Abstract
Ribonucleases with antitumor activity are mainly found in the oocytes and embryos of frogs, but the role of these ribonucleases in frog development is not clear. Moreover, most frog ribonuclease genes have not been cloned and characterized. In the present study, a group of ribonucleases were isolated from Rana catesbeiana (bullfrog). These ribonucleases in mature oocytes, namely RC-RNase, RC-RNase 2, RC-RNase 3, RC-RNase 4, RC-RNase 5 and RC-RNase 6, as well as liver-specific ribonuclease RC-RNase L1, were purified by column chromatographs and detected by zymogram assay and western blotting. Characterization of these purified ribonucleases revealed that they were highly conserved in amino acid sequence and had a pyroglutamate residue at their N-termini, but possessed different specific activities, base specificities and optimal pH values for their activities. These ribonucleases were cytotoxic to cervical carcinoma HeLa cells, but their cytotoxicities were not closely correlated to their enzymatic specific activities. Some other amino acid residues in addition to their catalytic residues were implicated to be involved in the cytotoxicity of the frog ribonucleases to tumor cells. Because the coding regions lack introns, the ribonuclease genes were cloned by PCR using genomic DNA as template. Their DNA sequences and amino acid sequences are homologous to those of mammalian ribonuclease superfamily, ∼50 and ∼25%, respectively.
INTRODUCTION
Ribonucleases are widely found in living organisms and have been suggested to be involved in RNA metabolism and regulation of gene expression (1). Several abundant ribonucleases have been isolated from tissues of various animals and have been well characterized. For example, ribonucleases have been purified from various bovine tissues, i.e. pancreas, liver, kidney, brain and seminal fluids. RNase A from bovine pancreas has been extensively characterized and is widely used in molecular biology (2). In humans, several ribonucleases have also been isolated, such as RNase 1 (human pancreatic RNase), RNase 2 (eosinophil-derived neurotoxin), RNase 3 (eosinophil cationic protein), RNase 4, RNase 5 (angiogenin) and RNase 6 (k6) (3). The occurrence of several homologous ribonucleases in different tissues of the same animal suggests that the existence of a family of homologous genes expresses in a tissue-specific fashion. Although these ribonucleases are abundant in some tissues and well characterized biochemically, their biological importance is still not clear (4,5). Recently, several proteins with known biological activities were found to have intrinsic ribonucleolytic activity, which is essential for the biological activities of the majority of these proteins. For example, angiogenin possesses both angiogenesis and ribonucleolytic activities (6). Eosinophil-derived neurotoxin and eosinophil cationic protein exert both neurotoxicity and ribonucleolytic activities (7,8). The S-RNases from the pistils of some flowering plants are involved in self-incompatibility through the degradation of ribosomal RNA of pollen tubes (9). Bovine seminal ribonuclease, a homodimer with two identical 124 amino acid residue subunits, exerts both antitumor and ribonucleolytic activities (10). However, most mammalian ribonucleases are not cytotoxic to tumor cells because they are recognized and inhibited by specific mammalian ribonuclease inhibitors (11).
Several oocyte ribonucleases from frog Rana spp. possess cytotoxicity toward tumor cells. Onconase, isolated from the oocytes of Rana pipiens, combined with tamoxifen or doxorubicin, is currently being evaluated in human phase III clinical trials for tumor therapy, and it is approaching maturity as a member of the arsenal of anticancer drugs (12). The sialic acid-binding lectin from the eggs of Rana catesbeiana (bullfrog) and Rana japonica exerts agglutination activity toward tumor cells (13). RC-RNase isolated from the oocytes of R.catesbeiana is identical to the sialic-acid-binding lectin and is also cytotoxic to several tumor cell lines (14–16).
There are several notable differences between frog and mammalian ribonucleases in addition to frog ribonucleases’ antitumor activity (17). First, the RNA substrate specificity of frog ribonucleases is pyrimidine–guanine and that of mammals is pyrimidine–adenine. Second, the ribonucleases from frogs are resistant to the ribonuclease inhibitor from human placenta, whereas those of mammals are susceptible to the inhibitor. Third, the location of the fourth disulfide bridge of frog ribonucleases is different from those of mammalian ribonucleases. Fourth, the abundance of ribonucleases in frog is present in oocytes and embryos but not in other organs (17). Although frog ribonucleases have novel properties compared with mammalian ribonucleases, the biologically functional differences between them are not well studied. To elucidate the functional roles of ribonucleases in frog development and to explore their potential applications in tumor therapy, in the present study we isolated a group of ribonucleases and cloned the gene family from bullfrog. We found that these purified ribonucleases exert cytotoxicity toward HeLa cells and that their cytotoxicity does not closely correlate to their specific activity. Due to the lack of introns in the coding regions, we cloned the ribonuclease genes from genomic DNA for the analysis of amino acid residues involved in the specific biological functions.
MATERIALS AND METHODS
Purification of ribonucleases from oocytes
Native bullfrogs (R.catesbeiana) were obtained from a local frog farm. The purification of oocyte ribonucleases was carried out at 4°C. Mature ovaries (60–100 g per frog) from mature female bullfrog were subjected to ultracentrifugation for 45 min at 100 000 g using a Beckman SW41 rotor. The yolk granules containing the majority of ribonucleases in the pellet were extracted by NaCl as described previously (14). The clear supernatant was applied onto a phosphocellulose column and eluted with a 0.09–0.7 M KCl gradient in buffer A (19 mM HEPES, pH 7.9, 0.1 mM EDTA). The fractions containing ribonucleases were identified by zymogram and western blotting, then collected and loaded onto subsequent column chromatographs and eluted with salt gradients in buffer A as demonstrated in Figure 1B. Total protein obtained from each purification step was determined by the Bradford method (18). Protein components were analyzed by 13.3% SDS–PAGE and Coomassie Brilliant blue R staining (19). For western blotting analysis, the membrane with transferred proteins was blocked by 5% skim milk in phosphate-buffered saline and incubated with rabbit anti-RNase antiserum (1:1000 dilution, prepared in this laboratory) and goat anti-rabbit antibodies conjugated with alkaline phosphatase. Ribonucleases were visualized by the addition of a bromochloroindolyl phosphate/nitro blue tetrazolium substrate (16). The amounts of purified ribonucleases on Coomassie blue-stained gels were estimated using bovine pancreatic RNase A as a standard.
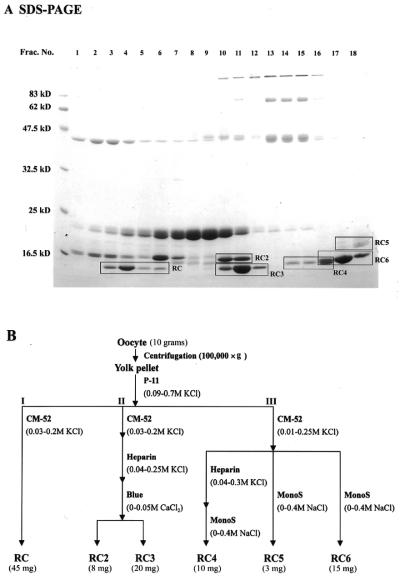
Figure 1.
Identification and purification scheme of multiple ribonucleases in bullfrog oocytes. (A) SDS–PAGE analysis of phosphocellulose column eluates. The ribonucleases in the yolk granules of oocytes were extracted by 0.09 M NaCl and subjected to fractionation by column chromatography on phosphocellulose and equal aliquots were taken for SDS–PAGE and Coomassie blue staining. (B) Purification scheme of ribonucleases from bullfrog oocytes. RC, RC 2, RC 3, RC 4, RC 5 and RC 6 represent RC-RNase, RC-RNase 2, RC-RNase 3, RC-RNase 4, RC-RNase 5 and RC-RNase 6, respectively. P-11, phosphocellulose (Whatman); CM52, carboxymethyl cellulose (Whatman); Heparin, heparin Sepharose 4B (Pharmacia); Blue, blue dextran Sepharose 4B (Pharmacia); Mono S, FPLC mono S (Pharmacia).
Amino acid sequence analysis and mass spectrometry
Purified RC-RNase, RC-RNase 2 and RC-RNase 3 were reduced with dithiothreitol and S-carboxymethylated with iodoacetate (20). These modified proteins were then cleaved with cyanogen bromide in 70% formic acid at room temperature and purified by reverse-phase HPLC on a column of Vydac C18 before sequence analysis (21). The purified RC-RNase 4, RC-RNase 5 and RC-RNase 6 were directly cleaved with cyanogen bromide in 70% formic acid, separated by SDS–PAGE and transferred to ProBlot™ Membranes (Applied Biosystems) for sequence analyses. For the determination of N-terminal amino acid sequences, the purified proteins were incubated with pfu pyroglutamate aminopeptidase (TaKaRa Biomedicals, Japan) at 70°C for 4 h to remove N-terminal pyroglutamate before sequence analysis. Automated Edman degradation was performed on a Gas/Liquid Phase Model 470A/900A sequencer (Applied Biosystems) equipped with an on-line Model 120A phenythiohydantoin-amino acid analyzer (21). The mass spectrometry analyses of ribonucleases were performed on a VG Quatoo-Bio-Q (Fisons Instruments, UG Biotech, Altrincham, UK), a triple quadrupole instrument with a mass range for singly charged ions of 4000 (22).
Ribonuclease activity assay
The ribonuclease activity of column fractions was determined by the ability for dinucleotide CpG cleavage, followed by thin layer chromatography (PEI-cellulose F Merck, Darmstadt, Germany) and UV illumination at 254 nm (23). The ribonuclease activity of each purified ribonuclease was determined by the release of acid-soluble nucleotides from bakers’ yeast total RNA after ribonuclease digestion. One unit of enzyme activity is defined as the amount of enzyme producing 1 U A260 acid-soluble material at 37°C for 15 min (23). The ribonuclease activity of the column fraction was also analyzed by zymogram assay on RNA-casting PAGE (24). Briefly, after electrophoresis the gel was washed twice with 25% isopropyl alcohol in 10 mM Tris–HCl, pH 7.5, to remove SDS for protein renaturation. The activity was visualized by incubating the gel at room temperature for 30 min in 10 mM Tris–HCl, pH 7.5, followed by 0.2% toluidine blue O staining.
Nucleotide base specificity of ribonuclease
The specific cleavage sites of ribonucleases were determined by incubation of ribonucleases with 5′-32P-labeled synthetic 18mer RNA with known sequence, 5′-AAGGUUAUCCGCACUGAA-3′, followed by denaturing gel electrophoresis and autoradiography (25). The kcat/Km of ribonucleases toward dinucleotides CpG, UpG and UpU was performed as follows. Briefly, dinucleotides were digested with ribonucleases at 37°C for 10 min in buffer containing 100 mM Mes, pH 6.0, 50 mM NaCl, 0.1 mg/ml bovine serum albumin and separated by reverse-phase HPLC using 1–5% acetonitrile (depending on products) in 0.1% TFA on a column of Vydac C18 with a Waters automated gradient controller.
cDNA cloning of upstream region of ribonuclease genes
Total RNA from bullfrog liver was isolated by the acid guanidinium thiocyanate/phenol/chloroform method (26). Briefly, 0.5 g of liver was frozen in liquid nitrogen and then ground by mortar. The powder was dissolved in guanidinium thiocyanate. After phenol/chloroform extraction, the RNA was precipitated with ice-cold isopropyl alcohol. Purification of mRNA (100 µg in total) was performed using a Quickprep Micro mRNA Purification Kit from Pharmacia Biotech (Uppsala, Sweden). The synthesis of double-stranded cDNA was performed using a Marathon™ cDNA Amplification Kit from Clontech (Palo Alto, CA) according to the manufacturer’s instructions. Adaptor (Ap1,2) containing oligonucleotides Ap1 and Ap2 was ligated to the 5′ end of these cDNAs.
Genomic DNA cloning of the downstream region of ribonuclease genes by Genome Walker
The g-DNA from blood cells was extracted by QIAamp Blood Kit (Qiagen, Hilden, Germany) and digested with EcoRI and filled in by Klenow DNA polymerase. The genomic DNA fragments ranging from 500 to 1000 bp were eluted and ligated with Genome Walker Adaptor Ap3,4 (Clontech Universal Genome Walker™ Kit).
Cloning of full-length bullfrog ribonuclease gene family
To clone the bullfrog ribonuclease gene family, degenerated primers (Perkin Elmer, Branchburg, NJ) were designed based on the highly conserved regions of the N- and C-terminal amino acid sequences of the frog ribonuclease including RC-RNase, onconase and lectin from R.japonica, liver ribonuclease from bullfrog, and from the DNA sequence of the RC-RNase gene (16). A 320 bp DNA fragment was amplified from genomic DNA by PCR and ligated to pGEM-T vector (Promega, Madison, WI). The cycle protocol for PCR was as follows: 95°C for 5 min; 30 cycles of 95°C for 40 s, 55°C for 40 s and 72°C for 2 min; 72°C for 2 min. RC-RNase 2 and RC-RNase 3 gene fragments were obtained from these clones.
The upstream region of the RC-RNase 2 gene was obtained from the above-metioned adaptor Ap1,2-tagged c-DNA by Nest PCR using oligonucleotides Ap1 and Ap2, and oligonucleotide from RC-RNase 2 internal DNA sequence as 5′ and 3′ end primers, respectively. The putative signal peptide sequences were included in these clones.
The downstream region of the RC-RNase 3 gene was obtained from Genome Walker adaptor Ap3,4 tagged g-DNA by Nest PCR using oligonucleotides from RC-RNase 3 internal DNA sequence and oligonucleotides (Ap3, Ap4) as 5′ and 3′ end primers, respectively. The full length members of the ribonuclease gene family, named rcr2, rcr3, rcr4 and rcr6 were obtained from bullfrog genomic DNA by PCR using oligonucleotide from the putative signal peptide sequence of rcr and rcr2 genes and oligonucleotide from the downstream region of RC-RNase 3 gene as 5′ and 3′ end primers, respectively. The full-length products of the coding region of RC-RNase L1, rcrl1, were obtained using oligonucleotides from above-mentioned putative signal peptide sequence and oligonucleotide from the downstream sequence of RC-RNase as 5′ and 3′ primers, respectively.
DNA sequence analysis
DNA sequencing was performed by the dideoxy chain termination method using a d-Rhodamine Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer Applied Biosystems). The nucleotide sequence was analyzed by a Perkin-Elmer 377 automated DNA sequencer.
Assay of cytotoxicity by ATP Lite-M measurement
ATP is a marker for cell viability because it is present in all metabolically active cells, and the concentration declines very rapidly when cells undergo necrosis or apoptosis. The ATP Lite-M assay system (Packard BioScience Company, The Netherlands) is based on the production of light caused by the reaction of ATP with the added luciferase and d-luciferin. The emitted light is proportional to the ATP concentration and was counted by a Packard Top Count Microplate Scintillation and Luminescence Counter. Briefly, cells (2–5 × 103) in 96-well plates in 100 µl DMEM were lyzed with 50 µl lysis buffer for 2 min followed by the addition of 50 µl substrate solution for 1 min, then transferred to a dark adapt plate for 10 min before luminescence counting.
Accession numbers
The nucleotide sequences reported in this paper have been submitted to GenBank with accession numbers AF039104 for rcr (RC-RNase), AF242553 for rcr2 (RC-RNase 2), AF242554 for rcr3 (RC-RNase 3), AF242555 for rcr4 (RC-RNase 4), AF242556 for rcr6 (RC-RNase 6) and AF288642 for rcrl1 (RC-RNase L1).
RESULTS
Purification of multiple ribonucleases from mature bullfrog oocytes
To characterize the RC-RNase immunoreactive proteins in the oocytes, which are the most ribonuclease-abundant tissues, the ovary extracts from mature bullfrogs were subjected to column chromatography on phosphocellulose. As shown in Figure 1A, ribonucleases were separated into three groups using KCl salt gradient elution and designated as group I, group II and group III, respectively, based on the elution order of the chromatography using zymogram and western blotting assays. A ribonuclease in group I, namely RC-RNase, was purified to homogeneity by further chromatography on carboxymethyl cellulose. Two ribonucleases in group II, namely RC-RNase 2 and RC-RNase 3, were further purified subsequently by carboxymethyl cellulose column chromatography, heparin Sepharose 4B and blue dextran Sepharose 4B column chromatography. Three ribonucleases in group III, namely RC-RNase 4, RC-RNase 5 and RC-RNase 6, were purified by carboxymethyl cellulose, heparin Sepharose 4B and/or FPLC mono S column chromatographies. Although RC-RNase 5 in column eluates was not detectable on zymogram and western blotting using anti-RC-RNase antibody, it was visible on Coomassie blue stained gel and defined as the ribonuclease family member by amino acid sequencing. The purification scheme is shown in Figure 1B. Approximately 45 mg of RC-RNase, ∼8 mg of RC-RNase 2, ∼20 mg of RC-RNase 3, ∼10 mg of RC-RNase 4, ∼3 mg of RC-RNase 5 and ∼15 mg of RC-RNase 6 were obtained from 10 g of matured ovary. The yield of RC-RNase 5 varied depending on the degree of maturation of ovary. The homogeneity of these purified oocyte ribonucleases was determined by SDS–PAGE (Fig. 2A). Bovine pancreatic ribonuclease, RNase A, and a liver-specific ribonuclease, RC-RNase L1, purified from bullfrog liver using a method similar to that for oocytic ribonuclease purification, were used for comparison. As shown in Figure 2B, most bullfrog ribonucleases except RC-RNase 5 and RNase A were recognized by anti RC-RNase antibody. RC-RNase, RC-RNase 2, RC-RNase 3, RC-RNase 4 and RC-RNase 6 were not recognized by anti RNase A and anti RC-RNase 5 antibodies (data not shown). Based on zymogram assay, RNase A was the most active, then RC-RNase, RC-RNase L1, RC-RNase 4 and RC-RNase 2, in descending order (Fig. 2C). Table 1 lists the relative specific activities of these purified ribonucleases which were in good agreement with those obtained from zymogram assay with the exception of RC-RNase 2. The low activity of RC-RNase 2 on the zymogram may have been due to inefficient renaturation of the ribonuclease in gel during the assay.
Figure 2.
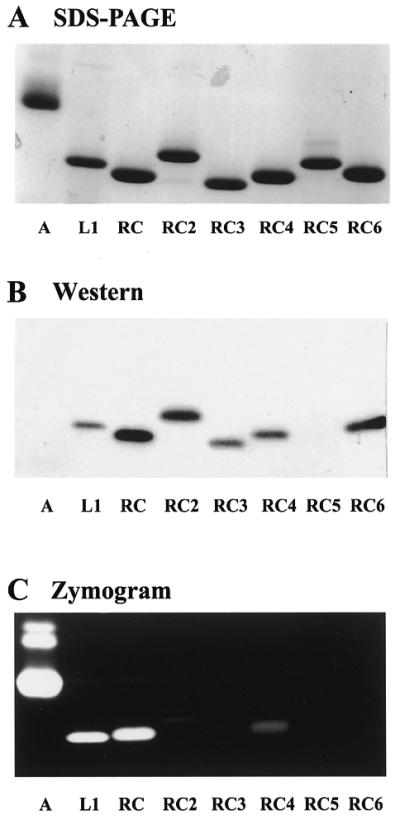
Analyses of multiple purified ribonucleases from bullfrog. (A) SDS–PAGE analysis. Purified ribonucleases (2 µg each) were separated by 13.3% SDS–PAGE and stained by Coomassie blue. (B) Western blotting. Purified ribonucleases (0.1 µg each) were separated by 13.3% SDS–PAGE, transferred to nitrocellulose membranes and probed with anti RC-RNase antibody. (C) Zymogram analysis. Ribonucleases (5 ng each) were separated by RNA-casting 13.3% non-reducing SDS–PAGE and stained by toluidine blue O. Lane A, bovine pancreatic ribonuclease A; lane L1, bullfrog liver-specific ribonuclease (RC-RNase L1); lanes RC, RC 2, RC 3, RC 4, RC 5 and RC 6 represent RC-RNase, RC-RNase 2, RC-RNase 3, RC-RNase 4, RC-RNase 5 and RC-RNase 6, respectively.
Table 1. Enzymatic properties of ribonuclease family.
| RNase A | RC-RNase L1 | RC-RNase | RC-RNase 2 | RC-RNase 3 | RC-RNase 4 | RC-RNase 5 | RC-RNase 6 | |
|---|---|---|---|---|---|---|---|---|
| Molecular mass (Da) |
– |
12 383 |
12 440 |
12 192 |
11 870 |
12 062 |
13 182 |
12157 |
| Specific activity (OD/µg)a |
1085.6 |
531.5 |
964.4 |
1.008 |
0.033 |
0.15 |
0.113 |
0.007 |
| Base preferencesb |
CA, UA |
CG |
CG, UG |
UG |
CU, UG |
CG |
UU, CG |
CA, CU |
|
kcat/Km for CpG (M–1min–1) |
2.6 × 107 |
7.2 × 107 |
7.7 × 107 |
1.2 × 104 |
4.6 × 103 |
5.7 × 106 |
2.0 × 103 |
5.1 × 102 |
|
kcat/Km for UpG (M–1min–1) |
1.7 × 107 |
1.4 × 107 |
7.8 × 107 |
5.6 × 105 |
4.0 × 104 |
3.8 × 105 |
2.7 × 103 |
8.3 ×102 |
|
kcat/Km for UpU (M–1min–1) |
1.5 × 106 |
2.8 × 105 |
8.6 × 105 |
6.8 × 103 |
6.8 × 102 |
ND |
ND |
ND |
| Optimal pHc |
8.0–8.5 |
6.5–7.5 |
7.5–8.0 |
6.5–8.0 |
4.5–5.0 |
5.5–6.0 |
6.0–7.5 |
5.0–5.5 |
| Optimal temperature (°C)d | 55 | 65 | 65 | 65 | 65 | 65 | 55 | 65 |
ND, non-detectable.
aResults obtained from acid-soluble method as described in Materials and Methods.
bResults obtained from the RNA sequence gel as shown in Figure 3.
cRelative activities were determined by acid-soluble method in 50 mM sodium acetate (pH 3.5–6.5) and 50 mM Tris–HCl (pH 6.5–9.5).
dRelative activities were determined by acid-soluble method in the buffer with respective optimal pH value.
Partial amino acid sequences of purified bullfrog ribonucleases
To further characterize the purified ribonucleases from bullfrog, the N-terminal amino acid sequence was determined by automatic amino acid sequencing. No signal was obtained by Edman degradation except after pfu pyroglutamate aminopeptidase treatment. Therefore, the N-terminal residues of bullfrog oocyte ribonucleases were assumed to be pyroglutamate. The N-terminal amino acid sequences were determined up to 18 residues after the N-terminal pyroglutamate had been enzymatically removed. N-terminal amino acid sequences of CNBr-derived fragments of these oocyte ribonucleases, as well as liver-specific RC-RNase L1, were also determined. The amino acid sequences of these ribonuclease fragments including RC-RNase5 were homologous to that of bullfrog RC-RNase and were identical to those obtained from respective DNA clones. These results suggest that these ribonucleases are derived from individual members of a gene family, as opposed to being differential posttranslational products of the same gene. It is of importance to note that residue 12 of RC-RNase L1 was Thr rather than Arg as reported by Nitta et al. (27).
Ribonucleotide base specificity of bullfrog ribonucleases
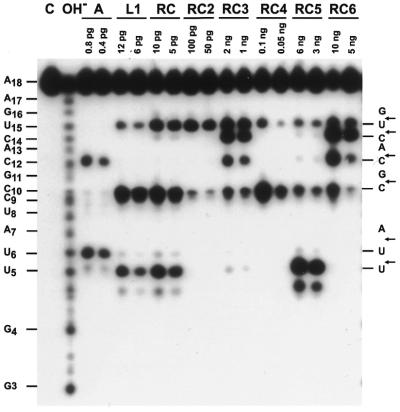
The base specificity of mammalian ribonucleases is pyrimidine–adenine, while that of some known frog ribonucleases is pyrimidine–guanine (12). As shown in Figure 3, using a 32P-end-labeled synthetic oligoribonucleotide as substrate and analyzing by denatured urea–PAGE and autoradiography, the base specificity of RNase A was CpA and UpA, whereas those of RC-RNase L1, RC-RNase and RC-RNase 5 were CpG, UpG and UpU as reported (13,23). However, the base preferences of RC-RNase 2 and RC-RNase 4 were UpG and CpG, respectively. Interestingly, the base specificities of RC-RNase 3 and RC-RNase 6 were quite broad because the bonds between CpA, CpU, CpG and UpG were cleaved by these two ribonucleases. These results are in good agreement with the kcat/Km values of those ribonucleases toward dinucleotides CpG and UpG (Table 1). In some instances, the relative kcat/Km values of ribonucleases for UpU, CpG and UpG did not closely correlate with the results on RNA sequencing gel, e.g. RC-RNase, RC-RNase L1 and RC-RNase 5, because the internal UpU bond of the primary product may have been further cleaved by the same ribonuclease.
Figure 3.
Base specificity of purified bullfrog ribonucleases toward a synthetic RNA substrate. A 5′-end-labeled oligoribonucleotide was partially digested with ribonucleases as indicated in Figure 2 at 30°C for 10 min, separated by 15% urea–PAGE and visualized by autoradiography. C, 5′-32P-labeled RNA substrate; OH–, RNA substrate treated with 0.05 M sodium bicarbonate-carbonate, pH 9.2, at 90°C for 4 min. The RNA sequence and cleavage sites are shown in the left and right margins, respectively.
Catalytic properties of bullfrog ribonucleases
Ribonucleases are abundant in oocytes and embryos of bullfrogs, and may be regulated by microenvironments to exert specific functions. Therefore, it was of interest to investigate factors that affect ribonuclease activity. The optimal pH values of RC-RNase 3, RC-RNase 6 and RC-RNase 4 were 4.5–5.0, 5.0–5.5 and 5.5–6.0, respectively, while those of other ribonucleases ranged from 6 to 8.5 (Table 1). In general, the optimal pH values of most ribonuclease activity seem broad, yet the optimal pH values of each individual ribonuclease ranged from acidic to basic condition. The optimal temperature of most bullfrog ribonuclease activities was 65°C, whereas that of RNase A and RC-RNase 5 was 50°C (Table 1). Divalent cations were not required for catalytic activity. Their ribonuclease activities were destroyed by reducing agents, e.g. dithiothreitol and 2-mercapto-ethanol, but were not inhibited by mammalian ribonuclease inhibitor from human placenta (data not shown).
Cloning of bullfrog ribonuclease gene family
On the basis of amino acid sequence homology among Rana spp. frog ribonucleases and the known DNA sequence of the RC-RNase gene (16), degenerated primers were used to clone ribonuclease fragments of the gene family by PCR using bullfrog genomic DNA as template, based on the lack of introns in the coding region of ribonuclease. Using a combination of 5′-RACE and 3′ Genome Walker, the coding regions of full-length mature proteins, except RC-RNase 5, in the oocyte ribonuclease family and the liver-specific RC-RNase L1 were cloned. The DNA sequences of pupative signal sequence of RC-RNase, RC-RNase 2 and RC-RNase 4 were obtained. Fragments of the 3′-untranslated regions of the gene family are also obtained. The length of the 3′-untranslated region varied with genes.
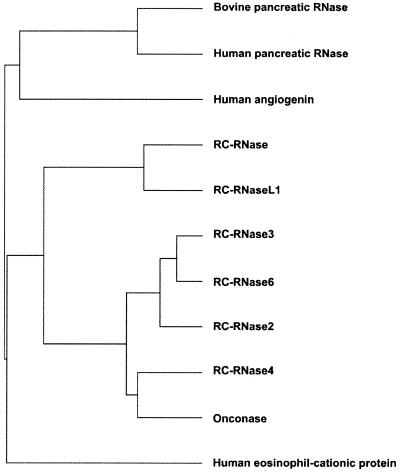
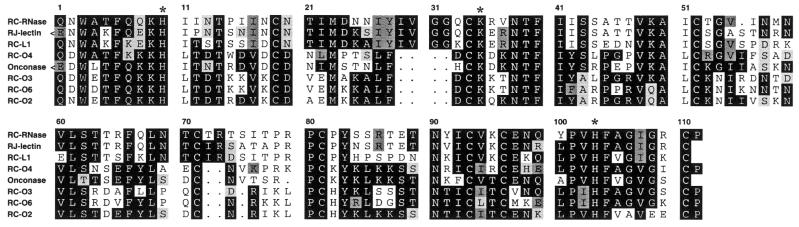
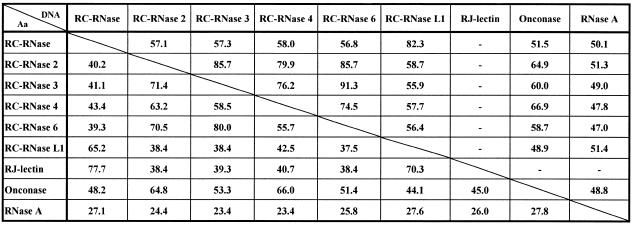
The DNA sequences of bullfrog ribonuclease coding region were compared with those of mammalian ribonucleases and expressed in a phylogenetic tree as shown in Figure 4. These ribonuclease genes belong to a distinct class in the ribonuclease gene superfamily and are evolutionarily far from mammalian eosinophil-cationic proteins, angiogenins and pancreatic ribonucleases. As shown in Figure 5, the deduced amino acid sequences of bullfrog ribonuclease genes were aligned with other frog ribonucleases, e.g. onconase from R.pipiens and sialic acid-binding lectin from R.japonica. Identical and similar amino acid residues are boxed in black and grey, respectively. The N-terminal amino acid residue of bullfrog ribonucleases deduced from DNA sequence was Gln, whereas it was pyroglutamate when obtained according to automatic amino acid sequence analyses, and molecular weight calculations by ANTHEPROT (28) and mass spectrometry analyses (Table 1). The conserved amino acid residues for catalytic activities of the ribonuclease family were marked by asterisks. The identities of both DNA and amino acid sequences of known frog ribonucleases and bovine RNase A were analyzed by pairwise sequence alignment and expressed as percentages as shown in Table 2. Frog ribonucleases have ∼50% identity in nucleic acid sequence and ∼25% identity in amino acid sequence when compared to bovine RNase A.
Figure 4.
Phylogenetic tree of ribonuclease genes. The DNA sequences of the following ribonuclease genes were clustered under the pileup and figure commands in the GCG program: bullfrog oocyte ribonucleases (RC-RNase, RC-RNase 2, RC-RNase 3, RC-RNase 4, RC-RNase 6), onconase from R.pipiens (32), human eosinophil-derived neurotoxin (34), bovine pancreatic RNase (35), human pancreatic RNase (36) and human angiogenin (37).
Figure 5.
Amino acid sequence alignment of ribonucleases from Rana spp. frogs. RJ-lectin, the lectin or ribonuclease from oocytes of R.japonica (12); onconase, the ribonuclease from the oocytes of R.pipiens (12). The amino acid sequences were analyzed and aligned under the commands of pileup and prettybox in the GCG programs. Asterisks indicate conserved amino acid residues for catalytic activities of ribonuclease superfamily.
Table 2. Pairwise protein and nucleic acid sequence identity matrix for frog ribonucleases and bovine pancreatic RNase A (%a).
Aa, amino acid; DNA, nucleic acid; –, data not available.
aPercentages were obtained by the pairwise sequence alignment program in BCM Search Launcher (http://www.hgsc.bcm.tmc.edu/SearchLauncher/ ).
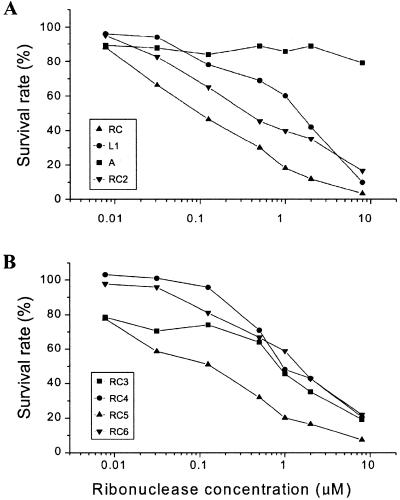
Cytotoxicity of bullfrog ribonucleases to HeLa cells
To investigate the potential biological functions of the bullfrog ribonuclease family in embryogenesis, which includes a period of intense proliferation and differentiation, the effects of these ribonucleases on cell cytotoxicity/proliferation were analyzed. The IC50 of these ribonucleases to human cervical carcinoma (HeLa) cell growth were determined. As shown in Figure 6A and B, RC-RNase (∼0.1 µM) was the most toxic, then in descending order: RC-RNase 5 (∼0.15 µM), RC-RNase 2 (∼0.3 µM), RC-RNase 3, RC-RNase 4, RC-RNase 6 and RC-RNase L1 (all ∼1–2 µM). Bovine RNase A was not cytotoxic to HeLa cells even at concentrations up to 8 µM. The IC50 of RC-RNase to human fibroblast cells WI38 was >128 µM by the same assay condition (data not shown). It is of importance to note that the cytotoxicity of a ribonuclease toward a tumor cell line does not closely correlate with its catalytic activity (Table 1; Figs 2C and 6). Therefore, other amino acid residues are likely involved in the cytotoxicity of ribonucleases to HeLa cells in addition to conventional catalytic residues, e.g. His10, Lys35 and His103 of RC-RNase.
Figure 6.
Concentration-dependent cytotoxicities of bullfrog ribonucleases toward HeLa S-3 cells. Cells (3 × 103) were seeded on a 96-well plate overnight and incubated with bullfrog ribonucleases and bovine pancreatic ribonuclease for 48 h (six wells per treatment). The survival rates of ribonuclease-treated HeLa S-3 cells were counted by ATP-Lite M (Packard BioScience Company) and expressed as a percentage.
DISCUSSION
To date, three oocytic ribonucleases and one liver ribonuclease have been purified from Rana spp. frogs (12–16,27). These ribonucleases are purified by column chromatography and assayed by the catalytic activities of each column eluate. Consequently, only one predominant ribonuclease was purified from each tissue, whereas other ribonucleases were ignored because of their low catalytic activities. In the present study, using both western blotting and catalytic activity assay, we purified four new novel ribonucleases as well as RC-RNase from bullfrog oocytes where these ribonucleases are stored (16,24). Conventional column chromatography was employed to purify these abundant ribonucleases to a high purity at a low cost within a short time period. The availability of the newly identified bullfrog ribonucleases would be helpful to the investigation of these ribonucleases as potential agents for tumor therapy, and to the studies of their biological functions and structure–function relationships (29–30).
One unique property of frog Rana spp. ribonucleases is the RNA substrate preference for pyrimidine–guanine rather than pyrimidine–adenine. From the analyses of base specificity, specific activity and kcat/Km of bullfrog ribonucleases toward dinucleotides, we found that some ribonucleases possess high specificity as well as strong catalytic activity, e.g. RC-RNase and RC-RNase L1 for CpG and UpG, while some ribonucleases, e.g. RC-RNase 3 and RC-RNase 6, have very weak catalytic activity and low base specificity. Based on the alignment of amino acid sequences of these ribonucleases and information from structural studies by NMR and X-ray crystallography (29–30), possible amino acid residues involved in these activities could be investigated. Furthermore, cloning of the bullfrog ribonuclease gene family in this study and site-directed mutagenesis of possible residues will provide valuable information for the study of the special biological properties, e.g. base specificity, catalytic activity as well as cytotoxicity.
The cytotoxicity of RC-RNase to HeLa cells was completely abolished if the catalytic activity was destroyed by H10A and H103A mutations (16). In contrast, the cytotoxicities of these native oocytic ribonucleases do not closely correlate with their relative catalytic activities. For example, RC-RNase 5 has 5000-fold less catalytic activity than RC-RNase, but they have similar cytotoxicity (IC50, ∼0.1–0.15 µM). Before its ribonuclease activity was found, RC-RNase was originally defined as a sialic acid-binding lectin which agglutinates tumor cells, but not neuramidase-treated tumor cells (13,17). Therefore, a specific domain of RC-RNase may be involved in the binding of ribonucleases to cell surface receptors and subsequent internalization into the cell rather than the catalytic active domain.
Onconase, a ribonuclease isolated from the oocytes of the frog R.pipiens, is currently being evaluated in human phase III clinical trials. The possible mechanisms of its antitumor activities are being studied, e.g. entry of ribonuclease through surface receptor binding into cytosol, degradation of specific RNA and induction of apoptosis in tumor cells (12,31), and the onconase gene was recently cloned (32), but the direct receptor/substrate of the ribonuclease and the detailed mechanism of cytotoxicity in tumor cells are still not clear. Preparation of multiple and large quantities of ribonucleases and cloning of these genes in this report will provide a good source for the studies of antitumor mechanisms at molecular and cellular level and for the clinical trial of human tumor therapy, e.g. identification of ribonuclease-binding proteins, ribonuclease entry routes and possible residues involved in cytotoxicity.
These ribonucleases were predominantly found in the oocyte and early embryos of bullfrog rather than in other developmental stages or other organs (24), but their possible biological functions on bullfrog embryonic development or host defence mechanism are still not clear. Therefore, large-scale preparations of the ribonuclease family from oocyte and preparation of specific antibodies and characterizations of these ribonucleases will help us in examining the biosynthesis and distribution of these ribonucleases during bullfrog development. Cloning of the ribonuclease gene family is a prerequisite to analyze the expression of each ribonuclease gene in frog development or oocyte maturation. Due to the involvement of apoptosis in Xenopus laevis early development (33) and the induction of apoptosis observed in onconase-treated HeLa cells (31) and RC-RNase-treated HeLa cells (unpublished results), it is suggested that these oocytic ribonucleases may play roles in embryonic development through their cytotoxic activities.
In conclusion, we have developed efficient methods to purify large quantities of novel oocyte ribonucleases which have the potential to be therapeutic agents for tumor therapy. We have cloned these ribonuclease genes to help us define the functional and structural relationships of these novel ribonucleases. The findings from these studies will enable us to understand the roles of novel ribonucleases in frog development, to elucidate the molecular mechanisms of their antitumor activities, and to make human–frog chimeric ribonucleases, which have low immunogeneity but possess high antitumor activity, for potential human tumor therapy.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Y. S. Lin, S. Lin-Chao and J. S. Huang for critical readings and helpful discussions of the manuscript, and research assistants L. J. Chang, Y. Y. Hsiao and S. M. Yu for assisting with ribonuclease preparation and gene cloning. This work was supported by Academia Sinica, the National Science Council of the Republic of China (NSC87-2316-B-001-002) and National Health Research Institutes.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +886 2 2789 9167; Fax: +886 2 2782 9142; Email: ydliao@ibms.sinica.edu.tw AF039104, AF242553–AF242556, AF288642
REFERENCES
- 1.D’Alessio G. (1993) Trends Cell Biol., 3, 106–109. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn P. and Moore,S. (1982) In Boyer,P.D. (ed.), The Enzymes, 3rd edn. Academic Press, New York, Vol. 15, pp. 317–433.
- 3.Rosenberg H.F. (1998) Cell. Mol. Life Sci., 54, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deutscher M.P. (1993) J. Biol. Chem., 268, 13011–13014. [PubMed] [Google Scholar]
- 5.Barnard E.A. (1969) Nature, 221, 340–344. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro R., Riordan,J.F. and Vallee,B.L. (1986) Biochemistry, 25, 3527–3532. [DOI] [PubMed] [Google Scholar]
- 7.Gleich G.J., Loegering,D.A., Bell,M.P., Chekel,J.L., Ackerman,S.J. and McKean,D.J. (1986) Proc. Natl Acad. Sci. USA, 83, 3146–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gullberg U., Widerner,B., Arnason,U., Egesten,A. and Olsson,I. (1986) Biochem. Biophys. Res. Commun., 139, 1239–1242. [DOI] [PubMed] [Google Scholar]
- 9.Parry S.K., Liu,Y.H., Clarke,A.E. and Newbigin,E. (1997) In D’Alessio,G. and Riordan,J.F. (eds), Ribonuclease: Structures and Functions. Academic Press, New York, pp. 192–211.
- 10.Vescia S., Tramontano,D., Augusti-Tocco,G. and D’Alessio,G. (1980) Cancer Res., 40, 3740–3744. [PubMed] [Google Scholar]
- 11.Leland P.A., Schultz,L., Kim,B.M. and Raines,R.T. (1998) Proc. Natl Acad. Sci. USA, 95, 10407–10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Youle R.J. and D’Alessio,G. (1997) In D’Alessio,G. and Riordan,J.F. (eds), Ribonuclease: Structures and Functions. Academic Press, New York, pp. 491–514.
- 13.Okabe Y., Katayama,N., Iwama, M, Watanabe,H., Ohgi,K., Irie,M., Nitta,K., Kawauchi,H., Takayanagi,Y., Oyama,F., Titani,K., Abe,Y., Okazaki,T., Inokuchi,N. and Koyama,T. (1991) J. Biochem., 109, 786–790. [DOI] [PubMed] [Google Scholar]
- 14.Liao Y.D., Huang,H.C., Chan,H.J. and Kuo,S.J. (1996) Protein Expr. Purif., 7, 194–202. [DOI] [PubMed] [Google Scholar]
- 15.Titani K., Takio,K., Kuwada,M., Nitta,K., Sakakibara,F., Kawauchi,H., Takayanagi,G. and Hakomori,S.I. (1987) Biochemistry, 26, 2189–2194. [DOI] [PubMed] [Google Scholar]
- 16.Huang H.C., Wang,S.C., Leu,Y.J., Lu,S.C. and Liao,Y.D. (1998) J. Biol. Chem., 273, 6395–6401. [DOI] [PubMed] [Google Scholar]
- 17.Irie M., Nitta,K. and Nonaka,T. (1998) Cell. Mol. Life Sci., 54, 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradford M.M. (1976) Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U.K. (1970) Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 20.Allen G. (1989) In Burdon,R.H. and van Knippenberg,P.H. (eds), Laboratory Techniques in Biochemistry and Molecular Biology, 2nd edn. Elsevier, Amsterdam, The Netherlands, Vol. 9, pp. 19–104.
- 21.Hewick R.M., Hunkapillar,M.W., Hood,L.E. and Dreyer,W.J. (1981) J. Biol. Chem., 256, 7990–7997. [PubMed] [Google Scholar]
- 22.Hong J.L., Liu,L.F., Wang,L.Y., Tsai,S.P., Hsieh,C.H., Hsiao,C.D. and Tam,M.F. (1994) Biochem. J., 304, 825–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y.D. (1992) Nucleic Acids Res., 20, 1371–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Y.D. and Wang,J.J. (1994) Eur. J. Biochem., 222, 215–220. [DOI] [PubMed] [Google Scholar]
- 25.Liao Y.D. (1995) Mol. Biol. Rep., 20, 149–154. [DOI] [PubMed] [Google Scholar]
- 26.Chomczynski P. and Sacchi,N. (1987) Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 27.Nitta R., Katayama,N., Okabe,Y., Iwama,M., Watanabe,H., Abe,Y., Okazaki,T., Ohgi,K. and Irie,M. (1989) J. Biochem., 106, 729–735. [DOI] [PubMed] [Google Scholar]
- 28.Geourjon C. and Deléage,G. (1995) ANTHEPROT 2.0: A three-dimensional module fully coupled with protein sequence analysis methods. J. Mol. Graph., 13, 209–212. [DOI] [PubMed] [Google Scholar]
- 29.Chen C., Hom,K., Huang,R.F., Chong,P.J., Liao,Y.D. and Huang,T.H. (1996) J. Biomol. NMR, 8, 331–344. [DOI] [PubMed] [Google Scholar]
- 30.Chang C.F., Chen,C., Chen,Y.C., Hom,K., Huang,R.F. and Huang,T.H. (1998) J. Mol. Biol., 283, 231–244. [DOI] [PubMed] [Google Scholar]
- 31.Iordanov M.S., Ryabinina,O.P., Wong,J., Dinh,T.H., Newton,D.L., Rybak,S.M. and Magun,B.E. (2000) Cancer Res., 60, 1983–1994. [PubMed] [Google Scholar]
- 32.Chen S.L., Le,S.Y., Newton,D.L., Maizel,J.V.,Jr and Rybak,S.M. (2000) Nucleic Acids Res., 28, 2375–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hensey C. and Gautier,J. (1998) Dev. Biol., 203, 36–48. [DOI] [PubMed] [Google Scholar]
- 34.Barker R.L., Loegering,D.A., Ten,R.M., Hamann,K.J., Pease,L.R. and Gleich,G.J. (1989) J. Immunol., 143, 952–955. [PubMed] [Google Scholar]
- 35.Carsana A., Confalone,E., Palmieri,M., Libonati,M. and Furia,A. (1988) Nucleic Acids Res., 16, 5491–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seno M., Futami,J., Kosaka,H., Seno,S. and Yamada,H. (1994) Biochim. Biophys. Acta, 1218, 466–468. [DOI] [PubMed] [Google Scholar]
- 37.Kurachi K., Davie,E.W., Strydom,D.J., Riordan,J.F. and Vallee,B.L. (1985) Biochemistry, 24, 5499–5502.4074710 [Google Scholar]