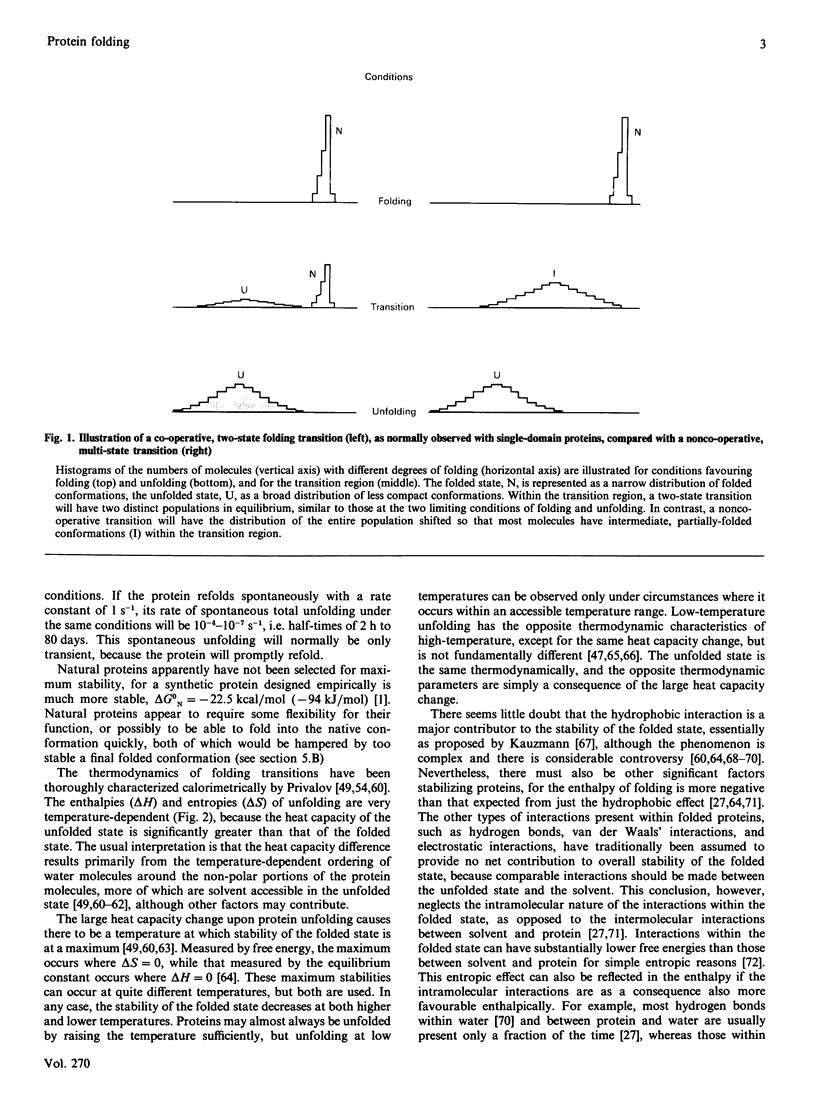
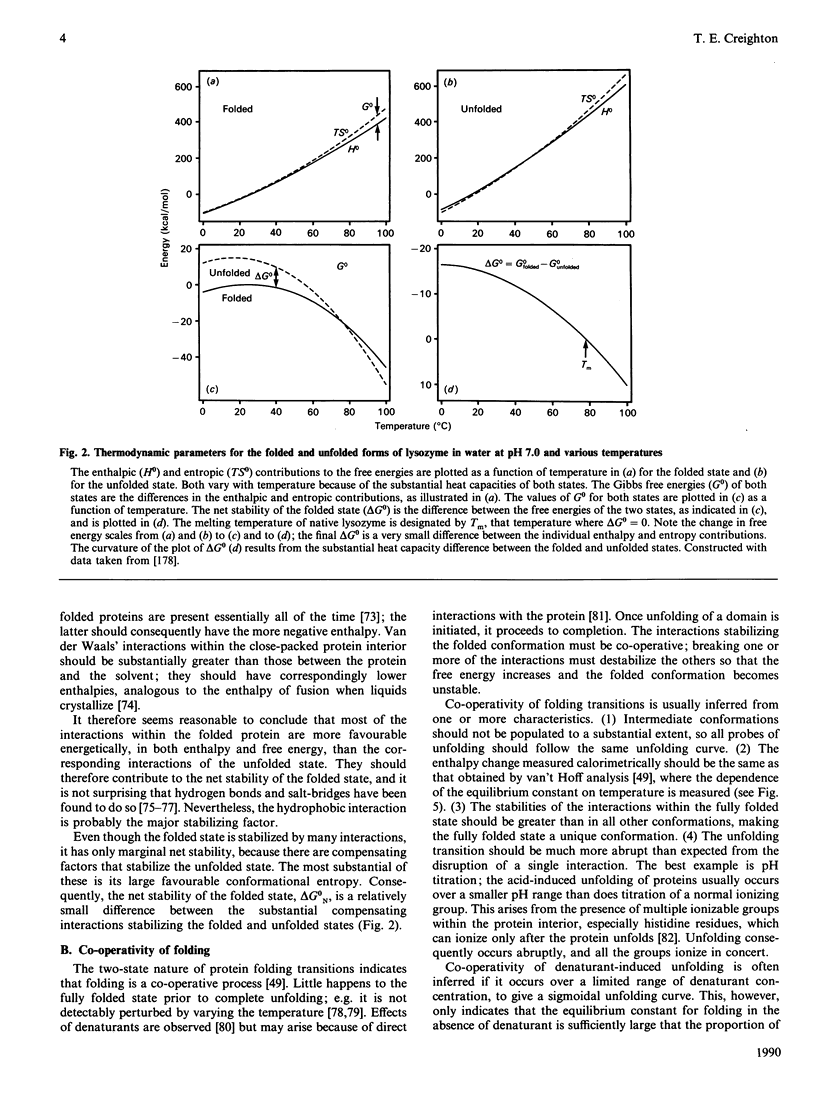
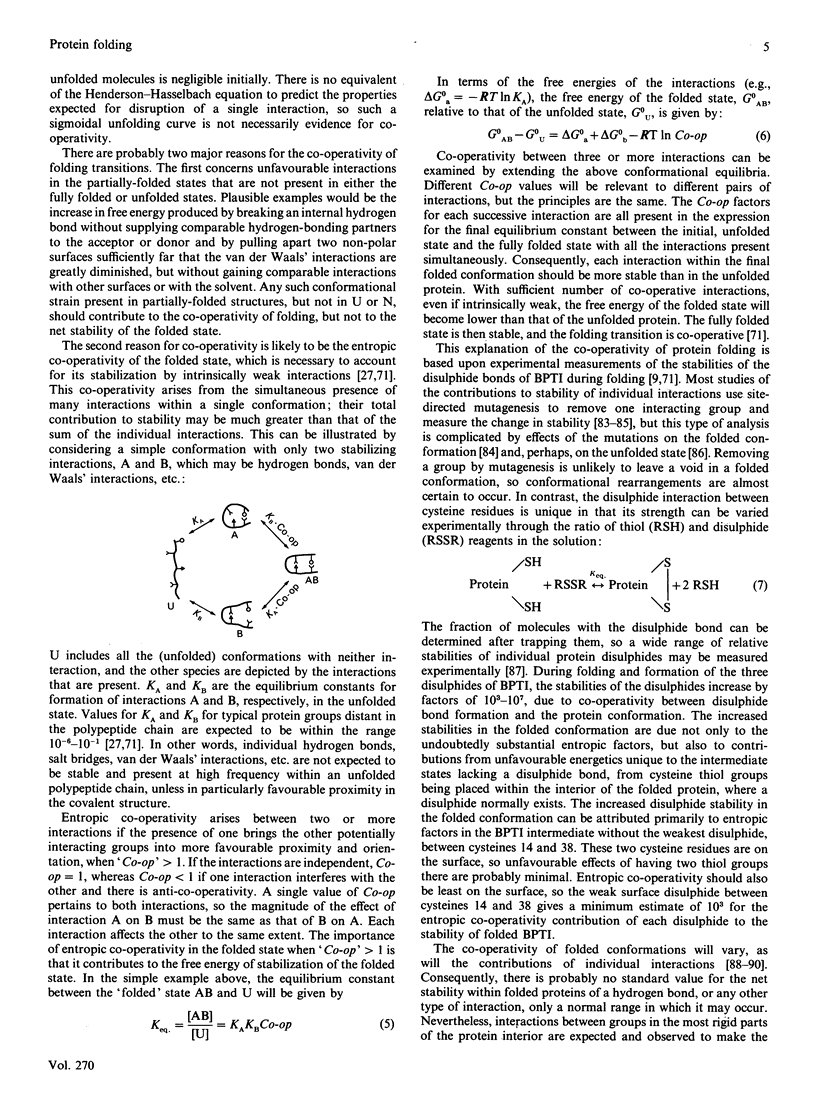
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Sun D. P., Nye J. A., Muchmore D. C., Matthews B. W. Temperature-sensitive mutations of bacteriophage T4 lysozyme occur at sites with low mobility and low solvent accessibility in the folded protein. Biochemistry. 1987 Jun 30;26(13):3754–3758. doi: 10.1021/bi00387a002. [DOI] [PubMed] [Google Scholar]
- Alber T., Sun D. P., Wilson K., Wozniak J. A., Cook S. P., Matthews B. W. Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature. 1987 Nov 5;330(6143):41–46. doi: 10.1038/330041a0. [DOI] [PubMed] [Google Scholar]
- Anderson D. E., Becktel W. J., Dahlquist F. W. pH-induced denaturation of proteins: a single salt bridge contributes 3-5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry. 1990 Mar 6;29(9):2403–2408. doi: 10.1021/bi00461a025. [DOI] [PubMed] [Google Scholar]
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Anfinsen C. B., Scheraga H. A. Experimental and theoretical aspects of protein folding. Adv Protein Chem. 1975;29:205–300. doi: 10.1016/s0065-3233(08)60413-1. [DOI] [PubMed] [Google Scholar]
- Bajaj M., Blundell T. Evolution and the tertiary structure of proteins. Annu Rev Biophys Bioeng. 1984;13:453–492. doi: 10.1146/annurev.bb.13.060184.002321. [DOI] [PubMed] [Google Scholar]
- Baker E. N., Hubbard R. E. Hydrogen bonding in globular proteins. Prog Biophys Mol Biol. 1984;44(2):97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- Baldwin R. L. How does protein folding get started? Trends Biochem Sci. 1989 Jul;14(7):291–294. doi: 10.1016/0968-0004(89)90067-4. [DOI] [PubMed] [Google Scholar]
- Baldwin R. L. Intermediates in protein folding reactions and the mechanism of protein folding. Annu Rev Biochem. 1975;44:453–475. doi: 10.1146/annurev.bi.44.070175.002321. [DOI] [PubMed] [Google Scholar]
- Baldwin R. L. Temperature dependence of the hydrophobic interaction in protein folding. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J., Dobson C. M., Evans P. A., Hanley C. Characterization of a partly folded protein by NMR methods: studies on the molten globule state of guinea pig alpha-lactalbumin. Biochemistry. 1989 Jan 10;28(1):7–13. doi: 10.1021/bi00427a002. [DOI] [PubMed] [Google Scholar]
- Becktel W. J., Schellman J. A. Protein stability curves. Biopolymers. 1987 Nov;26(11):1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- Bello J. Stability of protein conformation: internal packing and enthalpy of fusion of model compounds. J Theor Biol. 1977 Sep 7;68(1):139–142. doi: 10.1016/0022-5193(77)90232-6. [DOI] [PubMed] [Google Scholar]
- Bowie J. U., Reidhaar-Olson J. F., Lim W. A., Sauer R. T. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990 Mar 16;247(4948):1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- Brandts J. F., Brennan M., Lung-Nan Lin Unfolding and refolding occur much faster for a proline-free proteins than for most proline-containing proteins. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4178–4181. doi: 10.1073/pnas.74.10.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandts J. F., Halvorson H. R., Brennan M. Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry. 1975 Nov 4;14(22):4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- Brandts J. F., Hu C. Q., Lin L. N., Mos M. T. A simple model for proteins with interacting domains. Applications to scanning calorimetry data. Biochemistry. 1989 Oct 17;28(21):8588–8596. doi: 10.1021/bi00447a048. [DOI] [PubMed] [Google Scholar]
- Campbell I. D., Dobson C. M., Williams R. J. The study of conformational states of proteins by nuclear magnetic resonance. Biochem J. 1985 Oct 1;231(1):1–10. doi: 10.1042/bj2310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazin W. J., Kördel J., Drakenberg T., Thulin E., Brodin P., Grundström T., Forsén S. Proline isomerism leads to multiple folded conformations of calbindin D9k: direct evidence from two-dimensional 1H NMR spectroscopy. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2195–2198. doi: 10.1073/pnas.86.7.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. L., Baase W. A., Schellman J. A. Low-temperature unfolding of a mutant of phage T4 lysozyme. 2. Kinetic investigations. Biochemistry. 1989 Jan 24;28(2):691–699. doi: 10.1021/bi00428a042. [DOI] [PubMed] [Google Scholar]
- Chen B. L., Schellman J. A. Low-temperature unfolding of a mutant of phage T4 lysozyme. 1. Equilibrium studies. Biochemistry. 1989 Jan 24;28(2):685–691. doi: 10.1021/bi00428a041. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M., Dodson G. G., Hodgkin D. C. Transmission of conformational change in insulin. Nature. 1983 Apr 7;302(5908):500–505. doi: 10.1038/302500a0. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986 Apr;5(4):823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- Creighton E. T. Possible implications of many proline residues for the kinetics of protein unfolding and refolding. J Mol Biol. 1978 Nov 5;125(3):401–406. doi: 10.1016/0022-2836(78)90411-4. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Accessibilities and reactivities of cysteine thiols during refolding of reduced bovine pancreatic trypsin inhibitor. J Mol Biol. 1981 Sep 5;151(1):211–213. doi: 10.1016/0022-2836(81)90230-8. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. An empirical approach to protein conformation stability and flexibility. Biopolymers. 1983 Jan;22(1):49–58. doi: 10.1002/bip.360220110. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Disulfide bonds as probes of protein folding pathways. Methods Enzymol. 1986;131:83–106. doi: 10.1016/0076-6879(86)31036-x. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Disulphide bonds and protein stability. Bioessays. 1988 Feb-Mar;8(2):57–63. doi: 10.1002/bies.950080204. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Effects of urea and guanidine-HCl on the folding and unfolding of pancreatic trypsin inhibitor. J Mol Biol. 1977 Jun 25;113(2):313–328. doi: 10.1016/0022-2836(77)90144-9. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Experimental studies of protein folding and unfolding. Prog Biophys Mol Biol. 1978;33(3):231–297. doi: 10.1016/0079-6107(79)90030-0. [DOI] [PubMed] [Google Scholar]
- Creighton T. E., Goldenberg D. P. Kinetic role of a meta-stable native-like two-disulphide species in the folding transition of bovine pancreatic trypsin inhibitor. J Mol Biol. 1984 Nov 5;179(3):497–526. doi: 10.1016/0022-2836(84)90077-9. [DOI] [PubMed] [Google Scholar]
- Creighton T. E., Hillson D. A., Freedman R. B. Catalysis by protein-disulphide isomerase of the unfolding and refolding of proteins with disulphide bonds. J Mol Biol. 1980 Sep 5;142(1):43–62. doi: 10.1016/0022-2836(80)90205-3. [DOI] [PubMed] [Google Scholar]
- Creighton T. E., Kalef E., Arnon R. Immunochemical analysis of the conformational properties of intermediates trapped in the folding and unfolding of bovine pancreatic trypsin inhibitor. J Mol Biol. 1978 Aug 5;123(2):129–147. doi: 10.1016/0022-2836(78)90317-0. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Kinetic study of protein unfolding and refolding using urea gradient electrophoresis. J Mol Biol. 1980 Feb 15;137(1):61–80. doi: 10.1016/0022-2836(80)90157-6. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. On the relevance of non-random polypeptide conformations for protein folding. Biophys Chem. 1988 Aug;31(1-2):155–162. doi: 10.1016/0301-4622(88)80021-8. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. The single-disulphide intermediates in the refolding of reduced pancreatic trypsin inhibitor. J Mol Biol. 1974 Aug 15;87(3):603–624. doi: 10.1016/0022-2836(74)90106-5. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Toward a better understanding of protein folding pathways. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5082–5086. doi: 10.1073/pnas.85.14.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton T. Reactivities of the cysteine residues of the reduced pancreatic trypsin inhibitor. J Mol Biol. 1975 Aug 25;96(4):777–782. doi: 10.1016/0022-2836(75)90152-7. [DOI] [PubMed] [Google Scholar]
- Degrado W. F. Design of peptides and proteins. Adv Protein Chem. 1988;39:51–124. doi: 10.1016/s0065-3233(08)60375-7. [DOI] [PubMed] [Google Scholar]
- Denton J. B., Konishi Y., Scheraga H. A. Folding of ribonuclease A from a partially disordered conformation. Kinetic study under folding conditions. Biochemistry. 1982 Oct 12;21(21):5155–5163. doi: 10.1021/bi00264a008. [DOI] [PubMed] [Google Scholar]
- Dobson C. M., Evans P. A., Williamson K. L. Proton NMR studies of denatured lysozyme. FEBS Lett. 1984 Mar 26;168(2):331–334. doi: 10.1016/0014-5793(84)80273-2. [DOI] [PubMed] [Google Scholar]
- Evans P. A., Dobson C. M., Kautz R. A., Hatfull G., Fox R. O. Proline isomerism in staphylococcal nuclease characterized by NMR and site-directed mutagenesis. Nature. 1987 Sep 17;329(6136):266–268. doi: 10.1038/329266a0. [DOI] [PubMed] [Google Scholar]
- Evans P. A., Kautz R. A., Fox R. O., Dobson C. M. A magnetization-transfer nuclear magnetic resonance study of the folding of staphylococcal nuclease. Biochemistry. 1989 Jan 10;28(1):362–370. doi: 10.1021/bi00427a050. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. Dissection of the structure and activity of the tyrosyl-tRNA synthetase by site-directed mutagenesis. Biochemistry. 1987 Dec 15;26(25):8031–8037. doi: 10.1021/bi00399a001. [DOI] [PubMed] [Google Scholar]
- Fischer G., Schmid F. X. The mechanism of protein folding. Implications of in vitro refolding models for de novo protein folding and translocation in the cell. Biochemistry. 1990 Mar 6;29(9):2205–2212. doi: 10.1021/bi00461a001. [DOI] [PubMed] [Google Scholar]
- Fischer G., Wittmann-Liebold B., Lang K., Kiefhaber T., Schmid F. X. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature. 1989 Feb 2;337(6206):476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- Freedman R. B. Protein disulfide isomerase: multiple roles in the modification of nascent secretory proteins. Cell. 1989 Jun 30;57(7):1069–1072. doi: 10.1016/0092-8674(89)90043-3. [DOI] [PubMed] [Google Scholar]
- Garel J. R., Nall B. T., Baldwin R. L. Guanidine-unfolded state of ribonuclease A contains both fast- and slow-refolding species. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1853–1857. doi: 10.1073/pnas.73.6.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvey E. P., Matthews C. R. Effects of multiple replacements at a single position on the folding and stability of dihydrofolate reductase from Escherichia coli. Biochemistry. 1989 Mar 7;28(5):2083–2093. doi: 10.1021/bi00431a018. [DOI] [PubMed] [Google Scholar]
- Goldenberg D. P., Creighton T. E. Circular and circularly permuted forms of bovine pancreatic trypsin inhibitor. J Mol Biol. 1983 Apr 5;165(2):407–413. doi: 10.1016/s0022-2836(83)80265-4. [DOI] [PubMed] [Google Scholar]
- Goldenberg D. P., Frieden R. W., Haack J. A., Morrison T. B. Mutational analysis of a protein-folding pathway. Nature. 1989 Mar 9;338(6211):127–132. doi: 10.1038/338127a0. [DOI] [PubMed] [Google Scholar]
- Goldenberg D. P. Genetic studies of protein stability and mechanisms of folding. Annu Rev Biophys Biophys Chem. 1988;17:481–507. doi: 10.1146/annurev.bb.17.060188.002405. [DOI] [PubMed] [Google Scholar]
- Goodman E. M., Kim P. S. Folding of a peptide corresponding to the alpha-helix in bovine pancreatic trypsin inhibitor. Biochemistry. 1989 May 16;28(10):4343–4347. doi: 10.1021/bi00436a033. [DOI] [PubMed] [Google Scholar]
- Goraj K., Renard A., Martial J. A. Synthesis, purification and initial structural characterization of octarellin, a de novo polypeptide modelled on the alpha/beta-barrel proteins. Protein Eng. 1990 Mar;3(4):259–266. doi: 10.1093/protein/3.4.259. [DOI] [PubMed] [Google Scholar]
- Goto Y., Hamaguchi K. Formation of the intrachain disulfide bond in the constant fragment of the immunoglobulin light chain. J Mol Biol. 1981 Mar 5;146(3):321–340. doi: 10.1016/0022-2836(81)90391-0. [DOI] [PubMed] [Google Scholar]
- Griko YuV, Venyaminov SYu, Privalov P. L. Heat and cold denaturation of phosphoglycerate kinase (interaction of domains). FEBS Lett. 1989 Feb 27;244(2):276–278. doi: 10.1016/0014-5793(89)80544-7. [DOI] [PubMed] [Google Scholar]
- Griko Y. V., Privalov P. L., Venyaminov S. Y., Kutyshenko V. P. Thermodynamic study of the apomyoglobin structure. J Mol Biol. 1988 Jul 5;202(1):127–138. doi: 10.1016/0022-2836(88)90525-6. [DOI] [PubMed] [Google Scholar]
- Gruenewald B., Nicola C. U., Lustig A., Schwarz G., Klump H. Kinetics of the helix-coil transition of a polypeptide with non-ionic side groups, derived from ultrasonic relaxation measurements. Biophys Chem. 1979 Jan;9(2):137–147. doi: 10.1016/0301-4622(79)87008-8. [DOI] [PubMed] [Google Scholar]
- Hall J. G., Frieden C. Protein fragments as probes in the study of protein folding mechanisms: differential effects of dihydrofolate reductase fragments on the refolding of the intact protein. Proc Natl Acad Sci U S A. 1989 May;86(9):3060–3064. doi: 10.1073/pnas.86.9.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding M. W., Galat A., Uehling D. E., Schreiber S. L. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature. 1989 Oct 26;341(6244):758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- Harrison S. C., Durbin R. Is there a single pathway for the folding of a polypeptide chain? Proc Natl Acad Sci U S A. 1985 Jun;82(12):4028–4030. doi: 10.1073/pnas.82.12.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U., Neupert W. Protein sorting to mitochondria: evolutionary conservations of folding and assembly. Science. 1990 Feb 23;247(4945):930–938. doi: 10.1126/science.2406905. [DOI] [PubMed] [Google Scholar]
- Hawley S. A., Mitchell R. M. An electrophoretic study of reversible protein denaturation: chymotrypsinogen at high pressures. Biochemistry. 1975 Jul 15;14(14):3257–3264. doi: 10.1021/bi00685a036. [DOI] [PubMed] [Google Scholar]
- Hibbard L. S., Tulinsky A. Expression of functionality of alpha-chymotrypsin. Effects of guanidine hydrochloride and urea in the onset of denaturation. Biochemistry. 1978 Dec 12;17(25):5460–5468. doi: 10.1021/bi00618a021. [DOI] [PubMed] [Google Scholar]
- Hollecker M., Creighton T. E. Evolutionary conservation and variation of protein folding pathways. Two protease inhibitor homologues from black mamba venom. J Mol Biol. 1983 Aug 5;168(2):409–437. doi: 10.1016/s0022-2836(83)80026-6. [DOI] [PubMed] [Google Scholar]
- Hollecker M., Creighton T. E., Gabriel M. Preliminary circular dichroism study of the conformations of intermediates trapped during protein folding. Biochimie. 1981 Nov-Dec;63(11-12):835–839. doi: 10.1016/s0300-9084(82)80269-1. [DOI] [PubMed] [Google Scholar]
- Hollecker M., Larcher D. Conformational forces affecting the folding pathways of dendrotoxins I and K from black mamba venom. Eur J Biochem. 1989 Jan 15;179(1):87–94. doi: 10.1111/j.1432-1033.1989.tb14524.x. [DOI] [PubMed] [Google Scholar]
- Howarth O. W., Lian L. Y. Ribonuclease A: carbon-13 nuclear magnetic resonance assignments, binding sites, and conformational flexibility. Biochemistry. 1984 Jul 17;23(15):3515–3521. doi: 10.1021/bi00310a020. [DOI] [PubMed] [Google Scholar]
- Ikeguchi M., Kuwajima K., Mitani M., Sugai S. Evidence for identity between the equilibrium unfolding intermediate and a transient folding intermediate: a comparative study of the folding reactions of alpha-lactalbumin and lysozyme. Biochemistry. 1986 Nov 4;25(22):6965–6972. doi: 10.1021/bi00370a034. [DOI] [PubMed] [Google Scholar]
- Ikemura H., Takagi H., Inouye M. Requirement of pro-sequence for the production of active subtilisin E in Escherichia coli. J Biol Chem. 1987 Jun 5;262(16):7859–7864. [PubMed] [Google Scholar]
- Jaenicke R. Folding and association of proteins. Prog Biophys Mol Biol. 1987;49(2-3):117–237. doi: 10.1016/0079-6107(87)90011-3. [DOI] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Kato S., Okamura M., Shimamoto N., Utiyama H. Spectral evidence for a rapidly formed structural intermediate in the refolding kinetics of hen egg-white lysozyme. Biochemistry. 1981 Mar 3;20(5):1080–1085. doi: 10.1021/bi00508a006. [DOI] [PubMed] [Google Scholar]
- Kelley R. F., Richards F. M. Replacement of proline-76 with alanine eliminates the slowest kinetic phase in thioredoxin folding. Biochemistry. 1987 Oct 20;26(21):6765–6774. doi: 10.1021/bi00395a028. [DOI] [PubMed] [Google Scholar]
- Kellis J. T., Jr, Nyberg K., Fersht A. R. Energetics of complementary side-chain packing in a protein hydrophobic core. Biochemistry. 1989 May 30;28(11):4914–4922. doi: 10.1021/bi00437a058. [DOI] [PubMed] [Google Scholar]
- Kellis J. T., Jr, Nyberg K., Sali D., Fersht A. R. Contribution of hydrophobic interactions to protein stability. Nature. 1988 Jun 23;333(6175):784–786. doi: 10.1038/333784a0. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu Rev Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- Kosen P. A., Creighton T. E., Blout E. R. Circular dichroism spectroscopy of bovine pancreatic trypsin inhibitor and five altered conformational states. Relationship of conformation and the refolding pathway of the trypsin inhibitor. Biochemistry. 1981 Sep 29;20(20):5744–5754. doi: 10.1021/bi00523a017. [DOI] [PubMed] [Google Scholar]
- Kosen P. A., Creighton T. E., Blout E. R. Circular dichroism spectroscopy of the intermediates that precede the rate-limiting step of the refolding pathway of bovine pancreatic trypsin inhibitor. Relationship of conformation and the refolding pathway. Biochemistry. 1983 May 10;22(10):2433–2440. doi: 10.1021/bi00279a020. [DOI] [PubMed] [Google Scholar]
- Kosen P. A., Creighton T. E., Blout E. R. Ultraviolet difference spectroscopy of intermediates trapped in unfolding and refolding of bovine pancreatic trypsin inhibitor. Biochemistry. 1980 Oct 14;19(21):4936–4944. doi: 10.1021/bi00562a037. [DOI] [PubMed] [Google Scholar]
- Kuwajima K., Mitani M., Sugai S. Characterization of the critical state in protein folding. Effects of guanidine hydrochloride and specific Ca2+ binding on the folding kinetics of alpha-lactalbumin. J Mol Biol. 1989 Apr 5;206(3):547–561. doi: 10.1016/0022-2836(89)90500-7. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6(2):87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- Kuwajima K., Yamaya H., Miwa S., Sugai S., Nagamura T. Rapid formation of secondary structure framework in protein folding studied by stopped-flow circular dichroism. FEBS Lett. 1987 Aug 31;221(1):115–118. doi: 10.1016/0014-5793(87)80363-0. [DOI] [PubMed] [Google Scholar]
- Lang K., Schmid F. X., Fischer G. Catalysis of protein folding by prolyl isomerase. Nature. 1987 Sep 17;329(6136):268–270. doi: 10.1038/329268a0. [DOI] [PubMed] [Google Scholar]
- Lang K., Schmid F. X. Role of two proline-containing turns in the folding of porcine ribonuclease. J Mol Biol. 1990 Mar 5;212(1):185–196. doi: 10.1016/0022-2836(90)90314-C. [DOI] [PubMed] [Google Scholar]
- Lang K., Schmid F. X. Use of a trypsin-pulse method to study the refolding pathway of ribonuclease. Eur J Biochem. 1986 Sep 1;159(2):275–281. doi: 10.1111/j.1432-1033.1986.tb09864.x. [DOI] [PubMed] [Google Scholar]
- Levitt M., Chothia C. Structural patterns in globular proteins. Nature. 1976 Jun 17;261(5561):552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Sauer R. T. Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature. 1989 May 4;339(6219):31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- Lin L. N., Brandts J. F. Separation of the nativelike intermediate from unfolded forms during refolding of ribonuclease A. Biochemistry. 1988 Dec 13;27(25):9037–9042. doi: 10.1021/bi00425a023. [DOI] [PubMed] [Google Scholar]
- Liu W., Tsou C. L. Activity change during unfolding of bovine pancreatic ribonuclease A in guanidine. Biochim Biophys Acta. 1987 Dec 18;916(3):455–464. doi: 10.1016/0167-4838(87)90192-0. [DOI] [PubMed] [Google Scholar]
- Loebermann H., Tokuoka R., Deisenhofer J., Huber R. Human alpha 1-proteinase inhibitor. Crystal structure analysis of two crystal modifications, molecular model and preliminary analysis of the implications for function. J Mol Biol. 1984 Aug 15;177(3):531–557. [PubMed] [Google Scholar]
- Luger K., Hommel U., Herold M., Hofsteenge J., Kirschner K. Correct folding of circularly permuted variants of a beta alpha barrel enzyme in vivo. Science. 1989 Jan 13;243(4888):206–210. doi: 10.1126/science.2643160. [DOI] [PubMed] [Google Scholar]
- Lynn R. M., Konishi Y., Scheraga H. A. Folding of ribonuclease A from a partially disordered conformation. Kinetic study under transition conditions. Biochemistry. 1984 May 22;23(11):2470–2477. doi: 10.1021/bi00306a023. [DOI] [PubMed] [Google Scholar]
- Manning M. C., Woody R. W. Theoretical study of the contribution of aromatic side chains to the circular dichroism of basic bovine pancreatic trypsin inhibitor. Biochemistry. 1989 Oct 17;28(21):8609–8613. doi: 10.1021/bi00447a051. [DOI] [PubMed] [Google Scholar]
- Marqusee S., Robbins V. H., Baldwin R. L. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A., Kellis J. T., Jr, Serrano L., Fersht A. R. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989 Jul 13;340(6229):122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Becktel W. J., Matthews B. W. Hydrophobic stabilization in T4 lysozyme determined directly by multiple substitutions of Ile 3. Nature. 1988 Aug 4;334(6181):406–410. doi: 10.1038/334406a0. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Genetic and structural analysis of the protein stability problem. Biochemistry. 1987 Nov 3;26(22):6885–6888. doi: 10.1021/bi00396a001. [DOI] [PubMed] [Google Scholar]
- Mattice W. L. The beta-sheet to coil transition. Annu Rev Biophys Biophys Chem. 1989;18:93–111. doi: 10.1146/annurev.bb.18.060189.000521. [DOI] [PubMed] [Google Scholar]
- Murphy K. P., Privalov P. L., Gill S. J. Common features of protein unfolding and dissolution of hydrophobic compounds. Science. 1990 Feb 2;247(4942):559–561. doi: 10.1126/science.2300815. [DOI] [PubMed] [Google Scholar]
- Nall B. T., Baldwin R. L. Thermal unfolding transition of ribonuclease A measured by 2'-CMP binding. Biochemistry. 1977 Aug 9;16(16):3572–3576. doi: 10.1021/bi00635a011. [DOI] [PubMed] [Google Scholar]
- Oas T. G., Kim P. S. A peptide model of a protein folding intermediate. Nature. 1988 Nov 3;336(6194):42–48. doi: 10.1038/336042a0. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Creighton T. E. The disulphide folding pathway of ribonuclease T1. J Mol Biol. 1986 Apr 5;188(3):477–486. doi: 10.1016/0022-2836(86)90169-5. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Laurents D. V., Thomson J. A. pH dependence of the urea and guanidine hydrochloride denaturation of ribonuclease A and ribonuclease T1. Biochemistry. 1990 Mar 13;29(10):2564–2572. doi: 10.1021/bi00462a019. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S., Marqusee S., Ridgeway T., Laue T. M., Baldwin R. L. Relative helix-forming tendencies of nonpolar amino acids. Nature. 1990 Mar 15;344(6263):268–270. doi: 10.1038/344268a0. [DOI] [PubMed] [Google Scholar]
- Page M. I., Jencks W. P. Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1678–1683. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. Heat-shock proteins. Coming in from the cold. Nature. 1988 Apr 28;332(6167):776–777. doi: 10.1038/332776a0. [DOI] [PubMed] [Google Scholar]
- Perutz M. F. Mechanisms of cooperativity and allosteric regulation in proteins. Q Rev Biophys. 1989 May;22(2):139–237. doi: 10.1017/s0033583500003826. [DOI] [PubMed] [Google Scholar]
- Pfeil W., Privalov P. L. Thermodynamic investigations of proteins. III. Thermodynamic description of lysozyme. Biophys Chem. 1976 Jan;4(1):41–50. doi: 10.1016/0301-4622(76)80005-1. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Gill S. J. Stability of protein structure and hydrophobic interaction. Adv Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Griko YuV, Venyaminov SYu, Kutyshenko V. P. Cold denaturation of myoglobin. J Mol Biol. 1986 Aug 5;190(3):487–498. doi: 10.1016/0022-2836(86)90017-3. [DOI] [PubMed] [Google Scholar]
- Privalov P. L. Stability of proteins. Proteins which do not present a single cooperative system. Adv Protein Chem. 1982;35:1–104. [PubMed] [Google Scholar]
- Privalov P. L. Stability of proteins: small globular proteins. Adv Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- Privalov P. L. Thermodynamic problems of protein structure. Annu Rev Biophys Biophys Chem. 1989;18:47–69. doi: 10.1146/annurev.bb.18.060189.000403. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Tiktopulo E. I., Venyaminov SYu, Griko YuV, Makhatadze G. I., Khechinashvili N. N. Heat capacity and conformation of proteins in the denatured state. J Mol Biol. 1989 Feb 20;205(4):737–750. doi: 10.1016/0022-2836(89)90318-5. [DOI] [PubMed] [Google Scholar]
- Ptitsyn O. B., Pain R. H., Semisotnov G. V., Zerovnik E., Razgulyaev O. I. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 1990 Mar 12;262(1):20–24. doi: 10.1016/0014-5793(90)80143-7. [DOI] [PubMed] [Google Scholar]
- Puett D. The equilibrium unfolding parameters of horse and sperm whale myoglobin. Effects of guanidine hydrochloride, urea, and acid. J Biol Chem. 1973 Jul 10;248(13):4623–4634. [PubMed] [Google Scholar]
- Ramdas L., Nall B. T. Folding/unfolding kinetics of mutant forms of iso-1-cytochrome c with replacement of proline-71. Biochemistry. 1986 Nov 4;25(22):6959–6964. doi: 10.1021/bi00370a033. [DOI] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Unity in function in the absence of consensus in sequence: role of leader peptides in export. Science. 1989 Mar 3;243(4895):1156–1159. doi: 10.1126/science.2646712. [DOI] [PubMed] [Google Scholar]
- Regan L., DeGrado W. F. Characterization of a helical protein designed from first principles. Science. 1988 Aug 19;241(4868):976–978. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- Roder H., Elöve G. A., Englander S. W. Structural characterization of folding intermediates in cytochrome c by H-exchange labelling and proton NMR. Nature. 1988 Oct 20;335(6192):700–704. doi: 10.1038/335700a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs D. H., Schecter A. N., Eastlake A., Anfinsen C. B. Immunological distinction between the possible origins of enzymatic activity in a polypeptide fragment of staphylococcal nuclease. Nature. 1974 Sep 20;251(5472):242–244. doi: 10.1038/251242a0. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. The thermodynamic stability of proteins. Annu Rev Biophys Biophys Chem. 1987;16:115–137. doi: 10.1146/annurev.bb.16.060187.000555. [DOI] [PubMed] [Google Scholar]
- Scheraga H. A. Effect of side chain-backbone electrostatic interactions on the stability of alpha-helices. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5585–5587. doi: 10.1073/pnas.82.17.5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid F. X., Baldwin R. L. Detection of an early intermediate in the folding of ribonuclease A by protection of amide protons against exchange. J Mol Biol. 1979 Nov 25;135(1):199–215. doi: 10.1016/0022-2836(79)90347-4. [DOI] [PubMed] [Google Scholar]
- Schmid F. X., Grafl R., Wrba A., Beintema J. J. Role of proline peptide bond isomerization in unfolding and refolding of ribonuclease. Proc Natl Acad Sci U S A. 1986 Feb;83(4):872–876. doi: 10.1073/pnas.83.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid F. X. Proline isomerization during refolding of ribonuclease A is accelerated by the presence of folding intermediates. FEBS Lett. 1986 Mar 31;198(2):217–220. doi: 10.1016/0014-5793(86)80408-2. [DOI] [PubMed] [Google Scholar]
- Schmid F., Blaschek H. An early intermediate in the folding of ribonuclease A is protected against cleavage by pepsin. Biochemistry. 1984 May 8;23(10):2128–2133. doi: 10.1021/bi00305a004. [DOI] [PubMed] [Google Scholar]
- Schwarz F. P. Interaction of cytidine 3'-monophosphate and uridine 3'-monophosphate with ribonuclease a at the denaturation temperature. Biochemistry. 1988 Nov 1;27(22):8429–8436. doi: 10.1021/bi00422a020. [DOI] [PubMed] [Google Scholar]
- Segawa S., Kume K. Comparison between the unfolding rate and structural fluctuations in native lysozyme--effects of denaturants, ligand binding, and intrachain cross-linking on hydrogen exchange and unfolding kinetics. Biopolymers. 1986 Oct;25(10):1981–1996. doi: 10.1002/bip.360251012. [DOI] [PubMed] [Google Scholar]
- Segawa S., Sugihara M. Characterization of the transition state of lysozyme unfolding. I. Effect of protein-solvent interactions on the transition state. Biopolymers. 1984 Nov;23(11 Pt 2):2473–2488. doi: 10.1002/bip.360231122. [DOI] [PubMed] [Google Scholar]
- Segawa S., Sugihara M. Characterization of the transition state of lysozyme unfolding. II. Effects of the intrachain crosslinking and the inhibitor binding on the transition state. Biopolymers. 1984 Nov;23(11 Pt 2):2489–2498. doi: 10.1002/bip.360231123. [DOI] [PubMed] [Google Scholar]
- Semisotnov G. V., Rodionova N. A., Kutyshenko V. P., Ebert B., Blanck J., Ptitsyn O. B. Sequential mechanism of refolding of carbonic anhydrase B. FEBS Lett. 1987 Nov 16;224(1):9–13. doi: 10.1016/0014-5793(87)80412-x. [DOI] [PubMed] [Google Scholar]
- Serrano L., Fersht A. R. Capping and alpha-helix stability. Nature. 1989 Nov 16;342(6247):296–299. doi: 10.1038/342296a0. [DOI] [PubMed] [Google Scholar]
- Shortle D., Meeker A. K. Residual structure in large fragments of staphylococcal nuclease: effects of amino acid substitutions. Biochemistry. 1989 Feb 7;28(3):936–944. doi: 10.1021/bi00429a003. [DOI] [PubMed] [Google Scholar]
- Shortle D. Probing the determinants of protein folding and stability with amino acid substitutions. J Biol Chem. 1989 Apr 5;264(10):5315–5318. [PubMed] [Google Scholar]
- Siekierka J. J., Hung S. H., Poe M., Lin C. S., Sigal N. H. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature. 1989 Oct 26;341(6244):755–757. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- Silen J. L., Frank D., Fujishige A., Bone R., Agard D. A. Analysis of prepro-alpha-lytic protease expression in Escherichia coli reveals that the pro region is required for activity. J Bacteriol. 1989 Mar;171(3):1320–1325. doi: 10.1128/jb.171.3.1320-1325.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. L., Hendrickson W. A., Honzatko R. B., Sheriff S. Structural heterogeneity in protein crystals. Biochemistry. 1986 Sep 9;25(18):5018–5027. doi: 10.1021/bi00366a008. [DOI] [PubMed] [Google Scholar]
- Spolar R. S., Ha J. H., Record M. T., Jr Hydrophobic effect in protein folding and other noncovalent processes involving proteins. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8382–8385. doi: 10.1073/pnas.86.21.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J. P., Kim P. S. Role of a subdomain in the folding of bovine pancreatic trypsin inhibitor. Nature. 1990 Apr 12;344(6267):685–688. doi: 10.1038/344685a0. [DOI] [PubMed] [Google Scholar]
- States D. J., Creighton T. E., Dobson C. M., Karplus M. Conformations of intermediates in the folding of the pancreatic trypsin inhibitor. J Mol Biol. 1987 Jun 5;195(3):731–739. doi: 10.1016/0022-2836(87)90192-6. [DOI] [PubMed] [Google Scholar]
- Sturtevant J. M. Heat capacity and entropy changes in processes involving proteins. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Hayano T., Suzuki M. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature. 1989 Feb 2;337(6206):473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- Taniuchi H. Formation of randomly paired disulfide bonds in des-(121-124)-ribonuclease after reduction and reoxidation. J Biol Chem. 1970 Oct 25;245(20):5459–5468. [PubMed] [Google Scholar]
- Tweedy N. B., Hurle M. R., Chrunyk B. A., Matthews C. R. Multiple replacements at position 211 in the alpha subunit of tryptophan synthase as a probe of the folding unit association reaction. Biochemistry. 1990 Feb 13;29(6):1539–1545. doi: 10.1021/bi00458a027. [DOI] [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. NMR evidence for an early framework intermediate on the folding pathway of ribonuclease A. Nature. 1988 Oct 20;335(6192):694–699. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]
- Villafane R., King J. Nature and distribution of sites of temperature-sensitive folding mutations in the gene for the P22 tailspike polypeptide chain. J Mol Biol. 1988 Dec 5;204(3):607–619. doi: 10.1016/0022-2836(88)90359-2. [DOI] [PubMed] [Google Scholar]
- Wagner G. Characterization of the distribution of internal motions in the basic pancreatic trypsin inhibitor using a large number of internal NMR probes. Q Rev Biophys. 1983 Feb;16(1):1–57. doi: 10.1017/s0033583500004911. [DOI] [PubMed] [Google Scholar]
- Wearne S. J., Creighton T. E. Further experimental studies of the disulfide folding transition of ribonuclease A. Proteins. 1988;4(4):251–261. doi: 10.1002/prot.340040404. [DOI] [PubMed] [Google Scholar]
- West S. M., Kelly S. M., Price N. C. The unfolding and attempted refolding of citrate synthase from pig heart. Biochim Biophys Acta. 1990 Mar 1;1037(3):332–336. doi: 10.1016/0167-4838(90)90034-d. [DOI] [PubMed] [Google Scholar]
- West S. M., Price N. C. The unfolding and attempted refolding of mitochondrial aspartate aminotransferase from pig heart. Biochem J. 1990 Jan 1;265(1):45–50. doi: 10.1042/bj2650045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetlaufer D. B. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc Natl Acad Sci U S A. 1973 Mar;70(3):697–701. doi: 10.1073/pnas.70.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P. E., Dyson H. J., Lerner R. A. Conformation of peptide fragments of proteins in aqueous solution: implications for initiation of protein folding. Biochemistry. 1988 Sep 20;27(19):7167–7175. doi: 10.1021/bi00419a001. [DOI] [PubMed] [Google Scholar]
- Wüthrich K. Protein structure determination in solution by nuclear magnetic resonance spectroscopy. Science. 1989 Jan 6;243(4887):45–50. doi: 10.1126/science.2911719. [DOI] [PubMed] [Google Scholar]
- Yagisawa S. Two types of rate-determining step in chemical and biochemical processes. Biochem J. 1989 Nov 1;263(3):985–988. doi: 10.1042/bj2630985. [DOI] [PMC free article] [PubMed] [Google Scholar]