Abstract
Transcription factors (TFs) form homo- or hetero-dimeric DNA binding complexes along with associated co-regulators that can have transcriptional repressor or activator functions. Defining the specific composition of the complexes is therefore key to understanding their biological role. Here, we utilized bimolecular fluorescence complementation (BiFC) to visualize the formation of defined TF dimers and associated co-regulators derived from the activator protein-1 (AP-1) and myocyte enhancer factor 2 (MEF2) families. Firstly, BiFC signals were observed in cells co-expressing TFs tagged with complimentary combinations of the split fluorescent protein, demonstrating the engineered formation of defined dimer complexes. Next, we applied this approach and determined that defined AP-1 dimers localized at discrete sub-nuclear locations. Subsequently, a combination of BiFC coupled with GFP binding peptide (GBP)-nanotrap allowed observation of protein-protein interactions between a co-regulator, HDAC4, and defined BiFC-MEF2 engineered dimers. To determine transactivation properties of defined TF dimers in a cellular system, the Gal4-DNA binding domain fused to GBP was utilized to assess the transcriptional properties of the BiFC-TF dimers using a generically applicable Gal4/UAS luciferase reporter gene assay system. Here, we report efficacy of a BiFC/GBP-nanobody approach that allows engineering, visualization, and functional analysis of defined TF dimers.
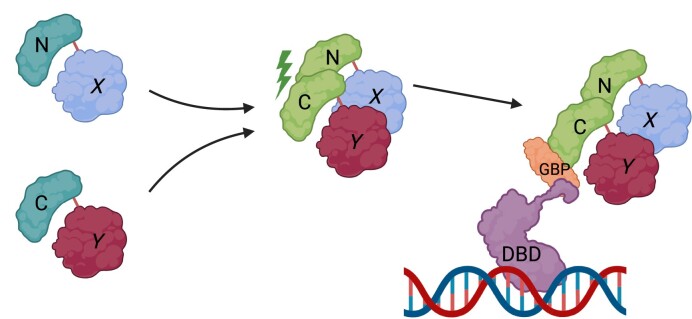
Graphical Abstract
Graphical Abstract.
Introduction
Tagging proteins of interest with fluorescent probes has revolutionized experimental approaches in molecular cell biology research. Attaching a green fluorescent protein (GFP) tag, for example, to a protein of interest has become a standard technique to determine protein sub-cellular localization and trafficking in a live cell context. Fluorescence tagging in combination with continually developing fluorescence microscopy technologies is thus a major tool in protein characterization (1,2). The expanding array of fluorescent proteins for tagging has allowed a multiplex approach to visualizing multiple proteins simultaneously within the cell (3). Since most if not all cellular activities require assembly of protein complexes, elucidating the function of protein-protein interactions (PPIs) is an important parameter for analyzing the biological role and regulation of PPIs (4). Although many different molecular tools have been developed, dissecting the function of specific protein: protein subunit interactions in defined dimer combinations or in the context of larger protein complexes still remains intractable. One strategy to achieve the goal of unambiguously assessing protein-protein interactions has been the implementation of bi-molecular complementation to engineer specific protein interactions. The principle of which is based on reconstitution of a split functional domain in which each half is attached to proteins of interest (4–7). Upon protein-protein interaction (PPI) of the tagged proteins, the separated N-terminal and C-terminal halves of the split domain are brought into register allowing reconstitution of function (6,8). Detection of the PPI occurring between the bimolecular fluorescence complementation (BiFC) tagged proteins of interest by visualizing and measuring the activity of the reconstituted functional domain thus allows a definitive readout of specific PPIs (9).
Dimer formation is a particularly common and important feature of many sequence-specific DNA binding transcription factors (TFs). Homo- and hetero-dimer formation of TF subunits has evolved to determine DNA binding specificity, signal pathway responsiveness and repressor or activator function, making it a crucial regulatory step in the control of gene expression (10). Thus, regulated dimer pairing of different TF combinations and their interaction with co-regulators is a critical determinant of the transcriptome and ultimately the generation of specialized cell types and adaptation to the cellular environment. It is therefore imperative to develop cell biological tools that allow visualization and functional analysis of defined TF PPIs in a cellular context.
In this study, we have utilized the previously described bimolecular fluorescence complementation (BiFC) technique (4,5,11) to engineer and visualize specific TF dimer combinations derived from the myocyte enhancer factor 2 (MEF2) TF, activator protein-1 (AP-1) TF family, and WW domain-containing transcriptional regulator protein-1 (WWTR1/TAZ). Interrogating the function of specific dimer pairs has been elusive because of the complexity and potential number of homo- and heterodimeric subunit combinations. We therefore reasoned that engineering defined dimer combinations using BiFC would allow us to unambiguously visualize specific TF dimer combinations at sub-nuclear compartments and also assess their properties in functional assay systems. Also, we document that reconstituted BiFC proteins are efficiently recognized by a GFP Binding Peptide nanobody (GBP). GBP is a recombinant single chain nanobody from Llama. Since nanobodies are small single chain antibodies (12–15 kDa), they can be efficiently expressed in cells and also used to capture reconstituted GFP moieties by biochemical affinity capture (12,13). This feature allowed us the utility of developing antibody-based protein dimer analysis in the context of living cells. Since GBP nanobodies recognize reconstituted BiFC, GFP derived proteins, a combination of the two techniques (BiFC coupled with GBP-nanotrap technology) allowed us to test the recruitment of a putative transcriptional co-regulator to specific dimer pairs visualized by BiFC using live cell imaging. In addition, we have further developed the system to determine the functional transactivation potential of an engineered dimer pair by integrating the use of the heterologous Gal4/UAS luciferase reporter gene system (14) in which the addition of a Gal4–DNA binding domain (Gal4–DBD) fused to GBP was utilized (Gal4–DBD–GBP) (13). This approach thus allows the recruitment of specific BiFC tagged TF dimers to the transcriptional machinery in the cell with a quantitative reporter gene readout allowing the transactivation properties of specific dimer combinations to be analyzed. Here, we report the efficacy of combining BiFC and nanobody technologies to characterize protein interactions.
Materials and methods
Cell culture
C2C12 myoblasts (CRL-1772), C3H10T1/2 (CCL-226) and HEK293T (CRL-1573) cells were obtained from the American Type Culture Collection. Cells were kept in growth media (GM) consisting of Dulbecco's modified Eagle's medium (DMEM: D6429, MilliporeSigma) and supplemented with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (516106, MilliporeSigma) in a moisturized incubator (Model 3120, ThermoFisher) at 5% CO2 and 37°C.
Antibodies
α HA (12CA5) antibody was obtained from Developmental Studies Hybridoma Bank (The University of Iowa, Department of Biology, Iowa City). Alexa Fluor 488 conjugated secondary antibodies for immunofluorescence analysis were obtained from ThermoFisher (goat α-rabbit AF-546).
Immunofluorescence analysis
C2C12 cells on polymer-coated glass-bottom dishes (#81158, ibidi) were fixed with 4% paraformaldehyde (PFA) in phosphate buffer saline (PBS: P3813, MilliporeSigma) on ice for 5 mins and for 10 mins at room temperature. The cells were then washed 3X with PBS and permeabilized with ice-cold 90% Me-OH for 1 min on ice. Cells were washed again 3× with PBS and incubated with blocking buffer (5% FBS in PBS) for 2 h at room temperature, and then incubated with the indicated primary antibodies in blocking buffer overnight at 4°C. After washed 3× with PBS, cells were incubated with α-rabbit AF-546 secondary antibody in the blocking buffer at room temperature for 1.5 h. The cells were washed 3× with 1XPBS and counter stained with Hoechst 33342 (B2261-25MG, MilliporeSigma) and subjected to confocal fluorescent imaging with a Zeiss Observer Z1 microscope equipped with a Yokogawa CSU-X1 spinning disk equipped with 100X objective lens (α Plan-APOCHROMAT 100X NA1.46 Oil DIC(UV) VIS-IR, 420792–9800, Zeiss). Images were recorded by AxioCam MRm camera (Zeiss) or Evolve camera (electron-multiplying CCD camera: Teledyne Photometrics) and processed using Zen 2.5 (blue edition) software (Zeiss).
Live-cell imaging
Cells on polymer-coated glass-bottom dishes (#81158, Ibidi) were transfected with indicated expression plasmids. Next day, the cell culture media was replaced with FluoBrite (A1896701, ThermoFisher) supplemented with 10%FBS (HyClone). The cells were maintained in a moisturized environmental chamber (5% CO2) at 37°C on the confocal fluorescent microscope (Zeiss Observer Z1) during imaging. Images were recorded by Evolve camera (electron-multiplying CCD camera: Teledyne Photometrics) or AxioCam MRm camera (Zeiss) and processed using Zen 2.5 (blue edition) software (Zeiss).
Transfections
Cells on polymer-coated glass-bottom dishes (#81158, Ibidi) were transfected with a mixture of Lipofectamine 2000 (#11668027, ThermoFisher) and the indicated mammalian expression plasmids (5–10 ng) diluted with Opti-MEM (#31985062, ThermoFisher) according to the manufacturer's protocol. Next day, transfected cells were transferred to FluoroBright DMEM (A189701 ThermoFisher) with 10%FBS for live-cell imaging.
For gene-reporter luciferase assays, 20 000–40 000 cells per well were seeded onto 12-well plates (CLS3516 Millipore Sigma) and transfected the next day after replenishment with fresh GM. 10ng of the appropriate firefly luciferase reporter, 10ng of the mammalian expression vector constructs and 3 ng of no-promoter renilla luciferase vector (E2271 Promega) were mixed in Opti-MEM, and the mixture was combined with polyethylenimine (PEI, Linear (MW 25000) (#23966 Polysciences), 1 μg/ml in milliQ water) at 1:3 (mass:mass) ratio diluted with Opti-MEM, After 15 min incubation at room temp, the mixture was added to the cell culture media and incubated overnight. Next day, the cells were transferred to fresh GM and harvested for the assay.
Reporter gene assay
After recovery in fresh GM, transfected cells were washed 3× with ice-cold 1XPBS and detached from the plate in 200 μl per well of luciferase lysis buffer (50 mM Tris–HCl at pH 7.4 with 1% Triton X-100) at 4°C with intense rocking for 30 min. Detached cells were collected into eppendorf tubes and further lysed by vigorous vortexing at 4°C for 15 min. Cellular debris was pelleted by centrifugation at 15 000 rpm at 4°C for 10 min and supernatant was subjected to luciferase gene reporter assay. Firefly and renilla luciferase activity was measured by a dual injection system luminometer (Berthold) with Firefly luciferase substrate (E1501 Promega) and Renilla luciferase substrate (E2820 Promega). Firefly luciferase activity values were normalized by Renilla luciferase activity values to account for transfection efficiency. The average of the normalized luciferase values in triplicate was calculated and fold-activation in relation to the control values was calculated and graphed. Error bars represent standard deviation (Stdev) of the triplicates.
Plasmids
The BiFC-HA-NV vector was constructed of the mVenus (1–155)-linker (GGGSX3) coding region which was PCR-amplified with primers (forward: agcaagcttgatttaggtgacactatagaatac, reverse: tagaattcgcttcccccgccgcttccgcctccgcttcctc) using pCS2plus-mVenus 1–155-I152L-GGGS as template (a kind gift from Marco Morsch (Addgene #162613)) (15). The resultant PCR-product was inserted at BamHI/EcoRI site of pcDNA3-HA vector previously described (16). The BiFC-HA-CC vector was constructed of the mCerulean (156–239)-linker (GGGSX3) coding region which was PCR-amplified with primers (forward: agcaagcttgatttaggtgacactatagaatac, reverse: tagaattcgcttcccccgccgcttccgcctccgcttcctc) using pCS2plus-mCerulean 156–239-GGGS (a kind gift from Marco Morsch (Addgene #162616)) (15) as template. The resultant PCR-product was inserted at BamHI/EcoRI sites of pcDNA3-HA vector previously described (16). MEF2A, MEF2D, c-Jun, and Fra2 ORFs were inserted into each of the BiFC-HA-NV or BiFC-HA-CC vectors at EcoRI/XhoI sites for BiFC-HA-NV- or BiFC-HA-CC-tagging. GBP-LaminB1 expression vector was described previously (17). pCAG-GBP1-10gly-Gal4DBD was a kind gift from Connie Cepko (Addgene #49438) (18).
Results
Determination of sub-nuclear localization of a specific protein dimer visualized by bimolecular fluorescence complementation (BiFC) tagging in live cells
One major advantage of the BiFC approach compared to conventional fluorescence protein tagging is that exclusive visualization of the intended protein-protein interaction (PPI) is possible (11). A large number of sequence specific DNA binding transcription factors (sTFs) form dimers, and homo- or hetero-dimer formation is often an essential and regulated property of their DNA binding site specificity and, thus, transactivation potential (10). We sought to utilize this method to definitively characterize the function of a defined dimer pair among the AP-1 family members. Conventional fluorescent tagging does not allow unambiguous dimer characterization due to the myriad possible dimer combinations between the protein products encoded by the 7 genes. In contrast, BiFC signals only occur when the fluorescence signal is reconstituted by the 2 separate GFP derivative proteins’ parts being brought together by the particular protein:protein interaction under study (6).
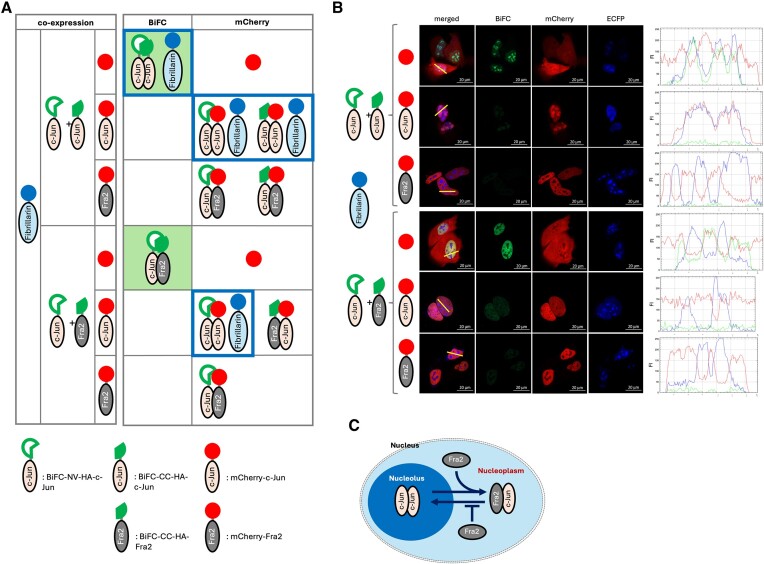
Previously, we reported that Jun proteins have nucleolar localization signal (NoLS), and ectopically expressed Jun proteins (c-Jun, JunB, and JunD) localized at the nucleolus (19). In contrast, Fra2 does not have NoLS, and ectopically expressed Fra2 exclusively localized at the nucleoplasm. Since Fra2 does not form homodimer but heterodimer with Jun proteins (20), we suggested that Fra2 heterodimerizes with c-Jun and regulate sub-nuclear localization of AP-1 complex (19). In this study, we took advantage of using BiFC system, by which we selectively visualized a specific dimer combination of AP-1 complex and eliminated fluorescent signals from unintended AP-1 dimers if tagged with conventional fluorescence proteins.
First, to test the efficacy of the BiFC system to detect AP-1 dimer formation, we fused the BiFC component tags to c-Jun and Fra2 (Supplementary Figure S1). In addition, mCherry or mCherry tagged c-Jun or Fra2 was included for a competitive dimer partner to demonstrate the specificity (Figure 1A). In agreement with previous observations, in the cells expressing BiFC tagged c-Jun, BiFC signals were observed indicating the localization of c-Jun/c-Jun homodimers in the nucleolus (co-localized with a nucleolar marker protein, Fibrillarin, tagged with ECFP) (Figure 1B and extended data in Supplementary Figure S2A and B). Co-expressed mCherry diffused across the whole cell but was predominantly excluded from the nucleolus (see fluorescence signal scanning graph). When mCherry-c-Jun was co-expressed with BiFC-c-Jun it resulted in diminished BiFC signal intensity and mCherry signals were observed in the nucleolus indicating mCherry-c-Jun formed dimers with BiFC tagged c-Jun in a competitive manner and resulted in reduction of the BiFC signal generation. In addition, importantly, the resultant dimer localized to the nucleolus. Therefore, regardless of the attached tag, c-Jun/c-Jun dimers largely localized within the nucleolus. As expected, when we co-expressed mCherry-Fra2, which forms a more stable heterodimer with c-Jun than c-Jun/c-Jun homodimers (20), with BiFC-c-Jun, the BiFC signal was strongly decreased indicative of mCherry-Fra2/BiFC-c-Jun heterodimerization, and mCherry signals were observed in the nucleoplasm but not in the nucleolus (see the graph for fluorescence signal intensity in which blue peaks (Fibrillarin) and red peaks (Fra2/c-Jun) were mutually exclusive).
Figure 1.
Visualization of different sub-nuclear localization of a specific AP-1 dimer pair. (A) A schematic of the experimental design to visualize different sub-nuclear localization of c-Jun/c-Jun homodimer (nucleolus) and c-Jun/Fra2 heterodimer (nucleoplasm). (B) C2C12 cells were transfected with indicated constructs and subjected to live cell confocal fluorescence microscopy analysis. The BiFC signal (green) was indicative of formation of AP-1 dimer pairing between BiFC tagged c-Jun or Fra2. CFP signal marks the nucleolus by fusing to the nucleolar protein, Fibrillarin. Red signal depicts the cellular localization of mCherry, mCherry-c-Jun, or mCherry-Fra2 which competes with BiFC tagged c-Jun or Fra2 for AP-1 dimer formation. Line scan analysis along with the yellow line on the merged micrograph indicates sub-nuclear localization (nucleolus or nucleoplasm) of a specific AP-1 dimer. (C) A schematic of the suggested model for different sub-nuclear localization of a specific dimer pairing such as c-Jun/c-Jun homodimer (nucleolus) or c-Jun/Fra2 heterodimer (nucleoplasm), in which Fra2 dictates localization of c-Jun by formation of the AP-1 dimer with c-Jun.
Next, c-Jun and Fra2 were BiFC tagged and co-expressed, and the reconstituted BiFC signal was indicative of c-Jun/Fra2 heterodimer formation in the nucleoplasm. In the nuclei, the BiFC signal (green) was colocalized with mCherry (red) and excluded from the nucleolus marked by Fibrillarin (blue peak in the graph). When mCherry-c-Jun was co-expressed, BiFC signals were mostly diminished and mCherry signals were observed broadly in the nucleoplasm with some in the nucleolus reflecting the previously known fact that the c-Jun/c-Jun homodimer is less stable than the c-Jun/Fra2 heterodimer (20). Finally, when mCherry-Fra2 was co-expressed, the BiFC signals were essentially eradicated, and mCherry signals were clearly excluded from the nucleolus demonstrating that mCherry-Fra2 heterodimerized with BiFC-c-Jun (Fra-2 does not homodimerize) and is localized in the nucleoplasm but not the nucleolus. Altogether, these data demonstrate c-Jun/c-Jun homodimer localization in the nucleolus and c-Jun/Fra2 heterodimer localization in the nucleoplasm, supporting our previous report that Fra2 regulates c-Jun sub-nuclear localization ((19) and Figure 1C). Thus, determination of the sub-nuclear localization of a specific protein dimer combination mediated by BiFC tagging is much more defined and unambiguous compared to conventional fluorescence protein tagging and co-localization analysis as we reported previously (19).
The authors want to clarify that the signal strength and frequency of complementation due to specific protein:protein interactions in most cases that we have documented far outweighs any BiFC signal from BiFC-NV/BiFC-CC components alone, even for the weakest interactions. However, some low-level complementation of BiFC-NV and BiFC-CC components can be detected, especially at long confocal exposure times, thus necessitating appropriate controls to be included in any BiFC based experimentation.
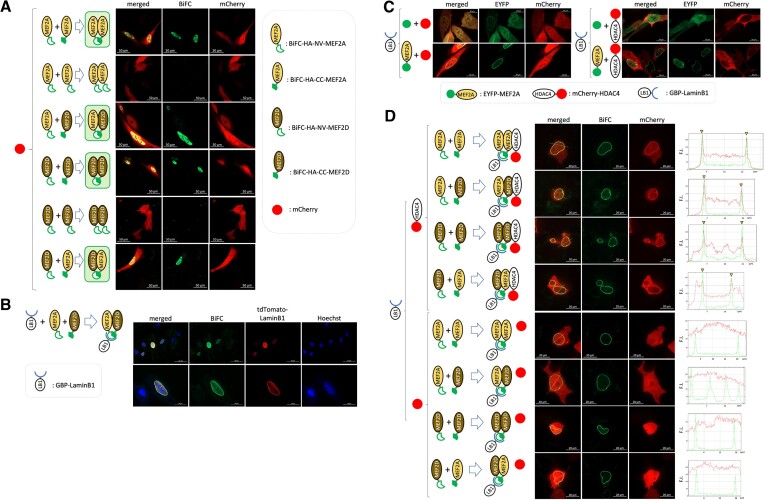
To further test the efficacy of the BiFC system to detect PPI, we next fused the BiFC component tags to the transcriptional regulators MEF2A and MEF2D (Supplementary Figures S3 and S4, Figure 2A extended data in Supplementary Figure S5) to monitor homo- and heterodimer formation in live cells. When transfected, BiFC signals were only observed in the nucleoplasm of the transfected (mCherry+) cells co-expressing a complementary combination of the BiFC-HA-NV and BiFC-HA-CC tagged MEF2A/D homodimers or heterodimers. Of note, we observe different nuclear distributions of the different MEF2 dimers (e.g. MEF2A dimers form condensates in some of the cells). We attribute this heterotypic pattern to be related to the cell state which determines both post-translational modifications (PTMs) of MEF2 and also protein interactions which collectively alter the valency and therefore the phase separation properties and subcellular localization. Further analysis of this point is ongoing.
Figure 2.
Visualization of a specific dimer pairing by BiFC technique and 3-way PPI using BiFC and GBP-nanotrap techniques. (A) C2C12 cells were transfected with a combination of indicated expression constructs in the left side panel. mCherry was included for monitoring transfection. Transfected cells were subjected to live cell imaging by confocal fluorescence microscopy. BiFC signal (green) generation due to complementation and mCherry (red) were depicted in represented micrographs. (B) HEK293T cells were transfected with GBP-LaminB1 (GBP-LB1), BiFC-NV-HA-MEF2A, and BiFC-CC-HA-MEF2D. Transfected cells were subjected to live cell imaging by confocal fluorescence microscopy technique. The nuclei (blue) were stained with Hoechst 33342. (C) C2C12 cells were transfected with a combination of the indicated expression constructs in the left side panel. Expression of EYFP or EYFP-MEF2A (green) and mCherry or mCherry-HDAC4 (red) was visualized by live cell confocal fluorescence microscopy. (D) C2C12 cells were transfected with a combination of the indicated expression constructs in the left side panel. Generation of BiFC signal (green) due to complementation of BiFC tags indicates a specific MEF2 dimer formation. Co-localization of BiFC signal and mCherry-HDAC4 but not mCherry was depicted by line scan analysis of the intensity of the fluorescence signal along the yellow line in the merged micrographs. Signal intensity peak matches are indicated by yellow triangle.
(Note: Expression of BiFC-HA fusion proteins was confirmed by immunofluorescence analysis on fixed transfected cells (Supplementary Figure S4)).
Nanobody recognition of reconstituted BiFC tagged protein dimers
Since some transcriptional co-regulators interact with specific TF dimer pairs, a specific dimer can differentially regulate transcription of its target genes through dimer dependent recruitment of different co-regulators (21). Therefore, detection of PPIs with a specific dimer combination is an important step to dissect the function of TFs. To determine whether we could visualize such a 3-way interaction, we used a combination of the GBP-nanotrap (22) and BiFC techniques. We have previously used the GBP-nanotrap to visualize PPIs (17,23). In brief, we engineer a GBP-nanotrap at a specific sub-cellular site by fusing GBP to an ‘anchor’ protein. Using this approach, we can then localize GFP-fusion protein (bait) at the trap site through the high affinity GBP-GFP interaction and then test if the putative interacting protein (prey) fused to mCherry, is enriched at the trap site by PPI between the bait and prey. We reasoned that this approach would then allow us to enrich a particular dimer combination at a defined sub-cellular site such as the nuclear lamina (e.g. reconstituted GFP captured at the lamina by GBP-lamin-B1) and then assess the recruitment of a co-regulator tagged with a different fluorescent moiety (e.g. mCherry).
We first tested whether GBP, a recombinant Llama single chain antibody, can recognize the complemented BiFC in live cells. We fused GBP to Lamin-B1 (LB1) (LB1-GBP), a component of the fibrous layer of the inner nuclear membrane (24), and co-expressed BiFC-HA-NV-MEF2A and BiFC-HA-CC-MEF2D (Figure 2B extended data in Supplementary Figure S6). Proper localization of LB1-GBP was monitored by co-expression of tdTomato-Lamin-B1. As predicted, the reconstituted BiFC signal, indicating the formation of a MEF2A/MEF2D heterodimer, was observed at the nuclear envelope and not in the nucleoplasm, where MEF2 proteins are normally localized (Supplementary Figure S7), indicating the robust nature of the protein interaction and the recruitment mechanism to GBP. Figure 2B documents BiFC-MEF2A/D dimers captured by the LB1-GBP anchored at the nuclear envelope. Therefore, GBP recognises reconstituted BiFC indicating the formation of binary PPI between the BiFC tagged proteins, in this case formation of the MEF2A/D dimer in live cells. Since defined dimer pairs are exclusively anchored to a defined subcellular location (specified here by the LB1-GBP trap), this allowed us to unambiguously detect/visualize a PPI between a specific MEF2 dimer pair along with a known co-regulator in live cells.
Protein–protein interaction of a defined protein dimer with a co-regulator can be visualized using combined BiFC/GBP-nanotrap technology
Previously, we documented PPIs in live cells using a GBP-nanotrap combined with fluorescent tagging and microscopy techniques (17,23). For, example, GFP tagged protein (bait) with co-expression of LB1-GBP and mCherry tagged putative interacting protein (prey) was successful in detecting the bait-prey interaction by means of an enrichment of mCherry tagged protein at the GBP-nanotrap anchored site. When GFP-MEF2A proteins were trapped by the GBP anchored at the periphery of the nucleus by LB1-GBP, mCherry-tagged HDAC4, but not mCherry proteins alone, were enriched at the periphery of the nucleus (Figure 2C also Supplementary Figure S7) indicating HDAC4 interaction with GFP-MEF2A captured by LB1-GBP (Figure 2C). Due to the nature of transient transfections, the expression levels of exogenous proteins are not perfectly controlled at a 1:1 ratio. Therefore, in some cases, an excess of one protein, such as unbound HDAC4 with MEF2 in our experiments, can result in some protein being localized at native sites as well as at the trap site (in this case the nuclear membrane). Therefore, the exogenous expression levels can affect the localization of the recruited protein to some degree. However, it is apparent in our data that HDAC4 did not localize at the nuclear membrane trap without the BiFC tagged MEF2 protein being appropriately trapped at the nuclear membrane by lamin B1-GBP (Supplementary Figure S7). In view of that, the trap at the nuclear membrane (in this case mediated by Lamin B1-GBP) still allows unambiguous documentation of protein interactions with the GBP trapped protein complex, even if there are other localization sites of exogenously transfected proteins within the cell. This also demonstrates that some empirical determination of the stoichiometry of the exogenously expressed component proteins can be fine-tuned for greater imaging resolution. Some nuclei expressing the GBP trap components can, in some cases, exhibit examples of atypical nuclear shape changes (Figure 2D and extended data in Supplementary Figure S8). However, extensive previous visualization of nuclei and nucleoli in these cell populations at high magnification does indicate that these shape changes typically reflect natural variations and features of these cells. However, it is also possible that exogenous expression of transcriptional regulators might result in nuclear shape changes.
Although this is an effective method to detect the HDAC4/MEF2A interaction, there is some ambiguity since we could not eliminate the possibility that heterologous dimer partners, for example endogenous MEF2D, are contributing to the HDAC4 recruitment in this assay. We used the interaction with HDAC4 primarily as a proof of principle since its interaction with MEF2 has been well documented in the literature (25). Therefore, to circumvent the ambiguity of endogenous proteins contributing to the dimer combination, we utilized the added specificity of BiFC tagging to assess the ability of a specific MEF2 dimer in combination with HDAC4 (26) (Figure 2D extended data in Supplementary Figure S8). In this assay system, since the intended dimer formation is a strict requirement of the generation of the BiFC signal due to the complementation and, importantly, recruitment to the GBP-nanotrap, the trapped dimer composition at the site is unambiguous. We first confirmed that no BiFC signals were generated by either BiFC-HA-NV- or BiFC-HA-CC-tagged MEF2A alone (Supplementary Figure S3). In addition, mCherry signals were enriched at the nuclear envelope where the MEF2A dimer was anchored only if mCherry was fused to HDAC4 (Figure 2D). Lastly, defined combinations of MEF2A and MEF2D BiFC were capable of recruiting HDAC4 in live cell analysis. These data are in agreement with the known HDAC4 interaction with the MADS-MEF2 domain that is highly conserved among the MEF2 family members (27). More importantly, it provides a proof of principle illustration of the utility of our approach in defining a specific TF dimer and its interaction with a known co-regulator. Therefore, this system can be used to test how a putative interacting protein associates with specific or multiple possible MEF2 dimer combinations in a live cell context.
Functional analysis of an engineered TF dimer using a combined BiFC/UAS-Gal4 reporter gene system
Since we observed that complemented BiFC can be recognized by GBP in live cells, we next considered whether GBP could be used to recruit a defined TF protein complex in a functional reporter gene assay by tethering GBP to a DNA binding domain (DBD) (13), which could then recruit the BiFC mediated defined dimer pair exclusively to a cellular reporter gene assay system.
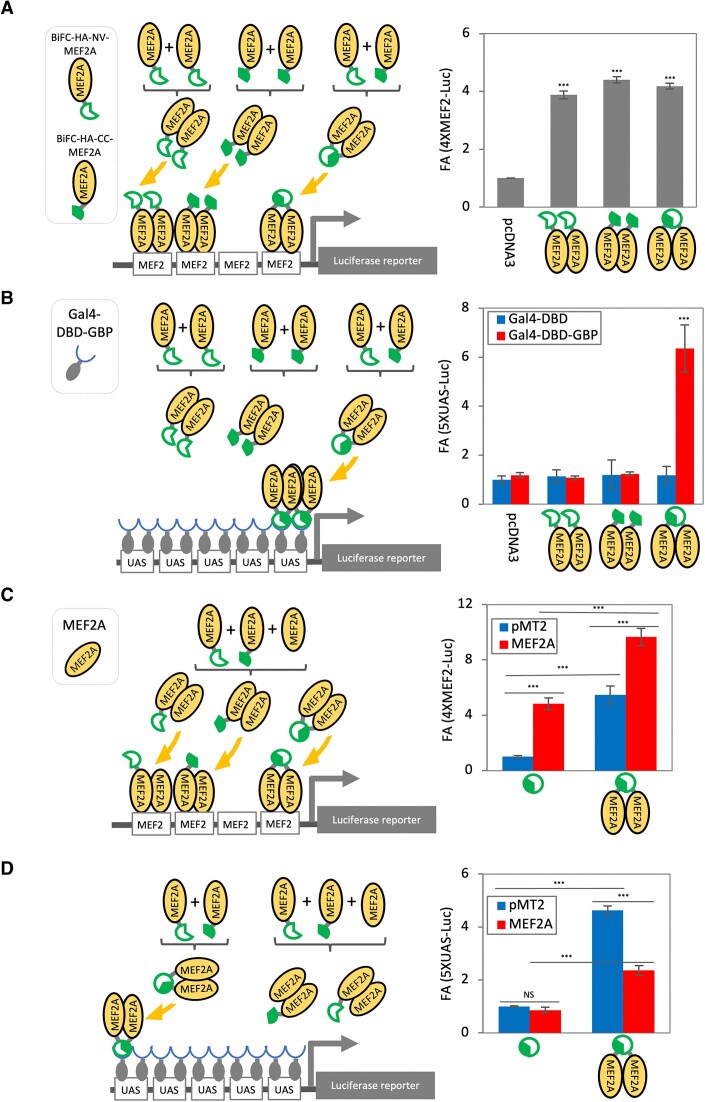
Firstly, we tested if addition of the BiFC tag to MEF2A influenced its transactivation properties using a synthetic multimerized MEF2-Luc reporter gene system (Figure 3A and C). Either in combination or alone, BiFC-HA-MEF2A and/or BiFC-HA-CC-MEF2A activated this reporter gene expression indicating that the BiFC tagged MEF2A is functional and can bind to the cis-regulatory elements (4XMEF2 binding sites) and potentiate transcription independent of complementation.
Figure 3.
Measurement of transcriptional potential of a specific dimer pair. C3H10T1/2 cells were transfected with the indicated constructs in addition to the promoter-less renilla luciferase construct in triplicate per condition. The cells were harvested and subjected to luciferase reporter gene assay. A constant volume of cell lysate was used to determine reporter gene activity (firefly luciferase) and transfection control (renilla luciferase), which was used for standardizing the transfection efficiency. An average of the standardized firefly luciferase values of each condition in triplicate was calculated and fold activation (FA) cpmpared to control condition was graphed. Error bar = standard deviation, n = 3.
Since our aim was to develop a reporter system that exclusively measures the transcriptional activation potential of specific dimer pairs and to eliminate the transcriptional contribution from endogenous MEF2s, we next implemented the heterologous Gal4/UAS-reporter gene assay system based on yeast Gal4, in which the Gal4–DNA binding domain (Gal4–DBD) binds the upstream activator sequence (UAS), driving luciferase reporter gene expression (Figure 3B). This ‘modular’ system has been extensively used previously to assess TF function by fusing the Gal4–DBD to the protein of interest. In this way PPI recruitment is separated from DNA binding and transactivation potential of recruited proteins can be assessed in a simple reporter gene system in which alterations in DNA binding are neutralized (14). In a further proof of principle experiment, we evaluated the transcriptional activation potential of a specific MEF2 dimer pair using the BiFC/GBP-nanotrap/Gal4/UAS-reporter gene assay.
To recruit complemented BiFC-MEF2A dimer but no other endogenous MEF2 dimers specifically to the UAS cis-regulatory elements, we used an expression vector for the expression of GBP fused to Gal4–DBD (Gal4–DBD–GBP) in live cells (18). This adaptor protein can therefore bind to the UAS through its GAL4-DBD while also recruiting any BiFC mediated dimer pair through the GBP moiety (Figure 3B and D). As we expected, without co-expression of Gal4–DBD–GBP, BiFC–MEF2A and GFP–MEF2A did not activate the UAS reporter compared to pcDNA3 empty vector control (Figure 3B), in contrast to the results from the 4XMEF2-luc reporter. However, in the presence of the Gal4–DBD–GBP, complemented BiFC–MEF2A, but not either BiFC-HA-VN- nor BiFC-HA-CC-MEF2A alone, robustly activated the reporter gene (Figure 3B). Since re-constitution of MEF2 dimers using BiFC is a strict requirement for the binding to GBP, this system provides an unequivocal readout of the transactivation potential of the engineered dimer. Therefore, in contrast to the conventional MEF2 reporter gene system which responds to any endogenous MEF2 dimer combinations, the combination of BiFC tagging and co-expression of Gal4–DBD–GBP allows measurement of the activity of a defined dimer pair. Next, we tested if the re-constituted MEF2A is signal responsive by the previously well characterized MKK6/p38MAPK signaling pathway (25). This BiFC tagged MEF2A dimer was robustly activated by co-expression of MKK6/p38MAPK indicating that the system is also responsive to kinase mediated regulation (Supplementary Figure S9A). We further documented here that activities of MEF2A/MEF2A and MEF2A/MEF2D dimers in this condition were comparable to each other, and a dimerization competent mutated form of MEF2A 1–91 (truncation of C-terminus) containing the MADS-MEF2 domain (responsible for dimer formation) inhibited MEF2A activity by formation of an unproductive dimer, consistent with its previously reported activity as a transdominant repressor of MEF2 function (28) (Supplementary Figure S9B).
We also tested whether this BiFC/GBP-Gal4–DBD/UAS-reporter system can be used for characterization of competitive binding. Since MEF2A dimer formation of BiFC tagged MEF2A can be monitored by generation of BiFC signals, we reasoned that addition of non-tagged MEF2A should compete for dimer formation with the BiFC-MEF2A and, at higher stoichiometries, extinguish the complementation (6). While co-expression of non-tagged MEF2A with BiFC tagged MEF2A further activated the 4XMEF2-luc reporter (Figure 3C), in the BiFC/GBP–Gal4–DBD/UAS-reporter system, addition of non-tagged MEF2A repressed the 5× UAS-luc reporter gene expression, supporting this idea (Figure 3D). Therefore, these data indicate that the reduction of BiFC complementation can be used to document competitive protein binding.
Phase separated biomolecular condensates can be visualized using BiFC
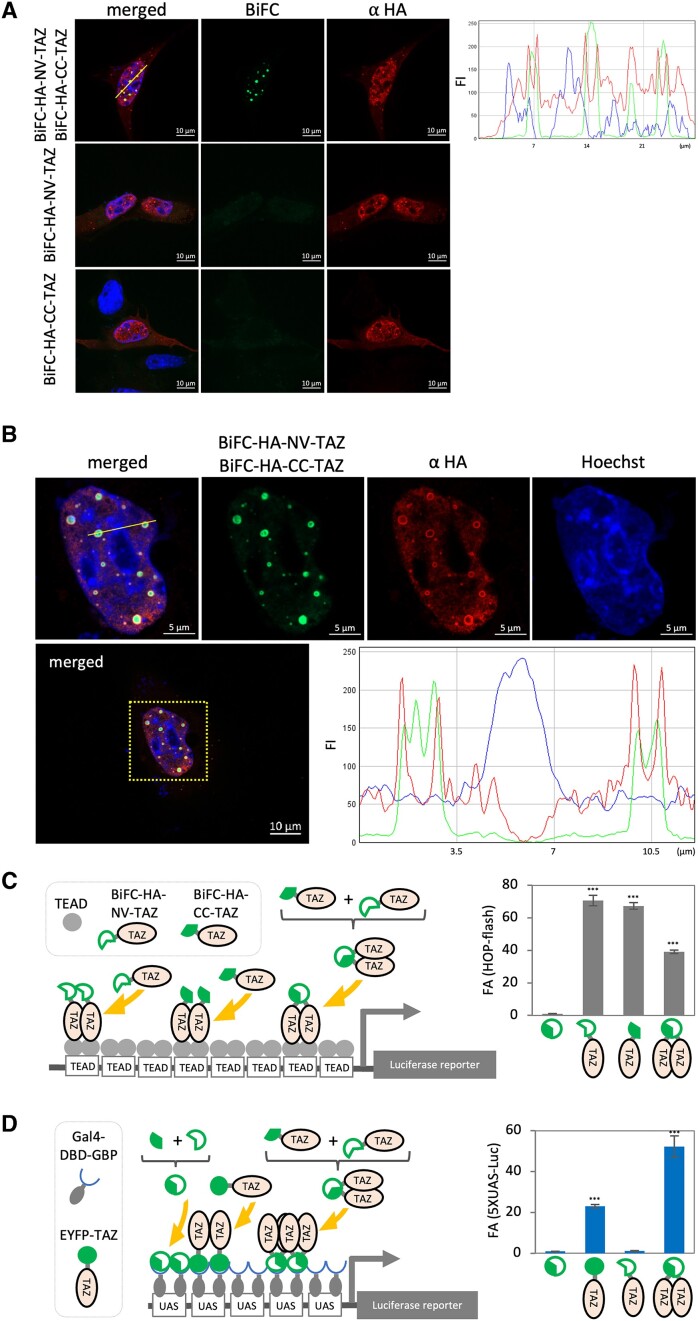
There is growing interest concerning the implications of phase separation by Liquid/Liquid phase separation (LLPS) in transcriptional control (29–31). We therefore wanted to test whether biomolecular condensates can be visualized using BiFC. For this we utilized the Hippo signaling pathway co-regulator, TAZ since it has been shown to robustly form LLPS condensates in cellular systems by homotypic multiple weak intra-/inter-interactions (32–34). To initially test efficacy of the BiFC system for the detection of TAZ, we compared the total expression of BiFC-HA-TAZ detected by immunofluorescence (IF) technique against BiFC signals generated by TAZ. First, we confirmed expression of BiFC-HA tagged TAZ by IF analysis for HA detection (Figure 4A). IF analysis indicates TAZ proteins form spherical condensates illustrating that the tags attached to TAZ allow LLPS formation (Figure 4A). We have observed spherical puncta of TAZ transcriptional regulators and fusion of puncta that is consistent with features of LLPS but we also recognize that these condensates could be reflective of a localized accumulation of protein complexes that does not necessarily conform to the requisite criteria for LLPS. We do acknowledge that the phenomena of phase separation and its molecular basis is still under debate in the scientific community (35–40). However, since TAZ has been well documented in terms of forming phase condensates and satisfying the criteria as such (33,34,41,42), these data indicate that the BiFC technique may be an effective method to define PPI’s involved in biomolecular condensates. Interestingly, we noted that peaks of BiFC signals and HA signals were not always matched, and BiFC signals were often restricted to the core of the LLPS (Figure 4B) suggesting dynamic interaction between TAZ at the surface of the condensates and condensed TAZ accumulation at the core possibly prevents recognition of the HA epitope by the antibody.
Figure 4.
LLPS condensate formation and co-activator functions of TAZ can be recorded by BiFC technique. (A) C2C12 cells were transfected with indicated constructs. The cells were fixed and subjected to IF analysis. The BiFC signal (green) was indicative of formation of TAZ dimers. HA signal (red) depicting distribution of the exogenous TAZ protein. BiFC and HA signal distribution were analyzed by line scan for green and red signal intensity along with the yellow line on the merged micrograph (B) One nucleus (yellow squared) was magnified and Green (BiFC), Red (HA), and Blue (Nucleus) signals were rendered by confocal fluorescence microscopy. Line scan analysis showed the distribution of BiFC (green) and HA (red) signals in the nucleus (blue). (C) C3H10T1/2 cells were transfected with indicated constructs and the promoter-less Renilla luciferase construct ws used to monitor transfection efficiency in triplicate per condition. The luciferase assay was performed using constant amount of total cell lysate. The firefly luciferase value was standardized by the corresponding renilla luciferase value. An average of the standardized firefly luciferase values of each condition in triplicate was calculated and fold activation (FA) compared to control condition was graphed. Complementation of BiFC tag independent (C) or dependent (D) reporter system. Error bar = standard deviation, n = 3.
Next, we examined if co-activator function can be quantified using a gene reporter system. First, we used a well-characterized YAP/TAZ reporter gene system, HOP-flash, which consists of multimerized TEAD binding cis-regulatory elements driving the expression of the luciferase reporter gene (Figure 4C). BiFC tagged TAZ subunits were active independent of the complementation status of the BiFC (Figure 4C) indicating that BiFC tagged TAZ can interact with TEAD and activate transcription. These data confirmed that addition of the BiFC-HA tag to TAZ does not prevent its interaction with TEAD or negate its transcriptional potential as a co-activator.
Next, we sought to test if the co-activator potential of TAZ could be assessed using BiFC combined with the GBP-Gal4–DBD/UAS-reporter system, thus by-passing TEAD binding to DNA. To by-pass TEAD binding, we used Gal4–DBD–GBP, which works as a synthetic GFP dependent TF (18) (Figure 4D). In addition, EYFP-TAZ was included as a positive control. As predicted, in the presence of both BiFC-HA-NV-TAZ and BiFC-HA-CC-TAZ, the reporter gene was robustly activated, but without complementation the reporter gene was silent. Therefore, these data indicate that the transactivation potential of a co-regulator that does not bind to DNA directly, in this case TAZ, can be assessed using this system.
Discussion
In this study, we outline a novel approach to dissect the function of defined protein dimer pairs in living cells using coupled GBP-nanotrap/BiFC techniques. Remarkably, the functional properties of defined protein dimers are retained in the BiFC complementation system. Moreover, the recognition of reconstituted BiFC by engineered nanobodies (GBP nanotrap), that can be expressed in cells, substantively expands the toolbox of techniques available to dissect protein interactions in living cells.
Previously, we documented PPI between GFP-fusion protein and mCherry tagged protein using a GBP-nanotrap (17,23). In the current study, we further develop this approach by replacing the intact GFP tag that we originally used with the BiFC split GFP system, allowing unambiguous assessment of PPIs between engineered homo- and hetero-dimer pairs as well as with a specific co-regulator. One of the considerable advantages of this system is the capability to monitor specific PPIs in live cells. This feature will allow a spectrum of PPI properties to be assessed. For example, the requirement of post-translational modification for PPIs can be assessed using live cell fluorescence microscopy combined with pharmacological manipulation of kinases or expression of phosphosite mutants.
A substantial avenue of transcription factor biology over several decades has been to assess the transactivation function of transcription factors and how their PPIs and post-translational modifications (PTMs) can modulate this function. However, definitive analysis has always been problematic due to the inseparable nature of DNA binding and transactivation function which can be regulated independently thus confounding the data, resulting in ambiguity in interpretation. Also, the contribution of endogenous undefined dimer partners in cellular based assays also complicates this analysis. In view of this, we utilized the heterologous UAS-luciferase reporter gene assay system with a Gal4–DBD–GBP adaptor peptide combined with BiFC complementation in order to circumvent these issues. In our experiments, since complementation of BiFC only occurs when BiFC tagged MEF2 subunits form defined dimers, the recruitment of the reconstituted dimer is required for recruitment to the UAS through the Gal4–DBD–GBP adaptor. Therefore, the reporter gene readout can only be a function of the defined dimer pair. This level of protein complex specification has not previously been possible in cellular reporter gene-based assay systems, allowing us to decisively assess the transactivation potential of a specific MEF2 dimer even in the presence of endogenous cellular MEF2 proteins.
Since transcriptional activation potential of MEF2 proteins is known to be regulated by a plethora of post-translational modifications (PTMs) we wanted to assess whether the defined BiFC dimer pairs can also be regulated by PTMs. For example, a variety of kinases differentially phosphorylate MEF2 isoforms on their less conserved TADs (43), controlling dimer pairing by BiFC tagging would therefore allow us to assess how PTMs regulate specific MEF2 dimer pairs. In our proof of principle experiments documented here, we report that the previously well characterized regulation of MEF2 by the MKK6/p38 MAPK pathway is conserved in our reconstitution system, thus highlighting the utility of this approach to characterize the effects of specific PTMs and kinases or other regulatory molecules on defined TF dimer pairs. Moreover, this general reporter gene strategy could be employed in drug screening approaches to identify small molecules that can modulate transcription factor properties in which the DNA binding properties are held constant due to the Gal4–DBD–GBP module. One caveat of our studies is that the dimer pairs engineered by BiFC may not necessarily reflect the properties of the natural dimer complexes. The aim is to unambiguously define properties of the dimer pairs using BiFC complementation which would then allow further experimentation to assess those properties in the natural dimer context. We also note that there is a possibility that the N-terminal and the C-terminal fragments of the BiFC system can generate low level fluorescence signals when co-expressed, generating some noise in this system (6,44–50). In our experiments (Supplementary Figure S1), there was no clear difference in complementation frequencies between BiFC-NV/BiFC-CC and the low affinity interaction between the BiFC-NV-c-Jun/BiFC-CC-Fra2 combination. This is, in this case, partially due to the ubiquitously expressed endogenous cellular AP-1 factors competing with the BiFC-tagged AP-1 factors for dimer formation which minimizes the frequency of specific complementation events (11,51). However, we would like to point out that the BiFC signal generation due to the BiFC-NV-c-Jun/BiFC-CC-Fra2 is a result of specific Jun/Fra2 heterodimer formation because ectopically expressed dimerization competitors (mCherry tagged c-Jun or Fra2 but not mCherry itself), greatly reduced the BiFC signal (Figure 1B and Supplementary Figure S2). Therefore, the BiFC signal generated by the BiFC-NV-c-Jun and BiFC-CC-Fra2 combination is primarily due to specific heterodimer formation between BiFC-NV-c-Jun and BiFC-CC-Fra2. Based on this competition analysis, generation of the BiFC signals does reflect the interaction of the intended test proteins. This example also highlights the generic requirement to perform a similar competition analysis as a control for the specificity of the interaction, particularly for low affinity PPIs.
An area of major interest in cell biology is the discovery and function of biomolecular condensates, the impact of which is still being evaluated. Since we, and others, have previously reported that the Hippo pathway effector, TAZ, has a very robust capacity for phase separation in living cells (32–34), we further extended our studies to determine if the approach highlighted in the current study could also be useful in analyzing phase separation properties in biological systems. As stated above, TAZ forms LLPS condensates in the nucleus and this is important for the co-activation function of TAZ protein. In support of this idea, we observed that BiFC GFP signal was observed in spherical condensates in the nucleus of BiFC-HA-TAZ transfected cells, and BiFC tagged TAZ showed transcriptional activation potential by binding to a Gal4–DBD–GBP in a Gal4 UAS based assay system. Collectively, these data indicate that BiFC reconstituted protein complexes can be observed in phase separated condensates in cells and also that BiFC TAZ retains its robust transactivation potential in functional reporter gene assays.
At this juncture, the generalizable application of these methods to many different types of protein interactions in different cellular contexts requires considerable empirical testing of other known interactions. However, the public availability of the system components and the widespread use of fluorescence based detection systems and luciferase based reporter gene assay systems should allow extensive analysis of the overall utility of this system to study defined protein:protein interactions in various cellular contexts.
In summary, we outline an experimental approach that allows engineering, cellular visualization and functional analysis of defined transcription factor dimer combinations using a coupled BiFC/GBP-nanobody based technique in living cells. This approach facilitates analysis of the transcriptional activation properties of defined TF dimer combinations and also constitutes a potentially more generalizable approach to assessing defined protein interactions.
Supplementary Material
Contributor Information
Tetsuaki Miyake, Department of Biology, York University, Toronto, ON, M3J 1P3, Canada; Muscle Health Research Centre (MHRC), York University, Toronto, ON, M3J 1P3, Canada; Centre for Research in Biomolecular Interactions (CRBI), York University, Toronto, ON, M3J 1P3, Canada.
John C McDermott, Department of Biology, York University, Toronto, ON, M3J 1P3, Canada; Muscle Health Research Centre (MHRC), York University, Toronto, ON, M3J 1P3, Canada; Centre for Research in Biomolecular Interactions (CRBI), York University, Toronto, ON, M3J 1P3, Canada.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Natural Sciences and Engineering Research Council of Canada [RGPIN-2018-05896]; Canadian Institutes of Health Research [PJT-159644]. Funding for open access charge: NSERC.
Conflict of interest statement. None declared.
References
- 1. Stearns T. Green fluorescent protein. The green revolution. Curr. Biol. 1995; 5:262–264. [DOI] [PubMed] [Google Scholar]
- 2. Zimmer M. Green fluorescent protein (GFP): applications, structure, and related photophysical behavior. Chem. Rev. 2002; 102:759–781. [DOI] [PubMed] [Google Scholar]
- 3. Matz M.V., Lukyanov K.A., Lukyanov S.A.. Family of the green fluorescent protein: journey to the end of the rainbow. Bioessays. 2002; 24:953–959. [DOI] [PubMed] [Google Scholar]
- 4. Kerppola T.K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 2008; 37:465–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kerppola T.K. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem. Soc. Rev. 2009; 38:2876–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kodama Y., Hu C.D.. Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. BioTechniques. 2012; 53:285–298. [DOI] [PubMed] [Google Scholar]
- 7. Ventura S. Bimolecular fluorescence complementation: illuminating cellular protein interactions. Curr. Mol. Med. 2011; 11:582–598. [DOI] [PubMed] [Google Scholar]
- 8. Miller K.E., Kim Y., Huh W.K., Park H.O.. Bimolecular Fluorescence Complementation (BiFC) Analysis: advances and Recent Applications for Genome-Wide Interaction Studies. J. Mol. Biol. 2015; 427:2039–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang T., Yang N., Liang C., Xu H., An Y., Xiao S., Zheng M., Liu L., Wang G., Nie L.. Detecting protein-protein interaction based on protein fragment complementation assay. Curr. Protein Pept. Sci. 2020; 21:598–610. [DOI] [PubMed] [Google Scholar]
- 10. Datta R.R., Rister J.. The power of the (imperfect) palindrome: sequence-specific roles of palindromic motifs in gene regulation. Bioessays. 2022; 44:e2100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu C.D., Chinenov Y., Kerppola T.K.. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002; 9:789–798. [DOI] [PubMed] [Google Scholar]
- 12. De Meyer T., Muyldermans S., Depicker A.. Nanobody-based products as research and diagnostic tools. Trends Biotechnol. 2014; 32:263–270. [DOI] [PubMed] [Google Scholar]
- 13. Harmansa S., Affolter M.. Protein binders and their applications in developmental biology. Development. 2018; 145:dev148874. [DOI] [PubMed] [Google Scholar]
- 14. Brand A.H., Perrimon N.. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993; 118:401–415. [DOI] [PubMed] [Google Scholar]
- 15. Don E.K., Maschirow A., Radford R.A.W., Scherer N.M., Vidal-Itriago A., Hogan A., Maurel C., Formella I., Stoddart J.J., Hall T.E.et al.. In vivo validation of bimolecular fluorescence complementation (BiFC) to investigate aggregate formation in amyotrophic lateral sclerosis (ALS). Mol. Neurobiol. 2021; 58:2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perry R.L., Yang C., Soora N., Salma J., Marback M., Naghibi L., Ilyas H., Chan J., Gordon J.W., McDermott J.C.. Direct interaction between myocyte enhancer factor 2 (MEF2) and protein phosphatase 1alpha represses MEF2-dependent gene expression. Mol. Cell. Biol. 2009; 29:3355–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pagiatakis C., Sun D., Tobin S.W., Miyake T., McDermott J.C.. TGFbeta-TAZ/SRF signalling regulates vascular smooth muscle cell differentiation. FEBS J. 2017; 284:1644–1656. [DOI] [PubMed] [Google Scholar]
- 18. Tang J.C., Szikra T., Kozorovitskiy Y., Teixiera M., Sabatini B.L., Roska B., Cepko C.L.. A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation. Cell. 2013; 154:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miyake T., McDermott J.C.. Nucleolar localization of c-Jun. FEBS J. 2022; 289:748–765. [DOI] [PubMed] [Google Scholar]
- 20. Rauscher F.J. 3rd, Voulalas P.J., Franza B.R. Jr., Curran T.. Fos and Jun bind cooperatively to the AP-1 site: reconstitution in vitro. Genes Dev. 1988; 2:1687–1699. [DOI] [PubMed] [Google Scholar]
- 21. Krasnov A.N., Mazina M.Y., Nikolenko J.V., Vorobyeva N.E.. On the way of revealing coactivator complexes cross-talk during transcriptional activation. Cell Biosci. 2016; 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirchhofer A., Helma J., Schmidthals K., Frauer C., Cui S., Karcher A., Pellis M., Muyldermans S., Casas-Delucchi C.S., Cardoso M.C.et al.. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 2010; 17:133–138. [DOI] [PubMed] [Google Scholar]
- 23. Ehyai S., Miyake T., Williams D., Vinayak J., Bayfield M.A., McDermott J.C.. FMRP recruitment of beta-catenin to the translation pre-initiation complex represses translation. EMBO Rep. 2018; 19:e45536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin F., Worman H.J.. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics. 1995; 27:230–236. [DOI] [PubMed] [Google Scholar]
- 25. Cornwell J.D., McDermott J.C.. MEF2 in cardiac hypertrophy in response to hypertension. Trends Cardiovasc. Med. 2022; 33:204–212. [DOI] [PubMed] [Google Scholar]
- 26. Miska E.A., Karlsson C., Langley E., Nielsen S.J., Pines J., Kouzarides T.. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999; 18:5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu J., McKinsey T.A., Nicol R.L., Olson E.N.. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:4070–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ornatsky O.I., Andreucci J.J., McDermott J.C.. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 1997; 272:33271–33278. [DOI] [PubMed] [Google Scholar]
- 29. Manning S.A., Kroeger B., Harvey K.F.. The regulation of Yorkie, YAP and TAZ: new insights into the Hippo pathway. Development. 2020; 147:dev179069. [DOI] [PubMed] [Google Scholar]
- 30. Mir M., Bickmore W., Furlong E.E.M., Narlikar G.. Chromatin topology, condensates and gene regulation: shifting paradigms or just a phase?. Development. 2019; 146:dev182766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Watson M., Stott K.. Disordered domains in chromatin-binding proteins. Essays Biochem. 2019; 63:147–156. [DOI] [PubMed] [Google Scholar]
- 32. Yu M., Peng Z., Qin M., Liu Y., Wang J., Zhang C., Lin J., Dong T., Wang L., Li S.et al.. Interferon-gamma induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol. Cell. 2021; 81:1216–1230. [DOI] [PubMed] [Google Scholar]
- 33. Franklin J.M., Guan K.L.. YAP/TAZ phase separation for transcription. Nat. Cell Biol. 2020; 22:357–358. [DOI] [PubMed] [Google Scholar]
- 34. Tripathi S., Miyake T., Kelebeev J., McDermott J.C.. TAZ exhibits phase separation properties and interacts with Smad7 and beta-catenin to repress skeletal myogenesis. J. Cell Sci. 2022; 135:jcs259097. [DOI] [PubMed] [Google Scholar]
- 35. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shin Y., Chang Y.C., Lee D.S.W., Berry J., Sanders D.W., Ronceray P., Wingreen N.S., Haataja M., Brangwynne C.P.. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2019; 176:1518. [DOI] [PubMed] [Google Scholar]
- 37. Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M.et al.. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018; 175:1842–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shimobayashi S.F., Ronceray P., Sanders D.W., Haataja M.P., Brangwynne C.P.. Nucleation landscape of biomolecular condensates. Nature. 2021; 599:503–506. [DOI] [PubMed] [Google Scholar]
- 39. Gibson B.A., Doolittle L.K., Schneider M.W.G., Jensen L.E., Gamarra N., Henry L., Gerlich D.W., Redding S., Rosen M.K.. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019; 179:470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shin Y., Berry J., Pannucci N., Haataja M.P., Toettcher J.E., Brangwynne C.P.. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell. 2017; 168:159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cai D., Feliciano D., Dong P., Flores E., Gruebele M., Porat-Shliom N., Sukenik S., Liu Z., Lippincott-Schwartz J.. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 2019; 21:1578–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu Y., Wu T., Gutman O., Lu H., Zhou Q., Henis Y.I., Luo K.. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 2020; 22:453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cox D.M., Du M., Marback M., Yang E.C., Chan J., Siu K.W., McDermott J.C.. Phosphorylation motifs regulating the stability and function of myocyte enhancer factor 2A. J. Biol. Chem. 2003; 278:15297–15303. [DOI] [PubMed] [Google Scholar]
- 44. Kerppola T.K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006; 1:1278–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kerppola T.K. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 2006; 7:449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vidi P.A., Przybyla J.A., Hu C.D., Watts V.J.. Visualization of G protein-coupled receptor (GPCR) interactions in living cells using bimolecular fluorescence complementation (BiFC). Curr. Protoc. Neurosci. 2010; Chapter 5:Unit 5.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Waadt R., Kudla J.. In Planta Visualization of Protein Interactions Using Bimolecular Fluorescence Complementation (BiFC). CSH Protoc. 2008; 2008:pdb prot4995. [DOI] [PubMed] [Google Scholar]
- 48. Shyu Y.J., Hiatt S.M., Duren H.M., Ellis R.E., Kerppola T.K., Hu C.D.. Visualization of protein interactions in living Caenorhabditis elegans using bimolecular fluorescence complementation analysis. Nat. Protoc. 2008; 3:588–596. [DOI] [PubMed] [Google Scholar]
- 49. Hu C.D., Grinberg A.V., Kerppola T.K.. Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis. Curr. Protoc. Protein Sci. 2005; Chapter 19:19.10.1–19.10.21. [DOI] [PubMed] [Google Scholar]
- 50. Hu C.D., Grinberg A.V., Kerppola T.K.. Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis. Curr. Protoc. Cell Biol. 2006; Chapter 21:Unit 21.23. [DOI] [PubMed] [Google Scholar]
- 51. Yuan Z., Gong S., Luo J., Zheng Z., Song B., Ma S., Guo J., Hu C., Thiel G., Vinson C.et al.. Opposing roles for ATF2 and c-Fos in c-Jun-mediated neuronal apoptosis. Mol. Cell. Biol. 2009; 29:2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.