Abstract
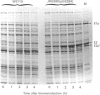
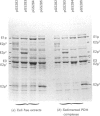
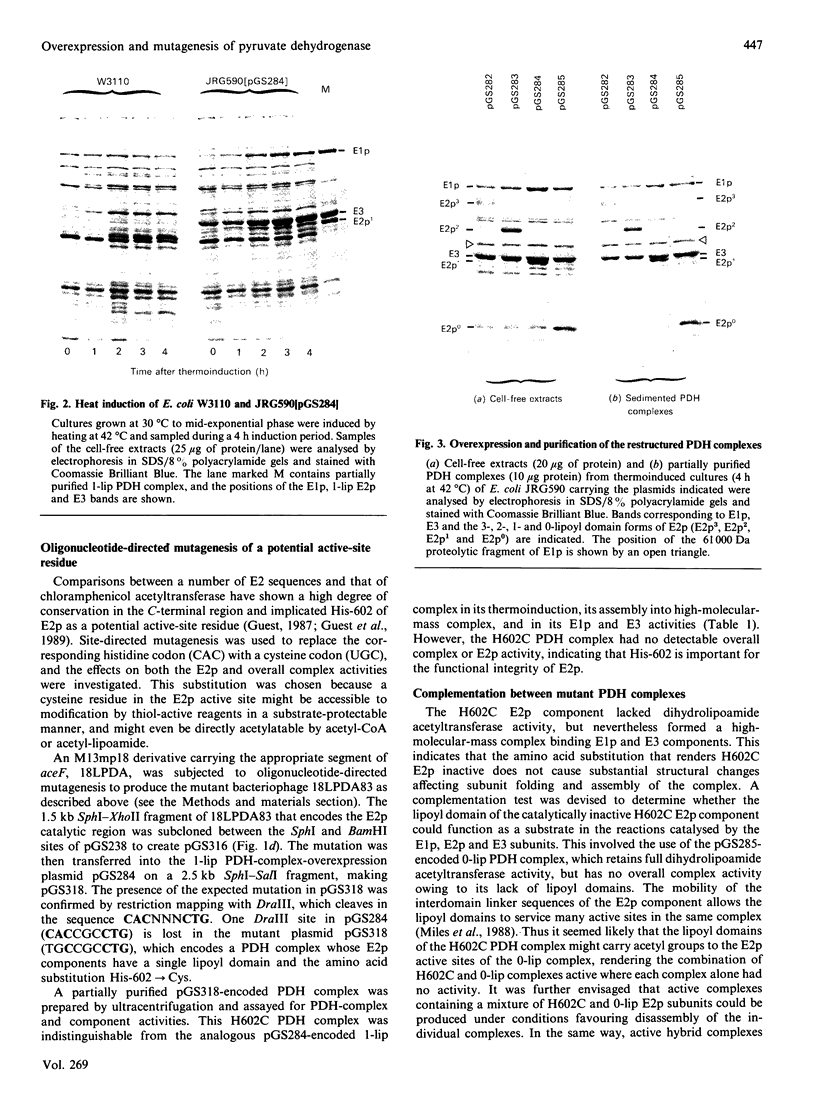
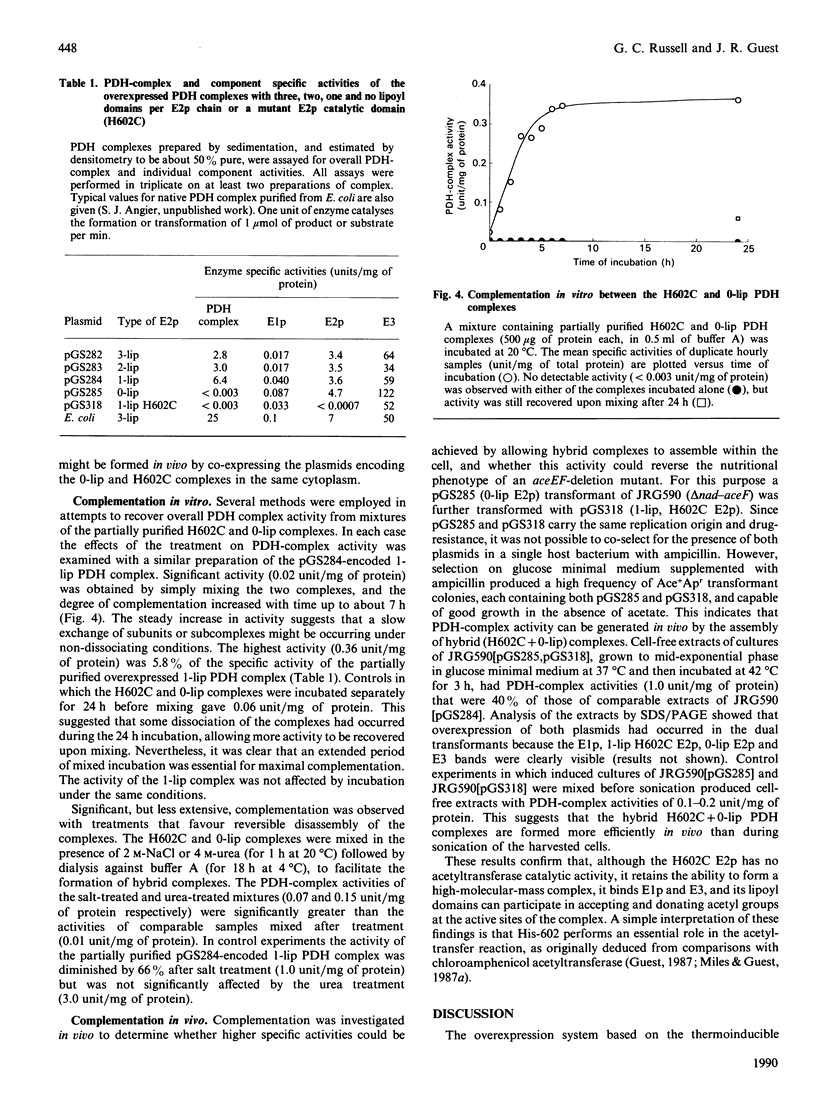
The aceEF-lpd operon of Escherichia coli encodes the pyruvate dehydrogenase (E1p), dihydrolipoamide acetyltransferase (E2p) and dihydrolipoamide dehydrogenase (E3) components of the pyruvate dehydrogenase multienzyme complex (PDH complex). A thermoinducible expression system was developed to amplify a variety of genetically restructured PDH complexes, including those containing three, two, one and no lipoyl domains per E2p chain. Although large quantities of the corresponding complexes were produced, they had only 20-50% of the predicted specific activities. The activities of the E1p components were diminished to the same extent, and this could account for the shortfall in overall complex activity. Thermoinduction was used to express a mutant PDH complex in which the putative active-site histidine residue of the E2p component (His-602) was replaced by cysteine in the H602C E2p component. This substitution abolished dihydrolipoamide acetyltransferase activity of the complex without affecting other E2p functions. The results support the view that His-602 is an active-site residue. The inactivation could mean that the histidine residue performs an essential role in the acetyltransferase reaction mechanism, or that the reaction is blocked by an irreversible modification of the cysteine substituent. Complementation was observed between the H602C PDH complex and a complex that is totally deficient in lipoyl domains, both in vitro, by the restoration of overall complex activity in mixed extracts, and in vivo, from the nutritional independence of strains that co-express the two complexes from different plasmids.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison N., Williams C. H., Jr, Guest J. R. Overexpression and mutagenesis of the lipoamide dehydrogenase of Escherichia coli. Biochem J. 1988 Dec 15;256(3):741–749. doi: 10.1042/bj2560741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. L., Danson M. J., Hale G., Hooper E. A., Perham R. N. Self-assembly and catalytic activity of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Nature. 1977 Jul 28;268(5618):313–316. doi: 10.1038/268313a0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleile D. M., Hackert M. L., Pettit F. H., Reed L. J. Subunit structure of dihydrolipoyl transacetylase component of pyruvate dehydrogenase complex from bovine heart. J Biol Chem. 1981 Jan 10;256(1):514–519. [PubMed] [Google Scholar]
- Chin D. T., Goff S. A., Webster T., Smith T., Goldberg A. L. Sequence of the lon gene in Escherichia coli. A heat-shock gene which encodes the ATP-dependent protease La. J Biol Chem. 1988 Aug 25;263(24):11718–11728. [PubMed] [Google Scholar]
- Danson M. J., Perham R. N. Evidence for two lipoic acid residues per lipoate acetyltransferase chain in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochem J. 1976 Dec 1;159(3):677–682. doi: 10.1042/bj1590677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussey S. P., Guest J. R., James O. F., Bassendine M. F., Yeaman S. J. Identification and analysis of the major M2 autoantigens in primary biliary cirrhosis. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8654–8658. doi: 10.1073/pnas.85.22.8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest J. R., Angier S. J., Russell G. C. Structure, expression, and protein engineering of the pyruvate dehydrogenase complex of Escherichia coli. Ann N Y Acad Sci. 1989;573:76–99. doi: 10.1111/j.1749-6632.1989.tb14988.x. [DOI] [PubMed] [Google Scholar]
- Guest J. R., Creaghan I. T. Further studies with lipoamide dehydrogenase mutants of Escherichia coli K12. J Gen Microbiol. 1974 Mar;81(1):237–245. doi: 10.1099/00221287-81-1-237. [DOI] [PubMed] [Google Scholar]
- Guest J. R., Lewis H. M., Graham L. D., Packman L. C., Perham R. N. Genetic reconstruction and functional analysis of the repeating lipoyl domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Mol Biol. 1985 Oct 20;185(4):743–754. doi: 10.1016/0022-2836(85)90059-2. [DOI] [PubMed] [Google Scholar]
- Kleanthous C., Cullis P. M., Shaw W. V. 3-(Bromoacetyl)chloramphenicol, an active site directed inhibitor for chloramphenicol acetyltransferase. Biochemistry. 1985 Sep 24;24(20):5307–5313. doi: 10.1021/bi00341a006. [DOI] [PubMed] [Google Scholar]
- Kleanthous C., Shaw W. V. Analysis of the mechanism of chloramphenicol acetyltransferase by steady-state kinetics. Evidence for a ternary-complex mechanism. Biochem J. 1984 Oct 1;223(1):211–220. doi: 10.1042/bj2230211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langley D., Guest J. R. Biochemical genetics of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: genetic characterization and regulatory properties of deletion mutants. J Gen Microbiol. 1978 May;106(1):103–117. doi: 10.1099/00221287-106-1-103. [DOI] [PubMed] [Google Scholar]
- Leslie A. G., Moody P. C., Shaw W. V. Structure of chloramphenicol acetyltransferase at 1.75-A resolution. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4133–4137. doi: 10.1073/pnas.85.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J. S., Guest J. R. Molecular genetic aspects of the citric acid cycle of Escherichia coli. Biochem Soc Symp. 1987;54:45–65. [PubMed] [Google Scholar]
- Miles J. S., Guest J. R., Radford S. E., Perham R. N. Investigation of the mechanism of active site coupling in the pyruvate dehydrogenase multienzyme complex of Escherichia coli by protein engineering. J Mol Biol. 1988 Jul 5;202(1):97–106. doi: 10.1016/0022-2836(88)90522-0. [DOI] [PubMed] [Google Scholar]
- Miles J. S., Guest J. R. Subgenes expressing single lipoyl domains of the pyruvate dehydrogenase complex of Escherichia coli. Biochem J. 1987 Aug 1;245(3):869–874. doi: 10.1042/bj2450869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., VanBogelen R. A., Vaughn V. The genetics and regulation of heat-shock proteins. Annu Rev Genet. 1984;18:295–329. doi: 10.1146/annurev.ge.18.120184.001455. [DOI] [PubMed] [Google Scholar]
- Packman L. C., Hale G., Perham R. N. Repeating functional domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. EMBO J. 1984 Jun;3(6):1315–1319. doi: 10.1002/j.1460-2075.1984.tb01969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packman L. C., Perham R. N. Chain folding in the dihydrolipoyl acyltransferase components of the 2-oxo-acid dehydrogenase complexes from Escherichia coli. Identification of a segment involved in binding the E3 subunit. FEBS Lett. 1986 Oct 6;206(2):193–198. doi: 10.1016/0014-5793(86)80979-6. [DOI] [PubMed] [Google Scholar]
- Packman L. C., Perham R. N., Roberts G. C. Cross-linking and 1H n.m.r. spectroscopy of the pyruvate dehydrogenase complex of Escherichia coli. Biochem J. 1982 Aug 1;205(2):389–396. doi: 10.1042/bj2050389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perham R. N., Packman L. C., Radford S. E. 2-Oxo acid dehydrogenase multi-enzyme complexes: in the beginning and halfway there. Biochem Soc Symp. 1987;54:67–81. [PubMed] [Google Scholar]
- Radford S. E., Laue E. D., Perham R. N., Miles J. S., Guest J. R. Segmental structure and protein domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Genetic reconstruction in vitro and 1H-n.m.r. spectroscopy. Biochem J. 1987 Nov 1;247(3):641–649. doi: 10.1042/bj2470641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. J., Pettit F. H., Eley M. H., Hamilton L., Collins J. H., Oliver R. M. Reconstitution of the Escherichia coli pyruvate dehydrogenase complex. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3068–3072. doi: 10.1073/pnas.72.8.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schauder B., Blöcker H., Frank R., McCarthy J. E. Inducible expression vectors incorporating the Escherichia coli atpE translational initiation region. Gene. 1987;52(2-3):279–283. doi: 10.1016/0378-1119(87)90054-0. [DOI] [PubMed] [Google Scholar]
- Spencer M. E., Darlison M. G., Stephens P. E., Duckenfield I. K., Guest J. R. Nucleotide sequence of the sucB gene encoding the dihydrolipoamide succinyltransferase of Escherichia coli K12 and homology with the corresponding acetyltransferase. Eur J Biochem. 1984 Jun 1;141(2):361–374. doi: 10.1111/j.1432-1033.1984.tb08200.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur J Biochem. 1983 Jul 1;133(3):481–489. doi: 10.1111/j.1432-1033.1983.tb07490.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the pyruvate dehydrogenase component. Eur J Biochem. 1983 Jun 1;133(1):155–162. doi: 10.1111/j.1432-1033.1983.tb07441.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Lewis H. M., Darlison M. G., Guest J. R. Nucleotide sequence of the lipoamide dehydrogenase gene of Escherichia coli K12. Eur J Biochem. 1983 Oct 3;135(3):519–527. doi: 10.1111/j.1432-1033.1983.tb07683.x. [DOI] [PubMed] [Google Scholar]
- Willms C. R., Oliver R. M., Henney H. R., Jr, Mukherjee B. B., Reed L. J. Alpha-keto acid dehydrogenase complexes. VI. Dissociation and reconstitution of the dihydrolipoyl transacetylase of Escherichia coli. J Biol Chem. 1967 Mar 10;242(5):889–897. [PubMed] [Google Scholar]
- Yeaman S. J. The 2-oxo acid dehydrogenase complexes: recent advances. Biochem J. 1989 Feb 1;257(3):625–632. doi: 10.1042/bj2570625. [DOI] [PMC free article] [PubMed] [Google Scholar]