Abstract
Genomic SELEX is a method for studying the network of nucleic acid–protein interactions within any organism. Here we report the discovery of several interesting and potentially biologically important interactions using genomic SELEX. We have found that bacteriophage MS2 coat protein binds several Escherichia coli mRNA fragments more tightly than it binds the natural, well-studied, phage mRNA site. MS2 coat protein binds mRNA fragments from rffG (involved in formation of lipopolysaccharide in the bacterial outer membrane), ebgR (lactose utilization repressor), as well as from several other genes. Genomic SELEX may yield experimentally induced artifacts, such as molecules in which the fixed sequences participate in binding. We describe several methods (annealing of oligonucleotides complementary to fixed sequences or switching fixed sequences) to eliminate some, or almost all, of these artifacts. Such methods may be useful tools for both randomized sequence SELEX and genomic SELEX.
INTRODUCTION
As genomic sequences from a variety of organisms are becoming available, it is important to develop rapid methods for identifying the regulatory network of interactions between nucleic acids and proteins. Genomic SELEX is an in vitro selection–amplification method proposed for identification of biologically important nucleic acid–protein interactions (1–4). Like SELEX [systematic evolution of ligands by exponential enrichment (5,6)] of randomized sequence nucleic acids, genomic SELEX consists of repeated rounds of binding a library of nucleic acids to the target protein, separating the bound nucleic acids from the unbound ones and amplifying the bound ones for the next round. The starting libraries in genomic SELEX, unlike in randomized sequence SELEX, are derived from the genome of the organism of interest. At the end of the selection, the resulting high affinity nucleic acid molecules are cloned and sequenced.
Genomic SELEX is conceptually related to other high-throughput methods of functional analysis of the genomes, such as the one-hybrid, two-hybrid and three-hybrid systems (7–10). It can be used to construct nucleic acid–protein linkage maps, similar to protein–protein linkage maps (11,12). Genomic SELEX was first tested on Escherichia coli DNA, with a known double-stranded DNA-binding protein, MetJ. Many of the reported genomic sites plus some previously unreported, but biologically plausible, sites were isolated (Y.Y.He, D.Brown, C.Workman, G.D.Stormo and L.Gold, personal communication).
In this study, genomic SELEX was tested on RNA, which because of its structural complexity might yield more unexpected results than double-stranded DNA. SELEX was performed with a library made of E.coli genomic DNA that was transcribed in vitro with T7 RNA polymerase. Because whole genomic DNA was used for the library, transcribing it probably yields a number of fragments not normally transcribed in E.coli. The library was constructed using a novel method (13) and was shown to contain most of the genomic sequences from E.coli. The RNA from this library was selected for its ability to bind bacteriophage MS2 coat protein (MS2 CP). Advantages of this system include the known genome of E.coli and the known sequence requirements for RNA that binds to MS2 CP, which make it possible to predict binding based on the sequence of the selected RNAs.
MS2 CP has two known functions. Its first function is to form the phage coat and its second function is to repress the translation of the phage replicase gene by binding its mRNA at the ribosome binding site (14,15). The RNA elements that are important for this interaction have been determined from the comprehensive biochemical analysis of mutants (14–16). These binding data have been supported both by the crystal structure of MS2 CP complexed with the RNA (17,18) and by the randomized sequence SELEX (19) with closely related phage R17 coat protein. However, it was not known whether MS2 CP binds any E.coli RNA and what role such interactions might play. It is possible that MS2 regulates gene expression in the host cell, as some other viruses do, for example oncogenic adenovirus (20). The potential MS2/E.coli interactions raise interesting biological questions and may reveal unknown aspects of host–parasite interactions.
Thus, the primary goal of this study was to test genomic SELEX on an RNA library from a well-studied organism and a protein with a known RNA binding specificity. The secondary goal was to find out if MS2 CP interacts with E.coli RNA in any interesting way that may prompt further investigations into its potential other biological roles.
MATERIALS AND METHODS
Regular SELEX
A non-aggregating MS2 CP variant V75E;A81G with RNA-binding properties identical to wild-type was purified as previously described (21). Any endogenous E.coli RNA was removed in the purification process, as indicated by the binding stoichiometry and the UV absorbance spectrum (21).
SELEX was carried out essentially as described before (19,22). Detailed protocols can be obtained from http://mcdb.colorado.edu/labs/gold_lab/ . SELEX was initiated with 1 nmol of RNA, transcribed from the E.coli genomic DNA library (13). In each round of selection, 1 nmol of RNA was denatured in TE at 95°C for 1 min, quickly chilled on ice and incubated on ice for 10 min. Binding buffer was added to give a final concentration of 100 mM HEPES–KOH pH 7.5, 80 mM KCl, 10 mM MgCl2. The mixture was pre-filtered through nitrocellulose to reduce the fraction of the nitrocellulose-binding RNA. The volume was adjusted with the binding buffer to make up for the loss on the filter. MS2 CP was added to the final concentration of 100 nM of the dimer in a 0.1 ml reaction. Binding proceeded for 45 min at room temperature (22–24°C).
The binding reaction was vacuum manifold-filtered through nitrocellulose and washed with 5 ml of the binding buffer. The bound RNA was eluted from the filters, precipitated with ethanol and amplified for the next round of SELEX. Primer B (5′-tcccgctcgtcgtctg-3′) served for reverse transcription and primers B and A (5′-gaaattaatacgactcactatagggaggacgatgcgg-3′; T7 promoter underlined) served for PCR. In this PCR, as well as in all others in this study, the relatively low concentrations of MgCl2 (3 mM) and dNTPs (50 µM each) were used to decrease the error rate.
SELEX with annealing of the complementary oligonucleotides
Instead of 1 nmol of RNA, 0.1 nmol of RNA and 0.4 nmol of each of the two complementary oligonucleotides were used (primer B and primer cA: 5′-ccgcatcgtcctccc-3′). An extra 10 min incubation at room temperature was introduced directly after the addition of the binding buffer to allow oligonucleotides to anneal. The annealed oligonucleotides decreased the yield of the full-length reverse transcription product by only 10%. Otherwise, this SELEX was identical to the regular SELEX.
SELEX with switching the fixed sequences
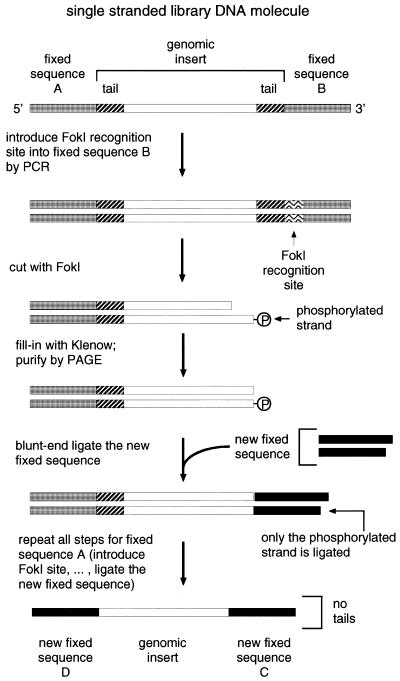
Step 1. Switching the 3′ fixed sequence: DNA purification and PCR. Regular SELEX was carried out for three rounds using the old fixed sequence primers A and B described above. Since the old fixed sequences did not contain FokI restriction sites, the sites had to be introduced by PCR (in future, the sites might be easier to introduce during library construction). DNA product from either the reverse transcription reaction or from PCR after round 3, was purified from primer B. Primer B interferes with the subsequent steps if not completely removed. Reverse transcription product was purified on a Microcon-30 filter (Amicon, Beverly, MA), by centrifugation three times with 0.2 ml of TE buffer for 10 min at 16 000 g. PCR product, since it contains more primer B, had to be purified, instead of Microcon-30, by native polyacrylamide gel electrophoresis (PAGE) with ethidium bromide staining, followed by crush-and-soak elution for 30 min at 37°C.
Purified DNA was amplified by PCR (Fig. 3) using primer A and primer B+FokI (5′-tcccgctcgtGgATGg-3′). Primer B+FokI introduces the FokI recognition site (GgATG, shown in boldface) into the old 3′ fixed sequence and differs from primer B where indicated by the uppercase nucleotides (G, ATG). The amplified DNA was extracted with chloroform/phenol, twice with chloroform, then ethanol-precipitated and resuspended in water (unpurified PCR product inhibits subsequent FokI digestion).
Figure 3.
Outline of the strategy of switching the fixed sequences (see Materials and Methods).
Step 2. FokI digestion. Purified DNA product of 0.1 ml PCR was incubated with FokI (New England Biolabs, Beverly, MA) at a ratio of >1.5 U/µg PCR product (the ratio was estimated assuming that <100% of the primers were converted into the PCR product). This ratio had to be optimized with every new DNA preparation. FokI digestion was carried out in 40 µl at 37°C for 1 h in the manufacturer’s buffer with 0.1% of Tween-20 detergent to decrease the exonuclease activity.
Step 3. Klenow extension. dNTPs (final concentration 0.5 mM each) and the Klenow fragment of E.coli DNA polymerase I (US Biochemicals, Cleveland, OH; final concentration 370 U/ml) were added directly to the FokI digest. Extension proceeded for 15 min at 37°C. The FokI-digested and Klenow-extended library DNA was then purified from the other digestion fragments by native PAGE as in Step 1. Of the input DNA, 60% was cut by FokI as expected, 40% was degraded non-specifically and a negligible fraction was left uncut.
Step 4. Ligation. The purified library DNA was resuspended in water and blunt-end ligated to the new fixed sequence. The new fixed sequence was a duplex of two DNA oligonucleotides: C (5′-ggtgcggcagttcggt-3′) and its complement, cC (5′-accgaactgccgcacct-3′). The duplex was formed by incubation of the mixture of 200 pmol of each oligonucleotide in 4 µl of TE at 95°C for 1 min, followed by 60°C for 10 min and room temperature for 10 min. The duplex was added to the purified DNA and incubated with 2 U of T4 DNA ligase (Boehringer Mannheim, Indianapolis, IN) in the manufacturer’s buffer in 20 µl for 1 h at 30°C. The ligation yield was 50%, estimated by Molecular Dynamics (Sunnyvale, CA) phosphorimager quantification of 32P-labeled DNA separated by PAGE. The overall yield of all steps was 10% relative to the input DNA at the beginning of Step 2. The relatively high blunt-end ligation yield was achieved by keeping all DNAs as concentrated as possible (more than a few µM), since the Km of ligase for blunt ends is 50 µM (23). To reduce ligation of the library DNA molecules to each other, an excess of oligonucleotides over the library DNA was used (>2-fold excess, estimated by assuming that <100% of the primers was converted into the library PCR product and that <100% of product was recovered after gel purification). The length of the ligation products was verified by PAGE.
Step 5. PCR. One-third of the ligation product was amplified by PCR with the new 3′ fixed sequence primer C (sequence shown above) and primer A+FokI (which introduces the FokI site into the old 5′ fixed sequence and relates to primer A as B+FokI relates to B). Higher concentrations of the primers were used in this PCR (10 µM each, instead of 1 µM as in all other PCRs in this study). This served to provide an excess of primer C over cC (cC is complementary to C and was carried over from the ligation in Step 4).
Only one of the two major ligation products can be amplified in PCR with primers C and A+FokI, namely, the product in which the duplex of oligonucleotides C and cC has been ligated in one of the two possible orientations with respect to the FokI-digested library DNA molecule. A single T was added to the 3′-end of oligonucleotide cC, as shown above, in order to create a single base overhang in the C–cC duplex. Under the experimental conditions, adding a single overhanging T directs ligation more toward the desired orientation: blunt end to blunt end, as opposed to the undesired orientation of overhanging end to blunt end (data not shown).
Step 6. Switching the 5′ fixed sequence. DNA was purified as in the last paragraph of Step 1 and then Steps 2–5 for 3′ fixed sequence were essentially repeated for the 5′ fixed sequence. For the PCR in Step 5, primers for the new fixed sequences were used: C (Step 4) and D (5′-gaaattaatacgactcactatagggaaagcccacgcc-3′). The resulting molecules had both fixed sequences replaced with the new ones, with both tails removed entirely. One such molecule in its RNA form is shown in Figure 1D. After switching the fixed sequences, SELEX proceeded as in ‘Regular SELEX’, with 1 µM RNA and 100 nM MS2 CP. In the SELEX experiment where the new fixed sequences were chosen without the help of the STOGEN computer program (see below), the primers that correspond to C and D were, respectively, 5′-atgtcgggccgccgaa-3′ and 5′-gaaattaatacgactcactatagggcccggcgcataa-3′.
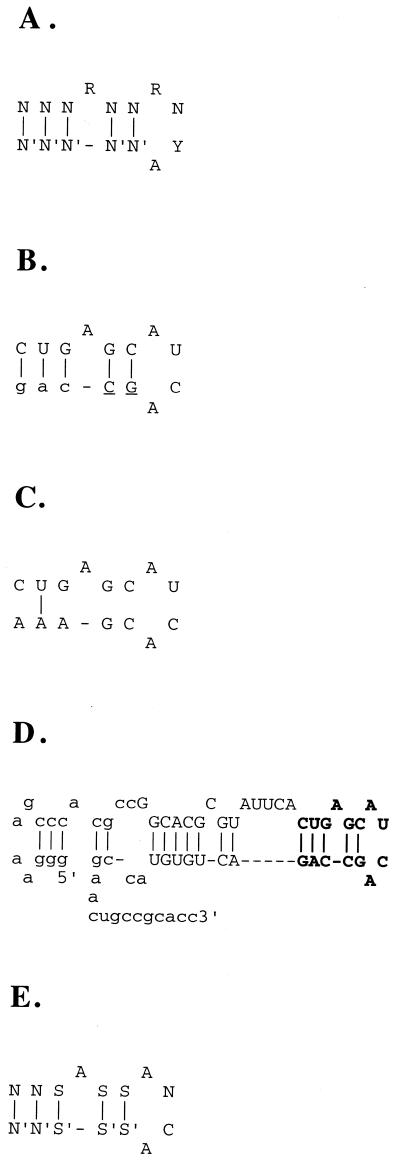
Figure 1.
MS2 CP binding sites. (A) The consensus binding site of MS2 CP. NN′ is any base pair, R is either A or G, Y is either U or C. (B) A frequent selection artifact. The fixed sequence is shown in lowercase, the genomic insert in uppercase, the tail is underlined. (C) The actual genomic sequence (from GenBank) that corresponds to the artifact shown in (B). It is shown ‘folded’ only for comparison with (A) and (B) and is not predicted to bind MS2 CP. (D) The predicted structure of the major isolate (rffG) from SELEX with switching the fixed sequences. The genomic insert nucleotides are in uppercase, fixed sequence (starting with ggg at the 5′-end) in lowercase. The consensus binding site is shown in boldface. (E) The SELEX consensus binding site of MS2 CP. SS′ is either GC or CG base pair. The first two NN′ base pairs must have at least one SS′.
Binding analysis
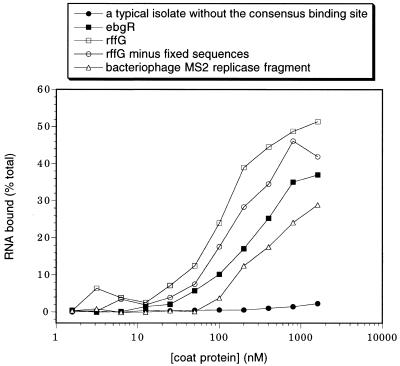
The MS2 replicase fragment (the natural MS2 CP binding site; Fig. 4) was chemically synthesized, amplified by PCR and labeled by in vitro transcription. The resulting RNA molecule contained the fragment of the original MS2 sequence (as published in GenBank) with the same fixed sequences (for primers C and D) attached to it as in the real SELEX isolates. The molecule also matched the SELEX isolates in length (70 nt, with most isolates being 60–80 nt). The MS2 CP binding site was positioned approximately in the middle of the molecule.
Figure 4.
Binding of RNA to MS2 CP. SELEX isolates with the consensus binding site, rffG and ebgR, bind well. To test whether the fixed sequences contribute to binding of the rffG isolate, the RNA fragment that corresponds exactly to its insert was obtained from E.coli genomic DNA using PCR and in vitro transcription. This RNA fragment (rffG minus fixed sequences) binds only marginally worse than the original rffG SELEX isolate. For comparison, the natural MS2 CP binding site (bacteriophage MS2 replicase fragment) binds more weakly than all of the SELEX isolates with the consensus binding site (Table 1), except the secY isolate (data not shown). A typical SELEX isolate without the consensus binding site does not appreciably bind MS2 CP and neither does the starting library (data not shown).
The RNAMOT site nos 8, 12 and 14 (Table 2) were amplified from E.coli genomic DNA template by PCR. The transcribed RNA molecules had the MS2 CP binding sites positioned approximately in the middle. The molecules also had the same fixed sequences, and were 70 nt long.
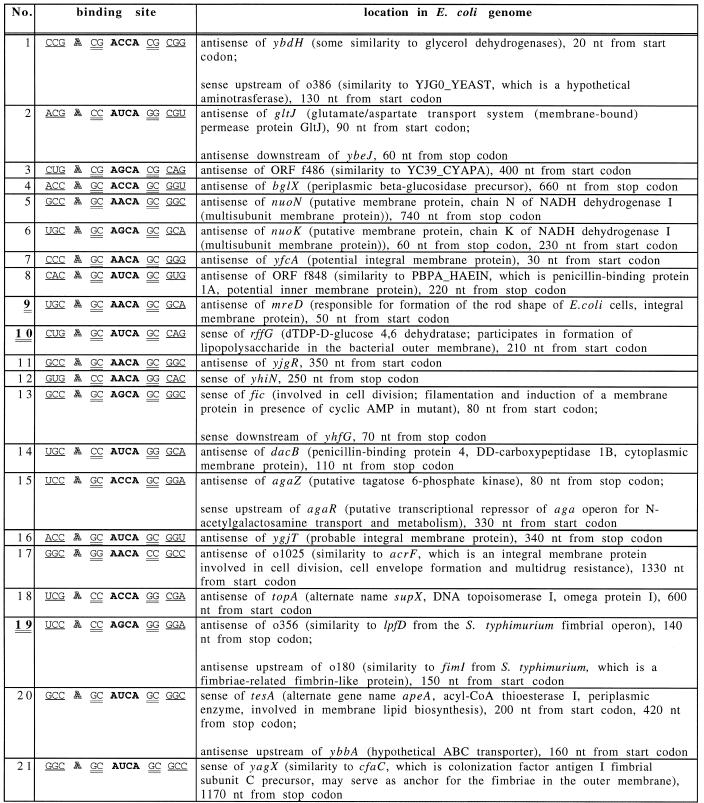
Table 2. Matches found in the E.coli genome to the structure of SELEX consensus MS2 CP binding site, using the RNAMOT program.
The sites shown double-underlined and in bold (9, 10 and 19) correspond to SELEX isolates 5, 1 and 3 in Table 1. The consensus binding site elements (Fig. 1E) are separated by spaces. The ANCA loop is in bold, the bulged A is outlined, the 2 nt stem is double-underlined and the 3 nt stem is underlined.
Labeled RNA (∼0.1 nM) was bound to MS2 CP in variable excess concentrations as during SELEX, but without pre-filtering, at 24–25°C. Each binding reaction was filtered through nitrocellulose and washed with 0.5 ml of the binding buffer.
STOGEN: a computer program to choose new fixed sequences
To reduce the possible influence of the fixed sequences on the outcome of SELEX, a computer program to design new fixed sequences was developed. The program (available by anonymous ftp from ftp://beagle.colorado.edu/pub/stogen ) takes the old fixed sequences as input. It generates possible candidates for the new fixed sequences, using four heuristic rules with user-adjustable parameters (see below). For computational efficiency, the program does not generate and test all possible sequences of a given length, but rather randomly generates a subset of sequences, tests them and repeats the process again, until it arrives at sequences that conform to all of the rules (hence the name of the program, STOGEN: stochastic generator).
The STOGEN rules are that the new fixed sequences should (i) have approximately the same annealing temperatures to the primers as the old fixed sequences, in order to facilitate amplification; (ii) form among themselves as little secondary structure as possible; (iii) share as little similarity as possible to the old ones; (iv) have minimal potential to form secondary structure with the genomic insert.
RESULTS
Binding sites from the regular MS2 CP SELEX agree with the known consensus structure
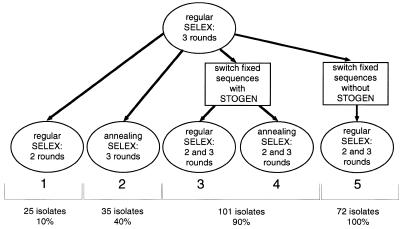
A library of genomic DNA was prepared from E.coli B by random primer extension (13). The library contained ∼65 nt genomic inserts flanked by fixed sequences, which serve as primer annealing sites for amplification. ‘Insert’ refers to the genomic sequence located in the library molecule between the two fixed sequences. In each round of SELEX, the transcribed library was allowed to bind MS2 CP and then the bound RNA was amplified. In SELEX experiment 1, the DNA was cloned and sequenced after five rounds, when the optimal binding was observed.
Out of 25 isolates sequenced, 12 had the predicted consensus (15) binding site (Fig. 1A), which could be identified either by folding by hand, or by computerized Zuker–Turner folding (24–26). Of the 12 isolates, 10 were found in GenBank, which included the complete E.coli sequence, by the BLAST search (27) using the network service at the NCBI (http://www.ncbi.nlm.nih.gov ). The remaining two isolates did not have significant similarities to any sequences in GenBank and might have resulted from contamination of the starting genomic library DNA.
Surprisingly, the genomic sequences obtained from GenBank that corresponded to 9 out of the 10 isolates with the consensus binding site, did not contain this site. Thus, they were not predicted to bind MS2 CP. In other words, 9 out of the 10 isolates were experimentally-induced artifacts.
One of the frequent artifacts is shown in Figure 1B and C. In this isolate, the fixed sequence participates in forming the binding site. This isolate also has several mutations in the insert that participate in forming the binding site. All of the mutations are at the junction between the insert and the fixed sequence. This junction is much more prone to mutations than the rest of the genomic insert because of the random sequence introduced when the randomized primer misannealed during the construction of the library (13). The mutated region of the genomic insert at its junction with the fixed sequence is termed the ‘tail’.
In the isolate shown in Figure 1B and C, as well as in most other isolates, the tail and the fixed sequence both participate in forming the binding site. The corresponding genomic sequences from GenBank were very different from the tails and, obviously, from the fixed sequences and thus were unable to form the binding site.
Annealing of oligonucleotides complementary to the fixed sequences reduces the fraction of SELEX artifacts
The first method to solve the problem of artifacts was designed to reduce the participation in binding of only the fixed sequences, but not the tails. The genomic SELEX described above (termed regular genomic SELEX) was carried out for three rounds. In the three subsequent rounds, two DNA oligonucleotides complementary to the two fixed sequences were annealed to RNA prior to its binding to MS2 CP. SELEX with annealing was carried out for three rounds (Fig. 2, SELEX experiment 2). Switching from ‘no annealing’ to ‘annealing’, rather than doing all six rounds ‘annealing’, should reduce the fraction of isolates that require annealing for binding to MS2 CP.
Figure 2.
Outline of the genomic SELEX experiments 1–5, with the number of isolates sequenced at the end of each SELEX and (for isolates found in GenBank) percentage of the isolates in which the MS2 CP binding site was present in the genomic sequence.
Out of 35 sequenced isolates from ‘annealing’ SELEX, 25 had the consensus binding site and 16 of those 25 were found in GenBank. Of these 16 isolates, 7 (40%) had a consensus binding site present in the corresponding genomic sequence from GenBank and the rest were artifacts as described above. The fraction of artifacts in which fixed sequences, but not tails, participated in binding, decreased only ∼2-fold.
Switching fixed sequences eliminates most of the SELEX artifacts
The second, and more efficient, method of reducing the artifacts consists of switching the fixed sequences halfway through the course of SELEX, replacing them with entirely new fixed sequences and at the same time eliminating the ‘tails’ altogether (Fig. 3).
FokI endonuclease was used to cut the 3′ fixed sequence and the tail of the library DNA after round 3 of regular SELEX. FokI cuts at a specific distance (9–13 nt, regardless of their sequence) away from its recognition site, which was introduced in the fixed sequence near its junction with the genomic insert. After digestion with FokI, the overhang at the cut end of the library DNA was extended with the Klenow fragment of E.coli DNA polymerase, and blunt-end ligated to the new 3′ fixed sequence, which was a duplex of synthetic oligonucleotides. The ligation product was amplified by PCR and the whole procedure was repeated—this time to switch the 5′ fixed sequence. The new fixed sequences were chosen using a specially developed computer program, STOGEN, but may also be chosen manually, with careful consideration of all the important factors involved (see Materials and Methods).
After switching the 5′ and 3′ fixed sequences, SELEX was performed in two different ways in parallel: (i) with annealing of the DNA oligonucleotides complementary to the new fixed sequences and (ii) without any oligonucleotides, as in regular SELEX (Fig. 2, SELEX experiments 3 and 4). Both SELEX experiments gave virtually identical results. After two and three rounds of SELEX with the new fixed sequences, 101 isolates were sequenced. Out of 101 isolates, 77 had the consensus binding site and 76 out of these 77 were found in GenBank.
The fixed sequences never made up any part of the consensus binding site. In a few cases we believe that the FokI treatment did not completely remove the tails. One characteristic of the tails is their higher mutation frequency and their proximity to the fixed sequences. Nineteen out of 76 molecules had this characteristic and, out of these, one had a mutation that made a part of the consensus binding site. Internal, rather than tail, mutations caused four artifacts.
All of the SELEX isolates with the MS2 CP consensus binding site (Table 1) had higher-affinity RNCA tetraloop instead of the lower-affinity RNUA tetraloop of the natural MS2 CP binding site on MS2 mRNA (28,29). As expected, these SELEX isolates bound better than the natural binding site, as measured by the nitrocellulose filter-binding assay (Fig. 4). Twenty-four of 101 sequenced isolates did not contain the consensus binding site and the negligible binding of one of them is shown in Figure 4. None of the F6-like, 3 nt loop variants, which bind more weakly than the RNCA tetraloop site (30), have been found in the genomic SELEXes.
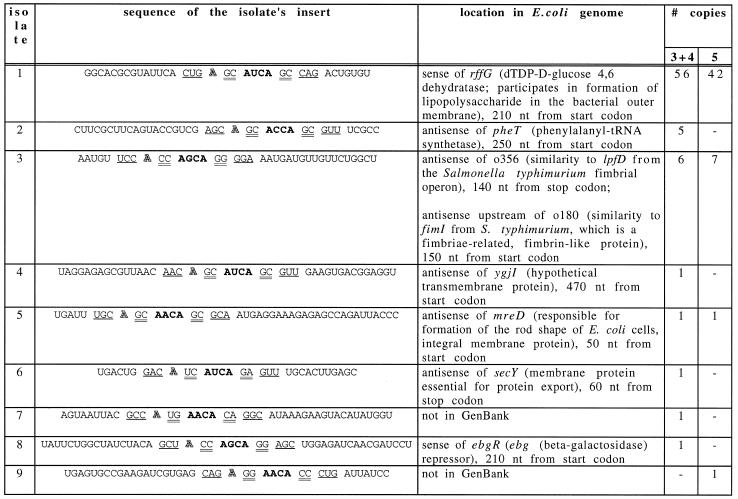
Table 1. Isolates with the consensus binding site from SELEXes with switching fixed sequences.
The consensus binding site elements (Fig. 1A) are separated by spaces. The RNYA loop is in bold, the bulged A is outlined, the 2 nt stem is double-underlined and the 3 nt stem is underlined. Isolate No. 1 is shown folded in Figure 1D. ‘# copies’ indicates in how many copies a particular isolate was found, out of the total 101 isolates sequenced in SELEX experiments 3 and 4 and out of 72 isolates in SELEX experiment 5 (Fig. 2).
The consensus of the isolates in Table 1, with consideration of their frequencies in the selected pool, is shown in Figure 1E. Most of the differences between this SELEX consensus site and the consensus site in Figure 1A, should make the binding tighter or the binding structure more stable (14,15). This SELEX consensus also agrees with the data from the randomized sequence SELEX (19).
Fifty-six isolates were from the sense strand of mRNA of the rffG gene (Fig. 1D). These molecules were not only the most frequent, but also the tightest binders among the SELEX isolates. The corresponding genomic fragment (without the fixed sequences) also bound MS2 CP well (Fig. 4). The rffG open reading frame, o355, was mistakenly labeled rffE in the GenBank version 90.0 (31). The enzyme dTDP-d-glucose-4,6-dehydratase is encoded by rffG. This enzyme participates in formation of O-specific polysaccharide or O antigen, which, joined together with lipid A via core oligosaccharide, forms lipopolysaccharide in the bacterial outer membrane (32). The enzyme also participates in formation of the polysaccharide part of the enterobacterial common antigen, a cell surface glycolipid (33). Other isolates that bind MS2 CP were located in six distinct genomic sites.
Comparison of the MS2 CP binding sites predicted by the computer search with the sites found by SELEX
Other sites in the E.coli genome that bind to MS2 CP were much less frequent than rffG, in these (Table 1) and prior rounds of SELEX (data not shown). This raised the question about the efficiency of finding all potentially biologically important binding sites. To check if any other binding sites were missed by SELEX, a search was performed for the MS2 CP consensus binding site (Fig. 1A) in the complete E.coli genome, using the RNAMOT program (34). The search revealed 412 matches to the consensus binding site, each of which theoretically binds the coat protein as well as, or better than, the wild-type MS2 mRNA. Only 280 sites were expected by chance.
To narrow this list to only the tightest binding sites, the SELEX consensus binding site (Fig. 1E) was searched for. It is based on the genomic SELEX isolates and is more restrictive than the consensus binding site, which is based on the studies of mutants. RNAMOT found 21 such SELEX consensus binding sites (Table 2). Only three sites were expected to be found at random.
Three binding sites were found both by SELEX and by the RNAMOT program, including the major (rffG) SELEX isolate. Most of the minor SELEX isolates were not found by RNAMOT. Some of these did not fit the SELEX consensus used for searching the database. For example, had G:U pairs been allowed in the consensus, these sites would have been found, too. Others, like isolate nos 7 and 9 from Table 1, were not in the database.
Most of the RNAMOT sites were not found by SELEX. It is possible that some of them were under-represented in the starting library, that they were poorly amplifiable in SELEX or that they bound MS2 CP weakly because RNA folds into alternate, non-binding structures within the context of a larger molecule.
An interesting feature of the major rffG isolate, relative to other SELEX isolates, was a fairly long stem that supported the binding site (Fig. 1D). Perhaps a longer stem provides extra stability to the correct binding site structure and thus reduces the fraction of molecules folded into other, non-binding, structures. In the randomized sequence SELEX for R17 coat protein binders, Schneider et al. also found mostly long (7 bp) and stable (mostly G:C or C:G base pairs) stems (19). Also, in the regular MS2 CP genomic SELEX (without switching the fixed sequences or annealing of oligonucleotides; Fig. 2, experiment 1) many isolates used fixed sequences not only to form the consensus binding site, but also to extend its stem past the minimum 5 bp. In SELEX experiments 3 and 4 the fixed sequences, while not forming the consensus binding site, sometimes extended the stem to longer than the minimal 5 bp. In SELEX experiment 5, the new fixed sequences were chosen without STOGEN and their potential to base-pair to each other and to the insert was accidentally overlooked. In most of the isolates from this SELEX, the fixed sequences extended the minimal stem by an additional 12 bp.
However, some RNAMOT sites (nos 8, 12 and 14, Table 2) are also predicted to have long stems, just like the most frequent genomic SELEX isolates (nos 1, 2 and 3, Table 1) and yet they were not found in SELEX. They fit the randomized sequence SELEX consensus (19) as well or better than the most frequent genomic SELEX isolates. They also bind MS2 CP rather well. Sites nos 8 and 14 bind MS2 CP with affinities between those of SELEX isolate nos 1 and 2 (data not shown). Thus, these sites were not isolated in SELEX for reasons other than their affinity to MS2 CP. Site No. 12 binds MS2 CP with affinity only slightly weaker than SELEX isolate No. 6.
DISCUSSION
Genomic SELEX: libraries and selection methods
Genomic SELEX for E.coli RNA that binds to phage MS2 CP showed that fixed sequences and tails of the library molecules can influence the outcome of SELEX, with the result that most of the isolates turn out to be artifacts (Fig. 1). The fraction of artifacts can be decreased ∼2-fold by annealing of DNA oligonucleotides complementary to the fixed sequences. If the fixed sequences are switched halfway through the SELEX rounds and tails are eliminated (Figs 2 and 3), the fraction of artifacts becomes only a few percent of the total isolates. The new fixed sequences must be chosen with consideration of several important factors, which was done using a specially designed computer program, STOGEN.
The genomic sites isolated by SELEX have markedly different frequencies in the final pool (Tables 1 and 2), which may be influenced by several factors, such as affinity to MS2 CP, frequency in the starting library and ability to be amplified in SELEX. To identify potentially interesting, but less frequent isolates, one can deplete the pool from the major isolates, for instance by using complementary biotinylated oligonucleotides and immobilized streptavidin.
It is possible that the starting libraries constructed using other methods will have a more uniform distribution of genomic fragments. Libraries made by mechanical fragmentation and blunt-end ligation (35) are good candidates to test in future experiments. Perhaps a mixture of several libraries, each constructed independently by a different method, would be more uniform than any of the individual libraries alone.
Proteins other than MS2 CP may have significantly longer binding sites and thus may require longer library inserts. It would also be interesting to test whether genomic SELEX with longer inserts yields fewer artifacts, as the fixed sequences and tails become smaller relative to the insert.
Like other methods of investigating nucleic acid–protein interactions, genomic SELEX has its limitations. Genomic SELEX is biased toward strong interactions. Some RNA molecules may be bound weakly but, because they are abundant, engage in significant interactions. It is also expected that a sizeable fraction of interactions identified in genomic SELEX may be biologically irrelevant, due to competition for binding to multiple proteins, folding of RNA into alternate structures, the absence of RNA expression and other factors. Because of the inherent complexity of the problem and the fact that the strengths of different methods often complement each other, genomic SELEX is best used in combination with other methods (both computational and experimental) of studying nucleic acid–protein interactions.
Locations of MS2 CP binding sites
The observed binding site locations within the genes do not follow any obvious pattern. In particular, the sites are not located close to the predicted ribosome binding sites. This was determined by comparing the GenBank sequences with the translation initiation site consensus (36) using GCG fitconsensus program (26).
MS2 CP binding may affect, for example, mRNA stability, as is the case with many protein–RNA interactions (37–40). MS2 is known to specifically repress the synthesis of some E.coli proteins (41). Since bacterial mRNA synthesis at the same time is affected relatively little (42,43), repression of the protein synthesis must be post-transcriptional, for example, at the RNA level, as suggested by the genomic SELEX results.
Some binding sites isolated by SELEX are located on the antisense RNA strand. They may still have a biological role. There are many known cases in which the antisense RNA negatively regulates the sense strand gene expression (44). Often, the mechanism involves RNaseIII cleavage of the sense–antisense RNA duplex. It is possible that MS2 CP binding prevents the antisense strand from hybridizing to the sense strand and thus positively regulates the sense strand gene expression. Indeed, there are several known examples of proteins that bind to the antisense strand RNA and thus affect the sense strand gene expression (39,40,45,46).
All of the antisense sites listed in Table 1 are close (within a few hundred nucleotides) to promoter-like sequences. This was determined using computerized promoter search (47). Of course, the antisense molecules can also be transcribed by read-through from the upstream genes on the antisense strand.
The functions of the genes that contain MS2 CP binding sites
Most of the E.coli genes containing MS2 CP binding sites have something to do with the cell surface (Table 1). It is possible that the phage changes the cell surface through the interaction of the coat protein with the E.coli RNA. MS2 CP–RNA interactions may play several possible roles:
MS2 CP–RNA interactions may benefit the phage, e.g. by facilitating its budding [a non-lytic mode of MS2 replication (48–51)] or by changing the E.coli adhesion properties so that the infected cell could wander away to infect more cells.
Escherichia coli may use the same MS2 CP–RNA interactions to reverse these processes—to the benefit of itself or other, uninfected cells.
Both phage and bacteria may benefit from the interactions, if it prevents other phages from entering the cell or if bacterial virulence is positively affected.
The examples below illustrate these possible scenarios. They do not in any way prove that the MS2 CP–RNA interactions identified by genomic SELEX have a specific biological role (most of them probably do not). The examples serve only to point out some of the many possible mechanisms in which these interactions can be utilized by the phage and the bacteria.
The potential interaction between MS2 CP and rffG RNA (isolate No. 1, Table 1) may affect the cell surface by changing the O antigen, whose synthesis is rffG-dependent. Phages are well-known to cause changes in the O antigen (52–54). In some cases, these changes protect both the bacteria and the resident phage from subsequent infection by other phages (55,56), the process in which lipopolysaccharide and its O antigen part play an active role (57,58).
The potential interaction between MS2 CP and o356 or o180 genes (isolate No. 3, Table 1), which display similarities to fimbriae-related genes lpfD and fimI, may also change the bacterial surface to make the cell resistant to other phages. Indeed, there are known examples of phages that require another fimbrial gene (fimU) for infection (59).
The potential binding of MS2 CP to secY (isolate No. 6) may affect budding of the phage, since SecY is known to interact with phage coat proteins (60). The binding may also make the cell resistant to other phages, because secY is necessary for export of many surface proteins, such as phage λ receptor (61).
In addition to affecting bacterial gene expression, MS2 CP–RNA interactions may be used by the phage to assess the physiological state of the cell and thus to adjust the phage life cycle accordingly. The E.coli RNAs may compete with MS2 replicase site for binding to MS2 CP, decreasing the translational repression of the replicase and thus influencing the number of phage particles produced. The concentrations of the E.coli RNAs will vary depending on the state of the bacterial cell. For example, the level of rffG mRNA, through its binding to MS2 CP, may indicate to the phage the state of the cell surface. Having MS2 CP bound to many different RNAs encoding functionally-related proteins would be a good way to produce more phage replicase when needed. The fact that RNA fragments from related genes were isolated in genomic SELEX supports this hypothesis.
Many bacteriophages are known to adapt their life cycle to variations in the physiological state of the bacterial cell. For example, MS2 lysis is dependent on the availability of nutrients (51), temperature (48), bacterial growth phase (49,50), pH of the media and the state of the cell surface (62). Lysis of cells infected by phage φX174 is dependent on the media composition and the function of a number of bacterial genes related to the cell surface, including envC and dacB (62).
If desired, the presence and the effects of the interactions between the E.coli genes and MS2 CP in vivo can be tested in a separate study using well-established methods. The results of the present study merely point to an interesting connection between bacteriophage MS2 and the cell surface of the bacteria that it infects. Genomic SELEX appears capable of uncovering surprising interactions even in a relatively simple and well-studied system. One can expect that for other proteins, or mixtures of proteins, this method will also frequently reveal previously unrecognized biological regulatory loops (1–3).
MS2 CP genomic SELEX results suggest that to reduce the fraction of experimentally-induced artifacts among the isolates, selection should be carried out with replacement of the fixed sequences. Annealing of oligonucleotides complementary to the fixed sequences is not as effective by itself and, when coupled with replacement of the fixed sequences, has no additional effect. The improvements in genomic SELEX methodology resulting from the present study can be useful for the future genomic and randomized sequence SELEX applications.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Ken Blount (University of Colorado, Boulder) for providing us with MS2 coat protein and P. Allen, D. Brown, D. Burke, T. Cech, C. Chapon, H. Chen, S. Dutcher, Y. Y. He, K. Krauter, L. Leinwand, S. Ringquist, G. Stormo, M. Wecker, M. Yarus and S. Zimmerman for helpful ideas and discussions. H.E.J. was supported by a post-doctoral fellowship from the Swedish Natural Science Research Council. This work was supported by NIH grant GM19963 to L.G. and by funds from NeXstar Pharmaceuticals.
REFERENCES
- 1.Gold L., Polisky,B., Uhlenbeck,O. and Yarus,M. (1995) Annu. Rev. Biochem., 64, 763–797. [DOI] [PubMed] [Google Scholar]
- 2.Gold L. (1995) J. Biol. Chem., 270, 13581–13584. [DOI] [PubMed] [Google Scholar]
- 3.Gold L., Brown,D., He,Y.-Y., Shtatland,T., Singer,B.S. and Wu,Y. (1997) Proc. Natl Acad. Sci. USA, 94, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gold L., Singer,B., He,Y.Y. and Brody,E. (1997) Curr. Opin. Genet. Dev., 7, 848–851. [DOI] [PubMed] [Google Scholar]
- 5.Tuerk C. and Gold,L. (1990) Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 6.Ellington A.D. and Szostak,J.W. (1990) Nature, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 7.Chien C.T., Bartel,P.L., Sternglanz,R. and Fields,S. (1991) Proc. Natl Acad. Sci. USA, 88, 9578–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J.J. and Herskowitz,I. (1993) Science, 262, 1870–1874. [DOI] [PubMed] [Google Scholar]
- 9.SenGupta D.J., Zhang,B., Kraemer,B., Pochart,P., Fields,S. and Wickens,M. (1996) Proc. Natl Acad. Sci. USA, 93, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Putz U., Skehel,P. and Kuhl,D. (1996) Nucleic Acids Res., 24, 4838–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel P.L., Roecklein,J.A., SenGupta,D. and Fields,S. (1996) Nature Genet., 12, 72–77. [DOI] [PubMed] [Google Scholar]
- 12.Fromont-Racine M., Rain,J.C. and Legrain,P. (1997) Nature Genet., 16, 277–282. [DOI] [PubMed] [Google Scholar]
- 13.Singer B.S., Shtatland,T., Brown,D. and Gold,L. (1997) Nucleic Acids Res., 25, 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhlenbeck O.C., Carey,J., Romaniuk,P.J., Lowary,P.T. and Beckett,D. (1983) J. Biomol. Struct. Dyn., 1, 539–552. [DOI] [PubMed] [Google Scholar]
- 15.Witherell G.W., Gott,J.M. and Uhlenbeck,O.C. (1991) Prog. Nucleic Acids Res. Mol. Biol., 40, 185–220. [DOI] [PubMed] [Google Scholar]
- 16.Stockley P.G., Stonehouse,N.J., Murray,J.B., Goodman,S.T., Talbot,S.J., Adams,C.J., Liljas,L. and Valegård,K. (1995) Nucleic Acids Res., 23, 2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valegård K., Murray,J.B., Stockley,P.G., Stonehouse,N.J. and Liljas,L. (1994) Nature, 371, 623–626. [DOI] [PubMed] [Google Scholar]
- 18.Valegård K., Murray,J.B., Stonehouse,N.J., van den Worm,S., Stockley,P.G. and Liljas,L. (1997) J. Mol. Biol., 270, 724–738. [DOI] [PubMed] [Google Scholar]
- 19.Schneider D., Tuerk,C. and Gold,L. (1992) J. Mol. Biol., 228, 862–869. [DOI] [PubMed] [Google Scholar]
- 20.Brockmann D. and Esche,H. (1995) Curr. Top. Microbiol. Immunol., 199, 81–112. [DOI] [PubMed] [Google Scholar]
- 21.LeCuyer K.A., Behlen,L.S. and Uhlenbeck,O.C. (1995) Biochemistry, 34, 10600–10606. [DOI] [PubMed] [Google Scholar]
- 22.Tuerk C. (1997) Methods Mol. Biol., 67, 219–230. [DOI] [PubMed] [Google Scholar]
- 23.Sugino A., Goodman,H.M., Heyneker,H.L., Shine,J., Boyer,H.W. and Cozzarelli,N.R. (1977) J. Biol. Chem., 252, 3987–3994. [PubMed] [Google Scholar]
- 24.Serra M.J. and Turner,D.H. (1995) Methods Enzymol., 259, 242–261. [DOI] [PubMed] [Google Scholar]
- 25.Zuker M. (1989) Science, 244, 48–52. [DOI] [PubMed] [Google Scholar]
- 26. Genetics Computer Group. (1994) Program Manual for the Wisconsin Package, 8th Ed., Madison, WI.
- 27.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 28.Johansson H.E., Dertinger,D., LeCuyer,K.A., Behlen,L.S., Greef,C.H. and Uhlenbeck,O.C. (1998) Proc. Natl Acad. Sci. USA, 95, 9244–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowary P.T. and Uhlenbeck,O.C. (1987) Nucleic Acids Res., 15, 10483–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Convery M.A., Rowsell,S., Stonehouse,N.J., Ellington,A.D., Hirao,I., Murray,J.B., Peabody,D.S., Phillips,S.E. and Stockley,P.G. (1998) Nature Struct. Biol., 5, 133–139. [DOI] [PubMed] [Google Scholar]
- 31.Marolda C.L. and Valvano,M.A. (1995) J. Bacteriol., 177, 5539–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raetz C.R.H. (1996) In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd Edn. ASM Press, Washington, DC, pp. 1035–1063.
- 33.Rick P.D. and Silver,R.P. (1996) In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd Edn. ASM Press, Washington, DC, pp. 104–122.
- 34.Gautheret D., Major,F. and Cedergren,R. (1990) Comput. Applic. Biosci., 6, 325–331. [DOI] [PubMed] [Google Scholar]
- 35.Kinzler K.W. and Vogelstein,B. (1989) Nucleic Acids Res., 17, 3645–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalphin M.E., Brown,C.M., Stockwell,P.A. and Tate,W.P. (1997) Nucleic Acids Res., 25, 246–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glatz E., Nilsson,R.P., Rutberg,L. and Rutberg,B. (1996) Mol. Microbiol., 19, 319–328. [DOI] [PubMed] [Google Scholar]
- 38.Bovy A., de Kruif,J., de Vrieze,G., Borrias,M. and Weisbeek,P. (1993) Plant Mol. Biol., 22, 1047–1065. [DOI] [PubMed] [Google Scholar]
- 39.van Biesen T. and Frost,L.S. (1994) Mol. Microbiol., 14, 427–436. [DOI] [PubMed] [Google Scholar]
- 40.Lee S.H., Frost,L.S. and Paranchych,W. (1992) Mol. Gen. Genet., 235, 131–139. [DOI] [PubMed] [Google Scholar]
- 41.Sugiyama T. and Stone,H.O.,Jr (1968) J. Mol. Biol., 36, 91–105. [DOI] [PubMed] [Google Scholar]
- 42.Berzin V., Rosenthal,G. and Gren,E.J. (1974) Eur. J. Biochem., 45, 233–242. [DOI] [PubMed] [Google Scholar]
- 43.Yamazaki H. (1969) Virology, 37, 429–436. [DOI] [PubMed] [Google Scholar]
- 44.Simons R.W. and Kleckner,N. (1988) Annu. Rev. Genet., 22, 567–600. [DOI] [PubMed] [Google Scholar]
- 45.Tasheva E.S. and Roufa,D.J. (1995) Genes Dev., 9, 304–316. [DOI] [PubMed] [Google Scholar]
- 46.Eguchi Y. and Tomizawa,J. (1990) Cell, 60, 199–209. [DOI] [PubMed] [Google Scholar]
- 47.Hertz G.Z. and Stormo,G.D. (1996) Methods Enzymol., 273, 30–42. [DOI] [PubMed] [Google Scholar]
- 48.Engelberg H. and Soudry,E. (1971) J. Virol., 8, 257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haywood A.M. (1974) J. Gen. Virol., 22, 431–435. [Google Scholar]
- 50.Ricciuti C.P. (1972) J. Virol., 10, 162–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Propst-Ricciuti B. (1976) J. Gen. Virol., 31, 323–330. [DOI] [PubMed] [Google Scholar]
- 52.Verma N.K., Brandt,J.M., Verma,D.J. and Lindberg,A.A. (1991) Mol. Microbiol., 5, 71–75. [DOI] [PubMed] [Google Scholar]
- 53.Clark C.A., Beltrame,J. and Manning,P.A. (1991) Gene, 107, 43–52. [DOI] [PubMed] [Google Scholar]
- 54.Lindberg A.A. (1977) In Sutherland,I. (ed.), Surface Carbohydrates of the Prokaryotic Cell. Academic Press, London, pp. 289–356.
- 55.Barksdale L. and Arden,S.B. (1974) Annu. Rev. Microbiol., 28, 265–299. [DOI] [PubMed] [Google Scholar]
- 56.Kuzio J. and Kropinski,A.M. (1983) J. Bacteriol., 155, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Ley P., de Graaff,P. and Tommassen,J. (1986) J. Bacteriol., 168, 449–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Datta D.B., Arden,B. and Henning,U. (1977) J. Bacteriol., 131, 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alm R.A. and Mattick,J.S. (1996) J. Bacteriol., 178, 3809–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters E.A., Schatz,P.J., Johnson,S.S. and Dower,W.J. (1994) J. Bacteriol., 176, 4296–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swidersky U.E., Rienhofer-Schweer,A., Werner,P.K., Ernst,F., Benson,S.A., Hoffschulte,H.K. and Muller,M. (1992) Eur. J. Biochem., 207, 803–811. [DOI] [PubMed] [Google Scholar]
- 62.Young R. (1992) Microbiol. Rev., 56, 430–481. [DOI] [PMC free article] [PubMed] [Google Scholar]