Abstract
Centrosomes are the canonical microtubule organizing centers (MTOCs) of most mammalian cells, including spermatocytes. Centrosomes comprise a centriole pair within a structurally ordered and dynamic pericentriolar matrix (PCM). Unlike in mitosis, where centrioles duplicate once per cycle, centrioles undergo two rounds of duplication during spermatogenesis. The first duplication is during early meiotic prophase I, and the second is during interkinesis. Using mouse mutants and chemical inhibition, we have blocked centriole duplication during spermatogenesis and determined that non-centrosomal MTOCs (ncMTOCs) can mediate chromosome segregation. This mechanism is different from the acentriolar MTOCs that form bipolar spindles in oocytes, which require PCM components, including gamma-tubulin and CEP192. From an in-depth analysis, we identified six microtubule-associated proteins, TPX2, KIF11, NuMA, and CAMSAP1-3, that localized to the non-centrosomal MTOC. These factors contribute to a mechanism that ensures bipolar MTOC formation and chromosome segregation during spermatogenesis when centriole duplication fails. However, despite the successful completion of meiosis and round spermatid formation, centriole inheritance and PLK4 function are required for normal spermiogenesis and flagella assembly, which are critical to ensure fertility.
Keywords: Centriole, Centrosome, Meiosis, Spermatogenesis, PLK4
Subject terms: Cell Cycle
Synopsis
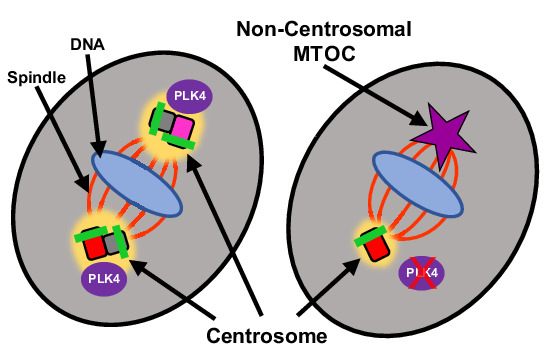
PLK4 is required for centriole duplication and maturation during mammalian spermatogenesis. However, when duplication and maturation fail, spermatocytes form non-centrosomal microtubule organizing centers to complete chromosome segregation during meiosis I and II.
PLK4 is the master regulator of centriole duplication and maturation during mammalian spermatogenesis.
Spermatocytes are essential for the formation of mature spermatozoa that harbor a flagella.
Spermatocytes can rely on a non-centrosomal microtubule organizing center (ncMTOC) to mediate chromosome segregation during meiosis I and II.
PLK4 is required for centriole duplication and maturation during mammalian spermatogenesis. However, when duplication and maturation fail, spermatocytes form non-centrosomal microtubule organizing centers to complete chromosome segregation during meiosis I and II.
Introduction
Infertility remains a public health issue that affects around 15% of couples worldwide (Sun et al, 2019). A major cause of infertility can arise from errors during meiosis (Potapova and Gorbsky, 2017). Meiotic cells undergo one round of DNA replication, followed by two successive rounds of chromosome segregation, termed meiosis I and meiosis II. In meiosis I, homologous chromosome pairs, which are linked via crossover recombination events, are segregated. The sister chromatid pairs, which had remained associated with one another during meiosis I, segregate during meiosis II. The accurate separation of the chromosomes via bipolar spindles during meiosis is critical, as aneuploidy events can lead to infertility, spontaneous abortion, and the development of genetic disease in the resulting offspring (Potapova and Gorbsky, 2017). The mechanism of bipolar spindle formation is sexually dimorphic in mammals. In oocytes, bipolar spindles are formed via an acentriolar microtubule organizing center (aMTOC), whereas spermatocytes rely on a canonical centriole-containing centrosome for bipolar spindle formation (Lerit and Poulton, 2016; Gruhn and Hoffmann, 2022; So et al, 2019; Schatten and Sun, 2009). While the importance of the centrosome is well established in mitotic cells, very little is known about the regulation of centrosome biogenesis during spermatogenesis.
A centrosome consists of two centrioles and associated proteins known as the pericentriolar matrix (PCM). Centrioles are cylindrical complexes made up of nine triplicate microtubules and exist in pairs orthogonally oriented to each other (Jana, 2021). The older centriole, termed the mature parent centriole, has additional maturation markers known as subdistal and distal appendages that are required for the recruitment of PCM to the centrioles and centriole membrane docking during ciliogenesis, respectively (Tanos et al, 2013; Mazo et al, 2016). During mitosis and meiosis, the PCM acts as the MTOC by concentrating γ-tubulin ring complexes (γ-TuRC) that serve as nucleation sites for the assembly of bipolar spindles required for chromosome segregation (Bornens, 2002). Following meiosis II, haploid spermatids undergo spermiogenesis, where the centrioles become the foundation for the microtubule structure responsible for flagella formation (Avidor-Reiss et al, 2020; Bettencourt-Dias et al, 2005).
Polo-like kinase 4 (PLK4) is considered the “master regulator” of centriole duplication during interphase in mitotically dividing cells (de Cárcer et al, 2011; Zitouni et al, 2014). PLK4-mediated centriole duplication in mitotic cells is tightly regulated to ensure this duplication event occurs only once per cell cycle (Bettencourt-Dias et al, 2005; Habedanck et al, 2005; Slevin et al, 2012; de Cárcer et al, 2011; Sillibourne and Bornens, 2010; Klebba et al, 2013, 2015; Cunha-Ferreira et al, 2013; Park et al, 2019). Two PLK4 proteins dimerize via crypto polo box interactions and activate each other via trans-autophosphorylation of their downstream regulatory element domain during the G1 phase (Klebba et al, 2015). The active PLK4 dimer induces centriole duplication during the G1 to S phase transition, and PLK4 then undergoes proteasomal degradation (Nigg and Raff, 2009; Rogers et al, 2009; Hatch et al, 2010). This self-degradation mechanism of PLK4 helps to prevent overduplication of centrioles. PLK4 misregulation is known to promote tumorigenesis and is associated with multiple hallmarks of cancer, such as resisting cell death, genome instability and mutations, and sustained proliferative signaling (Zhang et al, 2021; Wang et al, 2019; Ko et al, 2005; Holland and Cleveland, 2014)
Orthologs of PLK4 have been previously assessed during meiosis in both Drosophila melanogaster and Caenorhabditis elegans. In these model organisms, PLK4 has been found to be important for centriole duplication during spermatogenesis (Peters et al, 2010; Bettencourt-Dias et al, 2005). However, the function of PLK4 during mammalian spermatogenesis had not been determined. Centriole duplication during mammalian meiosis differs markedly from mitosis in that there are two centriole duplication events that are not linked to the G1 to S phase transition (Wellard et al, 2021; Alfaro et al, 2021). Instead, centrioles are duplicated during early prophase (leptotene stage) of meiosis I and again in interkinesis before entering meiosis II (Wellard et al, 2021; Alfaro et al, 2021). Herein, we report the localization pattern of PLK4 during mouse and human spermatogenesis, and we demonstrate the importance of PLK4 regarding centriole duplication and maturation, centrosome biogenesis, and spermiogenesis. We also show that if centriole duplication is prevented, mouse spermatocytes can still proceed through meiosis I and II and form haploid spermatids. This relies on spermatocytes forming non-centrosomal MTOCs (ncMTOCs) that mediate bipolar spindle formation.
Results
PLK4 localizes to the centrosome throughout mammalian spermatogenesis
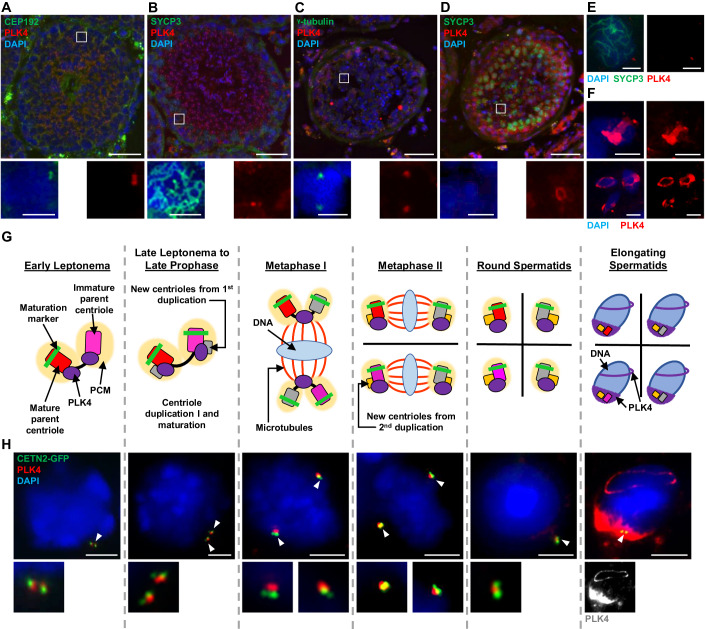
In mitotically dividing cells, PLK4 protein levels are tightly regulated to ensure centriole duplication only occurs at the G1 to S transition (Nigg and Holland, 2018). While the detailed mechanics behind PLK4 activation at the G1 to S transition have not been fully elucidated, this regulation is mediated by PLK4 dimerization and trans-autophosphorylation, which triggers PLK4 polyubiquitination and subsequent proteolysis (Cunha-Ferreira et al, 2013; Holland et al, 2010; Klebba et al, 2013; Holland et al, 2012; Guderian et al, 2010). As a result, it is challenging to visualize PLK4 via western blot analysis (Byrne et al, 2020). In contrast, PLK4 was reported to be detected via western blot throughout the first wave of spermatogenesis, suggesting higher levels of PLK4 are needed during spermatogenesis compared to mitotically dividing cells (Jordan et al, 2012). We performed a series of immunofluorescence (IF) microscopy approaches to assess the localization pattern of PLK4 during spermatogenesis. Immunolabelled cryosections of seminiferous tubules from adult wild-type (WT) mice demonstrated that PLK4 is present as two foci during meiotic prophase that colocalized with centrosomal marker CEP192 (Fig. 1A,B). PLK4 signal remained present following meiotic divisions in round spermatids and during all stages of spermiogenesis (Fig. 1A,B). To test the conservation of the PLK4 localization pattern, we immunolabelled cryosections of human testes with an antibody against PLK4. PLK4 colocalized with the centrosome component γ-tubulin in primary spermatocytes (Fig. 1C). Furthermore, PLK4 localization was observed as two foci in SYCP3-positive human primary spermatocytes (Fig. 1E). As observed in mouse seminiferous tubules, PLK4 remained present in round and elongating spermatids (Fig. 1D,F).
Figure 1. Assessment of PLK4 localization in murine and human spermatocytes throughout spermatogenesis.
(A) Cryosection (16 µm thick) of an adult WT (26 weeks) mouse testis immunolabeled against CEP192 (green) and PLK4 (red) and stained with DAPI (blue). Scale bar = 50 µm. Zoomed images of PLK4 localization with CEP192 are shown directly below. Scale bar = 5 µm. (B) Cryosection (16 µm thick) of an adult WT (26 weeks) mouse testis immunolabeled against SYCP3 (green) and PLK4 (red) and stained with DAPI (blue). Scale bar = 50 µm. Zoomed images of PLK4 localization in SYCP3-positive cells are shown directly below. Scale bar = 5 µm. (C) Cryosection (16 µm thick) of an adult (24 years) human testis immunolabeled against γ-tubulin (green) and PLK4 (red) and stained with DAPI (blue). Scale bar = 50 µm. Zoomed images of γ-tubulin and PLK4 co-localization are shown directly below. Scale bar = 5 µm. (D) Cryosection (16 µm thick) of an adult (24 years) human testis immunolabeled against SYCP3 (green) and PLK4 (red) and stained with DAPI (blue). Scale bar = 50 µm. Zoomed images of PLK4 localization at the perinuclear ring are shown directly below. Scale bar = 5 µm. (E) Cryosection (16 µm thick) of an adult (24 years) human testis immunolabeled against SYCP3 (green) and PLK4 (red) and stained with DAPI (blue). Images demonstrate PLK4 localization in prophase I spermatocytes. Scale bars = 5 µm. (F) Cryosections (16 µm thick) of an adult (24 years) human testis immunolabeled against PLK4 (red) and stained with DAPI (blue). Images demonstrate PLK4 localization in elongating spermatids. Scale bars = 5 µm. (G) Diagram of the PLK4 localization pattern during spermatogenesis aligned with cells from the corresponding stages of spermatogenesis captured from tubule squash preparations performed on WT control mice (H). In the diagram, red rectangle with rounded corners = mature parent centriole, pink rectangle with rounded corners = immature parent centriole, green bar = maturation marker, the yellow oval = PCM, purple shapes = PLK4, gray rectangle with rounded corners = new centrioles from first centriole duplication, yellow rectangle with rounded corners = new centriole from second centriole duplication, blue oval = DNA, red lines = microtubules. (H) Control spermatocytes harboring CETN2-GFP (green) were immunolabeled against PLK4 (red) and stained with DAPI (blue). The white arrowheads indicate the centrosome. Zoomed images of the centrioles (or PLK4 staining at the elongating spermatid stage) are shown directly below the corresponding image. Scale bars = 5 µm.
To closely assess the localization of PLK4 in spermatocytes at different stages, we took advantage of the first semi-synchronous wave of spermatogenesis (Fig. 1G,H). In prior work, we established that the first of two rounds of centriole duplication occurs during the leptotene sub-stage of meiotic prophase (Wellard et al, 2021; Alfaro et al, 2021). We determined that PLK4 localizes to both parent centrioles prior to centriole duplication (Fig. 1G,H). Following the first round of centriole duplication, PLK4 localizes in between the parent centrioles of each centriole pair. When centrosomes separate during late prophase, PLK4 continues to be detected between centriole pairs, and this remains the case during the first meiotic division. Following meiosis I, centrioles duplicate again at interkinesis, and PLK4 signal persists between the centriole pairs during meiosis II and in the resulting round spermatids (Fig. 1G,H). During the transformation of round spermatids towards spermatozoa (spermiogenesis), PLK4 localization broadens around the centrioles at the base of the spermatid, where the flagella will form (Fig. 1G,H). In addition, PLK4 signal can be detected as a ring that surrounds the spermatid nucleus (Fig. 1G,H). Collectively, this localization pattern is reminiscent of the manchette and perinuclear ring structures, respectively, which are important for the elongation of the sperm nucleus and the formation of functional spermatozoa (Lehti and Sironen, 2016; Kato et al, 2004). PLK4 was also observed as a ring-like structure and at the manchette in elongating human spermatids (Fig. 1D,F). These observations indicate that PLK4 localization patterns during spermatogenesis are conserved between mice and humans.
Taken together, we demonstrate that unlike what has been described for mitotically dividing cells, PLK4 remains present at the centrosome following both rounds of centriole duplication during meiosis. In addition, PLK4 localization during spermiogenesis mimics components of the perinuclear ring and manchette, indicating that PLK4 has roles during spermatid to spermatozoa morphogenesis.
Overexpression of PLK4 in primary spermatocytes leads to centriole overduplication in meiosis I
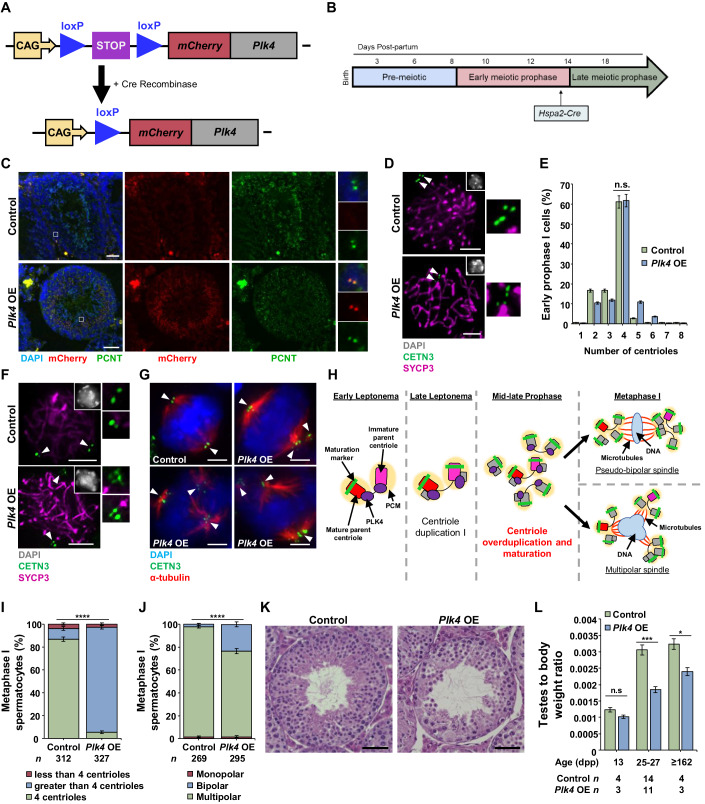
If PLK4 is overexpressed or not degraded prior to the next mitotic cycle, centrioles undergo amplification, which can result in the formation of multipolar spindles that cause chromosome missegregation (Moyer et al, 2015; Holland et al, 2012). Therefore, it was surprising to see that PLK4 remains associated with the centrioles throughout meiosis, especially considering that centriole duplication occurs at two defined times (Fig. 1F) (Wellard et al, 2021; Alfaro et al, 2021). We wondered whether the function of PLK4 in spermatocytes was negatively regulated following centriole duplication and if we could override this by stimulating the overexpression of PLK4. We accomplished this using a conditional Plk4 overexpression (OE) mouse model (Fig. 2A,B; see “Methods”) (Marthiens et al, 2013). An mCherry-Plk4 transgene driven by a CAG promoter was conditionally expressed when a stop cassette, flanked by loxP sites, was removed via Cre-mediated recombination (Fig. 2A). To stimulate overexpression following the first centriole duplication, we used the Hspa2 promoter-driven Cre recombinase, which is expressed during mid-prophase I in spermatocytes (Fig. 2B) (Inselman et al, 2010).
Figure 2. PLK4 OE leads to centriole overduplication by late prophase in mouse spermatocytes.
(A) Diagram of the Plk4 OE allele before and after Cre-mediated recombination. The non-excised allele harbors the CAG promoter (yellow box and arrow) upstream of two loxP sites (blue triangle), flanking a stop cassette (purple box). Further downstream is a non-endogenous copy of Plk4 fused with an mCherry tag (red and gray boxes). The stop codon was excised via Cre recombination, leaving a single loxP site and resulting in Plk4 OE. (B) The Hspa2 promoter used to drive Cre recombinase expression in the Plk4 OE mouse model is expressed in mid-prophase I at ~14 dpp. (C) Cryosections (16 µm thick) of an adult WT and Plk4 OE (26 weeks) mouse testis immunolabeled against PCNT (green) and mCherry (red) and stained with DAPI (blue). Zoomed images of mCherry and PCNT localization are shown to the right of their respective image. Scale bars = 50 µm. (D) Representative images of mid-prophase spermatocytes in control and Plk4 OE mice immunolabeled against SYCP3 (purple) and CETN3 (green) and stained with DAPI (gray inset in the upper right corner). The white arrowheads indicate the centrosome. Zoomed images of the centrioles are shown to the right of each image. Scale bars = 5 µm. (E) Quantification of CETN3 foci observed during early prophase I in both control and Plk4 OE spermatocytes. Immunolabeling was performed on three biological replicates with ≥36 spermatocytes quantified per replicate. The total number of cells quantified for control and Plk4 OE mice was 144 and 220, respectively. (F) Representative images of late prophase spermatocytes in control and Plk4 OE mice immunolabeled against SYCP3 (purple) and CETN3 (green) and stained with DAPI (gray inset in the upper right corner). The white arrowheads indicate the centrosome. Zoomed images of the centrioles are shown to the right of each image. Scale bars = 5 µm. (G) Representative images of metaphase I spermatocytes in control and Plk4 OE mice immunolabeled against α-tubulin (red) and CETN3 (green) and stained with DAPI (blue). The white arrowheads indicate a centrosome. Scale bars = 5 µm. (H) Diagram of the effect of Plk4 OE on centrioles during mammalian spermatogenesis. Plk4 OE spermatocytes underwent centriole overduplication in mid-to-late prophase, which led to pseudobipolar or multipolar spindle formation at metaphase I. Red rectangle with rounded corners = mature parent centriole, pink rectangle with rounded corners = immature parent centriole, green bar = maturation marker, yellow oval = PCM, purple oval = PLK4, gray rectangle with rounded corners = new centrioles from centriole duplication events, blue oval = DNA, red lines = microtubules. (I) Quantification of CETN3 foci per metaphase I spermatocyte. Immunolabeling was performed on three biological replicates with ≥23 spermatocytes quantified per replicate. The total number of cells quantified for control and Plk4 OE mice was 312 and 327, respectively. P value = <0.0001. (J) Quantification of the spindle orientation in metaphase I spermatocytes. Immunolabeling was performed on five biological replicates, with ≥19 spermatocytes quantified per replicate. The total number of cells quantified for control and Plk4 OE mice was 269 and 295, respectively. P value = <0.0001. (K) H&E staining of 5 µm thick testis sections of 95 dpp control or Plk4 OE mice. Scale bar = 50 µm. (L) Quantification of the average testis to body weight ratio of control and Plk4 OE mice. Measurements were performed using ≥3 mice for each age group. P values for 13 dpp, 25–27 dpp, and ≥162 dpp were 0.0751, 0.0009, and 0.0252, respectively. Data information: For all graphs (E, I, J, L), error bars show mean ± standard error of the mean (SEM). P values were obtained from two-tailed Student’s t test. n.s. (not significant), *P < 0.05 **P < 0.01, ***P < 0.0001, ****P < 0.0001.
Through immunohistochemistry of cryosectioned seminiferous tubules, we demonstrated that mCherry-PLK4 localized to the centrosome in spermatocytes (Fig. 2C), which complemented our observations of PLK4 localization in WT spermatocytes (Fig. 1A,B). From the assessment of centriole duplication during prophase I, we observed that the first round of centriole duplication was unaffected in the Plk4 OE spermatocytes, which was expected given that this process occurs before Hspa2-Cre recombinase expression (Fig. 2D,E) (Inselman et al, 2010). However, by mid-prophase I, after Hspa2-Cre recombinase expression occurs, centriole overduplication was observed in Plk4 OE spermatocytes (Fig. 2F). The majority of spermatocytes that progressed to metaphase I exhibited centriole overduplication, which was a rare occurrence to be observed in control metaphase I spermatocytes (Fig. 2G–I). Centriole numbers in Plk4 OE metaphase I spermatocytes ranged from 5 up to 29. Of these Plk4 OE metaphase I-staged spermatocytes, 23% contained multipolar spindles compared to 2% of spermatocytes with multipolar spindles observed in controls (Fig. 2J). From our analysis of histological preparations of adult Plk4 OE seminiferous tubule cross sections, we observed that the aberrancies in centriole number, spindle assembly, and chromosome alignment led to defects in spermatogenesis and germ cell depletion (Fig. 2K). Consequently, Plk4 OE mice had reduced testis to body weight ratios compared to controls (Fig. 2L). Furthermore, fertility tests demonstrated that Plk4 OE male mice were infertile (no litters were produced when four Plk4 OE males were each bred to two fertile WT females for a minimum of 8 weeks).
Taken together, these observations highlight several key points related to PLK4 function and regulation during male meiosis. First, PLK4 is a crucial regulator of centriole duplication during spermatogenesis, as Plk4 OE leads to centriole overduplication. Second, an unknown regulatory mechanism prevents PLK4 from triggering centriole overduplication in WT spermatocytes, which is overcome by an increase in PLK4 levels in the Plk4 OE model. Lastly, an excess of PLK4 alone was sufficient to induce extreme centriole overduplication without the need to increase the levels of other centriole components. This indicates that not only is PLK4 responsible for centriole duplication, but it is also the limiting factor of centriole duplication during spermatogenesis, which is also the case for mitotically dividing cells (Moyer et al, 2015; Holland et al, 2012).
Inhibition of PLK4 leads to centriole duplication failure
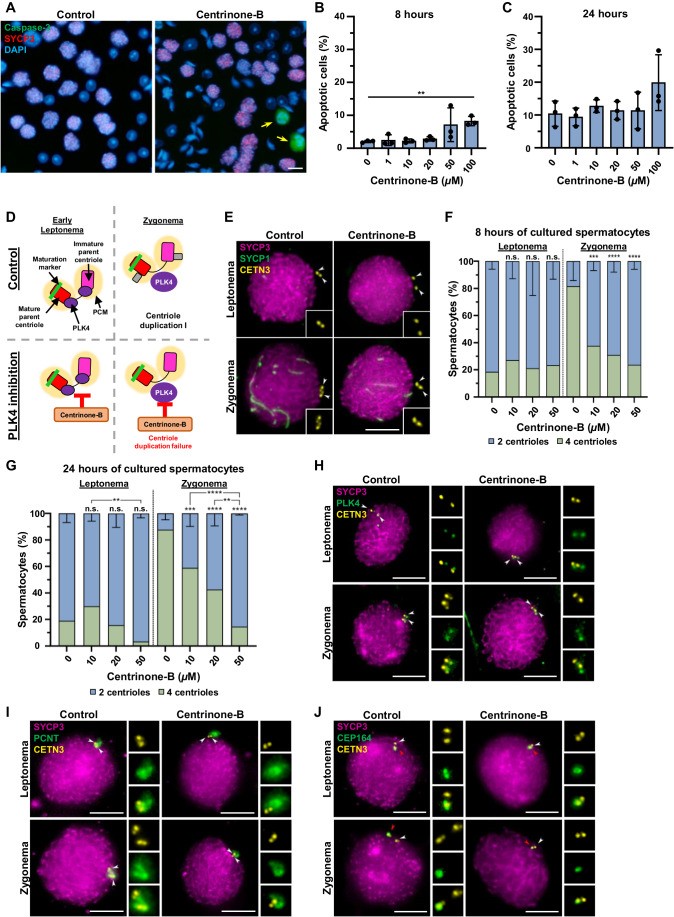
Having demonstrated that PLK4 overexpression during spermatogenesis promotes centriole amplification, we wanted to further examine the role of PLK4 by abrogating its function. It is known that the kinase activity of PLK4 is critical for centriole duplication in mitotically dividing cells (Holland and Cleveland, 2014; Holland et al, 2010). Therefore, we used a PLK4-specific kinase inhibitor, centrinone-B (Wong et al, 2015), on organotypic cultures of mouse seminiferous tubules to determine whether PLK4 kinase activity is required for the first meiotic centriole duplication event at leptonema. In somatic cell lines, the optimal, efficient concentration used was 500 nM (Tkach et al, 2022). However, higher concentrations of centrinone-B were required in our experiments because inhibition of PLK4 relied on media diffusion into fragments of seminiferous tubules that were embedded in agarose gel partially submerged in culture media (Sato et al, 2011; Alfaro et al, 2021). We tested increasing concentrations of centrinone-B from 10 to 100 µM for 8 and 24 h, first assessing cell viability (Fig. 3A–C). Cell viability was predominantly affected after 8 h of culture with 100 µM of centrinone-B, which was the highest concentration assessed (Fig. 3B). Next, we assessed centriole duplication using concentrations that did not significantly affect spermatocyte viability (0, 10, 20, and 50 µM) following 8 and 24 h of culture. Chromosome synapsis, interpreted by SYCP1 and SYCP3 localization, was used to determine spermatocyte prophase stages. We focused on centriole duplication during the first meiotic division, which occurs prior to the leptotene-to-zygotene transition (Wellard et al, 2021; Alfaro et al, 2021). Results revealed a significant reduction in the number of zygotene stage cells that successfully duplicated their centrioles after 8 or 24 h of 10–50 µM centrinone-B treatments (Fig. 3D–G). Moreover, the number of cells with non-duplicated centrioles increased in higher centrinone-B concentrations (Fig. 3F,G). As the 24-h treatment of 50 µM centrinone-B was the most effective for inhibiting centriole duplication in spermatocytes, all the subsequent analyses were performed using this culture condition.
Figure 3. PLK4 inhibition leads to centriole duplication failure in vitro.
(A) Representative images of control and centrinone-B-treated mouse spermatocytes immunolabelled against caspase-3 (green) and SYCP3 (red) and stained with DAPI (blue). Centrinone-B was added to spermatocytes for 8 h. The yellow arrows indicate apoptotic spermatocytes. Scale bars = 10 µm. (B) Quantification of apoptotic spermatocytes in cultures of seminiferous tubules after treatment of vehicle (control) or 10, 20, 50, or 100 μM of centrinone-B for 8 h. The percentage of apoptotic cells was calculated by counting spermatocytes, excluding Sertoli cells and spermatids. Data represents the average percentage of apoptotic spermatocytes for three biological replicates. P values for apoptotic cells treated for 8 h with 10, 20, 50, or 100 μM of centrinone-B compared to the control were 0.6966, 0.6686, 0.1256, 0.1602, and 0.0018, respectively. P values for apoptotic cells treated for 8 h with 10, 20, 50, or 100 μM of centrinone-B compared to the control were 0.7347, 0.4042, 0.7382, 0.0814, and 0.1539, respectively. (C) Quantification of apoptotic spermatocytes in cultures of seminiferous tubules after treatment of vehicle (control) or 10, 20, 50, or 100 μM of centrinone-B for 24 h. The percentage of apoptotic cells was calculated by counting spermatocytes, excluding Sertoli cells and spermatids. Data represents the average percentage of apoptotic spermatocytes for three biological replicates. (D) Diagram illustrating how the inhibition of PLK4 in vitro by centrinone-B leads to centriole duplication failure during early prophase. Red rectangle with rounded corners = mature parent centriole, pink rectangle with rounded corners = immature parent centriole, green bar = maturation marker, yellow oval = PCM, purple oval = PLK4, gray rectangle with rounded corners = new centrioles from first centriole duplication, orange box = centrinone-B, red T shape = indicates inhibition. (E) Representative images of mouse spermatocytes under control and 50 µM centrinone-B treatment conditions for 24 h. Spermatocytes were immunolabeled against SYCP1 (green), SYCP3 (magenta), and CETN3 (yellow). The white arrowheads indicate the centrioles. Zoomed images of the centrioles are inset on each corresponding image. Scale bar = 10 µm. (F) Quantification of CETN3 foci observed during leptonema and zygonema in spermatocytes under control and centrinone-B treatment conditions. Immunolabeling was conducted in three different biological replicates and with increasing concentrations of centrinone-B (10, 20, and 50 µM) for 8 h. P values for leptotene stage spermatocytes treated for 8 h with 10, 20, and 50 µM of centrinone-B compared to the control were 0.909, 0.9969, and 0.9802, respectively. P values for zygotene stage spermatocytes treated for 8 h with 10, 20, and 50 µM of centrinone-B compared to the control were 0.0002, <0.0001, and <0.0001, respectively. (G) Quantification of CETN3 foci observed during leptonema and zygonema in spermatocytes under control and centrinone-B treatment conditions. Immunolabeling was conducted in three different biological replicates and with increasing concentrations of centrinone-B (10, 20, and 50 µM) for 24 h. P values for leptotene stage spermatocytes treated for 24 h with 10, 20, and 50 µM of centrinone-B compared to the control were 0.2775, 0.9424, and 0.0753, respectively. P values for zygotene stage spermatocytes treated for 24 h with 10, 20, and 50 µM of centrinone-B compared to the control were 0.0639, <0.0001, and 0.0012, respectively. (H) Representative images of mouse spermatocytes under control and 50 µM centrinone-B treatment conditions for 24 h. Spermatocytes were immunolabeled against PLK4 (green), SYCP3 (magenta), and CETN3 (yellow). The white arrowheads indicate the centrioles. Zoomed images of the centrioles are shown to the right of each corresponding image. Scale bars = 10 µm. (I) Representative image of mouse spermatocytes under control and 50 µM centrinone-B treatment conditions for 24 h. Spermatocytes were immunolabeled against PCNT (green), SYCP3 (magenta), and CETN3 (yellow). The white arrowheads indicate the centrioles. Zoomed images of the centrioles are shown to the right of each corresponding image. Scale bars = 10 µm. (J) Representative images of mouse spermatocytes under control and 50 µM centrinone-B treatment conditions for 24 h. Spermatocytes were immunolabeled against CEP164 (green), SYCP3 (magenta), and CETN3 (yellow). The white arrowheads indicate the centrioles. The red arrowheads indicate the mature centrioles. Zoomed images of the centrioles are shown to the right of each corresponding image. Scale bars = 10 µm. Data information: For all graphs (B, C, F, G), bars and error bars represent mean ± standard deviation (SD). For graphs (B, C), P values were obtained from one-way ANOVA with Holm–Sidak’s comparisons test. For graphs (F, G), P values were obtained from two-way ANOVA with Turkey’s multiple comparisons test. n.s. (not significant), **P < 0.01, ***P < 0.001, ****P < 0.0001.
Next, we assessed PLK4 localization to address whether centrinone-B treatment affected PLK4 distribution and its kinase function. We hypothesized that PLK4 localization would not be affected based on observations previously reported for somatic cells (Wong et al, 2015). For this, we analyzed PLK4, synaptonemal complex formation (SYCP3), and centrioles (CENT3) and determined that PLK4 was detected as a small signal near each centriole (Fig. 3H). This PLK4 distribution was identical regardless of whether centriole duplication had occurred or not in both untreated and centrinone-B-treated spermatocytes. Furthermore, the PLK4 localization pattern observed in these assays corroborates with the localization data shown in Fig. 1H.
Because centrinone-B treatment inhibited centriole duplication, we then asked whether this could also alter centrosome integrity. We did not observe any alterations in the PCM revealed by pericentrin (PCNT) immunostaining in treated spermatocytes compared to the control (Fig. 3I). In addition, we studied CEP164, a marker for the mature parent centriole in mouse spermatocytes (Wellard et al, 2021; López-Jiménez et al, 2022). No alteration of CEP164 distribution was observed, as the protein was located at the mature parent centriole before and after centriole duplication in control and with unduplicated centrioles following centrinone-B treatment (Fig. 3J).
Collectively, these data demonstrated that PLK4 is the master regulator of centriole duplication during meiosis I in mouse spermatocytes, and its kinase activity is critical for this process. These observations align well with what has been reported for mitotically dividing cells (Neitzel et al, 2022). However, there are differences between spermatocytes and mitotic cells regarding PLK4 levels, localization pattern, and centriole duplication, where PLK4 remains stable and localizes to the centrosome throughout spermatogenesis, and centrioles duplicate twice during meiosis (Fig. 1F). While pleiotropic effects of centrinone-B cannot be fully excluded, the culture conditions and drug treatment used for PLK4 inhibition did not significantly compromise cell viability and showed no effect on chromosome synapsis, centrosome integrity, or PLK4 localization in spermatocytes. Therefore, to further our understanding of PLK4 function during spermatogenesis, we next performed in vivo studies using knockout mice.
Conditional mutation of Plk4 causes male infertility
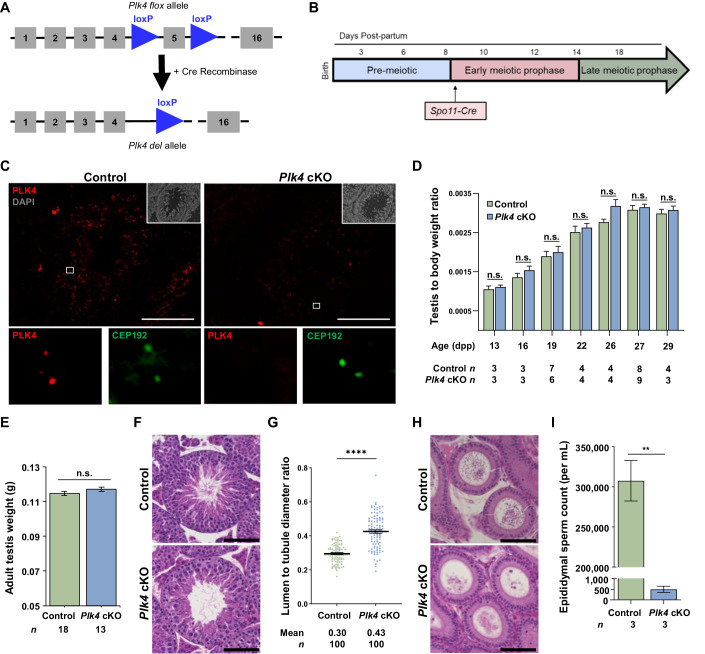
PLK4 is essential during embryonic development (Harris et al, 2011; Rosario et al, 2010). Therefore, to assess the effects of PLK4 depletion on centriole duplication and avoid embryonic lethality, we used a conditional knockout (cKO) allele for Plk4 that harbors loxP sites that flank exon 5, which encodes the kinase domain of PLK4 (LoMastro et al, 2022). To excise the floxed region of Plk4, we first tested the Stra8-Cre recombinase that conditionally knocked out Plk4 in spermatogonia (Sadate-Ngatchou et al, 2008). However, the germ cell population was depleted prior to meiotic entry, leading to a 2.7-fold reduction in the testis to body weight ratio in adult mice. This has also been reported for cKOs of other essential kinases when using the Stra8-Cre transgene, including PLK1, Aurora A, and Aurora B (Wellard et al, 2020, 2021). For the remainder of the study, we utilized the Spo11-Cre recombinase transgene, which expresses Cre recombinase in spermatocytes at 10 days post-partum (dpp), corresponding to the preleptotene/leptotene stage of meiosis I (Lyndaker et al, 2013; Wellard et al, 2020; Hwang et al, 2018a). Mice homozygous for the Plk4 flox allele and hemizygous for the Spo11-Cre transgene (Plk4 flox/flox, Spo11-Cre tg/0) were termed Plk4 cKO (see Fig. 4A,B and “Methods”).
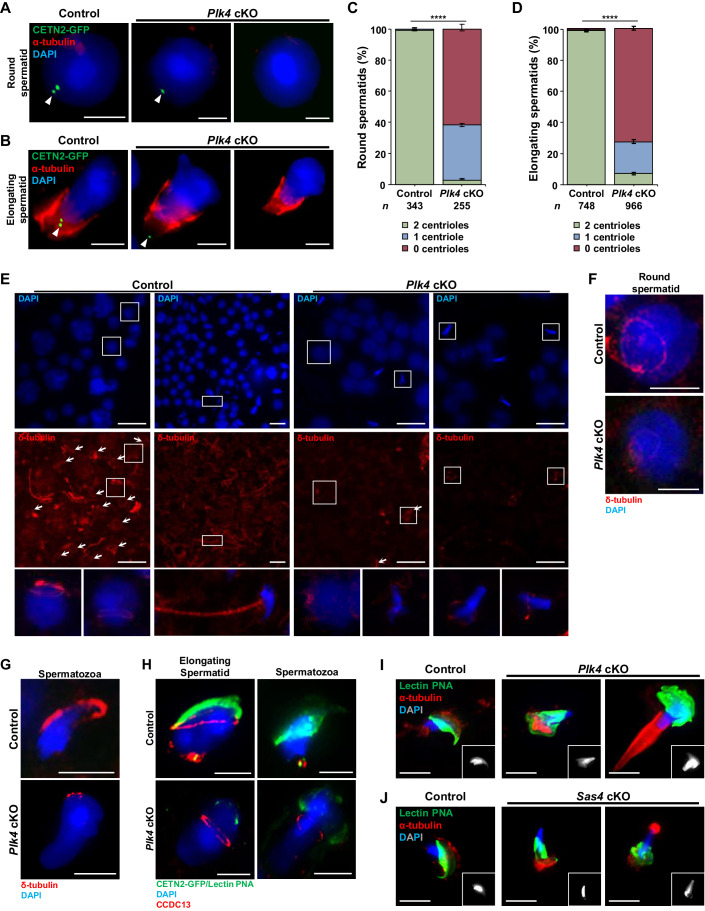
Figure 4. Basic characterization of the testes and epididymides from Plk4 cKO mice.
(A) Diagram of the Plk4 cKO allele before and after Cre-mediated recombination. The Plk4 flox allele harbors two loxP sites (blue triangle) flanking exon 5 (gray box). Excision of exon 5 by Cre recombinase results in the Plk4 deletion allele (Plk4 del). (B) The Spo11 promoter used to drive Cre recombinase expression in the Plk4 cKO mouse model is expressed in early meiotic prophase at ~9 dpp. (C) Cryosections (16 µm thick) of adult control and Plk4 cKO (26 weeks) mouse testis immunolabeled against PLK4 (red) and stained with DAPI (gray inset in the upper right corner). Zoomed images of PLK4 and CEP192 (green) localization are shown below each corresponding image. Scale bars = 100 µm. (D) Quantification of the average testis to body weight ratio of control and Plk4 cKO mice. Measurements were performed using ≥3 mice for 13, 16, 19, 22, 26, 27, and 29 dpp. P values for 13 dpp, 16 dpp, 19, 22, 26, and 29 dpp were 0.5928, 0.2907, 0.6027, 0.5581, 0.0817, and 0.5912, respectively. (E) Quantification of the average testis to body weight ratio of control and Plk4 cKO mice. Measurements were performed using ≥3 adult mice ages 60 to 173 dpp. P value = 0.47. (F) H&E staining of 5 µm thick testis sections of 65 dpp control and Plk4 cKO mice. Scale bar = 50 µm. (G) Quantification of the average lumen diameter to tubule diameter ratio in control and Plk4 cKO mice. The average tubule diameter was 412.7 µm and 422.0 µm in control and Plk4 cKO mice, respectively. The average tubule lumen diameter was 122.6 µm and 180.5 µm in control and Plk4 cKO mice, respectively. Quantification was performed in three biological replicates with ≥33 tubules quantified per replicate. The total number of tubules measured for the control and Plk4 cKO was 100 each. P value = <0.0001. (H) H&E staining of 5 µm thick epididymide sections of 95 dpp control and Plk4 cKO mice. Scale bar = 50 µm. (I) Epididymal sperm count in control and Plk4 cKO mice, respectively. The average sperm count was 307,500 sperm/mL and 500 sperm/mL in control and Plk4 cKO epididymides, respectively. Quantification was performed using three biological replicates. P value = 0.0067. Data information: For all graphs (D, E, G, I), error bars show mean ± SEM. P values were obtained from two-tailed Student’s t test. n.s. (not significant), **P < 0.01, and ****P < 0.0001.
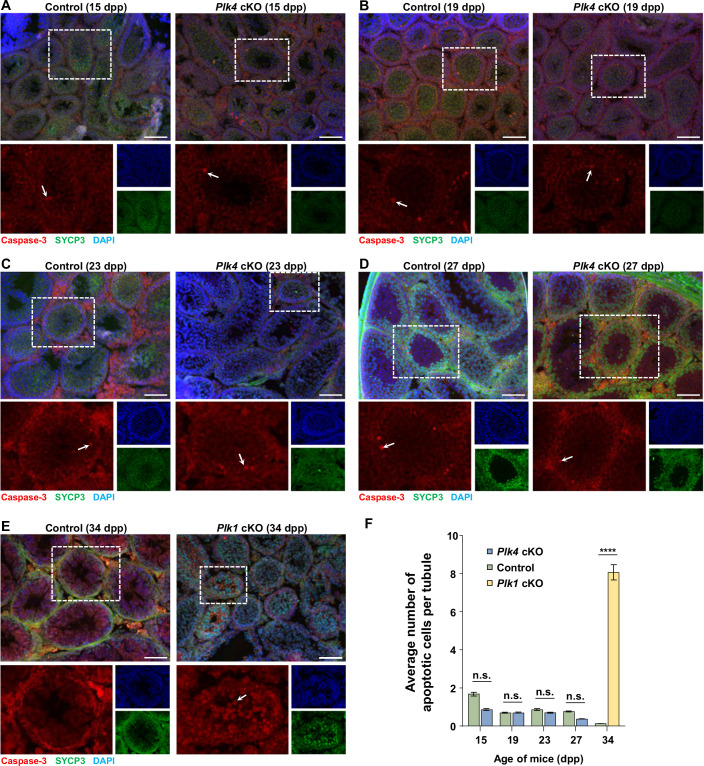
Immunostaining of testis sections showed that the PLK4 signal was not detected at the centrosome in Plk4 cKO testicular cells (Fig. 4C). There was no significant difference in the testis to body weight ratios of control and Plk4 cKO mice ranging from 13 to 29 days post-partum (dpp) or adults (Fig. 4D,E). Furthermore, based on the assessment of cleaved caspase-3, we did not observe a significant difference in the incidence of apoptosis between control and Plk4 cKO (Fig. EV1A–F). However, Plk4 cKO mice were infertile (no litters were produced when four Plk4 cKO males were each bred to two fertile WT females for a minimum of 8 weeks). Assessment of hematoxylin and eosin (H&E) stained testis cross sections revealed that Plk4 cKO mouse testes had a significantly larger lumen to seminiferous tubule diameter ratio than the control testes (Fig. 4F,G). The larger lumen diameter was because the Plk4 cKO spermatozoa lacked flagella (Fig. 4F). Assessment of epididymitis cross sections indicated that there were very few spermatozoa detected for the Plk4 cKO, and those that were present appeared abnormal compared to the control (Fig. 4H). Epidydimal sperm counts revealed that Plk4 cKOs had approximately 500 times fewer spermatozoa than control mice (Fig. 4I).
Figure EV1. Assessment of apoptosis during spermatocytes.
(A–D) Cryosections (16 µm thick) of control and Plk4 cKO mouse testis at 15 (A), 19 (B), 23 (C), and 27 (D) dpp, immunolabeled against SYCP3 (green), caspase-3 (red) and stained with DAPI (blue). Zoomed images of individual tubules are shown directly below the corresponding images. White arrows indicate an example of an apoptotic spermatocyte. Scale bar = 100 µm. (E) The Plk1 cKO (Plk1 flox/flox, Spo11-Cre tg/0) mouse model has been reported previously to exhibit increased levels of apoptosis in primary spermatocytes and was used as a positive control (Wellard et al, 2021). Cryosections (16 µm thick) of control and Plk1 cKO (34 dpp) mouse testis immunolabeled against SYCP3 (green), caspase-3 (red) and stained with DAPI (blue). Zoomed images of individual tubules are shown directly below the corresponding images. White arrows indicate an example of an apoptotic spermatocyte. Scale bar = 100 µm. (F) Quantification of apoptotic spermatocytes in control, Plk4 cKO and Plk1 cKO spermatocytes. Quantification was performed in ≥26 tubules. Error bars show mean ± SEM. P values were obtained from two-tailed Student’s t test. n.s. (not significant) and ****P < 0.0001. P value = <0.0001.
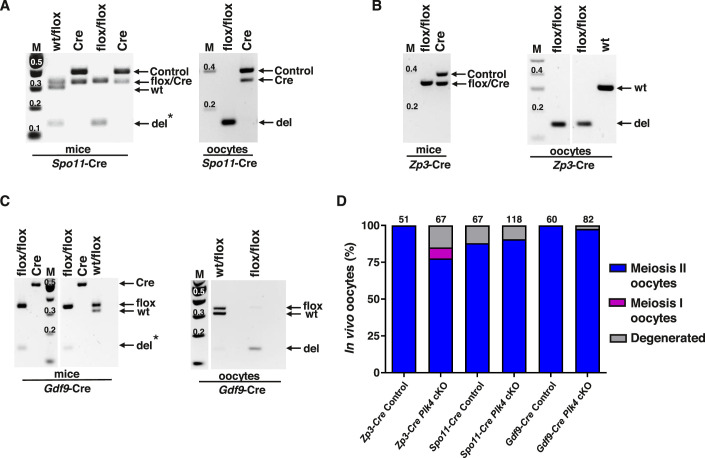
To determine whether PLK4 is required for oogenesis, we excised the floxed Plk4 allele in preleptotene oocytes, primordial follicles, and primary follicles with Spo11-Cre, Gdf9-Cre, and Zp3-Cre, respectively (Lan et al, 2004; Lyndaker et al, 2013; Wellard et al, 2020; Hwang et al, 2017). Unlike what was observed in males, all three Plk4 cKO female mouse models were fertile (four Plk4 cKO females for each Cre driver were bred to fertile WT males, and all produced three litters of 5–8 pups over a 12-week period). Genotyping of isolated oocytes demonstrated that the Plk4 flox allele was successfully excised when Cre recombinase was expressed (Fig. EV2A–C). Nevertheless, Plk4 cKO oocytes successfully formed bipolar spindles and showed no delay or error in their ability to progress from the germinal vesicle (GV) stage to the metaphase II (MII) arrest stage when compared to controls (Fig. EV2D). These assessments indicate that PLK4 is not essential for chromosome segregation in oocytes or female fertility.
Figure EV2. Assessment of Plk4 cKO female mice.
(A–C) PCR analysis of mouse and oocyte DNA showing the absence of the flox allele and presence of the del allele in oocytes after the expression of Cre recombinase driven by the Spo11 (A), Zp3 (B), and Gdf9 (C) promoters. WT Plk4 band 307 bp, flox floxed Plk4 band 341 bp, del deletion band 143 bp, M DNA marker, kb kilobases, Cre Cre recombinase band 355 bp for Spo11-Cre and Zp3-Cre, and 500 bp for Gdf9-Cre, Cont internal control band for Spo11-Cre and Zp3-Cre recombinases 420 bp. *Non-tissue-specific Cre recombinase expression. (D) Oocytes from all Plk4 cKO mice successfully progressed to MII stage in vivo. Mouse age 45-77 days. Oocytes from 3 superovulated mice were analyzed for each genotype. The total number of oocytes quantified from Zp3-cre control, Zp3-cre Plk4 cKO, Spo11-cre control, Spo11-cre Plk4 cKO, Gdf9-cre control and Gdf9-cre Plk4 cKO mice were 51, 67, 67, 118, 60, and 82, respectively.
Unlike spermatocytes, oocytes do not develop flagella, which requires centrioles during formation (Nigg and Raff, 2009). In addition, the spindle assembly and organization processes during meiotic divisions vary between spermatogenesis and oogenesis. While spermatocytes use a canonical centrosome containing two centrioles for spindle assembly and organization, oocytes lack centrioles (Manandhar et al, 2005). Instead, oocytes rely on forming acentriolar microtubule organizing centers (aMTOCs) that consist of PCM components and liquid-like spindle domain proteins, which are critical for successful chromosome segregation following meiotic resumption (So et al, 2019). Thus, we postulate that PLK4 depletion in oocytes does not have the same effects on fertility as observed in spermatocytes due to the sexually dimorphic nature of MTOCs.
Conditional mutation of Plk4 in primary spermatocytes leads to centriole duplication and maturation failure
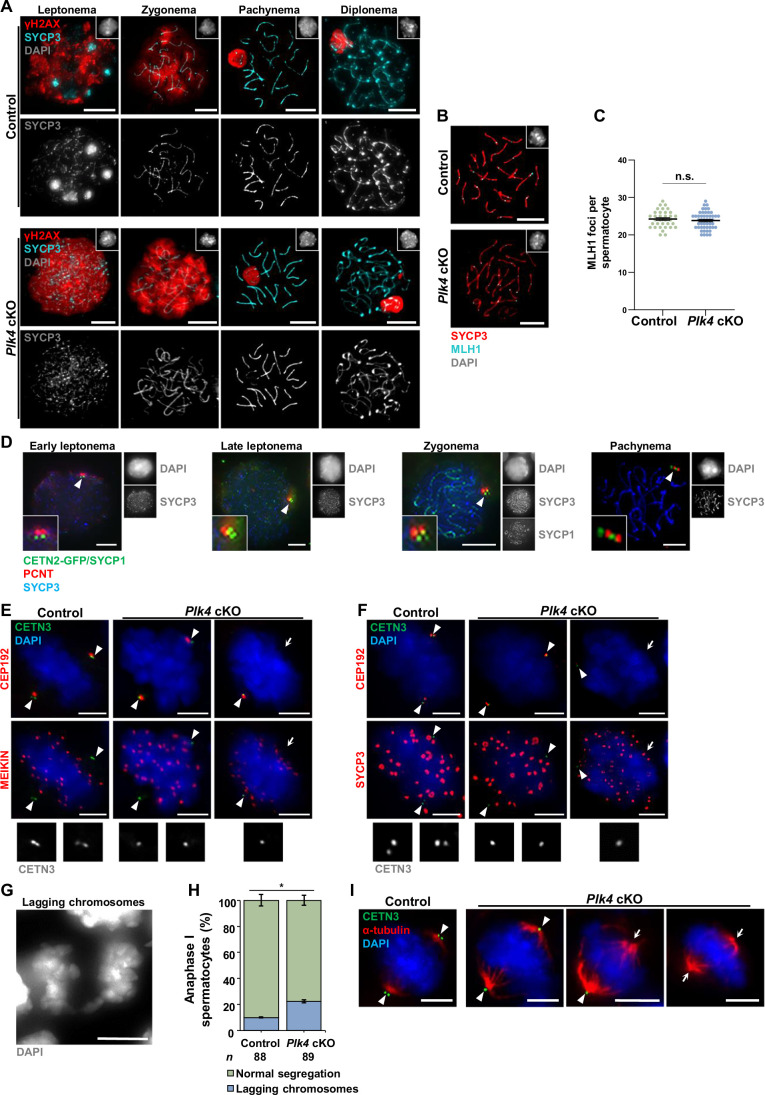
As the floxed region of the Plk4 cKO allele was excised during the preleptotene/leptotene stage of meiosis I via Spo11-Cre transgene expression, we closely assessed features of meiotic prophase progression. A previous report showed that a gene-trap mutation of Cep63, which encodes for a pericentriolar component, resulted in homologous recombination and chromosome synapsis defects that led to germ cell apoptosis during meiotic prophase (Marjanović et al, 2015). In contrast, we observed no apparent difference in double-strand break formation, repair, or sex body formation when comparing control and Plk4 cKO prophase I spermatocyte chromatin spreads immunolabelled for γH2AX (Fig. EV3A). Axial element formation, homologous chromosome synapsis, and desynapsis in Plk4 cKO spermatocytes were equivalent to that of the control (Fig. EV3A). In addition, we found no difference in MLH1 foci, which mark the majority (90–95%) of crossover recombination sites, in pachytene stage spermatocytes (Fig. EV3B,C) (Anderson et al, 1999). Lack of meiotic recombination and synapsis defects (Fig. EV3A–C) or increased germ cell apoptosis (Fig. EV1) differs from what was reported for mice harboring the Cep63 gene-trap allele (Marjanović et al, 2015). This may be explained by the fact that the Cep63 gene-trap mutation was present within all mouse cells, whereas the Plk4 cKO only occurred in primary spermatocytes. For instance, the Cep63 gene-trap mutation leads to microcephaly (Marjanović et al, 2015), which could affect the regulation of the hypothalamus–pituitary gonadal axis (Cerbone et al, 2020).
Figure EV3. Additional characterization of meiotic progression and centriole biogenesis during prophase I to metaphase II.
(A) Prophase I control and Plk4 cKO spermatocytes from 14 and 20 dpp mice were immunolabeled against SYCP3 (cyan), γ-tubulin (red), and stained with DAPI (gray inset in the upper right corner). Zoomed images of the centrioles are inset on the corresponding images. Scale bars = 10 µm. (B) Representative images of mid-prophase I spermatocytes in 20 dpp control and Plk4 cKO mice immunolabeled against SYCP3 (red), MLH1 (cyan), and stained with DAPI (gray inset in the upper right corner). Scale bars = 10 µm. (C) Quantification of MLH1 foci observed along SYCP3 stretches during pachynema in both control and Plk4 cKO spermatocytes. Immunolabeling was performed on 3 biological replicates with ≥ 10 spermatocytes quantified per replicate. The total number of cells quantified for control, and Plk4 cKO mice were 34 and 49, respectively. P value = 0.4386. (D) Representative images of early leptotene stage, late leptotene stage, zygotene stage, and pachytene stage control spermatocytes expressing CETN2-GFP (green) from 10 dpp mice immunolabeled against SYCP1 (green), PCNT (red), SYCP3 (blue), and stained with DAPI (gray outset). The white arrowheads indicate the centrosome. Individual gray outset channel of SYCP3 and SYCP1 are displayed to the right of the corresponding images. Zoomed images of the centrioles and inset on corresponding images. Scale bars = 5 µm. (E) Characterization of metaphase I compared to metaphase II spermatocytes by observing DNA size (stained with DAPI) was confirmed with MEIKIN and SYCP3 immunostaining. Both SYCP3 and MEIKIN staining are present in metaphase I spermatocytes, but no longer in metaphase II spermatocytes (Kim et al, 2015). Representative image of metaphase I control and Plk4 cKO spermatocytes from 27 dpp mice immunolabeled against CETN3 (green), MEIKIN (red) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate the ncMTOC. Zoomed images of the centrioles (gray) are displayed below the corresponding images. Scale bars = 5 µm. (F) Representative image of metaphase I control and Plk4 cKO spermatocytes harboring the CETN2-GFP transgene (green) from 27 dpp mice immunolabeled against SYCP3 (red) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate the ncMTOC. Zoomed images of the centrioles (gray) are displayed below the corresponding images. Scale bars = 5 µm. (G) Representative image of chromosome missegregation event during metaphase I stained with DAPI. Scale bars = 5 µm. (H) Quantification of lagging chromosomes in anaphase I spermatocytes. Immunolabeling was performed on 5 biological replicates with ≥ 10 spermatocytes quantified per replicate. The total number of cells quantified for control and Plk4 cKO mice were 89 and 89, respectively. P value = 0.0278. (I) Representative images of metaphase II spermatocytes in 27 dpp control and Plk4 cKO mice expressing CETN2-GFP (green) immunolabeled against α-tubulin (red) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate the ncMTOC. Scale bars = 5 µm. Data information: For all graphs (C, H), error bars show mean ± SEM. P values were obtained from two-tailed Student’s t test. n.s (not significant), *P < 0.05.
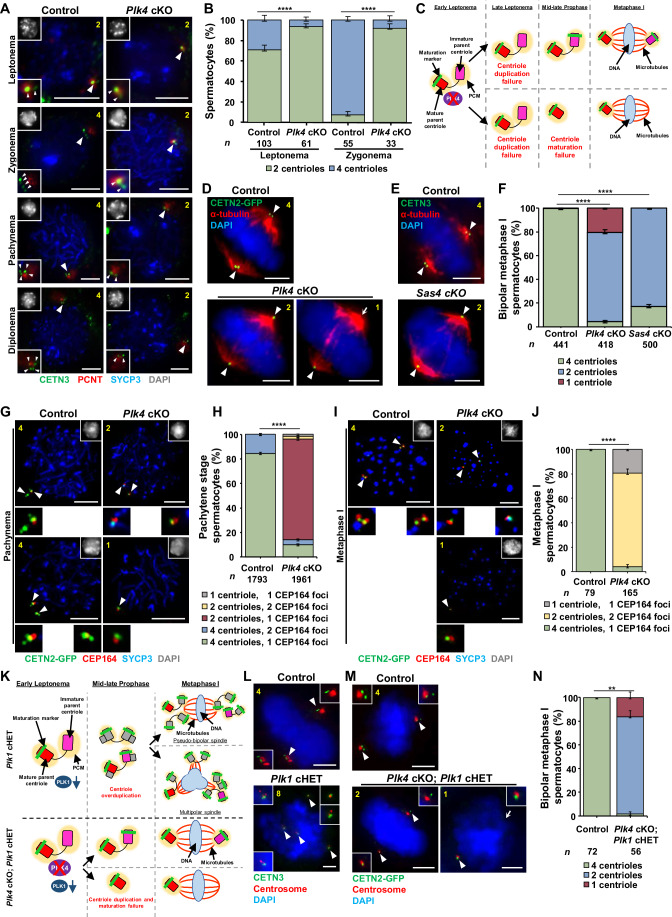
We next sought to determine whether PLK4 depletion specifically affected centriole biogenesis in spermatocytes. We first assessed centriole numbers during meiotic prophase in control spermatocytes. We assessed centrioles using a CETN3 antibody or via expression of CETN2-GFP (Higginbotham et al, 2004). We confirmed that centriole duplication occurred normally in spermatocytes from mice harboring the Cetn2-Gfp transgene (Fig. EV3D). In agreement with previous reports (Wellard et al, 2021; Alfaro et al, 2021), control spermatocytes entered meiosis with two centrioles and underwent centriole duplication in late leptonema (Fig. 5A,B). Control spermatocytes harbored four centrioles throughout the subsequent sub-stages of meiosis I (Fig. 5A,B). While Plk4 cKO spermatocytes entered meiosis at leptonema with two centrioles like control spermatocytes, the majority of them failed to undergo centriole duplication during late leptonema, and by early zygonema, spermatocytes continued to harbor only two centrioles (Fig. 5A,B). At metaphase I, 75.4% of spermatocytes contained only two centrioles (Fig. 5C–F). In addition, it was observed that 20.3% of Plk4 cKO spermatocytes in metaphase I harbored only one centriole (Figs. 5C–F and EV3E,F). Centriole duplication failure during meiotic prophase was not completely penetrant in the Plk4 cKO. We hypothesized that this was due to the overlap in timing of the Spo11-Cre expression and the first round of centriole duplication during spermatogenesis. Nevertheless, the Plk4 cKO centriole biogenesis defects were pronounced and significantly discernable from the control.
Figure 5. Assessment of centriole duplication and maturation in Plk4 cKO mice.
(A) Prophase I control and Plk4 cKO spermatocytes from 13 to 27 dpp mice were immunolabeled against CETN3 (green), PCNT (red), and SYCP3 (blue) and stained with DAPI (gray inset in upper left corner). The white arrowheads indicate the centrosome. Zoomed images of the centrioles are inset on the corresponding images. White arrowheads indicate the centrosome. The yellow number indicates the total number of centrioles per cell. Scale bars = 5 µm. (B) Quantification of centriole foci observed during leptonema and zygonema in both control and Plk4 cKO spermatocytes. Immunolabeling was performed on three biological replicates with ≥10 spermatocytes quantified per replicate. The total number of cell quantified for control and Plk4 cKO mice were 158 and 99, respectively. P values for leptonema and zygonema were XXX and XXXX, respectively. (C) Diagram illustrating how Plk4 cKO leads to both centriole duplication and maturation failure. As a result, metaphase I spermatocytes were observed to harbor either one centriole at each bipolar spindle pole, or a single centriole at one of the two bipolar spindle poles. Red rectangle with rounded corners = mature parent centriole, pink rectangle with rounded corners = immature parent centriole, green bar = maturation marker, yellow oval = PCM, purple oval = PLK4, red X = indicates depletion, blue oval = DNA, red lines = microtubules. (D) Metaphase I control and Plk4 cKO spermatocytes harboring CETN2-GFP (green) were immunolabeled against α-tubulin (red) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrow indicates the acentriolar spindle pole. The yellow number indicates the total centrioles per cell. Scale bars = 5 µm. (E) Metaphase I control and Sas4 cKO spermatocytes were immunolabeled against CETN3 (green), α-tubulin (red) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The yellow number indicates the total centrioles per cell. Scale bars = 5 µm. (F) Quantification of centrin foci per control, Plk4 cKO, and Sas4 cKO metaphase I spermatocytes. Immunolabeling was performed on three biological replicates, with ≥100 spermatocytes quantified per replicate. The total number of cells quantified for control, Plk4 cKO, and Sas4 cKO mice was 441, 418, and 500, respectively. P values for Plk4 cKO and Sas4 cKO were <0.0001 and <0.0001, respectively. (G) Pachytene stage control and Plk4 cKO spermatocytes harboring CETN2-GFP (green) were immunolabeled against CEP164 (red), SYCP3 (blue) and stained with DAPI (gray inset in the upper right corner) Zoomed images of the centrioles are shown below the corresponding images. The white arrowheads indicate the centrosome. The yellow number indicates the total centrioles per cell. Scale bars = 5 µm. (H) Quantification and localization of CEP164 foci in relation to centrioles in control and Plk4 cKO pachytene stage spermatocytes. Immunolabeling was performed on three biological replicates with ≥323 spermatocytes quantified per replicate. The total number of cells quantified for control, and Plk4 cKO mice were 1793 and 1961, respectively. P value = <0.0001. (I) Metaphase I stage control and Plk4 cKO spermatocytes expressing CETN2-GFP were immunolabeled against CEP164 (red) and SYCP3 (blue) and stained with DAPI (gray inset in the upper right corner). Zoomed images of the centrioles are shown below the corresponding images. The white arrowheads indicate the centrosome. The yellow number indicates the total number of centrioles per cell. Scale bars = 5 µm. (J) Quantification and localization of CEP164 foci in relation to centrioles in control and Plk4 cKO metaphase I spermatocytes. Immunolabeling was performed on three biological replicates, with ≥12 spermatocytes quantified per replicate. The total number of cells quantified for control and Plk4 cKO mice was 79 and 165, respectively. P value = <0.0001. (K) Diagram comparing centriole duplication in Plk1 cHET and Plk4 cKO; Plk1 cHET spermatocytes. Plk1 cHET spermatocytes undergo centriole overduplication, which leads to metaphase I spermatocytes with either pseudobipolar or multipolar spindle poles. In contrast, Plk4 cKO; Plk1 cHET spermatocytes undergo centriole duplication and maturation failure, in line with what is observed in Plk4 cKO spermatocytes. Plk4 cKO; Plk1 cHET metaphase I spermatocytes harbor either one centriole at each bipolar spindle pole, or a single centriole at one of the two bipolar spindle poles. Red rectangle with rounded corners = mature parent centriole, pink rectangle with rounded corners = immature parent centriole, green bar = maturation marker, yellow oval = PCM, purple oval = PLK4, red X = indicates depletion, teal oval = PLK1, teal arrow = indicates reduction, gray rectangle with rounded corners = new centrioles from first centriole duplication, blue oval = DNA, red lines = microtubules. (L) Metaphase I control and Plk1 cHET spermatocytes were immunolabeled against CETN3 (green) and CEP164 (red) and stained with DAPI (blue). Zoomed images of the centrioles are inset on the corresponding images. The white arrowheads indicate the centrosome. The yellow number indicates the total number of centrioles per cell. Scale bars = 5 µm. (M) Metaphase I control and Plk4 cKO; Plk1 cHET spermatocytes were immunolabeled against CETN3 (green) and CEP192 (red), and DNA was stained with DAPI (blue). Zoomed images of the centrioles are inset on the corresponding images. The white arrowheads indicate the centrosome. The white arrow indicates the acentriolar spindle pole. The yellow number indicates the total number of centrioles per cell. Scale bars = 5 µm. (N) Quantification of centrin foci per control and Plk4 cKO; Plk1 cHET metaphase I spermatocytes. Immunolabeling was performed on 3 biological replicates with ≥15 spermatocytes quantified per replicate. The total number of cells quantified for control and Plk4 cKO; Plk1 cHET mice was 72, and 56, respectively. P value = 0.002. Data information: For all graphs (B, F, H, J, N), error bars show mean ± SEM. P values were obtained from two-tailed Student’s t test. **P < 0.01, ****P < 0.0001.
Based on research with mammalian cell lines, centrioles that fail to undergo maturation cannot serve as an independent MTOC, become unstable, and are disassembled (Wang et al, 2017; Karasu et al, 2022). Therefore, we hypothesized that the depletion of PLK4 caused a proportion of the centrioles to remain immature and degenerate (Fig. 5C). To further examine this hypothesis, we probed for CEP164, a distal appendage component of the centriole that associates with centrioles during maturation (Nigg and Holland, 2018). WT spermatocytes enter meiosis with two centrioles, the mature parent centriole that harbors distal and subdistal appendages and the immature parent centriole that does not contain any of these maturation markers (Wellard et al, 2021; López-Jiménez et al, 2022). At pachynema, cells with a single maturation marker colocalizing with the mature parent centriole and cells with two maturation markers colocalizing with both parent centrioles were observed (Fig. 5G,H). By metaphase I, CEP164 colocalized with all four centrioles in control spermatocytes (Fig. 5I,J). These results correspond with our previous characterization of centriole maturation (Wellard et al, 2021; López-Jiménez et al, 2022). Analysis of Plk4 cKO spermatocytes at pachynema and metaphase I indicated that a population of spermatocytes had delayed and defective centriole maturation, which led to the degeneration of the immature parent centriole (Fig. 5G–J).
We next assessed whether the centriole maturation defect and subsequent centriole loss were due to the inability to build new centrioles or specifically due to the loss of PLK4. To address this, we utilized male mice that harbored a Sas4 (CenpJ) cKO allele (Sas4 flox/flox, Spo11-Cre tg/0; termed Sas4 cKO) (Bazzi and Anderson, 2014). SAS4 is a structural component of the centriole and is required during the assembly of new centrioles (McIntyre et al, 2012; Leidel and Gönczy, 2003). In Sas4 cKO mice, 82.8% of spermatocytes failed to duplicate centrioles during meiotic prophase, leading to the formation of spermatocytes containing only two centrioles (Fig. 5E,F). However, we never observed Sas4 cKO spermatocytes that only had a single centriole. This suggests that centrioles still undergo centriole maturation despite centriole duplication failure. Taken together, these results indicate that PLK4 is directly responsible for centriole duplication and maturation processes during spermatogenesis.
PLK4 functions upstream of PLK1 during centriole duplication
We had previously observed that another PLK family member, PLK1, was essential for centrosome biogenesis (Wellard et al, 2021). Conditional heterozygous (cHet) mutation of Plk1 resulted in more than one round of centriole duplication during meiotic prophase (Fig. 5K,L). This indicates that PLK1 is also important for regulating centriole duplication during spermatogenesis. To determine whether the centriole overduplication phenotype in Plk1 cHet spermatocytes is dependent on PLK4 function, we created mutant mice that harbored compound mutations of Plk1 cHet and Plk4 cKO (Plk4 flox/flox, Plk1 +/flox, Spo11-cre tg/0; termed Plk4 cKO; Plk1 cHET). We assessed meiosis I spermatocytes and observed that centriole duplication was blocked in Plk4 cKO; Plk1 cHET mice (Fig. 5K–N). This demonstrated that PLK4 function is required to stimulate centriole amplification when PLK1 levels are reduced. Thus, PLK4 functions upstream of PLK1, and PLK4 is the master regulator of centriole biogenesis during mammalian spermatogenesis.
Non-centriolar spindle poles form in Plk4 cKO spermatocytes during meiosis I
Despite Plk4 cKO spermatocytes failure to duplicate and mature centrioles during meiotic prophase, they still progressed to form a bipolar spindle at metaphase I (Fig. 5D,F). This was intriguing because centrioles are a core centrosome component, and the centrosome is the primary MTOC in spermatocytes (Chemes, 2012). However, there are other cases where the centrioles are not required for bipolar spindle pole formation in mammalian cells (Meunier and Vernos, 2016; So et al, 2019; Coquand et al, 2021). For instance, oocytes rely on aMTOCs, which comprise PCM components that coalesce to organize the metaphase I and II bipolar spindles (So et al, 2019). Based on this alternative method of spindle organization, we hypothesized that spermatocytes may use a similar aMTOC mechanism as a backup to ensure that chromosome segregation successfully takes place. Therefore, we assessed the localization of PCM proteins that are known to be components of aMTOCs in oocytes.
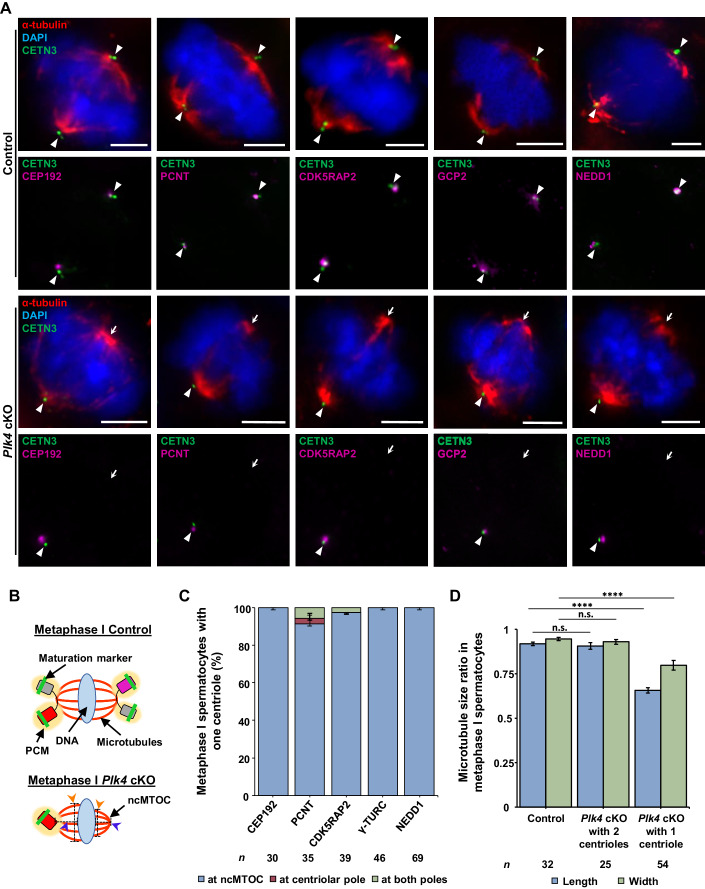
Assessment of PCM components CEP192, CDK5RAP2, PCNT, NEDD1, and components of the γ-TURC complex (γ-tubulin, GCP2, and GCP4), in metaphase I control spermatocytes demonstrated they all colocalized with the centrioles at both spindle poles (Figs. 6A and EV4A–E). However, in Plk4 cKO spermatocytes containing a single centriole, PCM components colocalized with the centriole but were not present at the acentriolar spindle pole (Figs. 6A–C and EV4A–E). Among these observations, the most striking to us was the absence of the γ-TURC components, which is canonically required for microtubule nucleation (Roostalu and Surrey, 2017; Sulimenko et al, 2022). The absence of γ-TURC and PCM components indicated that the spindle pole was not only acentriolar but also lacked major centrosomal components. These observations demonstrate that spermatocytes are capable of forming a non-centrosomal spindle pole when centriole duplication and maturation fail.
Figure 6. PCM components only localize to the spindle pole containing centrioles.
(A) Metaphase I control and Plk4 cKO spermatocytes were immunolabeled against CETN3 (green), α-tubulin (red), CEP192, PCNT, CDK5RAP2, GCP2, and NEDD1 (purple) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate the ncMTOC. Scale bars = 5 µm. (B) Diagram illustrating the ncMTOC and its reduced spindle width and length in Plk4 cKO metaphase I spermatocytes. Red rectangle with rounded corners = mature parent centriole, pink rectangle with rounded corners = immature parent centriole, green bar = maturation marker, yellow oval = PCM, gray rectangle with rounded corners = new centrioles from first centriole duplication, blue oval = DNA, red lines = microtubules, orange arrowheads and black dashed line = indicate spindle width measurement, blue arrowheads and black dashed line = indicate spindle length measurement. (C) Quantification of PCM component foci localization in metaphase I spermatocytes with only one centriole in Plk4 cKO mice (24–27 dpp). Immunolabeling was performed on ≥3 biological replicates with ≥10 spermatocytes quantified per replicate. The total number of cells quantified and immunolabelled for CEP192, PCNT, CDK5RAP2, γ-TURC, and NEDD1 were 30, 35, 39, 46, and 69, respectively. (D) Quantification of spindle length and width ratios in control and Plk4 cKO metaphase I spermatocytes. Measurement of spindle length and width were performed in metaphase I spermatocytes immunolabeled against α-tubulin on ≥3 biological replicates with ≥8 spermatocytes quantified per replicate. The total number of cells quantified for control spermatocytes, Plk4 cKO spermatocytes with 2 centrosomes, and Plk4 cKO spermatocytes with one centrosome were 32, 25, and 54, respectively. P values for Plk4 cKO with 2 centrioles spindle length and width were 0.5453 and 0.4161, respectively. P values for Plk4 cKO with 1 centriole spindle length and width were <0.0001 and <0.0001, respectively. Data information: For all graphs (C, D), error bars show mean ± SEM. For graph (D), P values were obtained from a two-tailed Student’s t test. n.s. (not significant), and ****P < 0.0001.
Figure EV4. Assessment of microtubule-associated factors during prophase I.
Representative image of prophase I (A–L) control and Plk4 cKO spermatocytes from 24–27 dpp mice expressing CETN2-GFP (green) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate. Scale bars = 5 µm. (A) Immunolabeled against α-tubulin (red) and CEP192 (purple). (B) Immunolabeled against α-tubulin (red) and PCNT (purple). (C) Immunolabeled against α-tubulin (red) and CDK5RAP2 (purple). (D) Immunolabeled against α-tubulin (red) and GCP2 (purple). (E) Immunolabeled against α-tubulin (red) and NEDD1 (purple). (F) Immunolabeled against α-tubulin (red) and TPX2 (purple). (G) Immunolabeled against α-tubulin (red) and CAMSAP1 (purple). (H) Immunolabeled against CAMSAP2 (red and purple). (I) Immunolabeled against CAMSAP3 (red and purple). (J) Immunolabeled against NuMA (red and purple). (K) Immunolabeled against α-tubulin (red) and KIF11 (purple). (L) Immunolabeled against α-tubulin (red) and dynein (purple).
To further characterize aspects of the non-centrosomal spindle pole, we compared the length and width of spindle poles in both control and Plk4 cKO metaphase I spermatocytes (Fig. 6B and “Methods”). The spindle pole on the non-centrosomal side of Plk4 cKO metaphase I spermatocytes was reduced in both length and width compared to the canonical spindle pole in metaphase I spermatocytes (Fig. 6B,D). We also observed a 2.2-fold increase in the number of chromosome missegregation events in Plk4 cKO anaphase I spermatocytes compared to controls (Fig. EV3J,K). Nevertheless, Plk4 cKO spermatocytes progressed to metaphase II harboring 2, 1, or 0 centrioles and formed spermatids without a significant increase in apoptosis (Figs. 4F–H, EV1 and EV3L), indicating that non-centrosomal MTOCs (ncMTOCs) are proficient in mediating chromosome segregation.
ncMTOCs in spermatocytes consist of a subset of microtubule-stabilizing and organizing proteins
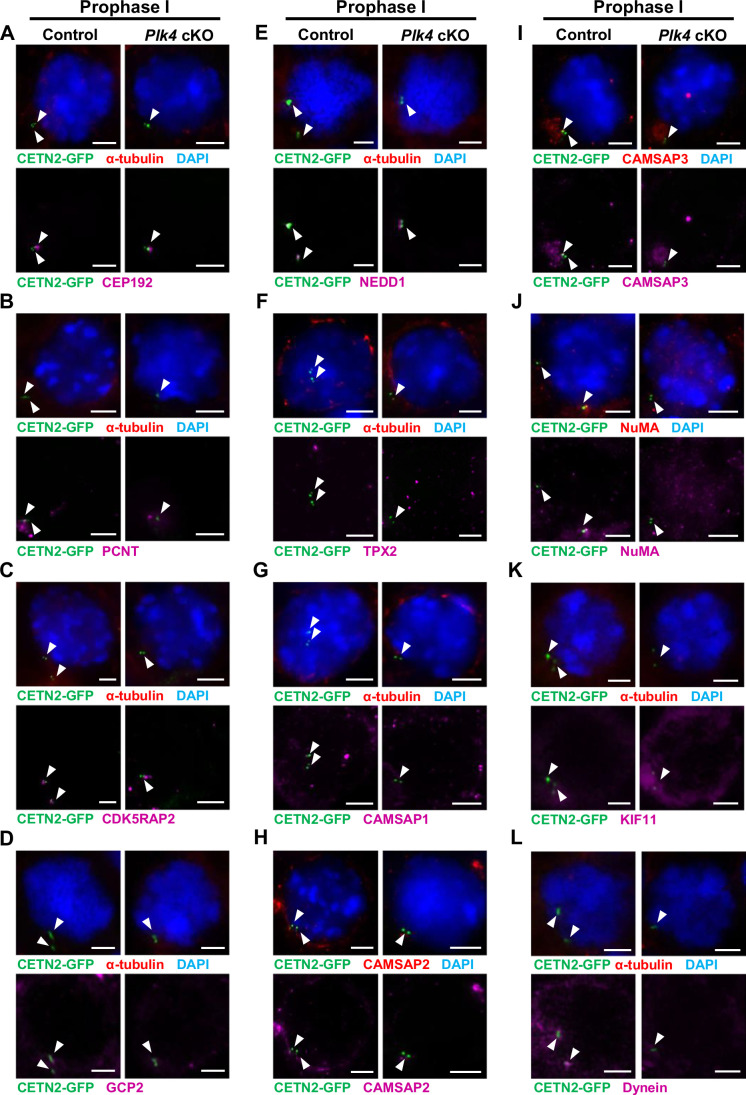
Our next aim was to determine what factors contribute to the formation of the non-centrosomal spindle pole in the Plk4 cKO spermatocytes. We identified proteins that were good candidates for aiding in microtubule nucleation, organization, and stabilization and evaluated their localization in control and Plk4 cKO metaphase I spermatocytes (Fig. 7A). One class of proteins assessed were microtubule motors, including dynein and the kinesins KIF2A, KIF18a, and KIFC1 (Mountain et al, 1999; Norris et al, 2018; Roostalu et al, 2018; Ganem and Compton, 2004; Ferenz et al, 2009; Weaver et al, 2011; van der Voet et al, 2009). We also examined ch-TOG and HURP, both of which are known microtubule stabilizers in mitotic cells, as well as microtubule severing proteins KATNB1, KATNA1, and KATNAL1 (Dudka et al, 2019; De Luca et al, 2008; Booth et al, 2011; Dunleavy et al, 2021). However, none of these molecular motors, microtubule stabilizers, and microtubule severing enzymes localized to the non-centrosomal spindle pole upon assessment via IF microscopy (Fig. 7A; Appendix Fig. S1A–W).
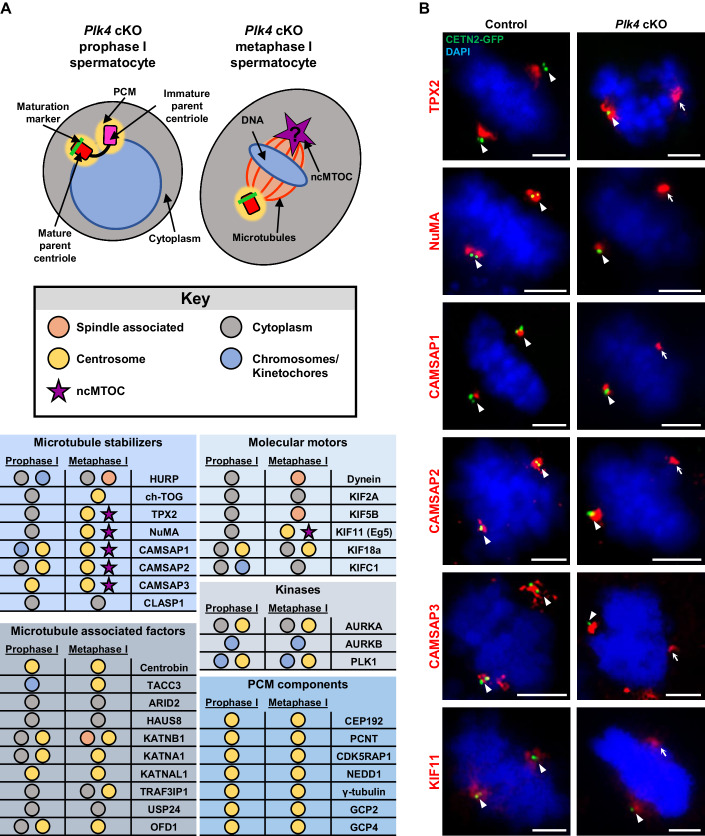
Figure 7. Identification of microtubule-associated proteins that localized to the ncMTOC.
(A) Diagram illustrating a Plk4 cKO prophase I and metaphase I spermatocyte. Red rectangle with rounded corners = mature parent centriole, pink rectangle with rounded corners = immature parent centriole, green bar = maturation marker, yellow oval = PCM, purple star = ncMTOC, blue circle/oval = DNA, red lines = microtubules, orange shading between red lines = spindle associated region, gray circle/oval = cytoplasm. The localization of microtubule-associated proteins was assessed and reported in the table presented below the diagram key. Observed localization corresponds to the diagram and is outlined in the key as follows: spindle associated (orange circle), cytoplasm (gray circle), centrosome (yellow circle), chromosomes/kinetochores (blue circle), and ncMTOC (purple star). See Fig. EV4 for representative images of prophase I and metaphase I control and Plk4 cKO spermatocytes immunostained for all microtubule-associated proteins listed in the table. (B) Metaphase I spermatocytes expressing CETN2-GFP were immunolabeled against TPX2, NuMA, CAMSAP1, CAMSAP2, CAMSAP3, and KIF11 (red) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate the ncMTOC. Scale bars = 5 µm.
From our extensive analyses, however, we did identify six proteins that localized to the ncMTOC in spermatocytes: TPX2 (TPX2 microtubule nucleation factor), CAMSAP1-3 (calmodulin regulated spectrin associated proteins), KIF11 (Eg5; Kinesin Family Member 11), and NuMA (nuclear mitotic apparatus protein1) (Figs. 7B and EV4F–K). All three CAMSAP proteins are minus-end microtubule stabilizers that prevent minus-end microtubule catastrophe and promote microtubule nucleation (Mao et al, 2019; Imasaki et al, 2022; Jiang et al, 2014; Khalaf-Nazzal et al, 2022; Coquand et al, 2021). TPX2 promotes microtubule nucleation and stabilizes along the lengths of the microtubules by binding within the grooves of the alpha and beta-microtubule subunits to help prevent unwanted microtubule catastrophe (Roostalu et al, 2015; Zhang et al, 2017a). KIF11 is a plus-end-directed microtubule motor protein that is known to aid in bipolar spindle orientation (She et al, 2020, 2022; Kapitein et al, 2005). NuMA plays an important role in the formation and organization of the metaphase spindle by helping to orient microtubule minus ends to a unified location and facilitating connections between the spindle pole and the cellular membrane to ensure proper spindle orientation (Gallini et al, 2016; van der Voet et al, 2009).
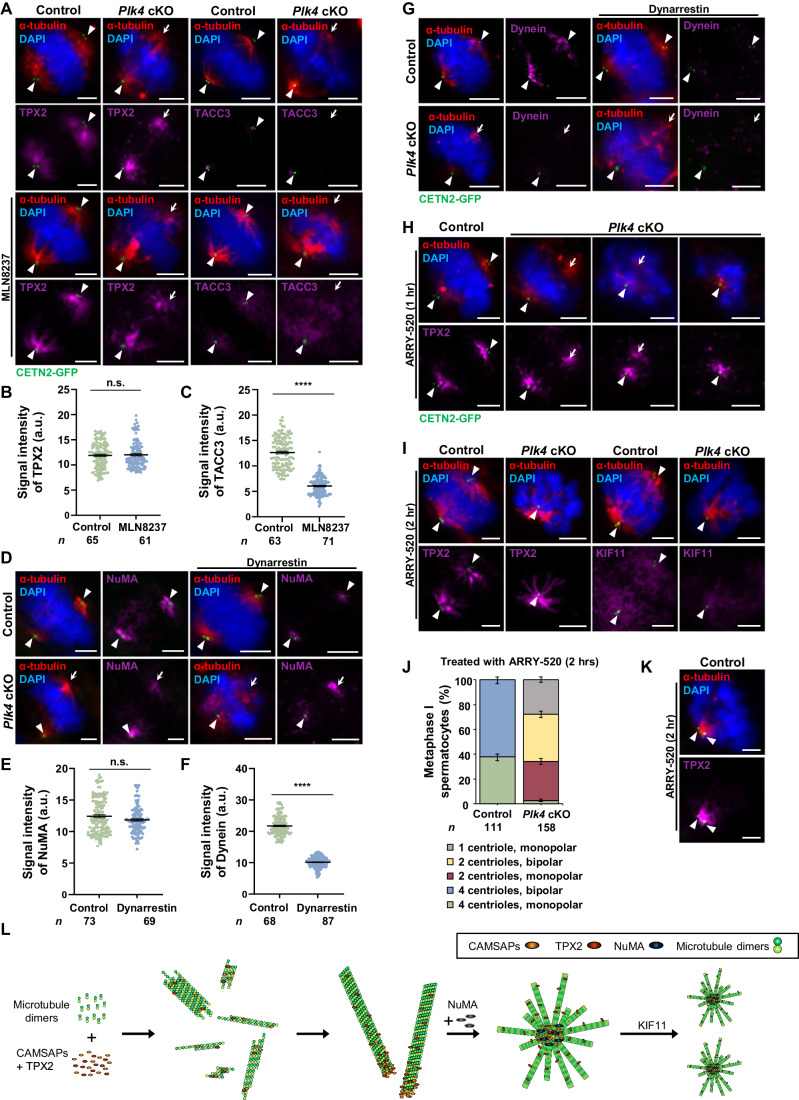
TPX2 is known to be a binding partner of Aurora A kinase (AURKA), which has been implicated in spindle assembly, γ-TuRC activation, and centrosome separation (Zhang et al, 2017b; Wellard et al, 2020). We assessed multiple antibodies against AURKA to determine its localization in Plk4 cKO spermatocytes. From these assessments, we concluded that AURKA does not localize to the ncMTOC (Appendix Fig. S1M). However, to further assess the role of AURKA in Plk4 cKO spermatocytes, we treated seminiferous tubules with an AURKA inhibitor, MLN8237 (see “Methods”). MLN8237 treatment did not disrupt TPX2 localization or bipolar spindle assembly in spermatocytes (Fig. 8A). Plk4 cKO spermatocytes with a single centriole successfully formed bipolar spindles and TPX2 localized to both the centrosome and ncMTOC (Fig. 8A). As a control, we assessed the localization of another known AURKA binding partner and substrate, TACC3. Inhibition of AURKA led to the disruption of TACC3 localization to the spindle poles in mouse spermatocytes at metaphase I, as previously reported (Fig. 8A–C) (Simerly et al, 2024). This indicated that AURKA inhibition was successful, and AURKA kinase activity is not required for TPX2 localization and ncMTOC formation in Plk4 cKO spermatocytes.
Figure 8. ncMTOC spindle formation in Plk4 cKO spermatocytes.
(A) Control and MLN8237 treated (2 h) metaphase I spermatocytes expressing CETN2-GFP were immunolabeled against α-tubulin (red), TPX2 (purple), and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate the ncMTOC. Scale bars = 5 µm. (B) Quantification of the signal intensity of TPX2 at the centrosome in control and MLN8237 treated metaphase I spermatocytes. Immunolabeling was performed on three biological replicates, with ≥20 spermatocytes quantified per replicate. The total number of cells quantified for control and MLN8237 treated spermatocytes was 65 and 61, respectively. P value = 0.7421. (C) Quantification of the signal intensity of TACC3 at the centrosome in control and MLN8237 treated metaphase I spermatocytes. Immunolabeling was performed on three biological replicates, with ≥20 spermatocytes quantified per replicate. The total number of cells quantified for control and MLN8237 treated spermatocytes was 63 and 71, respectively. P value = <0.0001. (D) Control and dynarrestin-treated (2 h) metaphase I spermatocytes expressing CETN2-GFP were immunolabeled against α-tubulin (red) and NuMA (purple) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate the ncMTOC. Scale bars = 5 µm. (E) Quantification of the signal intensity of NuMA at the centrosome in control and dynarrestin-treated metaphase I spermatocytes. Immunolabeling was performed on three biological replicates, with ≥20 spermatocytes quantified per replicate. The total number of cells quantified for control and dynarrestin-treated spermatocytes was 73 and 69, respectively. P value = 0.0504. (F) Quantification of the signal intensity of dynein at the centrosome in control and dynarrestin-treated metaphase I spermatocytes. Immunolabeling was performed on three biological replicates, with ≥20 spermatocytes quantified per replicate. The total number of cells quantified for control and dynarrestin-treated spermatocytes was 68 and 87, respectively. P value = <0.0001. (G) Control and dynarrestin-treated (2 h) metaphase I spermatocytes expressing CETN2-GFP were immunolabeled against α-tubulin (red) and KIF11 (purple) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate the ncMTOC. Scale bars = 5 µm. (H) Metaphase I spermatocytes treated with ARRY-520 (1 h) expressing CETN2-GFP were immunolabeled against α-tubulin (red) and TPX2 (purple) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate the ncMTOC. Scale bars = 5 µm. (I) Metaphase I spermatocytes treated with ARRY-520 (2 h) expressing CETN2-GFP were immunolabeled against α-tubulin (red) and TPX2 (purple) and stained with DAPI (blue). The white arrowheads indicate the centrosome. The white arrows indicate the ncMTOC. Scale bars = 5 µm. (J) Quantification of the spindle orientation in metaphase I spermatocytes treated with ARRY-520 for 2 h. Immunolabeling was performed on one biological replicate. The total number of cells quantified for control and Plk4 cKO mice was 111 and 158, respectively. (K) Metaphase I spermatocytes treated with ARRY-520 (2 h) expressing CETN2-GFP were immunolabeled against α-tubulin (red) and TPX2 (purple) and stained with DAPI (blue). The white arrowheads indicate the centrosome. Scale bars = 5 µm. (L) Diagram illustrating the proposed roles of NuMA, TPX2, CAMSAP1-3, and KIF11 during non-centrosomal spindle assembly. TPX2 and CAMSAP1-3 aid in microtubule nucleation and stabilization. NuMA helps to coordinate microtubule organization, focusing microtubules into an organized spindle pole. KIF11 is necessary for MTOC separation and bipolar spindle formation. Green circles = microtubule dimers, orange oval = CAMSAP1-2, red oval = TPX2, blue oval = NuMA. Data information: For all graphs (C–F, J), error bars show mean ± SEM. P values were obtained from two-tailed Student’s t test. n.s. (not significant), ****P < 0.0001.
We also performed additional assessments examining the effects of inhibiting microtubule motor proteins Dynein and KIF11. While we did not observe the minus-end-directed motor protein dynein at the ncMTOC through IF studies (Appendix Fig. S1A–F), it has been implicated in NuMA localization (He et al, 2023; Merdes et al, 1996, 2000). In spermatocytes, the meiosis I specific DYNLRB2-containing dynein complex is responsible for interacting with NuMA and successfully delivering it to the spindle poles (He et al, 2023). When we treated control and Plk4 cKO spermatocytes with the dynein inhibitor, Dynarrestin, we observed an increase in instances of disordered chromatin organization at the metaphase plate (Fig. 8D). However, NuMA localization to centrosomes or ncMTOCs was not perturbed (Fig. 8E,F). Dynein localization indicated that the inhibitor treatment was successful, as the protein no longer colocalized with the spindle (Fig. 8F,G). While dynein is not observed at the ncMTOCs in Plk4 cKO spermatocytes, its motor function is likely important for microtubule movements contributing to spindle assembly and metaphase plate formation in spermatocytes.
Treatment of spermatocytes with the KIF11 inhibitor, ARRY-520, revealed that KIF11 is critical for maintaining a bipolar spindle during spermatogenesis. We observed control and Plk4 cKO spermatocytes after one and two hours of ARRY-520 treatment to assess bipolar spindle assembly. After one hour, there was an increase in Plk4 cKO spermatocytes with ncMTOCs that failed to fully form bipolar spindles (Fig. 8H). After two hours, all Plk4 cKO spermatocytes harboring a single centriole were monopolar (Fig. 8I,J). There was also an increase in monopolar spindles in control metaphase I spermatocytes (Fig. 8J,K). Moreover, KIF11 no longer colocalized with the spindle poles, indicating ARRY-520 treatment was successful (Fig. 8I). Taken together, this suggests that KIF11 is critical for both centrosomal and non-centrosomal bipolar spindle formation in mammalian spermatocytes.
Based on these findings, we propose that CAMSAP1-3 and TPX2 serve as the base for microtubule nucleation, promoting polymerization in the absence of γ-tubulin while helping stabilize the microtubule minus ends and preventing catastrophe at the base of the microtubules (Fig. 8L). TPX2 further contributes to microtubule stabilization along the lengths of the microtubules. KIF11 is critical for MTOC separation and the formation of a bipolar spindle. NuMA organizes the microtubules by focusing the minus ends at a unified pole and orienting that pole with the cellular membrane, ensuring the spindle is in a bipolar arrangement. Based on their localization to the centrosome and ncMTOC in spermatocytes, CAMSAP1-3, TPX2, NuMA, and KIF11 are likely key factors in MTOC regulation during spermatogenesis. Their importance is made more apparent when centriole duplication fails.
Plk4 cKO spermatocytes fail to successfully complete spermiogenesis
Spermiogenesis is the process by which round spermatids undergo the necessary restructuring to become spermatozoa. In mice, this includes nuclear remodeling involving chromosome compaction and nuclear polarization to one side of the cell in addition to drastic changes in the cell structure, such as the formation of the flagella and the hooked sperm head, a process in part mediated by the manchette (O’Donnell, 2014). The manchette is a microtubule-based structure that forms between the perinuclear ring at the base of the acrosome and the axoneme (Lehti and Sironen, 2016). The manchette facilitates major cell morphological changes during spermiogenesis by serving as a system for protein trafficking (Lehti and Sironen, 2016). Manchette microtubules are anchored to the perinuclear ring complex and it is thought that microtubules nucleate from this location (Lehti and Sironen, 2016; Yoshida et al, 1994).
Despite having a non-centrosomal spindle pole, Plk4 cKO spermatocytes still progress through spermatogenesis without evidence of increased apoptosis (Fig. EV1). We observed that a majority of Plk4 cKO round and elongating spermatids harbored no centrioles (61.6% and 72.7%, respectively) compared to control spermatids that canonically have two centrioles (Fig. 9A–D). Our assessments of PLK4 localization in elongating spermatids indicated that PLK4 localized to the perinuclear ring and manchette structures (Fig. 1H). To assess the integrity of the perinuclear ring in Plk4 cKO spermatocytes, we probed for δ-tubulin, a known component of the perinuclear ring structure (Fig. 9E–H) (Kato et al, 2004). δ-tubulin also localized to the flagella of spermatozoa (Fig. 9E). We also analyzed the coiled-coil domain-containing protein 13, CCDC13. This centriolar satellite protein has been identified in mitotically dividing cells as a critical component during ciliogenesis (Staples et al, 2014), but has not been previously assessed in meiotic cells. CCDC13 localized to the perinuclear ring and the centrin signal in control elongating spermatids (Fig. 9H). However, in Plk4 cKO spermatocytes, δ-tubulin-positive flagella were absent, and there were defects in the perinuclear ring structure and its connection to the base of the acrosome (Fig. 9E–H). Based on α-tubulin localization, we observed no change in manchette morphology in Plk4 cKO elongating spermatids (Fig. 9B,I). However, the manchette was remained present in Plk4 cKO spermatozoa when it was disassembled in control spermatozoa (Fig. 9B,I). Together, the PLK4 localization pattern and Plk4 cKO defects observed during spermiogenesis indicated that PLK4 has multiple functions during spermatid morphogenesis that are critical for the formation of elongated spermatozoa harboring functional flagella.
Figure 9. Assessment of spermiogenesis in Plk4 cKO mice.
(A) Control and Plk4 cKO round spermatids expressing CETN2-GFP (green) were immunolabeled against α-tubulin (red) and stained with DAPI (blue). The white arrowheads indicate the centrosome. Scale bars = 5 µm. (B) Control and Plk4 cKO elongating spermatids expressing CETN2-GFP (green) were immunolabeled against α-tubulin (red) and stained with DAPI (blue). The white arrowheads indicate the centrosome. Scale bars = 5 µm. (C) Quantification of centrin foci in control and Plk4 cKO round spermatids. Immunolabeling was performed on three biological replicates with ≥20 spermatocytes quantified per replicate. The total number of cells quantified for control, and Plk4 cKO mice were 343 and 255, respectively. P value = <0.0001. (D) Quantification of centrin foci in control and Plk4 cKO elongating spermatids. Immunolabeling was performed on three biological replicates, with ≥36 spermatocytes quantified per replicate. The total number of cells quantified for control and Plk4 cKO mice was 748 and 966, respectively. P value = <0.0001. (E) Representative image of adult control and Plk4 cKO spermatocytes from mice immunolabeled against δ-tubulin (red) and stained with DAPI (blue). The white arrows indicate perinuclear ring structure. Zoomed image of spermatozoa and tail inset on the corresponding image. Scale bars = 25 µm. (F) Control and Plk4 cKO round spermatids from mice immunolabeled against δ-tubulin (red) and stained with DAPI (blue). Scale bars = 5 µm. (G) Control and Plk4 cKO spermatozoa from mice immunolabeled against δ-tubulin (red) and stained with DAPI (blue). Scale bars = 5 µm. (H) Elongating spermatid and spermatozoa control and Plk4 cKO spermatocytes harboring CETN2-GFP (green) were immunolabeled against lectin–PNA (green), CCDC13 (red), and stained with DAPI (blue). Scale bars = 5 µm. (I) Control and Plk4 cKO spermatozoa were immunolabeled against lectin–PNA (green), α-tubulin (red), and stained with DAPI (blue, gray inset in lower right corner). Scale bars = 5 µm. (J) Control and Sas4 cKO spermatozoa were immunolabeled against lectin–PNA (green), α-tubulin (red), and stained with DAPI (blue, gray inset in lower right corner). Scale bars = 5 µm. Data information: For all graphs (C, D), error bars show mean ± SEM. P values were obtained from two-tailed Student’s t test. ****P < 0.0001.
Control spermatids form flagella and hooked sperm heads during spermiogenesis to become spermatozoa, whereas Plk4 cKO spermatids failed to complete this structural remodeling (Fig. 9I). Instead, Plk4 cKO spermatozoa exhibited misshapen elongated configurations, never successfully forming the hooked head structure or flagella, including Plk4 cKO spermatids that inherited a single centriole (Fig. 9I). Similar spermiogenesis defects were observed for the Sas4 cKO mice (Fig. 9J). Overall, these observations indicate that PLK4 and centriole inheritance during meiosis are critical for mediating spermiogenesis. Furthermore, the defects during spermiogenesis are likely the primary cause of reduced epididymal sperm counts and infertility observed for the Plk4 cKO male mice.
Discussion
Spermatocytes can utilize a non-centrosomal spindle pole to mediate chromosome segregation during meiosis
We determined that mouse spermatocytes are capable of mediating bipolar chromosome segregation in the absence of centriole and centrosome components. We discovered that CAMSAP1-3, TPX2, NuMA, and KIF11 localize to ncMTOCs and are likely key components that ensure microtubule nucleation and bipolar spindle formation (Tsuchiya and Goshima, 2021). TPX2 and CAMSAP1-3 are microtubule-stabilizing proteins that have been observed to promote microtubule nucleation in the absence of γ-tubulin (Tsuchiya and Goshima, 2021; Meunier and Vernos, 2016). CAMSAP1-3 are microtubule minus-end stabilizers that bind with microtubules to prevent catastrophe at the sites closest to microtubule nucleation (Mao et al, 2019; Jiang et al, 2014). CAMSAP1 and 2 have also been shown to serve as nucleation sites for microtubules in cells undergoing mitotic division (Coquand et al, 2021; Imasaki et al, 2022). Additionally, in glial cells, microtubules were observed to nucleate from foci of GFP-labeled CAMSAP1 and 2 within the basal process region where γ-tubulin was undetectable (Coquand et al, 2021). TPX2 is also a microtubule stabilizer that binds more evenly along the lengths of microtubules to help prevent microtubule catastrophe, in addition to helping promote microtubule nucleation in the absence of γ-tubulin (King and Petry, 2020; Schatz et al, 2003; Zhang et al, 2017a; Roostalu et al, 2015). NuMA is responsible for helping orient the spindle in relation to the cell membrane and aids in the focusing of microtubule minus ends to a unified point (Gallini et al, 2016; van der Voet et al, 2009). Lastly, KIF11 has been shown to be required for centrosome separation and bipolar spindle orientation in mitotic and meiotically dividing cells (She et al, 2020). This, plus-end-directed microtubule motor protein, provides an outward pushing force on MTOCs through the cross-linking of antiparallel microtubules (She et al, 2022; Raaijmakers et al, 2012). Together, these six factors play key roles in successful spindle pole formation and function (Fig. 8L). Future assessments will be conducted to determine how these, and additional factors yet to be identified, coordinate ncMTOC formation during spermatogenesis.
Plk4 cKO mice are infertile due to defects during spermiogenesis
Despite being able to successfully complete meiosis I and II in the absence of centriole duplication, Plk4 cKO mice were still infertile. We attribute this phenotype to a failure during spermiogenesis. During spermiogenesis, spermatids must undergo major structural reconfiguration, forming both a head and tail structure needed for spermatozoa function during fertilization. Similar to other ciliary structures, the centriole acts as the base structure for the microtubule axoneme to polymerize and extend to form the flagella (Avidor-Reiss et al, 2019, 2020). In mouse and human spermatocytes, the two centrioles are oriented perpendicular to one another in order to successfully anchor to what will become the head of the spermatozoa and serve as the base structure for the axoneme to extend from (Chemes, 2012; Manandhar et al, 1998). Both Plk4 and Sas4 cKO result in the formation of spermatids that do not harbor a centriole pair. Therefore, it was expected that Plk4 and Sas4 cKO spermatocytes undergoing spermiogenesis failed to form flagella. Plk4 cKO spermatids also exhibited defects in sperm head morphogenesis. We found that PLK4 localizes to the pericentriolar ring and the manchette structures (Fig. 1H). Due to this observed localization beyond the centrosome, PLK4 is likely involved in additional aspects of cellular restructuring during spermiogenesis beyond flagella formation. Although centrioles are disassembled in rodents during spermiogenesis, we show that centrioles are required for flagella formation and successful execution of spermiogenesis, just as they are in human spermatozoa (Manandhar et al, 1998, 1999).
Centriole biogenesis in non-mammalian organisms
PLK4 localization to the centrosome throughout mammalian spermatogenesis suggests that the kinase is regulated differently in spermatocytes compared to mitotic cells. A difference in PLK4 regulation between mitotic and meiotic cell types is further supported by research on the Caenorhabditis elegans ortholog of PLK4, ZYG-1. A truncated version of ZYG-1 that was no longer capable of localizing to mitotic centrosomes was still able to localize to the centrosome in spermatocytes (Peters et al, 2010). This suggests that PLK4 localization to the centrosome is mediated differently in meiotic vs mitotic cell types. Identifying spermatocyte-specific interaction partners of PLK4 may shed light on how it remains localized to the centrosome throughout both meiotic divisions.
It has been previously reported that the Drosophila melanogaster PLK4 ortholog, SAK, is required for centriole duplication in pre-meiotic and meiotic cells during spermatogenesis (Bettencourt-Dias et al, 2005). When SAK was depleted via RNAi, centriole duplication failed but cells progressed through spermatogenesis (Bettencourt-Dias et al, 2005). It was shown that centrosomal components, D-PLP (PCNT) and γ-tubulin, were absent from spindle poles that lacked centrioles (Bettencourt-Dias et al, 2005). Also, in line with our observations, SAK-depleted spermatocytes were unable to successfully complete spermiogenesis, resulting in sperm lacking flagella (Bettencourt-Dias et al, 2005). Despite the similarities between our work and the reported findings in D. melanogaster, there are some key differences. First, depletion of SAK resulted in centriole duplication failure in the germ line, prior to entering meiosis (Bettencourt-Dias et al, 2005). They observed meiosis I spermatocytes that were devoid of centrioles, indicating duplication failure had taken place prior to meiotic entry (Bettencourt-Dias et al, 2005). Therefore, the role of SAK in relation to centriole duplication was not assessed in strictly a meiotic context. Secondly, centriole duplication only occurs once during D. melanogaster spermatogenesis at the beginning of the first prophase, whereas centrioles duplicate twice in mouse and human spermatocytes (Riparbelli et al, 2018; Bettencourt-Dias et al, 2005; Wellard et al, 2021; Breslow and Holland, 2019). D. melanogaster spermatids canonically complete spermiogenesis after inheriting a single centriole. Mammalian spermatids each inherit two centrioles. Our results demonstrate that both centrioles are required for successful spermiogenesis, as Plk4 and Sas4 cKO spermatids that harbor less than two centrioles failed to form flagella (Fig. 9I). Lastly, the expression patterns of PLK4 during spermatogenesis also vary between mice and D. melanogaster. While we observed PLK4 at the centrosome throughout spermatogenesis in mice (Fig. 1H), SAK levels are reduced as spermatogenesis progresses in D. melanogaster (Khire et al, 2015). This reduction in SAK is a requirement for successful D. melanogaster embryo development after fertilization (Khire et al, 2015; Varmark et al, 2007; Sullenberger et al, 2023; Hatch et al, 2010) The differences in centriole duplication processes, PLK4/SAK expression levels, and PLK4/SAK localization patterns between D. melanogaster and mouse spermatogenesis highlight the importance of assessing the roles of PLK4 in mammalian spermatocytes.
Centrioles and human disease
Previously, the role of mammalian PLK4 had only been assessed in the context of mitotic division. Our work demonstrated that PLK4 is critical for successful centriole duplication and maturation during spermatogenesis and was also essential for normal sperm morphogenesis. We showed similar PLK4 localization patterns in mouse and human spermatocytes, which suggests that the function of PLK4 is conserved between species. Importantly, PLK4 has been linked with cases of human infertility. It has been reported that individuals with a 13 base pair deletion or a missense mutation in PLK4 exhibit a combination of non-obstructive azoospermia and Sertoli cell only (SCO) syndrome (Miyamoto et al, 2016; Tang et al, 2022; Muranishi et al, 2024). Our characterization of PLK4 in mice can contribute to developing better diagnostic and treatment plans for patients with PLK4 and centrosome-related fertility defects.
Conditions resulting from centriole aberrancies, or ciliopathies, are most commonly associated with respiratory system deficiencies or other organs in the body that rely heavily on multiciliate cell types (Inaba and Mizuno, 2016). However, ciliopathies can manifest in the context of flagella formation and, thus, can result in fertility issues (Linck et al, 2016). Conditions such Bardet-Biedl and Kartagener syndromes are examples of such conditions where subfertility is an observed phenotype in patients (Lindstrand et al, 2016; Sha et al, 2014; Halbert et al, 1997). Patients with these conditions experience primary ciliary dyskinesia or the abnormal/absent cilia motion that has detrimental impacts on not only the flagella of spermatozoa in males, but also on the ciliated cells along the female reproductive tract (Newman et al, 2023). Characterization of PLK4 in the context of spermatogenesis, as well as the identification of the ncMTOC are key findings that can help contribute to developing better treatment for patients experiencing these kinds of subfertility or infertility issues. As we observed round spermatids in Plk4 and Sas4 cKO mice, procedures such as round spermatid injection (ROSI) or other reproductive interventions could be used to overcome infertility caused by centriole biogenesis defects (Tanaka and Watanabe, 2023).
Conclusion
We have demonstrated that PLK4 is required for centriole duplication and maturation during mammalian spermatogenesis. Despite a lack of centrioles in Plk4 cKO spermatocytes, these cells successfully complete chromosome segregation by utilizing a ncMTOC mechanism. The exact mechanism by which PLK4 is regulated during spermatogenesis remains unknown. In mitotic cells, PLK4 is regulated by undergoing autophosphorylation and proteasomal degradation in order to prevent centriole overduplication. However, PLK4 is present at the centrosome throughout spermatogenesis, suggesting that PLK4 is regulated differently during meiosis. We hypothesize that PLK4 is regulated through functional inhibition during spermatogenesis. When PLK4 was overexpressed in spermatocytes we observed centriole overduplication. This indicates that excess PLK4 overcomes this predicted canonical inhibitory mechanism. Future research aimed at determining how PLK4 and, thus, centriole duplication, is regulated during mammalian spermatogenesis will be a critical next step.
Methods
Ethics statement
All mice were bred at Johns Hopkins University (JHU, Baltimore, MD), the Uniformed Services University of Health Sciences (USUHS, Bethesda, MD), and Universidad Autónoma de Madrid (UAM, Madrid, Spain). Protocols for their care and use were approved by the Institutional Animal Care and Use Committees (IACUC) of JHU, USUHS, and UAM. All research was performed in accordance with the National Institutes of Health and U.S. Department of Agriculture (JHU and USUHS) and European Union (UAM) guidelines. Studies involving deidentified donor testes tissues have been reviewed and designated by Johns Hopkins University Bloomberg School of Public Health and USUHS IRBs as “not human subjects research” (IRB No.: 00006700 and DBS.2024.685) and approved for use by The USU Human Anatomical Materials Review Committee (HAMRC; #71-22-034).
Mice
Mice harboring the Sas4 cKO allele (C57BL/6N; Cenpjtm1a(EUCOMM)Wtsi) and mice harboring a Plk1 cKO allele (C57BL/6N; Plk1 tm1c(EUCOMM)Hmgu) were previously described (Bazzi and Anderson, 2014; Phan et al, 2021; Little and Jordan, 2020; Wellard et al, 2021). Within the Sas4 allele are two loxP sites flanking the 5th exon of Sas4 (McIntyre et al, 2012). Within the Plk1 allele are two loxP sites flanking the 3rd exon of Plk1. The Plk4 cKO allele (B6;SJL-Plk4em1Ahol/J) contains two loxP sites that flank the 5th exon of Plk4 (LoMastro et al, 2022). Mice harboring the Plk4-mCherry OE allele were obtained from the Institute Curie, Paris. The Plk4-mCherry OE allele contains a floxed stop cassette upstream of the Plk4-mCherry sequence, preventing Plk4-mCherry expression prior to excision by Cre recombinase (Marthiens et al, 2013). Mice harboring the Centrin-GFP transgene (CB6-Tg(CAG-EGFP/CETN2)3-4Jgg/J) were obtained from the Jackson Laboratory (Higginbotham et al, 2004). Conditional mutation was achieved by crossing in a hemizygous Cre recombinase transgene that is under the control of a meiosis-specific promoter. The Spo11 (Tg(Spo11-cre)1Rsw), Stra8 (Tg(Stra8-iCre)1Reb/LguJ), and Hspa2 (Tg(Hspa2-cre)1Eddy/J) promoters were used to drive Cre recombinase expression in this study (Lyndaker et al, 2013; Inselman et al, 2010; Sadate-Ngatchou et al, 2008).
Oocyte harvesting and culture
Oocytes were collected as previously reported (Hwang et al, 2018b). Female mice were intraperitoneally injected with 5IU of pregnant mare serum gonadotropin (PMSG; Sigma) to prime ovarian follicles. After 44–48 h germinal vesicle-staged oocytes were harvested from the ovaries into M2 medium supplemented with 5% fetal bovine serum (FBS; Life Technologies), 3 mg/mL bovine serum albumin (BSA), and 10 mM milrinone (Sigma). Oocyte cumulus complexes were treated for 1–2 min in 300 IU/mL hyaluronidase (Sigma) in M2 medium supplemented with 3 mg/mL BSA to denude oocytes of the surrounding cumulus cells. Oocytes were then washed and cultured for 24 h in M2 media supplemented with 5% FBS and 3% BSA at 37 °C. Oocytes were visually assessed for progression to metaphase I and metaphase II. Metaphase II oocytes were collected for genotyping by intraperitoneally injecting female mice with 5IU of PMSG, followed by human chorionic gonadotropin (HCG; Sigma) 48 h later. Oocytes were collected from the ampulla, and ~20 denuded oocytes were lysed in 10 μL of PBND buffer that was heated for 30 min at 56 °C, 10 min at 95 °C, then brought to room temperature (Scavizzi et al, 2015).
PCR genotyping
PCR genotyping was performed using AccuStart II PCR SuperMix (Quanta BioSciences). Primers used during this study are described in Appendix Table S1. PCR conditions: 94 °C for 2 min; 34–38 cycles of 94 °C for 20 s; 58 °C for 30 s; and 72 °C for 1 min, then a final extension of 72 °C for 10 min.
Human testes
Deidentified decedent adult human donor testes (HT10, age 24) were obtained through the Washington Regional Transplant Consortium (WRTC, Annandale, VA, USA), which are now known as Infinite Legacy (Halethorpe, MD, USA).
Tubule squash preparations
Mouse tubule squash preparations were performed as previously described (Wellard et al, 2018). Primary and secondary antibodies and dilutions used are presented in Appendix Table S2. Tubule squash preparations were mounted in Vectashield + DAPI (4’, 6-diamidino-2-phenylindole) medium (Vector Laboratories). Full Z-stack captured images were utilized to manually identify centrioles, centrosomes, spindle morphology, and chromosome structure.
Chromatin spread preparations
Mouse chromatin spread preparations were performed as previously described (Wellard et al, 2020). Primary and secondary antibodies and dilutions used are presented in Appendix Table S2. Chromatin spread preparations were immunolabeled and mounted in Vectashield + DAPI (Vector Laboratories). Full Z-stack captured images were utilized to manually identify chromosome structure.
Inhibitor treatment of seminiferous tubules
Seminiferous tubules were prepared for inhibitor treatment as previously reported (Simerly et al, 2024). Testes from 24 to 27 dpp mice were removed into KRB enriched with 10 mM HEPES (eKRB). Tubules were transferred into 50-mL falcon tubes and allowed to settle for 5 min in eKRB at 34 °C. The supernatant was then decanted, and tubules were resuspended in 10 mL of 1 mg/ml collagenase IV in eKRB and agitated for 5 min at 37 °C. Following agitation, tubules were allowed to settle by gravity sedimentation and washed twice in eKRB. After the second wash, they were resuspended in eKRB containing either 10 µM of MLN8237 (AURKA inhibitor; Cayman Chemicals), 10 µM of ARRY-520 (KIF11 inhibitor, Cayman Chemicals), or 10 µM of DYNARRESTIN (Dynein inhibitor, Cayman Chemicals) and incubated at 34 °C for 1–2 h. Following incubation, tubules underwent tubule squash preparation as described above.
Culture of seminiferous tubules and centrinone-B treatment
Culture of seminiferous tubules was performed as previously described (Alfaro et al, 2021; Sato et al, 2011). Testes from adult C57BL/6 (WT) mice were removed, detunicated, and fragments of seminiferous tubules were cultured at 34 °C in an atmosphere with 5% CO2 onto agarose gel half-soaked in MEMα culture medium (Gibco A10490-01) supplemented with KnockOut Serum Replacement (KOSR) (Gibco 10828-010), antibiotics (penicillin/streptomycin; Biochrom AG, A2213), and 1–100 µm of centrinone-B (Tocris, LCR-323). Controls were kept in MEMα culture medium without Centrinone-B. After 8 and 24 h, control and inhibitor-treated seminiferous tubules were subjected to the squashing technique described above.
Histology and cryosectioning
For histological assessment, mouse testis tissue was fixed in bouins fixative (Ricca Chemical Company) prior to paraffin embedding. Serial sections, 5 µm thick, were mounted onto slides and stained with hematoxylin and eosin. For cryosectioning testis tissue was embedded in O.C.T. compound (Fisher) and frozen at −80 °C. Serial sections, 16 µm thick, were cut using Cryo3 (Sakura Tissue-Tek, Torrance, CA, USA) and mounted onto Selectfrost Adhesion microscope slides (Fisher). Slides were then fixed with 2.5% formalin in PBS for 10 min at room temperature and washed in PBS. Slides were then immunolabeled as previously described (Atkins et al, 2020) with primary and secondary antibodies (see Appendix Table S2).
Epididymal sperm count
Mouse epididymides (one per mouse) were dissected and placed into PBS, then cut into several smaller pieces and incubated for 30 min at 30 °C to allow time for sperm to be released from the tissue. The sperm-containing solution was then transferred to a fresh tube, separating released sperm from epididymal tissue, and diluted as necessary before counting sperm with a hemocytometer.
Microscope image acquisition
Tubule squash and chromatin spread images were captured using a Zeiss CellObserver Z1 linked to an ORCA-Flash 4.0 CMOS camera (Hamamatsu). Histology and cryo-sectioning images were captured using a Keyence BZ-800 and BZ-X800 Viewer and Analyzer software. Images were processed using ZEN 2012 blue edition imaging software (Zeiss) or BZ-X800 Viewer and Analyzer software (Keyence). Images were analyzed using the Zeiss ZEN 2012 blue edition image software, and Photoshop (Adobe) was used to prepare the figure images.
Microtubule length and width measurements
Spindle measurements were made using ZEN 2012 blue edition imaging software (Zeiss). The spindle length was obtained by measuring from where microtubule nucleation began at an MTOC to the aligned chromosomes on the metaphase I spindle. The width of the spindle was measured proximal to aligned chromosomes on the metaphase I spindle. The width and length of both spindles of a metaphase I spermatocyte were measured and reported as a ratio of A:B, with A being the smaller of the two length or width measurements (Fig. 6B,D).
Data and statistical analysis
All graphics and statistical analyses were performed with GraphPad Prism 9.0 software. Each figure legend provides details of cell and animal counts and statistical analyses.
Supplementary information
Acknowledgements
The authors thank Dr. Masatoshi Takeichi for the CAMSAP3 antibody, Dr. Hisham Bazzi for Sas4 flox/flox mice (Cenpjtm1c(EUCOMM)Wtsi), and Dr. Véronique Marthiens for mCherry-Plk4 mice (Tg(CAG-mCherry/Plk4)#Reba). The authors thank Infinite Legacy and the deidentified testis donor posthumously. This work was funded by NIGMS grant to PWJ (R01GM11755), NICHD grant to PWJ (R01HD114180), and training grant fellowships (NCI, NIH; CA009110; NICHD, NIH; F31HD111265) to MWS.
Expanded view
Author contributions
Marnie W Skinner: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Visualization; Methodology; Writing—original draft; Writing—review and editing. Carter J Simington: Data curation; Formal analysis; Validation; Investigation; Visualization; Writing—review and editing. Pablo López-Jiménez: Data curation; Formal analysis; Validation; Investigation; Visualization; Methodology; Writing—original draft; Writing—review and editing. Kerstin A Baran: Data curation; Formal analysis; Validation; Visualization; Writing—review and editing. Jingwen Xu: Formal analysis; Validation; Investigation; Visualization; Writing—review and editing. Yaron Dayani: Formal analysis; Validation; Investigation; Visualization; Writing—review and editing. Marina V Pryzhkova: Data curation; Formal analysis; Validation; Investigation; Visualization; Methodology; Writing—review and editing. Jesús Page: Supervision; Writing—review and editing. Rocío Gómez: Conceptualization; Formal analysis; Supervision; Funding acquisition; Investigation; Methodology; Writing—original draft; Writing—review and editing. Andrew J Holland: Resources; Formal analysis; Investigation; Visualization; Writing—review and editing. Philip W Jordan: Conceptualization; Resources; Data curation; Formal analysis; Supervision; Funding acquisition; Investigation; Visualization; Methodology; Writing—original draft; Project administration; Writing—review and editing.
Source data underlying figure panels in this paper may have individual authorship assigned. Where available, figure panel/source data authorship is listed in the following database record: biostudies:S-SCDT-10_1038-S44319-024-00187-6.
Data availability
Request for data should be addressed to Philip Jordan (philip.jordan@usuhs.edu). Source data can be accessed through this link: https://www.ebi.ac.uk/biostudies/bioimages/studies/S-BIAD1185?key=a7d044f4-198e-4c8c-82ea-85edda37a38c.
The source data of this paper are collected in the following database record: biostudies:S-SCDT-10_1038-S44319-024-00187-6.
Disclosure and competing interests statement
Philip Jordan is on the scientific advisory board of Gameto, Inc. The remaining authors declare no competing interests.
Supplementary information
Expanded view data, supplementary information, appendices are available for this paper at 10.1038/s44319-024-00187-6.
References
- Alfaro E, López-Jiménez P, González-Martínez J, Malumbres M, Suja JA, Gómez R (2021) PLK1 regulates centrosome migration and spindle dynamics in male mouse meiosis. EMBO Rep 22:e51030 10.15252/embr.202051030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LK, Reeves A, Webb LM, Ashley T (1999) Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151:1569–1579 10.1093/genetics/151.4.1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins A, Xu MJ, Li M, Rogers NP, Pryzhkova MV, Jordan PW (2020) SMC5/6 is required for replication fork stability and faithful chromosome segregation during neurogenesis. eLife 9:e61171 10.7554/eLife.61171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T, Carr A, Fishman EL (2020) The sperm centrioles. Mol Cell Endocrinol 518:110987 10.1016/j.mce.2020.110987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T, Mazur M, Fishman EL, Sindhwani P (2019) The role of sperm centrioles in human reproduction—the known and the unknown. Front Cell Dev Biol 7:188 10.3389/fcell.2019.00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H, Anderson KV (2014) Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci USA 111:E1491–500 10.1073/pnas.1400568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM (2005) SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol 15:2199–2207 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Booth DG, Hood FE, Prior IA, Royle SJ (2011) A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J 30:906–919 10.1038/emboj.2011.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M (2002) Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 14:25–34 10.1016/S0955-0674(01)00290-3 [DOI] [PubMed] [Google Scholar]
- Breslow DK, Holland AJ (2019) Mechanism and regulation of centriole and cilium biogenesis. Annu Rev Biochem 88:691–724 10.1146/annurev-biochem-013118-111153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne DP, Clarke CJ, Brownridge PJ, Kalyuzhnyy A, Perkins S, Campbell A, Mason D, Jones AR, Eyers PA, Eyers CE (2020) Use of the Polo-like kinase 4 (PLK4) inhibitor centrinone to investigate intracellular signalling networks using SILAC-based phosphoproteomics. Biochem J 477:2451–2475 10.1042/BCJ20200309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerbone M, Katugampola H, Dattani MT (2020) Hypothalamic and pituitary disorders affecting the fetus and neonate. In: Kovacs CS, Deal CL (eds) Maternal-fetal and neonatal endocrinology. Elsevier, pp 709–734
- Chemes HE (2012) Sperm centrioles and their dual role in flagellogenesis and cell cycle of the zygote. In: Schatten H (ed) The centrosome. Humana Press, Totowa, NJ, pp 33–48
- Coquand L, Victoria GS, Tata A, Carpentieri JA, Brault J-B, Guimiot F, Fraisier V, Baffet AD (2021) CAMSAPs organize an acentrosomal microtubule network from basal varicosities in radial glial cells. J Cell Biol 220:e202003151 10.1083/jcb.202003151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Bento I, Pimenta-Marques A, Jana SC, Lince-Faria M, Duarte P, Borrego-Pinto J, Gilberto S, Amado T, Brito D et al (2013) Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr Biol 23:2245–2254 10.1016/j.cub.2013.09.037 [DOI] [PubMed] [Google Scholar]
- de Cárcer G, Manning G, Malumbres M (2011) From Plk1 to Plk5: functional evolution of polo-like kinases. Cell Cycle 10:2255–2262 10.4161/cc.10.14.16494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Brunetto L, Asteriti IA, Giubettini M, Lavia P, Guarguaglini G (2008) Aurora-A and ch-TOG act in a common pathway in control of spindle pole integrity. Oncogene 27:6539–6549 10.1038/onc.2008.252 [DOI] [PubMed] [Google Scholar]
- Dudka D, Castrogiovanni C, Liaudet N, Vassal H, Meraldi P (2019) Spindle-length-dependent HURP localization allows centrosomes to control kinetochore-fiber plus-end dynamics. Curr Biol 29:3563–3578.e6 10.1016/j.cub.2019.08.061 [DOI] [PubMed] [Google Scholar]
- Dunleavy JEM, O’Connor AE, Okuda H, Merriner DJ, O’Bryan MK (2021) KATNB1 is a master regulator of multiple katanin enzymes in male meiosis and haploid germ cell development. Development 148:dev199922 10.1242/dev.199922 [DOI] [PubMed] [Google Scholar]
- Ferenz NP, Paul R, Fagerstrom C, Mogilner A, Wadsworth P (2009) Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr Biol 19:1833–1838 10.1016/j.cub.2009.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallini S, Carminati M, De Mattia F, Pirovano L, Martini E, Oldani A, Asteriti IA, Guarguaglini G, Mapelli M (2016) NuMA phosphorylation by Aurora-A orchestrates spindle orientation. Curr Biol 26:458–469 10.1016/j.cub.2015.12.051 [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Compton DA (2004) The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol 166:473–478 10.1083/jcb.200404012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn JR, Hoffmann ER (2022) Errors of the egg: the establishment and progression of human aneuploidy research in the maternal germline. Annu Rev Genet 56:369–390 10.1146/annurev-genet-072820-033609 [DOI] [PubMed] [Google Scholar]
- Guderian G, Westendorf J, Uldschmid A, Nigg EA (2010) Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J Cell Sci 123:2163–2169 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof Y-D, Wilkinson CJ, Nigg EA (2005) The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7:1140–1146 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Halbert SA, Patton DL, Zarutskie PW, Soules MR (1997) Function and structure of cilia in the fallopian tube of an infertile woman with Kartagener’s syndrome. Hum Reprod 12:55–58 10.1093/humrep/12.1.55 [DOI] [PubMed] [Google Scholar]
- Harris RM, Weiss J, Jameson JL (2011) Male hypogonadism and germ cell loss caused by a mutation in Polo-like kinase 4. Endocrinology 152:3975–3985 10.1210/en.2011-1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T (2010) Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol 191:721–729 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Gillies JP, Zang JL, Córdoba-Beldad CM, Yamamoto I, Fujiwara Y, Grantham J, DeSantis ME, Shibuya H (2023) Distinct dynein complexes defined by DYNLRB1 and DYNLRB2 regulate mitotic and male meiotic spindle bipolarity. Nat Commun 14:1715 10.1038/s41467-023-37370-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Bielas S, Tanaka T, Gleeson JG (2004) Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res 13:155–164 10.1023/B:TRAG.0000026071.41735.8e [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW (2014) Polo-like kinase 4 inhibition: a strategy for cancer therapy? Cancer Cell 26:151–153 10.1016/j.ccr.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Zhu Q, Bauer M, Verma IM, Nigg EA, Cleveland DW (2012) The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev 26:2684–2689 10.1101/gad.207027.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW (2010) Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol 188:191–198 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G, Sun F, O’Brien M, Eppig JJ, Handel MA, Jordan PW (2017) SMC5/6 is required for the formation of segregation-competent bivalent chromosomes during meiosis I in mouse oocytes. Development 144:1648–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G, Verver DE, Handel MA, Hamer G, Jordan PW (2018a) Depletion of SMC5/6 sensitizes male germ cells to DNA damage. Mol Biol Cell 29:3003–3016 10.1091/mbc.E18-07-0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang GH, Hopkins JL, Jordan PW (2018b) Chromatin spread preparations for the analysis of mouse oocyte progression from prophase to metaphase II. J Vis Exp 132:e56736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imasaki T, Kikkawa S, Niwa S, Saijo-Hamano Y, Shigematsu H, Aoyama K, Mitsuoka K, Shimizu T, Aoki M, Sakamoto A et al (2022) CAMSAP2 organizes a γ-tubulin-independent microtubule nucleation centre through phase separation. eLife 11:e77365 10.7554/eLife.77365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Mizuno K (2016) Sperm dysfunction and ciliopathy. Reprod Med Biol 15:77–94 10.1007/s12522-015-0225-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inselman AL, Nakamura N, Brown PR, Willis WD, Goulding EH, Eddy EM (2010) Heat shock protein 2 promoter drives Cre expression in spermatocytes of transgenic mice. Genesis 48:114–120 10.1002/dvg.20588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana SC (2021) Centrosome structure and biogenesis: variations on a theme? Semin Cell Dev Biol 110:123–138 10.1016/j.semcdb.2020.10.014 [DOI] [PubMed] [Google Scholar]
- Jiang K, Hua S, Mohan R, Grigoriev I, Yau KW, Liu Q, Katrukha EA, Altelaar AFM, Heck AJR, Hoogenraad CC et al (2014) Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev Cell 28:295–309 10.1016/j.devcel.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Jordan PW, Karppinen J, Handel MA (2012) Polo-like kinase is required for synaptonemal complex disassembly and phosphorylation in mouse spermatocytes. J Cell Sci 125:5061–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF (2005) The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435:114–118 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Karasu OR, Neuner A, Atorino ES, Pereira G, Schiebel E (2022) The central scaffold protein CEP350 coordinates centriole length, stability, and maturation. J Cell Biol 221:e202203081 10.1083/jcb.202203081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Nagata Y, Todokoro K (2004) Delta-tubulin is a component of intercellular bridges and both the early and mature perinuclear rings during spermatogenesis. Dev Biol 269:196–205 10.1016/j.ydbio.2004.01.026 [DOI] [PubMed] [Google Scholar]
- Khalaf-Nazzal R, Fasham J, Inskeep KA, Blizzard LE, Leslie JS, Wakeling MN, Ubeyratna N, Mitani T, Griffith JL, Baker W et al (2022) Bi-allelic CAMSAP1 variants cause a clinically recognizable neuronal migration disorder. Am J Hum Genet 109:2068–2079 10.1016/j.ajhg.2022.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khire A, Vizuet AA, Davila E, Avidor-Reiss T (2015) Asterless reduction during spermiogenesis is regulated by Plk4 and is essential for zygote development in Drosophila. Curr Biol 25:2956–2963 10.1016/j.cub.2015.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Ishiguro K, Nambu A, Akiyoshi B, Yokobayashi S, Kagami A, Ishiguro T, Pendas AM, Takeda N, Sakakibara Y et al (2015) Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature 517:466–471 10.1038/nature14097 [DOI] [PubMed] [Google Scholar]
- King MR, Petry S (2020) Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat Commun 11:270 10.1038/s41467-019-14087-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba JE, Buster DW, McLamarrah TA, Rusan NM, Rogers GC (2015) Autoinhibition and relief mechanism for Polo-like kinase 4. Proc Natl Acad Sci USA 112:E657–66 10.1073/pnas.1417967112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba JE, Buster DW, Nguyen AL, Swatkoski S, Gucek M, Rusan NM, Rogers GC (2013) Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr Biol 23:2255–2261 10.1016/j.cub.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MA, Rosario CO, Hudson JW, Kulkarni S, Pollett A, Dennis JW, Swallow CJ (2005) Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet 37:883–888 10.1038/ng1605 [DOI] [PubMed] [Google Scholar]
- Lan Z-J, Xu X, Cooney AJ (2004) Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol Reprod 71:1469–1474 10.1095/biolreprod.104.031757 [DOI] [PubMed] [Google Scholar]
- Lehti MS, Sironen A (2016) Formation and function of the manchette and flagellum during spermatogenesis. Reproduction 151:R43–54 10.1530/REP-15-0310 [DOI] [PubMed] [Google Scholar]
- Leidel S, Gönczy P (2003) SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell 4:431–439 10.1016/S1534-5807(03)00062-5 [DOI] [PubMed] [Google Scholar]
- Lerit DA, Poulton JS (2016) Centrosomes are multifunctional regulators of genome stability. Chromosome Res 24:5–17 10.1007/s10577-015-9506-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linck RW, Chemes H, Albertini DF (2016) The axoneme: the propulsive engine of spermatozoa and cilia and associated ciliopathies leading to infertility. J Assist Reprod Genet 33:141–156 10.1007/s10815-016-0652-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrand A, Frangakis S, Carvalho CMB, Richardson EB, McFadden KA, Willer JR, Pehlivan D, Liu P, Pediaditakis IL, Sabo A et al (2016) Copy-number variation contributes to the mutational load of Bardet-Biedl syndrome. Am J Hum Genet 99:318–336 10.1016/j.ajhg.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little TM, Jordan PW (2020) PLK1 is required for chromosome compaction and microtubule organization in mouse oocytes. Mol Biol Cell 31:1206–1217 10.1091/mbc.E19-12-0701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoMastro GM, Drown CG, Maryniak AL, Jewett CE, Strong MA, Holland AJ (2022) PLK4 drives centriole amplification and apical surface area expansion in multiciliated cells. eLife 11:e80643 10.7554/eLife.80643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Jiménez P, Pérez-Martín S, Hidalgo I, García-Gonzalo FR, Page J, Gómez R (2022) The male mouse meiotic cilium emanates from the mother centriole at zygotene prior to centrosome duplication. Cells 12:142 10.3390/cells12010142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndaker AM, Lim PX, Mleczko JM, Diggins CE, Holloway JK, Holmes RJ, Kan R, Schlafer DH, Freire R, Cohen PE et al (2013) Conditional inactivation of the DNA damage response gene Hus1 in mouse testis reveals separable roles for components of the RAD9-RAD1-HUS1 complex in meiotic chromosome maintenance. PLoS Genet 9:e1003320 10.1371/journal.pgen.1003320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar G, Schatten H, Sutovsky P (2005) Centrosome reduction during gametogenesis and its significance. Biol Reprod 72:2–13 10.1095/biolreprod.104.031245 [DOI] [PubMed] [Google Scholar]
- Manandhar G, Simerly C, Salisbury JL, Schatten G (1999) Centriole and centrin degeneration during mouse spermiogenesis. Cell Motil Cytoskeleton 43:137–144 [DOI] [PubMed] [Google Scholar]
- Manandhar G, Sutovsky P, Joshi HC, Stearns T, Schatten G (1998) Centrosome reduction during mouse spermiogenesis. Dev Biol 203:424–434 10.1006/dbio.1998.8947 [DOI] [PubMed] [Google Scholar]
- Mao B-P, Li L, Ge R, Li C, Wong CKC, Silvestrini B, Lian Q, Cheng CY (2019) CAMSAP2 Is a microtubule minus-end targeting protein that regulates BTB dynamics through cytoskeletal organization. Endocrinology 160:1448–1467 10.1210/en.2018-01097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjanović M, Sánchez-Huertas C, Terré B, Gómez R, Scheel JF, Pacheco S, Knobel PA, Martínez-Marchal A, Aivio S, Palenzuela L et al (2015) CEP63 deficiency promotes p53-dependent microcephaly and reveals a role for the centrosome in meiotic recombination. Nat Commun 6:7676 10.1038/ncomms8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R (2013) Centrosome amplification causes microcephaly. Nat Cell Biol 15:731–740 10.1038/ncb2746 [DOI] [PubMed] [Google Scholar]
- Mazo G, Soplop N, Wang W-J, Uryu K, Tsou MFB (2016) Spatial control of primary ciliogenesis by subdistal appendages alters sensation-associated properties of cilia. Dev Cell 39:424–437 10.1016/j.devcel.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre RE, Lakshminarasimhan Chavali P, Ismail O, Carragher DM, Sanchez-Andrade G, Forment JV, Fu B, Del Castillo Velasco-Herrera M, Edwards A, van der Weyden L et al (2012) Disruption of mouse Cenpj, a regulator of centriole biogenesis, phenocopies Seckel syndrome. PLoS Genet 8:e1003022 10.1371/journal.pgen.1003022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW (2000) Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol 149:851–862 10.1083/jcb.149.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW (1996) A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87:447–458 10.1016/S0092-8674(00)81365-3 [DOI] [PubMed] [Google Scholar]
- Meunier S, Vernos I (2016) Acentrosomal microtubule assembly in mitosis: the where, when, and how. Trends Cell Biol 26:80–87 10.1016/j.tcb.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Bando Y, Koh E, Tsujimura A, Miyagawa Y, Iijima M, Namiki M, Shiina M, Ogata K, Matsumoto N et al (2016) A PLK4 mutation causing azoospermia in a man with Sertoli cell-only syndrome. Andrology 4:75–81 10.1111/andr.12113 [DOI] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA (1999) The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol 147:351–366 10.1083/jcb.147.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ (2015) Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J Cell Biol 209:863–878 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranishi Y, Kobori Y, Katoh-Fukui Y, Tamaoka S, Hattori A, Osaka A, Okada H, Nakabayashi K, Hata K, Kawai T et al (2024) Systematic molecular analyses for 115 karyotypically normal men with isolated non-obstructive azoospermia. Hum Reprod 39:1131–1140 10.1093/humrep/deae057 [DOI] [PubMed] [Google Scholar]
- Neitzel H, Varon R, Chughtai S, Dartsch J, Dutrannoy-Tönsing V, Nürnberg P, Nürnberg G, Schweiger M, Digweed M, Hildebrand G et al (2022) Transmission ratio distortion of mutations in the master regulator of centriole biogenesis PLK4. Hum Genet 141:1785–1794 10.1007/s00439-022-02461-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman L, Chopra J, Dossett C, Shepherd E, Bercusson A, Carroll M, Walker W, Lucas JS, Cheong Y (2023) The impact of primary ciliary dyskinesia on female and male fertility: a narrative review. Hum Reprod Update 29:347–367 10.1093/humupd/dmad003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Holland AJ (2018) Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat Rev Mol Cell Biol 19:297–312 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Raff JW (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139:663–678 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Norris SR, Jung S, Singh P, Strothman CE, Erwin AL, Ohi MD, Zanic M, Ohi R (2018) Microtubule minus-end aster organization is driven by processive HSET-tubulin clusters. Nat Commun 9:2659 10.1038/s41467-018-04991-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell L (2014) Mechanisms of spermiogenesis and spermiation and how they are disturbed. Spermatogenesis 4:e979623 10.4161/21565562.2014.979623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-E, Zhang L, Bang JK, Andresson T, DiMaio F, Lee KS (2019) Phase separation of Polo-like kinase 4 by autoactivation and clustering drives centriole biogenesis. Nat Commun 10:4959 10.1038/s41467-019-12619-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters N, Perez DE, Song MH, Liu Y, Müller-Reichert T, Caron C, Kemphues KJ, O’Connell KF (2010) Control of mitotic and meiotic centriole duplication by the Plk4-related kinase ZYG-1. J Cell Sci 123:795–805 10.1242/jcs.050682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TP, Maryniak AL, Boatwright CA, Lee J, Atkins A, Tijhuis A, Spierings DC, Bazzi H, Foijer F, Jordan PW et al (2021) Centrosome defects cause microcephaly by activating the 53BP1-USP28-TP53 mitotic surveillance pathway. EMBO J 40:e106118 10.15252/embj.2020106118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova T, Gorbsky GJ (2017) The consequences of chromosome segregation errors in mitosis and meiosis. Biology (Basel) 6:12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RGHP, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME (2012) Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J 31:4179–4190 10.1038/emboj.2012.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli MG, Persico V, Callaini G (2018) A transient microtubule-based structure uncovers a new intrinsic asymmetry between the mother centrioles in the early Drosophila spermatocytes. Cytoskeleton 75:472–480 10.1002/cm.21503 [DOI] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL (2009) The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol 184:225–239 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Cade NI, Surrey T (2015) Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat Cell Biol 17:1422–1434 10.1038/ncb3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Rickman J, Thomas C, Nédélec F, Surrey T (2018) Determinants of polar versus nematic organization in networks of dynamic microtubules and mitotic motors. Cell 175:796–808.e14 10.1016/j.cell.2018.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Surrey T (2017) Microtubule nucleation: beyond the template. Nat Rev Mol Cell Biol 18:702–710 10.1038/nrm.2017.75 [DOI] [PubMed] [Google Scholar]
- Rosario CO, Ko MA, Haffani YZ, Gladdy RA, Paderova J, Pollett A, Squire JA, Dennis JW, Swallow CJ (2010) Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci USA 107:6888–6893 10.1073/pnas.0910941107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE (2008) Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46:738–742 10.1002/dvg.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Katagiri K, Gohbara A, Inoue K, Ogonuki N, Ogura A, Kubota Y, Ogawa T (2011) In vitro production of functional sperm in cultured neonatal mouse testes. Nature 471:504–507 10.1038/nature09850 [DOI] [PubMed] [Google Scholar]
- Scavizzi F, Ryder E, Newman S, Raspa M, Gleeson D, Wardle-Jones H, Montoliu L, Fernandez A, Dessain M-L, Larrigaldie V et al (2015) Blastocyst genotyping for quality control of mouse mutant archives: an ethical and economical approach. Transgenic Res 24:921–927 10.1007/s11248-015-9897-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten H, Sun Q-Y (2009) The role of centrosomes in mammalian fertilization and its significance for ICSI. Mol Hum Reprod 15:531–538 10.1093/molehr/gap049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz CA, Santarella R, Hoenger A, Karsenti E, Mattaj IW, Gruss OJ, Carazo-Salas RE (2003) Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J 22:2060–2070 10.1093/emboj/cdg195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Y-W, Ding L, Li P (2014) Management of primary ciliary dyskinesia/Kartagener’s syndrome in infertile male patients and current progress in defining the underlying genetic mechanism. Asian J Androl 16:101–106 10.4103/1008-682X.122192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Z-Y, Zhong N, Wei Y-L (2022) Kinesin-5 Eg5 mediates centrosome separation to control spindle assembly in spermatocytes. Chromosoma 131:87–105 10.1007/s00412-022-00772-5 [DOI] [PubMed] [Google Scholar]
- She Z-Y, Zhong N, Yu K-W, Xiao Y, Wei Y-L, Lin Y, Li Y-L, Lu M-H (2020) Kinesin-5 Eg5 is essential for spindle assembly and chromosome alignment of mouse spermatocytes. Cell Div 15:6 10.1186/s13008-020-00063-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne JE, Bornens M (2010) Polo-like kinase 4: the odd one out of the family. Cell Div 5:25 10.1186/1747-1028-5-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simerly C, Robertson E, Harrison C, Ward S, George C, Deleon J, Hartnett C, Schatten G (2024) Male meiotic spindle poles are stabilized by TACC3 and cKAP5/chTOG differently from female meiotic or somatic mitotic spindles in mice. Sci Rep 14:4808 10.1038/s41598-024-55376-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin LK, Nye J, Pinkerton DC, Buster DW, Rogers GC, Slep KC (2012) The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication. Structure 20:1905–1917 10.1016/j.str.2012.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C, Seres KB, Steyer AM, Mönnich E, Clift D, Pejkovska A, Möbius W, Schuh M (2019) A liquid-like spindle domain promotes acentrosomal spindle assembly in mammalian oocytes. Science 364:eaat9557 10.1126/science.aat9557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples CJ, Myers KN, Beveridge RDD, Patil AA, Howard AE, Barone G, Lee AJX, Swanton C, Howell M, Maslen S et al (2014) Ccdc13 is a novel human centriolar satellite protein required for ciliogenesis and genome stability. J Cell Sci 127:2910–2919 [DOI] [PubMed] [Google Scholar]
- Sulimenko V, Dráberová E, Dráber P (2022) γ-Tubulin in microtubule nucleation and beyond. Front Cell Dev Biol 10:880761 10.3389/fcell.2022.880761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenberger C, Kong D, Avazpour P, Luvsanjav D, Loncarek J (2023) Centrosomal organization of Cep152 provides flexibility in Plk4 and procentriole positioning. J Cell Biol 222:e202301092 10.1083/jcb.202301092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Gong T-T, Jiang Y-T, Zhang S, Zhao Y-H, Wu Q-J (2019) Global, regional, and national prevalence and disability-adjusted life-years for infertility in 195 countries and territories, 1990-2017: results from a global burden of disease study, 2017. Aging 11:10952–10991 10.18632/aging.102497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Watanabe S (2023) How to improve the clinical outcome of round spermatid injection (ROSI) into the oocyte: correction of epigenetic abnormalities. Reprod Med Biol 22:e12503 10.1002/rmb2.12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Li K, Geng H, Xu C, Lv M, Gao Y, Wang G, Yu H, Shao Z, Shen Q et al (2022) Identification of deleterious variants in patients with male infertility due to idiopathic non-obstructive azoospermia. Reprod Biol Endocrinol 20:63 10.1186/s12958-022-00936-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos BE, Yang H-J, Soni R, Wang W-J, Macaluso FP, Asara JM, Tsou M-FB (2013) Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev 27:163–168 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach JM, Philip R, Sharma A, Strecker J, Durocher D, Pelletier L (2022) Global cellular response to chemical perturbation of PLK4 activity and abnormal centrosome number. eLife 11:e73944 10.7554/eLife.73944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya K, Goshima G (2021) Microtubule-associated proteins promote microtubule generation in the absence of γ-tubulin in human colon cancer cells. J Cell Biol 220:e202104114 10.1083/jcb.202104114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voet M, Berends CWH, Perreault A, Nguyen-Ngoc T, Gönczy P, Vidal M, Boxem M, van den Heuvel S (2009) NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat Cell Biol 11:269–277 10.1038/ncb1834 [DOI] [PubMed] [Google Scholar]
- Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C (2007) Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr Biol 17:1735–1745 10.1016/j.cub.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Wang J, Zuo J, Wang M, Ma X, Gao K, Bai X, Wang N, Xie W, Liu H (2019) Polo-like kinase 4 promotes tumorigenesis and induces resistance to radiotherapy in glioblastoma. Oncol Rep 41:2159–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Kong D, Hoerner CR, Loncarek J, Stearns T (2017) Centriole triplet microtubules are required for stable centriole formation and inheritance in human cells. eLife 6:e29061 10.7554/eLife.29061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LN, Ems-McClung SC, Stout JR, LeBlanc C, Shaw SL, Gardner MK, Walczak CE (2011) Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol 21:1500–1506 10.1016/j.cub.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellard SR, Hopkins J, Jordan PW (2018) A seminiferous tubule squash technique for the cytological analysis of spermatogenesis using the mouse model. J Vis Exp 132:e56453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellard SR, Schindler K, Jordan PW (2020) Aurora B and C kinases regulate chromosome desynapsis and segregation during mouse and human spermatogenesis. J Cell Sci 133:jcs248831 10.1242/jcs.248831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellard SR, Zhang Y, Shults C, Zhao X, McKay M, Murray SA, Jordan PW (2021) Overlapping roles for PLK1 and Aurora A during meiotic centrosome biogenesis in mouse spermatocytes. EMBO Rep 22:e51023 10.15252/embr.202051023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW et al (2015) Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348:1155–1160 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Ioshii SO, Imanaka-Yoshida K, Izutsu K (1994) Association of cytoplasmic dynein with manchette microtubules and spermatid nuclear envelope during spermiogenesis in rats. J Cell Sci 107(Pt 3):625–633 10.1242/jcs.107.3.625 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li G, Zhang L, Sun X, Zhang D, Lu J, Ma J, Yan J, Chen Z-J (2017a) Maternal common variant rs2305957 spanning PLK4 is associated with blastocyst formation and early recurrent miscarriage. Fertil Steril 107:1034–1040.e5 10.1016/j.fertnstert.2017.01.006 [DOI] [PubMed] [Google Scholar]
- Zhang R, Roostalu J, Surrey T, Nogales E (2017b) Structural insight into TPX2-stimulated microtubule assembly. eLife 6:e30959 10.7554/eLife.30959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang L, Huang L, Liu G, Nie X, Zhang X, Xing X (2021) SUN5 interacting with Nesprin3 plays an essential role in sperm head-to-tail linkage: research on Sun5 gene knockout mice. Front Cell Dev Biol 9:684826 10.3389/fcell.2021.684826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitouni S, Nabais C, Jana SC, Guerrero A, Bettencourt-Dias M (2014) Polo-like kinases: structural variations lead to multiple functions. Nat Rev Mol Cell Biol 15:433–452 10.1038/nrm3819 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Request for data should be addressed to Philip Jordan (philip.jordan@usuhs.edu). Source data can be accessed through this link: https://www.ebi.ac.uk/biostudies/bioimages/studies/S-BIAD1185?key=a7d044f4-198e-4c8c-82ea-85edda37a38c.
The source data of this paper are collected in the following database record: biostudies:S-SCDT-10_1038-S44319-024-00187-6.