Abstract
MicroRNAs (miRNAs) are essential regulators of gene expression, defined by their unique biogenesis, which requires the precise excision of the small RNA from an imperfect fold-back precursor. Unlike their animal counterparts, plant miRNA precursors exhibit variations in sizes and shapes. Plant MIRNAs can undergo processing in a base-to-loop or loop-to-base direction, with DICER-LIKE1 (DCL1) releasing the miRNA after two cuts (two-step MIRNAs) or more (sequential MIRNAs). In this study, we demonstrate the critical role of the miRNA/miRNA* duplex region in the processing of miRNA precursors. We observed that endogenous MIRNAs frequently experience suboptimal processing in vivo due to mismatches in the miRNA/miRNA* duplex, a key region that fine-tunes miRNA levels. Enhancing the interaction energy of the miRNA/miRNA* duplex in two-step MIRNAs results in a substantial increase in miRNA levels. Conversely, sequential MIRNAs display distinct and specific requirements for the miRNA/miRNA* duplexes along their foldback structure. Our work establishes a connection between the miRNA/miRNA* structure and precursor processing mechanisms. Furthermore, we reveal a link between the biological function of miRNAs and the processing mechanism of their precursors with the evolution of plant miRNA/miRNA* duplex structures.
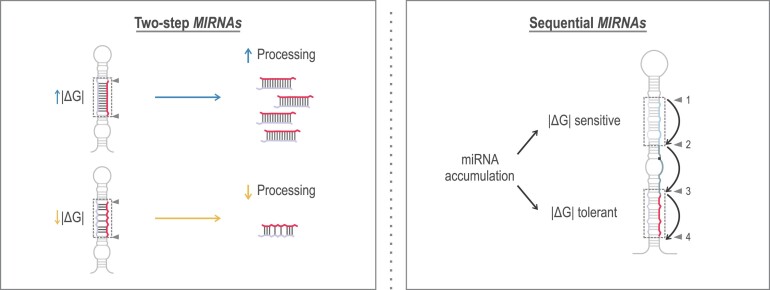
Graphical Abstract
Graphical Abstract.
Introduction
Small RNAs are essential regulators of gene expression in plants. Among these, miRNAs are a crucial class defined by their precise excision from a fold-back precursor, playing key roles as post-transcriptional regulators of plant genes. Evolutionarily conserved miRNAs significantly contribute to diverse plant processes, such as the regulation of transcription factors controlling plant development, hormone signalling, small RNA pathways and the response to nutritional deficiencies. MiRNAs are initially transcribed as longer transcripts by RNA polymerase II, referred to as primary miRNAs (pri-miRNAs), which are capped, polyadenylated and occasionally spliced [reviewed in (1–3)]. In the nucleus, the type III ribonuclease DCL1 aided by several proteins, primarily HYPONASTIC LEAVES1 (HYL1) and SERRATE (SE), processes the stem-loop precursor found within the primary transcript [reviewed in (1–3)]. This DCL1 complex cleaves the precursor to release two paired RNA strands of approximately 21 nucleotides (nt) with two 3′ nt overhanging (miRNA/miRNA*). One of these strands is incorporated into an ARGONAUTE (AGO) protein, generally AGO1 in plants, to regulate the miRNA target [reviewed in (4–6)]. That many proteins participate in all the steps of miRNA biogenesis, including MIRNA gene transcription, pri-miRNA stabilization, processing, as well as the fact that DCL1, SE and HYL1 activity are precisely regulated highlights the pathway's complexity [reviewed in (1–3)].
MIRNA precursors are a collection of diverse structures and lengths and their recognition and processing by the DCL1 complex may unfold through various mechanisms. Precursors can be processed by two DCL1 cuts (two-step MIRNA) or by three or four sequential cleavages, which produce several small RNA duplexes with the biologically relevant small RNA being released in the final two cuts (sequential MIRNAs) (7–15). The miRNA precursor harbours a double-stranded RNA (dsRNA) segment that can be below or above the miRNA/miRNA* region and which determines the position of DCL1’s first cut and therefore the processing direction; from base-to-loop or from loop-to-base, respectively (7,9–16). After the first cleavage, DCL1 generates a subsequent cut approximately 21 nt away, releasing the miRNA/miRNA* (17,18).
Studies of the precursor primary structure indicate that a GC-rich sequence signature favours their processing (19). Furthermore, recent work has shown that the identity of mismatches in miRNA precursors with imperfect fold-backs plays a role in their processing (20). Notably, C-C mismatches strongly disrupt the precursor processing, implying that this pair should be avoided in structural and functional analyses of plant miRNA precursors (20). Finally, several reports have described the importance of the miRNA/miRNA* structure on AGO sorting (21–26). Both AGO1 and AGO2 load miRNA duplexes with paired bases at 15th position however the pairing or unpairing of the 11th position of the miRNA/miRNA*, results in AGO2 or AGO1 differential loading (23). Other AGOs exclusively bind one miRNA, such as AGO7 loading miR390 duplex by recognizing the 5ʹ-A and a G^A mismatch at position 11th (21,27) and AGO10 selectively loading miR165/166 due to their extensive mismatches surrounding the 12th and 13th positions of the duplex (22,25).
Here, we focus on understanding the significance of the miRNA/miRNA* duplex region during the processing of Arabidopsis miRNA precursors. Our findings indicate that the structure of this duplex plays a crucial role, albeit with varying requirements depending on the precursor's processing mechanism. Interestingly, we found that most endogenous miRNA precursors undergo suboptimal processing, a situation that can be improved by adjusting the energy levels of the miRNA/miRNA* region in two-step MIRNAs. However, sequential MIRNAs have different miRNA/miRNA* requirements. We found that the miRNA/miRNA*, generated in the last two cuts by DCL1, is the most stable duplex from sequential precursors. In contrast, the other small RNAs have several unpaired bases on their central region, resulting in duplexes with low interacting energy and reduced final levels. Furthermore, we found that the distinct roles of the miRNA/miRNA* duplex regions in the processing of MIRNAs correlate with their sequence conservation during evolution.
Materials and methods
Plant material
All the experiments were done in wild-type (wt) Arabidopsis thaliana plants, Col-0 accession. Plant transformation was performed via the floral dip method (28). Transgenic plants were selected on plates containing Murashige and Skoog media with 50 μg/ml of kanamycin. Plants were grown at 16 hours light/8 hours dark photoperiod at 22°C and 100 μmol photons m−2 s−1 of light intensity.
Transgenes
MIR164a (AT2G47585), MIR171b (AT1G11735), MIR172a (AT2G28056), MIR172c (AT3G11435), MIR319a (AT4G23713), MIR319c (AT2G40805), MIR394a (AT1G20375) and MIR397a (AT4G05105) were cloned from Arabidopsis genomic DNA. Mutations were introduced by site-directed mutagenesis (11). All wt and mutant precursors were cloned in pCHF3 binary vector under a 35S viral promoter (29). See Supplementary Table S1 for the expressed sequences of each vector.
RNA expression analysis
Each sample corresponded to 15 pooled ten-day-old seedlings or inflorescences from independent primary transgenic plants. Plant material was collected and processed with TRI Reagent (MRC) following the manufacturer's instructions. For small RNA blots four to eight micrograms of total RNA were resolved in 17% (w/v) polyacrylamide denaturing gels (7M urea). RNA was transferred to a Nytran SPC membrane (GE Healthcare) by semi-dry blotting and UV-crosslinked. Probes listed on Supplementary Table S2 were 5′ end labelled with [γ -32P] ATP using T4 polynucleotide kinase (Thermo Scientific) and hybridized as described previously (11). Images were obtained with Typhoon FLA 7000 (GE Healthcare). U6 hybridization and the ethidium bromide (EB) staining were used as transference and RNA integrity controls respectively. The signal intensity of each band was determined using FIJI (30). The intensity values of the bands detected with the miRNA probes were normalized to the signal obtained with the U6 snRNA probe. In several figures featuring small RNA blots, the miRNA corresponding to the empty vector sample was not detectable. This is because the endogenous miRNA levels were low compared to the miRNAs derived from the transgenic precursors ectopically expressed from the 35S promoter in the analyzed tissues. The Appendix I contains the original images of every small RNA blot and biological replicates from Figures 1–5.
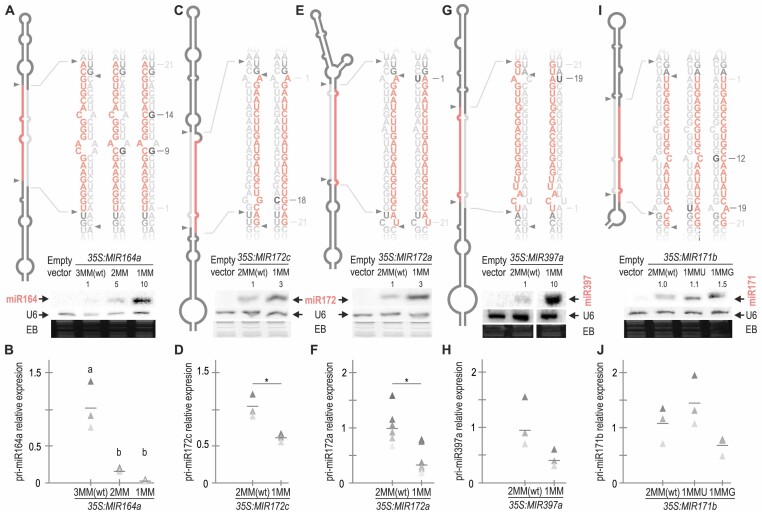
Figure 1.
Improving miRNA/miRNA* pairing increases miRNA accumulation. (A, C, E, G and I) Small RNA blots of Arabidopsis plants expressing different precursors from the 35S promoter. Each sample corresponds to 15 pooled ten-day-old seedlings from independent primary transgenic plants. The U6 hybridization and ethidium bromide (EB) staining of each gel are shown below. Numbers above small RNA blots correspond to miRNA levels quantified relative to the wt precursor. Top panels show a schematic representation of the precursor analysed according to the mfold secondary structure prediction (see Appendix II). MiRNA is shown in red and miRNA* in grey. Black letters specify the closed mismatches and numbers to the right indicate their position counting from the 5′end of the miRNA. Supplementary Figure S1E shows the original image of the miR397a blot. (B, D, F, H and J) Detection of pri-miRNA levels by RT-qPCR in at least three biological replicates that are shown as triangles in different shades of grey, lines represent the mean value for each MIRNA. Different letters in (B) indicate statistically significant differences, according to ANOVA (P-value = 0.0011) followed by Tukey's multiple comparison test (p < 0.05). Asterisks in D and F indicate statistically significant differences according to Student t-test (two-tailed), p < 0.05 (*). No statistically significant differences were observed in H and J. See Supplementary Table S4 for the statistical analysis conducted in all the samples.
Figure 5.
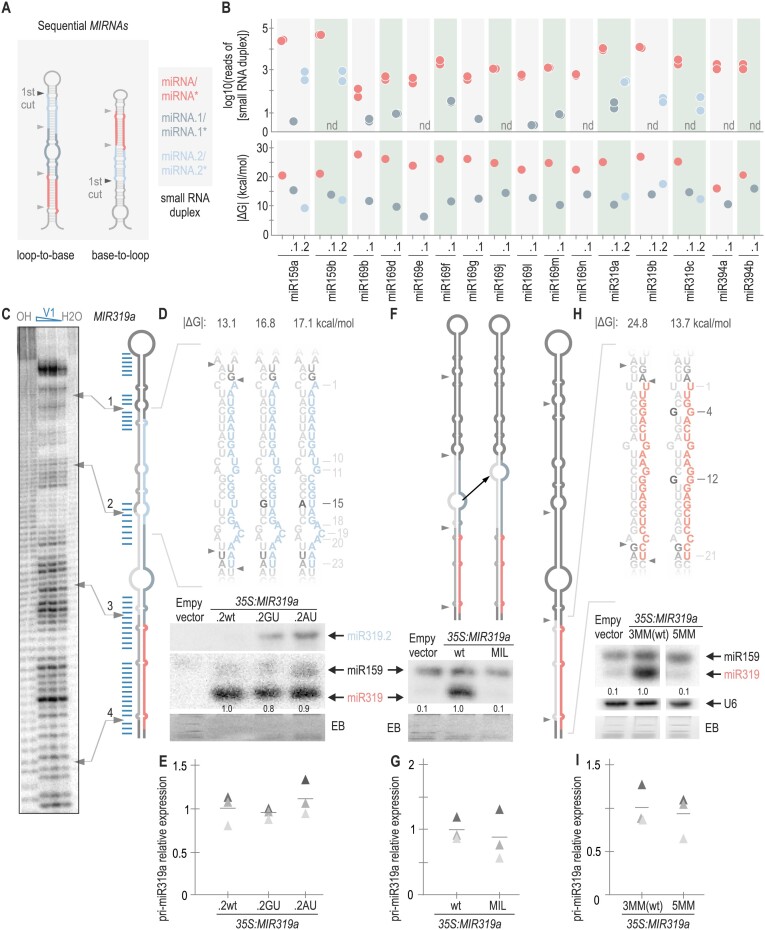
Sequentially processed MIRNAs generate several small RNA duplexes with different structural requirements. (A) Scheme of a sequential MIRNA. The first DCL1 cut is highlighted with a black arrow. Small RNAs derived from sequential precursors. MiRNA is shown in red, distal miRNA duplex (miRNA.2) is depicted in light blue and proximal miRNA duplex (miRNA.1) is indicated in dark grey (right). (B) Top panel: logarithm in base 10 of number of reads of small RNA duplex (n.d.: not detected). See Supplementary Table S3 for the number of reads detected for each duplex. Bottom panel: dot plot representing |ΔG| calculated for each small RNA duplex from evolutionary conserved sequentially processed MIRNAs. The colour code of the small RNA duplexes is shown in (A). (C) Structural analysis of MIR319a. Denaturing 8% (w/v) polyacrylamide gels shown: OH−: alkaline hydrolysis; V1: RNAse V1 in decreasing concentrations; H2O: incubation with water (control). Blue lines indicate the double-stranded bases obtained after V1 digestion as determined from polyacrylamide gels. The arrows correlate the position of DCL1 cuts in the precursor (right) with the experimental determination of the secondary structure (left). See Supplementary Figure S4BC for the original image of the polyacrylamide gel. (D, F and H) Small RNA blots of transgenic lines expressing different precursors from the 35S promoter. Each sample corresponds to 15 pooled inflorescences from independent primary transgenic plants. Numbers above small RNA blots correspond to miRNA levels quantified relative to the wt precursor. The EB staining of each gel is shown below. Top panels show a schematic representation of MIR319a including the wt and modified sequences of (D) miR319.2 (miR319a.2GU and miR319a.2AU) (F) MIR319a-MIL where a black arrow indicates how the central internal loop in miRNA.1 duplex was moved closer to DCL1 second cut (F); and MIR319a-3MMwt and MIR319a-5MM (H). These depictions represent the mfold secondary structure prediction for each precursor (see Appendix II). Black letters specify the closed mismatches, and the numbers indicate their position counting from the 5′end of the miRNA or miRNA.2. MiRNA is shown in red and miRNA* in grey, miRNA.2 is shown in light blue, and miRNA.2* is shown in grey. |ΔG| is shown above each small RNA sequence. (E, G and I) RT-qPCR of pri-miRNA determined in three biological replicates are shown as triangles in different shades of grey. Lines represent the mean value for each MIRNA. No statistically significant differences in E, G and I. See Supplementary Table S4 for the statistical analysis conducted in all the samples.
RT-qPCR
Pri-miRNA transcript levels were determined by RT-qPCR. 500 ng of total RNA were treated with RQ1 RNAse-free DNase (Promega). The first strand cDNA synthesis was carried out with M-MLV reverse transcriptase (Invitrogen) using dTV oligonucleotide for polyadenylated RNAs. Quantitative (q) PCR reactions were performed in Aria Mx Real-time PCR System (Agilent) using SYBR Green I (Roche) to monitor double-stranded (ds) DNA synthesis. At least three biological replicates were used in every experiment. The gene RPS26E (AT3G56340) was used to normalize the expression of pri-miRNA levels. Expression levels were relativized to the wt precursor. In Supplementary Figure S3, the expression levels were relativized to the empty vector since the expression of two different precursors was being compared and the oligonucleotides used hybridized in the transcribed regions of pCHF3. See Supplementary Table S2 for a complete list of the oligonucleotides used.
Analysis of small RNA libraries
We analysed publicly available and deposited data at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Small RNA libraries from seedlings used in Figures 5, 6 and Supplementary Figure S5 were obtained from ((11), accession number GSE116330). Small RNA libraries from leaves and inflorescences used in Supplementary Figure S4 and S5 were extracted from ((31), accession number GSM506656-8 and GSM506662-4). Only sequential MIRNAs with at least 100 reads in two libraries of seedlings or 100 reads in four libraries of leaves and inflorescences were considered for the analysis. The reads were mapped to different small RNAs from sequential precursors: miRNA, miRNA*, miRNA.1, miRNA.1*, miRNA.2 and miRNA.2*. The reads of each duplex were calculated as the sum of the reads for each small RNA composing the duplex. For example, to calculate the reads of miRNA/miRNA* duplex we add the reads of miRNA and miRNA*. Finally, we obtained the logarithm in base ten of the reads of each duplex. See Supplementary Table S3 for the number of reads used to calculate small RNA duplex accumulation from sequential MIRNAs.
Figure 6.
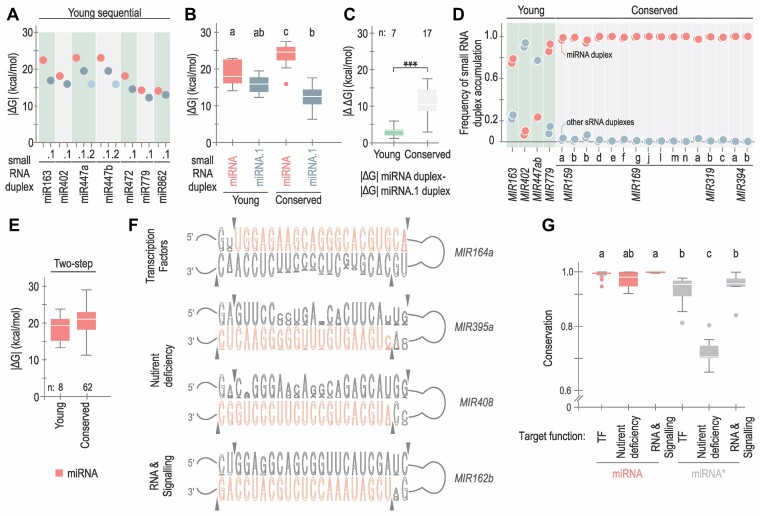
Conservation and divergence of small RNA duplexes. (A) Dot plot representing |ΔG| calculated for each small RNA duplex derived from evolutionary young sequentially processed MIRNAs. Duplexes are shown in red (miRNA/miRNA*), dark grey (miRNA.1/miRNA.1*) and light blue (miRNA.2/miRNA.2*). (B) Box plots representing |ΔG| values of miRNA (red) and miRNA.1 (dark grey) duplexes from conserved and young sequential MIRNAs. Different letters indicate statistically significant differences, according to ANOVA (P-value < 0.0001) followed by Tukey's multiple comparison test (P < 0.05). (C) Box plots representing differential interacting energy (|ΔΔG|) between the miRNA/miRNA* and the miRNA.1/miRNA.1* from each evolutionarily young (green) or conserved (light grey) sequential MIRNA. Asterisks indicate statistically significant differences according to Wilcoxon signed-rank test (two-sided), P < 0.001 (***). (D) Frequency of small RNA duplex accumulation. The colour is described in (A). (E) Box plots representing |ΔG| of miRNA duplexes from evolutionary young (left) and conserved (right) MIRNAs processed with two DCL1 cuts, no statistically significant differences were observed. See Supplementary Table S4 for all statistical tests performed. (F) Conservation of the miRNA/miRNA* region of MIR164a, MIR395a, MIR408 and MIR162b region. Sequence logos were generated using WebLogo (v 3.7.12). Arrows indicate the sites of DCL1 cuts. (G) Box plot showing the conservation of miRNA (red) and miRNA* (grey) using phastCons (v 1.5) for miRNA and miRNA* sequences according to the function of the miRNA’s targets: transcription factors (TF, 25 miRNAs, 651 orthologues), nutrient deficiency (nine miRNAs, 221 orthologues) and RNA metabolism and hormone signalling (RNA & Signalling, six miRNAs, 155 orthologues). Different letters indicate statistically significant differences (Kruskal–Wallis multiple comparison test, P-value < 0.05).
In vitro transcription
Pri-miR319a and pri-miR394a were transcribed in vitro using T7 RNA polymerase as described before (11). The transcription products were purified from a denaturing 6% (w/v) polyacrylamide gel and precipitated (Tris 0.01 M pH 8, EDTA 0.001 M pH 8, NaCl 0.6 M and three volumes of absolute ethanol). Using a T4 polynucleotide kinase (Thermo Fisher Scientific), 100 pmol of dephosphorylated pri-miR394a and pri-miR319a transcripts were 5′ end labelled with [γ -32P] ATP (10 μCi/μl). The products were purified from a denaturing 8% (w/v) polyacrylamide gel using autoradiography and precipitated as described above.
Nuclease digestions
Radioactively labelled products of in vitro transcription were partially digested using T1 RNAse (Fermentas, denaturing conditions), V1 RNase (Ambion, native conditions) and S1 nuclease (Fermentas, native conditions) as described before (11). Three different V1 concentrations were used 0.001, 0.005 and 0.01 U/μl. Three different S1 concentrations were used 0.25, 0.5 and 1 U/μl. Alkaline hydrolysis was included as a ruler, and an incubation with water as integrity control. The digested products were separated in an 8% (w/v) denaturing gel. The results were obtained by phosphor-imaging using a Typhoon FLA 7000 (GE Healthcare).
Statistical analysis
Statistical tests with statistically significant results are detailed in the Figure captions including p-values. All statistical tests are included and detailed in Supplementary Table S4. Their respective tables as well as details of sample sizes (n) and plant tissue used are included in Supplementary Table S4. RT-qPCR quantification, flowering time phenotype, |ΔG| of small RNA duplexes, and reads of small RNA duplexes were analyzed with InfoStat software (32) (http://www.infostat.com.ar).
Bioinformatics analysis
MIRNA secondary structure was predicted using the mfold web server for RNA folding (33). See Appendix II for the secondary structure of the wild-type and mutant precursors used here. Note that, as determined by mfold, the mutations in the miRNA/miRNA* duplexes do not affect the folding of the overall structures. ΔG values and secondary structure of small RNA duplexes were predicted using the Two-state-Melting version of mfold (http://www.unafold.org/mfold/applications/rna-folding-form.php) and (http://www.unafold.org/Dinamelt/applications/two-state-melting-hybridization.php, using energy rules: RNA (2.3).
Sequence conservation analysis
We employed a comprehensive approach to assess the conservation of miRNA and miRNA* sequences. We aligned sequences of miRNA and miRNA* from the orthologs of MIR164a, MIR395a, MIR408 and MIR162b from dicots species previously identified in (16). Then, using these aligned sequences, we generated a WebLogo (v 3.7.12) to visually represent the conservation patterns. To quantify the conservation of miRNA and miRNA* sequences, we utilized PhastCons to compute base-by-base conservation scores of aligned miRNA precursors. The alignment included orthologs present in at least 20 dicots species from (16), allowing us to evaluate the degree of conservation in both miRNA and miRNA* regions.
Results
The processing of two-step plant MIRNAs is suboptimal
The dsRNA region in the precursor that contains the miRNA has a variable number of mismatches. To assess their significance, we closed them in different DCL1 two-step MIRNAs through site-directed mutagenesis. Given that the structural determinants governing processing modes are located outside the miRNA/miRNA* region, specifically as additional dsRNA segments of 15–17 nt above or below the duplex (11), we do not expect these mutations to affect the processing mode. First, we cloned MIR164a-wt precursor, which contains three mismatches in the miR164/miR164* duplex (MIR164a-3MMwt) and introduced a point mutation in miR164a* to close a mismatch at position 9 (MIR164a-2MM) and two-point mutations to close the mismatches at position 9 and 14 (MIR164a-1MM) of the miRNA/miRNA* (Figure 1A). Next, we expressed MIR164a-3MMwt, MIR164a-2MM, and MIR164a-1MM from the 35S promoter in Arabidopsis (Figure 1A). Then, we analysed miRNA accumulation by small RNA blots in ten-day-old seedlings, with each sample being a pool of 15 independent primary transgenic plants. We found that closing one and two mismatches increased miR164 accumulation by 5- and 10-fold, respectively. Using RT-qPCR, we determined the pri-miR164 levels of these precursors. We found that MIR164a-2MM and MIR164a-1MM had a ∼6- and 10-fold reduction compared to MIR164a-wt (Figure 1B). The relative increase in miR164 and the decrease in pri-miR164 levels indicate that closing mismatches enhances the precursor processing. MIR164 overexpression is known to interfere with the meristem function and cause important developmental defects (34). We analysed the phenotypic defects in plants overexpressing MIR164a-2MM and MIR164a-1MM and found an increased frequency of these defects compared to the wt precursor, demonstrating that the increased levels in miR164 were biologically functional (Supplementary Figure S1A).
Next, we closed mismatches at position 18 of the miRNA/miRNA* of MIR172c and at position 1 of MIR172a by mutating the miRNA* sequence (Figure 1C and E). This led to a 3-fold increase in miR172 levels in both cases. Overexpression of miR172 is known to cause flower developmental defects (35), and we confirmed that the increased levels of miR172 by these mutant precursors also resulted in more pronounced defects (Supplementary Figure S1BC) and a decrease of their pri-miRNA transcripts (Figure 1D, F). We also introduced a mutation in MIR397a to close a mismatch in position 19 (Figure 1G, H). In this case, we observed a 10-fold increase in the miRNA levels (Figure 1G).
MIR164a, MIR172c, MIR172a and MIR397a are processed in two steps in a base-to-loop direction (9,11). To study a pri-miRNA processed by DCL1 in two steps in a loop-to-base direction, we assessed MIR171b. We generated two different mutants closing one mismatch at position 19 or at position 12 (MIR171b-1MMU and MIR171b-1MMG, respectively) and expressed them in Arabidopsis under the 35S promoter. In this case, we observed that a single change at position 19 almost did not affect the accumulation of the mature miRNA. However, with one change at position 12 in the miRNA/miRNA* duplex, we observed a 50% increase in the accumulation of miR171 (Figure 1I) with more pronounced developmental defects that are known to be caused by miR171 overexpression (12) (Supplementary Figure S1D). The latter mutant also accumulated less pri-miRNA, although the changes were not statistically significant (Figure 1J). In summary, our results indicate that reducing the number of mismatches in the miRNA/miRNA* regardless of the nt identities in precursors processed in two steps increases the production of miRNAs, which causes stronger developmental defects, while simultaneously reducing the levels of their primary transcripts. Taken together, our results show that the processing of several plant two-step MIRNAs is suboptimal in vivo.
Quantitative regulation of miRNA processing by the miRNA/miRNA* duplex
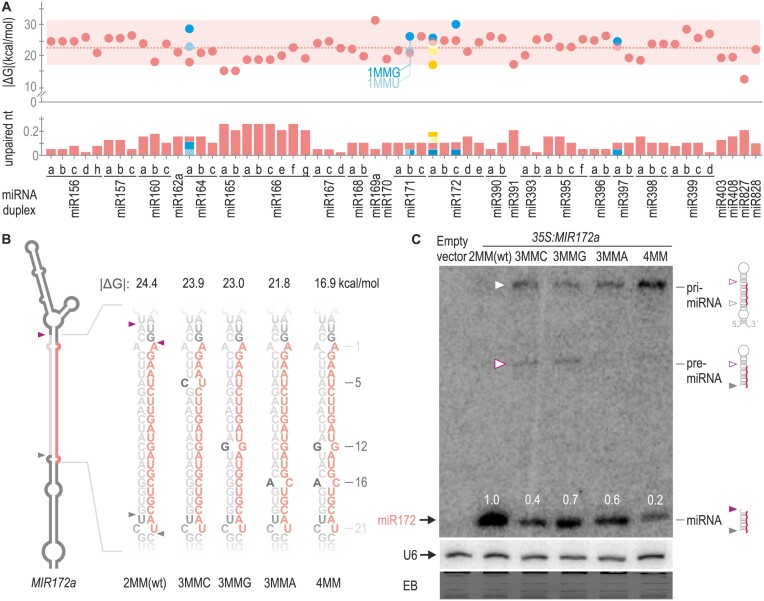
The observation that closing mismatches regardless of positions and nt identities led to increased processing efficiency prompted us to perform a systematic study of miRNA duplexes. We calculated the Gibbs free energy (ΔG) with the Two State Melting web server (33) and obtained the absolute value of ΔG (|ΔG|). Higher values of |ΔG| correspond to thermodynamically more stable miRNA/miRNA* (Supplementary Table S5). We analysed 62 Arabidopsis MIRNAs evolutionary conserved and experimentally validated to be processed by two cuts (11). Of these duplexes, 95% displayed interacting energy levels within the range of ∼17 to 31 kcal/mol (Figure 2A top panel, red box). Additionally, we assessed the frequency of unpaired nt in the duplex (including mismatches or bulges) which ranged from 0.05 to 0.26 (Figure 2A, bottom panel, Supplementary Table S5). All mutants tested in Figure 1 were within these limits (Figure 2A, blue dots and bars).
Figure 2.
Quantitative regulation of MIRNA processing efficiency by the miRNA/miRNA* secondary structure. (A) Dot plot representing the |ΔG| of 62 miRNA duplexes from two-step MIRNAs shown as red dots. A dotted red line shows the mean |ΔG|. A red box encloses 95% of the duplexes which have an interacting energy between ∼17 and 31 kcal/mol. Unpaired nt frequency is shown as red bars on the bottom panel. Blue and yellow dots and bars indicate modifications in the |ΔG| and unpaired nt frequency of miRNAs duplex from Figures 1 and 2B, respectively. (B) Schematic representation of MIR172a, the sequence of the wt and mutant miRNA duplexes is shown to the right. These depictions are based on mfold secondary structure prediction (see Appendix II). Black letters specify the introduced mismatches, and the numbers indicate their position counting from the 5′end of the miRNA. MiRNA is shown in red and miRNA* in grey. |ΔG| of the duplex are shown above each small RNA sequence. (C) Small RNA blot for miR172 transgenic lines expressing different precursors from the 35S promoter. Each sample corresponds to 15 pooled 10-day-old seedlings from independent primary transgenic plants. Numbers above small RNA blots correspond to miRNA levels quantified relative to the wt precursor. Schematic representation of the detected MIR172 processing intermediates is shown on the right. A grey empty arrow and purple empty arrow indicate the pri-miRNA and pre-miRNA detected in the blot, respectively. Both U6 hybridization and EB staining are shown at the bottom.
To investigate the impact of reducing miRNA duplex stability on miRNA accumulation, we generated three single mutants of MIR172a. These mutations included opening: an A–U pair at position 5 by replacing it with a C-U (MIR172a-3MMC), a G–C pair at position 12 with a G–G (MIR172a-3MMG), and a G–C pair at position 16 with an A–C (MIR172a-3MMA) (Figure 2A yellow dots and bars and Figure 2B). When these precursors were expressed in plants, all three mutants exhibited reduced levels of miRNA, approximately 40–70% of the wt precursor (Figure 2C). A similar effect, irrespective of the identity of the opened pair (A–U or G–C) or its position, was observed for all three mutants. Next, we combined both mutations at positions 12 and 16 (MIR172a-4MM), resulting in cumulative effects that further reduced miR172 levels to 20% (Figure 2C).
In the miR172a blot (Figure 2C) we observed RNA species of larger size also detected with the miRNA probe. In plants overexpressing MIR172a-wt, only the miRNA was detected. However, in the precursor mutants with additional mismatches, we observed the accumulation of pri-miR172 (Figure 2C, grey arrow). Furthermore, the MIR172a-4MM mutant accumulated increased levels of the pri-miR172 compared to the mutants with three mismatches, concurrently with decreased miR172 levels (Figure 2C). Interestingly, in two of the mutants (MIR172a-3MMC and MIR172a-3MMG) we detected an additional band corresponding to pre-miR172 (Figure 2C, purple arrow, see schematic representation to the right of the blot). These results support the observation that the overall number of mismatches in the miRNA/miRNA* quantitatively affects processing efficiency, and that lower processing efficiencies cause the accumulation of miRNA precursor intermediates, especially when the interacting energy is below the ∼17 kcal/mol threshold found for most two-step MIRNAs (Figure 2A, top panel).
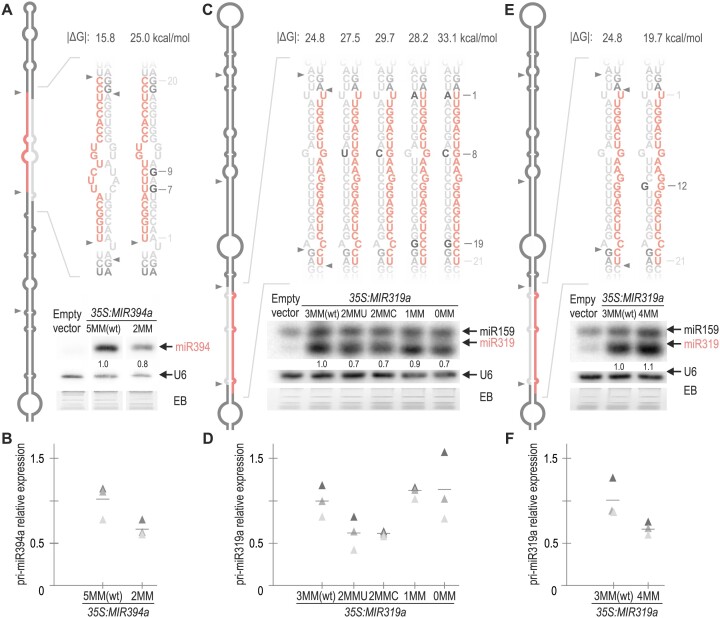
Processing of sequential MIRNAs is largely insensitive to variations in the miRNA/miRNA* region
In Arabidopsis, nearly 25% of experimentally validated processed MIRNAs require more than two DCL1 cuts to release the miRNA (11). The base-to-loop sequential MIR394a stands out by presenting the miRNA/miRNA* with the lowest interacting energy (Supplementary Table S5). To assess the impact of this low interaction energy, we introduced point mutations into miR394a*, increasing the interacting energy from 15.8 to 25 kcal/mol in MIR394a-2MM. In this case, small RNA blots revealed that the mutant precursor generated slightly reduced amounts of miR394 with minor changes in pri-miR394 levels (Figure 3A, B and Supplementary Figure S2A), indicating that the increase in the miR394/miR394* interaction energy does not result in a higher accumulation of the mature miRNA. Next, we sought to enhance the duplex stability of another sequentially processed MIRNA, the loop-to-base MIR319a. We introduced point mutations to progressively close the three mismatches within the miR319a duplex, creating MIR319a-2MMU, MIR319a-2MMC, MIR319a-1MM and MIR319a-0MM (Figure 3C). Small RNA blots showed that miR319 levels remained similar or were slightly reduced across all MIR319a versions (Figure 3C) without changes in pri-miRNA levels (Figure 3D).
Figure 3.
The biogenesis of sequential MIRNAs is tolerant to changes in the miRNA/miRNA*. (A,C and E) Small RNA blots of transgenic plants expressing different precursors from the 35S promoter. Each sample corresponds to 15 pooled inflorescences from independent primary transgenic plants. Numbers above small RNA blots correspond to miRNA levels quantified relative to the wt precursor. The U6 hybridization and the EB staining of each gel are shown below. Supplementary Figure S2A shows the original image of miR394a blot autoradiography. Note that the probe against miR319 also detects miRNA miR159, due to its high sequence similarity, and the abundance of endogenous miR159. Despite this, both miRNAs are easily distinguishable by their different electrophoretic mobilities (36). Top panels show a schematic representation of (A) MIR394a and (C and E) MIR319a precursor and the sequence of the wt and mutant miRNA duplexes according to the mfold secondary structure prediction (see Appendix II). Black letters specify the introduced mutations, and the numbers indicate their positions counting from the 5′end of the miRNA. MiRNA is shown in red and miRNA* in grey. |ΔG| of the duplex are shown above each small RNA sequence. (B,D and F) RT-qPCR analysis of pri-miRNA levels determined in three biological replicates are shown as triangles in different shades of grey. Lines represent the mean value for each MIRNA. No statistically significant differences were observed in B, D and F. See Supplementary Table S4 for the detailed statistical analysis conducted in all the samples.
Finally, we introduced a point mutation to generate a fourth mismatch in miR319a/miR319a*, reducing the duplex interacting energy (MIR319a-4MM). Small RNA blots showed MIR319a-3MMwt and MIR319a-4MM miR319 levels were comparable, as well as their pri-miRNA levels determined by RT-qPCR (Figure 3E, F). Phenotypic analysis of MIR319a-wt and all mutant versions agreed with the small RNA blot results (Supplementary Figure S2B–D). Most conspicuously, the production of miR319 was similar in a wide range of interaction energies from 19.7 to 33.1 kcal/mol, showing a ∼13 kcal/mol tolerance (Figure 3); in contrast to our observations in two-step MIRNAs, whose processing could be considerably modified by changes of 2 kcal/mol (Figures 1 and 2). Altogether, our findings suggest that while the processing of two-step MIRNAs is susceptible to changes in the miRNA duplex, sequential MIRNAs processing is largely insensitive to variations in the interacting energy of their miRNA/miRNA*, irrespective of their processing direction.
Two-step and sequential MIRNAs have differential miRNA/miRNA* requirements for their biogenesis
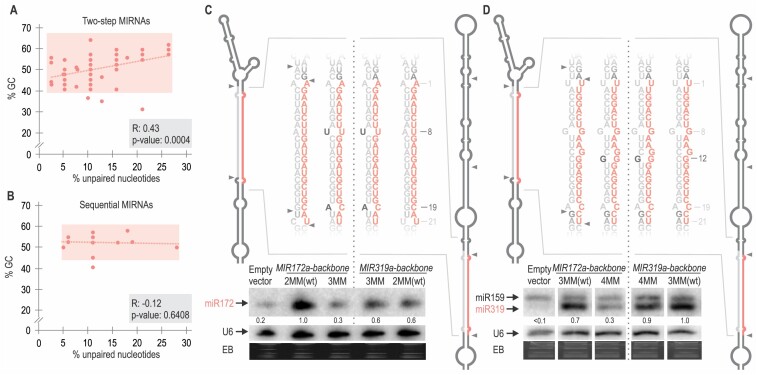
We sought to compare side-by-side the miRNA/miRNA* from two-step and sequential MIRNAs. We calculated the percentage of GC and percentage of unpaired nts for two-step (Figure 4A) and sequential (Figure 4B) MIRNAs (Supplementary Table S5). We observed that for two-step MIRNAs an increase in GC content correlated with an increase in unpaired positions (Figure 4A), suggesting a tendency to maintain the overall interacting energy of the miRNA/miRNA* within certain limits, as expected from its key role in processing efficiency. Conversely, we did not observe any correlation for the sequential MIRNAs (Figure 4B), in agreement with the low impact of this region on the processing efficiency of these precursors.
Figure 4.
Chimeric precursors expressing the same miRNAs show differential miRNA/miRNA* requirements depending on their processing mechanism. (A and B) Dot plot and correlation analysis of the percentage of GC nt versus percentage of unpaired nt in the miRNA/miRNA* of (A) two-step and (B) sequential MIRNAs shown as red dots. A red dotted line shows the linear regression curve obtained from the plotted data. For two-step miRNAs, there is a significant positive correlation, Spearman's correlation test (see Supplementary Table S4). (C and D) Small RNA blot of transgenic plants expressing different precursors from the 35S promoter. Each sample corresponds to 15 pooled inflorescences from independent primary transgenic plants. Numbers above small RNA blots correspond to miRNA levels quantified relative to the wt precursor. U6 hybridization and EB staining are shown at the bottom. Supplementary Figure S3C shows the original image of (D) blot. Schematic representation of the expressed MIRNA and their sequences: (C) miR172a-2MMwt and miR172a-3MM expressed from two-step MIR172a (left) and from the sequential MIR319a (right). (D) miR319a-3MMwt and miR319a-4MM expressed from MIR319a (right) and from MIR172a (left). These depictions represent the mfold secondary structure prediction for each precursor (see Appendix II). Black letters specify the introduced mismatches, and the numbers indicate their position counting from the 5′end of the miRNA. MiRNA is shown in red and miRNA* in grey.
To confirm that the miRNA/miRNA* requirements depend on the processing mechanism and not the primary sequence, we express the same miRNAs within the identical miRNA/miRNA* using a two-step (MIR172a) and a sequential (MIR319a) precursor backbone. First, we over-expressed miR172 sequence from MIR172 and MIR319 backbones using the wt secondary structure of miR172a/miR172a*, which has two mismatches, and introduced a third mismatch in the miRNA/miRNA* (Figure 4C, left). Small RNA blots revealed that MIR172 was more efficient than MIR319 backbone in expressing miR172 from the endogenous miRNA/miRNA* structure with two mismatches. However, introducing a third mismatch almost completely abolished miR172 production from the MIR172 backbone, while the production of miR172 from MIR319 was mostly unaffected (Figure 4C, right). Moreover, expression of miR172 from MIR172 backbone with 3MM in the miRNA/miRNA* failed to induce early flowering time in Arabidopsis, while either variant of MIR319 expressing miR172 produced early flowering time (Supplementary Figure S3A). Note that all precursors were expressed to similar levels (Supplementary Figure S3B).
Next, we expressed miR319a from MIR172 and MIR319 backbones using the same miRNA/miRNA* (Figure 4D). Small RNA blots showed that miR319a/miR319a* with 3MM produces similar amounts of miRNA from either MIR172a or MIR319a backbone (Figure 4D). However, introducing an additional mismatch (4MM) significantly affected miR319 from the MIR172a, but did not affect its biogenesis from the MIR319a backbone (Figure 4D). These results are in agreement with our previous observations indicating that the two-step MIRNAs are more sensitive to changes in the energy of the miRNA/miRNA* duplex than the sequential MIRNAs.
Distinct properties of small RNA duplexes from sequential MIRNAs
DCL1 processing of sequential MIRNAs releases the miRNA/miRNA* in the last two cuts. The other small RNA duplexes produced during the processing will be referred to as miRNA.1/miRNA.1* and miRNA.2/miRNA.2* according to their relative distance to the miRNA/miRNA* (Figure 5A, dark grey, and light blue lines, respectively).
We examined the relative accumulation of all small RNA duplexes derived from evolutionary conserved sequential MIRNAs: miRNA/miRNA*, miRNA.1/miRNA.1* and miRNA.2/miRNA.2* using publicly available small RNA libraries (11) (Supplementary Table S3). As expected from the definition of miRNAs (37), the data showed that miRNA/miRNA* had the highest number of small RNA reads in all the detected sequential MIRNAs, while miRNA.1/miRNA.1* and miRNA.2/miRNA.2* were detected at much lower levels (Figure 5B, top panel). This accumulation pattern was consistent across various small RNA libraries (Supplementary Figure S4A). We calculated the |ΔG| of the miRNA/miRNA* (red), miRNA.1/miRNA.1* (dark grey) and miRNA.2/miRNA.2* (light blue) (Figure 5B, lower panel and Supplementary Table S5). In all cases, the miRNA/miRNA* presented the highest |ΔG| when compared to miRNA.1 and miRNA.2 duplexes, thus showing that sequential MIRNAs mostly accumulate the small RNA duplex with the highest stability. Moreover, the interacting energy of most of the miRNA.1/miRNA.1* and miRNA.2/miRNA.2* fell within the 7–15 kcal/mol range (Figure 5B), which is lower than the miRNA/miRNA* energy of the sequential MIRNAs (15–27 kcal/mol) (Figure 5B) or the two-step MIRNAs (17–31 kcal/mol, Figure 2B).
To gain experimental insights into the secondary structure of these sequential precursors, we performed in vitro structural mapping of MIR319a (Figure 5C). The results showed that the most stable regions of miRNA.1/miRNA.1* and miRNA.2/miRNA.2* were located at the ends of the duplexes (Figure 5C, blue lines) (Supplementary Figure S4BC). Consequently, the structural features of these long sequential precursors rely on unstable small RNA duplexes (miRNA.1/miRNA.1* and miRNA.2/miRNA.2*), with internal mismatches and paired ends that generate suitable DCL1 cleavage sites (20). On the other hand, the miRNA/miRNA* presented an overall better pairing (Figure 5C, blue lines) as expected from its higher interacting energy and consequent accumulation.
To validate this model, we introduced changes in the miR319a.2/miR319a.2* region of MIR319a (13.1 kcal/mol) to match the minimal interaction energy of two-step miRNA/miRNA* of ∼17 kcal/mol (Figure 2A). Specifically, we closed one mismatch at position 15 of miR319a.2* by generating either a G:U (MIR319a.2GU, 16.8 kcal/mol) or A:U (MIR319a.2AU, 17.1 kcal/mol) base pair. Small RNA blots showed that MIR319a.2GU and MIR319a.2AU accumulated miR319a.2 while MIR319a.2wt did not (Figure 5D). We also mutated the miR319c.2/miR319c.2* region of MIR319c to increase the interacting energy yielding similar results (Supplementary Figure S4D). Next, we altered the relative position of a 12 unpaired nt internal loop found in the middle region of miR319a.1/miR319a.1*, from 9 nt below DCL1 second cut, to 2 nt below the cleavage site (Figure 5F). As predicted, the mutated precursor (MIR319a-MIL) failed to accumulate miR319. Finally, we introduce point mutations in miR319* to lower the interacting energy of miR319 duplex similar to a miR319a.2 duplex, MIR319a-5MM, with a |ΔG| of 13.7 kcal/mol. Small RNA blot showed that MIR319a-5MM failed to accumulate miRNA (Figure 5H). We did not observe significative changes in the pri-miRNA levels for any variant, suggesting that the changes in miRNA level were not due to the impairment of DCL1 first cleavage (Figure 5E, G and I). Overall, these results suggest that small RNA accumulation from a sequential MIRNA requires a minimal interacting energy independently of its relative position within the precursor.
Conservation and divergence of miRNA/miRNA*
Our results indicate distinct roles and structural requirements for the miRNA/miRNA*, depending on the precursor processing mechanism. We decided to analyse evolutionarily conserved and young MIRNAs. First, we examined the miRNA/miRNA* and miRNA.1/miRNA.1* regions of young sequential MIRNAs and calculated the |ΔG| for the different small RNA duplex (Supplementary Table S5). In young MIRNAs, the miRNA/miRNA* was more stable than the miRNA.1/miRNA.1* (Figure 6A and Supplementary Figure S5A), albeit the difference was not statistically significant when analysed as a group (Figure 6B). In conserved sequential MIRNAs, this difference was larger, due to both an increase in the interacting energy of the miRNA/miRNA* and a decrease in the interaction energy of the miRNA.1/miRNA.1* (Figure 6B). Considering each MIRNA individually, the energy difference (|ΔΔG|) between the miRNA/miRNA* and miRNA.1/miRNA.1* was on average approximately 11 kcal/mol for conserved MIRNAs and only 2–3 kcal/mol for the young ones (Figure 6C). Moreover, the accumulation pattern of small RNAs derived from young sequential MIRNAs did not have a clear preference for the miRNA as observed in the conserved ones (Figure 6D, Supplementary Table S3). Furthermore, it should be noted that miR402 and miR447 (miRbase v22, (38)) were not the most abundant small RNAs from their precursors in the libraries analysed.
We also compared the interacting energy of miRNA/miRNA* from evolutionarily young and conserved two-step precursors (Supplementary Table S5). In this case, while the interaction energy was higher in the conserved ones, this difference was not statistically significant (Figure 6E). Given the strong influence of miRNA/miRNA* interacting energy on the processing efficiency of two-step MIRNAs, we analysed the conservation of this region across different species. We detected a varying conservation of miRNA/miRNA* along MIRNA orthologs from different species (Figure 6F). For example, the sequence and therefore the secondary structure of MIR164a and MIR162b miRNA/miRNA* were rather conserved in different species, compared to MIR408 and MIR395a (Figure 6F). Interestingly, the miRNA* region was more variable in certain MIRNAs (Figure 6F). An inspection of the biological roles of these MIRNAs revealed that many of them participate in the response to nutrient deficiency. We then classified the miRNAs into three groups according to the biological roles of their targets: miRNAs that regulate transcription factors (TF), nutrient deficiency, and RNA processes and hormone signalling (RNA & Signalling) and calculated the conservation of the miRNA and miRNA* sequences using phastCons. We found that the miRNA sequences were equally conserved for the three groups (Figure 6G). On the other hand, we found that the miRNA* sequences were less conserved than the miRNAs, as expected (Figure 6G). However, the miRNA* sequences corresponding to the MIRNAs that participate in the response to nutrient deficiency were largely more variable than the others (Figure 6G). Therefore, we observed a correlation between the miRNA function and the conservation of the miRNA/miRNA* in two-step precursors.
Discussion
In contrast to their stereotypical animal counterparts, plant miRNA precursors comprise a collection of fold-back structures with largely variable shapes and sizes (7). To cope with this structural variation, the DCL1 complex processes MIRNAs in diverse ways, guided by different structural determinants beyond the miRNA/miRNA*. In certain cases, the processing complex identifies a 15–17 base pair ds RNA segment below the miRNA/miRNA* region that determines the position of the first cut at the base of this duplex (10,12–14,18). After this initial cleavage, DCL1 performs a second cut 21 nt away from the precursor end (17,39), proceeding in base-to-loop direction. However, only a fraction of the miRNA precursors harbours this structural determinant. In other cases, a dsRNA region above the miRNA/miRNA* region and below a small terminal loop guides the DCL1 complex to perform the first cut, resulting in a loop-to-base processing mechanism (9,11,15,16). In addition, approximately a quarter of the precursors in plants are processed sequentially by more than two DCL1 cuts, generating several small RNA duplexes (7–9,11,40). Here, we systematically analysed the role of the miRNA/miRNA* structure in miRNA biogenesis and found a link between the MIRNA processing mechanism and the structural features of this small RNA duplex.
Quantitative regulation of miRNA processing by the pairing of the miRNA/miRNA* in two-step precursors
It has been demonstrated that the identity of the bases at mismatched positions in the precursor influences miRNA precursor processing, with C–C mismatches being severely deleterious for MIRNA processing as they particularly increase the flexibility of the precursor stem (20). In addition, a previous study showed that a natural variant of MIR164c with a single nucleotide polymorphism (SNP) producing a G–C to G–U change resulted in reduced miRNA levels (41) and that a GC-signature formed by G-C pairs at specific positions favours the precursor processing (19). Therefore, we conducted studies modifying both G–C and A–U pairs, as well as different positions such as 1, 5, 8, 9, 12, 14, 16, 18 and 19 counting from the miRNA 5′ end and avoided introducing C-C mismatches (Figures 1, 2 and 4) in several two-step endogenous MIRNA precursors (MIR164a, MIR171b, MIR172c, MIR172a and MIR397a). We found that these changes in the miRNA/miRNA* affected miRNA biogenesis quantitatively. Our results show that the overall interaction energy and likely the rigidity of the stem quantitatively regulate the efficiency of processing in these precursors.
Most notably, we observed that these MIRNA precursors are processed at a sub-optimal efficiency in vivo. Therefore, it is plausible that this is a common feature of many plant MIRNA precursors processed in two steps. Furthermore, their biogenesis can be improved by increasing the miRNA/miRNA* pairing. We found that a minimal interacting energy of ∼17 kcal/mol is required for a functional miRNA/miRNA* in these precursors. Interestingly, the quantitative effect the miRNA/miRNA* allows the differential production of a miRNA in a wide dynamic range by adjusting its structure. The biogenesis of miR172 can be quantitatively up or downregulated by increasing (Figure 1) or decreasing (Figure 2) the paring of the miR172/miR172*, respectively, resulting in an expression range for the miRNA of more than one order of magnitude. This feature might be useful for the precise expression of artificial miRNAs designed to fine-tune the expression of key genes of interest.
In certain cases, the precise expression of a miRNA might be of biological significance, a possibility further supported by the fact that an increased number of mismatches in the miRNA/miRNA* tends to be compensated by an increase in the number of GC content (Figure 4). This might be of particular importance for those miRNAs involved in regulatory feedback loops, such as miR172, whose transcription is negatively regulated by its targets, AP2 transcription factors (42–44). In turn, the pairing of the miRNA/miRNA* could be targeted in vivo to inactivate miRNA expression, as shown by the ATPase subunit of the switch/sucrose non-fermentable (SWI/SNF) complex, CHR2, impairing MIR164b processing by increasing the unpaired bases on the miRNA/miRNA* region (45). In the same direction, m6A MIRNA methylation, prevents the miRNA/miRNA* to adopt an unpaired conformation, which reduces HYL1 binding and processing efficiency (46). Interestingly, we observed that miRNA/miRNA* from miRNAs regulating transcription factors and RNA & Signalling pathways were more conserved across evolution than those whose transcription is strongly activated during stress conditions, such as those miRNAs regulating enzymes, transporters and metal-binding proteins that become repressed during nutrient deficiency. We hypothesize that the expression of the latter miRNAs is predominantly regulated at the transcriptional level, suggesting that the biological regulation and function of the miRNA might be linked to the pattern of evolution of the miRNA/miRNA* region.
Processing of sequential MIRNA
The processing of sequential miRNA precursors, which generate at least two small RNA duplexes, harbours different structural determinants. Usually, the region that mostly accumulates a small RNA, which has an evolutionarily conserved target gene and is generated by the last two DCL1 cuts, also has the highest interaction energy. The other small RNA duplexes generally act as bridges allowing the processivity of the sequential precursor. These duplexes have a |ΔG| usually less than ∼15 kcal/mol, with a prevalence of interacting base pairs in the DCL1 cleavage sites. Increasing the interaction energy above 17 kcal/mol results in an increased accumulation of these small RNAs, while moving the mismatches near the DCL1 cleavage site abolishes the precursor processing. However, we cannot discard that these additional small RNA duplexes can have specific biological roles, as it has been reported that miR319b.2 can guide the cleavage of an additional target gene in Arabidopsis (47).
The miRNA/miRNA*, which is released by the last two DCL1 cuts, results quite impervious to the changes in the interaction energy. Changes in the range of ∼14 kcal/mol in the interaction of miR319/miR319* resulted in variations of up to 30% in accumulation (Figure 3–5). In contrast, changes of 2–3 kcal already produced significant modifications in the biogenesis of two-step miRNAs, which can be up to ∼0.1–10 times (Figures 1 and 2). Similar results were obtained after expressing the same miRNAs with identical miRNA/miRNA* using the two-step MIR172 and the sequential MIR319, being sensitive and tolerant to an increased number of mismatches, respectively. Interestingly, the data also revealed that the two-step MIR172 precursor was more effective than the sequential MIR319 when using a stable miRNA/miRNA* (Figure 4). This is in agreement with detailed studies of the two-step MIR390, which has been developed as a system to efficiently produce artificial miRNAs in plants (48–50).
Evolution of plant MIRNAs
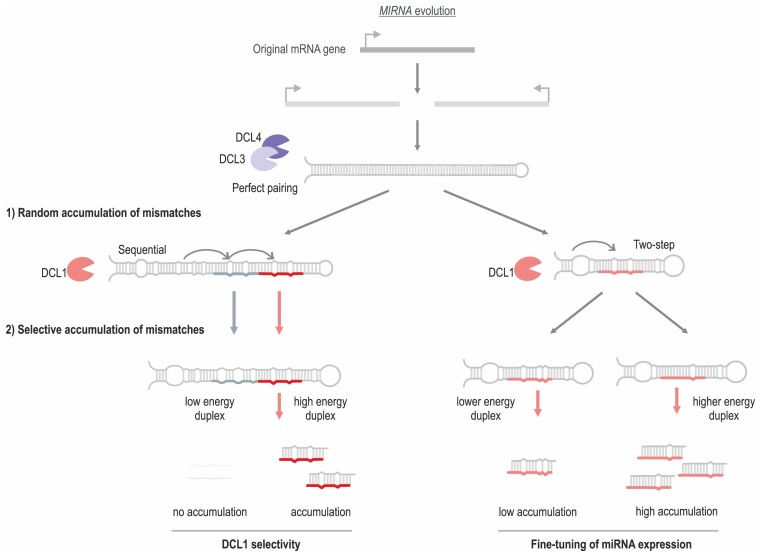
One hypothesis about the origin of miRNAs proposes that they were generated as inverted duplications of protein-coding genes (51,52). The result would be a perfectly paired stem-loop RNA that could be processed by diverse DCL proteins such as DCL3 and DCL4, in a process that generates diverse small RNAs potentially capable of regulating the gene of origin. Over time, this proto-MIRNA would accumulate mutations, giving rise to MIRNA with an increased number of mismatches along the stem of the precursor, resulting in a fold-back precursor preferentially processed by DCL1 (51–53) (Supplementary Figure S5B).
We propose that the accumulation of mutations in the fold-back structures proceeds through different phases (Figure 7). As a result of the first phase, the proto-MIRNA becomes a fold-back that is recognized by DCL1 and generates phased small RNA duplexes with relatively high precision (Figure 7). In the second stage, the accumulation of mutations in the miRNA/miRNA* promotes the accumulation of the miRNA over other small RNAs. Evolutionarily young sequential MIRNAs produce several small RNA duplexes that have similar energies, and accumulate to similar degrees (Figures 6 and 7), while evolutionary ancient sequential MIRNAs present additional mutations in the passenger small RNA duplexes and generate mainly a single miRNA (Figures 5–7). For example, the evolutionarily young MIR163 is processed sequentially by DCL1 generating two small RNAs that are easily detected in vivo (17). The energy difference between these two small RNA duplexes is less than 6 kcal/mol, in contrast to the 11 kcal/mol average difference found in evolutionarily conserved sequential MIRNAs. We propose that, in the case of sequential precursors, mutations provide specificity to generate a single miRNA (Figure 7, left).
Figure 7.
Proposed model of MIRNA evolution. A protein-coding gene undergoes an inverted duplication, generating a hairpin that is processed by DCL3 and/or DCL4. The processing of this proto-MIRNA generates a diverse mixture of small RNAs. In the next stage, the accumulation of mutations determines that the fold-back is processed by DCL1 and establishes the structural determinants for its first cut. In a subsequent stage, mutations in the small RNA duplex region of the precursor provide specificity for selected miRNAs in sequential MIRNAs, while mutations in the two-step precursors determine the quantitative levels of miRNA biogenesis.
On the other hand, the accumulation of mutations in the miRNA/miRNA* of two-step MIRNAs contributes to regulating the amount of the produced miRNA. We propose that for MIRNAs involved in complex gene regulatory networks, a precise amount of miRNA needs to be produced to meet the physiological demands. This might explain the conservation of the miRNA* and the fine-tuning of the miRNA level by the structure of miRNA/miRNA* in certain MIRNAs such as those involved in the regulation of transcription factors and RNA metabolism and signalling.
Supplementary Material
Acknowledgements
We thank Rodolfo Rasia, Carla Schommer, Ramiro Rodriguez, Julia Baulies and members of the J.F.P. lab for comments and discussions, and Gordon Rotherham for support during the writing of this manuscript. We thank Diego Aguirre for the technical support on this paper.
Contributor Information
Santiago Rosatti, Instituto de Biología Molecular y Celular de Rosario (IBR), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Universidad Nacional de Rosario, Rosario, Santa Fe, 2000, Argentina.
Arantxa M L Rojas, Instituto de Biología Molecular y Celular de Rosario (IBR), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Universidad Nacional de Rosario, Rosario, Santa Fe, 2000, Argentina.
Belén Moro, Instituto de Biología Molecular y Celular de Rosario (IBR), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Universidad Nacional de Rosario, Rosario, Santa Fe, 2000, Argentina; Centre for Research in Agricultural Genomics (CRAG), CSIC-IRTA-UAB-UB, Campus UAB, Barcelona 08193, Spain.
Irina P Suarez, Instituto de Biología Molecular y Celular de Rosario (IBR), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Universidad Nacional de Rosario, Rosario, Santa Fe, 2000, Argentina.
Nicolas G Bologna, Centre for Research in Agricultural Genomics (CRAG), CSIC-IRTA-UAB-UB, Campus UAB, Barcelona 08193, Spain.
Uciel Chorostecki, Faculty of Medicine and Health Sciences, Universitat Internacional de Catalunya, Sant Cugat del Vallès, Catalunya 08195, Spain.
Javier F Palatnik, Instituto de Biología Molecular y Celular de Rosario (IBR), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Universidad Nacional de Rosario, Rosario, Santa Fe, 2000, Argentina; Centro de Estudios Interdisciplinarios, Universidad Nacional de Rosario, Rosario, Sante Fe, 2000, Argentina.
Data availability
All data presented in this work is available in the manuscript as Supplementary Tables, Figures and Appendices. We also analyse publicly available and deposited data at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Small RNA libraries from seedlings used in Figures 5, 6 and Supplementary Figure S5 were obtained from ((11), accession number GSE116330). Small RNA libraries from leaves and inflorescences used in Supplementary Figures S4 and Figure S5 were extracted from ((31); accession numbers GSM506656-8 and GSM506662-4).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Consejo Nacional de Investigaciones Científicas y Técnicas [to B.M., A.M.L.R., S.R. and I.P.S.; J.F.P. is a member of the same institution]; Agencia Nacional de Promoción Científica y Técnica [PICT-2021-I-A-00513 and PICT-2019-2019-02619 to J.F.P.]. The open-access publication charge for this paper has been waived by Oxford University Press – NAR.
Conflict of interest statement. None declared.
References
- 1. Xu Y., Chen X.. microRNA biogenesis and stabilization in plants. Fundam. Res. 2023; 3:707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhan J., Meyers B.C.. Plant small RNAs: their biogenesis, regulatory roles, and functions. Annu. Rev. Plant Biol. 2023; 74:21–51. [DOI] [PubMed] [Google Scholar]
- 3. Mencia R., Gonzalo L., Tossolini I., Manavella P.A.. Keeping up with the miRNAs: current paradigms of the biogenesis pathway. J. Exp. Bot. 2023; 74:2213–2227. [DOI] [PubMed] [Google Scholar]
- 4. Ma Z., Zhang X.. Actions of plant argonautes: predictable or unpredictable?. Curr. Opin. Plant Biol. 2018; 45:59–67. [DOI] [PubMed] [Google Scholar]
- 5. Fang X., Qi Y.. RNAi in plants: an argonaute-centered view. Plant Cell. 2016; 28:272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin-Merchan A., Moro B., Bouet A., Bologna N.G.. Domain organization, expression, subcellular localization, and biological roles of ARGONAUTE proteins in Arabidopsis. J. Exp. Bot. 2023; 74:2374–2388. [DOI] [PubMed] [Google Scholar]
- 7. Bologna N.G., Mateos J.L., Bresso E.G., Palatnik J.F.. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009; 28:3646–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang W., Gao S., Zhou X., Xia J., Chellappan P., Zhou X., Zhang X., Jin H.. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol. 2010; 11:R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bologna N.G., Schapire A.L., Zhai J., Chorostecki U., Boisbouvier J., Meyers B.C., Palatnik J.F.. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 2013; 23:1675–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mateos J.L., Bologna N.G., Chorostecki U., Palatnik J.F.. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 2010; 20:49–54. [DOI] [PubMed] [Google Scholar]
- 11. Moro B., Chorostecki U., Arikit S., Suarez I.P., Hobartner C., Rasia R.M., Meyers B.C., Palatnik J.F.. Efficiency and precision of microRNA biogenesis modes in plants. Nucleic Acids Res. 2018; 46:10709–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song L., Axtell M.J., Fedoroff N.V.. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 2010; 20:37–41. [DOI] [PubMed] [Google Scholar]
- 13. Werner S., Wollmann H., Schneeberger K., Weigel D.. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 2010; 20:42–48. [DOI] [PubMed] [Google Scholar]
- 14. Cuperus J.T., Montgomery T.A., Fahlgren N., Burke R.T., Townsend T., Sullivan C.M., Carrington J.C.. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Nat. Acad. Sci. U.S.A. 2010; 107:466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim W., Kim H.E., Jun A.R., Jung M.G., Jin S., Lee J.H., Ahn J.H.. Structural determinants of miR156a precursor processing in temperature-responsive flowering in Arabidopsis. J. Exp. Bot. 2016; 67:4659–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chorostecki U., Moro B., Rojas A.L., Debernardi J.M., Schapire A.L., Notredame C., Palatnik J.F.. Evolutionary footprints reveal insights into plant microRNA biogenesis. Plant Cell. 2017; 29:1248–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kurihara Y., Watanabe Y.. Arabidopsis micro-RNA biogenesis through dicer-like 1 protein functions. Proc. Nat. Acad. Sci. U.S.A. 2004; 101:12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu H., Zhou Y., Castillo-Gonzalez C., Lu A., Ge C., Zhao Y.T., Duan L., Li Z., Axtell M.J., Wang X.J.et al.. Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat. Struct. Mol. Biol. 2013; 20:1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Narjala A., Nair A., Tirumalai V., Hari Sundar G.V., Shivaprasad P.V. A conserved sequence signature is essential for robust plant miRNA biogenesis. Nucleic Acids Res. 2020; 48:3103–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rojas A.M.L., Drusin S.I., Chorostecki U., Mateos J.L., Moro B., Bologna N.G., Bresso E.G., Schapire A., Rasia R.M., Moreno D.M.et al.. Identification of key sequence features required for microRNA biogenesis in plants. Nat. Commun. 2020; 11:5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Endo Y., Iwakawa H.O., Tomari Y.. Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 2013; 14:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu H., Hu F., Wang R., Zhou X., Sze S.H., Liou L.W., Barefoot A., Dickman M., Zhang X.. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011; 145:242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X., Niu D., Carbonell A., Wang A., Lee A., Tun V., Wang Z., Carrington J.C., Chang C.E., Jin H.. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat. Commun. 2014; 5:5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dalmadi A., Miloro F., Balint J., Varallyay E., Havelda Z.. Controlled RISC loading efficiency of miR168 defined by miRNA duplex structure adjusts ARGONAUTE1 homeostasis. Nucleic Acids Res. 2021; 49:12912–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao Y., MacRae I.J.. The molecular mechanism of microRNA duplex selectivity of Arabidopsis ARGONAUTE10. Nucleic Acids Res. 2022; 50:10041–10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iki T., Clery A., Bologna N.G., Sarazin A., Brosnan C.A., Pumplin N., Allain F.H.T., Voinnet O.. Structural flexibility enables alternative maturation, ARGONAUTE sorting and activities of miR168, a global gene silencing regulator in plants. Mol. Plant. 2018; 11:1008–1023. [DOI] [PubMed] [Google Scholar]
- 27. Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C.. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008; 133:128–141. [DOI] [PubMed] [Google Scholar]
- 28. Clough S.J., Bent A.F.. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- 29. Jarvis P., Chen L.J., Li H., Peto C.A., Fankhauser C., Chory J.. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998; 282:100–103. [DOI] [PubMed] [Google Scholar]
- 30. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia-Ruiz H., Takeda A., Chapman E.J., Sullivan C.M., Fahlgren N., Brempelis K.J., Carrington J.C.. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell. 2010; 22:481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Di Rienzo J., Casanoves F., Balzarini M., Gonzalez L., Tablada M., Robledo C.. InfoStat Versión 2018. 2020; Argentina: Centro de Transferencia InfoStat, FCA, Universidad Nacional de Córdoba. [Google Scholar]
- 33. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laufs P., Peaucelle A., Morin H., Traas J.. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004; 131:4311–4322. [DOI] [PubMed] [Google Scholar]
- 35. Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004; 303:2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palatnik J.F., Wollmann H., Schommer C., Schwab R., Boisbouvier J., Rodriguez R., Warthmann N., Allen E., Dezulian T., Huson D.et al.. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell. 2007; 13:115–125. [DOI] [PubMed] [Google Scholar]
- 37. Axtell M.J., Meyers B.C.. Revisiting criteria for plant MicroRNA annotation in the era of big data. Plant Cell. 2018; 30:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei X., Ke H., Wen A., Gao B., Shi J., Feng Y.. Structural basis of microRNA processing by Dicer-like 1. Nat. Plants. 2021; 7:1389–1396. [DOI] [PubMed] [Google Scholar]
- 40. Addo-Quaye C., Snyder J.A., Park Y.B., Li Y.F., Sunkar R., Axtell M.J.. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA. 2009; 15:2112–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Todesco M., Balasubramanian S., Hu T.T., Traw M.B., Horton M., Epple P., Kuhns C., Sureshkumar S., Schwartz C., Lanz C.et al.. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature. 2010; 465:632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sang Q., Vayssieres A., O’Maoileidigh D.S., Yang X., Vincent C., Bertran Garcia de Olalla E., Cerise M., Franzen R., Coupland G.. MicroRNA172 controls inflorescence meristem size through regulation of APETALA2 in Arabidopsis. New Phytologist. 2022; 235:356–371. [DOI] [PubMed] [Google Scholar]
- 43. Wollmann H., Mica E., Todesco M., Long J.A., Weigel D.. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development. 2010; 137:3633–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D.. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005; 8:517–527. [DOI] [PubMed] [Google Scholar]
- 45. Wang Z., Ma Z., Castillo-Gonzalez C., Sun D., Li Y., Yu B., Zhao B., Li P., Zhang X.. SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature. 2018; 557:516–521. [DOI] [PubMed] [Google Scholar]
- 46. Bhat S.S., Bielewicz D., Gulanicz T., Bodi Z., Yu X., Anderson S.J., Szewc L., Bajczyk M., Dolata J., Grzelak N.et al.. Mrna adenosine methylase (Mta) deposits M(6)A on Pri-Mirnas to modulate Mirna biogenesis In Arabidopsis Thaliana. Proc. Nat. Acad. Sci. U.S.A. 2020; 117:21785–21795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sobkowiak L., Karlowski W., Jarmolowski A., Szweykowska-Kulinska Z.. Non-canonical processing of arabidopsis pri-miR319a/b/c generates additional microRNAs to target one RAP2.12 mRNA isoform. Front. Plant Sci. 2012; 3:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cisneros A.E., Martin-Garcia T., Primc A., Kuziuta W., Sanchez-Vicente J., Aragones V., Daros J.A., Carbonell A.. Transgene-free, virus-based gene silencing in plants by artificial microRNAs derived from minimal precursors. Nucleic Acids Res. 2023; 51:10719–10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carbonell A., Fahlgren N., Mitchell S., Cox K.L. Jr, Reilly K.C., Mockler T.C., Carrington J.C. Highly specific gene silencing in a monocot species by artificial microRNAs derived from chimeric miRNA precursors. Plant J. 2015; 82:1061–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carbonell A., Takeda A., Fahlgren N., Johnson S.C., Cuperus J.T., Carrington J.C.. New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 2014; 165:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C.. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 2004; 36:1282–1290. [DOI] [PubMed] [Google Scholar]
- 52. Fahlgren N., Jogdeo S., Kasschau K.D., Sullivan C.M., Chapman E.J., Laubinger S., Smith L.M., Dasenko M., Givan S.A., Weigel D.et al.. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell. 2010; 22:1074–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vazquez F., Blevins T., Ailhas J., Boller T., Meins F. Jr. Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res. 2008; 36:6429–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this work is available in the manuscript as Supplementary Tables, Figures and Appendices. We also analyse publicly available and deposited data at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Small RNA libraries from seedlings used in Figures 5, 6 and Supplementary Figure S5 were obtained from ((11), accession number GSE116330). Small RNA libraries from leaves and inflorescences used in Supplementary Figures S4 and Figure S5 were extracted from ((31); accession numbers GSM506656-8 and GSM506662-4).