Abstract
The human herpesvirus entry mediator C (HveC), also known as the poliovirus receptor-related protein 1 (PRR1) and as nectin-1, allows the entry of herpes simplex virus type 1 (HSV-1) and HSV-2 into mammalian cells. The interaction of virus envelope glycoprotein D (gD) with such a receptor is an essential step in the process leading to membrane fusion. HveC is a member of the immunoglobulin (Ig) superfamily and contains three Ig-like domains in its extracellular portion. The gD binding site is located within the first Ig-like domain (V domain) of HveC. We generated a panel of monoclonal antibodies (MAbs) against the ectodomain of HveC. Eleven of these, which detect linear or conformational epitopes within the V domain, were used to map a gD binding site. They allowed the detection of HveC by enzyme-linked immunosorbent assay, Western blotting, and biosensor analysis or directly on the surface of HeLa cells and human neuroblastoma cell lines, as well as simian Vero cells. The anti-HveC V-domain MAbs CK6, CK8, and CK41, as well as the previously described MAb R1.302, blocked HSV entry. Their binding to soluble HveC was blocked by the association of gD with the receptor, indicating that their epitopes overlap a gD binding site. Competition assays on an optical biosensor showed that CK6 and CK8 (linear epitopes) inhibited the binding of CK41 and R1.302 (conformational epitopes) to HveC and vice versa. Epitope mapping showed that CK6 and CK8 bound between residues 80 and 104 of HveC, suggesting that part of the gD binding site colocalizes in the same region.
Among the 11 envelope glycoproteins of herpes simplex virus (HSV), glycoprotein D (gD) plays an essential role during viral entry into mammalian cells (14). gD binds specifically to one of several cell surface receptors during the pH-independent process that leads to fusion of the HSV envelope with the cell plasma membrane (13). Other essential glycoproteins such as gB and the gH-gL heterodimer also participate in the fusion event in ways that remain to be elucidated (9, 35, 38).
Several HSV gD receptors have been identified. Herpesvirus entry mediator A (HveA; also known as HVEM and TNFRSF14) is a member of the tumor necrosis factor receptor family which binds gD and allows the entry of most HSV-1 and HSV-2 strains (25, 41). HveB (nectin-2) and HveC (nectin-1) are members of the immunoglobulin (Ig) superfamily that are closely related to the poliovirus receptor (PVR; also known as CD155) and to the newly discovered nectin-3 (8, 21, 22, 33). Whereas the activity of HveB is limited to certain HSV-2 strains and some laboratory strains of HSV-1 (rid1 and ANG) and pseudorabies virus (PRV) (20, 39), HveC allows the entry of all the HSV-1 and HSV-2 strains tested as well as PRV and bovine herpesvirus 1 (10). Poliovirus receptor does not function as an HSV receptor but can be used by PRV and bovine herpesvirus 1 (10). A specific type of heparan sulfate modified by d-glucosaminyl-3-O-sulfotransferase 3 can substitute for HveA or HveC and binds to gD to allow the entry of HSV-1 KOS into cells (34).
HveB and HveC appear to be involved in cell-cell interaction and were named nectin-2 and nectin-1, respectively, according to their newly discovered function (1, 19, 37). In this paper, we will refer to them according to their viral usage (i.e., HveB and HveC).
Recently, mutations in the HveC gene (named PVRL1 in that study) were linked to a form of cleft lip/palate-ectodermal dysplasia in humans (36).
Although they have different structures, HveA and HveC bound to HSV-1 gD with similar affinity (17, 42). Using antibody competition and mutagenesis, the binding sites for HveC and HveA were mapped to common and distinct regions of gD (16, 28, 40). Reciprocally, the gD binding site on HveC has been localized to the first and most distal of the three Ig-like domains (or V domain) of its extracellular portion (4, 17). This V domain alone purified as a soluble protein was able to bind gD with full affinity and efficiently inhibited HSV infection (17). Moreover a monoclonal antibody (R1.302) could bind to the purified V domain of HveC and block HSV infection (4, 5). In addition, the V domain, when directly anchored on the cell surface through its natural transmembrane region, could mediate HSV entry, albeit with reduced capability (5). The precise location of the gD binding site within the V domain is yet to be defined. Monoclonal antibodies (MAbs) are useful tools to map functional sites on proteins such as cell surface receptors. Epitopes of MAbs able to interfere with ligand binding often colocalize with sites involved in such interactions (3, 15, 18, 30). Similarly, epitope mapping of virus-neutralizing MAbs provides useful indications about the location of receptor binding or functional sites on viral proteins (26, 27). For example, neutralizing anti-HSV-1 gD MAbs from group Ia, Ib, or VII directly interfered with gD binding to HveC and/or HveA but nonneutralizing MAbs did not interfere (16, 28). To apply this strategy to HveC, we raised 40 independent MAbs (numbered CK1 to CK41) against the ectodomain of HveC [HveC(346t)] expressed from a recombinant baculovirus. Eleven of these MAbs recognized epitopes within the V domain and were examined in more detail by enzyme-linked immunosorbent assay (ELISA), optical biosensor analysis, and native and denaturing Western blotting. We carried out epitope mapping studies using HveC truncations and synthetic peptides. Using optical biosensor technology, we showed competition between MAbs recognizing linear and conformational epitopes. Three MAbs, CK6, CK8, and CK41, recognized HveC on the cell surfaces of several cell lines and were also able to block HSV entry. Moreover, gD bound to soluble HveC prevented the binding of these MAbs to HveC. The information gathered about these MAbs has helped to define a specific site on HveC, which is important for virion gD binding.
MATERIALS AND METHODS
Cells and viruses.
IMR5 and SY5Y (human neuroblastoma cell lines) were maintained in Dulbecco's modified Eagle's medium plus 10% fetal calf serum (FCS). Vero and HeLa cells were grown in Dulbecco's modified Eagle's medium plus 5% FCS. M3A cells have been generated by transfecting CHO-IEβ8 cells (25) with pBG38, a plasmid expressing full-length human HveC under the control of a cytomegalovirus promoter (10). These cells express the β-galactosidase gene under the control of the viral ICP4 promoter induced upon infection. They were grown in Ham's F12 medium plus 5% FCS, 250 μg of G418 per ml, and 150 μg of puromycin per ml. CHO-IEβ8 cells were cultured in Ham's F12 medium plus 5% FCS and 150 μg of puromycin per ml. HSV-1 KOS tk12 (39) was grown and subjected to titer determination on Vero cells and was purified as described previously (11).
Protein production.
A version of HveC(346t) (16) without the C-terminal histidine tag [HveC(346t)hisless] was produced in the baculovirus system by the procedure described for HveC(346t) but using the downstream primer CGGTGATCAATGTTCGGGAGGAGACGGGGTGTA during PCR amplification (16). HveC(346t)hisless was purified from the supernatant of baculovirus-infected cells by affinity chromatography using the anti-HveC MAb XIV207 (R. J. Geraghty and P. G. Spear, unpublished data) and was eluted with 0.1 M glycine–0.5 M NaCl (pH 2.5), neutralized, dialyzed against phosphate buffer (pH 8.0) (150 mM NaCl, 0.1 M sodium phosphate), and concentrated. HveC(346t)hisless was used for immunization only; HveC(346t) was used for screening. The production and purification of HveC(346t), HveC(245t), HveC(143t), and gD(285t) were described previously (16, 17, 32).
Ab production and IgG purification.
Mice were immunized with HveC(346t)hisless until suitable titers of serum Abs were achieved. Hybridoma fusion was performed by a standard procedure. Hybridoma cells secreting anti-HveC(346t) Ig were subcloned twice. Igs were purified from mouse ascites using HiTrap protein G columns (Amersham Pharmacia) as specified by the manufacturer. IgGs were eluted using 2 ml of 0.1 M glycine (pH 2.7) and immediately neutralized with 60 μl of 1 M Tris (pH 9.0) prior to dialysis against phosphate-buffered saline (PBS). Typing was performed using a mouse hybridoma subtyping kit (Roche) as specified by the supplier. All the CK MAbs are of the IgG1 isotype with kappa light chains. The anti-tetrahistidine MAb was purchased from Qiagen Inc. The anti-HveC MAb R1.302 was kindly provided by S. McClellan (Beckman/Coulter). Anti-HveC rabbit polyclonal sera R154 against HveC and R7 against gD were described previously (12, 16).
Western blots.
Nondenaturing and nonreducing polyacrylamide gel electrophoresis PAGE have been described previously (6). Reduction and alkylation of HveC(346t) were performed as described previously (31).
ELISA. (i) Screening of MAbs secreted by hybridomas.
Plates were coated with HveC(346t) diluted to 10 μg/ml in PBS, blocked with 5% milk in PBS–0.1% Tween 20 (PBS-T milk), and incubated with hybridoma supernatants overnight at 4°C. Bound Ig was detected with goat anti-mouse IgG coupled with horseradish peroxidase (HRP). 2,2′-Azinobis(3-ethylbenzthazolinesulfonic acid) (ABTS) (Moss Inc.) was used as the substrate, and the absorbance was read at 405 nm.
(ii) Binding of IgG to HveC truncations.
The same procedure as above was used, except that dilutions of purified Ig were used to probe immobilized truncated forms of HveC, i.e., HveC(346t), HveC(245t), and HveC(143t).
Fluorescence-activated cell sorter (FACS) analysis.
Adherent cells were detached with 0.2 g of EDTA per liter in PBS (Versene; Gibco BRL). They were stained with saturating concentrations of anti-HveC MAbs (100 μg/ml; CK41 and R1.302 at 5 μg/ml) in PBS plus 3% bovine serum albumin (BSA), washed with PBS plus 3% BSA, and incubated with fluorescein isothiocyanate-conjugated goat anti-mouse in PBS plus 3% BSA. After a PBS wash, the cells were fixed with 1% paraformaldehyde in PBS.
Blocking assay.
Cells were grown to confluence in 96-well plates in their respective medium and chilled for 20 min at 4°C prior to the addition of Ig, which was serially diluted in cold medium containing 30 mM HEPES. After a 90-min incubation at 4°C, HSV-1 KOS tk12 carrying the β-galactosidase gene (39), also diluted in medium, was added to cells at a multiplicity of infection (MOI) of 2 to 4 PFU/cell. The cells were placed at 37°C and incubated for 6 h before being lysed by the addition of NP-40 to a final concentration of 0.5%. A 50-μl volume of cell lysate was mixed with an equal volume of β-galactosidase substrate (chlorophenol red-β-d-galactopyranoside; Roche). The level of entry was monitored by reading the absorbance at 595 nm for 50 min to record enzymatic activity, which is expressed as the change in absorbance per hour. Blocking activity of purified Ig is expressed as the percentage of virus entry into cells under test conditions as compared to viral infection in the absence of inhibitor (100%).
Immunoprecipitation.
Reaction mixtures (50 μl) containing 300 ng of HveC(346t) with or without 1.5 μg of gD(285t) were incubated in binding buffer (10 mM Tris [pH 8.0], 100 mM NaCl, 0.1% Nonidet P-40, 0.05% BSA, 0.05% chicken egg albumin) on ice for 1 h at 4°C. Purified Ig (500 ng) was added for 1 h at 4°C, followed by 50 μl of protein A-agarose (50%) for 1 h at 4°C. Beads were washed three times with high-salt buffer (10 mM Tris [pH 8.0], 500 mM NaCl, 0.1% Nonidet P-40, 0.05% BSA, 0.05% chicken egg albumin) and then boiled in 2× sodium dodecyl sulfate (SDS) sample buffer (28). Following SDS-PAGE (10% polyacrylamide), Western blots were probed with rabbit anti-HveC R154 (16) and anti-gD R7 (12) polyclonal sera followed by HRP-coupled anti-rabbit IgG.
Peptide mapping.
Synthetic 15-mer peptides (v1 to v11) covering the HveC(143t) domain were generated so that they overlapped by 5 amino acids (v1, Q31 to G44 with an additional D at the N terminus; v2, Y40 to A54; v3, H50 to Q64; v4, V60 to S74; v5, S70 to S84; v6, I80 to R94; v7, L90 to F104; v8, L100 to L114; v9, R110 to C124; v10, G120 to R134; v11, P130 to M143). Peptides were synthesized on a polyethylene glycol-modified cellulose membrane as described previously (29). Membrane strips were probed overnight at 4°C with anti-HveC Ig (5 μg/ml). Detection was performed using an HRP-coupled goat anti-mouse Ig followed by Supersignal detection substrate (Pierce).
Optical biosensor analysis.
Experiments were carried out on a Biacore X optical biosensor (Biacore AB) at 25°C. The running buffer was HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% polysorbate 20) (pH 7.4), the flow rate was set to 5 μl/min, and the data collection rate was 1 measurement/s.
(i) Binding properties of Ig.
To test the binding properties of any Ig, purified HveC(143t) was captured via its C-terminal histidine tag on a biosensor chip. First, an anti-histidine Ig (Qiagen Inc.) was directly coupled via primary amines to the surface of a CM5 chip using a standard procedure (BIAapplications handbook; Biacore AB, Uppsala, Sweden). The immobilization resulted in 4,100 and 3,600 resonance units (RU) of anti-histidine IgG immobilized on flow cell 1 (Fc1) and Fc2, respectively. At the beginning of each cycle, either 35 or 150 RU of HveC(143t) was captured on Fc2 only. Purified CK IgG (20 μg/ml) was then injected, and the association and dissociation of the complex were monitored for 2 min. The chip surface was regenerated by brief pulses of 0.2 M Na2CO3 (pH 10) until the response signal returned to baseline; then a new cycle was started.
(ii) Ig competition.
The same anti-histidine chip was used for competition studies. At the beginning of each cycle, 35 or 150 RU of HveC(143t) was captured via the C-terminal six-histidine tag on Fc2 only. An Ig, known to bind to HveC, was injected at a concentration of 20 μg/ml, for 10 min at 5 μl/min, on both Fc1 and Fc2 to saturate their epitope (primary Ig). Finally, the Ig to be tested (20 μg/ml) was injected on both Fc1 and Fc2 for 2 min to monitor its binding to HveC(143t). To regenerate the anti-His Ig chip surface, brief pulses of 0.2 M Na2CO3 (pH 10) (alternating with brief pulses of 10 mM glycine [pH 1.8] when necessary) were injected until the response signal returned to baseline. To determine the total amount of binding of each test Ig, no primary Ig was injected during the 10 min preceding the injection of the test Ig.
Sensorgrams were corrected for nonspecific binding and refractive index changes by subtracting the value obtained for the control sensorgram (Fc1) from the values obtained for the HveC surface sensorgram (Fc2). Data were analyzed with BIAevaluation software version 3.0 (Biacore AB). The amount of binding was determined as the RU measured 110 s after injection minus the RU measured at the time of injection of the test Ig. The binding of each test Ig in the absence of primary Ig was considered to be 100% (RUcontrol). Blocking of each tested Ig by itself resulted in residual binding (RUself). The binding of a given test MAb after preinjection of each primary MAb (Ign) was measured as RUIgn. Data are represented as percentage of binding of each tested Ig after blocking by the primary Ign, using the formula (RUIgn − RUself) × 100/(RUcontrol − RUself).
RESULTS
Generation of anti-HveC MAbs.
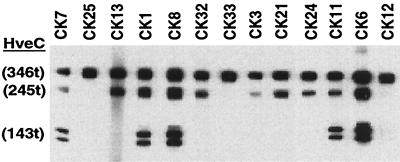
Forty independent hybridoma lines were isolated after two rounds of clonal selection. Each hybridoma secretes a MAb that was able to detect the soluble ectodomain of HveC [HveC(346t)] as shown by ELISA (data not shown). Initial mapping of the epitopes was performed by Western blot analysis of proteins electrophoresed under nonreducing and nondenaturing conditions. A mixture of three purified HveC truncated proteins representing the V domain alone [HveC(143t)], the V domain and the first C domain [HveC(245t)], or the complete ectodomain [HveC(346t)] (Fig. 1) (17) was probed with hybridoma culture supernatants. The results for a representative group of 13 MAbs are shown in Fig. 2. Several MAbs, such as CK1, CK6, CK7, CK8, and CK11, detected all three forms of HveC, indicating that their epitopes are located in the V domain of HveC. Another set, such as CK3, CK13, CK21, CK24, and CK32 recognized HveC(346t) and HveC(245t) but not HveC(143t), suggesting that their epitopes are located in the second Ig domain. Lastly, CK12, CK25, and CK33 recognized HveC(346t) only, indicating that their epitopes required the third Ig-like domain.
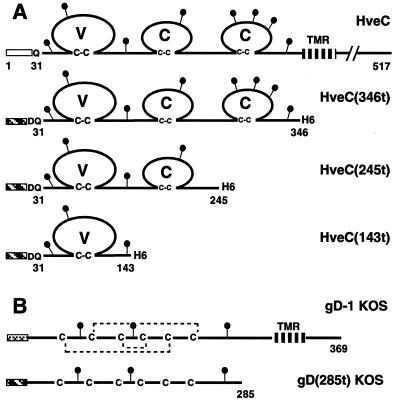
FIG. 1.
Schematic representation of recombinant proteins. (A) HveC constructs. Full-length HveC is shown as a solid line with amino acids numbered from the initial methionine. Three truncated forms of HveC were generated in the baculovirus system. Baculovirus-expressed proteins are truncated (t) at the indicated amino acid prior to the transmembrane region (TMR). The open box indicates the HveC natural signal peptide. The hatched box represents the mellitin signal peptide used in the baculovirus constructs. (B) gD constructs. Full-length gD from HSV-1 KOS and the truncated soluble gD(285t) produced in the baculovirus system are represented (32). Amino acids are numbered from the N terminus of the mature gD after cleavage of the gD signal peptide (cross-hatched box). Disulfide bonds are indicated by dotted lines. The black lollipops represent putative N-linked carbohydrates. H6 indicate the presence of a six-histidine tag at the C terminus of recombinant proteins with the exception of gD(285t).
FIG. 2.
Screening of hybridoma supernatant by native Western blot analysis. Three purified forms of truncated HveC [HveC(346t), HveC(245t), and HveC(143t)] were loaded on a gel under nonreducing and nondenaturing conditions, transferred to nitrocellulose, and probed with undiluted hybridoma culture supernatants. CK numbers correspond to the individual hybridoma clones. Discrete bands of HveC(143t) represent N-linked glycosylation variants (17).
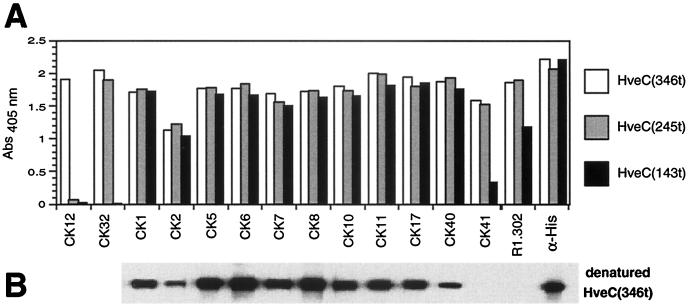
We previously showed that gD bound to HveC(143t) with full affinity, indicating that its binding site is entirely in the V domain of HveC (17). Therefore we decided to focus on the 11 MAbs whose epitopes were localized to this region. Igs (CK1, CK2, CK5, CK6, CK7, CK8, CK10, CK11, CK17, CK40, and CK41) were purified from ascites by protein G affinity chromatography and tested by ELISA (Fig. 3A). As expected from the native-gel Western blot results in Fig. 2, CK12 recognized only HveC(346t) and CK32 recognized both HveC(346t) and HveC(245t); the epitopes of these MAbs are localized in the third and second Ig-like domains, respectively. Each of the anti-V-domain CK MAbs recognized all three truncated forms efficiently (Fig. 3A); however, CK2 gave a consistently lower signal in all assays, presumably because of a lower affinity for HveC. The previously described MAb R1.302 (4) and an anti-histidine tag MAb were used as positive controls for conformational and linear epitopes, respectively, and both reacted with all three forms of HveC by ELISA. The anti-V-domain MAbs were further tested on a Western blot against HveC(346t) that had been previously reduced and alkylated. All of the V-domain MAbs, with the exception of CK41 and R1.302, were able to detect denatured HveC(346t). These last two MAbs did not efficiently detect HveC(143t) by ELISA, presumably because this truncated form of HveC underwent partial denaturation upon adsorption to the ELISA plate (17). Thus, 10 of the anti-V-domain MAbs are directed at linear epitopes whereas CK41 and R1.302 recognize conformational epitopes.
FIG. 3.
Characterization of anti-HveC V-domain Ig. (A) ELISA. Truncated forms of HveC [HveC(346t), HveC(245t), and HveC(143t)] were immobilized on a 96-well plate and incubated with purified Ig (10 μg/ml). Bound Anti-HveC Ig was detected with anti-mouse Ig-HRP and ABTS substrate. Absorbance was read at 405 nm. (B) Western blot. Reduced and alkylated HveC(346t) was loaded on a 16% polyacrylamide gel under reducing and denaturing conditions, transferred to nitrocellulose, and probed with anti-HveC Ig (0.1 μg/ml).
Detection of HveC on the cell surface.
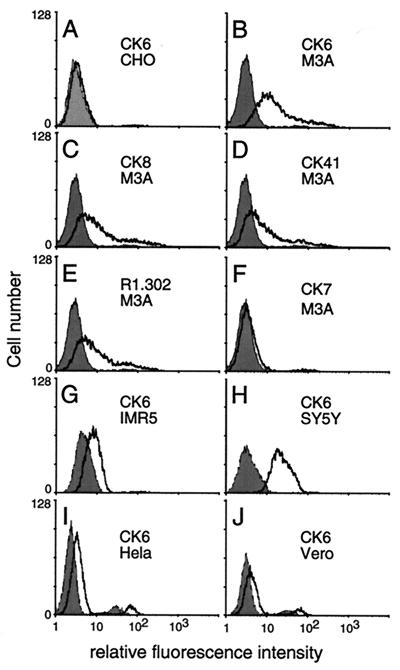
We used FACS analysis to test the ability of each CK MAb to detect HveC on CHO cells stably transfected with full-length human HveC cDNA (M3A cells) (Fig. 4B to F). Nontransfected CHO cells, which are refractory to HSV entry, were used as negative controls (Fig. 4A). MAbs CK6, CK8, CK41, and R1.302 gave a positive signal, indicating they bound to epitopes of HveC exposed on the surface of M3A cells (Fig. 4B to E). The remaining CK MAbs to the V domain, exemplified by CK7, were considered negative or weakly positive (Fig. 4F and data not shown). Using CK6 as a test MAb, we observed that the intensity of the FACS signal varied widely from cell line to cell line (Fig. 4G to J). Similar results were obtained with CK41 and R1.302 (data not shown), suggesting that variations of signal among the human cell lines reflect quantitative differences in cell surface expression of HveC rather than differences in epitope accessibility. The human neuroblastoma cell line SY5Y expressed a high level of surface HveC, whereas the IMR5 line displayed a more limited expression. HeLa cells expressed low levels of surface HveC. We also screened cell lines of animal origin commonly used to grow HSV, such as Vero, BHK, and L cells. Vero cells expressed low levels of HveC (Fig. 4J), whereas BHK and L cells were negative (data not shown). BHK cells were reported to be negative for HveC mRNA expression (4). Although murine HveC mRNA was detected in L cells (23), these cells did not express on their surface a form of HveC that has epitopes recognized by murine antibodies to human HveC.
FIG. 4.
Detection of HveC on the cell surface by FACS analysis using anti-HveC MAbs. (A) Nonpermissive CHO cells were tested as the negative controls for the nonspecific signal. (B to F) Susceptible M3A cells (CHO cells constitutively expressing human HveC) were tested for surface expression of HveC. (G to J) HSV-permissive cell lines were probed with the anti-HveC CK6 MAb. The gray shade represents fluorescence of cells in the absence of anti-HveC Ig but after incubation with the secondary antibody (goat anti-mouse–fluorescein isothiocyanate). The black line represents the fluorescence due to the anti-HveC MAb-specific binding on the cell surface.
Blocking virus infection.
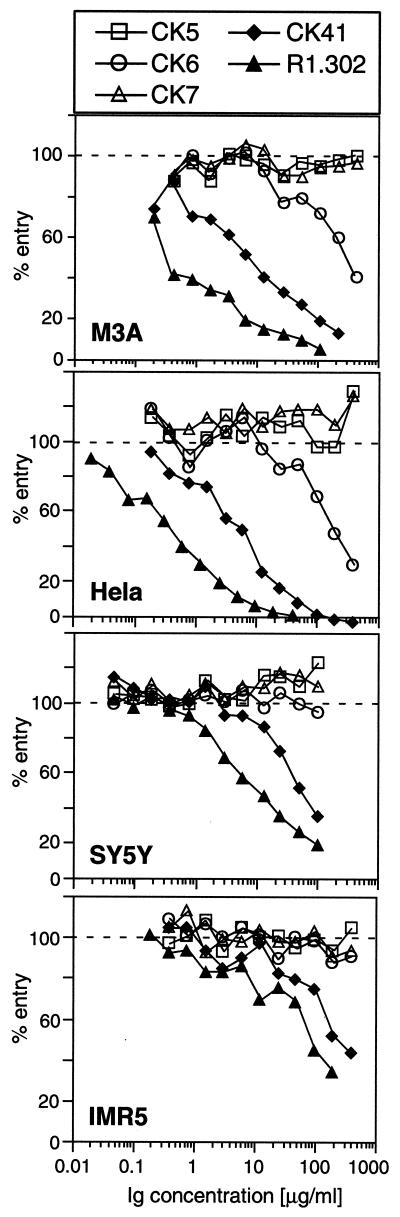
Since MAb R1.302 blocked the entry of HSV-1 into HeLa cells (4, 5), we determined whether any of the CK MAbs directed at the V domain had blocking capacity. We addressed this question using the cells and MAbs found to be positive by FACS analysis (Fig. 4). First we tested transfected CHO cells expressing the human HveC as the only HSV receptor (M3A cells). Entry of HSV-1 KOS into M3A cells was efficiently inhibited by the conformation-dependent MAbs R1.302 and CK41 (Fig. 5). Critical for this study, CK6 (Fig. 5) and CK8 (data not shown), which recognize linear epitopes, also blocked virus entry into M3A cells, albeit less efficiently. The same MAbs also protected HeLa cells from infection with high efficacy. When we tested the human neuroblastoma cell lines SY5Y and IMR5, we found that the blocking efficiency of R1.302 and CK41 was decreased and that CK6 did not block virus entry. The difficulty in blocking HSV entry into these cells might be due to the high level of HveC expressed by these cells or to the presence of other receptors.
FIG. 5.
Blocking of HSV infection with anti-HveC MAbs. M3A (CHO cells transfected with the human HveC), HeLa, SY5Y, and IMR5 cells were preincubated with increasing concentrations of purified anti-HveC Ig and infected with HSV-1 KOS tk12 at a MOI of 2 to 5 PFU/cell in the presence of Ig. The cells were lysed 6 h postinfection, and β-galactosidase activity was measured. 100% entry corresponds to the β-galactosidase activity induced following infection with HSV-1 KOS tk12 at a similar MOI in the absence of soluble Ig. Open symbols are used for MAbs with linear epitopes, and solid symbols are used for MAbs with conformational epitopes.
Blocking of anti-HveC MAb binding to HveC by gD.
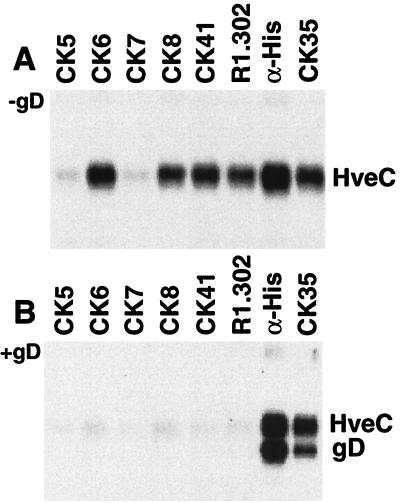
Binding of CK6, CK8, CK41, and R1.302 to HveC on the cell surface prevented HSV infection (Fig. 5) and blocked soluble gD binding to the cell surface (data not shown). We next used an in vitro assay to show that bound gD interfered with the recognition of HveC by these MAbs. We reasoned that an anti-HveC MAb could immunoprecipitate HveC with gD only if the MAb epitope was not masked as a result of the interaction with gD. When HveC(346t) was subjected to immunoprecipitation in the absence of gD, CK6, CK8, CK41, and R1.302 were able to immunoprecipitate native HveC(346t) (Fig. 6A). This correlates with their ability to detect HveC on the cell surface. The other anti-V-domain MAbs, such as CK5 and CK7, failed to immunoprecipitate HveC(346t) efficiently, suggesting that their epitopes were not accessible on soluble HveC(346t). When the high-affinity gD(285t) (32) is mixed with HveC(346t), a stable complex is formed in solution (16, 17). This complex can be precipitated with an anti-histidine tag MAb, which is specific for the C terminus of HveC(346t) (gD has no His tag) (Fig. 6B), or with an anti-HveC MAb (CK35), which recognizes an epitope in the second Ig-like domain of HveC, indicating that the bound gD did not cover their epitopes. In contrast, the anti-HveC V-domain MAbs were unable to precipitate the gD-HveC complex (Fig. 6B). This suggests that the epitopes of CK6, CK8, CK41, and R1.302 on HveC were inaccessible as a result of the gD interaction.
FIG. 6.
Immunoprecipitation of HveC and the HveC-gD complex. HveC(346t) alone (A) or mixed with an excess of gD(285t) (B) was subjected to immunoprecipitation with anti-HveC Ig. The immunoprecipitated proteins were separated on a 10% polyacrylamide gel and transferred to a nitrocellulose membrane. Both blots were probed simultaneously with rabbit polyclonal sera against HveC (R154) and gD (R7) followed by HRP-conjugated anti-rabbit Ig and enhanced chemiluminescence substrate. An anti-histidine tag MAb was used as a positive control to coprecipitate the complex. HveC(346t) carries a C-terminal histidine tag, while gD(285t) has no tag.
Mapping of linear epitopes on the HveC V domain.
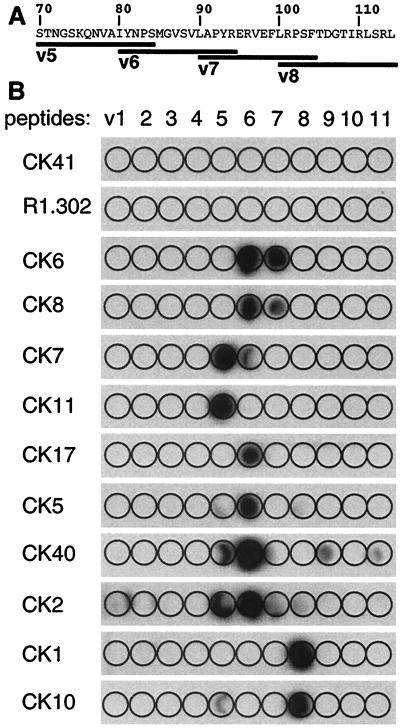
The capacity of CK6 and CK8, directed against linear epitopes, to inhibit HSV infection and to be blocked by gD bound to HveC prompted us to localize linear epitopes more accurately. We mapped them with a set of overlapping peptides spanning the V domain (peptides v1 to v11 [see Materials and Methods]). The peptides, which were directly synthesized on cellulose paper, were probed with each MAb. Figure 7 shows that only a limited number of peptides were recognized, suggesting that the most antigenic region of the human HveC V domain in mice is located between Ser70 and Leu114. This region contains a number of amino acid variations between the human HveC and mouse HveC sequences (see Fig. 10A), which are highly conserved overall (23). Each immunogenic peptide contains at least one nonconserved amino acid between mouse and human HveC. No MAbs were obtained against the conserved regions at the N terminus of HveC and at the C terminus of the V domain.
FIG. 7.
Peptide mapping of anti-HveC V-domain MAbs. (A) A partial sequence of the human HveC V-domain (aa 70 to 114) is indicated. Overlapping peptides v5 to v8 are shown as black bars. (B) Eleven peptides (v1 to v11) spanning the V domain of HveC (aa 31 to 143) were synthesized directly on a cellulose membrane. These strips were probed with the anti-HveC Ig (5 μg/ml). Due to the low binding of CK2, its autoradiogram was exposed for a longer time than the others were.
FIG. 10.
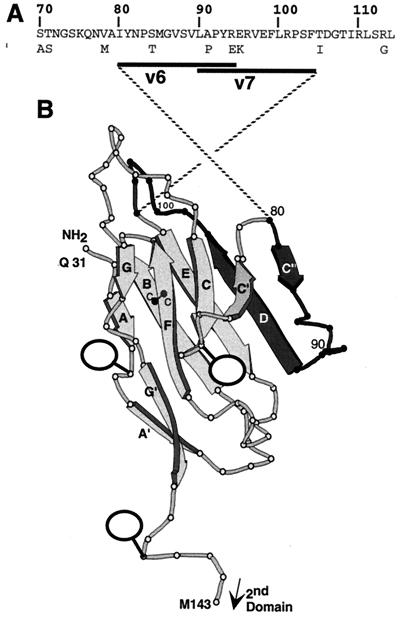
Location of CK6 and CK8 epitopes on the HveC V domain. (A) Partial sequence of the human HveC V domain (aa 70 to 114). The positions and amino acid variations of the mouse HveC sequence are also printed below the human sequence. Peptides v6 (aa 80 to 94) and v7 (aa 90 to 104), which are recognized by CK6 and CK8, are shown as black bars. (B) Three-dimensional model of the HveC V domain (based on a model of the poliovirus receptor V domain by Wimmer et al. [43]). The dark portion (aa 80 to 104) represents the position of peptides v6 and v7, encompassing the CK6 and CK8 epitopes. β-Sheets are named with bold letters. The disulfide bond between cysteines (c) located on β-sheets B and F is indicated. Balloons indicate the positions of N-linked carbohydrates.
CK6 and CK8 both detected peptides v6 and v7, suggesting that they reacted with the same or a closely related epitope between amino acids (aa) 80 and 104. CK6 and CK8 are the only MAbs that recognized peptide v7, although they do so with low efficiency. Likewise, the other MAbs could be paired according to the peptide(s) they recognized: CK7 with CK11, CK2 with CK40, CK5 with CK17, and CK1 with CK10. As expected, MAbs CK41 and R1.302 did not detect any synthetic peptides, reflecting the conformation-dependent nature of their epitopes.
Competition of MAbs analyzed by the optical biosensor.
To further define the anti-V-domain MAbs and to relate the conformational and linear epitopes, we performed Ab competition using an optical biosensor. To preserve the V-domain structure and to orient the molecules similarly, HveC(143t) was captured on the chip surface via its C-terminal His tag. First, the anti-V-domain IgGs were flowed across the chip and their binding was recorded. Seven MAbs (CK6, CK7, CK8, CK11, CK17, CK41, and R1.302) bound significantly to captured HveC(143t), although the binding of CK7, CK11, and CK17 was slightly lower (data not shown). The other MAbs (CK1, CK2, CK5, CK10, and CK40) failed to bind the HveC V domain in its native condition on the chip, suggesting that their epitopes were not exposed (data not shown).
Competition between the positive MAbs was performed to determine if they bound to overlapping epitopes. A primary (or blocking) Ab was bound to the captured HveC(143t) for 10 min, and the second (or test) Ab was then injected. The association of the test MAb with HveC(143t) was monitored for 2 min. As examples, Fig. 8 shows the results using CK6, CK11, and R1.302 as the test MAbs. The binding of a test antibody in the absence of a primary MAb served as a reference (Fig. 8, Control) and corresponded to 100% binding. Each antibody blocked itself, leaving only a residual binding, and this was considered background. The three MAbs presented in Fig. 8 displayed different patterns of competition, reflecting the fact that they belong to different groups. For example, binding of CK6 was blocked by CK8, CK41, and R1.302 but not by CK7, CK11, and CK17 (Fig. 8). Competition was then carried out in a reciprocal fashion between pairs of MAbs (Fig. 9). The percent binding of the test MAb, after 10 min of binding of the primary antibody, relative to its binding in the absence of primary MAb is shown in each square. Each interaction was categorized as an interfering interaction (black squares), a noninterfering interaction (white squares), or a partially interfering interaction (gray squares).
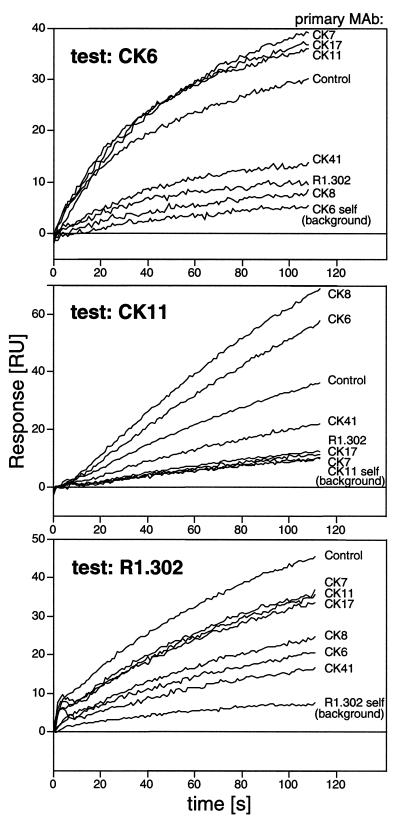
FIG. 8.
MAb competition measured on a biosensor. Overlaid sensorgrams of the binding of test MAbs CK6, CK11, and R1.302 to HveC(143t) after blocking with various primary MAbs are shown. The primary MAbs are indicated on the right. The sensorgrams are aligned at the time of injection of the test MAb. The binding of the test MAb to the captured HveC(143t) in the absence of a primary MAb is labeled “Control” and corresponds to 100% binding. Blocking of each test MAb by itself (self) resulted in residual binding, which was considered to represent background. A 35-RU amount of HveC(143t) was captured to analyze the binding of CK6 and R1.302, and 150 RU of HveC(143t) was used to bind CK11.
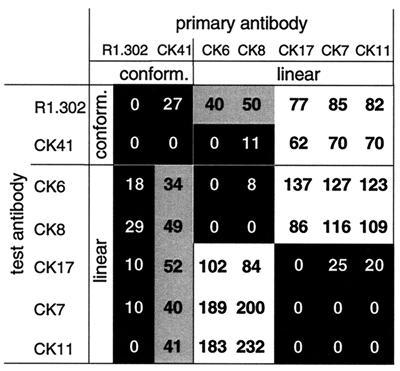
FIG. 9.
Competition between MAbs. The diagram indicates the amount of binding of each tested MAb (labeled on the left) after injection of the primary MAb (labeled at the top). The formula used to calculate the percentage of binding is (RUIgn − RUself) × 100/(RUcontrol − RUself), where RUcontrol represents the binding in the absence of primary MAb and RUself represents residual binding after self-blocking. This formula considers the RUcontrol to represent 100% binding and RUself to represent background normalized to zero (as on the diagonal of the diagram). A black background indicates low binding (0 to 30%), a gray background indicates binding from 30 to 60%, and a white background indicates binding greater than 60% of the binding measured in the absence of primary MAb.
The two conformation-dependent MAbs, CK41 and R1.302, interfered with each other in binding to HveC in a reciprocal manner, suggesting that their epitopes are overlapping. Similarly, CK6 and CK8 completely blocked the binding of each other, which was not unexpected since they bound to the same peptides (Fig. 7). A relationship between the pair CK41 and R1.302 and the pair CK6 and CK8 also emerged from this competition assay. CK6 and CK8 interfered to some extent with the binding of R1.302 and completely blocked the binding of CK41 to HveC(143t). These latter data are consistent with the idea that the conformational epitopes of R1.302 and CK41 are distinct, although both overlap each other and overlap the linear epitopes of CK6 and CK8. Another interference group consists of CK7, CK11, and CK17. Both CK7 and CK11 bound to the same peptide, but CK17 bound to an adjacent peptide. MAbs in this group were blocked by the conformation-dependent MAbs CK41 and R1.302, but the blocking was not reciprocal. The lack of reciprocity might be caused by differences in the affinity of each MAb involved or by conformational and oligomeric heterogeneity of the antigen on the chip (7).
We also noted that binding of CK6 or CK8 enhanced the binding of CK7 and CK11 twofold (Fig. 8 and 9). However, this enhancement was not observed for CK17. This suggests that binding of CK6 or CK8 modified the HveC(143t) structure in such a way that the epitopes of CK7 and CK11 were more exposed. This observation raised the possibility that the binding of CK6 or CK8 might induce a conformational change altering the epitopes of R1.302 and CK41, thus preventing the binding of the conformation-dependent MAbs.
DISCUSSION
Antibodies are useful tools to map functional sites involved in receptor-ligand interactions (15, 18, 26). In such cases, the functional site either overlaps the epitope or is disrupted upon antibody binding. We generated anti-HveC V-domain MAbs that interfered with gD-HveC interactions and then mapped their epitopes. Analysis of the epitopes covered by these blocking MAbs helped delineate a putative gD binding site on HveC.
gD and anti-HveC MAbs with blocking activities have a common binding site.
Overall, the most interesting anti-HveC MAbs were CK6, CK8, CK41, and R1.302, which were able to block infection and to prevent gD binding to the receptor on the cell surface and were inhibited by gD bound to HveC in vitro (Table 1). These MAbs could also compete with each other for binding to the V domain of HveC. Their epitopes appeared to be the most highly exposed on a native V domain (by biosensor analysis), on native HveC(346t) (by immunoprecipitation), and on HveC at the cell surface (by FACS). The common linear binding site of CK6 and CK8 within residues 80 to 104 (Fig. 10) probably overlaps the conformational epitopes of CK41 and R1.302. Our data also suggest that these residues form part of the gD binding site. However, gD binding is most likely to involve additional elements of HveC since the native HveC structure is required for gD binding (17). Because CK6 and CK8 detect two overlapping peptides, v6 (residues 80 to 94) and v7 (residues 90 to 104), we speculate that their epitopes are probably limited to residues 84 to 97. Although HveC seems to be particularly conserved among species, this small region contains four residues which differ between human and mouse HveC (Fig. 10A) (23). Murine HveC is also able to mediate HSV entry (23), although it remains to be seen if gD binding occurs with the same or a different affinity.
TABLE 1.
Properties of MAbs to the HveC V-domain
| MAb | Blocking of HSV-1 entrya | FACS resultb | Immunoprecipitationc | Blocked by gDd | Biosensor resulte | Peptide recognizedf |
|---|---|---|---|---|---|---|
| R1.302 | + | ++ | + | + | ++ | Noneg |
| CK41 | + | ++ | + | + | ++ | None |
| CK6 | + | ++ | + | + | ++ | v6(–v7)h |
| CK8 | + | ++ | + | + | ++ | v6(–v7) |
| CK11 | − | − | − | − | + | v5 |
| CK7 | − | − | − | − | + | v5(–v6) |
| CK17 | − | − | − | − | + | v6 |
| CK5 | − | − | − | − | − | v6 |
| CK40 | − | ± | − | − | − | (v5–)v6 |
| CK2 | − | + | − | − | − | (v5–v6) |
| CK1 | − | − | − | − | − | v8 |
| CK10 | − | − | − | − | − | v8 |
Blocking of HSV-1 KOS tested on M3A cells (CHO cells expressing human HveC).
Surface expression tested on transfected cells.
Immunoprecipitation of soluble HveC(346t).
Blocking by gD(285t) binding to HveC(346t) in a coimmunoprecipitation assay.
Detection of captured HveC(143t).
Peptides v5 (aa 70 to 84), v6 (aa 80 to 94), v7 (aa 90 to 104), and v8 (aa 100 to 114).
Conformational epitope; no peptides were detected.
Parentheses indicate a lower level of detection.
The antigenic region recognized by CK6 and CK8 (aa 80 to 104) is highlighted on a model of the HveC V domain (adapted from the model of the V domain of CD155 by Wimmer et al. [43]) (Fig. 10). This region mapped to the C" and D β-sheets and to the loop in between. The location of this loop at the periphery is consistent with the exposure of this region on the surface of the cell, where it can be accessible to virion gD and MAbs. Interestingly, the homologous region on the PVR V domain is involved in binding poliovirus particles (2, 3). Also, this particular region of the fourth Ig-like domain of CD4 plays a role in HIV gp120 binding (24).
Interestingly, binding of CK6 or CK8 seemed to enhance the binding of CK7 and CK11 but not CK17 as measured by the biosensor. This suggests that the nearby epitopes of CK7 and CK11 on β-sheet C′ became more exposed upon binding of CK8 or CK6, due to an induced conformational change of HveC(143t). Binding of CK6 and CK8 also prevented the binding of CK41 and reduced the binding of R1.302. As pointed out above, these latter antibodies rely on the HveC conformation for binding. This inhibition might have two causes: either a direct hindrance due to overlapping epitopes or a conformational alteration induced by CK6 and CK8. Although this experiment suggested that CK6 and CK8 slightly altered the HveC(143t) conformation, there is no evidence that it occurred on the surface of cells, nor that it would be sufficient to prevent the binding of gD and HSV entry. In such a case, however, the conformational gD binding site might be distinct from the CK6 or CK8 epitope but be modified upon MAb binding. This explanation appears unlikely since binding of gD prevents the detection of a gD-HveC complex by CK6 or CK8. These reciprocal data strongly suggest that CK6, CK8, and gD bind to the same region.
Location of anti-HveC MAb epitopes which lack blocking activity.
Aside from the blocking MAbs, three MAbs (CK7, CK11, and CK17) could be grouped by using a competition assay, which was performed with an optical biosensor. These three MAbs blocked each other in a reciprocal manner, suggesting that their epitopes overlap, at least partially. Since CK17 detected an overlapping peptide adjacent to the CK7 and CK11 binding site, it suggests that the epitope of CK17 is different. Binding of CK17, as well as CK5, on peptide v6, which was also detected by CK6 and CK8, suggests that this region of HveC is highly immunogenic and contains various epitopes. Although CK7, CK11, and CK17 bound to the native V domain, as shown by the biosensor, their epitopes were only poorly detected or not detected on the cell surface by FACS. These epitopes were not exposed on HveC(346t) in solution (inefficient immunoprecipitation), suggesting that they were hidden by other domains of HveC itself when present as a soluble tetramer (17). Thus, even though protein conformation and orientation can be retained on a biosensor chip, it does not entirely mimic the HveC V-domain presentation on a complex cell surface.
Inhibition of gD binding and blocking of HSV entry.
As shown above, soluble gD blocked the binding of CK6, CK8, CK41, and R1.302 to soluble HveC. These MAbs also blocked HSV entry into cells. Thus, the anti-HveC MAbs probably prevented HSV infection of mammalian cells by occupying the gD binding site, at least partially.
Both CK41 and R1.302, prevented gD binding to HveC on cells; however, in contrast to R1.302, CK41 failed to block binding of gD to purified HveC(346t) immobilized on an ELISA plate (data not shown). This suggests that the binding residues for these MAbs are slightly different. Overall, CK41 and R1.302 were more efficient than MAbs recognizing linear epitopes (CK6 and CK8) in their ability to block gD binding on cells and to block HSV infection. We presume that this enhanced efficacy correlates with their ability to better bind membrane-bound HveC and to achieve saturation of the cell surface receptors. FACS analysis showed that cultured cells express various amounts of HveC on their surface. The amount and the presentation of HveC on human cells might affect the inhibitory potential of anti HveC MAbs. For instance, HeLa cells, which express smaller amounts of HveC than IMR5 and SY5Y cells do, were more efficiently protected by anti-HveC MAbs. However, the quantity of HveC detected on human cell lines cannot be the only factor involved. For example, IMR5 cells expressed less HveC on their surface than SY5Y cells did, yet it was more difficult to block HSV entry into IMR5 cells than into SY5Y cells. The presence of other receptors, particularly HveA, might also explain why it is difficult to block HSV entry into IMR5 or SY5Y cells with anti-HveC MAbs (J. C. Whitbeck et al., unpublished data). However, the virus entry-blocking activity of R1.302 on SY5Y reached more than 80%, suggesting that HveC is the major receptor for HSV-1 KOS at the surface of these cells and that the contribution of HveA, or 3-O-S-heparan sulfate (34), is limited.
In summary, by using anti-HveC MAbs, we showed that surface expression of HveC is prevalent on cultured human cell lines. More direct quantification of HveC on the surface of various cells will help correlate the level of expression with susceptibility to infection or sensitivity to MAb blocking. Most of the cell lines can be protected from infection by several MAbs directed against the V domain of HveC. Fine epitope mapping of these blocking MAbs provided strong evidence that the region between residues 80 and 104 is involved in HveC binding to HSV gD. Usage of antibodies detecting this region demonstrated that this domain is well exposed on the cell surface, as expected for a viral binding site.
ACKNOWLEDGMENTS
This investigation was supported by Public Health Service grants NS-30606 and NS-36731 from the National Institute of Neurological Diseases and Stroke (to R.J.E. and G.H.C.) and by grant AI-18289 from the National Institute of Allergy and Infectious Diseases (to R.J.E. and G.H.C.). C.K. was supported by a fellowship (823A-053464) from the Swiss National Science Foundation. We thank the Schools of Dental and Veterinary Medicine of the University of Pennsylvania for the purchase of the Biacore X instrument.
We thank Patricia Spear for the XIV207 MAb and S. McClellan (Beckman/Coulter) for the R1.302 MAb. We are grateful to Laszlo Otvos for peptide synthesis and to Bruce Shenker for FACS assistance. Hybridoma production was performed at the Cell Center of the University of Pennsylvania. We thank Sharon Willis, Ann Rux, Sarah Connolly, Robert Geraghty, and Richard Milne for helpful discussions.
REFERENCES
- 1.Aoki J, Koike S, Asou H, Ise I, Suwa H, Tanaka T, Miyasaka M, Nomoto A. Mouse homolog of poliovirus receptor-related gene 2 product, mPRR2, mediates homophilic cell aggregation. Exp Cell Res. 1997;235:374–384. doi: 10.1006/excr.1997.3685. [DOI] [PubMed] [Google Scholar]
- 2.Aoki J, Koike S, Ise I, Sato-Yoshida Y, Nomoto A. Amino acid residues on human poliovirus receptor involved in interaction with poliovirus. J Biol Chem. 1994;269:8431–8438. [PubMed] [Google Scholar]
- 3.Bernhardt G, Harber J, Zibert A, deCrombrugghe M, Wimmer E. The poliovirus receptor: identification of domains and amino acid residues critical for virus binding. Virology. 1994;203:344–356. doi: 10.1006/viro.1994.1493. [DOI] [PubMed] [Google Scholar]
- 4.Cocchi F, Lopez M, Menotti L, Aoubala M, Dubreuil P, Campadelli-Fiume G. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc Natl Acad Sci USA. 1998;95:15700–15705. doi: 10.1073/pnas.95.26.15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attribute of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen G H, Isola V J, Kuhns J, Berman P W, Eisenberg R J. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J Virol. 1986;60:157–166. doi: 10.1128/jvi.60.1.157-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daiss J L, Scalice E R. Epitope mapping on BIAcore: theoretical and practical considerations. Methods Companion Methods Enzymol. 1994;6:143–156. [Google Scholar]
- 8.Eberlé F, Dubreuil P, Mattei M-G, Devilard E, Lopez M. The human PRR2 gene, related to the poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 9.Fuller A O, Lee W C. Herpes simplex virus type 1 entry through a cascade of virus-cell interactions requires different roles of gD and gH in penetration. J Virol. 1992;66:5002–5012. doi: 10.1128/jvi.66.8.5002-5012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geraghty R J, Krummenacher C, Eisenberg R J, Cohen G H, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 11.Handler C G, Eisenberg R J, Cohen G H. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol. 1996;70:6067–6075. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isola V J, Eisenberg R J, Siebert G R, Heilman C J, Wilcox W C, Cohen G H. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J Virol. 1989;63:2325–2334. doi: 10.1128/jvi.63.5.2325-2334.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson D C, Burke R L, Gregory T. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson D C, Ligas M W. Herpes simplex viruses lacking glycoprotein D are unable to inhibit virus penetration: quantitative evidence for virus-specific cell surface receptors. J Virol. 1988;62:4605–4612. doi: 10.1128/jvi.62.12.4605-4612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koike S, Ise I, Nomoto A. Functional domains of the poliovirus receptor. Proc Natl Acad Sci USA. 1991;88:4104–4108. doi: 10.1073/pnas.88.10.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krummenacher C, Rux A H, Whitbeck J C, Ponce de Leon M, Lou H, Baribaud I, Hou W, Zou C, Geraghty R J, Spear P G, Eisenberg R J, Cohen G H. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity while the third domain is involved in oligomerization of HveC. J Virol. 1999;73:8127–8137. doi: 10.1128/jvi.73.10.8127-8137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee B, Sharron M, Blanpain C, Doranz B J, Vakili J, Setoh P, Berg E, Liu G, Guy H R, Durell S R, Parmentier M, Chang C N, Price K, Tsang M, Doms R W. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 19.Lopez M, Aoubala M, Jordier F, Isnardon D, Gomez S, Dubreuil P. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood. 1998;92:4602–4611. [PubMed] [Google Scholar]
- 20.Lopez M, Cocchi F, Menotti L, Avitabile E, Dubreuil P, Campadelli-Fiume G. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J Virol. 2000;74:1267–1274. doi: 10.1128/jvi.74.3.1267-1274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez M, Eberlé F, Mattei M-G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 22.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 23.Menotti L, Lopez M, Avitabile E, Stefan A, Cocchi F, Adelaide J, Lecocq E, Dubreuil P, Campadelli-Fiume G. The murine homolog of human nectin 1 delta serves as a species non-specific mediator for entry of human and animal alphaherpesviruses in a pathway independent of detectable binding to gD. Proc Natl Acad Sci USA. 2000;97:4867–4872. doi: 10.1073/pnas.97.9.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moebius U, Clayton L K, Abraham S, Harrison S C, Reinherz E L. The human immunodeficiency virus gp120 binding site on CD4: delineation by quantitative equilibrium and kinetic binding studies of mutants in conjunction with a high-resolution CD4 atomic structure. J Exp Med. 1992;176:507–517. doi: 10.1084/jem.176.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 26.Moore J P, Sattentau Q J, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muggeridge M I, Chiang H-Y, Cohen G H, Eisenberg R J. Mapping of a functional site on herpes simplex virus glycoprotein D. J Cell Biochem Suppl. 1992;16C:132. [Google Scholar]
- 28.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on the herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otvos L, Paese A M, Bokonyi K, Giles-Davis W, Rogers M E, Hintz P A, Hoffmann R, Ertl H C. In situ stimulation of a T-helper cell hybridoma with a cellulose-bound peptide antigen. J Immunol Methods. 2000;233:95–105. doi: 10.1016/s0022-1759(99)00194-5. [DOI] [PubMed] [Google Scholar]
- 30.Peterson A, Seed B. Genetic analysis of monoclonal antibody and HIV binding sites on the human lymphocyte antigen CD4. Cell. 1988;54:65–72. doi: 10.1016/0092-8674(88)90180-8. [DOI] [PubMed] [Google Scholar]
- 31.Rux A H, Moore W T, Lambris J D, Abrams W R, Peng C, Friedman H M, Cohen G H, Eisenberg R J. Disulfide bond structure determination and biochemical analysis of glycoprotein C from herpes simplex virus. J Virol. 1996;70:5455–5465. doi: 10.1128/jvi.70.8.5455-5465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rux A H, Willis S H, Nicola A V, Hou W, Peng C, Lou H, Cohen G H, Eisenberg R J. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpes virus entry mediator. J Virol. 1998;72:7091–7098. doi: 10.1128/jvi.72.9.7091-7098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satoh-Horikawa K, Nakanishi H, Takahashi K, Miyahara M, Nishimura M, Tachibana K, Mizoguchi A, Takai Y. Nectin-3, a new member of immunoglobulin-like cell adhesion molecules that shows homophilic and heterophilic cell-cell adhesion activities. J Biol Chem. 2000;275:10291–10299. doi: 10.1074/jbc.275.14.10291. [DOI] [PubMed] [Google Scholar]
- 34.Shukla D, Liu J, Blaiklock P, Shworak N W, Bai X, Esko J D, Cohen G H, Eisenberg R J, Rosenberg R D, Spear P G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 35.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 201–232. [Google Scholar]
- 36.Suzuki K, Hu D, Bustos T, Zlotogora J, Richieri-Costa A, Helms J A, Spritz R A. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nat Genet. 2000;25:427–430. doi: 10.1038/78119. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warner M S, Martinez W, Geraghty R J, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by herpes simplex virus type 2, mutants of herpes simplex virus type 1 and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 40.Whitbeck J C, Muggeridge M I, Rux A, Hou W, Krummenacher C, Lou H, van Geelen A, Eisenberg R J, Cohen G H. The major neutralizing antigenic site on herpes simplex virus glycoprotein D overlaps a receptor-binding domain. J Virol. 1999;73:9879–9890. doi: 10.1128/jvi.73.12.9879-9890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the TNFR superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willis S H, Rux A H, Peng C, Whitbeck J C, Nicola A V, Lou H, Hou W, Salvador L, Cohen G H, Eisenberg R J. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wimmer E, Harber J J, Bibb J A, Gromeier M, Lu H H, Bernhardt G. Poliovirus receptors. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 101–127. [Google Scholar]