Abstract
We have developed a rapid-turnover culture system where the life span of a human immunodeficiency virus type 1-infected cell is controlled by periodic addition of a cytotoxic agent, mitomycin C. These mitomycin C-exposed cells are cocultured with a constant number of uninfected cells as new targets for the virus. Passage of the virus-infected cells under these conditions led to the emergence of a viral variant that was able to replicate efficiently in this culture system. After biologic and molecular cloning, we were able to identify a single frameshift mutation in the vpu open reading frame that was sufficient for growth of the mutant virus in the rapid-turnover assay. This virus variant spread more efficiently by cell-to-cell transfer than the parental virus did. Electron micrographs of cells infected with the Δvpu virus revealed a large number of mature viral capsids attached to the plasma membrane. The presence of these mature virus particles on the cell surface led to enhanced fusion and formation of giant syncytia with uninfected cells. Enhanced cell-to-cell transfer of the Δvpu virus provides an explanation for the survival of this mutant virus in the rapid-turnover culture system. The in vitro rapid-turnover culture system is a good representation of the in vivo turnover kinetics of infected cells and their continual replacement by host lymphopoietic mechanisms.
Human immunodeficiency virus type 1 (HIV-1) replication is continuous and occurs vigorously in infected individuals (4, 17, 28, 42). Primary acute HIV-1 infection is characterized by extremely high levels of plasma viremia, with values in excess of 106 copies of viral RNA/ml of blood (7). Resolution of the acute phase of HIV-1 infection correlates well with the appearance of robust cytotoxic T-cell responses to the virus (18, 21, 24, 34) and is followed by a variable period of clinical latency. The viral titer rapidly decreases to a new steady state that varies among individuals (the plasma HIV-1 RNA levels are typically in the range of 102 to 105 copies/ml) and is ultimately predictive of the subsequent rate of disease progression (23). Although the asymptomatic phase of infection is characterized by an absence of clinical symptoms, there is persistent replication of virus throughout the lymphoid system, especially in the germinal centers of peripheral lymph nodes (6, 25, 29). Remarkably, during this phase, the level of HIV-1 RNA in the plasma is reasonably stable in a given individual and reflects a quasi-steady state in which virus production equals virus clearance (4, 7).
Most of the plasma virus detected comes from recently infected CD4+ lymphocytes. Some studies have estimated that as many as one-third of peripheral and lymphoid CD4+ lymphocytes are HIV DNA positive, with a small proportion of them (0.1 to 1%) expressing viral RNA at any given time (3, 6, 25). These virus-infected T cells in vivo are turned over rapidly and have a short half-life (t1/2 ≈ 2 days) (1, 17, 28, 31, 32, 42). The rapid turnover of these infected CD4+ lymphoblasts is probably due to both virus-induced cytopathic effects and the host cytolytic effector mechanisms. Current in vitro assays for viral replication do not accurately represent this situation because they do not take into account the short half-life of infected cells in vivo. Therefore, in vitro culture systems used to analyze HIV-1 gene function do not have the same selective constraints as those present in vivo.
In this study, we have designed an in vitro assay system that mimics the short life span of infected T cells and the constant replenishment of uninfected target cells (the rapid-turnover assay). HIV-1-infected Jurkat T cells were killed every 3 days by the addition of a cytocidal agent and then cocultured with fresh uninfected Jurkat T cells. Sustained high level of viral replication was not achieved under rapid-turnover assay conditions following infection with HIVLai. However, continued propagation of the virus-infected cells led to the emergence of a new viral variant that could replicate under the selective conditions of rapid cell turnover. Spread of virus under the rapid-turnover conditions was correlated with a change in phenotype of the virus (increased numbers and sizes of syncytia). The virus was molecularly cloned, and the region responsible for the replication in the rapid-turnover assay was mapped by analysis of viral chimeras. The region of the virus that conferred this new phenotype mapped to a frameshift mutation in the vpu open reading frame (ORF) that was shown to be sufficient for survival and growth in the rapid-turnover assay. Moreover, the vpu mutation alone was responsible for converting the virus to one that spreads predominately by cell-to-cell fusion. Since viral replication in a system with rapid cell turnover kinetics depends on cell-to-cell transfer of virus, our data support the hypothesis that cell-to-cell spread of HIV is the predominant route of viral spread in vivo.
MATERIALS AND METHODS
Rapid-turnover assay.
Plasmid pLai is an infectious molecular clone of the T-cell-tropic isolate, Lai (26). Jurkat T cells were infected with HIVLai, and infected cell cultures were exposed to mitomycin C (50 μg/ml) 3 days postinfection and every third day after the initial mitomycin C exposure (see Fig. 1A). For mitomycin C treatment, cells were washed twice with phosphate-buffered saline (PBS) and then resuspended for 2 h at room temperature in the dark in PBS containing mitomycin C (50 μg/ml). Following incubation, the cells were washed twice with RPMI medium containing 10% fetal bovine serum (RPMI-FBS) and resuspended in 1 ml of medium containing fresh Jurkat T cells (106). After the next passage, dead cells were removed from the culture by Ficoll density gradient separation. This procedure was repeated every 3 days. Replication of virus was monitored by quantitation of p24 release into the supernatant.
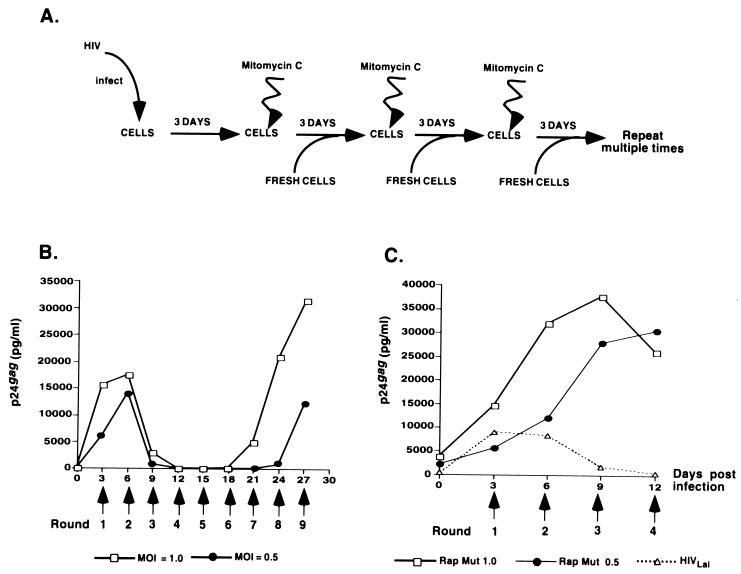
FIG. 1.
Establishment of a rapid-turnover assay system. (A) HIV-1Lai-infected Jurkat cells, 3 days postinfection, were treated with mitomycin C (50 μg/ml) for 120 min in the dark at room temperature and then cocultured with 106 uninfected Jurkat cells. Treatment with mitomycin C and coculture with uninfected cells were repeated every third day for several passages. (B) Jurkat cells were infected with HIVLai at an MOI of 1 or 0.5, and viral replication was monitored by measuring the amount of p24gag released into the supernatant at periodic intervals. The periodicity and number of exposures to mitomycin C are indicated below the graph. Note that by passage 3 (day 9 postinfection), Lai replication is suppressed to negligible levels. Infections were allowed to proceed until the appearance of a new actively replicating isolate. Viral supernatants from passage 9 (day 27 of culture), designated Rap Mut 1.0 or Rap Mut 0.5, were collected and used for characterization of the mutant and subsequent infections. (C) Jurkat cells were infected with viral supernatants from day 27 cultures from panel B (Rap Mut 1, Rap Mut 0.5), or with HIVLai (used as a control). The infected cultures were subjected to rapid-turnover assay conditions, as described above. Viral replication was monitored by measuring the p24gag content of the cell-free supernatants by ELISA.
Molecular cloning of rapid HIV-1 variants.
On day 27 postinfection (passage 9) (see Fig. 1A), cell-free supernatant was used for infection of fresh Jurkat T cells, and the rapid-turnover assay was performed every third day (Fig. 1B), as described above. At 12 days postinfection, infected cells were collected and extrachromosomal DNA was isolated by Hirt extraction and used for cloning of the proviruses. Briefly, 200 ng of DNA was amplified by PCR using the Expand Long Template kit (Boehringer-Mannheim) and the primer set 5′-AAATCTCTAGCAGTGGCGCCCGAACAG-3′ (sense) (+623 to +649) and 5′-GCACTCAAGGCAAGCTTTATTGAGGCT-3′ (antisense) (+9632 to +9606). The template was denatured for 5 min at 95°C, and this was followed by three cycles of denaturation (95°C for 10 s), annealing (55°C for 30 s), and extension (68°C for 10 min) and then by an additional 22 cycles in which only the extension time was changed (to 8 min). The resulting 9-kb product was subcloned into pGEM-T (Promega) by using T/A overhangs. Full-length infectious proviral clones were generated by digestion of the pGEM-T proviral clones with BssHII and AatII (position 714 in the provirus to the polylinker in the vector backbone). The BssHII-AatII fragment was subcloned into pLai that had been digested with the same restriction enzymes. The resulting full-length proviral clone contained the amplified region of the mutant virus ligated to the 5′ long terminal repeat (LTR) of Lai and was named pRap1 (to designate the rapid phenotype). To map the genetic component of the mutant virus that conferred the ability to grow within the parameters of the rapid-turnover assay, Lai/Rap1 chimeras were constructed. Initially, a 2,691-bp SalI-BamHI fragment (positions 5789 to 8480) of pRap1 was cloned into the corresponding sites of pLai to create plasmid pRap2. The reciprocal swap was also created by cloning the SalI-BamHI fragment (positions 5789 to 8480) of pLai into the similarly digested pRap1 to create pRap3. Also, a 584-bp BglII fragment (positions 7041 to 7628) carrying the env V3 of pRap1 was cloned into the similarly digested pLai to create plasmid pRap4. Finally, a 1,283-bp NdeI fragment (positions 5125 to 6408) of plasmid pRap1 was cloned into the corresponding sites of pLai to create plasmid pRap5. Nucleotide positions are numbered according to the HXB2R clone sequence (20). All viral constructs and the presence of mutations were confirmed by sequencing with an automated sequencer (ABI Prism).
Cells, transfection, and infection.
Jurkat T cells were cultured in RPMI-FBS. Jurkat-LTR-luc cells, which contain a luciferase gene under the control of the HIV-1 LTR (14), were maintained in RPMI-FBS containing 0.2 mg of hygromycin per ml. 293-T cells were propagated in Dulbecco's modified Eagle's medium containing 10% FBS. Virus stocks were prepared by calcium phosphate transfections of 293-T cells with plasmid DNAs of individual molecular clones, as described previously (14). At 2 days posttransfection, virus-containing supernatants were clarified by centrifugation (1,000 × g for 10 min) to remove cell debris. The virus stocks were assayed for their infectivity by the MAGI assay (41). Routinely, 106 Jurkat cells were infected at an equal multiplicity of infection (MOI) with different viruses. Following 2 h of adsorption, the cells were washed twice with PBS to remove residual input virus and then cultured at a density of 106/ml of RPMI-FBS. Spreading infections were maintained by replacing 75% of the cells and medium every third day. Cell supernatants were harvested, and the infectivity of cultures was monitored by a p24gag enzyme-linked immunosorbent assay ELISA (Coulter), as described previously (14).
Flow cytometry.
To determine the number of cells expressing p24gag in each of the infected cultures, cells were fixed in 1% paraformaldehyde, permeabilized in 0.5% Tween 20, and stained with a 1:50 dilution of murine anti-HIV-1 p24gag-fluorescein isothiocyanate (FITC) monoclonal antibody (KC57-FITC; Coulter). Flow cytometry was performed on a CALIBUR instrument (Becton Dickinson) with CellQuest (Becton Dickinson) data acquisition and analysis software.
Metabolic labeling.
For pulse-chase experiments, Jurkat cells were infected with either HIVLai or Rap5 virus. On day 3 postinfection, the cultures were washed once with PBS and incubated for 60 min at 37°C under 5% CO2 in methionine-deficient RPMI 1640 medium (Sigma). The cells were pulse-labeled with [35S]methionine-[35S]cysteine mix (New England Nuclear) for 60 min at 37°C under 5% CO2 and washed once in PBS, and equal portions were added to 500 μl of RPMI-FBS for each time point of the chase period and incubated at 37°C. At the indicated time points, cells were harvested and lysed in 500 μl of CHAPS buffer, containing 50 mM Tris-hydrochloride (pH 8.0), 5 mM EDTA, 100 mM NaCl, 0.5% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfate) (CHAPS), 0.2% (wt/vol) deoxycholate, and the protease inhibitors leupeptin, aprotinin, and phenylmethylsulfonyl fluoride (35). Cell debris was removed by centrifugation (12,000 × g, for 10 min at 4°C). For studies of virus particle release, cell supernatants were cleared of cell debris by centrifugation (1,000 × g, for 5 min). Virus particles were pelleted from the cell supernatants in a refrigerated microcentrifuge (14,000 × g for 100 min at 4°C) and then lysed in buffer containing 300 mM NaCl, 50 mM Tris-hydrochloride (pH 7.4), and 0.1% (vol/vol) Triton X-100 (35).
For steady-state analysis of envelope glycoprotein expression, Jurkat cells infected with HIVLai or Rap5 virus (72 h postinfection) were labeled with [35S]methionine-[35S]cysteine mix for 4 h. Cells and virus lysates were precleared by incubation with protein A-Sepharose preadsorbed with normal mouse serum for 30 min at 4°C. Viral proteins from the clarified lysates were immunoprecipitated with monoclonal antibodies to HIV-1 gp120 (2G12; AIDS Repository) (40) and HIV-1 gag polyprotein (76C; AIDS Repository) (37) preadsorbed to protein A-Sepharose (for 60 min at 4°C with rotation). Immunoprecipitated proteins were solubilized by boiling in sample buffer and separated on a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel. The gels were fixed for 20 min by incubation in 40% methanol–10% acetic acid, soaked in Amplify (Amersham Life Sciences) for 20 min, dried, and exposed to X-ray film (Kodak X-Omat AR). The densities of the visualized bands were quantitated by PhosphorImager analysis.
Electron microscopy.
Jurkat cells were infected at an MOI of 0.03 and fixed at 3 days postinfection. For fixation, the cells were washed three times with cold PBS and then fixed overnight in 4% paraformaldehyde–0.1% glutaraldehyde in 200 mM HEPES-KOH (pH 7.4) at 4°C. The cells were then washed and resuspended in 2% osmium tetroxide solution for 2 h on ice. The samples were dehydrated, embedded in Epon 812 resin, and sectioned.
Biotinylation and immunoprecipitation.
Sulfosuccinimidyl-6-biotinamidohexanoate (Sulfo-NHS-LC-biotin) (Pierce) was dissolved in dimethyl sulfoxide at 10 mg/ml. On day 3 postinfection, infected Jurkat cell cultures were washed three times with ice-cold PBS prior to biotinylation of cell surface proteins. Sulfo-NHS-LC-biotin was added to the cells in a volume of 1 ml PBS at a final concentration of 400 μg/ml for 60 min at room temperature. After biotinylation, the cells were washed three times with ice-cold PBS and then lysed in 500 μl of CHAPS lysis buffer (described above). Cell lysates containing equal numbers of p24gag+ cells were centrifuged at 12,000 × g for 10 min at 4°C to remove cell debris and then precleared by incubation at 4°C for 1 h with protein A-Sepharose beads (Pharmacia LKB Biotechnology) adsorbed with normal mouse serum. Viral envelope glycoproteins and a cell surface-associated receptor protein (Fas) were immunoprecipitated with a mouse anti-HIV-1 gp120 monoclonal antibody (2G12) and a mouse anti-Fas receptor monoclonal antibody (Santa Cruz) preadsorbed to protein A-Sepharose for 1 h at 4°C. The immunoprecipitated proteins were solubilized by boiling in sample buffer, separated on SDS–8% polyacrylamide gels, transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore), probed with horseradish peroxidase-conjugated streptavidin, and detected by enhanced chemiluminescence (ECL; Amersham Life Sciences).
Western blot analysis.
Jurkat cells (106) were infected with virus (Lai or Rap5) at an MOI of 0.03, as described above. On day 3 postinfection, the cells were washed with PBS and an aliquot of cells was used for fluorescence-activated cell sorter (FACS) analysis to determine the number of p24gag+ cells in culture, as described above. The rest of the cells were lysed in 200 μl of CHAPS lysis buffer. The lysates were centrifuged at 12,000 × g for 10 min to remove cell debris and boiled for 5 min in the presence of sample buffer (2% SDS, 1% 2-mercaptomethanol, 1% glycerol, 65 mM Tris-hydrochloride [pH 6.8]). Lysates from equal numbers of p24gag+ cells were loaded on SDS–8% polyacrylamide gels. Following electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes. The membranes were blocked for 60 min at room temperature with PBS containing 0.5% Tween 20 and 5% nonfat milk powder (Carnation) and incubated with a 1:1,000 dilution of a mouse anti-HIV-1 p55gag monoclonal antibody (76C) overnight at 4°C. The membranes were washed for 30 min in wash buffer (PBS containing 0.2% Tween 20) and then incubated with a 1:5,000 dilution of a horseradish peroxidase-conjugated anti-mouse monoclonal antibody (Jackson) for 60 min at room temperature. The membranes were washed three times for 30 min, and the bound antibody was detected with the ECL detection system.
Cell-to-cell fusion assay.
Jurkat cells that were mock infected, or infected with Lai or Rap5, were used as the source of viral envelope glycoproteins. To initiate cell-to-cell fusion, 1× 105 infected Jurkat cells (Lai or Rap5 infected) were cocultured with 2.5 × 105 Jurkat LTR-luc cells in 96-well plates. To inhibit subsequent rounds of viral replication, zidovudine (50 μM) was added to the cultures. After coincubation for the indicated times, the cells were washed and lysed in reporter lysis buffer and assayed for luciferase activity as specified by the manufacturer (Promega).
RESULTS
Generation of a rapid mutant in the short-half-life assay.
We set out to establish a model system which would allow us to ascertain the functions of accessory genes in cells subject to rapid turnover that would be akin to kinetic conditions in vivo, where infected cells have a short half-life (t1/2 ≈ 2 to 3 days) (4, 30–32). We did this by treating infected cells with mitomycin C, washing them, and then adding fresh uninfected cells (Fig. 1A). Treatment of Jurkat T cells with mitomycin C resulted in >95% cell death within 24 h (data not shown). However, no cells died for the first 8 h after treatment (data not shown), which should allow enough time for the mitomycin C-treated cells to transmit virus to the uninfected cells. This procedure was repeated every 3 days (Fig. 1A). Under these conditions, the life span of an infected cell is unlikely to exceed 3 days.
To our surprise, Jurkat cells infected with the HIVLai isolate (MOI = 1.0 or 0.5) and subjected to rapid-turnover conditions (Fig. 1A) were initially unable to sustain a viral infection. Rather, replication of the initial virus inoculum was nearly completely inhibited by the time of passage 3 (day 9 postinfection [Fig. 1B]). However, following six additional passages, virus emerged which was capable of surviving the rapid-turnover assay (Fig. 1B). To determine if the virus that grew out of the rapid-turnover culture passage (Fig. 1B) was a genetic variant, we compared the growth kinetics of the parental virus (HIVLai) with that of viral supernatants (named Rap Mut 1.0 and Rap Mut 0.5) derived from cells that had been subjected to nine passages in the presence of mitomycin C in the rapid-turnover assay. Indeed, virus that grew out of the first assay was able to replicate in the second rapid-turnover assay, while the replication of the parental virus, HIVLai, was again completely inhibited by day 9 postinfection (passage 3) (Fig. 1C). This indicates that while the parental HIVLai is not able to sustain a spreading infection under conditions where the life span of the infected cells is limited to 3 days, we were able to select for a viral variant that could.
Genotype of the rapid mutant.
To determine the molecular changes in the rapid mutant virus that were necessary for its survival in the rapid-turnover assay, we molecularly cloned the provirus from cells that were infected with Rap Mut 1.0 (viral supernatant containing the biologic clone, as described above) and had been subjected to four passages in the rapid-turnover assay. Extrachromosomal DNA was isolated from the infected culture by Hirt extraction and used as template for cloning the mutant provirus by long-range PCR. The resulting plasmid clone, pRap1, was a full-length viral clone containing the amplified region of the mutant virus (carrying all structural and accessory genes) ligated to the 5′-LTR of pLai (Fig. 2A).
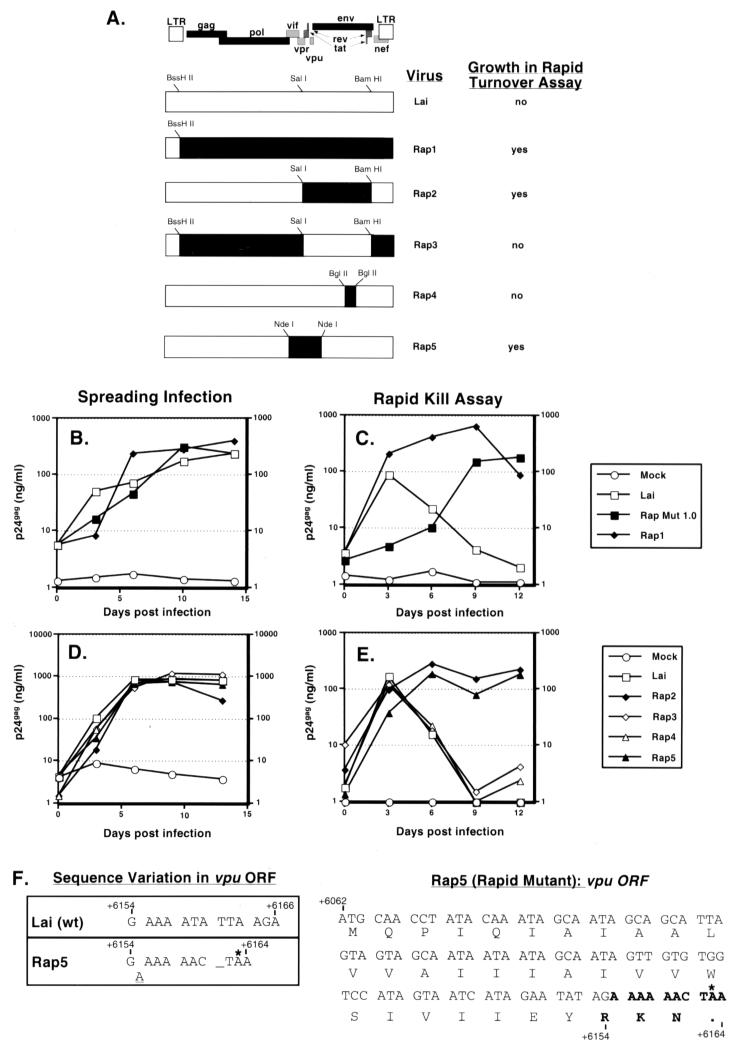
FIG. 2.
Characterization of mutations necessary for survival of the selected virus in the rapid-turnover assay. (A) Full-length HIVs with the indicated genomic composition were derived as described in Materials and Methods. Restriction sites used for cloning are indicated on each recombinant plasmid. Solid regions indicate sequences derived from the rapid mutant, while open boxes indicate sequences derived from Lai. The phenotypes of each of the chimeras in the rapid-turnover assay are indicated. Jurkat cells (106) were infected with the viral chimeras at an MOI of 0.03. (B to E) Cultures were subjected to the rapid-turnover assay protocol (C and E), as described in Materials and Methods, or infections were allowed to progress in the absence of mitomycin C (B and D). Cell-free culture supernatants were assayed for p24gag content by ELISA. Data shown are representative of two independent experiments. (F) The nucleotide and amino acid sequences of the wild-type (wt) and mutant vpu ORFs are shown and numbered according to the HXB2R clone sequence. The insertion of the A nucleotide in the mutant vpu ORF is indicated by a dash, and the termination codon indicated by an asterisk.
Sequencing of the full-length clone revealed multiple changes in the provirus. To determine the genetic components necessary for growth in the rapid-turnover assay, chimeras were constructed where portions of the pRap1 plasmid were exchanged with pLai (Fig. 2A). Infectious viruses were generated by transfection of 293-T cells and were analyzed for replication in the rapid-turnover assay (Fig. 2). All the molecular clones were infectious and were able to replicate to wild-type levels compared to the parental isolate, HIVLai, in spreading infections of Jurkat cells (Fig. 2B). In the rapid-turnover assay, Rap1, which contained the entire coding region of the rapid mutant, was able to replicate, as expected (Fig. 2C). In addition, the Rap2 virus, which contains the 2.69-kb SalI-BamHI fragment from the rapid mutant in the pLai backbone, was able to replicate in the rapid-turnover assay (Fig. 2E). This fragment carries the 3′ end of the vpr gene (nucleotides 5789 to 5850), the first exon of rev, the complete ORFs for tat and vpu, and the first 2,255 nucleotides of the env gene (6225 to 8480, which also includes the Rev response element). Conversely, the Rap3 clone, which contains the same 2.69-kb SalI-BamHI fragment from pLai cloned into the pRap1 backbone, failed to replicate in our rapid-turnover assay (Fig. 2E).
The sequence of the 2.69-kb SalI-BamHI fragment of the rapid mutant contained only two mutations. The first mutation, at nucleotide 6155, causes a frameshift in the vpu ORF, resulting in a premature stop codon, such that only the first 32 amino acids of Vpu are translated (Fig. 2F). The second difference, at nucleotide 7162, was a point mutation (C→A) in the V3 loop of the gp120 coding region, which results in a nonsynonymous amino acid change (proline to glutamate) (data not shown). To investigate the contribution of each of these mutations to the rapid-mutant phenotype, chimeras were constructed in the pLai backbone, such that the proviral clone expressed only one of the two mutations. The pRap4 plasmid contained the V3 region of the rapid mutant cloned into the pLai backbone (Fig. 2A), and the pRap5 plasmid contained sequences from the rapid mutant that included part of the vif ORF, the complete ORF for vpr, vpu, and the first 183 nucleotides of the env gene (nucleotides 5125 to 6408; Fig. 2A). Note that the only difference in sequence between pRap5 and the wild-type pLai plasmid is the frameshift mutation in the vpu ORF. The rapid-turnover assay was again performed with the Rap4 and Rap5 viruses, and the results are shown in Fig. 2E. The virus containing the vpu frameshift mutation (Rap5) was able to replicate in the rapid-turnover assay, while the virus containing the V3 mutation (Rap4) was completely inhibited, similar to the parental HIVLai clone (Fig. 2E). Note that all the viral chimeras are equally infectious in spreading infections (Fig. 2D). In addition, infectious virus molecularly cloned from the Rap Mut 0.5 biologic clone also contained the same frameshift mutation in the vpu ORF (data not shown). These results demonstrate that the vpu frameshift mutation is the only genetic change from the wild-type HIVLai virus that is required for survival in the rapid-turnover assay.
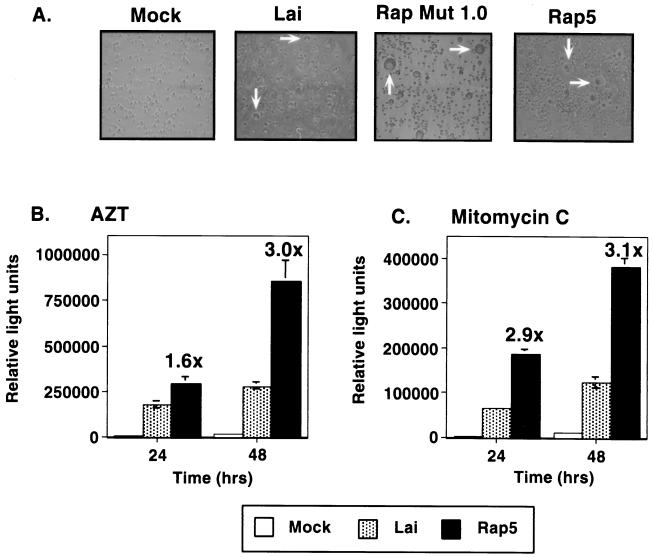
Enhanced fusogenicity observed in Rap5-infected cells.
Infection of Jurkat cells with the uncloned rapid virus mutant stock (Rap Mut 1.0) resulted in enlarged syncytia compared to infections with HIVLai (Fig. 3A). Similar results were also observed in infections with the Rap5 virus (Fig. 3A). Moreover, there were more of these syncytia in cultures infected with the Rap5 virus than in cultures infected with the parental isolate, HIV-1Lai. The large number of enlarged syncytia observed in the Rap5-infected cultures suggested to us the possibility that the rapid mutant virus was more capable of spreading by cell-to-cell fusion. To quantitate the fusion capability of each virus, fusion assays were performed in which cells were infected with either Lai or Rap5 virus. After 3 days of infection, equal numbers of p24gag+ cells from HIVLai- or Rap5-infected cultures were coincubated with uninfected Jurkat-LTR-luc cells (14) for various periods. Cocultures were carried out in the presence of the reverse transcriptase inhibitor zidovudine (50 μM), to prevent multiple rounds of infection. Cell-to-cell fusion between infected cells and the Jurkat-LTR-luc cells was quantified by measuring the luciferase activity of the lysed extracts. Indeed, the Rap5-infected cells initiated cell-to-cell fusion with a threefold-greater efficiency than the Lai-infected cells did (Fig. 3B). To confirm whether the enhanced fusion was also functional in the setting of the rapid-turnover assay, infected cultures were treated with mitomycin C (50 μg/ml) prior to coculture with Jurkat-LTR-luc cells. There was a similar threefold increase in the ability of the cells infected with the Rap5 virus to fuse with uninfected targets (Fig. 3C).
FIG. 3.
The cell-to-cell transfer of the Rap5 virus is greatly enhanced in a fusion assay. (A) Jurkat cells (106) were mock infected or infected with HIVLai, Rap Mut 1.0 (biologic clone), or Rap5 (molecular clone). Cultures were photographed on day 3 of infection. Representative phase micrographs of the cells infected with the indicated viruses are shown. The presence of syncytial cells in each of the infected cultures is indicated by white arrows. (B and C) At 3 days after infection with HIV-1Lai or Rap5 virus, 105 p24gag+ cells from each infected culture (as determined by FACS analysis) were mixed with Jurkat-LTR-luc cells (2.5 × 105) which contain a Tat-responsive LTR-luciferase reporter gene construct, in 96-well plates in triplicate. Fusion assays were performed in the presence of AZT (50 μM) to inhibit subsequent rounds of infection (B), or infected cells were exposed to mitomycin C (50 μg/ml) prior to coculture with Jurkat-LTR-luc cells (C). Fusion was quantified by measurement of the luciferase activity in the cell extracts after 24 and 48 h. The fold increase in luciferase activity (fusion) mediated by the Rap5 infection compared to the HIVLai infection is noted, and the standard deviations are indicated by error bars. Data shown are from one representative experiment performed in triplicate, which has been repeated multiple times.
The rapid mutant retains virus particles on the cell surface.
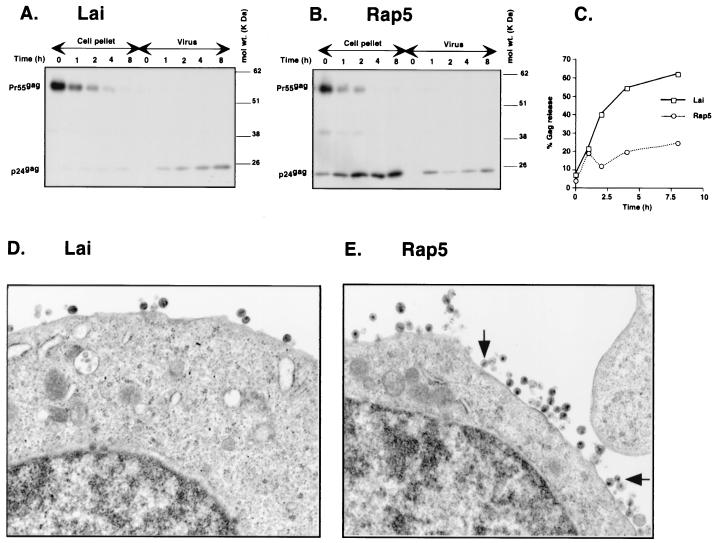
We wished to determine how the vpu frameshift mutation could lead to more efficient fusion and cell-to-cell transfer of virus. Since Vpu has been reported to affect particle release (11, 36, 38, 39), we first determined if this phenotype of virus-mediated enhanced cell-to-cell fusion was due to retention of viral particles on the plasma membrane of the producer cells. Jurkat cells were infected with either HIVLai or Rap5 virus, and the number of productively infected cells present in each culture was determined on day 3 postinfection by FACS analysis with an FITC-conjugated p24gag+ antibody. Cell cultures containing equal numbers of p24gag+ cells were pulse-labeled with [35S]methionine-[35S]cysteine for 1 h and chased for up to 8 h. At each time point, aliquots of cells and virions released into the supernatants were harvested, lysed, and processed for immunoprecipitation with a monoclonal antibody directed against the Gag protein (Fig. 4A and B). The release of virus particles into cell supernatants can be monitored by the accumulation of the p24gag protein in the cell-free viral supernatants. In cells infected with the parental isolate, HIVLai, there was an accumulation of Gag proteins in the cell-free virus supernatants over time (Fig. 4A). In contrast, infection with the Rap5 isolate resulted in an accumulation of the p24gag protein in the cell-associated fraction (Fig. 4B), especially at the later times. The relative amounts of p24gag and p55gag released from the cells producing HIVLai or Rap5 viruses were measured by image analysis with a PhosphorImager, and virus released into the supernatant was calculated as the percentage of Gag proteins present in the viral pellet relative to the sum of Gag proteins detected intra- and extracellularly. The results show that virus particle release from the Rap5-infected cells was three- to fourfold lower than that from the HIVLai-infected cells (Fig. 4C).
FIG. 4.
Inhibition of virus particle release in Rap5-infected Jurkat cells. (A and B) Jurkat cells (106) were infected with either Lai (A) or Rap5 (B) at an MOI of 0.03. At 72 h postinfection, the number of productively infected cells present in each infected culture was determined by FACS analysis (as described in Materials and Methods). Cultures containing equal numbers of p24gag+ cells (106) were labeled for 60 min with [35S]methionine-[35S]cysteine mix and chased for up to 8 h. Viral Gag proteins from the cell lysates or pelleted virions from cell-free viral supernatants were immunoprecipitated with a monoclonal antibody against the HIV-1 Gag polyprotein. The positions of p55gag and p24gag are indicated on the left of each panel, and the positions of the molecular mass markers in kilodaltons are indicated on the right. (C) Gag-specific proteins (p55gag and p24gag) were quantified by PhosphorImage analysis. The relative amounts of Gag proteins in the virus pellets (denoted as % Gag released) were calculated as the percentage of total p55gag and p24gag proteins for each time point and were plotted as a function of time. (D and E) Jurkat cells infected with Lai (D) or Rap5 (E) were processed for electron microscopy 72 h postinfection. Mature, electron-dense viral capsids left attached to the cell membrane in the Rap5-infected cell are indicated by black arrows.
Since the pulse-chase analysis suggested that the rapid mutant did not efficiently release viral particles, we next examined the infected cells by electron microscopy for the presence of virus particles in the cytoplasm and in the plasma membrane. Jurkat T cells infected with either HIVLai or Rap5 virus were processed for electron microscopy analysis 3 days postinfection. Viral capsids were clearly visible as they left the infected cell during infection with HIVLai (Fig. 4D). The cells infected with the Rap5 virus had many mature viral capsids left attached to the plasma membrane, suggesting impairment of virus particle release (Fig. 4E). Furthermore, these virus particles were mature, as suggested by the presence of the electron-dense cores. These results correlate well with the results of the pulse-chase analysis (Fig. 4A and B), where there was a three- to fourfold decrease in virus particle release in cells infected with Rap5 virus compared to those infected with Lai virus.
Effect of the vpu frameshift mutation on Env expression.
It has been previously reported that a point mutation in the translation initiation codon of vpu (which abolishes Vpu expression) has a positive effect on env expression (35). Since the enhanced cell-to-cell spread of Rap5 virus could be due to the presence of increased levels of Env glycoproteins on the cell surface, we decided to investigate the effects of the vpu frameshift mutation on Env expression in infected cells. To quantitate the relative levels of Env proteins, steady-state labeling of Jurkat cells infected with either HIVLai or Rap5 virus was performed (Fig. 5A).
FIG. 5.
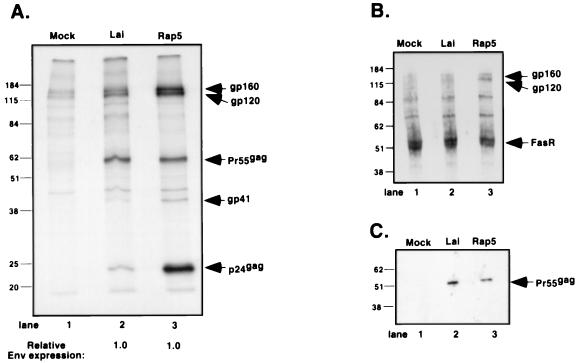
Effect of the vpu frameshift mutation on Env expression. (A) Jurkat cells (106) were infected with Lai or Rap at an MOI of 0.03. At 72 h postinfection, the cells were harvested and metabolically labeled with [35S]methionine-[35S]cysteine for 4 h. Viral proteins were immunoprecipitated with monoclonal antibodies against HIV-1 Env glycoproteins and HIV-1 Gag polyprotein. The amounts of gp160, gp120, gp41, p55gag, and p24gag were measured by PhosphorImage analysis, and the ratios of Env and Gag proteins relative to the Lai sample (lane 2) are indicated at the bottom of the fluorograph. (B) Jurkat cells (106) infected with Lai or Rap at an MOI of 0.03 were processed for cell surface biotinylation 72 h postinfection. Cultures (107 cells) were normalized for equal number of p24gag+ cells prior to biotinylation. Biotinylated extracts were immunoprecipitated with monoclonal antibodies against HIV-1 Env glycoproteins and cellular Fas receptor, electrophoresed on an SDS–10% polyacrylamide gel, blotted, and developed by ECL after incubation with streptavidin-horseradish peroxidase. (C) Western blot of extracts derived from Jurkat cells infected and processed as described for panel B. Solubilized proteins were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted with a monoclonal antibody against HIV-1 Gag polyprotein. Loading of samples was normalized according to the protein content of the extract. The positions of gp160, gp120, gp41, p55gag, p24gag, and Fas receptor are indicated on the right, and the positions of the molecular mass markers in kilodaltons are indicated on the left.
Determination of the amounts of Env glycoproteins (gp160, gp120, and gp41) present in each lane by image software analysis revealed a fourfold increase in the amounts of Env protein in the Rap5-infected cell lysate (Fig. 5A, lane 3) compared to those in the HIVLai-infected cell lysate (lane 2). However, the relative amounts of Env glycoproteins (the sum of gp160, gp120, and gp41) normalized to the level of total Gag proteins (the sum of p55gag and p24gag) present in the cells were similar. In fact, the Env-to-Gag protein ratios of each cellular lysate were identical (Fig. 5A), suggesting that the frameshift mutation in the vpu ORF did not have an effect on either the kinetics or the absolute level of Env expression. Therefore, the increased amounts of the Env glycoproteins (gp160, gp120, and gp41) present in the Rap5-infected cell lysate (Fig. 5A, lane 3) are a consequence of the greater number of virus particles left attached to the cell membrane (Fig. 4E). These data were also confirmed by Western blot analysis of Jurkat cells infected with HIVLai or Rap5 virus or transfected with Env expression constructs containing or lacking the Vpu frameshift mutation (data not shown). Note that there is no difference in the steady-state expression of the p55gag polyprotein (Fig. 5A, compare lanes 2 and 3). Rather, there is an accumulation of the p24gag protein in infected cells over time due to inhibition of Rap5 virus particle release.
We next wanted to determine whether this increase in total Env protein content in the cell was reflected in an increase in surface Env glycoprotein expression. To detect Env glycoproteins on the cell surface, we biotinylated proteins of infected Jurkat cells with Sulfo-NHS-LC-biotin, a compound that selectively biotinylates cell surface proteins. Equal numbers of p24gag+ cells from HIVLai- and Rap5-infected cultures were used for this experiment. The cells were lysed, and biotinylated Env glycoproteins were immunoprecipitated using Env-specific antisera. The biotinylated proteins on the blot were detected by conjugation to streptavidin-horseradish peroxidase followed by ECL, as described in Materials and Methods. As a control for biotinylation of cell surface proteins, endogenous Fas receptor molecules were also immunoprecipitated with an anti-Fas receptor monoclonal antibody. As an additional loading control, equal amounts (1/10) of the cell lysates were loaded on a 10% polyacrylamide gel and the amount of p55gag polyprotein present in each lysate was determined by Western blot analysis using Gag-specific mouse monoclonal antibodies (Fig. 5C). Note that there is an equal amount of p55gag in each infected cell lysate (Fig. 5C, lanes 2 and 3), suggesting that equal numbers of infected cells were used for immunoprecipitations. The results from a representative immunoprecipitation (Fig. 5B) demonstrate an approximately threefold increase in the surface presence of gp120 and gp160 in cells infected with Rap5 (Fig. 5B, lane 3) relative to that of biotinylated Fas receptor. This increase in the amount of surface Env is similar to that seen in Fig. 5A and reflects the presence of increased numbers of cell-associated virions in Rap5-infected cells.
From these experiments, we conclude that the frameshift mutation in the vpu ORF provides an unusual genetic advantage (permitting enhanced cell-to-cell transfer of virus without compromising its ability to replicate) to the virus in the rapid-turnover assay.
DISCUSSION
In this study, we describe a novel in vitro culture system that attempts to model the in vivo steady state of HIV-1 infection. This culture system utilizes the cell type (CD4+ T lymphocytes) that makes the most significant contribution to virus levels in vivo. Most in vitro culture systems do not take into account the short half-life of infected cells (31, 32), which in vivo is a consequence of host cytolytic effector mechanisms as well as of virus-induced lysis. Viral replication under such conditions could be significantly different from normal in vitro passage conditions where cells are under no selection pressure. To mirror the stresses that the host immune system places on infected cells in vivo, where infected CD4+ T lymphocytes have a shorter life than their uninfected counterparts, we set up a virus replication system where infected cells were killed every 3 days by addition of a cytotoxic agent, mitomycin C. To perpetuate the viral infection, fresh, uninfected cells were added every 3 days. Under such selective pressure, a new viral variant was isolated that was able to survive the rapid-turnover assay, while the parental isolate, HIV-1Lai, was unable to replicate under such culture conditions. The greater efficiency of cell-to-cell transfer mediated by the rapid mutant virus (Rap5) is a critical parameter for maintenance and active replication of virus in cells subject to a rapid turnover. We have also performed additional rapid-turnover experiments where the viral generation time has been limited to 2 days (infected cells were exposed to mitomycin C every 2 days), which is more reflective of the in vivo virus generation time (4, 28). The Rap5 virus was again able to survive and replicate under such culture conditions (data not shown).
The molecular change required for this new phenotype was a frameshift mutation in the vpu ORF that abolished Vpu expression. Vpu is an integral membrane phosphoprotein (81 amino acids) that has two known functions during the viral life cycle: enhancement of virus particle release from the infected cell surface (19, 36, 39), and down-regulation of CD4 antigen expression on the cell surface by targeting the nascent CD4 protein to ubiquitin-mediated proteolysis by the proteosomes (8, 22). The frameshift mutation in the vpu ORF affected both of its putative functions; that is, virus particle release was severely impaired in infected cells (Fig. 4) and CD4 antigen expression on the infected cell surface was twofold higher than in cells infected with the parental isolate, HIVLai (data not shown). The mechanism by which Vpu promotes virus particle release is still not clear.
The enhanced ability of the virus to mediate cell-to-cell transfer is probably responsible for the selection of this mutant in the rapid-turnover assay. Interestingly, in a recently published study, Hamm et al. report the isolation of an HIV-1 variant that was selected by in vitro passage of the virus in a CEM T-lymphoblastoid cell line (CEM/RevM10*) constitutively expressing a transdominant mutant of the Rev protein, RevM10 (16). This variant also had an insertion mutation in the vpu ORF that led to the introduction of a premature stop codon and truncation of the protein. Similar to our study, loss of Vpu expression probably allowed the virus variant to replicate in the CEM/RevM10* cells by mediating enhanced cell-to-cell transfer of virus.
The significance of the rapid-turnover culture system is best understood in the context of a simple steady-state model for virus infection (4). In the in vivo viral steady state, each infected cell produces enough virus particles in its lifetime to infect, on average, one other cell. Furthermore, the total number of cells is maintained at a constant level by replenishment of the dying cells by lymphopoietic mechanisms (Fig. 6A). Finally, although the time from infection to death is variable, it has been suggested that HIV-1-producing CD4+ T cells might die within 2 to 3 days of being infected (12, 27, 28). Since a persistent level of viral replication is predominantly observed in the lymph nodes (6, 15, 25), it can be assumed that the presence of large numbers and the proximity of virus-producing lymphocytes in the lymph nodes would promote intimate contacts between cells and favor cell-to-cell transmission of virus. In fact, it has been previously reported that cell-to-cell spread of virus was favored over infections with cell-free virus inocula (5, 33). In the in vivo setting, transmission of virus to T cells is most efficient in the context of antigen-presenting cells, such as dendritic cells (9). The presence of adhesion molecules such as DC-SIGN on the dendritic cell surface that capture HIV-1 virus particles and transmit virus allow the infection of replication-permissive T cells at a high efficiency (9, 10). This is probably why there have been isolated descriptions of primary isolates with vpu mutations that abolish Vpu expression and affect its particle release function (20). Hence, we would like to suggest that the vpu frameshift mutation conferred the ability on the virus to spread in vitro by cell-to-cell fusion, an infectious setting similar to the situation during in vivo virus transmission (2, 6, 13, 15, 25).
FIG. 6.
Modeling the steady state of HIV-1 infection in the rapid-turnover assay. For in vivo infection, kinetic modeling studies have established the average generation time of HIV-1 (defined as the time from release of a virion until it infects another CD4+ T cell and causes the release of a new generation of virus particles) as 2 to 3 days. The in vivo steady state makes the assumption that only one of these released virions would successfully infect another cell and produce the next generation of virus particles; that is, one productively infected cell leads to the productive infection of one other cell. It should be noted that in vivo, lymphocytes are much more likely to be infected by cell-associated HIV (chronically infected antigen-presenting cell such as macrophages or dendritic cells in the germinal centers) than by HIV in extracellular fluid. The number of CD4+ T cells is held constant by the production of new cells by the lymphopoietic mechanisms in face of cytotoxic T-lymphocyte (CTL)-mediated and virus-induced lysis of infected cells, such that infection, cell death, and cell replacement are in balance. In the in vitro rapid-turnover assay system, infection of Jurkat cells (activated CD4+ T cells) by Rap5 virus and the subsequent death of the infected cells either by virus-induced lysis or by addition of the cytotoxic agent mitomycin C are balanced by the addition of a constant number of uninfected Jurkat cells every third day. Steady state is achieved due to the unique ability of the Rap5 virus to mediate enhanced efficiency of cell-to-cell transfer. This unique phenotype allows the virus to survive the rapid-turnover assay in vitro and maintain high levels of viral replication over multiple generations.
New insights into virus population dynamics in vivo have been provided through a combination of experimental techniques and mathematical models (4, 27, 28). However, it has been hard to determine the roles of virus accessory genes in viral replication and disease progression due to the lack of a suitable in vitro replication system to provide a testable environment for novel therapeutics. Since the disease caused by HIV is a consequence of the accumulation of damage over the entire course of infection, any retardation of the process would be of significant help. In conclusion, we have developed an in vitro rapid-turnover assay system that mimics the turnover kinetics of infected T cells in vivo. This replication system will allow us to determine putative roles of viral accessory genes in the setting of an infected cell that is subjected to rapid turnover.
ACKNOWLEDGMENTS
We thank the FHCRC Flow Cytometry Laboratory, the EM Laboratory, Image Analysis, and the Biotechnology Facility for expert technical assistance, and we thank Wei Chun Goh, Steve Bartz, Marie Vodicka, Harmit Malik, Steve Dewhurst, and Maxine Linial for comments on the manuscript. The antibodies 76C and 2G12 were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, and this service is acknowledged.
This work was supported by NIH grant R01 AI30927 and the James B. Pendleton Fellowship.
REFERENCES
- 1.Cavert W, Notermans D W, Staskus K, Wietgrefe S W, Zupancic M, Gebhard K, Henry K, Zhang Z Q, Mills R, McDade H, Schuwirth C M, Goudsmit J, Danner S A, Haase A T. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 2.Cheynier R, Henrichwark S, Hadida F, Pelletier E, Oksenhendler E, Autran B, Wain-Hobson S. HIV and T cell expansion in splenic white pulps is accompanied by infiltration of HIV-specific cytotoxic T lymphocytes. Cell. 1994;78:373–387. doi: 10.1016/0092-8674(94)90417-0. [DOI] [PubMed] [Google Scholar]
- 3.Chun T W, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, Kuo Y H, Brookmeyer R, Zeiger M A, Barditch-Crovo P, Siliciano R F. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 4.Coffin J M. HIV viral dynamics. AIDS. 1996;10:S75–S84. [PubMed] [Google Scholar]
- 5.Dimitrov D S, Willey R L, Sato H, Chang L J, Blumenthal R, Martin M A. Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol. 1993;67:2182–2190. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embretson J, Zupancic M, Ribas J L, Burke A, Racz P, Tenner-Racz K, Haase A T. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature. 1993;362:359–362. doi: 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 7.Finzi D, Silliciano R F. Viral dynamics in HIV-1 infection. Cell. 1998;93:665–671. doi: 10.1016/s0092-8674(00)81427-0. [DOI] [PubMed] [Google Scholar]
- 8.Fujita K, Omura S, Silver J. Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J Gen Virol. 1997;78:619–625. doi: 10.1099/0022-1317-78-3-619. [DOI] [PubMed] [Google Scholar]
- 9.Geijtenbeek T B, Kwon D S, Torensma R, van Vliet S J, van Duijnhoven G C, Middel J, Cornelissen I L, Nottet H S, KewalRamani V N, Littman D R, Figdor C G, van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 10.Geijtenbeek T B, Torensma R, van Vliet S J, van Duijnhoven G C, Adema G J, van Kooyk Y, Figdor C G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 11.Gottlinger H G, Dorfman T, Cohen E A, Haseltine W A. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc Natl Acad Sci USA. 1993;90:7381–7385. doi: 10.1073/pnas.90.15.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossman Z, Feinberg M, Kuznetsov V, Dimitrov D, Paul W. HIV infection: how effective is drug combination treatment? Immunol Today. 1998;19:528–532. doi: 10.1016/s0167-5699(98)01353-x. [DOI] [PubMed] [Google Scholar]
- 13.Grossman Z, Feinberg M B, Paul W E. Multiple modes of cellular activation and virus transmission in HIV infection: a role for chronically and latently infected cells in sustaining viral replication. Proc Natl Acad Sci USA. 1998;95:6314–6319. doi: 10.1073/pnas.95.11.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gummuluru S, Emerman M. Cell cycle- and Vpr-mediated regulation of human immunodeficiency virus type 1 expression in primary and transformed T-cell lines. J Virol. 1999;73:5422–5430. doi: 10.1128/jvi.73.7.5422-5430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haase A T, Henry K, Zupancic M, Sedgewick G, Faust R A, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang Z Q, Dailey P J, Balfour H H, Jr, Erice A, Perelson A S. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science. 1996;274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 16.Hamm T E, Rekosh D, Hammarskjold M L. Selection and characterization of human immunodeficiency virus type 1 mutants that are resistant to inhibition by the transdominant negative RevM10 protein. J Virol. 1999;73:5741–5747. doi: 10.1128/jvi.73.7.5741-5747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 18.Jin X, Bauer D E, Tuttleton S E, Lewin S, Gettie A, Blanchard J, Irwin C E, Safrit J T, Mittler J, Weinberger L, Kostrikis L G, Zhang L, Perelson A S, Ho D D. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klimkait T, Strebel K, Hoggan M D, Martin M A, Orenstein J M. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korber B T, Kuiken C, Floey B, Hahn B, McCutchan F, Mellors J, Sodroski J. Human retroviruses and AIDS. Los Alamos, N.M: Los Alamos National Laboratory; 1998. [Google Scholar]
- 21.Kuroda M J, Schmitz J E, Charini W A, Nickerson C E, Lifton M A, Lord C I, Forman M A, Letvin N L. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J Immunol. 1999;162:5127–5133. [PubMed] [Google Scholar]
- 22.Margottin F, Bour S P, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 23.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 24.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 25.Pantaleo G, Graziosi C, Demarest J F, Butini L, Montroni M, Fox C H, Orenstein J M, Kotler D P, Fauci A S. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 26.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 27.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 28.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 29.Piatak M, Jr, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 30.Rodrigo A G, Shpaer E G, Delwart E L, Iversen A K, Gallo M V, Brojatsch J, Hirsch M S, Walker B D, Mullins J I. Coalescent estimates of HIV-1 generation time in vivo. Proc Natl Acad Sci USA. 1999;96:2187–2191. doi: 10.1073/pnas.96.5.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenzweig M, DeMaria M A, Harper D M, Friedrich S, Jain R K, Johnson R P. Increased rates of CD4(+) and CD8(+) T lymphocyte turnover in simian immunodeficiency virus-infected macaques. Proc Natl Acad Sci USA. 1998;95:6388–6393. doi: 10.1073/pnas.95.11.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachsenberg N, Perelson A S, Yerly S, Schockmel G A, Leduc D, Hirschel B, Perrin L. Turnover of CD4+ and CD8+ T lymphocytes in HIV-1 infection as measured by Ki-67 antigen. J Exp Med. 1998;187:1295–1303. doi: 10.1084/jem.187.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato H, Orenstein J, Dimitrov D, Martin M. Cell-to-cell spread of HIV-1 occurs within minutes and may not involve the participation of virus particles. Virology. 1992;186:712–724. doi: 10.1016/0042-6822(92)90038-q. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, Ghrayeb J, Forman M A, Montefiori D C, Rieber E P, Letvin N L, Reimann K A. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 35.Schubert U, Bour S, Willey R L, Strebel K. Regulation of virus release by the macrophage-tropic human immunodeficiency virus type 1 AD8 isolate is redundant and can be controlled by either Vpu or Env. J Virol. 1999;73:887–896. doi: 10.1128/jvi.73.2.887-896.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubert U, Clouse K A, Strebel K. Augmentation of virus secretion by the human immunodeficiency virus type 1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J Virol. 1995;69:7699–7711. doi: 10.1128/jvi.69.12.7699-7711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steimer K S, Puma J P, Power M D, Powers M A, George-Nascimento C, Stephans J C, Levy J A, Sanchez-Pescador R, Luciw P A, Barr P J, Hallewell R A. Differential antibody responses of individuals infected with AIDS-associated retroviruses surveyed using the viral core antigen p25gag expressed in bacteria. Virology. 1986;150:283–290. doi: 10.1016/0042-6822(86)90289-8. [DOI] [PubMed] [Google Scholar]
- 38.Strebel K, Klimkait T, Maldarelli F, Martin M A. Molecular and biochemical analyses of human immunodeficiency virus type 1 vpu protein. J Virol. 1989;63:3784–3791. doi: 10.1128/jvi.63.9.3784-3791.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger E F, Cohen E A, Lu Y C, Sodroski J G, Haseltine W A. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci USA. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vodicka M A, Goh W C, Wu L I, Rogel M E, Bartz S R, Schweickart V L, Raport C J, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 42.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]