Abstract
Although the green alga Chlamydomonas reinhardtii has long served as a reference organism, few studies have interrogated its role as a primary producer in microbial interactions. Here, we quantitatively investigated C. reinhardtii’s capacity to support a heterotrophic microbe using the established coculture system with Mesorhizobium japonicum, a vitamin B12-producing α-proteobacterium. Using stable isotope probing and nanoscale secondary ion mass spectrometry (nanoSIMS), we tracked the flow of photosynthetic fixed carbon and consequent bacterial biomass synthesis under continuous and diurnal light with single-cell resolution. We found that more 13C fixed by the alga was taken up by bacterial cells under continuous light, invalidating the hypothesis that the alga’s fermentative degradation of starch reserves during the night would boost M. japonicum heterotrophy. 15NH4 assimilation rates and changes in cell size revealed that M. japonicum cells reduced new biomass synthesis in coculture with the alga but continued to divide—a hallmark of nutrient limitation often referred to as reductive division. Despite this sign of starvation, the bacterium still synthesized vitamin B12 and supported the growth of a B12-dependent C. reinhardtii mutant. Finally, we showed that bacterial proliferation could be supported solely by the algal lysis that occurred in coculture, highlighting the role of necromass in carbon cycling. Collectively, these results reveal the scarcity of fixed carbon in this microbial trophic relationship (particularly under environmentally relevant light regimes), demonstrate B12 exchange even during bacterial starvation, and underscore the importance of quantitative approaches for assessing metabolic coupling in algal–bacterial interactions.
Keywords: rhizobia, chlorophyte, photoautotroph, diel, cobalamin, cocultivation, symbiosis, SIP
Introduction
Photosynthetic microbes are responsible for a large fraction of the primary productivity on our planet [1]. These organisms supply the microbial world with raw materials for life, thereby shaping communities and driving carbon cycling [2, 3]. Through exudation, lysis, and predation, photosynthetic microbes provide a variety of reduced carbon substrates with various diffusion rates and accessibility to heterotrophic bacteria. The primary consumers recycle carbon back to inorganic forms, closing the “microbial loop” that constitutes a major component of biogeochemical carbon cycling [2, 4, 5]. Heterotrophic bacteria can also influence algal productivity, for example, by changing the availability of critical micronutrients for algal growth and metabolism [6–8].
The green alga Chlamydomonas reinhardtii has served as a reference organism for discoveries in photosynthesis, cilia-based motility, and the eukaryotic cell cycle [9]. Extensive work on axenic (bacteria-free) cultures has provided detailed knowledge of the pathways and regulons for photosynthesis and primary metabolism. This chlorophyte has a flexible metabolic repertoire, with the capacity to grow by photoautotrophy alone or by acetate assimilation. Under diurnal cycles, C. reinhardtii’s metabolism is shaped by programmed waves of gene expression that accommodate diurnal changes in light and oxygen availability in natural environments [10, 11]. Photosynthesis fuels cell growth and starch synthesis during the day, and starch reserves are glycolytically degraded at night, providing a source of energy until the next dawn. Although extensive knowledge of its metabolism makes C. reinhardtii an attractive model organism to study microbial trophic interactions, relatively little is known about how C. reinhardtii contributes to its environment and influences its neighbors in the microbial world.
In its natural soil context, C. reinhardtii is surrounded by other microbes that exchange nutrients, compete for resources, and communicate with one another. Chlamydomonas reinhardtii can enrich for particular heterotrophic bacteria from bulk soil, demonstrating that the alga’s growth can influence the metabolisms and behaviors of various bacteria [12, 13]. Studies of Chlamydomonas–bacteria interactions have primarily focused on impacts to the alga’s behavior, in part due to technical challenges in probing bacterial physiology in mixed cultures. Bacteria are known to supply vitamins and fixed nitrogen, facilitate iron assimilation, and impart thermal tolerance to algal cultures [6, 7, 14–20]. Coculture of C. reinhardtii with various heterotrophic bacteria (e.g. Bradyrhizobium japonicum, Pseudomonas spp.) has been shown to stimulate algal H2 production, starch accumulation, and viability under sulfur deprivation by consuming O2 and alleviating accumulation of fermentation byproducts from the medium [21]. Bacterial secondary metabolites can also influence C. reinhardtii’s cell cycle and aggregation [22, 23]. In addition, interactions between C. reinhardtii and vitamin B12-producing rhizobia like Mesorhizobium japonicum (previously referred to as Mesorhizobium loti), Sinorhizobium meliloti, and Rhizobium leguminosarum have been used to study the evolution of vitamin dependency across the algal tree of life [6, 15, 24–28]. Chlamydomonas reinhardtii does not require vitamin B12 for growth, because it has retained a functional cobalamin-independent isoform of methionine synthase, METE1. However, when vitamin B12 is supplied, METE1 is repressed, and the vitamin is used as a cofactor for the cobalamin-dependent isoform, METH1. When C. reinhardtii was grown in the laboratory with a continuous supply of the vitamin, the alga acquired deleterious mutations in the repressed METE1 gene, yielding B12-requiring strains (mete1) in <500 generations [25].
The interaction between the C. reinhardtii mete1 mutant and M. japonicum is the most well-studied C. reinhardtii coculture system. It has been suggested to be facultatively mutualistic: the mutant alga receives the vitamin cofactor, and the bacterium is believed to receive reduced carbon compounds that support heterotrophic growth. However, the mechanism and magnitude of the flux of carbon transfer from C. reinhardtii to M. japonicum are unknown. Quantitative investigations of algal–bacterial interactions have been challenging due to technical barriers for studying eukaryotes and prokaryotes simultaneously. We have addressed this challenge by using spatiotemporal stable isotope tracing via nanoscale secondary ion mass spectrometry (nanoSIMS) to probe this simple model interaction—a technique that has typically been used for studying complex polycultures from the field. We quantified the transfer of fixed carbon from wild-type C. reinhardtii to M. japonicum in continuous and diurnal light. Although some photosynthetically fixed carbon was taken up by the bacterial population, M. japonicum growth was severely limited, resulting in reductive bacterial division. Thus, C. reinhardtii primary productivity yields a low carrying capacity for this bacterium, particularly under diurnal light. Nonetheless, M. japonicum provided adequate B12 to enable growth of the C. reinhardtii mete1 strain even when the bacterium itself exhibited signs of starvation. These results highlight that in natural contexts, bacteria may not proliferate to the high cell densities that are possible in nutrient-rich media often used in the laboratory, but they can still greatly influence the metabolism and evolution of their neighbors in the microbial world [25].
Materials and methods
Strains and culture conditions
Three strains of C. reinhardtii were used in this study. The mete1 strain generated through experimental evolution on 1000 ng/l vitamin B12 [25] and its parental wild-type strain were provided by Alison G. Smith (University of Cambridge, UK). According to its haplotype, this strain is closely related to strain S24- (SI Fig. 1) [29]. For experiments testing the impact of diurnal light, we used the cell-wall reduced strain CC-5390. This strain is amenable to synchronization under diurnal cycles, providing a system with exceptional signal-to-noise for resolving diurnal patterns [10]. Mesorhizobium japonicum strain MAFF303099 was also provided by Alison G. Smith.
All experiments were performed in amended high salt minimal (HSM) medium [30] with a modified trace element solution [31], 2 mM MgSO4·7H2O, 10 μg/l biotin, and 10 μM CoCl2 unless otherwise specified (Supplementary Data File S2: Media composition). Cultures were grown in either 250 ml Erlenmeyer flasks containing 100 ml medium and bubbled with filter-sterilized air (provided by an aquarium pump), 6-well plates with 3 ml medium, or 250 ml beakers with 50 ml medium. Algal cultures and cocultures were incubated at 28°C, agitated at 110–130 rpm, and illuminated with 200 μmol photons/m2/s cool white light (Sylvania Dulux L55WFT55DL/841) continuously or under diurnal 12-h-light/12-h-dark cycles.
Chlamydomonas reinhardtii cells were precultivated axenically in photoautotrophic conditions for at least three passages prior to experiments. The mete1 strain was precultivated with 200–250 ng/l vitamin B12 (cyanocobalamin). For experiments comparing coculture in continuous light versus diurnal light, C. reinhardtii CC-5390 cells were precultivated in flat-panel photobioreactors (Photon System Instruments, Drásov, Czechia) as previously described under the respective light regime to achieve adequate synchrony of the diurnal populations [10].
Bacterial cells were precultivated in 14 ml polystyrene round-bottom tubes with 3 ml amended HSM containing 0.2% sucrose, agitated at 200 rpm, and incubated at 28°C. When larger volumes were required for inoculation, cells were precultivated instead in 250 ml Erlenmeyer flasks containing 100 ml medium, continuously bubbled with air, and agitated at 110–130 rpm. Bacterial cells were collected by centrifugation at 8000 xg and washed three times in 0.85% NaCl to remove sucrose before use as inoculum. Optical density at 600 nm (OD600) was used to estimate the cell density of washed cell suspensions used for inoculation.
Cell number, size, and cytotoxicity measurements
Chlamydomonas reinhardtii cell density and size were determined using a hemocytometer and ImageJ or a Beckman Coulter Multisizer 3 with a 50 μm orifice (Beckman Coulter, CA, USA). Bacterial cell density was determined by counting colony-forming units (CFU) in 10 μl spots of serial dilutions on Typtone Yeast (TY) agar medium (Supplementary Data File S2: Media composition) after 4 d of incubation at 30°C in the dark.
Algal cellular integrity was estimated using CellTox Green Cytotoxicity Assay (Promega, WI, USA). Chlamydomonas reinhardtii sample densities were adjusted to 1 × 106 cells/ml. Triplicate killed control samples were prepared by heating cells at 90°C for 10 min. Fluorescence was measured in a black-walled 96-well plate using a SpectraMax iD3 plate reader (Molecular Devices, CA, USA). Background fluorescence of a medium blank was subtracted from all values. Results were reported relative to killed control samples.
Samples for fluorescence microscopy were fixed overnight in 4% paraformaldehyde at 4°C in the dark. Fixed samples were then collected by centrifugation at 10 000 xg, washed in phosphate-buffered saline (PBS) (pH 7), and stored in 1:1 ethanol:PBS at −20°C. For imaging, samples were spotted on 0.22 μm hydrophilic isopore polycarbonate filters and washed with sterile water to remove residual salts. Prepared filters were mounted on slides with 5 μg/ml 4',6-diamidino-2-phenylindole (DAPI) in Citifluor antifade mounting medium buffered with PBS and imaged using a Zeiss Axioimager M2 microscope with a DAPI filter set (Ex 350/50, Em 460/50). Bacterial cell lengths were measured manually using ImageJ Version 2.1.0/1.53c by drawing a line from one pole of the cell to the other along the longer axis.
Total organic carbon measurements
Total nonpurgeable organic carbon (NPOC) content of cells was determined using a TOC-L Shimadzu Total Organic Carbon Analyzer (Shimadzu, Kyoto, Japan). Ten- to forty-milliliters culture was collected by centrifugation at 8000 xg for 2 min, and the supernatant was collected and then stored at −20°C until analysis. Spent media were diluted 2-fold, and HCl was added to 27 mM. All samples were sparged to remove inorganic carbon. Some organic carbon may be purged from the sample by this method, so we report “nonpurgeable” organic carbon.
Stable isotope labeling and nanoSIMS
15NH4Cl (99% 15N, Cambridge Isotope Laboratories, MA, USA) was provided directly in the culture medium, replacing 50% of the NH4Cl in HSM. 13 h after inoculation, 13CO2 was provided to cultures by bubbling air through a solution of NaH13CO3 (99% 13C, Cambridge Isotope Laboratories) and then into the cultures as previously described [32]. Killed controls were generated by treating cells with 4% paraformaldehyde for 20 min in the dark. Coculture experiments were otherwise conducted as described above.
For nanoSIMS analyses, samples were fixed and deposited on filters as described above for fluorescence microscopy. Dry filter pieces were then mounted on 1 in round stubs using carbon tabs (Ted Pella, Inc., CA, USA) and gold-coated. Data were collected on a NanoSIMS 50 (CAMECA, Gennevilliers, France) using a 1.5 pA Cs+ primary ion beam. The secondary ion masses of 12C2−, 12C13C−, 12C14N−, 12C15N−, and 32S− were collected. Raw nanoSIMS images were processed using L’image (L. Nittler, Carnegie Institution of Washington, D.C., USA). Individual cell regions of interest (ROIs) were manually circled using size and high 12C14N signal to identify bacterial cells and high 12C13C signal along with the secondary electron image to identify algal cells. Isotope enrichment in atom percent enrichment (APE) for 12C13C−/12C2− and 12C15N−/12C14N− were calculated using standard ratios of 0.02247 and 0.00367, respectively. Enrichment data are also presented as net assimilation (Nnet and Cnet). The fraction of newly synthesized biomass relative to total biomass was calculated against an unlabeled biomass standard as previously described [33]:
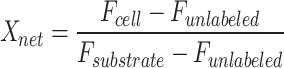 |
where F is the abundance of the heavy isotope (15N or 13C) over the total (14N + 15N or 12C + 13C, respectively). For 15N, net assimilation was calculated relative to the known isotopic composition of the initial source substrate added to the medium. For 13C, net bacterial assimilation was calculated relative to the average algal biomass isotopic composition in the respective experiment (continuous light or diurnal light) as a rough estimation of the source substrate.
Vitamin B12 bioassay
Vitamin B12 concentration was determined using the Escherichia coli B12 bioassay described previously [34, 35]. The bioassay uses two E. coli strains: the ∆metE∆metH control strain requires methionine for growth, and the ∆metE strain can grow when provided either methionine or B12. The amount of B12 in a sample is determined by subtracting the growth of the ∆metE∆metH control strain from the growth of the ∆metE strain after 24-h incubation in the sample and then comparing to a vitamin B12 standard curve.
Culture samples and vitamin B12 standards were boiled at 100°C for 10 min. Cell debris was removed by centrifugation at 16 000 xg for 2 min, and the supernatants were snap-frozen in liquid N2 and stored at −80°C until analysis. Escherichia coli strains were precultivated twice in polystyrene round-bottom tubes in 2 ml M9 medium (Supplementary Data File S2: Media composition) with 0.2% glucose and 1 mg/ml methionine at 37°C and 250 rpm agitation for 24 h. Escherichia coli inocula were then collected by centrifugation at 10 000 xg for 1 min and washed three times with M9 medium prior to use in the bioassay. Samples and standards were thawed at room temperature and diluted in 2× M9 medium with 0.4% glucose and water to a final volume of 2 ml in polystyrene round-bottom tubes, to which either the ∆metE strain or the ∆metE∆metH control strain was added to a starting density of 0.01 OD600. The bioassay was then incubated at 37°C and agitated at 250 rpm for 24 h, and growth was measured by OD600. Sample B12 concentration was determined relative to a standard curve.
Spent medium and cell lysate experiments
Wild-type C. reinhardtii was grown for 48 h in continuous light until density reached 1–4 × 106 cells/ml. Cells were collected by centrifugation in 500 ml polypropylene bottles for 4 min at 11 900 xg and 4°C. The supernatant was passed through a 0.22 μm filter to generate sterile spent medium. Cell pellets were resuspended in fresh medium to the original culture volume and then sonicated using a Fisherbrand Model 505 Sonic Dismembrator (Fisher Scientific, PA, USA) on ice for 45 min with alternating 10 s at 30% intensity and 15-s “off” steps. The supernatant of the resulting cell lysate was then passed through a 0.22 μm filter to generate sterile soluble cell lysate. Spent medium, soluble cell lysate, and parallel live C. reinhardtii cultures were then inoculated with M. japonicum as described above, and growth was monitored over another 48 h.
To determine the bacterial growth supported by substrates in C. reinhardtii cell lysate when these substrates are present at a relevant concentration, wild-type C. reinhardtii was grown for 48 h in continuous light, and spent medium and soluble cell lysate were prepared as described above. Then, the NPOC of the two culture fractions was measured, and the cell lysate was diluted to the same NPOC concentration as the spent medium using fresh medium. Three milliliters of the diluted cell lysate was inoculated with M. japonicum as described above in parallel with spent medium, fresh medium, and fresh medium with 150 μg/ml sucrose in polystyrene round-bottom tubes. Cultures were incubated at 30°C and agitated at 200 rpm, and growth was monitored for roughly 48 h.
Results
M. japonicum growth is severely limited in coculture with C. reinhardtii, especially under diurnal light
Previous work suggested that C. reinhardtii supports heterotrophic growth of the rhizobial bacterium M. japonicum when cocultured in minimal medium given light and atmospheric CO2 [25, 26, 28, 36]. As a first step in characterizing the transfer of fixed carbon in this relationship, we compared the growth of M. japonicum when incubated alone or together with wild-type C. reinhardtii in minimal medium with either no exogenous carbon source or with 150 μg/ml sucrose (Fig. 1). Coculture with the bacterium did not have a discernable impact on C. reinhardtii growth or physiology (Fig. 1B, SI Fig. 2, P > .05). Mesorhizobium japonicum grew considerably with sucrose and only marginally without it, but we found that coculture with C. reinhardtii did not significantly increase the maximum bacterial density in either condition (Fig. 1C, P > .05). Although the maximum density of M. japonicum in coculture without sucrose (~3 × 107–5 × 108 CFU/ml) was similar to what has been observed previously [28], we found that the bacterium was able to achieve a comparable density without the alga, in contrast to previous reports.
Figure 1.
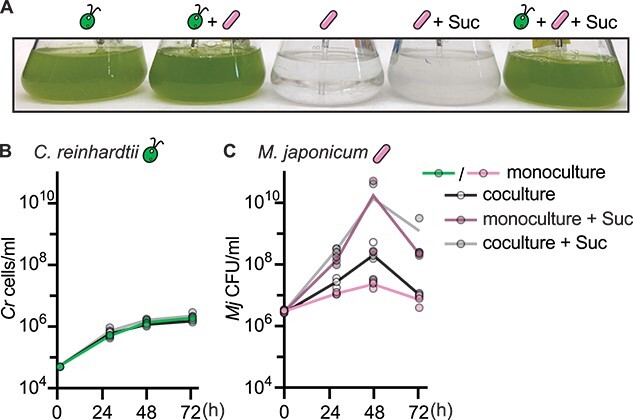
Wild-type C. reinhardtii does not significantly improve growth of M. japonicum. Triplicate cultures (n = 3) were inoculated with or without 150 μg/ml sucrose and maintained under continuous light, bubbled with air, and shaken at 110 rpm. (A) Representative image of cultures 72 h after inoculation. (B) Wild-type C. reinhardtii cell density in cultures over time after inoculation. (C) M. japonicum cell density in cultures over time after inoculation. The maximum bacterial density in coculture was not significantly different from the maximum bacterial density in monoculture with or without sucrose by two-tailed Student’s t-tests (P > .05).
As the growth of M. japonicum was not significantly stimulated by wild-type C. reinhardtii, it seemed that the alga did not supply adequate substrates for the bacterium’s heterotrophic metabolism under these conditions. This could be because C. reinhardtii released fixed carbon compounds in insufficient quantities or because M. japonicum is unable to metabolize the specific compounds generated by C. reinhardtii. We had initially illuminated cocultures with continuous light to maximize algal carbon fixation and growth rate, but we hypothesized that the alga’s nighttime metabolism could stimulate additional growth of M. japonicum. During the day, the alga fixes carbon to build biomass and stores photosynthate as starch [10]. After sufficient growth during the light phase, cells initiate DNA synthesis and mitosis in the dark, followed by a G0 phase during which starch reserves are degraded by glycolysis and genes for fermentation are expressed. Fermentation “waste products” released by algae in the night could serve as a readily available source of fixed carbon for cooccurring bacteria.
To test the impact of different light regimes on the interaction, we used the readily synchronized C. reinhardtii strain CC-5390. Synchronized populations allow excellent temporal separation of cells in distinct metabolic phases of the cell cycle and could reveal a diurnal pattern in bacterial growth resulting from the algal partner’s diurnal metabolism. We compared cocultures and monocultures under continuous and diurnal light over several days with high temporal resolution. Chlamydomonas reinhardtii’s doubling time is slower under diurnal light (cells only divide once per day), but we designed the experiment to achieve the same final amount of algal biomass under the two light regimes. Chlamydomonas reinhardtii strain CC-5390 density reached roughly 4 × 106 cells/ml under all conditions (Fig. 2A and B). This was roughly two times higher than the density reached by the wild-type strain used in Fig. 1, although the strains appeared to reach stationary phase in the same amount of time under continuous light (72 h). Algal growth was again unaffected by the bacterium. In contrast, we found that M. japonicum density increased significantly in the presence of CC-5390 under both light regimes (Fig. 2C–E). CC-5390 may support more growth of the bacterium than the wild-type strain because it is more prone to cell lysis and releases more NPOC into the spent medium (SI Fig. 3). However, we could not discern a clear temporal pattern in bacterial proliferation over the diurnal cycles, and contrary to our hypothesis, we found that continuously illuminated cocultures supported 3-fold more bacterial cells than did diurnally illuminated cocultures (Fig. 2E, P = .04). Spent medium from continuous light-grown C. reinhardtii had significantly more NPOC than spent medium from diurnal light-grown C. reinhardtii of the same culture density, suggesting that more dissolved organic carbon was available to the bacterium under continuous light (Fig. 2F, P < .05). Algal cell lysis was similar under the two light regimes, so differences in algal spent medium NPOC suggest that C. reinhardtii exudes more reduced carbon in continuous light than in diurnal light (SI Fig. 4, P > .05). Finally, the presence of M. japonicum did not significantly reduce the concentration of NPOC in the spent medium (Fig. 2F, Student’s t-test, P > .05). This could mean that either M. japonicum did not take up significant amounts of organic carbon from the medium or that it could not fully oxidize it to CO2 and instead released reduced carbon waste products back into the extracellular milieu.
Figure 2.
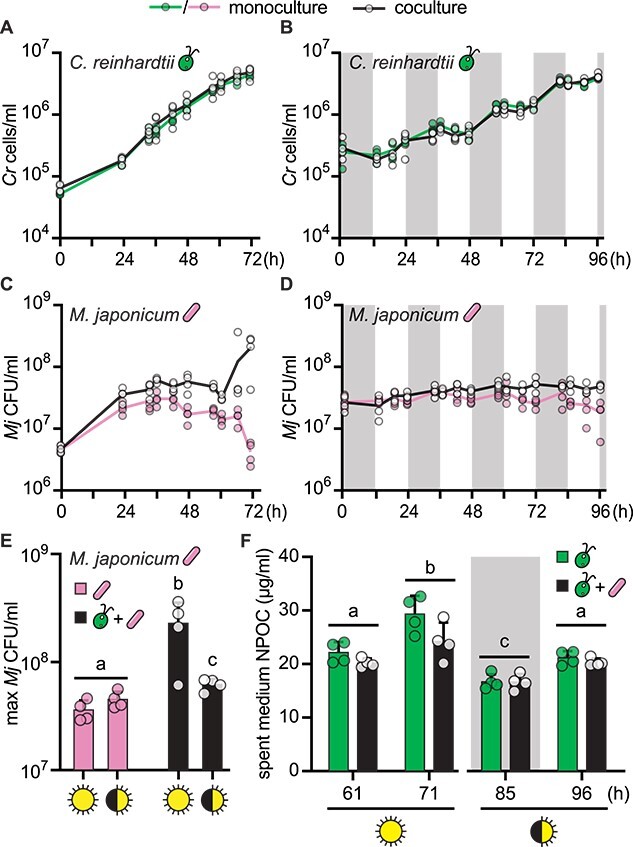
Coculture light regime impacts organic carbon release and M. japonicum growth. Quadruplicate (n = 4) monocultures (colors) and cocultures (black and white) were bubbled with air and shaken at 125 rpm. Cultures were inoculated to reach the same final algal cell density at the end of the experiment. (A, B) Growth of cell wall reduced strain CC-5390 in continuous light or diurnal light (12-h light/12-h dark), respectively, with and without M. japonicum. The grey backgrounds indicate sample timepoints that occurred during the dark phase. Under diurnal light, CC-5390 exhibits synchronous growth, doubling roughly once per day upon nightfall. (C, D) M. japonicum growth under the continuous and diurnal light regimes, respectively, with and without CC-5390. (E) Maximum bacterial density achieved under the various trophic conditions tested. (F) NPOC in spent medium from CC-5390 and cocultures sampled at the indicated times from the continuous and diurnal light regimes in (A) and (B), respectively. Lower-case letters indicate significantly different groups by paired two-tailed Student’s t-test (P < .05). Error bars represent the standard deviation from the mean.
Stable isotope probing reveals mismatch between bacterial biomass synthesis and division in coculture
To determine the amount of carbon fixed by C. reinhardtii that is taken up by M. japonicum, we conducted stable isotope probing experiments using 13CO2. Cocultures of CC-5390 and M. japonicum grown under continuous and diurnal light as in Fig. 2 were bubbled with air containing 13CO2. 15NH4Cl was added to the medium as an independent tracer of biomass synthesis, complementing measurements of cell density. We assayed label uptake with single-cell resolution using nanoSIMS.
We found substantial 13C enrichment of the algal cells, which remained at a similar concentration throughout the timepoints examined (Fig. 3A, SI Figs 5 and 6). In addition, we observed 13C enrichment of bacterial cells, which increased with time in cocultures under both light regimes (Fig. 3A, SI Figs 5 and 6, P < .05). Killed controls and monocultures of M. japonicum on various substrates demonstrated that this enrichment was not a result of nonspecific background or direct CO2 incorporation by the bacterium (i.e. through anapleurotic carboxylation reactions) (SI Figs 6 and 7). Thus, a meaningful amount of carbon derived from algal photoautotrophy was, in fact, incorporated into M. japonicum biomass in the cocultures.
Figure 3.
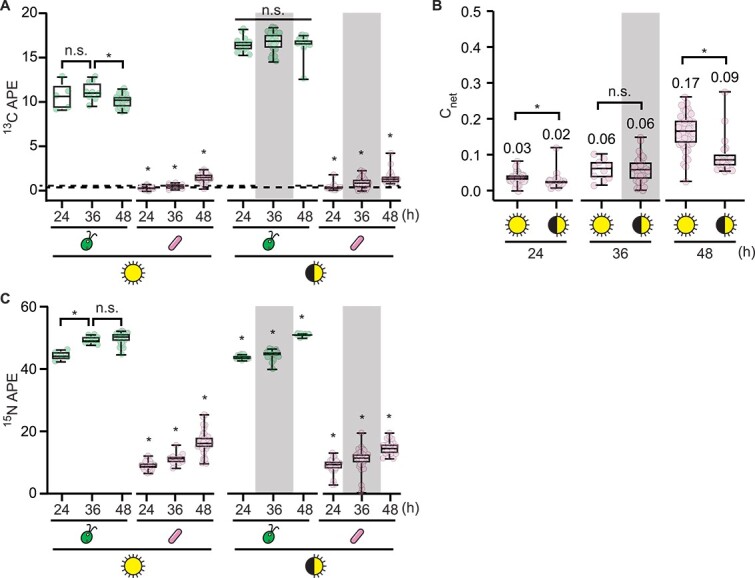
Stable isotope probing demonstrates carbon flux in continuous and diurnal light. Duplicate cocultures (n = 2) of CC-5390 and M. japonicum were grown as described in Fig. 2 but with 50% of their NH4+ provided as 15N and with 13CO2 added to the air starting 13 h after inoculation. Isotope enrichment was measured using nanoSIMS at the indicated times after inoculation. The grey backgrounds indicate sample timepoints that occurred during the dark phase. (A) 13C atom percent enrichment (APE) data of n ≥ 5 individual cells grown under continuous light or diurnal light. The dashed lines represent the maximum 13C APE that occurred in M. japonicum monoculture controls provided sucrose as a result of nonspecific background or direct CO2 incorporation during heterotrophic growth; see SI Fig. 6 for more details. Asterisks indicate significant differences over time in a given culture by two-tailed Student’s t-test (P < .05). (B) Cnet of M. japonicum cells (n ≥ 10), calculated relative to the algal biomass as the source. Median values are indicated, and asterisks indicate significant differences by two-tailed Student’s t-test (P < .05). (C) 15N APE data of n ≥ 5 individual cells grown under continuous light or diurnal light. Asterisks indicate significant differences over time in a given culture by two-tailed Student’s t-test (P < .05).
We compared the extent of carbon transfer under continuous and diurnal light by calculating the fraction of bacterial carbon that was derived from algal carbon, Cnet, using the average 13C content of algal biomass over the course of each experiment as a rough measure of the source carbon isotopic composition. We found that significantly more algal 13C was transferred to bacterial cells under continuous light over 48 h (0.17 median Cnet in continuous light, 0.09 median Cnet in diurnal light) (Fig. 3B, P = 6 × 10−6). This was consistent with our observation that spent medium NPOC and M. japonicum cell density were greater in cocultures exposed to continuous light than those exposed to diurnal light (Fig. 2, SI Fig. 8A).
Both the 13C and 15N enrichment in M. japonicum cells were lower than we had expected from the increase in bacterial CFUs observed in the experiment (>3-fold increase in CFUs during the first 24 h of coculture under continuous light), suggesting a mismatch between the change in cell density and the bacterial biomass synthesized (Fig. 3C, SI Fig. 8A). We quantified the magnitude of this mismatch using the 15N label, for which the source isotopic composition was better constrained, as opposed to that of the 13C label, which was delivered in the gas phase in an open system. Using the source 15N composition and the measured isotopic composition of bacterial cells at each timepoint, we calculated the fraction of newly synthesized biomass generated from the source substrate over the course of the experiment: Nnet [33]. We compared this to the expected Nnet that would be observed if each doubling in CFUs in the culture occurred upon a doubling in bacterial biomass (0.99 Nnet for cells grown on sucrose after 8–13 doublings, 0.63–0.97 Nnet for cells cocultivated with C. reinhardtii after 1–5 doublings) (SI Fig. 8B). Indeed, the measured Nnet of M. japonicum cells in the cocultures was much lower than the expected values at all measured timepoints (0.18–0.34 Nnet), whereas the measured values matched the expected values during logarithmic-phase growth on sucrose (Fig. 4A). This showed that in coculture with C. reinhardtii, M. japonicum cells reduced new biomass synthesis by 24 h and yet continued to divide—a hallmark of bacterial starvation often referred to as reductive division [37–39]. As nutrients available for a bacterium’s growth decrease, the cell volume added prior to cell division is reduced, leading to smaller daughter cells [40–43].
Figure 4.
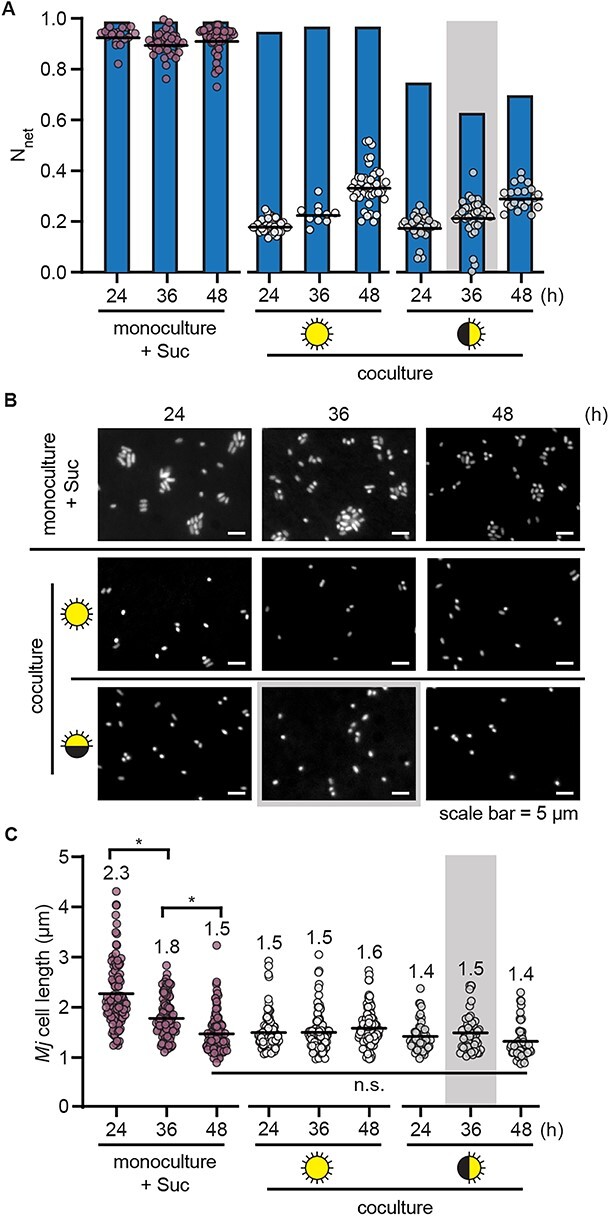
Mesorhizobium japonicum undergoes reductive division upon depletion of sucrose and during algal coculture. Duplicate cultures (n = 2) (monoculture with 150 μg/ml sucrose or coculture under continuous light or diurnal light) were grown as described in Fig. 2 but with 50% of their NH4+ provided as 15N and with 13CO2 added to the air starting 13 h after inoculation. Isotope enrichment was measured using nanoSIMS and cells were imaged by fluorescence microscopy at the indicated times after inoculation. The grey backgrounds indicate sample timepoints that occurred during the dark phase. (A) Nnet of M. japonicum cells (n ≥ 10) (circles) relative to the calculated expected Nnet (blue bars) if each doubling in CFU/ml represented a doubling in biomass (SI Fig. 8). Black lines represent mean values. (B) Representative fluorescence microscopy images of cells stained with DAPI. (C) Cell length of n ≥ 50 M. japonicum cells per culture type (n ≥ 25 cells from each of the duplicate cultures) measured using ImageJ. Mean cell length is indicated, and asterisks indicate significant differences by two-tailed Student’s t-test (P < .05).
If the deviation of the observed Nnet from the expected Nnet indeed reflected reductive division, bacterial cells in the coculture would be smaller than cells in the logarithmic phase of growth on sucrose, which are typically 2 μm in length. When we measured the size of the bacterial cells, we found that M. japonicum cells were indeed smaller in the cocultures: mean cell length was 1.4–1.5 μm after 24 h in coculture, whereas it was 2.3 μm after 24 h of growth on sucrose (Fig. 4B and C). When sucrose was presumably depleted from the medium over the subsequent day in the monoculture, mean cell length decreased to a similar extent, demonstrating that reductive division occurs upon carbon limitation in M. japonicum.
Taken together, these data show that photosynthetically fixed carbon was indeed transferred from C. reinhardtii to M. japonicum under both continuous and diurnal light and that although this amount was not sufficient for balanced logarithmic-phase growth of the bacterium, it allowed for a small amount of bacterial proliferation.
Reductively dividing bacteria still deliver sufficient vitamin B12 to support growth of the B12-requiring mete1 strain
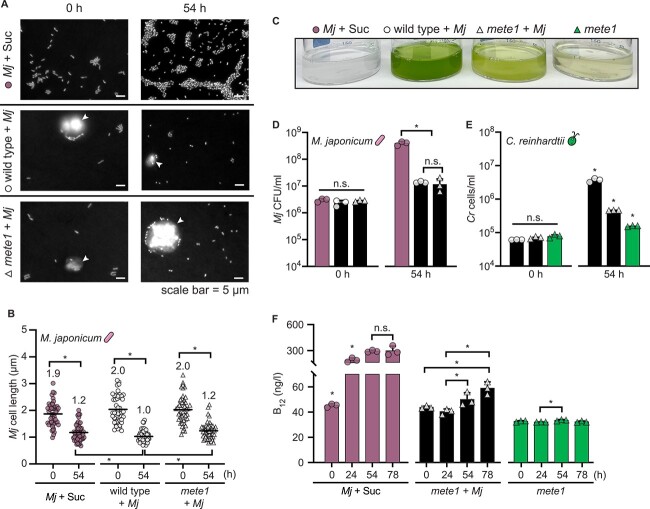
As reductive division had not previously been reported in the C. reinhardtii–M. japonicum interaction, we wondered whether the phenomenon also occurs during coculture with the B12-requiring mete1 strain of C. reinhardtii, and if so, whether the bacterium still synthesizes adequate B12 to support the mutant’s growth. We compared the change in bacterial cell size after growth on 150 μg/ml sucrose to changes during coculture with either wild-type C. reinhardtii or mete1. In all conditions, we observed a 37%–50% reduction in M. japonicum cell length over 54 h of cultivation (Fig. 5A and B). This suggested that bacterial carbon limitation occurs to a similar extent during coculture with the mete1 strain as with the wild type.
Figure 5.
Mesorhizobium japonicum delivers vitamin B12 to C. reinhardtii despite reductive division. Triplicate cultures (n = 3) were maintained under continuous light, shaken at 130 rpm, and assayed at the indicated times after inoculation. (A) Representative fluorescence microscopy images of M. japonicum with 150 μg/ml sucrose, in coculture with C. reinhardtii wild type, and in coculture with C. reinhardtii mete1. Cells were stained with DAPI, and C. reinhardtii cells are indicated with white arrowheads. (B) Length of M. japonicum cells (n = 50, 10–20 cells from each of the three triplicate cultures) measured using ImageJ. Mean cell length is indicated. (C) Representative image of cultures 54 h after inoculation. (D) M. japonicum density in monoculture with 150 μg/ml sucrose (pink bars) or in coculture (black bars) with wild-type (circles) and mete1 C. reinhardtii (triangles), estimated as CFU/ml. (E) Cell density of C. reinhardtii wild type (circles) or mete1 (triangles) in coculture (black bars) and of mete1 alone without added vitamin B12 (green bars). (F) Concentration of vitamin B12 in cultures measured using an E. coli bioassay. Error bars represent the standard deviation from the mean, and asterisks indicate significant differences by two-tailed Student’s t-test (P < .05).
To test whether reductively dividing M. japonicum still delivers vitamin B12 to C. reinhardtii, we compared the growth of the C. reinhardtii mete1 strain in coculture and monoculture without exogenous B12, and we monitored changes in vitamin B12 concentration overtime using a bioassay [34, 35]. mete1 growth was limited by B12 relative to the wild-type C. reinhardtii, but the B12-requiring strain grew significantly more in the presence of M. japonicum despite reductive bacterial division (Fig. 5C and E, P < .05). Furthermore, we found that M. japonicum produced significant vitamin B12 in coculture, but only after 24 h (Fig. 5F, P < .05), even though bacterial biomass synthesis and cell size have decreased substantially by that time (Fig. 4). This demonstrates that bacterial B12 synthesis and sharing occur despite the carbon limitation experienced during the trophic interaction with the alga.
Although there were roughly 10× more C. reinhardtii cells present in the wild-type coculture than the B12-limited mutant coculture, there was no difference in M. japonicum density between these two conditions (Fig. 5D, P > .05). Thus, greater algal density does not translate into greater M. japonicum growth, as noted previously [28]. We observed this phenomenon in cocultures inoculated at several starting relative abundances (SI Fig. 10). This suggests that carbon transfer from the alga to the bacterium could be indirect.
C. reinhardtii lysis can explain observed M. japonicum proliferation in coculture
To distinguish the source of the fixed carbon that supports M. japonicum proliferation in coculture, we compared M. japonicum growth with live C. reinhardtii to growth in C. reinhardtii spent medium and in the soluble fraction of C. reinhardtii cell lysate. Spent medium supported the same number of M. japonicum cells as did coculture, demonstrating that the observed increase does not require a direct interaction with the alga (Fig. 6A). Cell lysate from C. reinhardtii supported almost an order of magnitude more M. japonicum proliferation. Therefore, we hypothesized that the fixed carbon transferred in our cocultures could be sourced from a few lysed C. reinhardtii cells rather than from photosynthate exuded by live algal cells.
Figure 6.
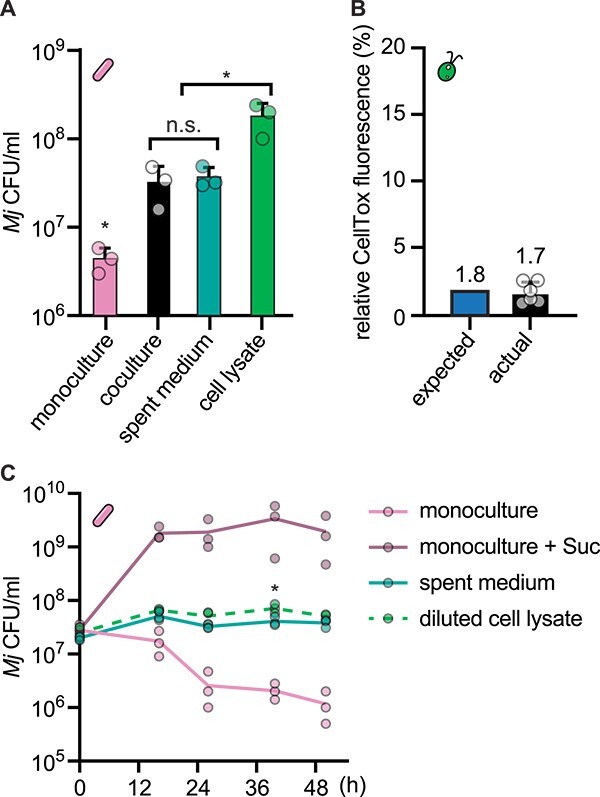
A small degree of algal lysis can explain the M. japonicum growth that occurs in coculture. Triplicate cultures (n = 3) of wild-type C. reinhardtii were grown under continuous light and shaken at 125 rpm for 24 h, reaching 1–4 × 106 cells/ml. These cultures were then split and used to generate parallel live algal cultures, algal spent media, and algal cell lysate, each then inoculated with M. japonicum in triplicate (n = 3). (A) M. japonicum density 45 h after inoculation in monoculture with no reduced carbon source, coculture with live C. reinhardtii cells, C. reinhardtii spent medium, or C. reinhardtii cell lysate. Asterisks indicate significant differences by two-tailed Student’s t-test (P < .05). (B) The approximate degree of C. reinhardtii cell lysis required to explain M. japonicum growth in coculture based on M. japonicum growth yield in C. reinhardtii cell lysate (expected), and the degree of C. reinhardtii cell lysis measured in the cultures used for the experiment in panel (A) by CellTox Green fluorescence, reported relative to a 100% killed control (actual). The expected value and the mean actual value are indicated. (C) M. japonicum density in triplicate cultures (n = 3) over time after inoculation in monoculture with or without 150 μg/ml sucrose, in C. reinhardtii spent medium, or in C. reinhardtii cell lysate when diluted to the same NPOC concentration as the spent medium. The asterisk indicates a significant difference in density between spent medium and diluted cell lysate 40 h after inoculation by Student’s t-test (P < .05); density was not significantly different between these two conditions at the other timepoints examined (P > .05). Error bars represent the standard deviation from the mean.
Having determined the increase in viable M. japonicum cells supported by lysate from a known density of C. reinhardtii cells (1.7 × 108 CFU/ml on average after 48 h in lysate from 1.6 × 106 C. reinhardtii cells/ml culture), we estimated the theoretical increase in M. japonicum CFUs supported by a single lysed C. reinhardtii cell to be 100 ± 43 CFU in 48 h. From that, we estimated how many C. reinhardtii cells would have to lyse in the coculture to enable the M. japonicum proliferation observed: roughly 1.1 × 105 C. reinhardtii cells/ml, which corresponds to 1.8% of the C. reinhardtii populations in those cocultures. To test the actual degree of algal lysis in our cultures, we measured algal cellular integrity using the CellTox Green Cytotoxicity Assay. This assay uses a membrane-impermeable DNA dye, resulting in fluorescence from cells whose plasma membrane has been compromised or from extracellular DNA [44]. We compared live culture fluorescence to killed control samples and found that 1%–3% of the wild-type C. reinhardtii population had compromised cellular integrity (Fig. 6B). Thus, the fraction of dead or dying C. reinhardtii cells measured by this assay is comparable to that needed to explain the observed M. japonicum proliferation.
Finally, we reasoned that if M. japonicum is primarily growing on algal cell lysate, then cell lysate should support the same amount of bacterial proliferation as spent medium when present at the same organic carbon content. Thus, we compared M. japonicum growth in spent medium and diluted cell lysate from wild-type C. reinhardtii when both were present at roughly 15 μg/ml NPOC. Indeed, algal cell lysate alone supported the same M. japonicum growth as spent medium when present at a relevant concentration: bacterial cell density was not significantly different between these two conditions at most timepoints examined (Fig. 6C, P > .05). The slight increase in M. japonicum density in the diluted cell lysate condition could be attributed to increased availability of micronutrients in the fresh medium used to dilute the algal cell lysate. Taken together, these data are consistent with the interpretation that M. japonicum growth in coculture is enabled by substrates that are released through C. reinhardtii cell lysis rather than by substrates exuded by live C. reinhardtii cells.
Discussion
Most studies of symbioses between C. reinhardtii and heterotrophic bacteria to date have focused on impacts on the alga, leaving open the question of how this model phototroph might influence bacterial growth and physiology. Here, we have investigated C. reinhardtii’s role as a primary producer in the established coculture system with M. japonicum. We have tied carbon cycling in the interaction to alterations in bacterial growth and physiology through quantitative, single-cell analyses. Under continuous light, we found that the alga increased the number of viable bacterial cells, albeit marginally so. We tested whether nighttime metabolism might increase the alga’s propensity for carbon sharing, but we found that diurnal illumination actually decreased the productivity of the partnership. Using nanoSIMS to visualize 13C enrichment with high spatial resolution, we quantitatively demonstrated transfer of carbon fixed by the model alga to the bacterium. Roughly 17% of the carbon in M. japonicum cells was derived from C. reinhardtii over 36 h under continuous light, whereas only ~9% of the M. japonicum carbon came from C. reinhardtii over the same amount of time under diurnal light. Bulk measurements showed that algal cultures of similar densities had more organic carbon in their spent medium after continuous illumination than after diurnal illumination. Together, these results suggest that the increased metabolic rate realized under continuous light may be more beneficial for M. japonicum (and potentially other bacteria) than fermentative catabolism of starch presumed to occur during the night. It is also possible that although C. reinhardtii expresses genes for fermentative metabolism and produces some lactate in the night [10], fermentative metabolites may not actually accumulate to high-enough levels in culture unless cells experience anoxia. Future research on the metabolic exchanges between C. reinhardtii and bacterial partners will help reveal the extent to which the results presented here are specific to M. japonicum or more broadly applicable.
With C. reinhardtii as the sole source of fixed carbon, M. japonicum exhibited signs of starvation, even under continuous light, highlighting the meagerness of the microbial trophic relationship. The quantitative stable isotope-probing experiments revealed that during coculture with C. reinhardtii, M. japonicum synthesized significantly less biomass than expected given the increase in viable bacterial cells. By direct measurement, we showed that M. japonicum divided into small daughter cells within 24 h of carbon limitation in coculture, a physiological change often referred to as reductive division. It is well known that as nutrients become limiting, bacterial size at birth and the amount of cell volume each bacterium adds prior to division decrease [40–43]. Studies on freshwater and marine bacteria have documented up to 90% reduction in cell volume during carbon limitation [45–48]. This phenomenon is often considered an adaptive strategy by which diverse bacteria survive starvation [37–39, 49]. By increasing cell number and reducing cell volume, bacteria increase the probability that a member of the population will encounter sufficient nutrients again and increase their surface area-to-volume ratio to improve nutrient uptake. Reductively dividing cells also exhibit changes in stress resistance and macromolecule synthesis. However, we found that despite its apparent starvation, M. japonicum provided enough vitamin B12 to support a B12-dependent C. reinhardtii mutant. Vitamin supply enables growth of auxotrophs, influences gene expression and metabolism, and can even give rise to vitamin dependency in algae [25, 50]. Thus, our results suggest that even under austere conditions with meager supply of growth substrates, bacteria can still influence algal physiology and evolution.
We also found that a direct interaction with C. reinhardtii was not required to achieve the M. japonicum proliferation observed in coculture: growth in algal spent medium yielded the same maximum density of viable M. japonicum cells as did coculture. Furthermore, mechanically lysed C. reinhardtii cells yielded considerably higher M. japonicum densities. This observation led us to hypothesize that perhaps this bacterium does not metabolize the exudates of live C. reinhardtii cells at all. Instead, perhaps the small amount of bacterial growth observed in coculture and in spent medium is due to the lysis of a small fraction of algal cells. It has long been appreciated that even during rapid exponential growth, microbial cultures experience a small death rate [51]. Furthermore, cell lysis is an important mechanism of nutrient transfer in natural environments, and microbial necromass is a major component of the soil organic carbon pool [52, 53]. For the experimental conditions examined here, we showed that ~2% of the C. reinhardtii population in our cultures are present as necromass. Based on the M. japonicum growth yields achieved in mechanically lysed C. reinhardtii cells, we calculated that this is almost exactly the amount of algal necromass required to explain the M. japonicum proliferation that occurred in coculture. When present at the same organic carbon concentration, algal cell lysate supported the same amount of M. japonicum growth as algal spent medium. Therefore, we propose that lysis is the primary mechanism of trophic transfer in this coculture system, rather than exudation or direct exchange.
Given that very little growth of M. japonicum was observed from an interaction with C. reinhardtii—even under conditions optimized for algal productivity—our results may suggest that this model alga exhibits a low carrying capacity for heterotrophic bacteria in the environment. Similar bacterial densities have been reported for other C. reinhardtii cocultures in minimal media, including those with Methylobacterium aquaticum [54, 55]. However, it is possible that other bacteria, such as those that may have coevolved with C. reinhardtii in its native environment, may possess greater capacity to metabolize compounds that C. reinhardtii releases during growth. In addition, other bacteria may achieve greater access to growth substrates through chemotaxis, interspecies signaling and direct exchange, or algicidal activity [3, 56–59]. The mode of cell lysis (i.e. mechanical vs. viral) could also greatly impact the algal-derived nutrients released to cooccurring heterotrophs [60]. Nonetheless, stable isotope-probing experiments on other algal–bacterial interactions and on seawater microbial communities have reported similar levels of trophic carbon transfer to those we observed here [56, 61, 62]. This suggests that although C. reinhardtii and M. japonicum may not commonly encounter one another in nature, this relationship may be representative of naturally occurring microbial phototroph–heterotroph interactions. Studying this model system has revealed important aspects of microbial ecology, and our quantitative approach can serve as a roadmap for the future work necessary to understand C. reinhardtii’s full potential as a primary producer in the microbial world.
Supplementary Material
Acknowledgements
We thank Dr. Alison G. Smith and her group at the University of Cambridge for providing strains and for their generous discussions with us. We thank Dr. Michiko E. Taga at UC Berkeley for her guidance on the work. We thank Dr. Denise Schichnes and the UC Berkeley Biological Imaging Facility for fluorescence microscopy support. We also thank Christina Ramon at Lawrence Livermore National Laboratory and Charles Perrino at UC Berkeley for analytical support.
Contributor Information
Sunnyjoy Dupuis, Department of Plant and Microbial Biology, University of California, Berkeley, CA 94720, United States; California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, CA 94720, United States.
Usha F Lingappa, California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, CA 94720, United States.
Xavier Mayali, Physical and Life Sciences Directorate, Lawrence Livermore National Laboratory, Livermore, CA 94550, United States.
Eve S Sindermann, California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, CA 94720, United States.
Jordan L Chastain, California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, CA 94720, United States; College of Chemistry, University of California, Berkeley, CA 94720, United States.
Peter K Weber, Physical and Life Sciences Directorate, Lawrence Livermore National Laboratory, Livermore, CA 94550, United States.
Rhona Stuart, Physical and Life Sciences Directorate, Lawrence Livermore National Laboratory, Livermore, CA 94550, United States.
Sabeeha S Merchant, Department of Plant and Microbial Biology, University of California, Berkeley, CA 94720, United States; California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, CA 94720, United States; Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, United States; Division of Environmental Genomics and Systems Biology, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, United States.
Conflicts of interest
None declared.
Funding
This work was supported by The Gordon and Betty Moore Foundation Symbiosis in Aquatic Systems Initiative Investigator Award GBML9203 (https://doi.org/10.37807/GBMF9203) to S.S.M., and the Lawrence Livermore National Laboratory μBiospheres Scientific Focus Area award SCW1039 to R.S. from the US Department of Energy Office of Biological and Environmental Research Genomic Sciences Program. S.D. acknowledges support from the US National Institute of Health T32 Genetic Dissection of Cells and Organisms training grant 1T32GM132022-01.
Data availability
All data underlying this article are available in Supplementary Data File S1.
References
- 1. Field CB, Behrenfeld MJ, Randerson JT et al. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 1979;281:237–40. 10.1126/science.281.5374.237 [DOI] [PubMed] [Google Scholar]
- 2. Azam F, Fenchel T, Field J et al. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 1983;10:257–63. 10.3354/meps010257 [DOI] [Google Scholar]
- 3. Seymour JR, Amin SA, Raina J-B et al. Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat Microbiol 2017;2:17065. 10.1038/nmicrobiol.2017.65 [DOI] [PubMed] [Google Scholar]
- 4. Pomeroy LR. The ocean’s food web, a changing paradigm. Bioscience 1974;24:499–504. 10.2307/1296885 [DOI] [Google Scholar]
- 5. Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol 2007;5:782–91. 10.1038/nrmicro1747 [DOI] [PubMed] [Google Scholar]
- 6. Croft MT, Lawrence AD, Raux-Deery E et al. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 2005;438:90–3. 10.1038/nature04056 [DOI] [PubMed] [Google Scholar]
- 7. Amin SA, Green DH, Hart MC et al. Photolysis of iron–siderophore chelates promotes bacterial–algal mutualism. Proc Natl Acad Sci 2009;106:17071–6. 10.1073/pnas.0905512106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hom EFY, Aiyar P, Schaeme D et al. A chemical perspective on microalgal-microbial interactions. Trends Plant Sci 2015;20:689–93. 10.1016/j.tplants.2015.09.004 [DOI] [PubMed] [Google Scholar]
- 9. Salomé PA, Merchant SS. A series of fortunate events: introducing Chlamydomonas as a reference organism. Plant Cell 2019;31:1682–707. 10.1105/tpc.18.00952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strenkert D, Schmollinger S, Gallaher SD et al. Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc Natl Acad Sci 2019;116:2374–83. 10.1073/pnas.1815238116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zones JM, Blaby IK, Merchant SS et al. High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 2015;27:2743–69. 10.1105/tpc.15.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Durán P, Flores-Uribe J, Wippel K et al. Shared features and reciprocal complementation of the Chlamydomonas and Arabidopsis microbiota. Nat Commun 2022;13:406. 10.1038/s41467-022-28055-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim B-H, Ramanan R, Cho D-H et al. Role of rhizobium, a plant growth promoting bacterium, in enhancing algal biomass through mutualistic interaction. Biomass Bioenergy 2014;69:95–105. 10.1016/j.biombioe.2014.07.015 [DOI] [Google Scholar]
- 14. Lőrincz Z, Preininger É, Kósa A et al. Artificial tripartite symbiosis involving a green alga (Chlamydomonas), a bacterium (Azotobacter) and a fungus (Alternaria): morphological and physiological characterization. Folia Microbiol (Praha) 2010;55:393–400. 10.1007/s12223-010-0067-9 [DOI] [PubMed] [Google Scholar]
- 15. Xie B, Bishop S, Stessman D et al. Chlamydomonas reinhardtii thermal tolerance enhancement mediated by a mutualistic interaction with vitamin B12-producing bacteria. ISME J 2013;7:1544–55. 10.1038/ismej.2013.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 2012;76:667–84. 10.1128/MMBR.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gobler C, Norman C, Panzeca C et al. Effect of B-vitamins (B1, B12) and inorganic nutrients on algal bloom dynamics in a coastal ecosystem. Aquat Microb Ecol 2007;49:181–94. 10.3354/ame01132 [DOI] [Google Scholar]
- 18. Sañudo-Wilhelmy SA, Gobler CJ, Okbamichael M et al. Regulation of phytoplankton dynamics by vitamin B12. Geophys Res Lett 2006;33:L04604. 10.1029/2005GL025046 [DOI] [Google Scholar]
- 19. Helliwell KE. The roles of B vitamins in phytoplankton nutrition: new perspectives and prospects. New Phytol 2017;216:62–8. 10.1111/nph.14669 [DOI] [PubMed] [Google Scholar]
- 20. Keshtacher-Liebso E, Hadar Y, Chen Y. Oligotrophic bacteria enhance algal growth under iron-deficient conditions. Appl Environ Microbiol 1995;61:2439–41. 10.1128/aem.61.6.2439-2441.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fakhimi N, Gonzalez-Ballester D, Fernández E et al. Algae-bacteria consortia as a strategy to enhance H2 production. Cells 2020;9:1353. 10.3390/cells9061353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krespach MKC, Stroe MC, Flak M et al. Bacterial marginolactones trigger formation of algal gloeocapsoids, protective aggregates on the verge of multicellularity. Proc Natl Acad Sci 2021;118:e2100892118. 10.1073/pnas.2100892118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Windler M, Stuart R, Deutzmann JS et al. Bacterial exometabolites influence Chlamydomonas cell cycle and double algal productivity. FEMS Microbiol Ecol 2022;98:fiac091. 10.1093/femsec/fiac091 [DOI] [PubMed] [Google Scholar]
- 24. Kazamia E, Czesnick H, Van NTT et al. Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol 2012;14:1466–76. 10.1111/j.1462-2920.2012.02733.x [DOI] [PubMed] [Google Scholar]
- 25. Helliwell KE, Collins S, Kazamia E et al. Fundamental shift in vitamin B12 eco-physiology of a model alga demonstrated by experimental evolution. ISME J 2015;9:1446–55. 10.1038/ismej.2014.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bunbury F, Helliwell KE, Mehrshahi P et al. Responses of a newly evolved auxotroph of Chlamydomonas to B12 deprivation. Plant Physiol 2020;183:167–78. 10.1104/pp.19.01375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Helliwell KE, Scaife MA, Sasso S et al. Unraveling vitamin B12 -responsive gene regulation in algae. Plant Physiol 2014;165:388–97. 10.1104/pp.113.234369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bunbury F, Deery E, Sayer AP et al. Exploring the onset of B12-based mutualisms using a recently evolved Chlamydomonas auxotroph and B12-producing bacteria. Environ Microbiol 2022;24:3134–47. 10.1111/1462-2920.16035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gallaher SD, Fitz-Gibbon ST, Glaesener AG et al. Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell 2015;27:2335–52. 10.1105/tpc.15.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sueoka N. Mitotic replication of deoxynucleic acid in Chlamydomonas reinhardi. Proc Natl Acad Sci 1960;46:83–91. 10.1073/pnas.46.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kropat J, Hong-Hermesdorf A, Casero D et al. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J 2011;66:770–80. 10.1111/j.1365-313X.2011.04537.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lingappa UF, Dupuis S, Mayali X et al. Isotopically labelled inorganic carbon delivered to algal cultures via bubbler bottle. PRO 2023. 10.17504/protocols.io.yxmvm3q8ol3p/v1 [DOI] [Google Scholar]
- 33. Dekas AE, Parada AE, Mayali X et al. Characterizing chemoautotrophy and heterotrophy in marine archaea and bacteria with single-cell multi-isotope nanoSIP. Front Microbiol 2019;10:10. 10.3389/fmicb.2019.02682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mok KC, Hallberg ZF, Taga ME. Purification and detection of vitamin B12 analogs. Methods Enzymol 2022;668:61–85. 10.1016/bs.mie.2021.11.023 [DOI] [PubMed] [Google Scholar]
- 35. Dupuis S, Schmollinger S, Merchant SS. Quantitative detection of vitamin B12 in algae by bioassay and ICP-MS/MS. PRO 2022. 10.17504/protocols.io.14egn7726v5d/v1 [DOI] [Google Scholar]
- 36. Laeverenz Schlogelhofer H, Peaudecerf FJ, Bunbury F et al. Combining SIMS and mechanistic modelling to reveal nutrient kinetics in an algal-bacterial mutualism. PLoS One 2021;16:e0251643. 10.1371/journal.pone.0251643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thorne SH, Williams HD. Adaptation to nutrient starvation in rhizobium leguminosarum bv. Phaseoli: analysis of survival, stress resistance, and changes in macromolecular synthesis during entry to and exit from stationary phase. J Bacteriol 1997;179:6894–901. 10.1128/jb.179.22.6894-6901.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shimaya T, Okura R, Wakamoto Y et al. Scale invariance of cell size fluctuations in starving bacteria. Commun Phys 2021;4:238. 10.1038/s42005-021-00739-5 [DOI] [Google Scholar]
- 39. Bergkessel M, Delavaine L. Diversity in starvation survival strategies and outcomes among heterotrophic proteobacteria. Microb Physiol 2021;31:146–62. 10.1159/000516215 [DOI] [PubMed] [Google Scholar]
- 40. Schaechter M, Maaløe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced growth of salmonella typhimurium. J Gen Microbiol 1958;19:592–606. 10.1099/00221287-19-3-592 [DOI] [PubMed] [Google Scholar]
- 41. Vadia S, Levin PA. Growth rate and cell size: a re-examination of the growth law. Curr Opin Microbiol 2015;24:96–103. 10.1016/j.mib.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Campos M, Surovtsev IV, Kato S et al. A constant size extension drives bacterial cell size homeostasis. Cell 2014;159:1433–46. 10.1016/j.cell.2014.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taheri-Araghi S, Bradde S, Sauls JT et al. Cell-size control and homeostasis in bacteria. Curr Biol 2015;25:385–91. 10.1016/j.cub.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haire TC, Bell C, Cutshaw K et al. Robust microplate-based methods for culturing and in vivo phenotypic screening of Chlamydomonas reinhardtii. Front. Plant Sci 2018;9:235. 10.3389/fpls.2018.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vrede K, Heldal M, Norland S et al. Elemental composition (C, N, P) and cell volume of exponentially growing and nutrient-limited bacterioplankton. Appl Environ Microbiol 2002;68:2965–71. 10.1128/AEM.68.6.2965-2971.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Troussellier M, Bouvy M, Courties C et al. Variation of carbon content among bacterial species under starvation condition. Aquat Microb Ecol 1997;13:113–9. 10.3354/ame013113 [DOI] [Google Scholar]
- 47. Holmquist L, Kjelleberg S. Changes in viability, respiratory activity and morphology of the marine vibrio sp. strain S14 during starvation of individual nutrients and subsequent recovery. FEMS Microbiol Ecol 1993;12:215–23. 10.1111/j.1574-6941.1993.tb00034.x [DOI] [Google Scholar]
- 48. Mårdén P, Tunlid A, Malmcrona-Friberg K et al. Physiological and morphological changes during short term starvation of marine bacterial islates. Arch Microbiol 1985;142:326–32. 10.1007/BF00491898 [DOI] [Google Scholar]
- 49. Nyström T, Larsson C, Gustafsson L. Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J 1996;15:3219–28. 10.1002/j.1460-2075.1996.tb00686.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sokolovskaya OM, Shelton AN, Taga ME. Sharing vitamins: Cobamides unveil microbial interactions. Science 2020;369:eaba0165. 10.1126/science.aba0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kelly CD, Rahn O. The growth rate of individual bacterial cells. J Bacteriol 1932;23:147–53. 10.1128/jb.23.2.147-153.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gude S, Pherribo GJ, Taga ME. Emergence of metabolite provisioning as a by-product of evolved biological functions. mSystems 2020;5:e00259-20. 10.1128/mSystems.00259-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sokol NW, Slessarev E, Marschmann GL et al. Life and death in the soil microbiome: how ecological processes influence biogeochemistry. Nat Rev Microbiol 2022;20:415–30. 10.1038/s41579-022-00695-z [DOI] [PubMed] [Google Scholar]
- 54. Ban S, Lin W, Wu F et al. Algal-bacterial cooperation improves algal photolysis-mediated hydrogen production. Bioresour Technol 2018;251:350–7. 10.1016/j.biortech.2017.12.072 [DOI] [PubMed] [Google Scholar]
- 55. Calatrava V, Hom EFY, Llamas Á et al. OK, thanks! A new mutualism between Chlamydomonas and methylobacteria facilitates growth on amino acids and peptides. FEMS Microbiol Lett 2018;365:fny021. 10.1093/femsle/fny021 [DOI] [PubMed] [Google Scholar]
- 56. Samo TJ, Kimbrel JA, Nilson DJ et al. Attachment between heterotrophic bacteria and microalgae influences symbiotic microscale interactions. Environ Microbiol 2018;20:4385–400. 10.1111/1462-2920.14357 [DOI] [PubMed] [Google Scholar]
- 57. Jackrel SL, Yang JW, Schmidt KC et al. Host specificity of microbiome assembly and its fitness effects in phytoplankton. ISME J 2021;15:774–88. 10.1038/s41396-020-00812-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aiyar P, Schaeme D, García-Altares M et al. Antagonistic bacteria disrupt calcium homeostasis and immobilize algal cells. Nat Commun 2017;8:1756. 10.1038/s41467-017-01547-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Raina J-B, Giardina M, Brumley DR et al. Chemotaxis increases metabolic exchanges between marine picophytoplankton and heterotrophic bacteria. Nat Microbiol 2023;8:510–21. 10.1038/s41564-023-01327-9 [DOI] [PubMed] [Google Scholar]
- 60. Pherribo GJ, Taga ME. Bacteriophage-mediated lysis supports robust growth of amino acid auxotrophs. ISME J 2023;17:1785–8. 10.1038/s41396-023-01452-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Arandia-Gorostidi N, Weber PK, Alonso-Sáez L et al. Elevated temperature increases carbon and nitrogen fluxes between phytoplankton and heterotrophic bacteria through physical attachment. ISME J 2017;11:641–50. 10.1038/ismej.2016.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. de-Bashan LE, Mayali X, Bebout BM et al. Establishment of stable synthetic mutualism without co-evolution between microalgae and bacteria demonstrated by mutual transfer of metabolites (NanoSIMS isotopic imaging) and persistent physical association (fluorescent in situ hybridization). Algal Res 2016;15:179–86. 10.1016/j.algal.2016.02.019 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data underlying this article are available in Supplementary Data File S1.