Abstract
Hybrids of cymbidium ringspot (CymRSV) and carnation Italian ringspot (CIRV) tombusviruses were used to identify viral symptom determinants responsible for the generalized necrosis in tombusvirus-infected plants. Surprisingly, symptoms of Nicotiana benthamiana infected with CymRSV/CIRV hybrids were distinctly different. It was demonstrated that not all chimeras expressing wild-type (wt) levels of p19 protein caused systemic necrosis as both parents CymRSV and CIRV did. We showed here that hybrids containing chimeric ORF1 were not able to induce lethal necrosis even if the viral replication of these constructs was not altered significantly. However, if a wt p33 (product of ORF1) of CymRSV was provided in trans in transgenic plants expressing p33 and its readthrough product p92, the lethal necrosis characteristic to tombusvirus infection was restored. In addition, the expression of p33 by a potato virus X viral vector in N. benthamiana caused severe chlorosis and occasionally necrosis, indicating the importance of p33 in wt symptoms of tombusviruses. Thus, our results provide evidence that elicitation of the necrotic phenotype requires the presence of the wt p33 in addition to the p19 protein of tombusviruses.
Plant viruses are responsible for severe diseases in plants, resulting in major losses in many important crops. Infection of plants with viruses usually results in different symptoms, which may vary greatly. The most common symptoms are perturbation in the growth of the plant, malformations, chlorotic or dark green spots on the leaves, and necrotic lesions or even generalized necrosis, leading to the death of the plant. The development of symptoms during a virus infection is likely to be the last step in a complex and little-understood process in which the virus interacts with the metabolism of the plant on many different levels (1). It is generally assumed that the functional and elicitation activities of viral proteins require specific interaction with host factors. These host factors can interact with individual viral genes, as demonstrated by several studies in which coding as well as noncoding regions were shown to be able to modulate the specific symptoms of a given virus infection (3, 10, 12, 15, 16, 30, 33).
Plant viruses that have a small RNA genome, such as tombusviruses, are well suited for studying the molecular bases of plant-virus interaction and symptom development. Tombusviruses have a broad experimental host range, and the members of the genus have been extensively studied at the molecular level. A number of infectious cDNA clones are also available (6, 14, 26). The genome of a tombusvirus is a linear, single-stranded monopartite RNA molecule of positive polarity, about 4,700 nucleotides long. The genome contains five open reading frames (ORFs) coding for proteins with approximate molecular masses of 33, 92, 22, and 19 kDa and for a 41-kDa coat protein (26). The genomic RNA acts as an mRNA for the translation of a 33-kDa protein (p33; ORF1), and a 92-kDa protein (p92; ORF2). The p92 protein is a product of readthrough of the amber termination codon of the p33 protein (26). Both p33 and p92 are required for viral replication (11, 17, 19, 20, 31). Using full-length hybrid infectious cDNA clones of the cymbidium ringspot (CymRSV) and carnation Italian ringspot (CIRV) viruses (Fig. 1), it has been shown (6) that the N-terminal half of ORF1 contained the determinants for the formation of vesiculated membraneous structures (multivesicular bodies [MVBs]), which are possible sites of tombusvirus replication (24). In addition, the p33 protein was localized by immunogold labeling to the periphery of vesiculated peroxisomes in CymRSV-infected cells (4) and was suggested to be a transmembrane protein.
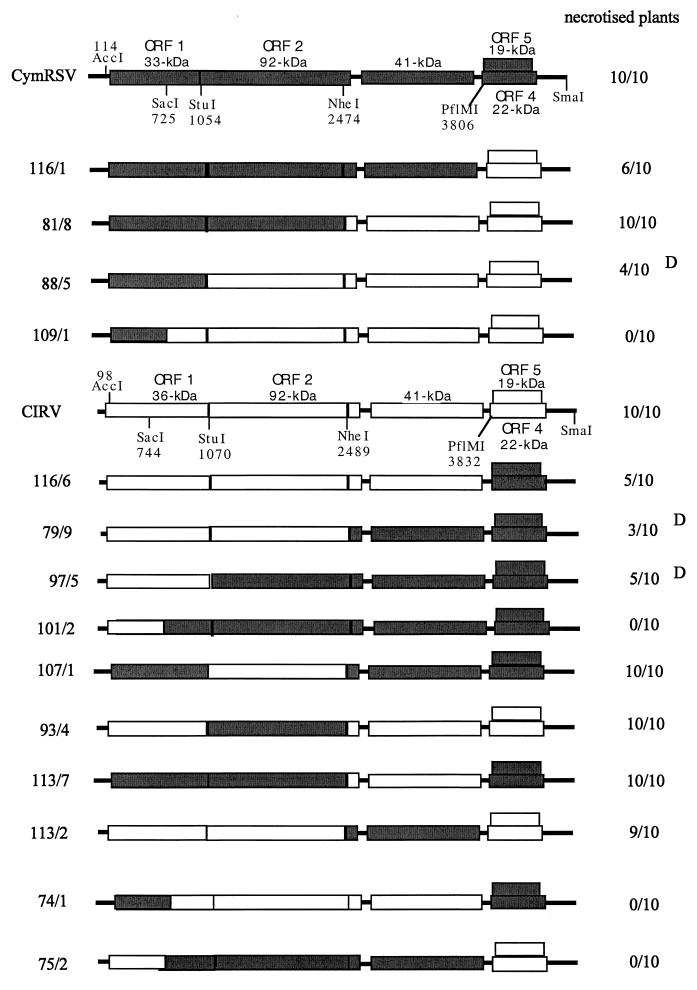
FIG. 1.
Schematic representation of and symptoms induced by the CymRSV/CIRV chimeras are shown with the ORFs. The approximate molecular masses of the proteins encoded by CymRSV and CIRV are shown. The common restriction endonuclease sites used for constructing chimeras are indicated. Numbers below the restriction sites show their positions in the viral genome. The number of plants showing typical necrotic symptoms for each construct is indicated on the right side. D, delayed symptoms. Ten plants were inoculated with each inoculum, and each experiment was repeated at least three times.
The 41-kDa coat protein is encoded by ORF3 and translated from subgenomic RNA 1 (sg1 RNA) (26). Two nested ORFs, ORF4 and ORF5, are located at the 3′ terminus of the virus genome and encode a 22-kDa protein (p22) and a 19-kDa protein (p19), respectively. Both p22 and p19 are translated from sg2 RNA (26). p22 is required for cell-to-cell movement (11, 23, 29, 30) and is also involved in symptom determination and the elicitation of resistance responses (8). Although the precise function of p19 has not been determined, it plays an important role in necrotic symptom development (11, 23, 30), and it was suggested to be a suppressor of posttranscriptional gene silencing (PTGS) (34). Previously, it was also suggested that p19 is solely responsible for the generalized necrosis of tombusvirus-infected plants (30) and participates in virus spread in a host-specific manner. However, symptoms of Nicotiana benthamiana plants infected with CymRSV/CIRV hybrids constructed previously (6) were surprisingly different from those of plants infected with CymRSV or CIRV. Moreover, not all chimeras expressing wild-type (wt) levels of p19 caused systemic necrosis (J. Burgyán, EMBO Workshop on Molecular Mechanisms in the Replicative Cycle of Viruses in Plants, 1997, abstr. 47). These preliminary observations suggested that viral factors other than p19 are also required to induce the systemic necrosis characteristic to N. benthamina and Nicotiana clevelandii systemically infected with known tombusviruses.
These conflicting results published on the role of p19 in inducing systemic necrosis (30;); J. Burgyán, abstr.) prompted us to analyze further the role of tombusvirus proteins in symptom development. In this report we show evidence using hybrid viruses that expression of the p19 protein of two tombusviruses is likely required with another viral protein(s) for virus-induced lethal necrosis. We also show that p33 (or p36 in CIRV) interacts directly or indirectly with p19 and that this interaction is required for development of systemic necrosis. Hybrid p33 genes did not support the development of generalized necrosis even if replication of these constructs was not altered significantly relative to wt virus. However, if wt p33 was provided by transgenic plants, the lethal necrosis characteristic to tombusvirus infection was restored.
MATERIALS AND METHODS
Plasmid constructs.
The infectious cDNA clones of CymRSV, Δ19 of CymRSV, CIRV, and most of the CymRSV/CIRV hybrid constructs (116/1, 81/8, 88/5, 109/1, 116/6, 79/9, 97/5, 101/2, 107/1, 93/4, 113/7, 113/2, 125/3, C80, and C80/G11) have been described previously (6, 11). Two additional chimeras were also prepared using PflMI and SmaI sites in the CIRV and CymRSV genomic sequences. The new constructs, designated 74/1 and 75/2, respectively, were obtained by replacing the coding region of the 3′ proximal nested genes (ORF4 and ORF5) and the 3′ noncoding region (UTR) between constructs 101/2 and 109/1. The potato virus X (PVX) vector (pP2C2S) used to express CymRSV proteins has been described previously (2, 7). The CymRSV cDNA fragments encoding p33 and p19 were PCR amplified from the G11 plasmid (11) using an oligonucleotide homologous to the first 22 and complementary to the last 21 nucleotides of ORF1 and ORF5, respectively. The PCR-amplified DNA fragments were cloned individually into EcoRV linearized PVX vector under the control of a duplicated PVX coat protein promoter.
In vitro transcription and plant inoculation.
Transcription of SmaI-linearized template DNA (CymRSV or CIRV derivatives) and inoculation of uncapped transcripts onto N. clevelandii and N. benthamiana plants were performed as described previously (11). PVX-derived plasmids were linearized with SpeI, and in vitro RNA transcripts were capped using a cap analogue (New England Biolabs) (7).
Protoplast preparation and inoculation.
Protoplasts were isolated from N. benthamiana plants and transfected with in vitro-synthesized transcripts of genomic RNA using the polyethylene glycol method as described (11).
RNA extraction, molecular hybridization, and protein analysis.
Samples of upper noninoculated leaves were taken 7 to 14 days after inoculation, when the first systemic symptoms had appeared. Inoculated plants that did not develop necrotic symptoms were kept for several weeks, and leaf samples were collected and examined periodically. Total nucleic acids were extracted from 50 mg of leaf tissue or 0.5 × 106 to 1 × 106 harvested protoplasts as described (35). Briefly, the homogenized plant material or pelleted protoplasts were resuspended in 600 μl of extraction buffer (0.1 M glycine–NaOH [pH 9.0] containing 100 mM NaCl, 10 mM EDTA, 2% sodium dodecyl sulfate [SDS], and 1% sodium lauroylsarcosine) and mixed with an equal volume of phenol. The aqueous phase was treated with equal volumes of phenol and chloroform, precipitated with ethanol, and resuspended in sterile water. The presence of virus-related RNA was assessed by Northern blot analysis using formaldehyde-agarose gels. Each RNA sample contained approximately 5 μg of total nucleic acids, and a 32P-labeled probe prepared by random priming (27) of a clone representing the 3′-terminal 60 nucleotides of CymRSV RNA was used for hybridization. This CymRSV sequence contains only one base mismatch compared to the corresponding sequence of the CIRV genome. The presence of viral gene products in infected plants was verified by Western blot analysis. About 100 mg of leaf tissue was rapidly ground in 2 volumes of Laemmli sample buffer, incubated at 100°C for 3 min, and fractionated by 0.1% SDS–12.5% polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to nitrocellulose membrane. The p33 and p19 proteins were detected by using antibodies raised in rabbits against purified p33 and p19, respectively (14, 17).
RESULTS
Symptoms and replication of CymRSV/CIRV hybrids.
In an effort to better understand the role of tombusvirus genes in virus-induced necrotic symptoms, 10 N. benthamiana plants were infected with a series of CymRSV/CIRV chimeras (Fig. 1). The symptoms obtained varied from lethal necrosis to attenuated mosaic and leaf distortion, even though virus accumulation in infected N. benthamiana plants did not change dramatically from that observed for the wt viruses (Fig. 2). The constructs in which ORF1 and 3′-proximal ORFs (4 and 5) were derived from the same parents caused typical wt necrotic symptoms, as indicated in Fig. 1. However, most of the constructs having terminal sequences deriving from different parents caused wt symptoms only in part of the inoculated plant, which were often delayed (Fig. 1). Construct 81/8 was an exception; it induced wt symptoms on both N. benthamiana and N. clevelandii. Symptom attenuation was particularly characteristic of two constructs (101/2 and 109/1), which never caused systemic necrosis on N. benthamiana (Fig. 1). These two constructs (101/2 and 109/1) were further analyzed on N. clevelandii. In contrast to symptom development on N. benthamiana, N. clevelandii plants inoculated with 101/2 showed the usual chlorotic lesions on the inoculated leaves, but the upper noninoculated leaves remained symptomless for several weeks, and no viral RNA could be detected in these leaves (not shown). Hybrid 109/1 showed a remarkable delay (7 to 10 days) in the development of systemic symptoms compared to the wt viruses on N. clevelandii. The upper, noninoculated leaves showed only leaf distortion and occasionally small necrotic lesions, but they failed to show the typical apical necrosis followed by death of the plant (not shown). These results indicated that the 5′ terminus of the viral genome plays a crucial role in determining wt (necrotic) symptoms and suggests that the altered 5′ terminus of viral genomes is not able to contribute to the elicitation of necrotic symptoms. Apart from symptoms caused by 81/8, an alternative explanation is that only the constructs having homologous (deriving from the same virus) 5′ and 3′ termini are able to elicit necrotic symptoms.
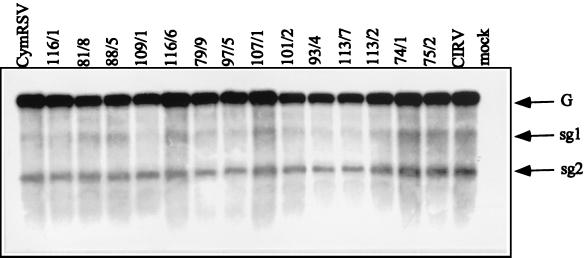
FIG. 2.
Northern blot analysis of RNA extracted from N. benthamiana plants inoculated with CymRSV/CIRV chimeras. Samples were taken from the first just-developed apical leaves showing systemic symptoms, which appeared 7 to 10 dpi. A 32P-labeled probe specific to the 3′ terminus of CymRSV was used for hybridization. G and sg1 and sg2 genomic and subgenomic RNAs, respectively.
To differentiate between these possibilities, two other constructs (74/1 and 75/2) were prepared by exchanging the two nested ORFs and 3′ UTRs between constructs 101/2 and 109/1 (Fig. 1). In order to address whether the terminal sequences deriving from the same virus can restore the wt symptoms on N. benthamiana, plants were infected with transcripts from 74/1 and 75/2. The attenuated symptoms (lack of necrosis) produced (Fig. 1) and viral RNA accumulation (Fig. 2) in these plants were the same as in the plants inoculated with 101/2 and 109/1. These results demonstrate that chimeras (101/2, 109/1, 74/1, and 75/2) having a hybrid ORF1 are not able to elicit wt symptoms regardless of the origin of the 3′ terminus.
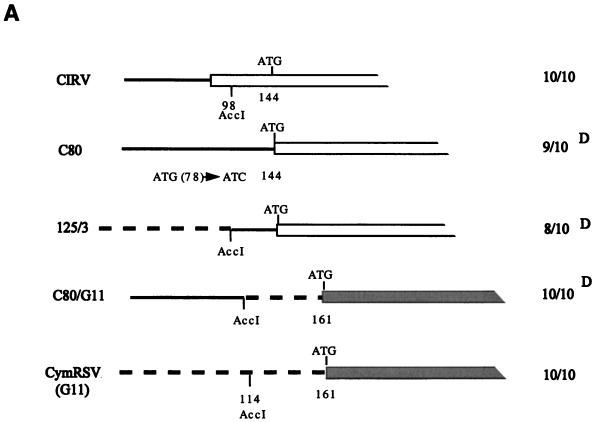
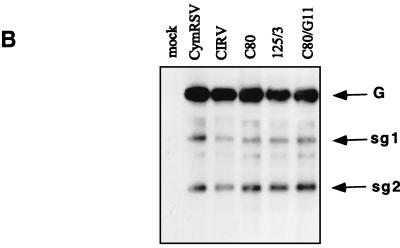
To find out whether the UTR and/or the N-terminal half of ORF1 is responsible for the attenuated symptoms, three other previously described (6) clones (C80, 125/3, and C80/G11) were tested (Fig. 3). In C80, the AUG initiation codon (positions 78 to 80) of ORF1 in wt CIRV was converted to AUC, so that the next available AUG was at positions 144 to 146. The protein product of ORF1 in C80 is 34 kDa instead of the wt 36 kDa. In clone 125/3, the first 98 nucleotides of CIRV were replaced with 114 nucleotides of the UTR of CymRSV, and in the sister clone C80/G11, the 114 nucleotides of the UTR of CymRSV were replaced with the first 98 nucleotides of C80. In vitro RNA transcripts derived from C80, 125/3, and C80/G11 were infectious on N. benthamiana and induced wt symptoms. While no significant differences could be observed in virus RNA accumulation in the upper noninoculated leaves (Fig. 3B), there was a delay of 4 to 5 days in symptom development (12 to 15 days postinoculation [dpi]) relative to wt viruses (Fig. 3A). Although the appearance of symptoms was delayed, these constructs (C80, 125/3, and C80/G11) induced the same necrotic phenotype as wt viruses. Therefore, it is unlikely that the UTR of tombusviruses has a significant role in symptom development.
FIG. 3.
(A) Diagrammatic representation of 5′-terminal mutants of CIRV (C80 and 125/3) and CymRSV (C80/G11). Only the 5′ leader sequence and a part of ORF1 are shown. Continuous and broken lines indicate the UTRs of CIRV and CymRSV, respectively. ATG indicates the initiation codon, and the number below shows the position in the viral genome. Solid and open boxes indicate ORF1 of CIRV and CymRSV, respectively. (B) Viral RNA accumulation in N. benthamiana plants inoculated with 5′-terminal mutants. The probe used for hybridization and definitions of symbols are the same as in Fig. 2.
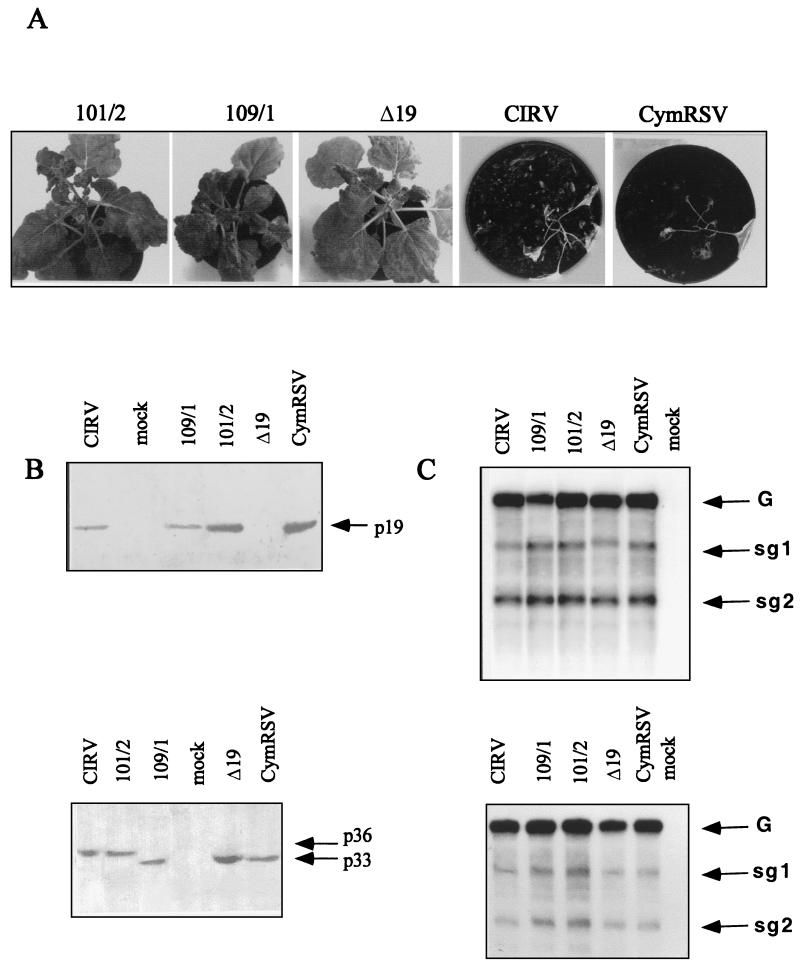
To better understand why the plants inoculated with constructs 101/2 and 109/1 show attenuated symptoms, we analyzed the level of p33 and p19 proteins and the corresponding sg RNAs, which are thought to play an important role in symptom development. N. benthamiana plants were inoculated again with 101/2, 109/1, Δ19 (in this clone the initiation codon of p19 was changed to CUG, not affecting the amino acid content of p22), and wt viruses. The apical leaves of N. benthamiana plants inoculated with CIRV and CymRSV necrotized shortly after the first systemic symptoms appeared (7 to 10 dpi) (Fig. 4A). Therefore, we were not able to follow virus accumulation in the new leaves of wt virus-infected plants because they died (12 to 15 dpi) before new leaves developed. In contrast, plants infected with 101/2, 109/1, and Δ19 developed less severe symptoms (Fig. 4A), and new leaves developed. For comparison, samples from upper leaves showing the first-appearing systemic symptoms (typically 7 to 10 dpi) were examined by Northern and Western analysis. The results did not show significant variation in the levels of genomic and subgenomic RNAs accumulated in inoculated plants or in transfected protoplasts (Fig. 4C). In addition, the expression of p33 and p19 proteins in plants infected with wt and hybrid constructs (101/2 and 109/1) was also similar (Fig. 4B). This evidence demonstrates that the wt level of p19 protein is not the only requirement for eliciting systemic necrosis in tombusvirus-infected plants. Interestingly, a marked decrease in the viral RNA level was observed in the newly developed leaves of plants infected with 101/2, 109/1, and Δ19. Figure 5 shows the accumulation of viral RNA in 101/2-infected plants, which was very similar to that detected in plants infected with 109/1 and Δ19 (not shown). It is important to note that no defective interfering (DI) RNA accumulation was detected, which could interfere with viral symptoms (26). This observation may suggest the activation of a plant defense mechanism (e.g., PTGS), which may inhibit the accumulation of viral RNA in the newly developed leaves.
FIG. 4.
Accumulation of viral RNAs and proteins in N. benthamiana plants and protoplasts inoculated with CymRSV/CIRV chimeras. (A) Symptoms of N. benthamiana inoculated with the viruses indicated. The photos were taken 4 wpi. (B) Western blot analysis of the accumulation of p19 (upper panel) and p33 (lower panel) in virus-infected plants. (C) Northern analysis of viral RNAs extracted from infected N. benthamiana plants (lower panel) and protoplasts (upper panel). Samples for protein and RNA extractions were taken from upper leaves showing the first appearing systemic symptoms (typically 7 to 10 dpi) and from protoplasts harvested 24 hpi.
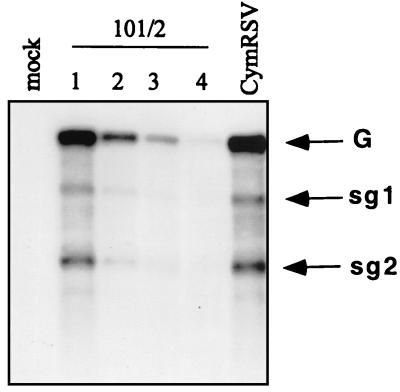
FIG. 5.
Relative level of virus-specific RNAs in 101/2- and CymRSV-infected N. benthamiana plants. Numbers above the lanes indicate the sequence of leaves developed after the first systemic symptoms in plants inoculated with 101/2. Note that the wt CymRSV-infected plants necrotized shortly after the first systemic symptoms appeared (7 to 10 dpi), and RNA samples were available only from the first apical leaves showing systemic symptoms.
Restoration of wt symptoms by 101/2 in transgenic plants expressing p33 and p92 of CymRSV.
The 101/2 construct was used to infect transgenic N. benthamiana plants (pol I) expressing biologically active p33 and p92 of CymRSV (17). Seven of 10 pol I plants inoculated with the 101/2 transcripts became infected and showed wt-like necrotic symptoms within 4 weeks postinoculation (wpi) in contrast to inoculated nontransgenic plants, all of which showed the typical attenuated symptoms caused by 101/2 (Fig. 6). It is worth noting that there were slight differences in the development of necrotic symptoms of wt virus-infected nontransgenic plants and pol I plants infected with 101/2 transcripts. The appearance of necrosis caused by the wt virus started on the small apical leaves after 7 to 10 dpi and culminated in the death of the plants, In the case of 101/2 inoculated pol I plants, partial necrosis appeared first on the petioles and the stem (15 to 20 dpi), extended slowly to the other parts of the plant, and was complete in 4 weeks. The Northern blot analysis of RNA extracts made from 101/2-inoculated pol I transgenic plants and from nontransgenic plants demonstrated that no significant alteration in the accumulation of viral RNA at 8 dpi was observed (not shown). These results suggest that CymRSV p33 (or/and p92) provided in trans is capable of restoring necrotic symptoms in the presence of the 101/2 hybrid virus. Sequence analysis of reverse transcription-PCR products made from the ORF1 region of 101/2 RNA extracted from infected pol I plants showed that no RNA recombination occurred between the transgene and the replicating challenge virus (not shown). Backinoculation of plant sap derived from pol I plants infected with 101/2 into nontransgenic plants resulted in attenuated symptoms, further supporting the conclusion that recombination had not occurred between 101/2 and the RNA of the transgene. For comparison, pol I transgenic plants were inoculated with other chimeras (109/1, 74/1 and 75/2), which were not able to elicit necrosis on nontransgenic plants. Pol I plants infected with 74/1 developed necrosis similarly to 101/2 infection. In contrast, 109/1 and 75/2 caused the same attenuated symptoms on pol I plants as on nontransgenic plants (not shown). These results underline the importance of compatibility between ORF1 and p19 suggested by the partially necrotic phenotype of genomes such as 116/1, 88/5, 116/6, 79/9, and 97/5.
FIG. 6.
Symptoms of 101/2-infected N. benthamiana nontransgenic (A), CymRSV p33- and p92-expressing transgenic plants inoculated with 101/2 (B), and mock-inoculated transgenic (C) plants. The photos were taken 5 wpi.
Expression of p33 and p19 proteins of CymRSV by PVX vector.
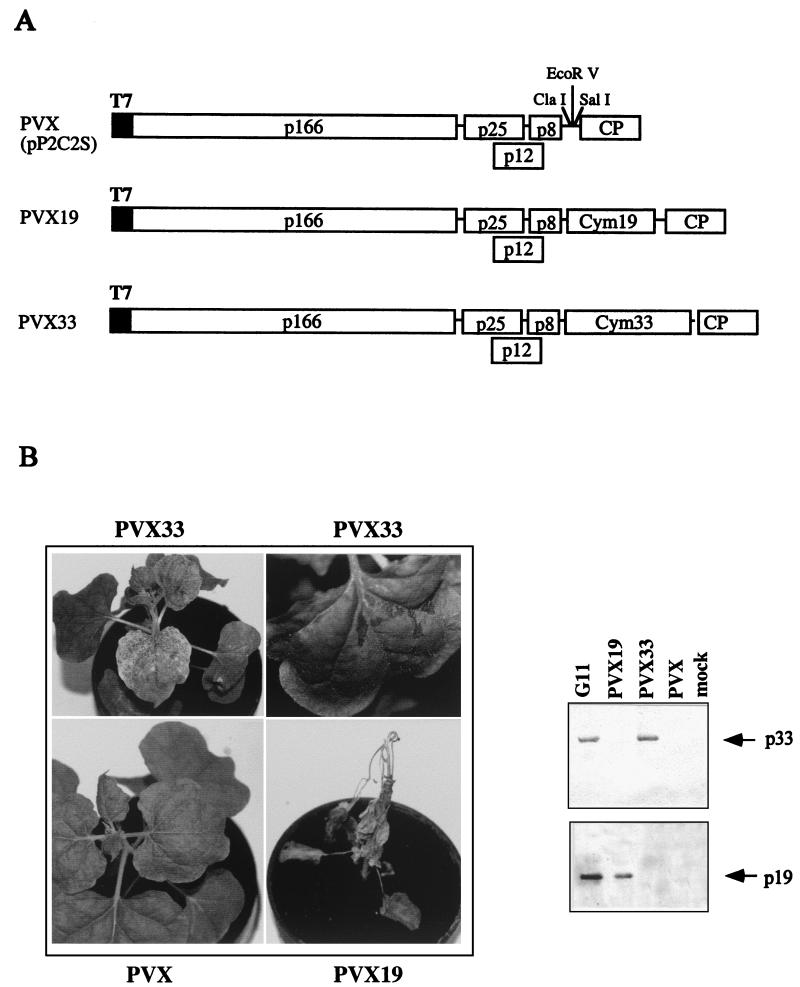
In order to complement the results obtained by gene exchange, the viral p33 and p19 proteins were individually expressed using a PVX vector, and the developing symptoms were monitored for up to 6 wpi. N. bethamiana plants were inoculated with PVX33 and PVX19 constructs, carrying the coding regions of p33 and p19, respectively (Fig. 7A). The presence of the appropriate CymRSV proteins in the upper noninoculated leaves of plants inoculated with PVX, PVX33, or PVX19 was verified by Western blot analysis (Fig. 7B). The secondary veins of systemically infected leaves of PVX33-inoculated plants became white and occasionally necrotized (Fig. 7B). These symptoms can be easily distinguished from the mild mosaic caused by PVX, suggesting an important role for the p33 protein in the viral symptoms. The majority (18 of 20) of the PVX19-infected plants showed systemic necrosis, which culminated in the death of the plants (Fig. 7B). Occasionally (2 of 20), PVX p19-inoculated plants failed to show generalized necrosis; instead, they showed mosaic and necrotic local lesions on the upper noninoculated leaves (not shown). These plants were not analyzed further.
FIG. 7.
(A) Diagram of PVX genome and derivatives expressing CymRSV p33 and p19 proteins. Boxes indicate ORFs, and the molecular mass of the encoded proteins is shown. (B) Symptoms of N. benthamiana plants and detection of CymRSV p33 and p19 proteins expressed by PVX vectors.
DISCUSSION
Hybrid tombusviruses carrying chimeric ORF1 are not able to induce wt symptoms.
We reported here that hybrid tombusviruses carrying chimera's of ORF1 derived from CymRSV and CIRV were not able to induce generalized necrosis on N. benthamiana compared to that caused by both parents. This was surprising, since both hybrids 101/2 and 109/1 were able to replicate and spread in the infected N. benthamiana plants at the wt level. We also showed that the replacement of another part of the genome between the two viruses (including the terminal noncoding regions) did not modify the induced symptoms significantly. Since the protein products of ORF1 and ORF2 are essential in virus replication (11, 17, 19, 20), it would not be surprising if the replication machinery of the ORF1 chimeras was also altered. We were particularly interested in the level of sg2, since it is well known from other reports (11, 23, 32) that p19 translated from sg2 is a pathogenicity determinant and plays a key role in causing severe symptoms in tombusvirus-infected plants. Moreover, it was suggested that the abundant expression of p19 protein of the closely related tomato bushy stunt virus (TBSV) was solely responsible for severe systemic necrosis (9, 30). However, we showed that the lack of systemic necrosis in plants infected with 101/2 and 109/1 was not due to an altered transcription level of genomic and both subgenomic RNAs, respectively. The efficient cell-to-cell movement of these hybrids in infected N. benthamiana plants was a further indication that there was no alteration in the level of sg2 RNA transcription because p22, responsible for the cell-to-cell movement, is translated from the same sg2 RNA as p19. Since the nucleotide sequences of the sg RNAs of 101/2 and 109/1 mutants were exactly the same as that of the wt viruses, the translational efficiency would be expected to be the same. In fact, Western blot analysis of plants infected with 101/2 and 109/1 showed a level of p19 accumulation similar to that in plants infected with wt viruses. These results were in contrast to the suggestion that p19 is the sole elicitor of the lethal necrotic symptom, and it is more likely that other viral factors are also involved in systemic lethal necrosis. Our results strongly suggest that the protein products of ORF1 (and/or the N terminus of ORF2, which is the readthrough product of ORF1) have an essential role in inducing the severe symptoms caused by tombusviruses. We showed that the chimeric protein products of ORF1 in 101/2 and 109/1 are replication competent, but these proteins are not able to induce lethal necrosis.
p33 protein plays an important role in the necrotic phenotype of CymRSV-infected plants.
The infection of transgenic plants expressing biologically active CymRSV p33 and p92 with 101/2 and 74/1 hybrids showed that wt systemic necrosis can be restored with p33 (and/or p92) provided in trans. Our results do not rule out a role for p92 in the elicitation of necrosis, but the expression of p33 by the PVX viral vector in N. benthamiana caused severe chlorosis and occasionally necrosis, indicating the importance of p33 in wt symptoms of tombusviruses. The relatively low level of p33 accumulation in transgenic plants could explain why pol I plants are asymptomatic even though they express p33 (17). Alternatively, other viral protein such as p19 are also involved in symptom development. Our evidence confirmed that the necrotic phenotype requires the presence of wt p33. We do not yet know wether there is any direct or indirect interaction between these proteins. However, the chimeras carrying the p33 and p19 genes from different parent viruses (116/1, 88/5, 116/6, 79/9, and 97/5) can necrotize only a part of the inoculated plants. Furthermore, the pol I plants expressing CymRSV p33 can only complement 101/2 and 74/1 genomes carrying CymRSV-derived p19 protein. These results indicate that the p33 and p19 proteins of different tombusviruses are not fully compatible and that their compatibility plays an important role in symptom development. These results coincide with the recent observation that the ability of heterologous DI RNA to protect virus-infected plants against systemic necrosis is determined by the 5′-proximal region (including the 5′ UTR and the coding region of ORF1) of the helper virus genome (14). In addition, it was suggested that DI RNA-mediated protection operates via a specific interference with viral products, perhaps preventing the interaction between p33 and p19 and thus the induction of necrotic symptoms. It was shown by biochemical analysis that the ORF1 protein products of both CymRSV (p33) and CIRV (p36) are integral membrane proteins (24), which are anchored to the membrane of modified peroxisomes or mitochondria. The expression of p36 in yeast cells resulted in membrane proliferation (25). The lack of lethal necrosis in 101/2 hybrid-infected plants could be the outcome of different compartmentalization of viral proteins derived from different parents. It was shown that 101/2 contains a signal which directs the virus replicase (including p33 and p92) to mitochondria and induces the formation of MVBs, where the virus probably replicates (6, 24). However, CymRSV, which represents 80% of the 101/2 genome, normally induces MVBs exclusively in peroxisomes (6). Therefore, it is possible that the viral factors that are required for the elicitation of wt symptoms accumulate in a different cell compartment, resulting in attenuated symptoms.
Role of p19 protein in the lethal necrosis caused by CymRSV.
The expression of pl9 of CymRSV in the PVX19-infected N. benthamiana plant resulted in lethal necrosis in most of the inoculated plants, in accordance with the previous observation on PVX expression of TBSV p19 (30). However, wt levels of p19 were not able to induce lethal necrosis in 101/2- and 109/1-infected plants. In addition, it was demonstrated recently that p19 of TBSV is a suppressor of PTGS (34). Therefore, p19 acts as virus-induced PTGS suppressor, and the severe symptoms caused by PVX19 are likely caused by the suppressing action of p19. Similar to this observation, it was shown recently that the HCPro protein of potato virus Y, which was one of the first-described suppressors of PTGS, is responsible for the severe symptoms caused by the PVX-HcPro construct (5). However, HCPro itself did not elicit necrosis, because transgenic plants expressing HCPro of tobacco etch virus are asymptomatic (22). A possible explanation for the necrotic symptoms induced by PVX19 is that p19, depending on the viral genetic background, can function as either a virulence (in tombusviruses) or an avirulence (in PVX19) determinant. Similar observation for the 2b PTGS suppressor protein of tomato aspermy virus has been described. It is a hypersensitive response elicitor (an avirulence determinant) when expressed by heterologous viruses (tobacco mosaic virus or PVX), but not when expressed by tomato aspermy virus itself (18).
The combined evidence strongly suggests that the systemic lethal necrosis of tombusviruses is induced by viral products, including p33, and the contribution of p19 is indirect (but essential), suppressing plant defense mechanisms. However, the precise role of p33 replication protein in viral symptoms remains to be determined. The observation that p33 has a crucial role in induced wt virus symptoms is not unique among plant viruses. It has been shown that the replication proteins of other viruses are also involved in the elicitation of viral symptoms (21; 16 and references therein;)).
ACKNOWLEDGMENTS
We thank David Baulcombe for generously supplying the PVX vector and Marcello Russo for providing a preprint of his article.
This research was supported by grants from the Hungarian OTKA (31929) and the Ministry of Education (FKFP0442/1999).
REFERENCES
- 1.Aranda M A, Maule A. Virus-induced host gene shutoff in animals and plants. Virology. 1998;243:261–267. doi: 10.1006/viro.1998.9032. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe D C, Chapman S, Santa Cruz S. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 1995;7:1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- 3.Berzal-Herranz A, de la Cruz A, Tenllado F, Diaz-Ruiz J R, Lopez L, Sanz A L, Vaquero C, Serra M T, Garcia-Luque L. The caosicum L3 gene-mediated resistance against the tobamoviruses is elicited by the coat protein. Virology. 1995;209:498–505. doi: 10.1006/viro.1995.1282. [DOI] [PubMed] [Google Scholar]
- 4.Bleve-Zacheo T, Rubino L, Melillo M T, Russo M. The 33K protein encoded by cymbidium ringspot tombusvirus localizes to modified peroxisomes of infected cells and uninfected transgenic plants. J Plant Pathol. 1997;79:197–202. [Google Scholar]
- 5.Brigneti G, Voinnet O, Li W X, Ji L H, Ding S W, Baulcombe D. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Burgyán J, Rubino L, Russo M. The 5′ terminal region of tombusvirus genome determines the origin of multivesicular bodies. J Gen Virol. 1996;77:1967–1974. doi: 10.1099/0022-1317-77-8-1967. [DOI] [PubMed] [Google Scholar]
- 7.Chapman S, Kavanagh T A, Baulcombe D C. Potato virus X as a vector for gene expression in plants. Plant J. 1992;2:549–557. doi: 10.1046/j.1365-313x.1992.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- 8.Chu M, Park J-W, Scholthof H B. Separate regions of tomato bushy stunt virus p22 protein mediate cell-to-cell movement versus elicitation of effective resistance responses. Mol Plant-Microbe Interact. 1999;12:285–292. [Google Scholar]
- 9.Chu M, Desvoyes B, Turina M, Noad R, Scholthof H B. Genetic dissection of tomato bushy stunt virus p19-protein-mediated host-dependent symptom induction and symptome invasion. Virology. 2000;266:79–87. doi: 10.1006/viro.1999.0071. [DOI] [PubMed] [Google Scholar]
- 10.Culver J N, Dawson W O. Tobacco mosaic virus elicitor coat protein genes produce a hypersensitive phenotype in transgenic Nicotiana sylvestris plant. Mol Plant Microbe Interact. 1991;4:458–463. [Google Scholar]
- 11.Dalmay T, Rubino L, Burgyán J, Kollár Á, Russo M. Functional analysis of cymbidium ringspot virus genome. Virology. 1993;194:697–704. doi: 10.1006/viro.1993.1310. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez I, Candresse T, le Gall O, Dunez J. The 5′ noncoding region of grapevine chrome mosaic Nepovirus RNA-2 triggers a necrotic response on three Nicotiana spp. Mol Plant Microbe Interact. 1999;12:337–344. doi: 10.1094/MPMI.1999.12.4.337. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton A J, Baulcombe D C. A novel species of small antisense RNA in post-transcriptional gene silencing. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 14.Havelda Z, Szittya Gy, Burgyán J. Characterization of the molecular mechanism of defective interfering RNA-mediated symptom attenuation in tombusvirus-infected plants. J Virol. 1998;72:6251–6256. doi: 10.1128/jvi.72.7.6251-6256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jupin I, Guilley H, Richards K E, Jonard G. Two proteins encoded by beet necrotic yellow vein virus influence symptome phenotype on leaves. EMBO J. 1992;11:479–488. doi: 10.1002/j.1460-2075.1992.tb05078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C-H, Palukaitis P. The plant defense response to cucumber mosai virus in cowpea is elicited by the viral polymerse gene and affects virus accumulation in single cells. EMBO J. 1997;16:4060–4068. doi: 10.1093/emboj/16.13.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kollár A, Burgyán J. Evidence that ORF 1 and 2 are the only virus encoded replicase gene of cymbidium ringspot tombusvirus. Virology. 1994;201:169–172. doi: 10.1006/viro.1994.1280. [DOI] [PubMed] [Google Scholar]
- 18.Li H-W, Lucy A P, Guo H-S, Li W-X, Ji L-H, Wong S-M, Ding S-W. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 1999;18:2683–2691. doi: 10.1093/emboj/18.10.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molinari P, Marusic C, Lucioli A, Tavazza R, Tavazza M. Identification of artichoke mottled crinkle virus (AMCV) proteins required for virus replication: complementation of AMCV p33 and p92 replication-defective mutants. J Gen Virol. 1998;79:639–647. doi: 10.1099/0022-1317-79-3-639. [DOI] [PubMed] [Google Scholar]
- 20.Oster S K, Wu B, White K A. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J Virol. 1998;72:584–551. doi: 10.1128/jvi.72.7.5845-5851.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padgett H S, Beachy R N. Analysis of a tobacco mosaic virus strain capable of overcoming N gene-mediated resistance. Plant Cell. 1993;5:577–586. doi: 10.1105/tpc.5.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruss G, Ge X, Shi X M, Carrington J C, Vance A B. Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell. 1997;6:859–868. doi: 10.1105/tpc.9.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rochon D M, Johnston J C. Infectious transcripts from cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic mRNA. Virology. 1991;181:656–665. doi: 10.1016/0042-6822(91)90899-m. [DOI] [PubMed] [Google Scholar]
- 24.Rubino L, Russo M. Membrane targeting sequences in tombusvirus infections. Virology. 1998;252:431–437. doi: 10.1006/viro.1998.9490. [DOI] [PubMed] [Google Scholar]
- 25.Rubino L, Di Franco A, Russo M. Expression of a plant virus non-structural protein in Saccharomyces cerevisiae causes membrane proliferation and altered mitochondrial morphology. J Gen Virol. 2000;81:279–286. doi: 10.1099/0022-1317-81-1-279. [DOI] [PubMed] [Google Scholar]
- 26.Russo M, Burgyán J, Martelli P G. The molecular biology of tombusviridae. Adv Virus Res. 1994;44:382–424. doi: 10.1016/s0065-3527(08)60334-6. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Scholthof H B, Desvoyes B, Kuecker J, Whitehead E. Biological activity of two Tombusvirus proteins translated from nested genes is influenced by dosage control via context-dependent leaky scanning. Mol Plant-Microbe Interact. 1999;12:670–679. [Google Scholar]
- 29.Scholthof H B, Morris T J, Jackson A O. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol Plant-Microbe Interact. 1993;6:309–322. [Google Scholar]
- 30.Scholthof H B, Scholthof K-B G, Jackson A O. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell. 1995;7:1157–1172. doi: 10.1105/tpc.7.8.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholthof K-B G, Scholthof H B, Jackson A O. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology. 1995;208:365–369. doi: 10.1006/viro.1995.1162. [DOI] [PubMed] [Google Scholar]
- 32.Scholthof K-B G, Scholthof H B, Jackson A O. The effect of defective interfering RNAs on the accumulation of tomato bushy stunt virus proteins and implications for disease attenuation. Virology. 1995;211:324–328. doi: 10.1006/viro.1995.1410. [DOI] [PubMed] [Google Scholar]
- 33.Taraporewala Z F, Culver J N. Identification of an elicitor active site within the three-dimensional structure of the tobacco mosaic tobamovirus coat protein. Plant Cell. 1996;8:169–178. doi: 10.1105/tpc.8.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voinnet O, Pinto Y, Baulcombe D C. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White J L, Kaper J M. A simple method for detection of viral satellite RNAs in small tissue samples. J Virol Methods. 1989;23:83–94. doi: 10.1016/0166-0934(89)90122-5. [DOI] [PubMed] [Google Scholar]