Abstract
Bovine leukemia virus (BLV) is associated with enzootic bovine leukosis and is closely related to human T-cell leukemia virus type 1 (HTLV-1). The Tax protein of BLV acts through the 5′ long terminal repeat (LTR) of BLV and activates the transcription of BLV. In this study, we amplified tax genes from BLV-infected cattle using PCR. We cloned the genes and monitored the transcriptional activities of the products. Seven independent mutant Tax proteins, with at least one amino acid substitution between residues 240 and 265, exhibited a markedly stronger ability to stimulate the viral LTR-directed transcription than the wild-type Tax protein. Analysis of chimeric Tax proteins derived from wild-type and mutant Tax proteins clearly demonstrated that a single substitution between residue 240 and 265 might be critical for the higher activities of the Tax mutant proteins. Furthermore, it appeared that transient expression of a Tax mutant protein was better able to increase the production of viral proteins and particles from a defective recombinant proviral clone of BLV than was wild-type Tax. Analysis of mutations within the U3 region of the LTR revealed that a cyclic AMP-responsive element in Tax-responsive element 2 might be sufficient for the enhanced activation mediated by the mutant proteins. In addition to the LTR of BLV, other viral enhancers, such as the enhancers of HTLV-1 and of mouse mammary tumor virus, which cannot be activated by wild-type BLV Tax protein, were activated by a Tax mutant protein. Our observations suggest that the transactivation activity and target sequence specificity of BLV Tax might be limited or negatively regulated by the region of the protein between amino acids 240 and 265.
Bovine leukemia virus (BLV) is the etiologic agent of enzootic bovine leukosis (EBL), which is the most common neoplastic disease of cattle, and it is often associated with persistent lymphocytosis, which is characterized by an increased number of normal B lymphocytes and the subsequent development of B-cell leukemia or lymphosarcoma after a long latency period (9). Sheep that are experimentally inoculated with BLV are readily infected, and some develop B-cell tumors at higher frequencies and after a shorter latency period than naturally inoculated cattle (3, 15). BLV is closely related to human T-cell leukemia virus type 1 (HTLV-1), which is the causative agents of adult T-cell leukemia and a chronic neurological disorder known as tropical spastic paraparesis or HTLV-1-associated myelopathy (10). BLV and HTLV constitute a unique subgroup within the retrovirus family, being characterized by similar genomic organizations, similar strategies for gene expression, and similar pathologies. In addition to the structural proteins Gag, Pol, and Env, these viruses encode at least two regulatory proteins, namely, Tax and Rex, in the pX region located between the env gene and the 3′ long terminal repeat (LTR). The Tax protein acts on a triplicate 21-bp motif known as the Tax-responsive element (TxRE) in the U3 region of the 5′ LTR, and it stimulates transactivation of the virus genome (13, 16, 20, 52). The TxRE consists of a cyclic AMP-response element (CRE)-like sequence, and it has been suggested that Tax binds indirectly to this element through cellular factors, such as members of the CREB/ATF family of basic-leucine zipper proteins which have been shown to bind to the CRE-like sequence (6, 43). The Tax protein of HTLV-1 is also known to modulate the expression of many cellular genes that are related to regulation of cell growth (61), but little is known about the Tax protein of BLV (27). The Tax proteins of BLV and HTLV-1 can cooperate with the Ha-Ras oncoprotein to induce the full transformation of primary rat embryo fibroblasts (34, 54). These findings indicate that the Tax protein is a key contributor to the oncogenic potential, as well as a key protein in the replication of the virus. The Rex protein interacts with the Rex-responsive element in the 3′ R regions of the BLV and HTLV-1 mRNAs and enhances the cytoplasmic accumulation of singly spliced and unspliced transcripts. This enhancement leads to an increase in the production of structural proteins and to a decrease in the level of the doubly spliced tax-rex mRNA (14, 42).
RNA viruses have high rates of variation in nucleotide sequence, as frequently observed in members of the lentivirus group, such as human immunodeficiency virus (HIV), and such variation is important for viral survival during immunological attack by the host's immune system. However, in BLV and HTLV-1, the genetic variability appears to be limited in vivo (12, 21, 57, 58). Moreover, it is difficult to detect transcripts of the BLV and HTLV genomes in fresh tumor cells or in fresh peripheral blood lymphocytes (PBL) from infected individuals (18, 29). These findings suggest that the BLV-HTLV subgroup might exploit a strict mechanism for control of the expression of viral proteins throughout the course of leukemogenesis in order to evade the host's immunosurveillance system. However, we do not yet know how viral expression is inhibited in vivo. The increased expression of BLV and HTLV-1 mRNAs can be induced by several activators of lymphocytes, such as fetal calf serum, lipopolysaccharides, and phorbol esters, after culture of lymphocytes in vitro (1, 24, 32, 33). Recent findings also indicate that interleukin-2 (IL-2) activates BLV mRNA and enhances levels of viral proteins, while IL-10 inhibits detection of BLV mRNA (39). Furthermore, phosphorylation of HTLV-1 Tax is critical for the transactivation function of Tax, and the extent of such phosphorylation in human lymphocytes is increased by treatment of cells with phorbol esters (5, 17). We showed previously that BLV-infected cattle retain a full-length proviral genome throughout the course of their disease (44), in sharp contrast to the high frequencies (30 to 50%) of deletions in proviruses in HTLV-1-induced tumors (30, 37, 46). These findings suggest that a signal transduction pathway that controls the activity of Tax in host cells, rather than any genetic change in the BLV proviral genome, might play an important role in regulating the activation and silencing of the virus.
In this study, we amplified tax genes from BLV-infected animals using PCR, and then we cloned and sequenced the genes and identified seven independent Tax mutants, which were associated with strikingly higher viral LTR-directed transcriptional activity than the wild type. Furthermore, we found that amino acid substitutions between amino acids 240 and 265 of Tax resulted in significantly increased transactivational activity that involved a CRE motif in the BLV LTR. Finally, we demonstrated that the mutant Tax protein also significantly activated the 21-bp enhancer of HTLV-1 and the LTR of mouse mammary tumor virus (MMTV) and was slightly effective with the LTRs of HIV type 1 (HIV-1) and of Moloney murine leukemia virus (M-MLV).
The mutant Tax proteins with elevated transactivation activity might help us to elucidate the mechanism for strict regulation of the expression of BLV.
MATERIALS AND METHODS
Amplification by PCR, cloning, and sequencing of the BLV tax gene.
To amplify the BLV tax gene by PCR, we used the oligonucleotide primers BtaxF (5′-ACCTCGAGATGGCAAGTGTTGTTGGTTGG-3′) and BtaxR (5′-AGTCTAGAGCTGACGTCTCTGTCTG-3′). The underlined sequences are restriction sites specific for XhoI and XbaI, respectively. Approximately 10 to 100 ng of genomic or plasmid DNA was amplified in a 30-μl reaction mixture that contained 1× EX Taq buffer (Takara Shuzo, Ohtsu, Japan) or 1× KOD-Plus buffer containing 1 mM MgSO4, (Toyobo, Kyoto, Japan), 0.25 mM each deoxynucleoside triphosphate, 0.33 μM each oligonucleotide primer, and 1 U of EX Taq (Takara Shuzo) or KOD-Plus™ (Toyobo) DNA polymerase. Amplification was achieved by 35 cycles of incubation at 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min 30 s. To obtain Tax-expressing plasmids, PCR products were introduced into the XhoI and XbaI sites of the expression plasmid pME18Neo (45). Several constructs were sequenced with a BigDye Terminator Cycle Sequencing Kit and a Genetic Analyzer (ABI PRISM 310; PE Applied Biosystems, Norwalk, Conn.) with the following primers: 7457R, 5′-GGGAACAACGTTCCATTGAG-3′; 7662R, 5′-GTTGTTCCAGGGAAGAAGGC-3′; 7855R, 5′-CTAGGGGTAGAATACAAGTAGC-3′; 8073R, 5′-CGTAGGGTCATGAAGAAGGAAGC-3′; and 8250R, 5′-CAGCTGACGTCTCTGTCTGGT-3′. To generate chimeric Tax-expressing plasmids that encoded chimeric proteins Chi1 through Chi4, we excised the XhoI-EcoRI fragment that contained the sequence corresponding to amino acids 1 to 228 of the Tax protein for Chi2 and Chi4 and the EcoRI-XbaI fragment that contained the sequence corresponding to amino acids 229 to 309 of the Tax protein for Chi1 and Chi3 from the expression plasmids that encoded the Tax mutants R73Q/V146I/S240T and A157V/T251A, respectively. We then replaced the various sequences with the sequence for an expression plasmid that encoded wild-type Tax.
Construction of plasmids.
To construct pGV-BLTR, we amplified the full-length BLV LTR by PCR in 1× KOD-Plus buffer, 0.2 mM each deoxynucleoside triphosphate, 1 mM MgSO4, and 1 U of KOD-Plus DNA polymerase with primers (0.33 μM) BLTRF (5′-TCCTGCAGTGTATGAAAGATCATGCCGAC-3′) and BLTRR (5′-AGTCTAGAATTGTTTGCCGGTCTCTC-3′), in which the underlined sequences correspond to restriction sites for PstI and XbaI, respectively, using the infectious BLV molecular clone pBLV-IF (23) as template DNA. The PCR product was treated with PstI, blunted with T4 DNA polymerase, digested with XbaI, and then ligated into the SmaI-NheI site of pGV(−), which is a plasmid that corresponds to pGV-P (Toyo Ink, Tokyo, Japan) without the simian virus 40 promoter sequence (BglII-HindIII region). To generate pGV-BLTRκB, two oligonucleotides, namely, BLTRκB1 (5′-TCGAGTGGCTAGAATCCCCGTACCTCCCCAACTTCCCCTTTCCCGAAAAATCCAC-3′) and BLTRκB2 (5′-TCGAGTGGATTTTTCGGGAAAGGGGAAGTTGGGGAGGTACGGGGATTCTAGCCA-3′), in which the underlined sequences correspond to restriction sites for XhoI, were annealed and ligated at the XhoI site of pGV-P. A clone with four copies of the oligonucleotide on the sense strand was selected and used for luciferase assays. pGV-TxRE2 was constructed by ligating an annealed oligonucleotide with TxRE2 sequences in the sense and antisense orientations (5′-CTGAGCTGGTGACGGCAGCTGGTGGCGCCACCAGCTGCCGTCACCAGC-3′; the underlined sequence corresponds to a restriction site for XhoI) at the XhoI site of pGV-P. To generate mutant reporter plasmids pGV-TxRE2m1 through pGV-TxRE2m5, we introduced point mutations into pGV-TxRE2 by PCR-based site-directed mutagenesis, as described previously (51), using KOD-Plus DNA polymerase and the following oligonucleotide primers: m1, 5′-GCCACCAGCTGACGTCACCAGCTC-3′; m2, 5′-GCCACCAGCACCCGTCACCAGCTC-3′; m3, 5′-GCCTGCAGCTGCCGTCACCAGCTC-3′; m4, 5′-GCCACCAGCTGCGGTGACCAGCTC-3′; and m5, 5′-GCCACGTGCTGCCGTCACCAGCTC-3′ (bases different from those in the wild-type sequence are shown in boldface). For construction of pGV-HL21, the WT/BL plasmid (a kind gift from J. Fujisawa, Kansai Medical University, Osaka, Japan), which contains five repeats of the HTLV-1 21-bp enhancer, was digested with XhoI, blunted, and then digested with XbaI. The fragment containing the 21-bp enhancers was ligated at the HindIII (blunted)-NheI sites of pGV-P. To generate pGV-HIV-1 LTR(U3R), the U3-to-R region of the LTR of HIV-1 was amplified by PCR as described above with primers HLF (5′-TGGGGTACCTGGAAGGGCTAATTTGGTG-3′; the underlined sequence corresponds to a restriction site for KpnI) and HLR (5′-CCGCTCGAGTATTGAGGCTTAAGCAGT-3′; the underlined sequence corresponds to a restriction site for XhoI) and the HIV-1 plasmid pNL432 as the template. The PCR product was subcloned into the KpnI-XhoI sites of pGV(−). To construct pGV-MMTV LTR, we ligated the BglII-HindIII fragment that contained the full-length MMTV LTR from a hybrid MMTV provirus plasmid (kindly donated by S. Yanagawa, Kyoto University, Kyoto, Japan) into the BamHI-HindIII sites of pBluescript II SK(+) (Stratagene, La Jolla, Calif.), and then the XbaI-HindIII fragment containing the full-length LTR of MMTV was subcloned into the NheI-HindIII sites of pGV(−). For construction of pGV-M-MLV LTR, the EcoRI-HindIII fragment of the full-length LTR of M-MLV, which had been cloned into pUC18 (a kind gift from A. Ishimoto, Kyoto University, Kyoto, Japan) was ligated into the EcoRI-HindIII sites of pBluescript II SK(+) and the SpeI-HindIII fragment containing the LTR of M-MLV was then subcloned into the NheI-HindIII sites of pGV(−). To obtain the HTLV-1 Tax-expressing plasmid pMEHtax, we subcloned the EcoRI-BamHI fragment that contained the tax sequence from pSGtax (a kind gift from M. Fujii, Niigata University, Niigata, Japan), into pBluescript II SK(+), and then the EcoRI-NotI fragment containing the tax sequence was subcloned into pME18Neo. The defective BLV infectious molecular clone pBLV-IFS240P was constructed by replacing the ClaI-Eco47III fragment that included the tax gene in pBLV-IF by the region of the mutant tax clone that encoded S240P. pRL-SV40 (Promega, Madison, Wis.) encodes a luciferase gene from Renilla and was used for normalization of the efficiencies of transfections.
Cells, extraction of DNA, and transfections.
PBL were separated from three BLV-infected but clinically and hematologically normal cows, and total chromosomal DNA was extracted from these cells as described previously (22). Tumor tissues were obtained from three BLV-infected cows with lymphoma, and genomic DNA was prepared from these tissues as described elsewhere (35). 293T cells, human embryonic kidney cells that express the large T antigen of simian virus 40, were maintained in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum.
For Western blotting, 293T cells (107) were transfected with 10 μg of a Tax expression plasmid or a BLV molecular clone and 1 μg of pRL-SV40 by electroporation in a Gene Pulser (Bio-Rad, Hercules, Calif.) operated at 975 μF and 290 V. For analysis of luciferase activity, 293T cells (105/well) were transfected with 1 μg of each reporter plasmid (which had an enhancer-promoter sequence upstream of the firefly gene for luciferase), 0.5 μg of a Tax-expressing plasmid, and 0.3 μg of pRL-SV40. Transfections were performed with the DOSPER reagent (Roche Molecular Biochemicals, Mannheim, Germany) as described previously (45).
Luciferase assay.
At 60 h after transfection, cells were harvested and subjected to the assay for luciferase activity as described elsewhere (45).
Western blotting analysis.
At 60 h after transfection, cells were divided into two aliquots; one aliquot was used for measurement of Renilla luciferase activity to monitor the efficiency of transfection, and the other aliquot of cells was lysed with lysis buffer (2% SDS and 2 mM phenylmethanesulfonyl fluoride plus Complete Protease Inhibitor Cocktail [Roche Molecular Biochemicals]). Proteins in lysates with equal Renilla luciferase activity were examined by Western blotting analysis as described previously (23). Virus particles were pelleted as described previously (23) and subjected to Western blotting as described above. We also examined the presence of Tax protein in cell lysates that were used for luciferase assays by Western blotting with BLV Tax-specific B1 polyclonal antibodies (a kind gift from M. Sakurai, National Institute of Animal Health, Tsukuba, Japan).
RESULTS
Ability of BLV Tax mutants to transactivate the BLV LTR.
To clarify the way in which individual animals infected with BLV progress from the asymptomatic stage to the disease stage, we investigated the correlation between Tax activity and the progression of lymphoma. We amplified tax genes from PBL and tumor tissues taken from animals at the asymptomatic stage (A63, A69, and A78) and the lymphoma stage (pr2170, pr2374, and pr2436), respectively, by PCR using EX Taq polymerase. We subcloned the PCR products into the mammalian cell expression vector pME18Neo. As a wild-type tax gene, a tax clone, IF-1, was similarly cloned from an infectious molecular clone of BLV, pBLV-IF (Table 1) (23).
TABLE 1.
Nucleotide changes and predicted amino acid substitutions in the BLV tax clones isolated in this studya
| tax clone type | Animal (source of clone) | Clone no. (clone name) | Codon no. | Codon change | Amino acid substitution |
|---|---|---|---|---|---|
| High transactivation activity | A63 | 3n (H261R) | 261 | CAC→CGC | H→R |
| 297 | TTA→TTG | Silent | |||
| 28n (A157V/T251A) | 45 | GAC→GAT | Silent | ||
| 157 | GCC→GTC | A→V | |||
| 251 | ACC→GCC | T→A | |||
| 297 | TTA→TTG | Silent | |||
| pr2374 | 2 (S265G) | 265 | AGT→GGT | S→G | |
| 297 | TTA→CTA | Silent | |||
| 9 (R73Q/V146I/S240T) | 45 | GAC→GAT | Silent | ||
| 73 | CGA→CAA | R→Q | |||
| 100 | CCC→CCT | Silent | |||
| 146 | GTC→ATC | V→I | |||
| 240 | TCT→ACT | S→T | |||
| 36n (H261Y) | 199 | ACT→ACC | Silent | ||
| 261 | CAC→TAC | H→Y | |||
| 284 | GTT→GTC | Silent | |||
| 297 | TTA→CTA | Silent | |||
| pr2436 | 5 (D258G) | 258 | GAC→GGC | D→G | |
| 18n (D247G) | 247 | GAC→GGC | D→G | ||
| Low transactivation activity | A63 | 1 (S240P) | 240 | TCT→CCT | S→P |
| 287 | AGA→AGG | Silent | |||
| 297 | TTA→TTG | Silent | |||
| Wild typeb | IF-1 | 297 | TTA→CTA | Silent |
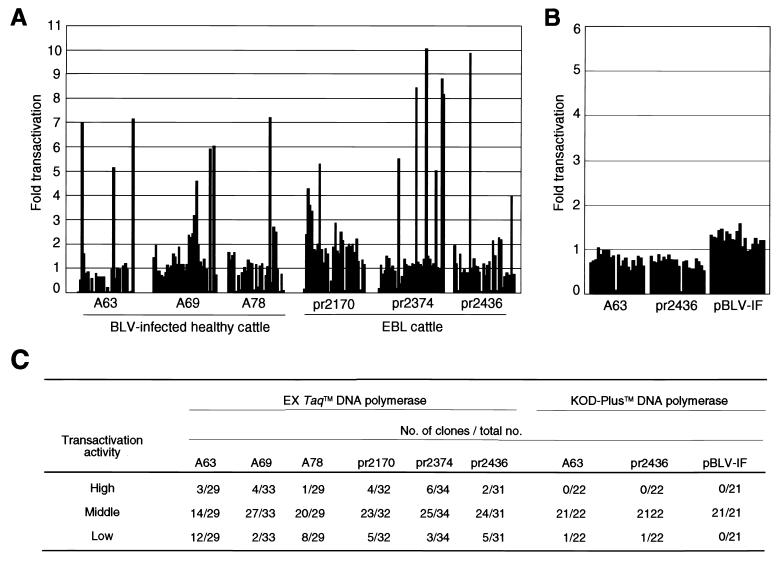
As shown in Fig. 1A and C, we obtained 91 and 97 tax clones from asymptomatic cattle and cattle with EBL, respectively. All 188 clones were introduced into 293T cells with the reporter plasmid pGV-BLTR, which included the firefly gene for luciferase driven by a BLV LTR, and then we performed luciferase analysis to compare the transactivation activities of these clones (Fig. 1). It was clear that various Tax proteins were isolated from the same individual in all cases, and in addition, the clones could be divided into three groups depending on transactivation capacity. The majority (approximately 71%) of clones had activities similar to that of the wild-type tax clone, irrespective of the stage of the disease. By contrast, some clones had higher or lower transactivation activity than the wild-type clone in the case of both the asymptomatic and the lymphoma stages. There was no significant difference in terms of the rate of isolation of Tax mutant clones with high, medium, and low transactivation activities between healthy cattle and those with EBL (chi-square test, 0.1 < P < 0.2).
FIG. 1.
Transactivation of the BLV LTR by various tax clones from BLV-infected cattle. (A and B) BLV tax genes were amplified by PCR with EX Taq (A) or KOD-Plus (B) DNA polymerase from the genomic DNAs of BLV-infected animals and from a molecular clone pBLV-IF and then subcloned into pME18Neo. 293T cells were transfected with the reporter plasmid pGV-BLTR, the reference plasmid pRL-SV40, and individual Tax-expressing plasmids. At 60 h after transfection, cells were recovered and the activities of firefly and Renilla luciferases were measured in lysates. For each sample, the firefly luciferase activity (pGV-BLTR) was normalized by reference to Renilla luciferase activity (pRL-SV40). The extent of transactivation (fold) was calculated by dividing the transactivation activity of the mutant Tax protein by that of wild-type Tax protein. (C) Summary of the results shown in panels A and B. The tax mutant clones were divided into three groups as follows, with wild-type activity being taken as 1: high, >3-fold transactivation; medium, 3- to 0.5-fold transactivation; low, <0.5-fold transactivation.
We also amplified the tax genes using KOD-Plus DNA polymerase, which has considerably higher fidelity than EX Taq polymerase (Fig. 1B and C). No mutant Tax clones with elevated transactivation activity were isolated from two BLV-infected animals and from pBLV-IF, indicating that the mutations that we identified were a result of errors made by EX Taq polymerase during the amplification of the tax gene by PCR.
Identification of the Tax mutants with elevated transactivation activity.
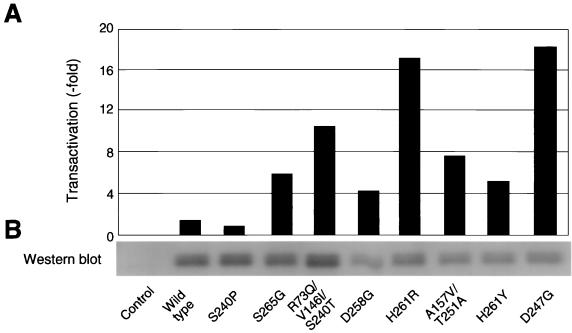
We focused on the Tax mutant proteins with significantly elevated transactivation activity in our effort to elucidate the mechanism that controls the activity of the Tax protein. We selected seven mutant tax clones with transactivation activity that was 4- to 18-fold higher than that of the wild-type Tax clone, and we determined the nucleotide sequences of these clones (Table 1). We first determined the nucleotide sequence of the wild-type tax clone IF-1 that we generated from the pBLV-IF provirus and compared it with the sequences determined by Sagata et al. (λBLV-1) (40). Of the 930 bp of the full-length tax gene that we sequenced, we identified only a single, silent point mutation at codon 297. In the case of the seven mutant tax clones, we found that each clone had at least one missense mutation between amino acids 240 and 265, namely, at codon 240 (S→T), at codon 247 (D→G), at codon 251 (T→A), at codon 258 (D→G), at codon 261 (H→R), at codon 261 (H→Y), and at codon 265 (S→G). Among the seven clones, only two clones (A63 28n [A157V/T251A] and pr2374 9 [R73Q/V146I/S240T]) also had missense mutations outside the region that included amino acids 240 to 265 of the Tax protein, namely, at codon 73 (R→Q), at codon 146 (V→I), and at codon 157 (A→V). The positions and amino acids after the various substitutions differed among the seven clones, with the exception of position 261 in clones H261R and H261Y. In addition, we used one clone with weak transactivation activity via the BLV LTR as a negative control in this study; this clone had a missense mutation at codon 240 (S→P).
To analyze the correlation between the expression of and the ability to activate the BLV LTR by the various Tax mutant proteins, we transiently transfected 293T cells with an expression vector that encoded wild-type Tax protein or a Tax mutant, together with pGV-BLTR and pRL-SV40 for normalization of the efficiency of transfection. A fraction of each cell lysate was used for analysis of expression of the reporter gene, and the remainder was used for detection of Tax protein by Western blotting with BLV Tax-specific polyclonal antibodies (Fig. 2). Two Tax mutant proteins, namely, R73Q/V146I/S240T and S265G, were specifically expressed at levels similar to that of the wild-type Tax protein, while the levels of expression of the other five mutant proteins, namely, D258G, A157V/T251A, H261R, H261Y, and D247G, were lower than that of the wild-type protein. The Tax mutant with weak transactivation activity, S240P, was synthesized at a level similar to that of the wild-type Tax protein. Thus, there was no obvious relationship between transactivation activity and the level of expression of the Tax protein, indicating that the increase in transactivation activity was due neither to an increase in amount nor to enhanced stability due to amino acid substitution.
FIG. 2.
Transactivation activity and expression of Tax mutant proteins. 293T cells were cotransfected with pME18Neo that encoded wild-type or mutant Tax protein or with the control plasmid pME18Neo together with pGV-BLTR and the reference plasmid pRL-SV40. At 60 h after transfection, cells were taken for measurements of Renilla luciferase activity. Lysates with equal Renilla luciferase activity were subjected to firefly luciferase assay (A) and Western blotting analysis (B). (A) Luciferase activities were monitored and is presented as described in the legend to Fig. 1. Each value represents the average obtained from two independent transfection experiments. (B) To analyze the expression of mutant Tax proteins, lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 10% polyacrylamide gel and then analyzed by Western blotting with polyclonal antibodies (B1) against BLV Tax. Similar results were obtained in two independent experiments.
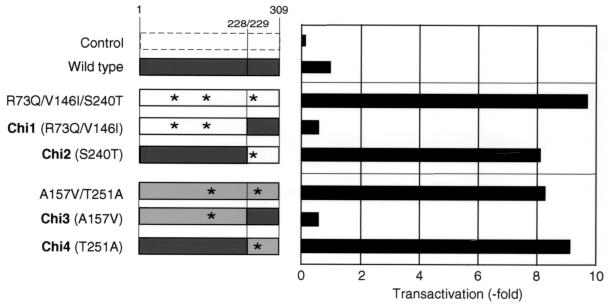
To evaluate whether only amino acid substitutions between amino acids 240 and 265 of the Tax protein might be involved in the elevated transactivation activity, we generated four chimeric proteins (Chi1 through Chi4) by replacing sequences that encoded each segment of wild-type Tax protein by sequences that corresponded to the amino-terminal region from amino acids 1 through 228 or to the carboxy-terminal region from amino acids 229 through 309 of two Tax mutants, R73Q/V146I/S240T and A157V/T251A. These Tax mutants have amino acid substitutions both beyond and between residues 240 and 265, as shown on the left in Fig. 3. Plasmids encoding chimeric parental-mutant or wild-type Tax protein were used to cotransfect 293T cells together with pGV-BLTR, and then transactivation activities were examined (Fig. 3). The luciferase activities of two chimeric proteins, Chi1 (R73Q/V146I) and Chi3 (A157V), with missense mutations outside residues 240 to 265 of the Tax protein had only approximately 10% of the activity of each parental mutant, even though they retained approximately 50% of the activity of the wild-type Tax protein. By contrast, the chimeras Chi2 (S240T) and Chi4 (T251A) each retained most of the transactivation activity of the respective parental mutant.
FIG. 3.
Transactivation of the BLV LTR by chimeric proteins derived from wild-type and mutant Tax proteins. Plasmids expressing chimeric proteins were created by replacing sequences that corresponded to the amino-terminal regions or the carboxy-terminal regions of the Tax mutants R73Q/V146I/S240T and A157V/T251A, with that of wild-type Tax protein, as shown on the left. 293T cells were transfected with pGV-BLTR, the reference plasmid pRL-SV40, and pME18Neo that encoded the wild-type or chimeric Tax protein or with the control plasmid pME18Neo. Luciferase activity was monitored and is presented as described in the legend to Fig. 1. Each value represents the average obtained from two independent transfection experiments. Asterisks indicate the positions of missense mutations in each Tax protein.
These results demonstrated that with a single amino acid substitution, such as S240T, D247G, T251A, D258G, H261R, H261Y, or S265G, between residues 240 and 265, the Tax protein of BLV is endowed with significantly increased ability to transactivate the BLV LTR. However, a loss-of-activity mutant of Tax, S240P, also has a missense mutation within this region. Thus, it is clear that not only the position of the missense mutation but also the particular amino acid inserted as a result might be an important factor in Tax activity.
A CRE, a cis element in TxRE2, is sufficient for the enhanced activation of the BLV LTR by the mutant Tax proteins.
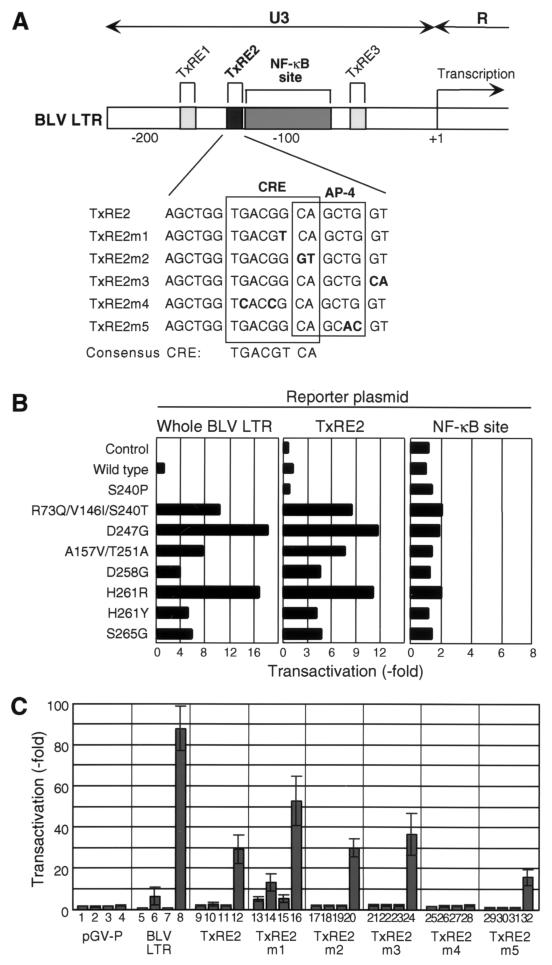
We attempted to identify the regions in the BLV LTR that might be required for the elevated transactivation activity. As shown in Fig. 4A, the U3 region of the BLV LTR contains three imperfect direct repeats known as TxREs (TxRE1, TxRE2, and TxRE3), which are critically important for the Tax-mediated activation of the LTR. An NF-κB-binding site has also been identified between TxRE2 and TxRE3 in the U3 region. It is associated with the strong activation of transcription of BLV in the presence of NF-κB and Tax in addition to a TxRE (7, 8).
FIG. 4.
Identification of the element in the BLV LTR that is involved in the enhanced transactivation activity of Tax mutants proteins. (A) Schematic illustration of the U3 region of the BLV LTR. The upper boxes represent the locations of cis-acting elements in the U3 region. The expanded region below the black box shows the sites of site-directed mutagenesis in TxRE2 in the experiment for which results are shown in panel C. The binding sites for transcription factors CREB (CRE) and AP-4 are indicated by open boxes. Bases different from the wild-type sequence are shown in boldface. The consensus cellular CRE sequence is also indicated. (B) Transactivation of the BLV LTR, TxRE2, and the NF-κB-binding site in the U3 region by Tax mutant proteins. The reporter plasmids (pGV-BLTR, pGV-TxRE2, and pGV-BLTRκB) were used for transfection together with pME18Neo that encoded wild-type or mutant Tax protein or with the control plasmid pME18Neo and the reference plasmid pRL-SV40. Luciferase activity was monitored and is presented as described in the legend to Fig. 1. The average values from two independent transfection experiments are shown. (C) Transactivation of wild-type and mutant forms of TxRE2 by Tax mutant proteins. The effector plasmid encoding wild-type Tax (bars 2, 6, 10, 14, 18, 22, 26, and 30), S240P (bars 3, 7, 11, 15, 19, 23, 27, and 31), or D247G (bars 4, 8, 12, 16, 20, 24, 28, and 32) or the control plasmid pME18Neo (bars 1, 5, 9, 13, 17, 21, and 25) was used for transfection together with the reporter plasmid that included the BLV LTR (bars 5 to 8), TxRE2 (wild type) (bars 9 to 12), TxRE2m1 (bars 13 to 16), TxRE2m2 (bars 17 to 20), TxRE2m3 (bars 21 to 24), TxRE2m4 (bars 25 to 28), or TxRE2m5 (bars 29 to 32), or the control plasmid pGV-P (bars 1 to 4), and the reference plasmid pRL-SV40. Luciferase activity was measured as described in the legend to Fig. 1. The results are presented as transactivation (fold) relative to the transactivation activity resulting from transfection with pGV-P and pME18Neo (bar 1). Average values from triplicate transfections with standard deviations (error bars) are shown.
We constructed two reporter plasmids in which the TxRE2 (pGV-TxRE2) or the NF-κB-binding site (pGV-BLTRκB) in the U3 region of the BLV LTR was linked to the upstream region of a luciferase gene. We transfected 293T cells with these reporter plasmids together with an expression vector that encoded wild-type Tax protein or a Tax mutant, or with the control vector pME18Neo, and then we monitored the transient expression of luciferase (Fig. 4B). We selected TxRE2 from among the three TxREs because it appears that TxRE2 is the most important element for the Tax-driven activation of the BLV LTR (27). When we used TxRE2 as an enhancer element, the transactivating activities of the seven independent Tax mutants with elevated activity remained similar to that observed with the full-length LTR. By contrast, the wild-type protein and four Tax mutant proteins, A157V/Y251A, D258G, H261Y, and S265G, failed to activate the NF-κB-binding site, while the remaining three mutant proteins, R73Q/V146I/S240T, D247G, and H261R, only weakly activated the NF-κB-binding site. The mutant protein S240P could not activate these enhancer sequences. Thus, it was clear that the TxRE2 site was the main site involved in the increased transactivation of BLV LTR by the Tax mutant proteins. In subsequent experiments, we used the Tax mutant protein D247G because this protein had the strongest activity.
We next examined the target element of the Tax mutant in further detail. TxRE2 consists of a CRE and a binding site for the transcription factor AP-4. We introduced point mutations into the CRE (to yield TxRE2m1 and TxRE2m4), into AP-4 (TxRE2m5), into the overlapping region of the two motifs (TxRE2m2), and outside the region of both motifs (TxRE2m3) in the TxRE2 region of the BLV LTR (Fig. 4A). The various constructs were used to transfect 293T cells together with an expression vector that encoded wild-type Tax, D247G, or S240P or the control vector pME18Neo, and then we performed the luciferase assay (Fig. 4C). In the presence of D247G, the luciferase activity associated with the cis-element mutants TxRE2m2 and TxRE2m3 was nearly identical to that associated with wild-type TxRE2, indicating that the overlapping and outside regions were not important in the increased transactivation mediated by the Tax mutant protein. By contrast, no transactivation activity was detected in cells transfected with all constructs when pGV-TxRE2m4, which has a disrupted CRE motif, was used as the reporter plasmid. TxRE2m1, which corresponded to the consensus CRE sequence, yielded elevated enhancer activity regardless of the effector plasmids used. The AP-4 mutant (TxRE2m5) was associated with transactivation activity at a level equivalent to approximately 50% of that associated with the wild-type TxRE2 but only in the presence of the mutant Tax protein D247G.
Taken together, the results indicate that the CRE motif in TxRE2 might be sufficient for the elevated transcription due to the Tax mutant proteins and that the AP-4 motif might play an auxiliary role. We failed to detect any transactivation activity with wild-type Tax and any variant of the TxRE2 sequence except TxRE2m1. Thus, it appears that wild-type Tax might need some other motif(s) in the LTR in addition to TxRE2.
Induction of the production of BLV structural proteins and virus particles from the defective molecular clone pBLV-IFS240P by the Tax mutant protein.
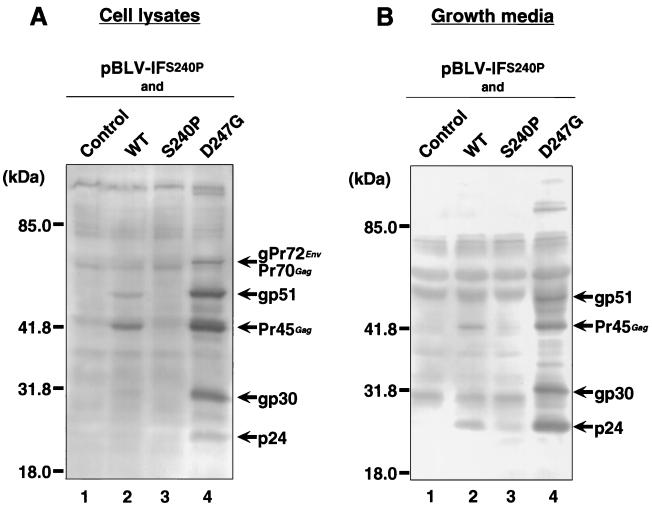
We next examined whether D247G could induce the production of viral proteins and virus particles from the defective recombinant provirus clone pBLV-IFS240P. The defective clone of BLV was made by replacing the tax gene of an infectious full-length molecular clone of BLV, pBLV-IF, by the gene for the transactivation-negative Tax mutant S240P. The defective clone was designated pBLV-IFS240P. As shown in Fig. 5, pBLV-IFS240P was unable to synthesize viral proteins by itself, even though it retained all of the normal viral genes apart from the tax sequence and the entire LTR sequence.
FIG. 5.
Induction of the expression of viral structural proteins and production of viral particles by Tax mutant proteins. A defective molecular clone, pBLV-IFS240P, was used to cotransfect 293T cells with a Tax-expressing plasmid that encoded wild-type (WT) Tax protein (lane 2), S240P (lane 3), or D247G (lane 4), or with the control plasmid pME18Neo (lane 1), and the reference plasmid pRL-SV40. At 60 h after transfection, cell lysates (A) and concentrated preparations of virus particles that had been released into the growth medium (B) were subjected to Western blotting analysis with serum from a BLV-infected sheep. The amounts of lysates and preparations of virus particles subjected to electrophoresis were varied by reference to the efficiency of transfection, which was assessed by monitoring Renilla luciferase activity. Arrows on the right indicate the positions of BLV structural proteins. Numbers on the left show molecular masses.
We transiently transfected 293T cells with pBLV-IFS240P, together with an expression vector that encoded wild-type Tax protein, D247G, or S240P or with the control vector, by electroporation and then analyzed the expression of BLV structural proteins by Western blotting with serum either from a BLV-infected sheep or from a control sheep (Fig. 5A). No detectable viral proteins were found in cells that had been cotransfected with pBLV-IFS240P and either the control plasmid or pME18Neo, which encoded the Tax mutant protein S240P. By contrast, bands corresponding to the structural proteins of cell-associated BLV, such as Gag and its precursors (p24, Pr45Gag, and Pr70Gag) and Env and its precursors (gp30, gp51, and gPr72env), were detected specifically in cells that had been cotransfected with both pBLV-IFS240P and an expression plasmid that encoded wild-type Tax or D247G. In the presence of D247G, the levels of expression of viral structural proteins from pBLV-IFS240P were much higher than those in the presence of the wild-type Tax protein.
We also examined the BLV particles that had been released from the cells (Fig. 5B). The intensities of the bands that corresponded to the structural protein in virions, such as p24, gp30, Pr45Gag, and gp51, obtained in the presence of D247G were higher than those obtained in the presence of the wild-type construct. Moreover, virus particles were not released from cells that had been cotransfected with pBLV-IFS240P and either the control plasmid or an expression plasmid that encoded S240P. No specific bands were detected in both types of analysis with the control serum from an uninfected sheep (data not shown).
Our results suggest that the specific amino acid substitutions between amino acids 240 and 265 in the Tax protein of BLV result in the considerably increased ability of Tax to activate the production of virus particles of BLV.
Tax mutant proteins with elevated transactivation activity also activate other retrovirus enhancers.
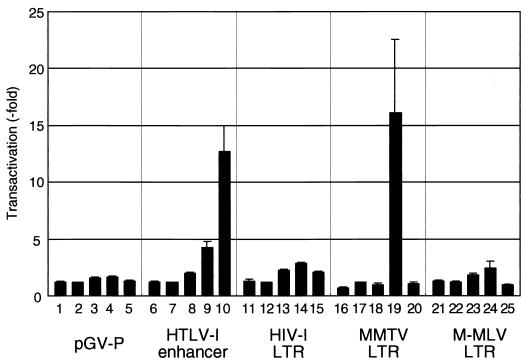
It has been reported that BLV Tax cannot activate the LTR of HTLV-1, which includes CRE motifs (19). However, the results in Fig. 4C showed that D247G was able to activate some modified CRE motifs, such as TxRE2m1 and TxRE2m2. The sequences of these two CRE motif (TGACGTCA and TGACGGGT) are more similar to that of the CRE of HTLV-1 (TGACGTGT) than to that of wild-type TxRE2 (TGACGGCA) (the bases different from those in the sequence of the CRE of HTLV-1 are shown in boldface). Our observations suggested that a Tax mutant protein with elevated transactivation activity might be able to activate the HTLV-1 enhancer. To examine the possibility, we cotransfected 293T cells with the reporter plasmid pGV-HL21, which encodes five tandemly repeated 21-bp enhancers of HTLV-1 that each contain a CRE motif, together with an effector plasmid that encodes wild-type Tax, D247G, S240P, or HTLV-1 Tax or the control vector pME18Neo, and then we analyzed the transactivation activity (Fig. 6). D247G exhibited considerable transactivation activity with the HTLV-1 enhancer, even though it was only 30% as effective as HTLV-1 Tax. The mutant Tax proteins H261R, R73Q/V146I/S240T, and A157V/T251A also have the ability to transactivate the HTLV-1 enhancer (data not shown).
FIG. 6.
Transactivation of the enhancer sequences of various retroviruses by Tax mutant proteins. The reporter plasmid pGV-P or pGV(−) with HL21 (HTLV-1 enhancer) (bars 6 to 10), the HIV-1 LTR (U3R) (bars 11 to 15), the MMTV LTR (bars 16 to 20), or the M-MLV LTR (bars 21 to 25), or the control plasmid pGV-P (bars 1 to 5), was used to cotransfect 293T cells together with the effector plasmid that encoded wild-type Tax protein (bars 2, 7, 12, 17, and 22), S240P (bars 3, 8, 13, 18, and 23), D247G (bars 4, 9, 14, 19, and 24), or HTLV-1 Tax protein (bars 5, 10, 15, 20, and 25), or with the control plasmid pME18Neo (bars 1, 6, 11, 16, and 21), and with the reference plasmid pRL-SV40. Luciferase activity was monitored as described in the legend to Fig. 1. The results are presented as the transactivation (fold) relative to transactivation activity observed after cotransfection of each reporter plasmid with the wild-type Tax-expressing plasmid. Average results from triplicate transfections with standard deviations (error bars) are shown.
We also examined the transactivation activity of D247G with three other retroviral enhancers, namely, the LTRs of HIV-1, MMTV, and M-MLV (Fig. 6). None of these three enhancers was activated by wild-type BLV Tax. D247G significantly transactivated the LTR of MMTV (16-fold enhancement of luciferase activity) and was slightly effective with the other two enhancers (2-fold enhancement of leuciferase activity in each case). No significant transactivation of the MMTV and M-MLV enhancers was evident in cells that expressed HTLV-1 Tax. This result demonstrates the possibility that mutants of BLV Tax, such as D247G, might be able to stimulate the expression of viral genes that are not activated by the wild-type Tax protein.
DISCUSSION
In this study, we have identified several mutant BLV Tax proteins that transactivated the LTR of BLV much more effectively than wild-type Tax (Fig. 7). These mutant proteins appeared to enhance the production of viral proteins and particles, as compared to the wild-type Tax protein, via the LTR of a cotransfected defective recombinant provirus clone of BLV. Nucleotide sequencing demonstrated that each mutant protein had at least one missense mutation between amino acids 240 and 265, namely, at codon 240, 247, 251, 258, 261, or 265, and some also had missense mutations outside this region, namely, at codon 73, 146, or 157. Analysis with chimeric proteins generated from wild-type and mutant Tax proteins clearly demonstrated that a single substitution between residues 240 and 265 was critical for the elevated transactivation activities of the Tax mutant proteins. The results of Western blotting analysis indicated that the increased transactivation activities were due to qualitative changes in the Tax protein and not to quantitative changes or increased stability of the Tax protein. Moreover, we examined the target of transactivation activity in detail and showed that a CRE motif was sufficient for transactivation by the Tax mutant proteins via the LTR of BLV. The mutant protein D247G also activated other retrovirus enhancers, such as the 21-bp enhancers of HTLV-1 and the LTRs of HIV-1, MMTV, and M-MLV, which were not activated by the wild-type Tax from BLV. However, the CRE motif was not necessarily essential for the activation of these retrovirus enhancers by the Tax mutants, since no CRE motif is present in the LTRs of HIV-1, MMTV, and M-MLV. Thus, our results suggest that a single substitution between amino acids 240 and 265 not only stimulates the transactivation activity of Tax protein but also increases the tolerance in terms of specificity for target sequences in other LTRs.
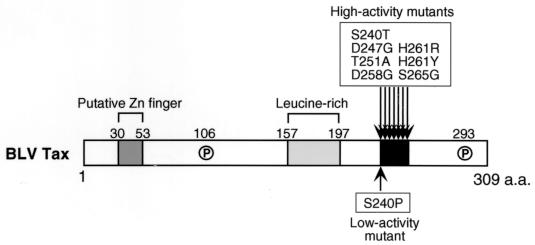
FIG. 7.
Schematic representation of the BLV Tax protein. The region between amino acids 240 and 265, in which missense mutations influenced the transactivation activity of the Tax protein, is shown by a black box. A putative zinc finger structure (dark-gray box; amino acids 30 to 53), a leucine-rich activation domain (light-gray box; amino acids 157 to 197), and sites of phosphorylation (amino acids 106 and 293) are also indicated.
Several Tax variants with activity markedly lower or higher than that of wild-type Tax protein were identified in BLV-infected animals by PCR with EX Taq polymerase. This result conflicts with previous reports by Willems et al. (57, 58) of extremely low levels of intrastrain variability among env genes and LTRs of BLV in vivo. To confirm the existence of tax variants in cattle, we also amplified the tax genes with KOD-Plus DNA polymerase, which has considerably higher fidelity than EX Taq polymerase. In contrast, no mutant Tax clones with elevated transactivation activity were identified from our BLV-infected animals and from pBLV-IF. Therefore, the Tax variants we identified in this study might not be seen in naturally BLV-infected cattle. However, further studies are required to determine whether or not our mutations are attributable to errors made by EX Taq polymerase during PCR.
The enhanced activity of Tax mutant proteins might have been due to alterations in the interaction between the Tax protein and cellular factors as a result of the amino acid substitutions between amino acids 240 and 265. Binding of Tax protein to cellular factors has been characterized in some detail, as follows. (i) HTLV-1 Tax protein cannot bind directly to its target cis-acting elements, such as the CRE motif and CArG box. However, it binds indirectly to these elements via cellular transcription factors such as CREB, ATF, and SRF (19, 43, 50). Similar findings were also obtained in a functional analysis of BLV Tax, but direct interactions of BLV Tax with specific cellular transcription factors have not yet been demonstrated (2). The BLV Tax mutant proteins with high transactivation activity might bind more effectively to these transcription factors. (ii) It has been reported that HTLV-1 Tax stimulates the binding of the basic-leucine zipper domain, which had been found in many transcription factors, including CREB and ATF, to the target motif of the Tax protein by stabilizing dimerization of the factors and by altering the relative affinity of the factors for different DNA-binding sites (4, 38, 50). It is possible that the mutations in Tax might lead to enhancement of these biochemical functions of Tax. (iii) HTLV-1 Tax has also been shown to interact with CREB-binding protein p300/CREB binding protein-associated factor (PCAF). These proteins are cellular coactivators with intrinsic histone acetyltransferase activity that bind to various transcription factors such as CREB. Both CREB-binding protein and PCAF are able to activate Tax-mediated transcription of HTLV-1 (25, 26, 31). The Tax mutant proteins with higher activity might have an enhanced ability to interact with these coactivators, and consequently, they might increase the ability of these coactivators and Tax to activate CRE-mediated transcription. (iv) An unknown cellular factor(s) might interact with Tax and regulate its function positively or negatively. Willems et al. (53, 55, 56, 59) performed a comprehensive analysis of the functional domains of the BLV Tax protein, and they identified a putative zinc finger motif (amino acids 30 to 53), a transactivating domain (amino acids 157 to 197), and two sites of phosphorylation (amino acids 106 and 293) (Fig. 7). In addition, they also showed the possibility that the Tax protein might be autoregulated by a region outside the transactivating domain, the exact location of which remains to be determined (55). The region between amino acids 240 and 265 of the Tax protein might act as an negative regulatory domain, and missense mutations in this region might enhance the transactivation activity of Tax. Identification of the factor(s) that might interact with Tax through amino acids 240 to 265 would help us to elucidate its role and the mechanism that controls the activity of Tax protein.
Why is the transactivation activity of the wild-type Tax protein lower than those of some of the Tax mutant proteins that we examined? The majority of tax clones isolated in this study had activity similar to that of wild-type tax irrespective of the lymphoma stage. Thus, we may also ask why it is that host individuals carry a great majority of BLV clones that encode Tax with only moderate transactivation activity and not high activity. BLV Tax with strong transactivation activity might be disadvantageous for the survival and expansion of BLV, and hence, the optimum form of Tax might be the Tax protein with transactivation activity suitable for both the induction and the repression of the expression of BLV. BLV-infected animals develop EBL after a long latency period. Moreover, during the latency period, the expression of viral proteins appears to be blocked at the transcriptional level (29, 32). This silencing is thought to be very important for escape of BLV from the host's immunosurveillance system. However, it is unknown how the expression of BLV is restricted in vivo to levels that are undetectable by conventional methods. Several candidate mechanisms for silencing have been proposed: (i) accumulation and export of unspliced viral mRNA by Rex protein (10), (ii) inactivation of a viral protein(s) caused by mutation or deletion of the proviral genome (29, 36, 48, 49), (iii) blockage of the transcription of the viral LTR by DNA methylation (11, 41), (iv) absence and/or inactivation of cellular factors that can activate LTR-directed transcription (2, 24, 28, 39), and (v) induction and/or activation of cellular factors that repress the viral transcription (39, 60). Recently, Van Den Broeke et al. (49) reported that the occurrence of a deficient Tax protein as a result of mutation may play an important role in the silenced phenotype observed in BLV-induced ovine B-cell tumors. However, the present results and previous findings indicate that BLV-infected animals retain a full-length BLV proviral genome, with functional tax and LTR sequences, throughout the course of the disease (reference 44 and our unpublished data). Furthermore, it appears that the extent of variations in LTR and env sequences is very limited in BLV-infected sheep (57). These results indicate that silencing of a BLV provirus in vivo is not necessarily associated with any deletion or mutation of the BLV proviral genome. Several lymphocyte activators, such as fetal calf serum, lipopolysaccharides, IL-2, and phorbol esters, can induce the expression of detectable amounts of viral mRNA in lymphocytes from BLV- and HTLV-1-infected individuals after culture in vitro (1, 24, 32, 33, 39). By contrast, IL-10 reduces the level of expression of the viral mRNA (39). Furthermore, phosphorylation of HTLV-1 Tax is critical for the transactivation function of Tax, and the phosphorylation of Tax in human lymphocytes is increased by treatment of these cells with phorbol esters (5, 17). Therefore, some signal transduction pathways in the host cell might regulate the activity of Tax through the region between amino acids 240 and 265 and, consequently, the activation or silencing of BLV.
The Tax protein also appears to be critical to the oncogenic potential of BLV and HTLV-1, and it induces the expression of many cellular genes, including c-fos (47, 54, 61). Therefore, we must now clarify the role of Tax mutant proteins with elevated transactivation activity in BLV-induced leukemogenesis.
ACKNOWLEDGMENTS
We thank K. Okada (Iwate University, Iwate, Japan) for kindly providing tumor tissue and peripheral blood from BLV-infected cattle and M. Sakurai (National Institute of Animal Health, Tsukuba, Japan) for Tax-specific antibodies. We also thank J. Fujisawa (Kansai Medical School, Osaka, Japan), A. Ishimoto (Kyoto University, Kyoto, Japan), S. Yanagawa (Kyoto University), and M. Fujii (Niigata University, Niigata, Japan) for kindly providing various plasmids.
This study was supported by Special Coordination Funds for the Promotion of Science and Technology from the Science and Technology Agency of the Japanese Government, by grants from the Ministry and Education, Science and Culture of Japan, by a President's Special Research Grant from RIKEN, and by a grant for a special postdoctoral researcher of RIKEN.
REFERENCES
- 1.Adam E, Kerkhofs P, Mammerickx M, Kettmann R, Burny A, Droogmans L, Willems L. Involvement of the cyclic AMP-responsive element binding protein in bovine leukemia virus expression in vivo. J Virol. 1994;68:5845–5853. doi: 10.1128/jvi.68.9.5845-5853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam E, Kerkhofs P, Mammerickx M, Burny A, Kettman R, Willems L. The CREB, ATF-1, and ATF-2 transcription factors from bovine leukemia virus-infected B lymphocytes activate viral expression. J Virol. 1996;70:1990–1999. doi: 10.1128/jvi.70.3.1990-1999.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aida Y, Miyasaka M, Okada K, Onuma M, Kogure S, Suzuki M, Minoprio P, Levy D, Ikawa Y. Further phenotypic characterization of target cells for bovine leukemia virus experimental infection in sheep. Am J Vet Res. 1989;50:1946–1951. [PubMed] [Google Scholar]
- 4.Baranger A M, Palmer C R, Hamm M K, Giebler H A, Brauweiler A, Nyborg J K, Schepartz A. Mechanism of DNA-binding enhancement by the human T-cell leukaemia virus transactivator Tax. Nature. 1995;376:606–608. doi: 10.1038/376606a0. [DOI] [PubMed] [Google Scholar]
- 5.Bex F, Murphy K, Wattiez R, Burny A, Gaynor R. Phosphorylation of the human T-cell leukemia virus type 1 transactivator Tax on adjacent serine residues is critical for Tax activation. J Virol. 1999;73:738–745. doi: 10.1128/jvi.73.1.738-745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boros I M, Tie F, Giam C Z. Interaction of bovine leukemia virus transactivator Tax with bZip proteins. Virology. 1995;214:207–214. doi: 10.1006/viro.1995.9939. [DOI] [PubMed] [Google Scholar]
- 7.Brooks P A, Nyborg J K, Cockerell G L. Identification of an NF-kappa B binding site in the bovine leukemia virus promoter. J Virol. 1995;69:6005–6009. doi: 10.1128/jvi.69.10.6005-6009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks P A, Cockerell G L, Nyborg J K. Activation of BLV transcription by NF-kappa B and Tax. Virology. 1998;243:94–98. doi: 10.1006/viro.1998.9035. [DOI] [PubMed] [Google Scholar]
- 9.Burny A, Cleuter Y, Kettmann R, Mammerickx M, Marbaix G, Portetelle D, van den Broeke A, Willems L, Thomas R. Bovine leukaemia: facts and hypotheses derived from the study of an infectious cancer. Vet Microbiol. 1988;17:197–218. doi: 10.1016/0378-1135(88)90066-1. [DOI] [PubMed] [Google Scholar]
- 10.Cann A J, Chen I S Y. Human T-cell leukemia virus types I and II. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1849–1880. [Google Scholar]
- 11.Cassens S, Ulrich U, Beimling P, Simon D. Inhibition of human T cell leukaemia virus type I long terminal repeat expression by DNA methylation: implications for latency. J Gen Virol. 1994;75:3255–3259. doi: 10.1099/0022-1317-75-11-3255. [DOI] [PubMed] [Google Scholar]
- 12.Daenke S, Nightingale S, Cruickshank J K, Bangham C R. Sequence variants of human T-cell lymphotropic virus type I from patients with tropical spastic paraparesis and adult T-cell leukemia do not distinguish neurological from leukemic isolates. J Virol. 1990;64:1278–1282. doi: 10.1128/jvi.64.3.1278-1282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derse D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J Virol. 1987;61:2462–2471. doi: 10.1128/jvi.61.8.2462-2471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derse D. trans-acting regulation of bovine leukemia virus mRNA processing. J Virol. 1988;62:1115–1119. doi: 10.1128/jvi.62.4.1115-1119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djilali S, Parodi A L, Levy D, Cockerell G L. Development of leukemia and lymphosarcoma induced by bovine leukemia virus in sheep: a hematopathological study. Leukemia. 1987;1:777–781. [PubMed] [Google Scholar]
- 16.Felber B K, Paskalis H, Kleinman-Ewing C, Wong-Staal F, Pavlakis G N. The pX protein of HTLV-I is a transcriptional activator of its long terminal repeats. Science. 1985;229:675–679. doi: 10.1126/science.2992082. [DOI] [PubMed] [Google Scholar]
- 17.Fontes J D, Strawhecker J M, Bills N D, Lewis R E, Hinrichs S H. Phorbol esters modulate the phosphorylation of human T-cell leukemia virus type I Tax. J Virol. 1993;67:4436–4441. doi: 10.1128/jvi.67.7.4436-4441.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franchini G, Wong-Staal F, Gallo R C. Human T-cell leukemia virus (HTLV-I) transcripts in fresh and cultured cells of patients with adult T-cell leukemia. Proc Natl Acad Sci USA. 1984;81:6207–6211. doi: 10.1073/pnas.81.19.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujii M, Tsuchiya H, Chuhjo T, Minamino T, Miyamoto K, Seiki M. Serum response factor has functional roles both in indirect binding to the CArG box and in the transcriptional activation function of human T-cell leukemia virus type I Tax. J Virol. 1994;68:7275–7283. doi: 10.1128/jvi.68.11.7275-7283.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujisawa J, Seiki M, Kiyokawa T, Yoshida M. Functional activation of the long terminal repeat of human T-cell leukemia virus type I by a trans-acting factor. Proc Natl Acad Sci USA. 1985;82:2277–2281. doi: 10.1073/pnas.82.8.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gessain A, Gallo R C, Franchini G. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J Virol. 1992;66:2288–2295. doi: 10.1128/jvi.66.4.2288-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes S H, Shank P R, Spector D H, Kung H J, Bishop J M, Varmus H E, Vogt P K, Breitman M L. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978;15:1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- 23.Inabe K, Ikuta K, Aida Y. Transmission and propagation in cell culture of virus produced by cells transfected with an infectious molecular clone of bovine leukemia virus. Virology. 1998;245:53–64. doi: 10.1006/viro.1998.9140. [DOI] [PubMed] [Google Scholar]
- 24.Jensen W A, Wicks-Beard B J, Cockerell G L. Inhibition of protein kinase C results in decreased expression of bovine leukemia virus. J Virol. 1992;66:4427–4433. doi: 10.1128/jvi.66.7.4427-4433.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang H, Lu H, Schiltz R L, Pise-Masison C A, Ogryzko V V, Nakatani Y, Brady J N. PCAF interacts with Tax and stimulates Tax transactivation in a histone acetyltransferase-independent manner. Mol Cell Biol. 1999;19:8136–8145. doi: 10.1128/mcb.19.12.8136. . (Erratum 20:1897, 2000.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashanchi F, Duvall J F, Kwok R P, Lundblad J R, Goodman R H, Brady J N. The coactivator CBP stimulates human T-cell lymphotrophic virus type I Tax transactivation in vitro. J Biol Chem. 1998;273:34646–34652. doi: 10.1074/jbc.273.51.34646. [DOI] [PubMed] [Google Scholar]
- 27.Katoh I, Yoshinaka Y, Ikawa Y. Bovine leukemia virus trans-activator p38tax activates heterologous promoters with a common sequence known as a cAMP-responsive element or the binding site of a cellular transcription factor ATF. EMBO J. 1989;8:497–503. doi: 10.1002/j.1460-2075.1989.tb03403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerkhofs P, Adam E, Droogmans L, Portetelle D, Mammerickx M, Burny A, Kettmann R, Willems L. Cellular pathways involved in the ex vivo expression of bovine leukemia virus. J Virol. 1996;70:2170–2177. doi: 10.1128/jvi.70.4.2170-2177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kettmann R, Deschamps J, Cleuter Y, Couez D, Burny A, Marbaix G. Leukemogenesis by bovine leukemia virus: proviral DNA integration and lack of RNA expression of viral long terminal repeat and 3′ proximate cellular sequences. Proc Natl Acad Sci USA. 1982;79:2465–2469. doi: 10.1073/pnas.79.8.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korber B, Okayama A, Donnelly R, Tachibana N, Essex M. Polymerase chain reaction analysis of defective human T-cell leukemia virus type I proviral genomes in leukemic cells of patients with adult T-cell leukemia. J Virol. 1991;65:5471–5476. doi: 10.1128/jvi.65.10.5471-5476.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok R P, Laurance M E, Lundblad J R, Goldman P S, Shih H, Connor L M, Marriott S J, Goodman R H. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 32.Lagarias D M, Radke K. Transcriptional activation of bovine leukemia virus in blood cells from experimentally infected, asymptomatic sheep with latent infections. J Virol. 1989;63:2099–2107. doi: 10.1128/jvi.63.5.2099-2107.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin H C, Dezzutti C S, Lal R B, Rabson A B. Activation of human T-cell leukemia virus type 1 tax gene expression in chronically infected T cells. J Virol. 1998;72:6264–6270. doi: 10.1128/jvi.72.7.6264-6270.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto K, Shibata H, Fujisawa J I, Inoue H, Hakura A, Tsukahara T, Fujii M. Human T-cell leukemia virus type 1 Tax protein transforms rat fibroblasts via two distinct pathways. J Virol. 1997;71:4445–4451. doi: 10.1128/jvi.71.6.4445-4451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKnight G S. The induction of ovalbumin and conalbumin mRNA by estrogen and progesterone in chick oviduct explant cultures. Cell. 1978;14:403–413. doi: 10.1016/0092-8674(78)90125-3. [DOI] [PubMed] [Google Scholar]
- 36.Ogawa Y, Sagata N, Tsuzuku-Kawamura J, Koyama H, Onuma M, Izawa H, Ikawa Y. Structure of a defective provirus of bovine leukemia virus. Microbiol Immunol. 1987;31:1009–1015. doi: 10.1111/j.1348-0421.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 37.Ohshima K, Kikuchi M, Masuda Y, Kobari S, Sumiyoshi Y, Eguchi F, Mohtai H, Yoshida T, Takeshita M, Kimura N. Defective provirus form of human T-cell leukemia virus type I in adult T-cell leukemia/lymphoma: clinicopathological features. Cancer Res. 1991;51:4639–4642. [PubMed] [Google Scholar]
- 38.Perini G, Wagner S, Green M R. Recognition of bZIP proteins by the human T-cell leukaemia virus transactivator Tax. Nature. 1995;376:602–605. doi: 10.1038/376602a0. [DOI] [PubMed] [Google Scholar]
- 39.Pyeon D, Splitter G A. Regulation of bovine leukemia virus tax and pol mRNA levels by interleukin-2 and -10. J Virol. 1999;73:8427–8434. doi: 10.1128/jvi.73.10.8427-8434.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sagata N, Yasunaga T, Tsuzuku-Kawamura J, Ohishi K, Ogawa Y, Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci USA. 1985;82:677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saggioro D, Forino M, Chieco-Bianchi L. Transcriptional block of HTLV-I LTR by sequence-specific methylation. Virology. 1991;182:68–75. doi: 10.1016/0042-6822(91)90649-v. [DOI] [PubMed] [Google Scholar]
- 42.Seiki M, Inoue J, Hidaka M, Yoshida M. Two cis-acting elements responsible for posttranscriptional trans-regulation of gene expression of human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1988;85:7124–7128. doi: 10.1073/pnas.85.19.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki T, Fujisawa J I, Toita M, Yoshida M. The trans-activator tax of human T-cell leukemia virus type 1 (HTLV-1) interacts with cAMP-responsive element (CRE) binding and CRE modulator proteins that bind to the 21-base-pair enhancer of HTLV-1. Proc Natl Acad Sci USA. 1993;90:610–614. doi: 10.1073/pnas.90.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tajima S, Ikawa Y, Aida Y. Complete bovine leukemia virus (BLV) provirus is conserved in BLV-infected cattle throughout the course of B-cell lymphosarcoma development. J Virol. 1998;72:7569–7576. doi: 10.1128/jvi.72.9.7569-7576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tajima S, Zhuang W Z, Kato M V, Okada K, Ikawa Y, Aida Y. Function and conformation of wild-type p53 protein are influenced by mutations in bovine leukemia virus-induced B-cell lymphosarcoma. Virology. 1998;243:735–746. [PubMed] [Google Scholar]
- 46.Tamiya S, Matsuoka M, Etoh K, Watanabe T, Kamihira S, Yamaguchi K, Takatsuki K. Two types of defective human T-lymphotropic virus type I provirus in adult T-cell leukemia. Blood. 1996;88:3065–3073. [PubMed] [Google Scholar]
- 47.Tanaka A, Takahashi C, Yamaoka S, Nosaka T, Maki M, Hatanaka M. Oncogenic transformation by the tax gene of human T-cell leukemia virus type I in vitro. Proc Natl Acad Sci USA. 1990;87:1071–1075. doi: 10.1073/pnas.87.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van den Broeke A, Cleuter Y, Chen G, Portetelle D, Mammerickx M, Zagury D, Fouchard M, Coulombel L, Kettmann R, Burny A. Even transcriptionally competent proviruses are silent in bovine leukemia virus-induced sheep tumor cells. Proc Natl Acad Sci USA. 1988;85:9263–9267. doi: 10.1073/pnas.85.23.9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Den Broeke A, Bagnis C, Ciesiolka M, Cleuter Y, Gelderblom H, Kerkhofs P, Griebel P, Mannoni P, Burny A. In vivo rescue of a silent Tax-deficient bovine leukemia virus from a tumor-derived ovine B-cell line by recombination with a retrovirally transduced wild-type tax gene. J Virol. 1999;73:1054–1065. doi: 10.1128/jvi.73.2.1054-1065.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner S, Green M R. HTLV-I Tax protein stimulation of DNA binding of bZIP proteins by enhancing dimerization. Science. 1993;262:395–399. doi: 10.1126/science.8211160. [DOI] [PubMed] [Google Scholar]
- 51.Weiner M P, Costa G L, Schoettlin W, Cline J, Mathur E, Bauer J C. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene. 1994;151:119–123. doi: 10.1016/0378-1119(94)90641-6. [DOI] [PubMed] [Google Scholar]
- 52.Willems L, Gegonne A, Chen G, Burny A, Kettmann R, Ghysdael J. The bovine leukemia virus p34 is a transactivator protein. EMBO J. 1987;6:3385–3389. doi: 10.1002/j.1460-2075.1987.tb02661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willems L, Chen G, Portetelle D, Mamoun R, Burny A, Kettmann R. Structural and functional characterization of mutants of the bovine leukemia virus transactivator protein p34. Virology. 1989;171:615–618. doi: 10.1016/0042-6822(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 54.Willems L, Heremans H, Chen G, Portetelle D, Billiau A, Burny A, Kettmann R. Cooperation between bovine leukaemia virus transactivator protein and Ha-ras oncogene product in cellular transformation. EMBO J. 1990;9:1577–1581. doi: 10.1002/j.1460-2075.1990.tb08277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willems L, Kettmann R, Burny A. The amino acid (157–197) peptide segment of bovine leukemia virus p34tax encompass a leucine-rich globally neutral activation domain. Oncogene. 1991;6:159–163. [PubMed] [Google Scholar]
- 56.Willems L, Kettmann R, Chen G, Portetelle D, Burny A, Derse D. A cyclic AMP-responsive DNA-binding protein (CREB2) is a cellular transactivator of the bovine leukemia virus long terminal repeat. J Virol. 1992;66:766–772. doi: 10.1128/jvi.66.2.766-772.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willems L, Thienpont E, Kerkhofs P, Burny A, Mammerickx M, Kettmann R. Bovine leukemia virus, an animal model for the study of intrastrain variability. J Virol. 1993;67:1086–1089. doi: 10.1128/jvi.67.2.1086-1089.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willems L, Kerkhofs P, Burny A, Mammerickx M, Kettmann R. Lack of LTR and ENV genetic variation during bovine leukemia virus-induced leukemogenesis. Virology. 1995;206:769–772. doi: 10.1016/s0042-6822(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 59.Willems L, Grimonpont C, Kerkhofs P, Capiau C, Gheysen D, Conrath K, Roussef R, Mamoun R, Portetelle D, Burny A, Adam E, Lefebvre L, Twizere J C, Heremans H, Kettmann R. Phosphorylation of bovine leukemia virus Tax protein is required for in vitro transformation but not for transactivation. Oncogene. 1998;16:2165–2176. doi: 10.1038/sj.onc.1201765. [DOI] [PubMed] [Google Scholar]
- 60.Xiao J, Buehring G C. In vivo protein binding and functional analysis of cis-acting elements in the U3 region of the bovine leukemia virus long terminal repeat. J Virol. 1998;72:5994–6003. doi: 10.1128/jvi.72.7.5994-6003.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshida M. Molecular biology of HTLV-I: recent progress. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:S63–S68. doi: 10.1097/00042560-199600001-00012. [DOI] [PubMed] [Google Scholar]