Abstract
Human herpesvirus 8 (HHV-8) or Kaposi's sarcoma-associated herpesvirus (KSHV) ORF59 protein (PF-8) is a processivity factor for HHV-8 DNA polymerase (Pol-8) and is homologous to processivity factors expressed by other herpesviruses, such as herpes simplex virus type 1 UL42 and Epstein-Barr virus BMRF1. The interaction of UL42 and BMRF1 with their corresponding DNA polymerases is essential for viral DNA replication and the subsequent production of infectious virus. Using HHV-8-specific monoclonal antibody 11D1, we have previously identified the cDNA encoding PF-8 and showed that it is an early-late gene product localized to HHV-8-infected cell nuclei (S. R. Chan, C. Bloomer, and B. Chandran, Virology 240:118–126, 1998). Here, we have further characterized PF-8. This viral protein was phosphorylated both in vitro and in vivo. PF-8 bound double-stranded DNA (dsDNA) and single-stranded DNA independent of DNA sequence; however, the affinity for dsDNA was approximately fivefold higher. In coimmunoprecipitation reactions, PF-8 also interacted with Pol-8. In in vitro processivity assays with excess poly(dA):oligo(dT) as a template, PF-8 stimulated the production of elongated DNA products by Pol-8 in a dose-dependent manner. Functional domains of PF-8 were determined using PF-8 truncation mutants. The carboxyl-terminal 95 amino acids (aa) of PF-8 were dispensable for all three functions of PF-8: enhancing processivity of Pol-8, binding dsDNA, and binding Pol-8. Residues 10 to 27 and 279 to 301 were identified as regions critical for the processivity function of PF-8. Interestingly, aa 10 to 27 were also essential for binding Pol-8, whereas aa 1 to 62 and aa 279 to 301 were involved in binding dsDNA, suggesting that the processivity function of PF-8 is correlated with both the Pol-8-binding and the dsDNA-binding activities of PF-8.
Kaposi's sarcoma (KS) is a vascular tumor frequently seen in human immunodeficiency virus type 1-infected people, especially homosexual AIDS patients (reviewed in reference 4). The tumor contains both inflammatory and angiogenic components that lead to formation of the signature of KS lesions: slit-like spaces surrounded with spindle cells that are thought to have originated from endothelial cells and monocytes (reviewed in reference 16). Human herpesvirus 8 (HHV-8), also known as KS-associated herpesvirus, is implicated in the pathogenesis of KS (reviewed in references 17 and 50). HHV-8 DNA has been detected in all epidemiological forms of KS (1, 8, 9, 15, 23, 36, 51) and in peripheral blood from patients prior to the onset of KS (1, 27, 37, 57). The lytic cycle of HHV-8 also seems to be important for KS development. Ganciclovir, which inhibits HHV-8 lytic replication in vitro (25, 33), reduces the risk of KS development in AIDS patients (19, 32, 34). Furthermore, high titers of antibodies against HHV-8 lytic antigens in AIDS patients are associated with increased risk for KS (46). Hence, it might be possible to delay the onset of KS with antiviral agents that specifically target the viral lytic cycle. In order to design antiviral drugs that are more specific for the HHV-8 lytic cycle and less toxic, it is essential to elucidate the molecular biology of HHV-8 DNA replication.
To date, little is known about the mechanism of, or the proteins involved in, HHV-8 DNA replication. The most extensively studied herpesvirus in this area is herpes simplex virus type 1 (HSV-1). HSV-1 encodes seven proteins that are required for viral DNA replication and for replication of origin-containing plasmid DNA (30, 49, 56, 58). These proteins include a DNA polymerase (Pol or UL30) (44), a processivity factor (UL42) (31, 41), an origin-binding protein (UL9) (39), a helicase-primase complex (composed of UL5, UL8, and UL52) (13), and a single-stranded DNA (ssDNA)-binding protein (ICP8) (42). UL42 is a processivity factor that enhances the affinity of the HSV-1 Pol for primer-template junctions (20, 21, 55). Hence, it increases the period of time Pol stays on the DNA template, resulting in long-chain DNA synthesis. UL42 is essential for HSV-1 DNA replication since a UL42 temperature-sensitive mutant or a UL42 null mutant is unable to support viral DNA synthesis and the subsequent production of infectious virions (24, 30).
HHV-8 encodes homologs of seven proteins required for DNA replication in other herpesviruses (38, 48). HHV-8 PF-8 (encoded by open reading frame 59 [ORF59]) is homologous to HSV-1 UL42, Epstein-Barr virus (EBV) BMRF1, herpesvirus saimiri ORF59 protein, human cytomegalovirus (HCMV) ICP36, HHV-6 p41, varicella-zoster virus gene 16 protein, and HHV-7 U27 (28). The cDNA encoding HHV-8 ORF59 protein was first identified by monoclonal antibody (MAb) 11D1 generated against body cavity-based B-cell lymphoma cell line BCBL-1 (5). BCBL-1 cells are latently infected with HHV-8, and the viral lytic cycle can be induced by TPA (12-O-tetradecanoylphorbol-13-acetate) (45). We reported that HHV-8 ORF59 encodes an early-late protein which localizes to HHV-8-infected cell nuclei and whose expression is induced by TPA treatment (5). Lin et al. (28) later renamed the ORF59 protein as PF-8 and showed that it interacted with HHV-8 DNA polymerase (Pol-8; encoded by ORF9) in vitro and stimulated DNA synthesis activity of Pol-8 on singly primed M13 template in DNA polymerase assays. They observed that only in the presence of PF-8 was Pol-8 able to synthesize full-length products and concluded that PF-8 is a processivity factor for HHV-8 Pol-8.
In this report, we extend the previous observations (5, 28) and present a further characterization of HHV-8 PF-8. Our studies demonstrate that PF-8 is phosphorylated, possesses double-stranded DNA (dsDNA)-binding activity, and associates with Pol-8 in vitro. These properties, together with the ability of PF-8 to enhance processivity of Pol-8, imply that by binding to dsDNA and Pol-8 at the same time, PF-8 might hold Pol-8 on the DNA template for an extended amount of time, so that Pol-8 can synthesize long stretches of DNA. This hypothesis was examined by elucidating the functional domains of PF-8. PF-8 truncation mutants were generated and assayed for stimulation of processive DNA synthesis by Pol-8, dsDNA-binding, and interaction with Pol-8. Our results show that amino acids (aa) 10 to 27 and 279 to 301 are critical for the processivity function of PF-8. In addition, while residues 10 to 27 of PF-8 are important for interacting with Pol-8, both the N terminus and residues 279 to 301 are required for binding dsDNA. These results demonstrate that the processivity function of PF-8 correlates with the physical interaction between PF-8 and Pol-8 and with that between PF-8 and dsDNA.
MATERIALS AND METHODS
Cells.
HHV-8-negative, EBV-negative BJAB cells and HHV-8-positive, EBV-negative BCBL-1 cells were maintained as previously described (5).
Antibodies.
The development and characterization of MAb 11D1, specific for HHV-8 PF-8, has been previously reported (5).
Plasmid constructs.
ORF59 was amplified from the pCD50 cDNA clone (5) by PCR, using primers 59A, 5′-GTG CGT CTA CGA ATT CAA TCA TGC CTG TGG-3′ (containing an EcoRI site), and 59B, 5′-CTC ACT GTC GCG GCC GCA CAT GGT GTC AAA TC-3′ (containing a NotI site). ORF9 was amplified from BCBL-1 DNA by PCR, using primers 9A, 5′-GCA GCG AAT TCC AGA TCA TGG ATT TTT TCA ATC-3′ (containing an EcoRI site), and 9B, 5′-AAG AGG AAA CTT TGC GGC CGC TGT TTC CG-3′ (containing a NotI site). Amplified fragments were purified and ligated into the EcoRI/NotI sites of pCI-neo (Promega, Madison, Wis.). All inserts were verified by sequencing at the Biotechnology Support Facility, University of Kansas Medical Center.
Construction of PF-8 truncation mutants.
All PF-8 truncation mutants were amplified from the pCD50 cDNA clone (5) by PCR, using the primers indicated below. For the C-terminal truncation mutants, the 5′ primer was the same as that used for amplifying full-length ORF59 (59A; see above). The 3′ primers were as follows: ΔC359–396, 5′-CCT TAG TTG CGG CCG CTG GGG GTC AGC TGG TGA C-3′; ΔC323–396, 5′-CTC CAA TTG CGG CCG CCT CCT TCA GTC CGG TAT AG-3′; ΔC302–396, 5′-TCC CGC GGC CGC CAC AGA TCA GTT TAC C-3′; ΔC279–396, 5′-CAG CCA GCG GCC GCA CCT TCC ACT TAT AAT ATT TCG-3′; ΔC234–396, 5′-GTA TGC ACC GCG GCC GCA TCC ACC TAC TTC TTC C-3′; and ΔC191–396, 5′-CTC GCT GGC GGC CGC AGT CAC CTA TTG GTC C-3′. The 3′ primers contain NotI sites. For the N-terminal truncation mutants, the 3′ primer was the same as that used for amplifying full-length ORF59 (59B; see above). The 5′ primers were as follows: ΔN1–9, 5′-TAT AGA ATT CAC CAT GAG GGT GGA CGT GAC CC-3′; ΔN1–27, 5′-GGG TCA ATG AAT TCA TTA TGA GTG CCA C-3′; ΔN1–62, 5′-GGC GTT CTG GAA TTC AGA ATG AAG AAT GCC C-3′; and ΔN1–127, 5′-CAA CCG GAA TTC GTC ATG ACC ACC ATT TCC-3′. The 5′ primers contain methionine start sites and EcoRI sites. All amplified fragments were ligated into the EcoRI/NotI sites of pCI-neo.
In vitro transcription-translation.
The TNT-coupled reticulocyte lysate system with T7 RNA polymerase (Promega) was used to transcribe and translate the ORF9, full-length ORF59, and ORF59 truncation mutants in the presence of [35S]Met according to the manufacturer's instructions.
λPPase treatment.
[35S]Met-labeled, in vitro translated (IVT) PF-8 polypeptide (20 μl) was incubated with 2,000 U of λ protein phosphatase (λPPase) at 30°C for 3 h according to the manufacturer's recommendation (New England Biolab, Beverly, Mass.). The mock-treated and λPPase-treated IVT PF-8 proteins were boiled with 2× sample buffer and resolved by sodium dodecyl sulfate–9% polyacrylamide gel electrophoresis (SDS–9% PAGE). The gel was amplified using Entensify (NEN, Boston, Mass.), dried, and exposed to Kodak (Rochester, N.Y.) XAR-5 film.
Radioimmunoprecipitation.
BJAB, BCBL-1, and TPA-induced BCBL-1 (20 ng of TPA [Sigma, St. Louis, Mo.] per ml in 10 ml of RPMI medium for 4 days) cells (107 cells each) were labeled with 1.5 mCi of [32P]orthophosphate (Amersham Pharmacia, Piscataway, N.J.) for 20 h. Identical samples were labeled with [35S]Met/Cys as per procedures described elsewhere (5). Lysis of 32P- or 35S-labeled cells and immunoprecipitation with MAb 11D1 was carried out as previously reported (5). The immunoprecipitates were dissolved in 2× sample buffer and resolved by SDS–9% PAGE.
DNA-cellulose chromatography.
DNA-cellulose chromatography was performed as described by Loh et al. (29). Briefly, 20 μl of [35S]Met-labeled IVT PF-8 polypeptide was treated with 2 μl of RNace-It Cocktail (Stratagene, La Jolla, Calif.) for 20 min at room temperature. The treated sample was diluted in 100 μl of binding buffer (10 mM Tris-HCl [pH 7.4], 0.5 mM EDTA, 1 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride) with 50 mM NaCl and centrifuged at 12,000 × g for 15 min at 4°C. The supernatant was then poured over 500 μl of equilibrated unmodified, ssDNA-, or dsDNA-cellulose (Sigma) in Poly-Prep columns (Bio-Rad, Hercules, Calif.). The columns were washed four times with two-bed volumes of binding buffer with 50 mM NaCl. Bound protein was eluted stepwise with 3 two-bed volumes of binding buffer with increasing concentrations of NaCl (0.1, 0.2, 0.3, 0.4, 0.5, 1, and 2 M). Three hundred microliters from the last wash fraction (50 mM NaCl) and from the first fraction of each high salt concentration was incubated with 2 volumes of acetone and 20 μg of bovine serum albumin (BSA) overnight at −20°C. The precipitated proteins were sedimented at 12,000 × g for 30 min at 4°C, solubilized with 35 μl of 2× sample buffer, resolved by SDS-PAGE, and visualized by fluorography. dsDNA-binding activities of PF-8 truncation mutants were examined in an identical manner.
Coimmunoprecipitation of IVT Pol-8 and IVT PF-8.
IVT Pol-8 and IVT PF-8 (20 μl of each) were incubated with 200 μl of MAb 11D1 tissue culture supernatant in the presence of 1 U of benzonase (endonuclease; Sigma) and 200 μl of binding buffer [100 mM (NH4)2SO4, 20 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 0.1 mM EDTA, 0.5 mM dithiothreitol, 4% glycerol, 0.1% NP-40, and 1 mM phenylmethylsulfonyl fluoride) for 2 h at 4°C. Protein A-Sepharose (100 μl; 25% [vol/vol]; Amersham Pharmacia) was then added to the mixture, which was incubated for another 2 h at 4°C. The immunoprecipitates were extensively washed with binding buffer, boiled in 2× sample buffer, and resolved by SDS–10% PAGE. Coimmunoprecipitation of PF-8 truncation mutants with Pol-8 was carried out in an identical manner.
Processivity assay.
Poly(dA):oligo(dT)16 template was prepared following the procedure described by Hamatake et al. (22). Oligo(dT)16 (Amersham Pharmacia) was end labeled with [γ-32P]ATP by T4 polynucleotide kinase according to the manufacturer's recommendation (Promega). Poly(dA) (Amersham Pharmacia) was mixed with labeled oligo(dT)16 at a ratio of 10:1 (wt/wt), which resulted in approximately one primer binding every 200 bp. The mixture was heated at 75°C for 5 min and cooled to room temperature for 30 min. Nonhybridized primers were removed by Micro Bio-Spin P30 chromatography (Bio-Rad). The DNA synthesis procedure was carried out as described before (28), with modifications. Briefly, 2 μl of IVT Pol-8 and 1, 2, or 5 μl of IVT PF-8 were incubated on ice for 15 min. An appropriate amount of TNT programmed with pCI-neo was added to bring the volume up to 7 μl. Eighteen microliters of buffer [100 mM (NH4)2SO4, 20 mM Tris-Cl (pH 7.5), 3 mM MgCl2, 0.1 mM EDTA, 0.5 mM dithiothreitol, 4% glycerol, 40 μg of BSA, 60 μM dTTP, and 1 μg of poly(dA):oligo(dT)16] was added, and reactions were carried out for 1 h at 37°C. To examine the kinetics of the stimulatory effect of PF-8, processivity reactions were set up with 2 μl of IVT Pol-8 and 5 μl of IVT PF-8 and were carried out for 10, 30, or 60 min. All reactions were quenched by the addition of 25 μl of 20 mM EDTA. DNA was extracted by phenol-chloroform followed by chloroform. Purified DNA products were mixed with denaturing PAGE loading dye (90% formamide, 10 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol), boiled for 5 min, and resolved by 7 M urea–15% PAGE.
To test the stability of PF-8 truncation mutants under the processivity assay conditions, reactions were set up as described above except without BSA, dTTP, and DNA template. Proteins were incubated for 1 h at 37°C and then resolved by SDS–12% PAGE.
RESULTS
HHV-8 PF-8 is a phosphoprotein.
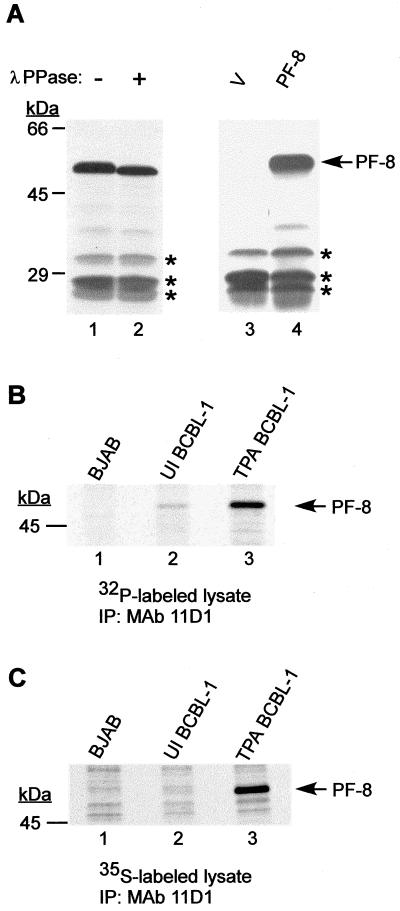
It has been reported that HSV-1 UL42, EBV BMRF1, HCMV ICP36, and HHV-6 p41 are phosphoproteins (7, 10, 18, 31, 47). To determine whether PF-8 (encoded by HHV-8 ORF59) is phosphorylated, [35S]methionine-labeled, IVT PF-8 polypeptide was mock treated, or treated with λPPase, which removes phosphates from serine, threonine, and tyrosine residues. The mock-treated and λPPase-treated samples were resolved by SDS-PAGE (Fig. 1A, lanes 1 and 2). The apparent molecular mass of the mock-treated PF-8 was 50 kDa (Fig. 1A, lane 1). The mobility of the λPPase-treated IVT PF-8 (Fig. 1A, lane 2) was faster than that of the mock-treated sample (Fig. 1A, lane 1), indicating that PF-8 was phosphorylated in vitro. The difference in the sizes of the phosphorylated and dephosphorylated forms of PF-8 was approximately 1 kDa. Since PF-8 was expressed from the PF-8-coding region carried by the pCI-neo expression vector, proteins synthesized due to the presence of the pCI-neo vector itself were considered nonspecific (Fig. 1A, lane 3). The mobilities of these nonspecific proteins did not change upon λPPase treatment (Fig. 1A, lanes 1 and 2), indicating that the shift in migration of PF-8 is due to the removal of phosphates.
FIG. 1.
Phosphorylation of HHV-8 PF-8. (A) IVT PF-8 is phosphorylated. PF-8 was expressed from pCI-neo expression vector with an in vitro transcription-translation (TNT) system in the presence of [35S]methionine (lane 4). TNT lysate programmed with pCI-neo vector (V) is shown in lane 3. [35S]Met-labeled IVT PF-8 was incubated with (lane 2) or without (lane 1) λPPase for 3 h. Samples were resolved by SDS–9% PAGE. Protein products synthesized by TNT due to the presence of the pCI-neo vector itself are indicated by asterisks. (B) PF-8 is phosphorylated in BCBL-1 cells. [32P]orthophosphate-labeled BJAB (lane 1), uninduced BCBL-1 (lane 2), and TPA-induced BCBL-1 (lane 3) lysates were immunoprecipitated with MAb 11D1 and resolved by SDS–9% PAGE. (C) PF-8 is expressed in BCBL-1 cells, but not in BJAB cells. [35S]Met/Cys-labeled BJAB (lane 1), uninduced BCBL-1 (lane 2), and TPA-induced BCBL-1 (lane 3) lysates were immunoprecipitated with MAb 11D1 and resolved by SDS–9% PAGE. The sizes of protein markers are shown to the left of each gel. The position of PF-8 is indicated at right. IP, immunoprecipitation.
To determine whether PF-8 was also phosphorylated in vivo, lysates from BJAB, uninduced, and TPA-induced BCBL-1 cells labeled with [32P]orthophosphate were immunoprecipitated with MAb 11D1, which is specific for HHV-8 PF-8 (5). An isotype control MAb (immunoglobulin G2b) against an HHV-6 glycoprotein did not react with PF-8 (data not shown), further demonstrating the specificity of MAb 11D1. There was no specific reaction between MAb 11D1 and the 32P-labeled HHV-8-negative BJAB cells (Fig. 1B, lane 1). However, MAb 11D1 recognized a phosphoprotein with an apparent molecular mass of 50 kDa from TPA-induced BCBL-1 cells, which is the expected size of PF-8 (Fig. 1B, lane 3). This phosphoprotein was also seen in uninduced BCBL-1 cells, albeit at a much lower level (Fig. 1B, lane 2). These results demonstrate that PF-8 expressed in BCBL-1 cells is phosphorylated.
To prove that the absence of the phosphorylated form of PF-8 in HHV-8-negative BJAB cells was due to the absence of expression of this protein, lysates from [35S]Met/Cys-labeled BJAB, uninduced, and TPA-induced BCBL-1 cells were immunoprecipitated with MAb 11D1. MAb 11D1 precipitated the 50-kDa PF-8 from TPA-induced BCBL-1 cells, but not from BJAB cells (Fig. 1C). The expression of PF-8 was induced by TPA treatment (Fig. 1C, lanes 2 and 3) as previously reported (5). The results of these experiments demonstrate that PF-8 is phosphorylated both in vitro and in vivo.
PF-8 has dsDNA-binding activity.
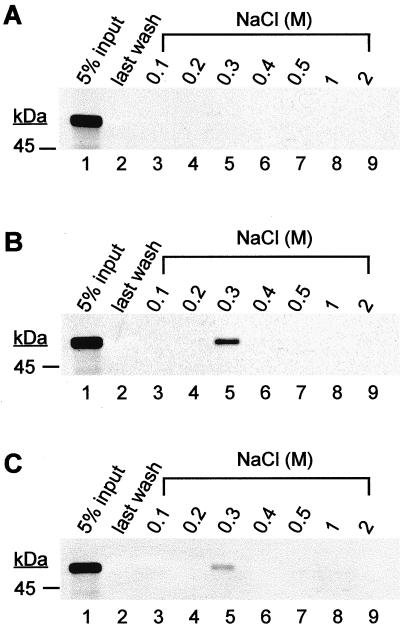
It has been established that HSV-1 UL42 and EBV BMRF1 have intrinsic dsDNA-binding activity (31, 41, 43, 53). To determine whether PF-8, a structural homolog of these viral processivity factors, was also capable of binding dsDNA, DNA-cellulose chromatography was performed. [35S]Met-labeled IVT PF-8 polypeptide was applied to an unmodified, ssDNA-, or dsDNA-cellulose column. The columns were extensively washed with buffer containing 50 mM NaCl. Increasing concentrations of NaCl were used to elute bound protein. Approximately 30% of the first fraction from each NaCl concentration was resolved by SDS-PAGE (Fig. 2). The last wash fraction was also analyzed by SDS-PAGE to ensure that the polypeptides eluted with high NaCl concentrations were not the result of insufficient washings (Fig. 2, lanes 2). As shown in Fig. 2A, PF-8 was not eluted from the unmodified cellulose even at a high salt concentration (2 M NaCl), indicating that there was no background binding between IVT PF-8 and the unmodified cellulose. In contrast, PF-8 was retained by both the dsDNA-cellulose column (Fig. 2B) and the ssDNA-cellulose column (Fig. 2C) at 50 mM NaCl and eluted in the presence of 0.3 M NaCl. The amount of PF-8 eluted from dsDNA was approximately fivefold more than that from ssDNA at 0.3 M NaCl as measured by densitometry (Fig. 2B and C, lane 5), suggesting that PF-8 has a higher affinity for dsDNA than for ssDNA.
FIG. 2.
dsDNA-binding activity of PF-8. IVT radiolabeled PF-8 was applied to 500 μl of equilibrated unmodified cellulose (A), dsDNA-cellulose (B), or ssDNA-cellulose (C) in columns. The columns were extensively washed (lane 2) and eluted stepwise with increasing concentrations of NaCl (0.1, 0.2, 0.3, 0.4, 0.5, 1, and 2 M in binding buffer). Thirty percent of the first fraction from each NaCl concentration was precipitated and resolved by SDS–9% PAGE (lanes 3 to 9). Five percent of column input is shown in lane 1. The sizes of protein markers are shown at left.
Coimmunoprecipitation of HHV-8 DNA polymerase and PF-8 in vitro.
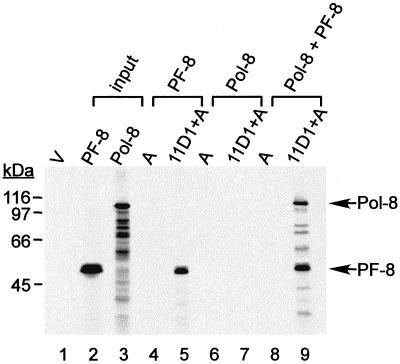
To investigate whether HHV-8 DNA polymerase (Pol-8) (encoded by ORF9) (Fig. 3, lane 3) interacted with PF-8 (Fig. 3, lane 2), we performed coimmunoprecipitation of [35S]Met-labeled IVT Pol-8 and PF-8 using MAb 11D1. Neither IVT protein bound protein A-Sepharose (Fig. 3, lanes 4, 6, and 8). MAb 11D1 precipitated the [35S]Met-labeled IVT PF-8 (Fig. 3, lane 5) but not Pol-8 (Fig. 3, lane 7), indicating that MAb 11D1 did not react with Pol-8 nonspecifically. Pol-8 was precipitated only in the presence of PF-8 (Fig. 3, lane 9), suggesting an association between Pol-8 and PF-8. Since an endonuclease was included in the binding buffer, this interaction between Pol-8 and PF-8 was not mediated by DNA. This result demonstrates that the HHV-8 DNA polymerase associates with PF-8 in vitro, independent of other viral proteins.
FIG. 3.
Coimmunoprecipitation of IVT Pol-8 and IVT PF-8 by MAb 11D1. Fifty percent PF-8 and 10% of Pol-8 used in the coimmunoprecipitation reactions are shown in lanes 2 and 3, respectively. PF-8 alone (lanes 4 and 5), Pol-8 alone (lanes 6 and 7), or PF-8 plus Pol-8 (lanes 8 and 9) was incubated with protein A-Sepharose alone (A) (lanes 4, 6, and 8) or with protein A-Sepharose and MAb 11D1 (lanes 5, 7, and 9). The immune complexes were extensively washed and resolved by SDS–10% PAGE. The sizes of protein markers are shown at left. The positions of Pol-8 and PF-8 are indicated at right. V, TNT lysate programmed with pCI-neo vector.
PF-8 increases processivity of Pol-8.
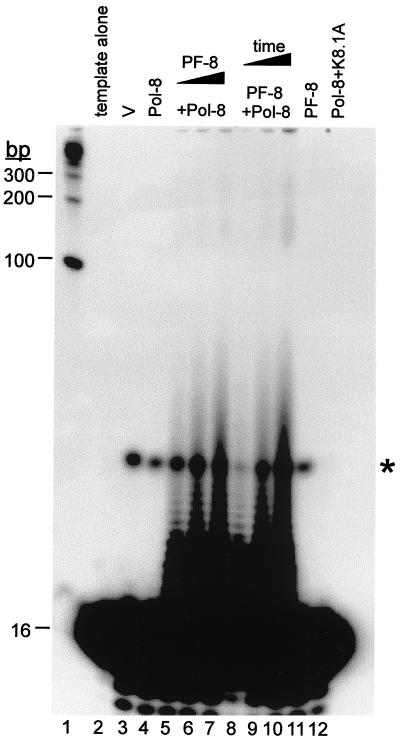
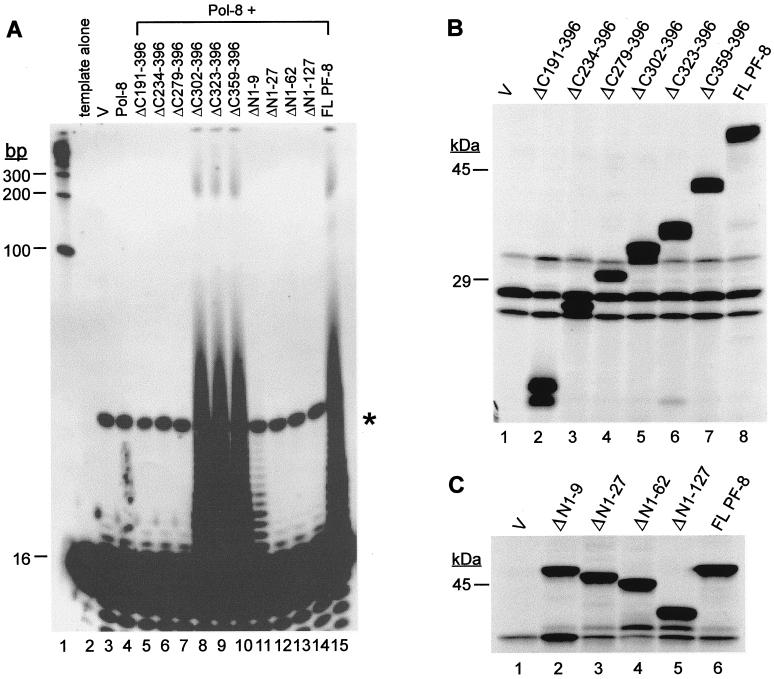
We next examined the effect of PF-8 on the processivity of HHV-8 Pol-8 on poly(dA):oligo(dT)16, a homopolymeric template. This template was used since, unlike the primed M13 template, it does not form any secondary structure or contain any sequence-specific high energy barrier (3, 54), so that the rate of production of elongated DNA is not affected by these pause sites. Oligo(dT)16 primer end labeled with [γ-32P]ATP was hybridized with poly(dA) template with an average length of 246 bases. Excess poly(dA):oligo(dT)16 was used to ensure that any Pol-8–PF-8 complex dissociated from a primer-template would not interact with the same primer-template again (3, 54). This is a rigorous assay for processivity since any DNA products synthesized result from a single processive cycle, but not from distributive elongation (3, 54). The DNA products were resolved by 15% denaturing PAGE (Fig. 4). An excess amount of nonelongated primers (16 bases in length) from the primed templates were seen near the bottom of the gel (Fig. 4), verifying that the extended primers were limited to one round of processive synthesis. Since the primer was end labeled, the intensity of each band is proportional to the number of elongated primer molecules with that particular DNA size. The observed bands correspond to the length of DNA that was synthesized prior to the dissociation of Pol-8 from the template.
FIG. 4.
Stimulation of the processivity of Pol-8 by PF-8 on poly(dA):oligo(dT)16 template. TNT lysate programmed with pCI-neo vector (V) (lane 3), Pol-8 (lane 4), Pol-8 plus increasing amounts of PF-8 (lanes 5 to 7), PF-8 (lane 11), or Pol-8 plus glycoprotein K8.1A (lane 12) were assayed for DNA synthesis on poly(dA):oligo(dT)16. Pol-8 and PF-8 were incubated with poly(dA):oligo(dT)16 for 10, 30, or 60 min (lanes 8, 9, or 10, respectively). DNA products were fractionated by 7 M urea–15% PAGE. Poly(dA):oligo(dT)16 template was electrophoresed in lane 2 to show the size of the primers (16 bases). Nonspecific DNA products synthesized by pCI-neo-programmed TNT lysate are indicated by an asterisk. The sizes of DNA markers (lane 1) are shown at left.
Pol-8 or PF-8 alone did not elongate any primer (Fig. 4, lanes 4 and 11, respectively). However, in the presence of Pol-8, increasing amounts of PF-8 enhanced the processivity of Pol-8 in a dose-dependent manner, indicated by the increasing quantities of labeled elongated DNA products and the appearance of DNA products greater than 200 bases long (Fig. 4, lanes 5 to 7). With constant amounts of Pol-8 and PF-8, the stimulation in processivity by PF-8 was also time dependent (Fig. 4, lanes 8 to 10). HHV-8 envelope glycoprotein K8.1A (6), used as a negative control, had no effect on the processivity of Pol-8 (Fig. 4, lane 12), demonstrating the specificity of the functional interaction between Pol-8 and PF-8. These results confirm that PF-8 is a processivity factor for Pol-8 in a more rigorous processivity assay than the DNA synthesis protocol utilized by Lin et al. (28).
Mapping the processivity domain of PF-8.
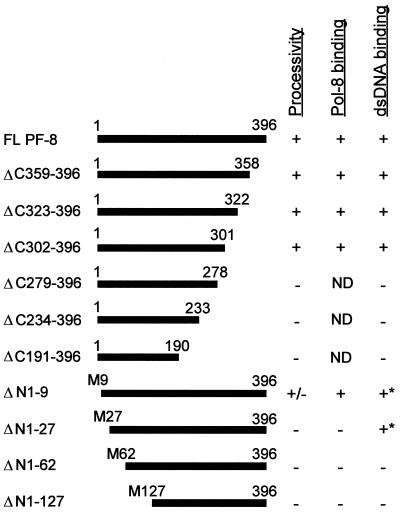
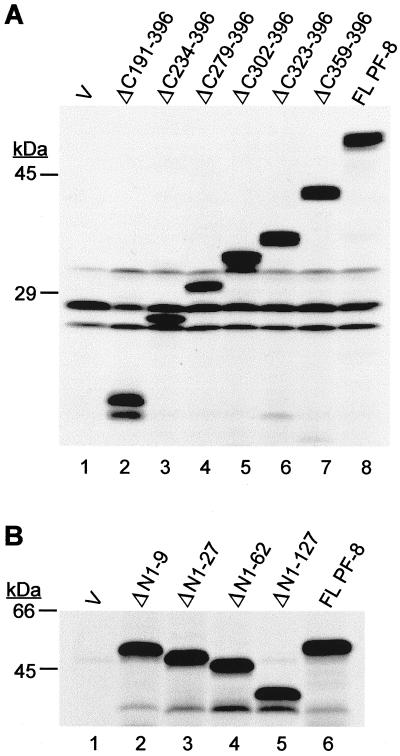
To examine the region(s) of PF-8 that is important for processivity function, PF-8 truncation mutants were generated. A schematic diagram of each mutant is shown in Fig. 5. PF-8 mutants were expressed by the in vitro transcription-translation system and fractionated by SDS-PAGE (Fig. 6). All mutants were resolved at their expected sizes and expressed at similar levels. PF-8 mutants were then tested for their abilities to stimulate processive DNA synthesis by Pol-8 on poly(dA):oligo(dT)16 template (Fig. 7A). C-terminal PF-8 mutants ΔC302–396, ΔC323–396, and ΔC359–396 were still functional in the processivity assay (Fig. 7A, lanes 8 to 10), indicating that aa 302 to 396 are dispensable for this function. In contrast, deletion of aa 279 to 396 totally abolished the processivity activity (Fig. 7A, lane 7), demonstrating the importance of aa 279 to 301 of PF-8 for processivity function. N-terminal mutant ΔN1–9 was not as efficient as the full-length PF-8 in enhancing the processivity of Pol-8 (Fig. 7A, lane 11), whereas mutants ΔN1–27, ΔN1–62, and ΔN1–127 were nonfunctional (Fig. 7A, lanes 12 to 14), suggesting that the N-terminal portion of PF-8 (aa 10 to 27) is essential. To ensure that the inability of some of the PF-8 mutants to function in the processivity assays was not due to degradation of the mutants, the integrity of the PF-8 mutants was monitored by SDS-PAGE over the course of the processivity assays. The result shows that all PF-8 truncation mutants were stable throughout the assay period (Fig. 7B and 7C).
FIG. 5.
Schematic representation and summary of properties of full-length PF-8 and PF-8 truncation mutants. Ability of full-length (FL) PF-8 and PF-8 mutants to enhance Pol-8 processivity and to interact with Pol-8 and dsDNA (as assessed in Fig. 7A, Fig. 8, and Fig. 9) are scored as follows: positive (+), negative (−), or weak (+/−). For dsDNA-binding activity, +∗ indicates an altered elution profile of the polypeptide from the dsDNA-cellulose column. Abbreviations: M, Met; ND, not determined.
FIG. 6.
Expression of IVT PF-8 truncation mutants. TNT lysate programmed with pCI-neo vector (V) (lane 1), C-terminal truncation mutants (panel A, lanes 2 to 7), N-terminal truncation mutants (panel B, lanes 2 to 5), and full-length PF-8 (panel A, lane 8, and panel B, lane 6) were resolved by SDS–12% PAGE. The apparent molecular masses of the following PF-8 mutants are as indicated: ΔC191–396, 20 kDa; ΔC234–396, 25 kDa; ΔC279–396, 30 kDa; ΔC302–396, 32 kDa; ΔC323–396, 34 kDa; ΔC359–396, 38 kDa; ΔN1–9, 49 kDa; ΔN1–27, 47 kDa; ΔN1–62, 45 kDa; ΔN1–127, 38 kDa. The sizes of protein markers are shown at left. FL, full-length.
FIG. 7.
Effects of PF-8 truncation mutants on the processivity of Pol-8. (A) TNT lysate programmed with pCI-neo vector (V) (lane 3), Pol-8 (lane 4), Pol-8 plus PF-8 mutants (lanes 5 to 14), or Pol-8 plus full-length PF-8 (lane 15) was incubated with components of the processivity assay for 1 h and DNA products were resolved by 7 M urea–15% PAGE. Poly(dA):oligo(dT)16 template alone is in lane 2. The sizes of DNA markers (lane 1) are shown at left. Nonspecific DNA products are indicated by an asterisk. (B and C) PF-8 mutants are stable under the processivity assay conditions. To monitor protein stability under assay conditions, processivity assays were carried out without the addition of BSA, dTTP, and template, and proteins were resolved by SDS–12% PAGE. (B) C-terminal truncation mutants. (C) N-terminal truncation mutants. The sizes of protein markers are shown at left.
The results of these experiments are summarized in Fig. 5; aa 10 to 27 and 279 to 301 of PF-8 are critical for its processivity function, but the C-terminal 95 aa (from residues 302 to 396) are dispensable.
Mapping the dsDNA-binding site on PF-8.
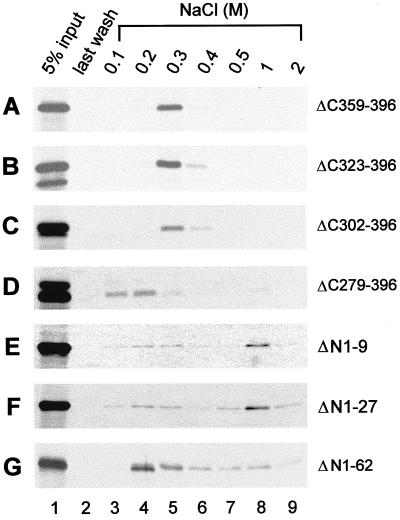
To map the region of PF-8 necessary for binding to dsDNA, the ability of the PF-8 truncation mutants to bind dsDNA-cellulose columns was examined. As shown in Fig. 8A to C, three C-terminal truncation mutants (ΔC359–396, ΔC323–396, and ΔC302–396) behaved like the full-length PF-8 (Fig. 2B), in that the 35S-labeled polypeptides were retained in the columns at 50 mM NaCl and eluted upon the addition of 0.3 M NaCl. Further deletion of the C terminus dramatically affected the elution profile of the polypeptide, such that most of the mutant ΔC279–396 polypeptides were eluted with 0.1 and 0.2 M NaCl (Fig. 8D). This result indicated that C-terminal truncation mutant ΔC279–396 had less affinity for dsDNA than the full-length PF-8, suggesting that residues 279 to 301 contribute to the dsDNA-binding activity of PF-8. Larger C-terminal deletions (mutants ΔC234–396 and ΔC191–396) also displayed weak binding to dsDNA, similar to the behavior of mutant ΔC279–396 (summarized in Fig. 5).
FIG. 8.
dsDNA-binding activities of PF-8 truncation mutants. [35S]Met-labeled IVT PF-8 mutants were applied to dsDNA-cellulose columns. DNA-cellulose chromatography was carried out as described in the legend to Fig. 2.
Deletion of the first 27 residues at the N terminus of PF-8 led to an alteration of elution profile. Mutants ΔN1–9 and ΔN1–27 were retained by the columns at 50 mM NaCl (Fig. 8E and F), but one population of the proteins was eluted from 0.1 to 0.3 M NaCl and one was eluted at 1 M NaCl. This biphasic property of fractionation suggested that even a small deletion of the N terminus of PF-8 was deleterious to the dsDNA-binding capacity of PF-8. Further truncation of the N terminus (ΔN1–62 and ΔN1–127) rendered PF-8 incapable of binding dsDNA with as high affinity as the full-length PF-8, since a majority of the proteins eluted at 0.2 M NaCl (Fig. 8G and Fig. 5).
The results of these tests are summarized in Fig. 5. The C-terminal 95 aa of PF-8 are dispensable for dsDNA-binding. However, residues 279 to 301 and the first 62 aa of the N terminus are critical for PF-8 binding to dsDNA.
Mapping the Pol-8-binding domain of PF-8.
Since the physical interaction with Pol-8 might also contribute to the processivity function of PF-8, we next examined whether the PF-8 truncation mutants associated with Pol-8 in vitro. We first determined the region of PF-8 recognized by MAb 11D1 by immunoprecipitating the 35S-labeled IVT PF-8 truncation mutants. All N-terminal truncation mutants could be recognized by MAb 11D1 (Fig. 9, lanes 7 to 10). In contrast, mutant ΔC302–396 was immunoprecipitated by MAb 11D1, but mutant ΔC279–396 was not (Fig. 9, lanes 3 and 4), demonstrating that deletion of aa 279 through 301 completely abolished the ability of MAb 11D1 to recognize PF-8. This suggests that either the epitope recognized by MAb 11D1 lies between aa 279 and aa 301 or the deletion of these 23 aa disrupts the conformation of PF-8 such that the epitope is no longer available.
FIG. 9.
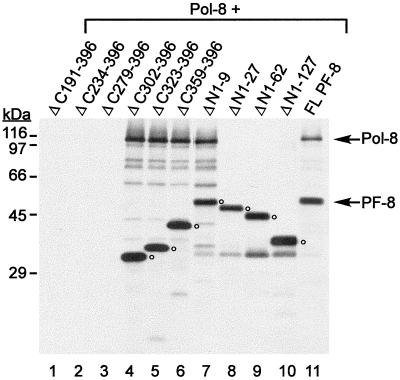
Coimmunoprecipitation of IVT Pol-8 and IVT PF-8 mutants by MAb 11D1. IVT C-terminal or N-terminal PF-8 truncation mutants were mixed with IVT Pol-8 and incubated with MAb 11D1 and protein A-Sepharose. The immune complexes were washed and resolved by SDS–11% PAGE. The sizes of protein markers are shown at left. The positions of Pol-8 and full-length PF-8 are indicated by arrows, and the positions of PF-8 mutants are indicated by open circles.
To map the region of PF-8 that interacts with Pol-8, coimmunoprecipitation of [35S]Met-labeled IVT Pol-8 and IVT PF-8 mutants with MAb 11D1 was carried out. Since mutants ΔC191–396, ΔC234–396, and ΔC279–396 could not be recognized by MAb 11D1 (Fig. 9, lanes 1 to 3), we are currently developing other methods to examine their abilities to physically interact with Pol-8. While ΔC302–396, ΔC323–396, ΔC359–396, and ΔN1–9 coprecipitated with Pol-8 (Fig. 9, lanes 4 to 7), ΔN1–27, ΔN1–62, and ΔN1–127 did not (Fig. 9, lanes 8 to 10), indicating that aa 10 to 27 of PF-8 are required for Pol-8-binding. In addition, these results confirm that MAb 11D1 does not bind Pol-8 nonspecifically, since MAb 11D1 did not precipitate Pol-8 in the presence of several of the PF-8 mutants (Fig. 9, lanes 1 to 3 and lanes 8 to 10). The results of the coimmunoprecipitation reactions are summarized in Fig. 5.
DISCUSSION
In this report, we have characterized HHV-8 PF-8 as a functional processivity factor and mapped regions of PF-8 important for activity. We observed that the 50-kDa PF-8 (encoded by ORF59) is phosphorylated in vitro and in vivo and that it binds strongly to dsDNA. PF-8 also coprecipitated with HHV-8 Pol-8 in vitro, confirming the work of Lin et al. (28). Using a stringent assay for processivity function, we showed that PF-8 enhances the processivity of Pol-8. These results not only demonstrate that PF-8 is a processivity factor, but also suggest a potential mechanism for its activity, enhancement of Pol-8 binding to primer-template junctions (see below). Additionally, we mapped the functional domains of PF-8 that are critical for enhancing the processivity of Pol-8, binding to dsDNA, or binding to Pol-8. Our results demonstrate that two regions of PF-8 are essential for processivity function, aa 10 to 27 and aa 279 to 301, and that these same regions are also important for PF-8 interaction with dsDNA and Pol-8.
PF-8 was phosphorylated both in an in vitro transcription-translation system and in BCBL-1 cells. The difference in the sizes of the phosphorylated and dephosphorylated forms of the IVT PF-8 was approximately 1 kDa, which is approximately equivalent to 13 phosphates (2). A search for common phosphorylation motifs using Motif Finder revealed that there are one potential cyclic AMP and cyclic GMP-dependent kinase phosphorylation site, six potential casein kinase II phosphorylation sites, and six potential protein kinase C phosphorylation sites in PF-8. This observation corresponds well with data demonstrating serine or threonine phosphorylation on other herpesviral polymerase accessory proteins, such as UL42 (HSV-1), BMRF1 (EBV), p41 (HHV-6), and ICP36 (HCMV) (7, 10, 18, 31, 47). A recent report shows that EBV BGLF4, a herpesvirus kinase, phosphorylates BMRF1 in vitro (12). Since HHV-8 ORF36 is homologous to EBV BGLF4 and the protein encoded by ORF36 has serine kinase activity (40), it will be interesting to examine whether the HHV-8 ORF36 protein can phosphorylate PF-8. However, in the in vitro transcription-translation system we used to express PF-8, PF-8 was phosphorylated not by viral protein, but by host protein kinase(s). The functional significance and in vivo regulation of the phosphorylation of these viral DNA polymerase accessory proteins are not known at present.
The dsDNA-binding activity of PF-8 was demonstrated using dsDNA-cellulose chromatography. Radiolabeled PF-8 was retained on a dsDNA column at low ionic strength and was displaced by increased salt concentration. The affinity of PF-8 for ssDNA was approximately fivefold less than that for dsDNA, consistent with the behavior of HSV-1 UL42 (55) and EBV BMRF1 (53). It has been proposed (20, 55) that this property of viral processivity factors may contribute to the enhanced specific binding of the viral polymerases toward primer-template DNA at the site of the growing 3′-OH end of newly synthesized DNA.
Our coimmunoprecipitation results demonstrated that PF-8 interacted with Pol-8 in the absence of other HHV-8 proteins and that this interaction was not mediated by DNA. The results also suggested that the PF-8 epitope recognized by MAb 11D1 is not on the Pol-8-binding interface, since MAb 11D1 did not interfere with the ability of the two proteins to interact. However, deletion of residues 279 to 301 of PF-8 abolished both the reactivity to MAb 11D1 and the processivity function. Further analysis is necessary to determine why MAb 11D1 does not inhibit the physical interaction between Pol-8 and PF-8 but requires a region within the processivity domain of PF-8 for reactivity.
In a more rigorous assay for processivity, PF-8 stimulated long-chain DNA synthesis by Pol-8 using poly(dA):oligo(dT)16 as a template. The use of excess poly(dA):oligo(dT)16 template ensured that the Pol-8–PF-8 complex did not associate with the same primer-template more than once so that single processive reactions were monitored (3, 54). This system is more rigorous because it minimizes production of elongated DNA products by distributive elongation—a process in which the polymerase dissociates and associates with the same primer-template repeatedly and hence does not depend on processivity of the polymerase.
Lin et al. (28) demonstrated physical and functional interactions between Pol-8 and PF-8 and enhancement of Pol-8 replicative activity by PF-8. Our results from the DNA chromatography assays, coimmunoprecipitation reactions, and processivity assays clearly demonstrate that PF-8 possesses properties common to other herpesviral processivity factors. In addition, our work suggests that the physical interaction between PF-8 and dsDNA could contribute to the stimulation of processivity of Pol-8 by increasing the affinity of Pol-8 for the primer terminus on the viral DNA template. Therefore, like HSV-1 UL42 (21), PF-8 might tether its polymerase (Pol-8) to the DNA primer terminus so that the polymerase can incorporate more nucleotides onto the growing chain of DNA without dissociating from the template.
To further examine this hypothesis, we generated PF-8 truncation mutants and tested their abilities to confer processivity. Our results demonstrated that aa 10 to 27 and aa 279 to 301 were critical for the processivity function of PF-8. This observation corresponds well with the predicted structure of the processivity domain of PF-8 (59). Zuccola et al. (59) compared the processivity domain of HSV-1 UL42 with other herpesvirus DNA polymerase accessory proteins using fold recognition methods and found that aa 10 to 300 of PF-8 could form a structure resembling the processivity domain of HSV-1 UL42. Our biochemical studies, which demonstrated the importance of aa 10 to 27 and aa 279 to 301 for PF-8 processivity function, support the algorithmic prediction of structural similarity by Zuccola et al. (59).
To correlate the processivity function of PF-8 with dsDNA- and Pol-8-binding activities, we also assayed PF-8 truncation mutants in DNA-cellulose chromatography assays and coimmunoprecipitation reactions with Pol-8. The C-terminal 95 aa were dispensable in all three functional assays (summarized in Fig. 5). Deletion of residues 279 to 301 dramatically rendered the polypeptide incapable of binding dsDNA. Since it was not possible to examine the physical interaction of mutants ΔC191–396, ΔC234–396, or ΔC279–396 with Pol-8 in the present system due to the absence of a MAb 11D1 epitope, the effect of deleting residues 279 to 301 on Pol-8-binding is not known at present. Nonetheless, the inability of mutant ΔC279–396 to bind dsDNA might contribute at least partially to the lack of processivity function of this mutant.
Although aa 1 to 9 of PF-8 are not included in the predicted processivity fold (59), we observed that deletion of these 9 aa dramatically reduced PF-8 processivity function, implying that this region might help to stabilize the processivity domain. Since mutant ΔN1–9 could precipitate Pol-8, and thus had an intact Pol-8-binding site, it is unlikely that this mutant polypeptide was globally misfolded. Interestingly, mutant ΔN1–9 had a biphasic dsDNA-binding property in that one population of the polypeptides had a higher affinity for dsDNA and the other had a lower affinity compared to the full-length PF-8. It is possible that this altered affinity for dsDNA affects the mobility of ΔN1–9 on DNA and thus reduces its processivity function. Deletion of aa 1 to 27 of PF-8 completely disrupted the physical association with Pol-8 and its processivity function. Its DNA-binding activity was also affected, indicated by the biphasic fractionation profile from the dsDNA-cellulose column. Since both mutant ΔN1–9 and ΔN1–27 had this altered elution profile, but only ΔN1–27 was nonfunctional in the processivity assay and incapable of binding Pol-8, it appears that the interaction between Pol-8 and PF-8 is critical for the processivity function of PF-8.
In summary, HHV-8 PF-8 is a phosphoprotein that binds dsDNA and Pol-8 in vitro. It enhances the processivity of Pol-8, and this function is closely associated with the ability to physically interact with Pol-8 and with dsDNA. This is consistent with the idea that both Pol-8-binding and dsDNA-binding is necessary for PF-8 to function as a processivity factor. The C-terminal region of PF-8 is dispensable for dsDNA binding, Pol-8 binding, and processivity of PF-8. These characteristics of PF-8 resemble those of HSV-1 UL42 (14, 22, 35, 52) and EBV BMRF1 (11, 26) despite only a 21.8 and a 28.5% aa identity to UL42 and BMRF1, respectively (28). Genetic and biochemical evidence collected to date suggests that there is a structural conservation among the herpesviral processivity factors without a high degree of amino acid sequence identity. Further understanding of the interaction between Pol-8 and PF-8 and the biology of HHV-8 DNA replication might contribute to the design of antiviral agents that could delay KS development.
ACKNOWLEDGMENTS
We thank Clark Bloomer at the Biotechnology Support Facility in KUMC for DNA sequencing.
This work was supported by Public Health Service grants CA75911 and CA82056 to B.C. and by a University of Kansas Medical Center Biomedical Research Training Program predoctoral fellowship to S.R.C.
REFERENCES
- 1.Ambroziak J A, Blackbourn D J, Herndier B G, Glogau R G, Gullett J H, McDonald A R, Lennette E T, Levy J A. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science. 1995;268:582–583. doi: 10.1126/science.7725108. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Bambara R A, Fay P J, Mallaber L M. Methods of analyzing processivity. Methods Enzymol. 1995;262:270–280. doi: 10.1016/0076-6879(95)62023-0. [DOI] [PubMed] [Google Scholar]
- 4.Beral V. Epidemiology of Kaposi's sarcoma. Cancer Surv. 1991;10:5–22. [PubMed] [Google Scholar]
- 5.Chan S R, Bloomer C, Chandran B. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology. 1998;240:118–126. doi: 10.1006/viro.1997.8911. [DOI] [PubMed] [Google Scholar]
- 6.Chandran B, Bloomer C, Chan S R, Zhu L, Goldstein E, Horvat R. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology. 1998;249:140–149. doi: 10.1006/viro.1998.9316. [DOI] [PubMed] [Google Scholar]
- 7.Chang C K, Balachandran N. Identification, characterization, and sequence analysis of a cDNA encoding a phosphoprotein of human herpesvirus 6. J Virol. 1991;65:2884–2894. doi: 10.1128/jvi.65.6.2884-2894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y, Ziegler J, Wabinga H, Katangole-Mbidde E, Boshoff C, Schulz T, Whitby D, Maddalena D, Jaffe H W, Weiss R A, Moore P S Uganda Kaposi's Sarcoma Study Group. Kaposi's sarcoma-associated herpesvirus and Kaposi's sarcoma in Africa. Arch Intern Med. 1996;156:202–204. [PubMed] [Google Scholar]
- 10.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, III C A H, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 11.Chen L-W, Lin L-S, Chang Y-S, Liu S-T. Functional analysis of EA-D of Epstein-Barr virus. Virology. 1995;211:593–597. doi: 10.1006/viro.1995.1443. [DOI] [PubMed] [Google Scholar]
- 12.Chen M-R, Chang S-J, Huang H, Chen J-Y. A protein kinase activity associated with Epstein-Barr virus BGLF4 phosphorylates the viral early antigen EA-D in vitro. J Virol. 2000;74:3093–3104. doi: 10.1128/jvi.74.7.3093-3104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crute J J, Tsurumi T, Zhu L, Weller S K, Olivo P D, Challberg M D. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc Natl Acad Sci USA. 1989;86:2186–2189. doi: 10.1073/pnas.86.7.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Digard P, Chow C S, Pirrit L, Coen D M. Functional analysis of the herpes simplex virus UL42 protein. J Virol. 1993;67:1159–1168. doi: 10.1128/jvi.67.3.1159-1168.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupin N, Grandadam M, Calvez V, Gorin I, Aubin J T, Havard S, Lamy F, Leibowitch M, Huraux J M, Escande J P, Agut H. Herpesvirus-like DNA sequences in patients with Mediterranean Kaposi's sarcoma. Lancet. 1995;345:761–762. doi: 10.1016/s0140-6736(95)90642-8. [DOI] [PubMed] [Google Scholar]
- 16.Ensoli B, Sturzl M. Kaposi's sarcoma: a result of the interplay among inflammatory cytokines, angiogenic factors and viral agents. Cytokine Growth Factor Rev. 1998;9:63–83. doi: 10.1016/s1359-6101(97)00037-3. [DOI] [PubMed] [Google Scholar]
- 17.Ganem D. KSHV and Kaposi's sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 18.Gibson W, Murphy T L, Roby C. Cytomegalovirus-infected cells contain a DNA-binding protein. Virology. 1981;111:251–262. doi: 10.1016/0042-6822(81)90669-3. [DOI] [PubMed] [Google Scholar]
- 19.Glesby M J, Hoover D R, Weng S, Graham N M H, Phair J P, Detels R, Ho M, Saah A J. Use of antiherpes drugs and the risk of Kaposi's sarcoma: data from the multicenter AIDS cohort study. J Infect Dis. 1996;173:1477–1480. doi: 10.1093/infdis/173.6.1477. [DOI] [PubMed] [Google Scholar]
- 20.Gottlieb J, Challberg M D. Interaction of herpes simplex virus type 1 DNA polymerase and the UL42 accessory protein with a model primer template. J Virol. 1994;68:4937–4945. doi: 10.1128/jvi.68.8.4937-4945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb J, Marcy A I, Coen D M, Challberg M D. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J Virol. 1990;64:5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamatake R K, Bifano M, Tenney D J, Hurlburt W W, Cordingley M G. The herpes simplex virus type 1 DNA polymerase accessory protein, UL42, contains a functional protease-resistant domain. J Gen Virol. 1993;74:2181–2189. doi: 10.1099/0022-1317-74-10-2181. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y Q, Li J J, Kaplan M H, Poiesz B, Katabira E, Zhang W C, Feiner D, Friedman-Kien A E. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet. 1995;345:759–761. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- 24.Johnson P A, Best M G, Friedmann T, Parris D S. Isolation of a herpes simplex virus type 1 mutant deleted for the essential UL42 gene and characterization of its null phenotype. J Virol. 1991;65:700–710. doi: 10.1128/jvi.65.2.700-710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kedes D H, Ganem D. Sensitivity of Kaposi's sarcoma-associated herpesvirus replication to antiviral drugs. Implications for potential therapy. J Clin Investig. 1997;99:2082–2086. doi: 10.1172/JCI119380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiehl A, Dorsky D I. Bipartite DNA-binding region of the Epstein-Barr virus BMRF1 product essential for DNA polymerase accessory function. J Virol. 1995;69:1669–1677. doi: 10.1128/jvi.69.3.1669-1677.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefrere J J, Meyohas M C, Mariotti M, Meynard J L, Thauvin M, Frotier J. Detection of human herpesvirus 8 DNA sequences before the appearance of Kaposi's sarcoma in human immunodeficiency virus (HIV)-positive subjects with a known date of HIV seroconversion. J Infect Dis. 1996;174:283–287. doi: 10.1093/infdis/174.2.283. [DOI] [PubMed] [Google Scholar]
- 28.Lin K, Dai C Y, Ricciardi R P. Cloning and functional analysis of Kaposi's sarcoma-associated herpesvirus DNA polymerase and its processivity factor. J Virol. 1998;72:6228–6232. doi: 10.1128/jvi.72.7.6228-6232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh L C, Britt W J, Raggo C, Laferte S. Sequence analysis and expression of the murine cytomegalovirus phosphoprotein pp50, a homolog of the human cytomegalovirus UL44 gene product. Virology. 1994;200:413–427. doi: 10.1006/viro.1994.1205. [DOI] [PubMed] [Google Scholar]
- 30.Marchetti M E, Smith C E, Schaffer P A. A temperature-sensitive mutation in a herpes simplex virus type 1 gene required for viral DNA synthesis maps to coordinate 0.609 through 0.614 in UL. J Virol. 1988;62:715–721. doi: 10.1128/jvi.62.3.715-721.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsden H S, Campbell M E M, Haarr L, Frame M C, Parris D S, Murphy M, Hope R G, Muller M T, Preston C M. The 65,000-Mr DNA-binding and virion trans-inducing proteins of herpes simplex virus type 1. J Virol. 1987;61:2428–2437. doi: 10.1128/jvi.61.8.2428-2437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin D F, Kuppermann B D, Wolitz R A, Palestine A G, Li H, Robinson C A the Roche Ganciclovir Study Group. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. N Engl J Med. 1999;340:1063–1070. doi: 10.1056/NEJM199904083401402. [DOI] [PubMed] [Google Scholar]
- 33.Medveczky M M, Horvath E, Lund T, Medveczky P G. In vitro antiviral drug sensitivity of the Kaposi's sarcoma-associated herpesvirus. AIDS. 1997;11:1327–1332. doi: 10.1097/00002030-199711000-00006. [DOI] [PubMed] [Google Scholar]
- 34.Mocroft A, Youle M, Gazzard B, Morcinek J, Halai R, Phillips A N. Anti-herpesvirus treatment and risk of Kaposi's sarcoma in HIV infection. AIDS. 1996;10:1101–1105. [PubMed] [Google Scholar]
- 35.Monahan S J, Barlam T F, Crumpacker C S, Parris D S. Two regions of the herpes simplex virus type 1 UL42 protein are required for its functional interaction with the viral DNA polymerase. J Virol. 1993;67:5922–5931. doi: 10.1128/jvi.67.10.5922-5931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore P S, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and those without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 37.Moore P S, Kingsley L A, Holmberg S D, Spira T, Gupta P, Hoover D R, Parry J P, Conley L J, Jaffe H W, Chang Y. Kaposi's sarcoma-associated herpesvirus infection prior to onset of Kaposi's sarcoma. AIDS. 1996;10:175–180. doi: 10.1097/00002030-199602000-00007. [DOI] [PubMed] [Google Scholar]
- 38.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olivo P D, Nelson N J, Challberg M D. Herpes simplex virus DNA replication: the UL9 gene encodes an origin binding protein. Proc Natl Acad Sci USA. 1988;85:5414–5418. doi: 10.1073/pnas.85.15.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J, Lee D, Seo T, Chung J, Choe J. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) open reading frame 36 protein is a serine protein kinase. J Gen Virol. 2000;81:1067–1071. doi: 10.1099/0022-1317-81-4-1067. [DOI] [PubMed] [Google Scholar]
- 41.Parris D S, Cross A, Haarr L, Orr A, Frame M C, Murphy M, McGeoch D J, Marsden H S. Identification of the gene encoding the 65-kilodalton DNA-binding protein of herpes simplex virus type 1. J Virol. 1988;62:818–825. doi: 10.1128/jvi.62.3.818-825.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powell K L, Littler E, Purifoy D J M. Nonstructural proteins of herpes simplex virus. II. Major virus-specific DNA-binding protein. J Virol. 1981;39:894–902. doi: 10.1128/jvi.39.3.894-902.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powell K L, Purifoy D J M. DNA-binding proteins of cells infected with herpes simplex virus type 1 and type 2. Intervirology. 1976;7:225–239. doi: 10.1159/000149955. [DOI] [PubMed] [Google Scholar]
- 44.Purifoy D J M, Lewis R B, Powell K L. Identification of the herpes simplex virus DNA polymerase gene. Nature. 1977;269:621–623. doi: 10.1038/269621a0. [DOI] [PubMed] [Google Scholar]
- 45.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 46.Rezza G, Andreoni M, Dorrucci M, Pezzotti P, Monini P, Zerboni R, Salassa B, Colangeli V, Sarmati L, Nicastri E, Barbanera M, Pristera R, Aiuti F, Ortona L, Ensoli B. Human herpesvirus 8 seropositivity and risk of Kaposi's sarcoma and other acquired immunodeficiency syndrome-related diseases. J Natl Cancer Inst. 1999;91:1468–1474. doi: 10.1093/jnci/91.17.1468. [DOI] [PubMed] [Google Scholar]
- 47.Roeckel D, Mueller-Lantzsch N. Biochemical characterization of two Epstein-Barr virus early antigen-associated phosphopolypeptides. Virology. 1985;147:253–263. doi: 10.1016/0042-6822(85)90128-x. [DOI] [PubMed] [Google Scholar]
- 48.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi's sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaffer P A, Aron G M, Biswal N, Benyesh-Melnick M. Temperature-sensitive mutants of herpes simplex virus type 1: isolation, complementation, and partial characterization. Virology. 1973;52:57–71. doi: 10.1016/0042-6822(73)90398-x. [DOI] [PubMed] [Google Scholar]
- 50.Schulz T F. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 51.Smith M S, Bloomer C, Horvat R, Goldstein E, Casparian M, Chandran B. Detection of human herpesvirus-8 DNA in Kaposi's sarcoma lesions and peripheral blood of HIV+ patients and correlation with serological measurements. J Infect Dis. 1997;176:84–93. doi: 10.1086/514043. [DOI] [PubMed] [Google Scholar]
- 52.Tenney D J, Hurlburt W W, Bifano M, Stevens J T, Micheletti P A, Hamatake R K, Cordingley M G. Deletions of the carboxy terminus of herpes simplex virus type 1 UL42 define a conserved amino-terminal functional domain. J Virol. 1993;67:1959–1966. doi: 10.1128/jvi.67.4.1959-1966.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsurumi T. Purification and characterization of the DNA-binding activity of the Epstein-Barr virus DNA polymerase accessory protein BMRF1 gene products, as expressed in insect cells by using the baculovirus system. J Virol. 1993;67:1681–1687. doi: 10.1128/jvi.67.3.1681-1687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.VonHippel P H, Fairfield F R, Dolejsi M K. On the processivity of polymerase. Ann NY Acad Sci. 1994;726:118–131. doi: 10.1111/j.1749-6632.1994.tb52803.x. [DOI] [PubMed] [Google Scholar]
- 55.Weisshart K, Chow C S, Coen D M. Herpes simplex virus processivity factor UL42 imparts increased DNA-binding specificity to the viral DNA polymerase and decreased dissociation from primer-template without reducing the elongation rate. J Virol. 1999;73:55–66. doi: 10.1128/jvi.73.1.55-66.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weller S K, Aschman D P, Sacks W R, Coen D M, Schaffer P A. Genetic analysis of temperature-sensitive mutants of HSV-1; the combined use of complementation and physical mapping for cistron assignment. Virology. 1983;130:290–305. doi: 10.1016/0042-6822(83)90084-3. [DOI] [PubMed] [Google Scholar]
- 57.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatzioannou T, Suggett F E A, Aldam D M, Denton A S, Miller R F, Weller I V D, Weiss R A, Tedder R S, Schulz T F. Detection of Kaposi's sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 58.Wu C A, Nelson N J, McGeoch D J, Challberg M D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zuccola H J, Filman D J, Coen D M, Hogle J M. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol Cell. 2000;5:267–278. doi: 10.1016/s1097-2765(00)80422-0. [DOI] [PubMed] [Google Scholar]