above a certain threshold (2:1 w/w ratio) that constitutes a molar excess of positive charges (from cationic liposomes) in the complexation mixture. Below this threshold (at a 1:1 w/w ratio, yielding complexes of net negative charge), uptake was severely impaired (data not shown).
Efficacy of ribozymes in co-transfection experiments in HeLa cells
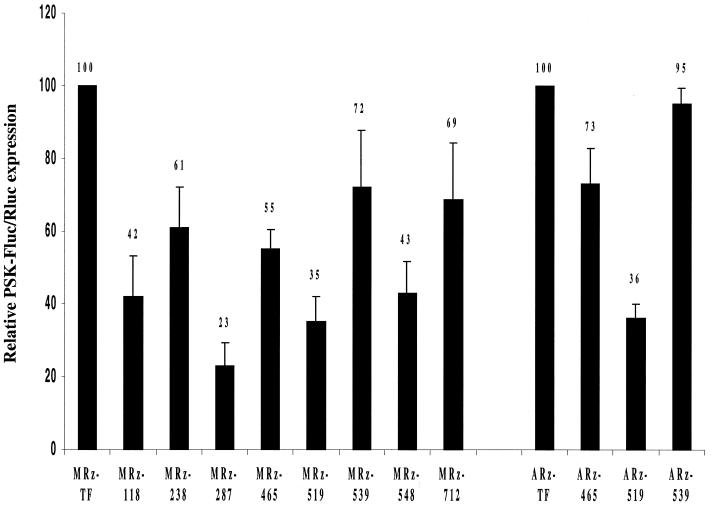
The in vivo efficiency of the selected ribozymes was evaluated in a cell culture assay in which 100 nM of ribozyme was co-transfected with a plasmid coding for a PSKH1–luciferase fusion protein. In order to correlate for transfection variability and improve experimental reproducibility, a plasmid coding for Renilla luciferase (Rluc) under the control of an EF1α-promoter was added to the transfection mixture as an internal control. This control proved to be essential as the general expression levels of Renilla luciferase varied up to 2–3-fold within the same experiment for different ribozymes (data not shown). Experiments were performed as far as possible with the full complement of ribozymes of identical chemistry so that in each experiment, the efficacies of ribozymes were compared under identical conditions. In each experiment, the relative firefly luciferase activity for all ribozymes was normalized to the levels for the control ribozyme (the expression of which was set at 100%). Normalization was always performed relative to the relevant control ribozyme. The data from experiments with both methylated and allylated ribozymes are summarized in Figure 4. The most effective methylated ribozyme in co-transfection assays proved to be MRz-287, targeting guc287. This ribozyme reduced the level (normalized) of luciferase expressed from the reporter gene to ~23% of the control levels, i.e. the levels in cells treated with irrelevant control ribozyme. The second most efficient ribozyme was MRz-519, which resulted in ~65% inhibition of expression. The ribozymes targeting sites guc118 and guc548, exhibited similar inhibitory effects, with residual expression levels just over 40%. At the other end of the spectrum we find the ribozymes targeting guc712 and guc539, which result in only ~30% inhibition.
Figure 4.
Lipofectamine-mediated co-transfection of HeLa cells with ribozyme, pTRE-PSK-Luc, and pRLuc as detailed in Materials and Methods. Eight methylated (MRz-) and three allylated (ARz-) PSKH1-specific ribozymes were analyzed together with their respective controls (MRz-TF and ARz-TF). PSKH1-dependent firefly luciferase expression was normalized to Renilla luciferase expression for each sample. Normalized expression in cells transfected with control ribozymes within each series was set at 100%. Data are averages of 4–7 independent experiments. Expression levels are indicated above the error bars (+SD) of each column.
Three of the ribozymes were synthesized also in the allylated version to evaluate any systematic differences in inhibitory capacity of methylated and allylated ribozymes. The effect of the allylated ribozymes varied over a relatively wide range, resulting in residual reporter gene expression ranging from ~35% for ARz-519 to 95% for ARz-539 (Fig. 4). Comparing the data for these ribozymes with those of their methylated counterparts, we observed the same activity ranking. For both types of modification a ribozyme targeting the guc519 cleavage site was superior to the ribozymes targeting the two other sites, gua465 and guc539, of which the latter site appeared to be least amenable to cleavage. This constitutes evidence that the observed inhibitory effects of these ribozymes are sequence specific. The difference in activity of methylated and allylated versions of otherwise identical ribozymes varies with the target of the ribozymes. While the two types of ribozymes targeting guc519 are equally active, the allylated ribozymes targeting gua465 and guc539 are less active than their methylated counterparts. The combined data suggest that methylation generally results in superior ribozymes and, further, that the allyl modification seems to be more detrimental to ribozyme efficacy when targeting poorly accessible sites. Reduced efficacy of allylated ribozymes is not due to differences in intracellular uptake, as the methylated and the allylated FITC-labeled ribozyme were internalized to the same level by lipofectamine-mediated transfection (see uptake experiments).
Ribozyme effects depend on ribozyme concentration and target gene promoter strength
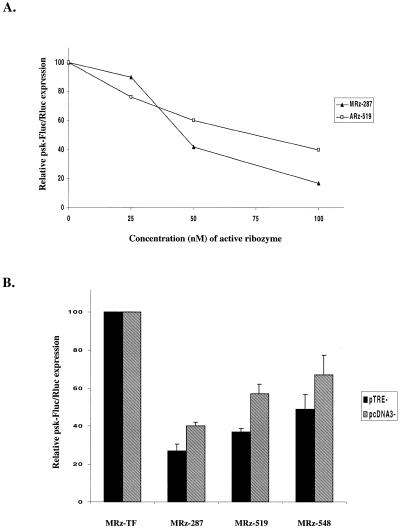
The concentration dependence of the ribozyme effect was investigated in co-transfection experiments in which the concentration of the active anti-PSKH1 ribozyme was varied and the total concentration of ribozyme adjusted to 100 nM with control ribozyme. For dose-dependence experiments, the most active allylated (ARz-519) and methylated (MRz-287) ribozymes were selected. These experiments confirmed the dose-dependence of the ribozyme effect (Fig. 5A). We next wanted to investigate whether increasing the steady-state target gene expression would reduce the specific inhibitory effect of ribozymes. Higher expression of the reporter was achieved by placing it under the transcriptional control of the CMV promoter in the pcDNA3 expression plasmid. This resulted in 20-fold enhancement of reporter expression (data not shown) compared to the weaker tetracycline-responsive promoter. In co-transfection experiments, this increase in reporter expression was accompanied by reduced apparent efficiencies of three of the most active PSKH1 ribozymes (Fig. 5B). The average levels of inhibition of target gene expression with the weak promoter were 73, 63 and 51% for ribozymes MRz-287, MRz-519 and MRz-548, respectively. With the strong promoter, yielding the 20-fold higher expression of reporter (measured in control ribozyme-treated cells), their inhibitory activities were 60, 43 and 33%, respectively. Thus the inhibitory effect of ribozymes, as expected, was dependent on the concentrations of both ribozyme and target, but the ribozymes were able to inhibit almost completely the 20-fold increase in the target mRNA.
Figure 5.
Dependence of ribozyme inhibitory activity on ribozyme concentration and reporter gene expression. (A) Transfection of HeLa cells with increasing concentrations of ARz-519 or MRz-287 ribozymes. Total concentration of ribozyme is adjusted to 100 nM with control ribozyme (ARz-TF or MRz-TF) and transfections performed after the standard protocol. Data for representative experiments are shown. (B) Parallel co-transfections of HeLa cells with selected ribozymes and different reporter constructs (pTRE-PSK-Luc and pcDNA3-PSK-Luc). Relative expression levels in control cells were 20-fold higher with the pcDNA3-PSK-Luc construct compared to pTRE-PSK-Luc.
Comparing ribozymes selected by MFold secondary structure prediction and in vitro accessibility assays
Ribozymes were ranked based on their inhibitory activity in co-transfection assays, with tied ranks given to ribozymes resulting in residual expression levels differing by <3% (Table 2). This classification can be used to assess the relative merits of the two methods of target site selection. The four ribozymes that were selected on the basis of library screening data (MRz-465, MRz-519, MRz-539, MRz-548) were compared to those selected on the basis of structural considerations (MRz-118, MRz-238, MRz-287, MRz-519, MRz-712) by Wilcoxon’s rank sum test on unpaired samples (50). The rank sums for the two groups of ribozymes were 18 (average rank = 4.5) and 20 (average rank = 5.0), respectively, which are higher than the critical rank sum of 11 required for a 5% significance level. From these data, ribozyme targets selected on the basis of in vitro accessibility are no more susceptible to ribozyme cleavage in vivo than those selected by structural prediction or other considerations.
Table 2. Rank correlations of ribozyme efficacy with various features of the predicted target site secondary structure.
| Ribozyme | Efficacy | ΔG (helix) | Stem | Stem+helix | Loop | ΔG (target) | ΔG (duplex) |
|---|---|---|---|---|---|---|---|
| MRz-287 | 1.0 | 1.0 | 1.0 | 1.0 | 4.0 | 1.0 | 1.0 |
| MRz-519 | 2.0 | 2.0 | 3.5 | 2.8 | 6.0 | 5.0 | 5.0 |
| MRz-118 | 3.5 (3.0) | 6.0 | 3.5 | 4.8 | 3.0 | 4.0 | 7.0 |
| MRz-548 | 3.5 (4.0) | 4.0 | 6.0 | 5.0 | 6.0 | 6.0 | 6.0 |
| MRz-465 | 5.0 | 3.0 | 2.0 | 2.5 | 2.0 | 3.0 | 4.0 |
| MRz-238 | 6.0 | 8.0 | 5.0 | 6.5 | 1.0 | 8.0 | 3.0 |
| MRz-712 | 7.5 (7.0) | 7.0 | 7.0 | 7.0 | 6.0 | 2.0 | 8.0 |
| MRz-539 | 7.5 (8.0) | 5.0 | 8.0 | 6.5 | 8.0 | 7.0 | 2.0 |
| Pearson’s correlation | 0.75 (0.69) | 0.77 (0.80) | 0.84 (0.83) | 0.16 | 0.35 | 0.18 | |
| coefficient (r) | |||||||
| Significance | 0.025 (<0.05) | <0.025 | 0.01 | >0.05 | >0.05 | >0.05 | |
| level (P) |
Ribozymes were ranked according to inhibitory activity in co-transfection assays (lowest rank for highest activity). Target sites were ranked according to free energy of limiting helix and target sequence (lowest rank for highest energy), stem length (lowest rank for shortest stems), loop length (lowest rank for largest loops), and the free energy of the ribozyme–RNA duplex (lowest rank for lowest enegy) (Table 1). Tied ranks were allotted to targets with the same length of stem and loop and to ribozymes resulting in residual expression levels differing by at most 3%. When considering stem and helix together, the average of the two individual ranks was used. Correlation coefficients have also been calculated for the case in which ribozymes were ranked without ties (numbers in parentheses). This did not significantly alter the results.
Correlation of ribozyme efficacy with the predicted target secondary structure
The two methods of target selection both resulted in ribozymes with very diverse inhibitory activities. We therefore decided to investigate whether there was any correlation between predicted structure of target sites and the in vivo efficacy of corresponding ribozymes. We approached this by attempting to break down the secondary structures of the target sequences in readily quantifiable parameters. In doing so we have disregarded certain factors that may influence hybridization efficiency but which cannot be easily quantified. Such factors include the positioning of single-stranded stretches within the target sequence and the ability of a specific hammerhead sequence to assume its proper catalytic three-dimensional structure. The presence of so-called free ends, the positioning of single-stranded stretches at the ends of target sequences, may be expected to be correlated with enhanced antisense binding (12), while elements of strong secondary structure in the neighborhood of the target sequence proper might impede ribozyme folding. We do not imply that these factors are unimportant, merely that as they cannot be easily quantified, other more readily quantifiable parameters should be investigated for predictive value. Three potentially important parameters for target site accessibility were local free energy of folding of the target sequence (11), the size of single-stranded stretches (loops) that may function as ‘hooks’ for nucleation of duplex formation (51,52), and the length and stability of stems and helices. Stems and helices may need to be opened up for full hybridization of ribozyme to occur and their length and stability may therefore influence hybridization efficiency. Consequently, the target sequences of all ribozymes were decomposed into length of the major loop, stem and helix, while free energies have been calculated both for the target sequence as a whole and for its major helical region (Table 1). Target sequence free energies were calculated for the energetically optimal folding of the RNA, by adding up the energy contributions from all base-pairings, stacking interactions, bulges and various loops that are contained within the target sequences, as indicated by the MFold program. MFold was also used to determine the free energy of the most stable (‘limiting’) helical region and to estimate the strength of the ribozyme-target hybrid (by folding the corresponding cis-acting ribozyme in which catalytic and substrate strands were connected through a 5 nt loop at stem I).
Spearman’s rank correlation test (53) was used to investigate the level of correlation of ribozyme efficacy ranking with various features of the predicted secondary structure of the target sequences (Table 2). No correlation was observed with the length of the major loop (Pearson’s correlation coefficient r = 0.16) or ribozyme–substrate duplex free energy (r = 0.18). The correlation with target sequence free energy was weak (r = 0.35) and not significant. Ribozyme efficacy was, however, significantly correlated (P <0.025) with both the length of the major base-paired stretch (r = 0.75) and the energy of the most stable helix (r = 0.77) within the target sequence (Table 2). Correlation was improved (r = 0.84) when considering stem length and helix stability together (for this analysis, the rank was taken as the average of the two individual ranks for stem and helix). Correlation coefficients were not significantly affected by ranking ribozyme activity without the use of ties (Table 2).
DISCUSSION
In this study we have analyzed DNA-armed chemically modified hammerhead ribozymes targeting eight GUC and GUA sites selected by two different methods, in vitro accessibility assays and MFold prediction. In a co-transfection controlled assay, the ribozymes resulted in residual luciferase reporter gene expression ranging from 23 (MRz-287) to 72% (MRz-539) (Fig. 4). This activity may in part be due to the presence of DNA arms which allow RNase H-mediated cleavage of DNA–RNA hybrids. Target gene expression normalized to the expression of a co-transfected non-target gene allowed the control of any non-target-specific sequence effects of the ribozymes. Non-specific effects are occasionally encountered in association with extended stretches of phosphorothioate (P=S) linkages (19). In an attempt to minimize such effects, no more than two consecutive P=S linkages were incorporated. Finally, ribozymes of different chemistry (with methylated RNA instead of DNA in the arms) against two of the targets (guc519 and guc548) were less efficient in inhibiting target gene expression than their DNA-armed counterparts (unpublished data). In combination, the above data make it unlikely that the observed effects are due to aspects of ribozyme sequence or chemistry unrelated to hybridization-specific activity.
The inhibitory effect of ribozymes was dependent on both the ribozyme concentration (Fig. 5A) and the steady-state expression levels of the reporter gene (Fig. 5B). A reduced inhibitory effect was observed for three selected methylated ribozymes when expressing the reporter gene from a stronger promoter resulting in 20-fold higher expression levels (Fig. 5B). However, the molar amount of target suppression when the stronger promoter was used increased to a degree almost matching the increase in the level of the exogenous target. This interesting fact suggests an enzymatic rather than a stoichiometric effect of the ribozymes on target gene expression. The enzymatic function may derive entirely from the ribozyme or include RNase H activity. It should, however, be noted that the increase in the total level of PSKH1 mRNA by expressing the exogenous transcript from the stronger promoter depends also on the unknown level of the endogenous transcript, which is another target for the ribozyme. The level of suppression achieved with the best ribozyme in this study (77%) is similar to that which has previously been reported for the best of several unmodified ribozymes in a similar luciferase reporter gene co-transfection assay (16). Other comparable studies have reported inhibition levels in the range of 40–80% for a set of 15 ribozymes (54), 50% inhibition with a pair of variously modified ribozymes (29), and 40–55% inhibition for a pair of 2′-F-pyrimidine modified ribozymes with phosphorothioates at both ends (27). Thus our results, achieved with generally more extensively modified ribozymes, compare favorably with previous reports.
We have also attempted to evaluate the relative effects of two commonly employed types of 2′-O-modifications on the activity of ribozymes. The same rank order of ribozymes targeting three selected sites was observed for both methylated and allylated DNA-armed ribozymes (Fig. 4). Allylated ribozymes were generally less efficient in inhibiting reporter gene expression in co-transfection assays compared to their methylated counterparts. The difference in activity of methylated and allylated versions of otherwise identical ribozymes appeared to correlate with the susceptibility of the target to inhibition by ribozyme. While the ribozyme targeting the least accessible of the dually target sites (guc539) apparently was most sensitive to the type of alkylation, ribozyme species of either modification were equally efficient when targeting the most susceptible site (guc519).
Predicted secondary structures of targets selected on the basis of the MFold program were very diverse and included a short stem–loop (guc287), a stem with a large internal loop (guc238), a bulged stem (guc519), and a hairpin structure (guc712), as well as a site near the translation initiation site that was presumed to be relatively unstructured (Fig. 2). In vitro accessibility assays with cleavage triplet-specific ODN-libraries identified three accessible GUC sites and one GUA site (Fig. 3A). In vitro cleavage assays with specific ODNs against all sites targeted by ribozymes confirmed that the sites that were selected by library screening were indeed more accessible in vitro than sites selected by alternative means (Fig. 3B). This confirms the utility of such semi-randomized libraries for identifying the most accessible sites in vitro. However, ribozymes targeting sites selected on the basis of in vitro accessibility assays were no more efficient in inhibiting target gene expression in a co-transfection assay than ribozymes targeting sites selected by theoretical means. In fact, the ribozyme targeting the most accessible site in vitro (MRz-465) ranked only as the fifth most active (Table 2), while the target site of the best ribozyme (MRz-287), was relatively inaccessible in vitro (Fig. 3B). The lack of correlation between in vitro accessibility and in vivo efficacy data is consistent with previous observations of other researchers (55–58). A possible explanation for the poor correlation is that the target mRNA is folded differently in vitro than in vivo. The secondary structure of the folded RNA may not be the energetically most stable (fast local folding events may prevent more energetically favorable interactions between distal regions). Structural features that promote hybridization in vivo and in vitro may also differ. Furthermore, the hybridization efficiency of 13mer antisense ODNs may not be entirely representative for the hybridization characteristics of longer (32mer) hammerhead ribozymes with varying degrees of secondary structure of their own. Finally, the generation of higher-order mRNA structures and modulation or masking of the mRNA secondary structure by RNA-binding proteins (9,10) or protein complexes (ribosomes) (59) in vivo may influence accessibility of target sites, although these factors would also tend to rule out the applicability of secondary structure predictions for target site selection. Recent data suggest that performing in vitro accessibility assays on an endogenous transcript in a protein environment (cell extracts) may improve the accuracy of the predictions (14,22).
Although in vitro accessibility assays proved to be of limited predictive value for the in vivo situation, correlative studies suggested that secondary structure predictions might have some merit. Significant correlation was found between ribozyme efficacy and the presence of short stems and energetically unstable helices within the ribozyme target sequence (Table 2). Ranking of ribozymes according to these two criteria, the relative efficacies of ribozymes were predicted nearly perfectly, the only significant discrepancy being the ribozyme targeting gua465. As well as being most accessible in vitro, this site also has a secondary structure that according to the above criteria should make it a significantly better target site than observed here. Other factors, such as the lack of single-stranded bases near the ends of the target sequence or a prohibitive environment for ribozyme folding, may explain the results. Notwithstanding this discrepancy, there is an apparently clear correlation between ribozyme efficacy and predicted target sequence secondary structure. Furthermore, the combined data from this study suggest that the previously reported correlation of ribozyme efficacy with local folding potential (11) may be incidental. Our data suggest that high target sequence free energy alone may not be sufficient for efficient ribozyme targeting. We propose that the above correlation is a consequence of the need to have some unpaired regions to facilitate fast nucleation of duplex formation (51,52), combined with short base-paired regions and helices of low stability that easily open up. Fulfillment of these criteria will in many cases result in a low local folding potential (high free energy) for the target site.
A recent study on the effect of varying RNA secondary structure on the efficiency of specific antisense ODNs concluded that target sequences located within regions designed to be unstructured were most effective, while targets within stable stem–loop structures were ineffective (13). Recent reports by Patzel and co-workers (12,14) described a theoretical approach for antisense ODN target site selection based on the prediction of large single-stranded stretches (loops) by MFold. Our data do not support a correlation of ribozyme activity with the length of loops, possibly because all our targets had shorter predicted loops than recommended in the above studies. Our hypothesis and the conclusions of the above studies are, however, not mutually exclusive. Target sites containing very large loops will have a good probability of also containing short helical regions, which we propose to be the limiting factor. In fact, applying our hypothesis of target site evaluation to three previously well characterized target sites (t351, t398, t498) (14,22) within mRNA for murine DNA methyl transferase, the same rank susceptibility of targets, in perfect accordance with actual inhibition data, is predicted by both theories.
In addition to the structure of the target site, the composition of the target sequence may also be of some importance. Sequences with a high G+C content will hybridize more efficiently with the complementary arms of their ribozymes and possibly increase the efficacy of the ribozyme. Although a general correlation between hybrid stability and ribozyme efficacy was not supported by our data (Table 2), it is worth noting that the most efficient ribozyme, MRz-287, has a substantially higher affinity for its target sequence than the other ribozymes (Table 1) due to an unusually high G+C content (13 out of 16 nt). In conclusion, the target sequence of MRz-287 represents the proposed desirable structural features for a good ribozyme/antisense target site. The target sequence consists of alternating short stretches of paired and unpaired bases, which limit stems and helical regions to no more than 3 bp. All other target sequences fold into secondary structures containing longer and more stable helical regions. One study has reported that hybridization accessibility for hammerhead ribozymes is correlated with the presence of unpaired bases near the cleavage triplet (60). Although our data do not suggest this to be a critical requirement for in vivo activity, the above criterium is also fulfiled for the guc287 target site, as the longest single-stranded stretch is situated around the cleavage triplet and includes the base preceding the scissile bond (Fig. 2). All of the above mentioned features of guc287 add up to a very effective ribozyme target site, in good agreement with its observed inhibitory capacity in HeLa cells.
In conclusion, our study indicates that there is a poor correlation between the apparent in vivo accessibility of a target and its accessibility in a completely cell-free in vitro assay as performed here. Thus such assays appear to be of limited value even for a preliminary selection of target sites. However, predictions by the MFold program suggest a correlation of certain features of the predicted secondary structures of target sequences, helical stability in particular, with ribozyme efficacy. The generality of these findings will, however, need to be investigated in an alternate test system. If these correlations should be confirmed, this would represent a significant improvement in the preliminary selection of candidate ribozymes. Ultimately, however, an empirical cell-based assay will still need to be performed to select the best of these candidates.
Acknowledgments
ACKNOWLEDGEMENTS
M.A. is a research fellow of the Norwegian Cancer Society. This work was supported by grants to H.P. from the Norwegian Cancer Society and the Norwegian Research Council.
DDBJ/EMBL/GenBank accession no. AJ272212
REFERENCES
- 1.Amarzguioui M. and Prydz,H. (1998) Cell. Mol. Life Sci., 54, 1175–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ludwig J., Blaschke,M. and Sproat,B.S. (1998) Nucleic Acids Res., 26, 2279–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaish N.K., Heaton,P.A., Fedorova,O. and Eckstein,F. (1998) Proc. Natl Acad. Sci. USA, 95, 2158–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milner N., Mir,K.U. and Southern,E.M. (1997) Nat. Biotechnol., 15, 537–541. [DOI] [PubMed] [Google Scholar]
- 5.Ho S.P., Bao,Y., Lesher,T., Malhotra,R., Ma,L.Y., Fluharty,S.J. and Sakai,R.R. (1998) Nat. Biotechnol., 16, 59–63. [DOI] [PubMed] [Google Scholar]
- 6.Brede G. (2000) EMBL accession no. AJ272212.
- 7.Zuker M., Mathews,D.H. and Turner,D.H. (1999) In Barciszewski,J. and Clark,B.F.C. (eds), Algorithms and Thermodynamics for RNA Secondary Structure Prediction: A Practical Guide in RNA Biochemistry and Biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 11–43.
- 8.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 9.Tsuchihashi Z., Khosla,M. and Herschlag,D. (1993) Science, 267, 99–102. [DOI] [PubMed] [Google Scholar]
- 10.Sioud M. (1994) J. Mol. Biol., 242, 619–629. [DOI] [PubMed] [Google Scholar]
- 11.Sczakiel G., Homann,M. and Rittner,K. (1993) Antisense Res. Dev., 3, 45–52. [DOI] [PubMed] [Google Scholar]
- 12.Patzel V., Steidl,U., Kronenwett,R., Haas,R. and Sczakiel,G. (1999) Nucleic Acids Res., 27, 4328–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vickers T.A., Wyatt,J.R. and Freier,S.M. (2000) Nucleic Acids Res., 28, 1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherr M., Rossi,J.J., Sczakiel,G. and Patzel,V. (2000) Nucleic Acids Res., 28, 2455–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimayama T., Nishikawa,S. and Taira,K. (1993) Nucleic Acids Res. Symp. Ser., 29, 177–178. [PubMed] [Google Scholar]
- 16.Sakamoto N., Wu,C.H. and Wu,G.Y. (1996) J. Clin. Invest., 98, 2720–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szymanski M., Fürste,J.P., Barciszewska,M.Z., Erdmann,V.A. and Barciszewski,J. (1997) Biochem. Mol. Biol. Int., 41, 439–447. [DOI] [PubMed] [Google Scholar]
- 18.Mir K.U. and Southern,E.M. (1999) Nat. Biotechnol., 17, 788–792. [DOI] [PubMed] [Google Scholar]
- 19.Jarvis T.C., Wincott,F.E., Alby,L.J., McSwiggen,J.A., Beigelman,L., Gustofson,J., DiRenzo,A., Levy,K., Arthur,M., Matulic-Adamic,J., Karpeisky,A., Gonzalez,C., Woolf,T.M., Usman,N. and Stinchcomb,D.T. (1996) J. Biol. Chem., 271, 29107–29112. [DOI] [PubMed] [Google Scholar]
- 20.Ho S.P., Britton,D.H., Stone,B.A., Behrens,D.L., Leffet,L.M., Hobbs,F.W., Miller,J.A. and Trainor,G.L. (1996) Nucleic Acids Res., 24, 1901–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birikh K.R., Berlin,Y.A., Soreq,H. and Eckstein,F. (1997) RNA, 3, 429–437. [PMC free article] [PubMed] [Google Scholar]
- 22.Scherr M. and Rossi,J.J. (1998) Nucleic Acids Res., 26, 5079–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matveeva O., Felden,B., Audlin,S., Gesteland,R.F. and Atkins,J.F. (1997) Nucleic Acids Res., 25, 5010–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoumadakis M. and Tabler,M. (1995) Nucleic Acids Res., 23, 1192–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieber A. and Strauss,M. (1995) Mol. Cell. Biol., 15, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hertel K.J., Pardi,A., Uhlenbeck,O.C., Koizumi,M., Ohtsuka,E., Uesugi,S., Cedergren,R., Eckstein,F., Gerlach,W.L., Hodgson,R. et al. (1992) Nucleic Acids Res., 20, 3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherr M., Grez,M., Ganser,A. and Engels,J.W. (1997) J. Biol. Chem., 272, 14304–14313. [DOI] [PubMed] [Google Scholar]
- 28.Parry T.J., Cushman,C., Gallegos,A.M., Agrawal,A.B., Richardson,M., Andrews,L.E., Maloney,L., Mokler,V.R., Wincott,F.E. and Pavco,P.A. (1999) Nucleic Acids Res., 27, 2569–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bramlage B., Alefelder,S., Marshall,P. and Eckstein,F. (1999) Nucleic Acids Res., 27, 3159–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuschl T. and Eckstein,F. (1993) Proc. Natl Acad. Sci. USA, 90, 6991–6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beigelman L., Karpeisky,A., Matulic-Adamic,J., Haeberli,P., Sweedler,D. and Usman,N. (1995) Nucleic Acids Res., 23, 4434–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyngstadaas S.P., Risnes,S., Sproat,B.S., Thrane,P.S. and Prydz,H.P. (1995) EMBO J., 14, 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu J.L., Veerapanane,D., Rossi,J., Natarajan,R., Thomas,L. and Nadler,J. (1995) Circ. Res., 77, 14–20. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q., Mullah,B., Hansen,C., Asundi,J. and Robishaw,J.D. (1997) J. Biol. Chem., 272, 26040–26048. [DOI] [PubMed] [Google Scholar]
- 35.Flory C.M., Pavco,P.A., Jarvis,T.C., Lesch,M.E., Wincott,F.E., Beigelman,L., Hunt,S.W.,III and Schrier,D.J. (1996) Proc. Natl Acad. Sci. USA, 93, 754–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sproat B.S., Rupp,T., Menhardt,N., Keane,D. and Beijer,B. (1999) Nucleic Acids Res., 27, 1950–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hendry P., McCall,M.J., Santiago,F.S. and Jennings,P.A. (1992) Nucleic Acids Res., 20, 5737–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimayama T., Nishikawa,F., Nishikawa,S. and Taira,K. (1993) Nucleic Acids Res., 21, 2605–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimayama T. (1994) Gene, 149, 41–46. [DOI] [PubMed] [Google Scholar]
- 40.Heidenreich O., Xu,X., Swiderski,P., Rossi,J.J. and Nerenberg,M. (1996) Antisense Nucleic Acid Drug Dev., 6, 111–118. [DOI] [PubMed] [Google Scholar]
- 41.Sinha N.D., Biernat,J., McManus,J. and Köster,H. (1984) Nucleic Acids Res., 12, 4539–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Usman N., Ogilvie,K.K., Jiang,M.-Y. and Cedergren,R.J. (1987) J. Am. Chem. Soc., 109, 7845–7854. [Google Scholar]
- 43.Green R., Szostak,J.W., Benner,S.A., Rich,A. and Usman,N. (1991) Nucleic Acids Res., 19, 4161–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polushin N.N., Pashkova,I.N. and Efimov,V.A. (1991) Nucleic Acids Res. Symp. Ser., 24, 49. [PubMed] [Google Scholar]
- 45.Polushin N.N., Morocho,A.M., Chen,B. and Cohen,J.S. (1994) Nucleic Acids Res., 22, 639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vinayak R., Andrus,A. and Hampel,A. (1995) Biomed. Pept. Proteins Nucleic Acids, 1, 227–230. [PubMed] [Google Scholar]
- 47.Wang F.S., Kobayashi,H., Liang,K.W., Holland,J.F. and Ohnuma,T. (1999) Hum. Gene Ther., 10, 1185–1195. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi H., Takemura,Y., Wang,F.S., Oka,T. and Ohnuma,T. (1999) Int. J. Cancer, 81, 944–950. [DOI] [PubMed] [Google Scholar]
- 49.Jankowsky E. and Schwenzer,B. (1996) Biochemistry, 35, 15313–15321. [DOI] [PubMed] [Google Scholar]
- 50.Wilcoxon F. (1945) Biomet. Bull., 6, 80. [Google Scholar]
- 51.Homann M., Rittner,K. and Sczakiel,G. (1993) J. Mol. Biol., 233, 7–15. [DOI] [PubMed] [Google Scholar]
- 52.Skripkin E., Paillart,J.C., Marquet,R., Blumenfeld,M., Ehresmann,B. and Ehresmann,C. (1996) J. Biol. Chem., 271, 28812–28817. [DOI] [PubMed] [Google Scholar]
- 53.Altman D.G. (1991) Practical Statistics for Medical Research. Chapman and Hall, London, UK, pp. 293–295.
- 54.Macejak D.G., Jensen,K.L., Jamison,S.F., Domenico,K., Roberts,E.C., Chaudhary,N., von Carlowitz,I., Bellon,L., Tong,M.J., Conrad,A., Pavco,P.A. and Blatt,L.M. (2000) Hepatology, 31, 769–776. [DOI] [PubMed] [Google Scholar]
- 55.L’Huillier P.J., Davis,S.R. and Bellamy,A.R. (1992) EMBO J., 11, 4411–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beck J. and Nassal,M. (1995) Nucleic Acids Res., 23, 4954–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Domi A., Beaud,G. and Favre,A. (1996) Biochimie, 78, 654–662. [DOI] [PubMed] [Google Scholar]
- 58.Ramezani A. and Joshi,S. (1996) Antisense Nucleic Acid Drug. Dev., 6, 229–235. [DOI] [PubMed] [Google Scholar]
- 59.Chen H., Ferbeyre,G. and Cedergren,R. (1997) Nat. Biotechnol., 15, 432–435. [DOI] [PubMed] [Google Scholar]
- 60.Campbell T.B., McDonald,C.K. and Hagen,M. (1997) Nucleic Acids Res., 25, 4985–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]