Abstract
Background
N6-methyladenosine (m6A) is the most common internal RNA modification and is involved in regulation of RNA and protein expression. AlkB family member 5 (ALKBH5) is a m6A demethylase. Given the important role of m6A in biological mechanisms, m6A and its regulators, have been implicated in many disease processes, including cancer. However, the contribution of ALKBH5 to invasive breast cancer (BC) remains poorly understood. The aim of this study was to evaluate the clinicopathological value of ALKBH5 in BC.
Methods
Publicly available data were used to investigate ALKBH5 mRNA alterations, prognostic significance, and association with clinical parameters at the genomic and transcriptomic level. Differentially expressed genes (DEGs) and enriched pathways with low or high ALKBH5 expression were investigated. Immunohistochemistry (IHC) was used to assess ALKBH5 protein expression in a large well-characterised BC series (n = 1327) to determine the clinical significance and association of ALKBH5 expression.
Results
Reduced ALKBH5 mRNA expression was significantly associated with poor prognosis and unfavourable clinical parameters. ALKBH5 gene harboured few mutations and/or copy number alternations, but low ALKBH5 mRNA expression was seen. Patients with low ALKBH5 mRNA expression had a number of differentially expressed genes and enriched pathways, including the cytokine-cytokine receptor interaction pathway. Low ALKBH5 protein expression was significantly associated with unfavourable clinical parameters associated with tumour progression including larger tumour size and worse Nottingham Prognostic Index group.
Conclusion
This study implicates ALKBH5 in BC and highlights the need for further functional studies to decipher the role of ALKBH5 and RNA m6A methylation in BC progression.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-024-01205-8.
Keywords: N6-methyladenosine, Epitranscriptomics, m6A, Prognosis, Breast cancer
Introduction
Breast cancer (BC) is the most commonly diagnosed cancer and the leading cause of cancer related mortality amongst women [1]. It is a heterogeneous group of diseases with distinct clinical, morphological, and molecular features between tumours that aid disease classification and inform treatment decision making [2].
N6-methyladenosine (m6A) is the most abundant internal mRNA modification and is dynamically regulated by a multiprotein complex of ‘writers’, ‘erasers’, and ‘readers’ that methylate, demethylate, and interpret the m6A mark, respectively [3]. The RNA methyltransferase complex is made up of methyltransferase-like 3 (METTL3), METTL14, and multiple adapter proteins [4]. AlkB family member 5 (ALKBH5) and fat-mass and obesity-associated protein (FTO) are currently the only identified m6A demethylases [5, 6].
The m6A modification is involved in a diverse set of mRNA transcription, splicing, translation, and stability functions [6–11]. Evidence is emerging that implicates the m6A epitranscriptomic modification in a variety of biological processes, including carcinogenesis [12–20]. Recent studies have also associated m6A regulators, including ALKBH5, in BC development, progression, and prognosis [21–27]. In BC, it has been reported that ALKBH5 expression is regulated by hypoxia inducible factors (HIFs), leading to increased expression of NANOG, thereby promoting the BC stem cell phenotype [28, 29]. ALKBH5 expression has also been shown to be increased in immortalised and transformed breast cell lines and tumour samples, and implicated in migration, invasion, and metastasis [22, 23, 27, 30–32]. However, the role of ALKBH5 in BC remains unclear. Therefore, this study aimed to investigate the relationship between ALKBH5 expression and clinicopathological factors in a large patient cohort and to relate this to mechanisms involving differential global gene expression identified using the TCGA-BRCA cohort of invasive BC cases stratified based on ALKBH5 mRNA expression.
Materials and methods
Cell line culture conditions
Human mammary epithelial cells (HMEC), MCF10A, MCF7, T-47D, MDA-MB-453, and MDA-MB-231 breast cells were utilised. HMEC, MCF-7, T47D, MDA-MB-231, and MDA-MB-453 were generously provided by Professor Lorraine Gudas (Weill Cornell Medicine). The MCF10A were a generous gift from Dr Cinzia Allegrucci (University of Nottingham). BC cell lines MCF7, T-47D and MDA-MB-231 were grown in phenol red containing RPMI-1640 medium with L-glutamine (Gibco) supplemented with 10% foetal bovine serum (FBS) (Sigma-Aldrich), 1% penicillin–streptomycin–glutamine (Gibco), and 1 mM sodium pyruvate (Gibco). MDA-MB-453 were maintained in DMEM media (Gibco) supplemented with FBS (Sigma-Aldrich) and 1% penicillin–streptomycin–glutamine (Gibco). HMEC cells grown in HuMEC media with the addition and HuMEC supplements (Gibco). MCF10A were grown in HuMEC media with HuMEC supplements (Gibco) and 100 ng/ml cholera toxin (Sigma-Aldrich). All cells were cultured at 5% CO2 at 37 °C.
Gene expression analysis
Cells were harvested for RNA using the GenElute™ Mammalian Total RNA Miniprep Kit (RTN70-1KT, Sigma-Aldrich), following manufacturer’s instructions. The qScript cDNA Synthesis Kit was used for complementary DNA (cDNA) synthesis (95047-100, Quantabio). For mRNA expression analysis, quantitative real-time polymerase chain reaction (qRT-PCR) was performed using ALKBH5 (Hs00539502_m1) and β-actin (Hs01060665_g1) Taqman probes (ThermoScientific) with LightCycler® 480 Probes Master (Roche Diagnostics) in a qRT-PCR machine (Bio-Rad) and the relative mRNA expression was determined by the Pfaffl method, as previously described [33].
Western blotting
Cell lysates in final sample buffer (100 mM Tris–HCl pH 6.8, 4% SDS and 20% glycerol) were used to assess protein expression of ALKBH5 in cell lines using western blotting (n = 3). The membrane was blocked using 5% bovine serum albumin or milk for 1 h at room temperature and probed with ALKBH5 antibody overnight at 4 °C (1:5000; Novus Biologicals, NBP1-82188) or β-actin antibody (1:10,000; Invitrogen, MA515739). For secondary antibodies, goat IgG HRP anti-rabbit or goat IgG HRP anti-mouse (1:10,000; Abcam, ab6721 and ab97023) were used for 1 h at room temperature, the signal was detected using Amersham™ ECL™ Prime reagent (GE Healthcare) and image captured using a ChemiDoc™ MP Imaging System (Bio-Rad). Full uncropped western blots are displayed in Supplementary Fig. 1.
Patient cohort
This study used a well-characterised retrospective cohort of patients diagnosed with primary invasive BC (Stage I–III, age (55, 20–87), tumour size (1.7250, 0.2–8)) at Nottingham University Hospitals NHS Trust—City Hospital Campus between 1998 and 2006 (n = 1327), as previously described [34]. The full patient demographics are described in Supplementary Table 1. This study was reviewed and approved by the Nottingham Research Ethics Committee, (approval # REC202313), and the research ethics committee of the University of Nottingham School of Veterinary Medicine and Science (approval # 2803 190814). The General Data Protection Regulation (GDPR) was applied, and informed consent obtained. The Helsinki Declaration of Human Rights was strictly observed. The Nottingham Prognostic Index (NPI) and hormone receptor status were used to inform patient management. Patients within the NPI excellent prognostic group (score ≤ 3.4) did not receive adjuvant therapy, but those patients with NPI > 3.4 received tamoxifen if ER-positive and were able to receive chemotherapy if ER-negative. Chemotherapy regimen included cyclophosphamide, methotrexate, and 5-flurouracil (CMF). Outcome data includes breast cancer specific survival (BCSS), disease free interval (DFI), and distant metastasis free survival (DMFS) [35].
Tissue microarrays and immunohistochemical staining
The cohort was arrayed using a tissue microarray (TMA) Grand Master (3D Histech), as previously described [36]. Immunohistochemical (IHC) staining was performed on 4-μm thick TMA sections using the Novolink polymer detection system (Leica Biosystems). Heat-induced antigen epitope retrieval was performed in citrate buffer (pH 6.0) for 20 min using a microwave oven. Sections were incubated with the primary ALKBH5 antibody (1:100; Novus Biologicals, NBP1-82188) diluted in Leica antibody diluent (Leica Biosystems) at room temperature for 1 h. Slides were washed and incubated with post primary block for 30 min. Novolink polymer was applied for 30 min followed by application of 3, 3′-diaminobenzidine (DAB) chromogen for 5 min. Slides were counterstained with Novolink haematoxylin for 6 min, dehydrated, and cover slipped.
Scoring of ALKBH5 protein expression
Stained TMA sections were scanned using a digital scanner (NanoZoomer, Hamamatsu Photonics) at × 20 magnification. High resolution images were viewed using Xplore (Phillips Pathology) to score ALKBH5 expression within the tumour cells. A modified histochemical score (H-score) was used to evaluate stained cells [37]. Staining intensity was assessed as follows: 0, negative; 1, weak; 2, moderate; 3, strong, and the percentage of the positively stained tumour cells was estimated subjectively. The final H-score was calculated by multiplying the percentage of positive cells (0–100) by the intensity (0–3), producing a total range of 0–300. Scoring was assessed independently by two researchers and an intraclass concordance of > 0.8 was confirmed.
ALKBH5 transcriptomic data
The cBioPortal for Cancer Genomics [38, 39] was used to investigate ALKBH5 copy number and mRNA expression alterations in BC patients utilising The Cancer Genome Atlas (TCGA) Firehose Legacy (n = 1108) [40] and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (n = 2509) [41, 42] cohorts. Kaplan Meier Plotter (KM-Plotter) was used to investigate the prognostic value of ALKBH5 for overall survival (OS), relapse free survival (RFS), and distant metastasis free survival (DMFS) [43] and the Breast Cancer Gene-Expression Miner v4.7 (bc-GenExMiner v4.7) database used to investigate ALKBH5 expression and clinical factors [44]. The UCSC Xena browser [45] was used to access the GDC TCGA BC RNA-sequencing dataset to determine ALKBH5 expression in normal (n = 113), primary tumour (n = 1097), and metastatic samples (n = 7). Utilising the METABRIC dataset, mRNA expression in primary patients were dichotomised into low and high ALKBH5 expression and correlated with clinical factors. Differential gene expression analysis was conducted on the primary tumour data from TCGA data set using DESeq2. Samples were dichotomised by quartile into lowest (< 4804.337) and highest (> 6911.127) expression of ALKBH5 and significantly differentially expressed genes (DEGs) identified (fold change ± 2 and FDR-corrected p-value < 0.05). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) [46] was used to investigate over representation analysis and enrichment of KEGG pathways with the up-and down-regulated DEGs.
Statistical analysis
Statistical analysis was performed using SPSS 24.0 statistical software (SPSS Inc.) or GraphPad Prism 8 (Dotmatics). A t-test was used to assess ALKBH5 mRNA expression in different cell lines (n = 5/6). ALKBH5 protein expression was dichotomised into low and high expression using the X-tile software, used to identify the optimal cut-off based on the association of the protein expression and patient outcome (BCSS) [47]. This has resulted in the following, for nuclear (0–300; low expression ≤ 135, high expression > 135), cytoplasmic (0–300; low expression ≤ 90, high expression > 90), and for nuclear and cytoplasmic staining combined (0–300 calculated but the scores were added together and dived by 2; low expression ≤ 88, high expression > 88). The chi-square test (χ2) was performed to analyse the relationships between expression and categorical variables. Survival curves were analysed by the KM and log rank test. The p-values ≤ 0.05 were considered significant. Data is reported in line with the REMARK guidance [48].
Results
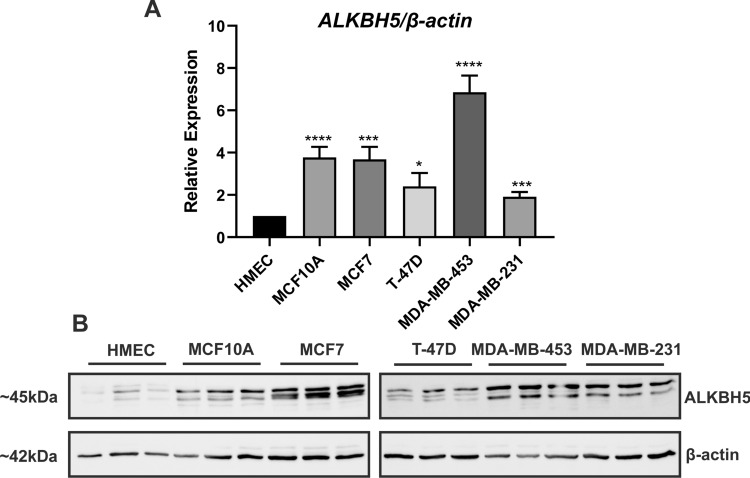
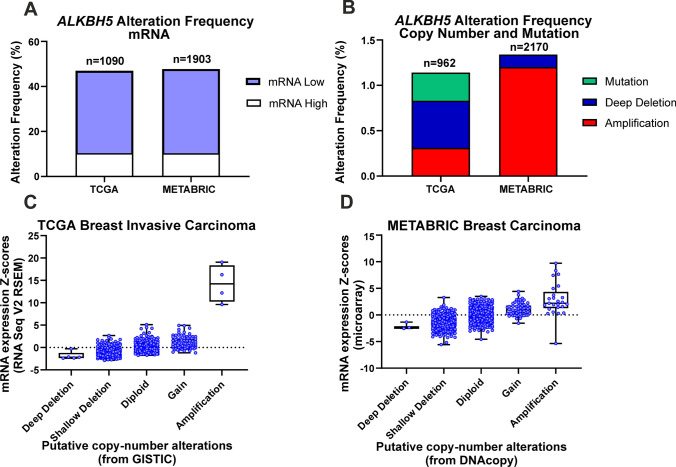
The basal expression of ALKBH5 was observed across breast cell lines at the mRNA and protein levels (Fig. 1A, B). In the TCGA dataset, 46.98% of samples showed ALKBH5 mRNA expression alterations (Fig. 2A). Similarly, 47.71% of samples in the METABRIC cohort had ALKBH5 mRNA alterations, the majority of which exhibited low mRNA expression, as compared with expression in the diploid samples (Fig. 2A). The TCGA dataset showed that 1.14% of patients harboured a copy number alteration (CNA), with 0.31% being amplification, 0.52% being deep deletion, and 0.31% being mutation (Fig. 2B). In the METABRIC cohort, 1.34% of patients had a CNA, with 1.2% being amplification, and 0.14% being deep deletion (Fig. 2B). Copy number gain was associated with high ALKBH5 mRNA expression (p < 0.001; Fig. 2C, D and Supplementary Table 2). ALKBH5 expression was lower in metastatic samples as compared to normal tissues (p < 0.05; Supplementary Fig. 2A).
Fig. 1.
ALKBH5 basal expression in breast cell lines. A ALKBH5 mRNA expression in primary human mammary epithelial cells (HMEC), non-malignant MCF10A and breast cancer MCF7, T-47D, MDA-MB-436, and MDA-MB-231 cell lines (n = 5/6). B Western blot showing ALKBH5 protein is expressed across the breast cell lines, β-actin was used as a loading control (n = 3). *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001 by t-test
Fig. 2.
Bioinformatic analysis of ALKBH5 in breast cancer datasets. The cBioPortal was used to investigate ALKBH5 mRNA and copy number alterations (A-D) in breast cancer patients from the TCGA (Firehose Legacy) and METABRIC datasets
The expression of ALKBH5 was then correlated with survival. Low expression of ALKBH5 was associated with shorter OS, RFS, and DMFS (p < 0.05; Fig. 3A–C). In the METABRIC dataset, low ALKBH5 mRNA expression was significantly associated with factors pertinent to poor prognosis including larger tumour size, high grade, and higher NPI (p < 0.05; Supplementary Table 3). It was also observed that low ALKBH5 expression was associated with shorter BCSS (p = 0.029; Supplementary Fig. 2B). Similar results were obtained utilising the bc-GenExMiner (Supplementary Table 4).
Fig. 3.
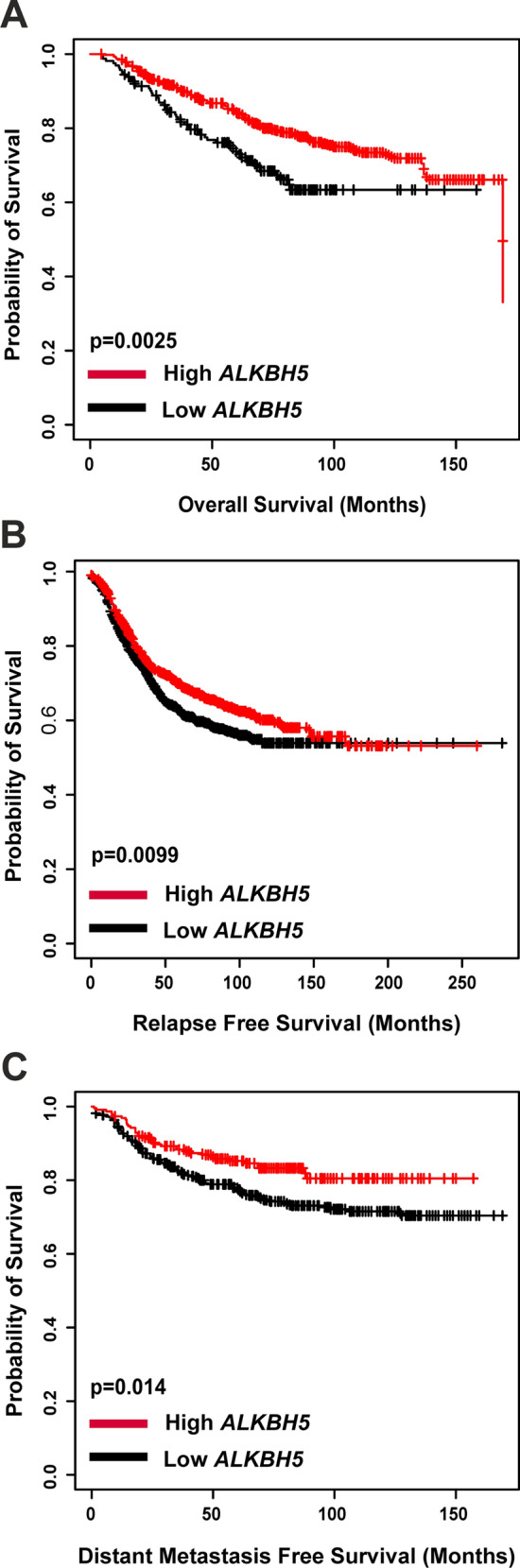
Kaplan–Meier plots was used to investigate ALKBH5 mRNA expression and (A) overall survival (n = 626), (B) relapse free survival (n = 1764), and (C) distant metastasis free survival (n = 664)
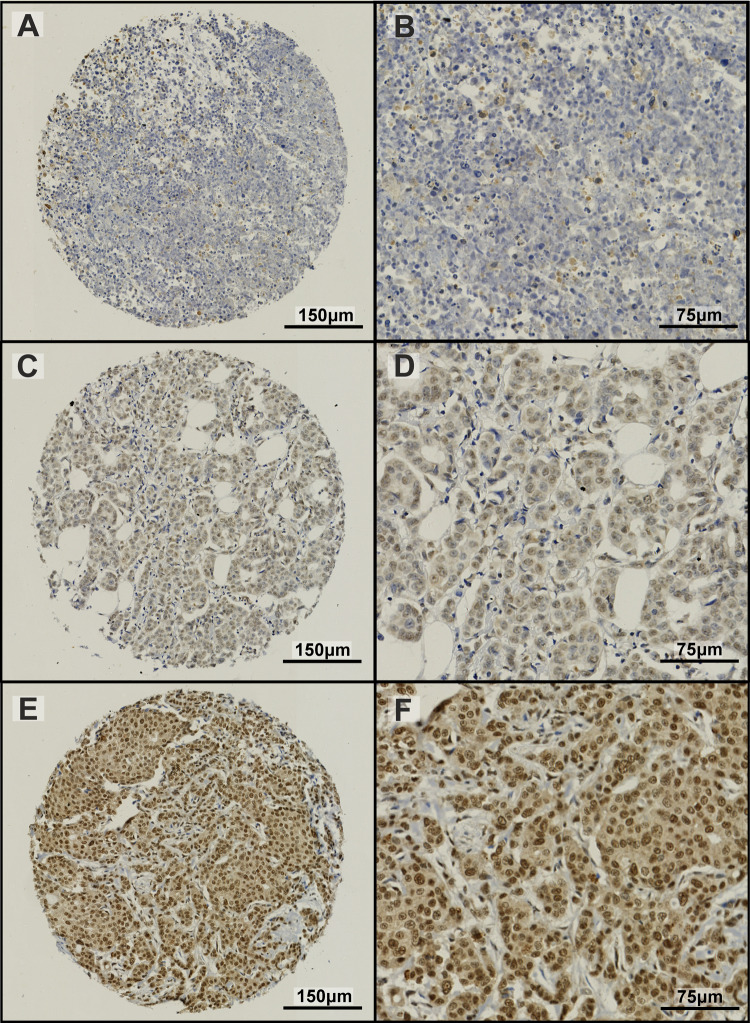
ALKBH5 protein expression in BC patient samples showed a range of staining in both the nuclei and cytoplasm of invasive tumour cells (Fig. 4). Supplementary Table 5 shows the association between ALKBH5 nuclear and cytoplasmic protein expression separately with clinicopathological parameters. Low ALKBH5 protein expression (combined nuclear and cytoplasmic expression) was significantly associated with a number of clinical parameters including larger tumour size, higher nodal stage, less tubule formation, presence of vascular invasion, hormone receptor negativity, and worse NPI prognostic group (p < 0.05, Table 1). Furthermore, low ALKBH5 protein expression was associated with worse prognosis (Fig. 5) and was a significant prognostic indicator, independent of other clinical parameters (Fig. 6, Supplementary Fig. 4 and Supplementary Table 6).
Fig. 4.
ALKBH5 immunohistochemical staining in the Nottingham Invasive BC TMA. A range of staining in the nuclear and cytoplasmic compartments was observed (A–F). Examples of weakly stained (A, B), moderately stained (C, D), and strongly stained (E, F) tumour samples are shown
Table 1.
Clinical associations with ALKBH5 combined protein expression in the Nottingham invasive breast carcinoma series
| Parameters | ALKBH5 protein Expression | ||
|---|---|---|---|
| Low (%) | High (%) | p-value | |
| Age | |||
| < 50 years | 52 (12.5) | 364 (87.5) | 0.609 |
| ≥ 50 years | 122 (13.5) | 780 (86.5) | |
| Tumour size | |||
| < 2 cm | 90 (11.6) | 688 (88.4) | 0.035 |
| ≥ 2 cm | 84 (15.6) | 456 (84.4) | |
| Grade | |||
| 1 | 18 (10.2) | 158 (89.8) | 0.04 |
| 2 | 59 (11.3) | 465 (88.7) | |
| 3 | 97 (15.7) | 521 (84.3) | |
| Stage | |||
| 1 | 91 (11.5) | 703 (88.5) | 0.012 |
| 2 | 53 (14.1) | 323 (85.9) | |
| 3 | 30 (20.3) | 118 (79.7) | |
| Tubule formation | |||
| 1 | 7 (10.1) | 62 (89.9) | < 0.001 |
| 2 | 32 (8.2) | 359 (91.8) | |
| 3 | 135 (15.7) | 723 (84.3) | |
| Pleomorphism | |||
| 1 | 0 (0) | 15 (100) | 0.315 |
| 2 | 48 (13.4) | 311 (86.6) | |
| 3 | 126 (13.3) | 818 (86.7) | |
| Mitosis | |||
| 1 | 69 (11.5) | 533 (88.5) | 0.194 |
| 2 | 37 (13.7) | 233 (86.3) | |
| 3 | 68 (15.2) | 378 (84.8) | |
| Multifocality | |||
| No | 124 (12.4) | 876 (87.6) | 0.066 |
| Yes | 50 (15.7) | 268 (84.3) | |
| Tumour type | |||
| NST | 117 (13.2) | 767 (86.8) | 0.016 |
| ILC, including lobular mixed | 20 (19.2) | 84 (80.8) | |
| Mixed NST and Iobular | 12 (14.8) | 69 (85.2) | |
| Mixed NST and special type | 4 (10.5) | 34 (89.5) | |
| Other Special tumour type including Mucinous, papillary, micropapillary, cribriform and adenoidcystic carcinoma | 1 (11.1) | 8 (88.9) | |
| Metaplastic carcinoma | 2 (66.7) | 1 (33.3) | |
| Tubular and tubular mixed | 18 (9) | 181 (91) | |
| Vascular invasion | |||
| Negative | 110 (12) | 806 (88) | 0.033 |
| Positive | 64 (15.9) | 338 (84.1) | |
| Associated DCIS | |||
| Negative | 22 (10.9) | 179 (89.1) | 0.173 |
| Positive | 152 (13.7) | 960 (86.3) | |
| LCIS | |||
| Negative | 148 (13.1) | 983 (86.9) | 0.252 |
| Positive | 26 (14.3) | 156 (85.7) | |
| Lymph node status | |||
| Negative | 91 (11.5) | 703 (88.5) | 0.015 |
| Positive | 83 (15.8) | 441 (84.2) | |
| ER | |||
| Negative | 55 (20.1) | 219 (79.9) | < 0.001 |
| Positive | 118 (11.3) | 925 (88.7) | |
| PgR | |||
| Negative | 91 (17) | 444 (83) | < 0.001 |
| Positive | 79 (10.3) | 691 (89.7) | |
| HER2 | |||
| Negative | 144 (12.7) | 990 (87.3) | 0.12 |
| Positive | 28 (15.5) | 153 (84.5) | |
| Triple negative | |||
| No | 135 (12.1) | 978 (87.9) | 0.009 |
| Yes | 36 (19) | 153 (81) | |
| Ki67 index groups | |||
| < 15 Hscore | 59 (12.1) | 430 (87.9) | 0.356 |
| ≥ 15 Hscore | 69 (14.1) | 422 (85.9) | |
| Molecular classes | |||
| Luminal types combined | 118 (11.3) | 925 (88.7) | < 0.001 |
| HER2 enriched | 17 (24.3) | 53 (75.7) | |
| TNBC | 36 (19) | 153 (81) | |
| Nottingham Prognostic Index | |||
| Good prognostic group | 41 (9.8) | 378 (90.2) | 0.006 |
| Moderate prognostic group | 92 (13.6) | 582 (86.4) | |
| Poor prognostic group | 41 (18.2) | 184 (81.8) | |
| Menopausal status | |||
| Pre | 56 (12) | 410 (88) | 0.135 |
| Post | 118 (13.8) | 734 (86.2) | |
| Chemotherapy | |||
| Non treated | 92 (11) | 745 (89) | 0.004 |
| Treated | 82 (17.1) | 398 (82.9) | |
| Endocrine therapy | |||
| Non treated | 70 (16.4) | 357 (83.6) | 0.013 |
| Treated | 104 (11.7) | 787 (88.3) | |
| Radiotherapy local | |||
| Non treated | 50 (13.4) | 322 (86.6) | 0.206 |
| Treated | 124 (13.1) | 822 (86.9) | |
| Radiotherapy LNs | |||
| Non treated | 123 (12.1) | 894 (87.9) | 0.02 |
| Treated | 51 (16.9) | 250 (83.1) | |
| Biological therapy | |||
| Non treated | 130 (13) | 873 (87) | 0.34 |
| Treated | 10 (14.3) | 60 (85.7) | |
Statistically significant associations are highlighted in bold
Fig. 5.
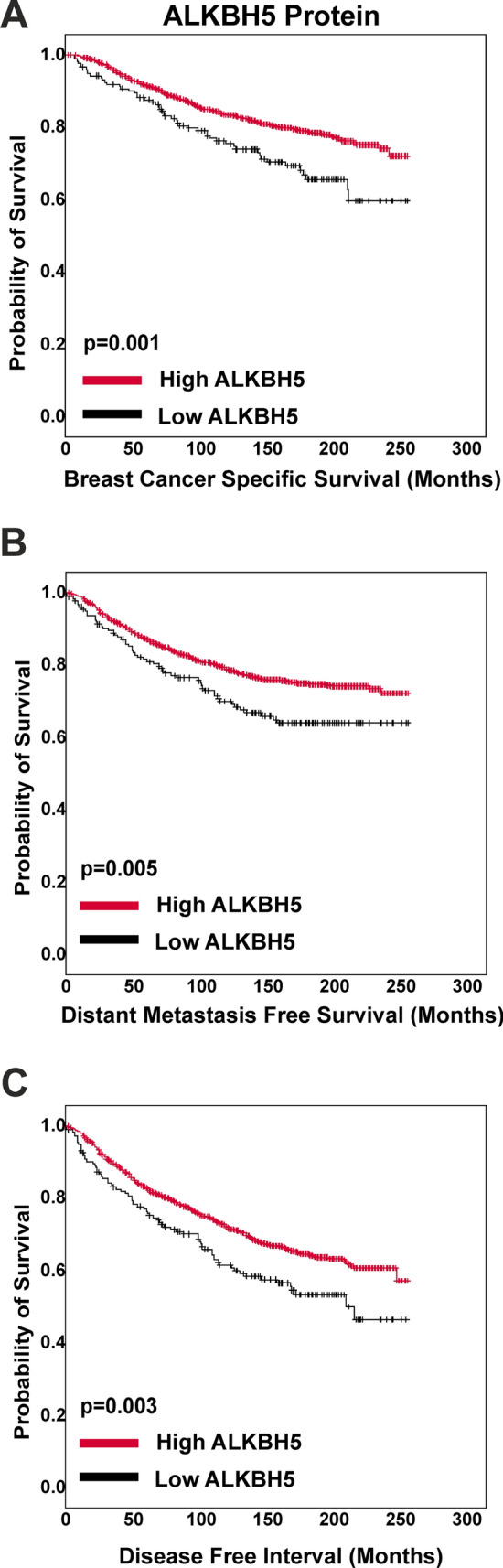
Kaplan–Meier plots was used to investigate ALKBH5 protein expression and (A) breast cancer specific survival, (B) distant metastasis free survival, and (C) disease free interval (n = 1318)
Fig. 6.
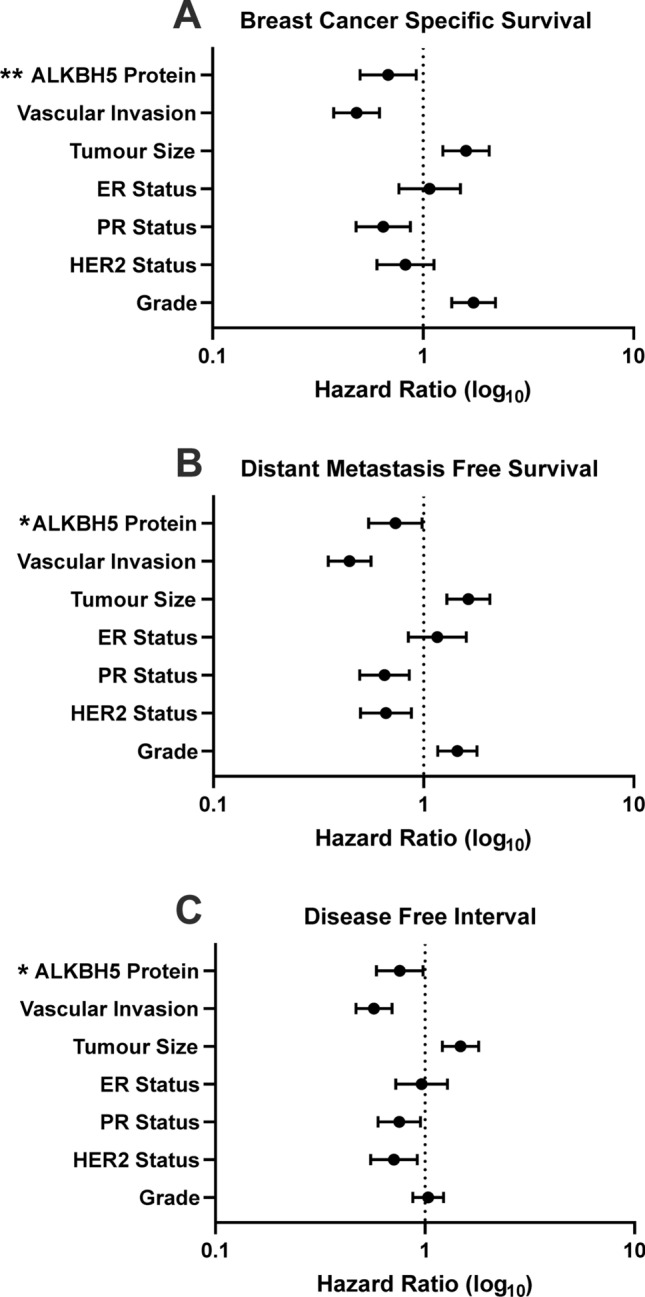
Forest plots showing the hazard ratios and 95% confidence interval of the multivariate survival analyses for ALKBH5 protein expression in the patient cohort for (A) breast cancer specific survival, (B) distant metastasis free survival, and (C) disease free interval. ALKBH5 protein expression was an independent prognostic factor
Given the evidence suggesting that low ALKBH5 expression is associated with worse outcome, the TCGA-BRCA cohort was stratified into low and high ALKBH5 expression, and DEGs identified. A total of 1964 DEGs were identified, 594 with lower expression, and 1321 more highly expressed in samples with low as compared to high ALKBH5 expression (Fig. 7 and Supplementary Table 7). Genes higher in tumours with low ALKBH5 expression were significantly enriched (FDR < 0.05) in the cytokine-cytokine receptor interaction pathway (Supplementary Table 8). In the lower DEGs in tumours with low ALKBH5 expression, 14 KEGG pathways were significantly enriched (FDR < 0.05). This included neural related pathways such as neuroactive ligand-receptor interaction, glutamatergic synapse, and dopaminergic synapse pathways (Supplementary Table 8).
Fig. 7.
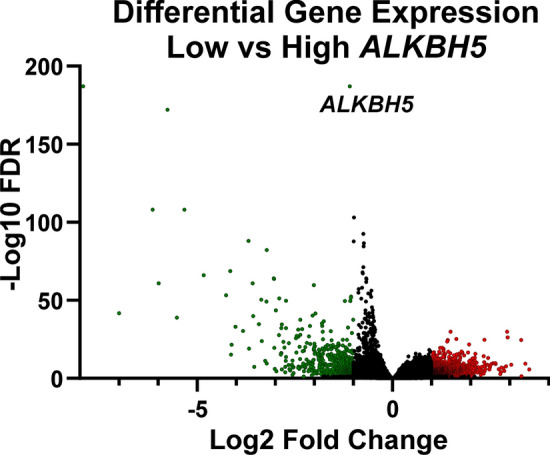
The TCGA RNA-seq dataset was stratified into low and high ALKBH5 expression by quartile, and the differentially expressed genes analysed using DeSeq2. Genes with significantly higher expression in low ALKBH5 are coloured red and genes significantly lower in low ALKBH5 are coloured green. Non-significantly differentially expressed genes are plotted in black. Significant gene expression: FC ± 2 and FDR < 0.05
Discussion
Whilst alterations in gene expression in BC have been extensively studied [49, 50], the contribution of covalent mRNA modifications such as m6A is still largely unknown. ALKBH5 is a m6A RNA demethylase [6], and the role of m6A components in cancer is only just being revealed. Several recent studies reported that reduced ALKBH5 expression in BC cell lines decreased viability, migration, invasion and tumour growth and metastasis in mouse models [29, 31, 32]. Mechanistically, hypoxia dependant expression of ALKBH5 promotes a BC stem cell phenotype [28, 29]. Despite the fundamental importance of m6A in cancer, the exact clinical relevance of ALKBH5 remains elusive as ALKBH5 protein and mRNA expression have been shown to be both increased [23, 31] or decreased [22, 31] in tumours compared to normal tissue. High ALKBH5 mRNA expression has previously been associated with ER-positive and PR-positive patients [31], suggesting that ALKBH5 may have distinct clinical significance depending on BC subtype. To address this, our study assessed the clinical association of ALKBH5 in a large well characterised cohort of BC patients.
ALKBH5 mRNA and protein expression was confirmed in malignant and non-malignant breast cell lines. Higher ALKBH5 mRNA expression was observed in transformed (MCF10A) and malignant cells compared to non-malignant primary HMEC at the mRNA level. ALKBH5 expression was increased, and m6A decreased, after immortalisation and oncogenic transformation of primary HMEC cells [30, 31], supporting a role for ALKBH5 in the progression and transformation of cells from a non-malignant to a malignant state.
Recent investigations into the prognostic value of ALKBH5 mRNA expression showed no clear association [23, 31, 51]. However, a recent study showed high ALKBH5 to be a predictor of poor survival in triple-negative BC (TNBC) patients [27]. At the mRNA level, the KM-Plotter, METABRIC dataset, and bc-GenExMiner revealed that low ALKBH5 was associated with unfavourable outcome. Additionally, the expression of ALKBH5 was found to be lower in metastatic samples compared to normal tissues, suggesting that lower ALKBH5 expression could lead to an increased chance of developing metastasis. Bioinformatic analysis using the cBioPortal for Cancer Genomics revealed that a small number of patients had CNA or mutation of ALKBH5 in both BC datasets investigated. In contrast to these modest changes in copy number, a large number of changes in ALKBH5 mRNA expression was observed with the majority of these resulting in lower mRNA expression. However, given the limited number of copy loss or loss of function mutations identified to date in BC patients, it is likely other mechanisms, such as epigenetic down-regulation of expression may also play a role in reduced ALKBH5 expression in BC.
Given the results on the mRNA level, ALKBH5 protein expression was assessed in a large cohort of well characterised BC patient samples. Consistent with previous studies in cancer, a range of ALKBH5 staining was identified in the nuclear and cytoplasmic compartments of cells [16, 23, 29, 52, 53]. Analysis of ALKBH5 nuclear and cytoplasmic staining separately revealed few associations with the clinicopathological parameters. However, low ALKBH5 combined protein expression was associated with parameters of poor prognosis in BC and worse survival.
To explore the functional role of ALKBH5 in BC, the TCGA BC primary tumour RNA-sequencing dataset was used to identify DEGs with low and high ALKBH5 expression. This revealed 1964 DEGS, 594 down-regulated, and 1321 DEGs up-regulated when ALKBH5 expression is lower. KEGG pathway revealed up-regulated DEGs were enriched in the cytokine-cytokine receptor interactions pathway. Cytokines in the tumour microenvironment play an important role in tumour pathogenesis, including in promoting metastasis [54]. Immune cells are attracted by oncogenic changes, and these cells secrete cytokines, chemokines, and growth factors to which the tumour responds leading to tumour development and progression [55]. Thus, suggesting tumours with low ALKBH5 have an increased cytokine signalling causing pro-survival and pro-metastatic signals. In addition, several studies have investigated the role of m6A RNA methylation and the immune system. The METTL3 m6A methyltransferase is important for T cell homeostasis and differentiation [56], and dendritic cell maturation and activation [57]. Recent investigations have implicated m6A in response to immunotherapy, an emerging and increasingly used therapy now being utilised in BC. Regulators of m6A influence the tumour immune microenvironment and response to anti-PD1 therapies [58–62]. Two matrix metalloproteinases (MMPs; MMP-1 and MMP-20) were identified as significantly up-regulated in tumours with low ALKBH5. MMPs play a pivotal role in cancer cell migration, invasion, and metastasis [63]. Multiple studies show MMP-1 plays a role in invasiveness by promoting local growth and the formation of metastasis [64–66]. Despite initial reports that MMP-20 expression was restricted to enamel, it is expressed in BC cell lines and tissue and promotes invasion in ovarian cancer [67, 68].
Fourteen significantly enriched pathways with down-regulated DEGs in tumours with low ALKBH5 expression were identified. The majority were related to neural signalling, including neuroactive ligand-receptor interactions, glutamatergic synapse, and dopaminergic synapse. Neuroactive ligand-receptor interaction was the most enriched pathway which has been shown to play a role in brain metastasis in TNBC [69]. Dopamine functions in many pathways through binding to its receptor. There is currently conflicting evidence on the role of dopamine receptor activation in cancers, including in BC [70, 71], however several studies have shown that stimulated dopamine signalling inhibits tumour growth [72–74]. Dopamine receptor D2 (DRD2) was down-regulated with low ALKBH5, and studies have shown DRD2 to be up or down-regulated in different cancer types [75–77]. A study has also implicated FTO in the control of DRD2 dependant signalling [78].
Altered metabolism is a widely accepted hallmark of cancer [79], and increased glutamine and glutamate signalling, including through up-regulation of receptors, increases cancer cell growth and proliferation [80]. Enrichment of this pathway suggests that glutamate signalling is down-regulated in these tumours. Whilst many studies have associated the increase of these receptors to be oncogenic, previous research has found that in cancer cells the inhibition of certain glutamate receptor subunits has led to the increased proliferation [81, 82]. Interestingly, glutamate metabotropic receptor 4 (GRM4) was down-regulated with low ALKBH5 expression. In BC high expression of GRM4 was associated with better prognosis in patients and furthermore may act as a tumour suppressor [83].
While this study presents promising findings of the potential role of ALKBH5 in invasive BC at both the mRNA and protein level using multiple large well-characterised cohorts. However, we acknowledge some limitations. Firstly, the protein expression BC cohort used in this study is a retrospective cohort. While these results were validated on additional publicly available BC transcriptomic cohorts, extending this study to include further patient cohorts, including ethnically diverse patient cohorts to further understand the prognostic value of ALKBH5 would be beneficial. In addition, in vitro and in vivo studies would allow for further insights to be gained into the mechanistic role of ALKBH5 in BC and provide a wider mechanistic context to its clinical relevance. Additionally, utilising a range of cell line and patient derived models representing different molecular subtypes of BC would allow for further characterisation of the role of ALKBH5 in BC.
Taken together, this study provides clinical evidence that ALKBH5 plays a role in BC. Low ALKBH5 mRNA and protein expression was shown to be associated with unfavourable clinical outcomes and worse prognosis. Further functional studies into the role of ALKBH5 and related mechanisms are therefore warranted to determine how reduced expression of ALKBH5 may contribute to poorer outcomes in BC. Given ALKBH5 functions as an m6A demethylase, the association of low ALKBH5 with poorer outcomes indicates that unopposed RNA m6A methylation mediated by METTL3 may promote BC progression. For this reason, a phase 1 clinical trial of the STC-15 METTL3 inhibitor (NCT05584111) is currently underway [84], and extending this to BC is justified [85].
Supplementary Information
Acknowledgements
We thank the Nottingham Health Science Biobank and Breast Cancer Now Tissue Bank for providing the tissue samples. We also gratefully acknowledge the financial support from the University of Nottingham, the BBSRC Doctoral Training Program (BB/I024291/1), and the School of Veterinary Medicine and Science, University of Nottingham. We acknowledge the support of the British Council ResearcherLinks program (RLWK10-458041157) and Kazan Federal University (2019–2021).
Author contributions
Conceptualisation: NPM, EAR, RGF, CSR. Data acquisition and curation: CLW, MA, MST, JLR, AEH, JNJ, NB, AAR, RRM, YAF. Formal analysis: CLW, MA, MST, JLR, AEH, JNJ, NB, AAR, RRM, YAF, SM, ARG, NPM. First manuscript draft: CLW. Funding acquisition: NPM, EAR. Supervision: NPM, CSR, RGF, EAR. Writing and editing draft: CLW, MA, MST, JLR, AEH, JNJ, NB, AAR, RRM, YAK, SM, ARG, CSR, RGF, EAR, NPM. All authors have read and agreed to the submitted version of the manuscript.
Funding
We gratefully acknowledge the funding from the School of Veterinary medicine and Science, University of Nottingham, the BBSRC Doctoral Training Program (BB/I024291/1), and Kazan Federal University (2019–2021). We also acknowledge the support of the British Council ResearcherLinks program (RLWK10-458041157).
Availability of data and materials
The transcriptomic and associated clinical data utilised in this study is publicly available from the cBioPortal for Cancer Genomics [38, 39], KM-Plotter [43], bc-GenExMiner v4.7 database [44], and the UCSC Xena browser [45]. The authors confirm the data is available on reasonable request.
Declarations
Ethics approval and consent to participate
This study was reviewed and approved by the Nottingham Research Ethics Committee, (approval # REC202313), and the research ethics committee of the University of Nottingham School of Veterinary Medicine and Science (approval # 2803 190814). The General Data Protection Regulation (GDPR) was applied, and informed consent obtained. The Helsinki Declaration of Human Rights was strictly observed.
Patient consent for publication
Not required.
Competing interests
The authors have no conflicts of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Corinne L. Woodcock, Email: corinne.woodcock1@nottingham.ac.uk
Nigel P. Mongan, Email: nigel.mongan@nottingham.ac.uk
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011;121(10):3786–8. 10.1172/JCI60534. 10.1172/JCI60534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Hsu PJ, Chen Y-S, Yang Y-G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616–24. 10.1038/s41422-018-0040-8. 10.1038/s41422-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao G, Li H-B, Yin Z, Flavell RA. Recent advances in dynamic m(6)A RNA modification. Open Biol. 2016;6(4): 160003. 10.1098/rsob.160003. 10.1098/rsob.160003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang Y-G, He C. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–7. 10.1038/nchembio.687. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. 10.1016/j.molcel.2012.10.015. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–20. 10.1038/nature12730. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–99. 10.1016/j.cell.2015.05.014. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adhikari S, Xiao W, Zhao YL, Yang YG. m(6)A: signaling for mRNA splicing. RNA Biol. 2016;13(9):756–9. 10.1080/15476286.2016.1201628. 10.1080/15476286.2016.1201628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540(7632):301–4. 10.1038/nature20577. 10.1038/nature20577 [DOI] [PubMed] [Google Scholar]
- 11.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu YC, Huttelmaier S, Skibbe JR, Su R, Deng X, Dong L, Sun M, Li C, Nachtergaele S, Wang Y, Hu C, Ferchen K, Greis KD, Jiang X, Wei M, Qu L, Guan JL, He C, Yang J, Chen J. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–95. 10.1038/s41556-018-0045-z. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Xiao J, Bai J, Tian Y, Qu Y, Chen X, Wang Q, Li X, Zhang Y, Xu J. Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol Cancer. 2019;18:137. 10.1186/s12943-019-1066-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, Qin X, Tang L, Wang Y, Hong GM, Wang X, Chen P, Gurbuxani S, Arnovitz S, Li Y, Li S, Strong J, Neilly MB, Larson RA, Jiang X, Zhang P, Jin J, He C, Chen J. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127–41. 10.1016/j.ccell.2016.11.017. 10.1016/j.ccell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bogler O, Majumder S, He C, Huang S. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591-606.e6. 10.1016/j.ccell.2017.02.013. 10.1016/j.ccell.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y, Zheng H, Li B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. 2019;512(3):479–85. 10.1016/j.bbrc.2019.03.093. 10.1016/j.bbrc.2019.03.093 [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res. 2019;38(1):163. 10.1186/s13046-019-1159-2. 10.1186/s13046-019-1159-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Meng S, Xu M, Wang S, He L, Xu X, Wang X, Xie L. Downregulation of N(6)-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N(6)-methyladenosine levels. Oncotarget. 2018;9(3):3752–64. 10.18632/oncotarget.23365. 10.18632/oncotarget.23365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang B, Yang Y, Kang M, Wang Y, Bi Y, He S, Shimamoto F. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19(1):3. 10.1186/s12943-019-1128-6. 10.1186/s12943-019-1128-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, Wong CC, Ng IO, Wong CM. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–70. 10.1002/hep.29683. 10.1002/hep.29683 [DOI] [PubMed] [Google Scholar]
- 20.Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H, Wu M, Liang Y, Zhu F, Zhang Y, Zhang X, Yuan Q, Li Y. The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019;38(19):3667–80. 10.1038/s41388-019-0683-z. 10.1038/s41388-019-0683-z [DOI] [PubMed] [Google Scholar]
- 21.Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, Liu Y, Zhang X, Zhang W, Ye L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–9. 10.1016/j.canlet.2017.11.018. 10.1016/j.canlet.2017.11.018 [DOI] [PubMed] [Google Scholar]
- 22.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, Wu X, Wan G. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18(1):46. 10.1186/s12943-019-1004-4. 10.1186/s12943-019-1004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, Ren F, Cui G, Sun R. N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J Cancer. 2019;10(22):5447–59. 10.7150/jca.35053. 10.7150/jca.35053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Y, Zheng C, Jin Y, Bao B, Wang D, Hou K, Feng J, Tang S, Qu X, Liu Y, Che X, Teng Y. Reduced expression of METTL3 promotes metastasis of triple-negative breast cancer by m6A methylation-mediated COL3A1 up-regulation. Front Oncol. 2020;10:1126. 10.3389/fonc.2020.01126. 10.3389/fonc.2020.01126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, Lu S, Xu D, Wu Y, Chen Q, Ding X, Guo H, Li Y, Fu B, Yao W, Wei M, Wu H. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020;39(31):5358–72. 10.1038/s41388-020-1338-9. 10.1038/s41388-020-1338-9 [DOI] [PubMed] [Google Scholar]
- 26.Zheng F, Du F, Qian H, Zhao J, Wang X, Yue J, Hu N, Si Y, Xu B, Yuan P. Expression and clinical prognostic value of m6A RNA methylation modification in breast cancer. Biomark Res. 2021;9(1):28. 10.1186/s40364-021-00285-w. 10.1186/s40364-021-00285-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Zou X, Chen Y, Cho WC, Zhou X. Effect of N6-methyladenosine regulators on progression and prognosis of triple-negative breast cancer. Front Genet. 2020;11: 580036. 10.3389/fgene.2020.580036. 10.3389/fgene.2020.580036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113(14):E2047–56. 10.1073/pnas.1602883113. 10.1073/pnas.1602883113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C, Zhi WI, Lu H, Samanta D, Chen I, Gabrielson E, Semenza GL. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7(40):64527–42. 10.18632/oncotarget.11743. 10.18632/oncotarget.11743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fry NJ, Law BA, Ilkayeva OR, Carraway KR, Holley CL, Mansfield KD. N(6)-methyladenosine contributes to cellular phenotype in a genetically-defined model of breast cancer progression. Oncotarget. 2018;9(58):31231–43. 10.18632/oncotarget.25782. 10.18632/oncotarget.25782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019;19(1):326. 10.1186/s12885-019-5538-z. 10.1186/s12885-019-5538-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, Abdelfattah N, Onyeagucha BC, Cui X, Lai Z, Mohammad TA, Gupta YK, Huang TH, Huang Y, Chen Y, Rao MK. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci Adv. 2018;4(10):eaar8263. 10.1126/sciadv.aar8263. 10.1126/sciadv.aar8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsson EM, Laursen KB, Whitchurch J, McWilliam A, Ødum N, Persson JL, Heery DM, Gudas LJ, Mongan NP. MiR137 is an androgen regulated repressor of an extended network of transcriptional coregulators. Oncotarget. 2015;6(34):35710–25. 10.18632/oncotarget.5958. 10.18632/oncotarget.5958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kariri YA, Joseph C, Kurozumi S, Toss MS, Alsaleem M, Raafat S, Mongan NP, Aleskandarany MA, Green AR, Rakha EA. Prognostic significance of KN motif and ankyrin repeat domains 1 (KANK1) in invasive breast cancer. Breast Cancer Res Treat. 2020;179(2):349–57. 10.1007/s10549-019-05466-8. 10.1007/s10549-019-05466-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El Ansari R, Craze ML, Miligy I, Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA, Green AR. The amino acid transporter SLC7A5 confers a poor prognosis in the highly proliferative breast cancer subtypes and is a key therapeutic target in luminal B tumours. Breast Cancer Res. 2018;20(1):21. 10.1186/s13058-018-0946-6. 10.1186/s13058-018-0946-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer. 2005;116(3):340–50. 10.1002/ijc.21004. 10.1002/ijc.21004 [DOI] [PubMed] [Google Scholar]
- 37.McCartyJr KS, Miller LS, Cox EB, Konrath J, McCartySr KS. Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol. 1985;109(8):716–21. [PubMed] [Google Scholar]
- 38.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. 10.1158/2159-8290.cd-12-0095. 10.1158/2159-8290.cd-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. 10.1126/scisignal.2004088. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400-416.e11. 10.1016/j.cell.2018.02.052. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale AL, Brenton JD, Tavaré S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52. 10.1038/nature10983. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, Tsui DW, Liu B, Dawson SJ, Abraham J, Northen H, Peden JF, Mukherjee A, Turashvili G, Green AR, McKinney S, Oloumi A, Shah S, Rosenfeld N, Murphy L, Bentley DR, Ellis IO, Purushotham A, Pinder SE, Borresen-Dale AL, Earl HM, Pharoah PD, Ross MT, Aparicio S, Caldas C. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. 10.1038/ncomms11479. 10.1038/ncomms11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–31. 10.1007/s10549-009-0674-9. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- 44.Jézéquel P, Gouraud W, Ben Azzouz F, Guérin-Charbonnel C, Juin PP, Lasla H, Campone M. bc-GenExMiner 4.5: new mining module computes breast cancer differential gene expression analyses. Database (Oxford). 2021. 10.1093/database/baab007. 10.1093/database/baab007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, Zhu J, Haussler D. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–8. 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199-w205. 10.1093/nar/gkz401. 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–9. 10.1158/1078-0432.Ccr-04-0713. 10.1158/1078-0432.Ccr-04-0713 [DOI] [PubMed] [Google Scholar]
- 48.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer. 2005;93(4):387–91. 10.1038/sj.bjc.6602678. 10.1038/sj.bjc.6602678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, Desmedt C, Ignatiadis M, Sengstag T, Schütz F, Goldstein DR, Piccart M, Delorenzi M. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10(4):R65. 10.1186/bcr2124. 10.1186/bcr2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alsaleem MA, Ball G, Toss MS, Raafat S, Aleskandarany M, Joseph C, Ogden A, Bhattarai S, Rida PCG, Khani F, Davis M, Elemento O, Aneja R, Ellis IO, Green A, Mongan NP, Rakha E. A novel prognostic two-gene signature for triple negative breast cancer. Mod Pathol. 2020;33(11):2208–20. 10.1038/s41379-020-0563-7. 10.1038/s41379-020-0563-7 [DOI] [PubMed] [Google Scholar]
- 51.Zhang B, Gu Y, Jiang G. Expression and prognostic characteristics of m(6) A RNA methylation regulators in breast cancer. Front Genet. 2020;11: 604597. 10.3389/fgene.2020.604597. 10.3389/fgene.2020.604597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thalhammer A, Bencokova Z, Poole R, Loenarz C, Adam J, O’Flaherty L, Schodel J, Mole D, Giaslakiotis K, Schofield CJ, Hammond EM, Ratcliffe PJ, Pollard PJ. Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1alpha (HIF-1alpha). PLoS ONE. 2011;6(1): e16210. 10.1371/journal.pone.0016210. 10.1371/journal.pone.0016210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu H, Yang X, Tang J, Si S, Zhou Z, Lu J, Han J, Yuan B, Wu Q, Lu Q, Yang H. ALKBH5 inhibited cell proliferation and sensitized bladder cancer cells to cisplatin by m6A-CK2α-mediated glycolysis. Mol Ther Nucleic Acids. 2021;23:27–41. 10.1016/j.omtn.2020.10.031. 10.1016/j.omtn.2020.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esquivel-Velázquez M, Ostoa-Saloma P, Palacios-Arreola MI, Nava-Castro KE, Castro JI, Morales-Montor J. The role of cytokines in breast cancer development and progression. J Interferon Cytokine Res. 2015;35(1):1–16. 10.1089/jir.2014.0026. 10.1089/jir.2014.0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–7. 10.1038/nature01322. 10.1038/nature01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, Bailis W, Cao G, Kroehling L, Chen Y, Wang G, Broughton JP, Chen YG, Kluger Y, Simon MD, Chang HY, Yin Z, Flavell RA. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548(7667):338–42. 10.1038/nature23450. 10.1038/nature23450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, Zhou Q, Cao X. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat Commun. 2019;10(1):1898. 10.1038/s41467-019-09903-6. 10.1038/s41467-019-09903-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, Aplin AE, Lu Z, Hwang S, He C, He YY. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10(1):2782. 10.1038/s41467-019-10669-0. 10.1038/s41467-019-10669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, Agrawal K, Gonzalez GM, Wang Y, Patel SP, Rana TM. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci USA. 2020;117(33):20159–70. 10.1073/pnas.1918986117. 10.1073/pnas.1918986117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, Huang X, Wang J, Dougherty U, Bissonnette MB, Shen B, Weichselbaum RR, Xu MM, He C. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566(7743):270–4. 10.1038/s41586-019-0916-x. 10.1038/s41586-019-0916-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yi L, Wu G, Guo L, Zou X, Huang P. Comprehensive analysis of the PD-L1 and immune infiltrates of m(6)A RNA methylation regulators in head and neck squamous cell carcinoma. Mol Ther Nucleic Acids. 2020;21:299–314. 10.1016/j.omtn.2020.06.001. 10.1016/j.omtn.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19(1):53. 10.1186/s12943-020-01170-0. 10.1186/s12943-020-01170-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278(1):16–27. 10.1111/j.1742-4658.2010.07919.x. 10.1111/j.1742-4658.2010.07919.x [DOI] [PubMed] [Google Scholar]
- 64.Liu H, Kato Y, Erzinger SA, Kiriakova GM, Qian Y, Palmieri D, Steeg PS, Price JE. The role of MMP-1 in breast cancer growth and metastasis to the brain in a xenograft model. BMC Cancer. 2012;12:583. 10.1186/1471-2407-12-583. 10.1186/1471-2407-12-583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakopoulou L, Giannopoulou I, Gakiopoulou H, Liapis H, Tzonou A, Davaris PS. Matrix metalloproteinase-1 and -3 in breast cancer: correlation with progesterone receptors and other clinicopathologic features. Hum Pathol. 1999;30(4):436–42. 10.1016/s0046-8177(99)90120-x. 10.1016/s0046-8177(99)90120-x [DOI] [PubMed] [Google Scholar]
- 66.McGowan PM, Duffy MJ. Matrix metalloproteinase expression and outcome in patients with breast cancer: analysis of a published database. Ann Oncol. 2008;19(9):1566–72. 10.1093/annonc/mdn180. 10.1093/annonc/mdn180 [DOI] [PubMed] [Google Scholar]
- 67.Kraus D, Reckenbeil J, Perner S, Winter J, Probstmeier R. Expression pattern of matrix metalloproteinase 20 (MMP20) in human tumors. Anticancer Res. 2016;36(6):2713–8. [PubMed] [Google Scholar]
- 68.Wang S, Jia J, Liu D, Wang M, Wang Z, Li X, Wang H, Rui Y, Liu Z, Guo W, Nie J, Dai H. Matrix metalloproteinase expressions play important role in prediction of ovarian cancer outcome. Sci Rep. 2019;9(1):11677. 10.1038/s41598-019-47871-5. 10.1038/s41598-019-47871-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou L, Gao HF, Liu DS, Feng JY, Gao DD, Xia W. Gene expression profiling of brain metastatic cell from triple negative breast cancer: understanding the molecular events. Gene. 2018;640:21–7. 10.1016/j.gene.2017.10.019. 10.1016/j.gene.2017.10.019 [DOI] [PubMed] [Google Scholar]
- 70.Sarkar C, Chakroborty D, Chowdhury UR, Dasgupta PS, Basu S. Dopamine increases the efficacy of anticancer drugs in breast and colon cancer preclinical models. Clin Cancer Res. 2008;14(8):2502–10. 10.1158/1078-0432.ccr-07-1778. 10.1158/1078-0432.ccr-07-1778 [DOI] [PubMed] [Google Scholar]
- 71.Jiang SH, Hu LP, Wang X, Li J, Zhang ZG. Neurotransmitters: emerging targets in cancer. Oncogene. 2020;39(3):503–15. 10.1038/s41388-019-1006-0. 10.1038/s41388-019-1006-0 [DOI] [PubMed] [Google Scholar]
- 72.Chakroborty D, Sarkar C, Mitra RB, Banerjee S, Dasgupta PS, Basu S. Depleted dopamine in gastric cancer tissues: dopamine treatment retards growth of gastric cancer by inhibiting angiogenesis. Clin Cancer Res. 2004;10(13):4349–56. 10.1158/1078-0432.ccr-04-0059. 10.1158/1078-0432.ccr-04-0059 [DOI] [PubMed] [Google Scholar]
- 73.Hoeppner LH, Wang Y, Sharma A, Javeed N, Van Keulen VP, Wang E, Yang P, Roden AC, Peikert T, Molina JR, Mukhopadhyay D. Dopamine D2 receptor agonists inhibit lung cancer progression by reducing angiogenesis and tumor infiltrating myeloid derived suppressor cells. Mol Oncol. 2015;9(1):270–81. 10.1016/j.molonc.2014.08.008. 10.1016/j.molonc.2014.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roney MSI, Park SK. Antipsychotic dopamine receptor antagonists, cancer, and cancer stem cells. Arch Pharm Res. 2018;41(4):384–408. 10.1007/s12272-018-1017-3. 10.1007/s12272-018-1017-3 [DOI] [PubMed] [Google Scholar]
- 75.Wu XY, Zhang CX, Deng LC, Xiao J, Yuan X, Zhang B, Hou ZB, Sheng ZH, Sun L, Jiang QC, Zhao W. Overexpressed D2 dopamine receptor inhibits non-small cell lung cancer progression through inhibiting NF-κB signaling pathway. Cell Physiol Biochem. 2018;48(6):2258–72. 10.1159/000492644. 10.1159/000492644 [DOI] [PubMed] [Google Scholar]
- 76.Jandaghi P, Najafabadi HS, Bauer AS, Papadakis AI, Fassan M, Hall A, Monast A, von Knebel Doeberitz M, Neoptolemos JP, Costello E, Greenhalf W, Scarpa A, Sipos B, Auld D, Lathrop M, Park M, Büchler MW, Strobel O, Hackert T, Giese NA, Zogopoulos G, Sangwan V, Huang S, Riazalhosseini Y, Hoheisel JD. Expression of DRD2 is increased in human pancreatic ductal adenocarcinoma and inhibitors slow tumor growth in mice. Gastroenterology. 2016;151(6):1218–31. 10.1053/j.gastro.2016.08.040. 10.1053/j.gastro.2016.08.040 [DOI] [PubMed] [Google Scholar]
- 77.Roy S, Lu K, Nayak MK, Bhuniya A, Ghosh T, Kundu S, Ghosh S, Baral R, Dasgupta PS, Basu S. Activation of D2 dopamine receptors in CD133+ve cancer stem cells in non-small cell lung carcinoma inhibits proliferation, clonogenic ability, and invasiveness of these cells. J Biol Chem. 2017;292(2):435–45. 10.1074/jbc.M116.748970. 10.1074/jbc.M116.748970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hess ME, Hess S, Meyer KD, Verhagen LA, Koch L, Bronneke HS, Dietrich MO, Jordan SD, Saletore Y, Elemento O, Belgardt BF, Franz T, Horvath TL, Ruther U, Jaffrey SR, Kloppenburg P, Bruning JC. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16(8):1042–8. 10.1038/nn.3449. 10.1038/nn.3449 [DOI] [PubMed] [Google Scholar]
- 79.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–37. 10.1038/nrc3038. 10.1038/nrc3038 [DOI] [PubMed] [Google Scholar]
- 80.Prickett TD, Samuels Y. Molecular pathways: dysregulated glutamatergic signaling pathways in cancer. Clin Cancer Res. 2012;18(16):4240–6. 10.1158/1078-0432.CCR-11-1217. 10.1158/1078-0432.CCR-11-1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luksch H, Uckermann O, Stepulak A, Hendruschk S, Marzahn J, Bastian S, Staufner C, Temme A, Ikonomidou C. Silencing of selected glutamate receptor subunits modulates cancer growth. Anticancer Res. 2011;31(10):3181–92. [PubMed] [Google Scholar]
- 82.Beretta F, Bassani S, Binda E, Verpelli C, Bello L, Galli R, Passafaro M. The GluR2 subunit inhibits proliferation by inactivating Src-MAPK signalling and induces apoptosis by means of caspase 3/6-dependent activation in glioma cells. Eur J Neurosci. 2009;30(1):25–34. 10.1111/j.1460-9568.2009.06804.x. 10.1111/j.1460-9568.2009.06804.x [DOI] [PubMed] [Google Scholar]
- 83.Xiao B, Chen D, Zhou Q, Hang J, Zhang W, Kuang Z, Sun Z, Li L. Glutamate metabotropic receptor 4 (GRM4) inhibits cell proliferation, migration and invasion in breast cancer and is regulated by miR-328-3p and miR-370-3p. BMC Cancer. 2019;19(1):891. 10.1186/s12885-019-6068-4. 10.1186/s12885-019-6068-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, Pilka ES, Aspris D, Leggate D, Hendrick AG, Webster NA, Andrews B, Fosbeary R, Guest P, Irigoyen N, Eleftheriou M, Gozdecka M, Dias JML, Bannister AJ, Vick B, Jeremias I, Vassiliou GS, Rausch O, Tzelepis K, Kouzarides T. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593(7860):597–601. 10.1038/s41586-021-03536-w. 10.1038/s41586-021-03536-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Achour C, Bhattarai DP, Groza P, Román CÁ, Aguilo F. METTL3 regulates breast cancer-associated alternative splicing switches. Oncogene. 2023. 10.1038/s41388-023-02602-z. 10.1038/s41388-023-02602-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomic and associated clinical data utilised in this study is publicly available from the cBioPortal for Cancer Genomics [38, 39], KM-Plotter [43], bc-GenExMiner v4.7 database [44], and the UCSC Xena browser [45]. The authors confirm the data is available on reasonable request.