Abstract
The combination of an amino acid deletion at codon 67 (Δ67) and Thr-to-Gly change at codon 69 (T69G) in the reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1) is associated with high-level resistance to multiple RT inhibitors. To determine the relative contributions of the Δ67 and T69G mutations on viral fitness, we performed a series of studies of HIV replication using recombinant variants. A high-level 3′-azido-3′-deoxythymidine (AZT)-resistant variant containing Δ67 plus T69G/K70R/L74I/K103N/T215F/K219Q in RT replicated as efficiently as wild-type virus (Wt). In contrast, the construct without Δ67 exhibited impaired replication (23% of growth of Wt). A competitive fitness study failed to reveal any differences in replication rates between the Δ67+T69G/K70R/L74I/K103N/T215F/K219Q mutant and Wt. Evaluation of proviral DNA sequences over a 3-year period in a patient harboring the multiresistant HIV revealed that the T69G mutation emerged in the context of a D67N/K70R/T215F/K219Q mutant backbone prior to appearance of the Δ67 deletion. To assess the impact of this stepwise accumulation of mutations on viral replication, a series of recombinant variants was constructed and analyzed for replication competence. The T69G mutation was found to confer 2′,3′-dideoxyinosine resistance at the expense of fitness. Subsequently, the development of the Δ67 deletion led to a virus with improved replication and high-level AZT resistance.
The emergence of antiretroviral drug-resistant forms of human immunodeficiency virus type 1 (HIV-1) has limited the efficacy of currently available therapeutic reverse transcriptase (RT) and protease inhibitors. Drug-resistant HIV-1 variants have been isolated from HIV-1-infected patients treated with monotherapy or combination therapy. The mutations associated with resistance to 3′-azido-3′-deoxythymidine (AZT) have been found to emerge in a specific order (7, 25). Markedly (>120-fold) higher levels of AZT resistance seem to coincide with the emergence of variants containing both M41L and T215Y changes and/or other multiple mutations in the HIV RT gene (7, 18, 21, 24, 25, 28–30). Following the cessation of therapy, and thus removal of the selective pressure of AZT, these drug-resistant mutants persist for a period of time (3, 8, 35) and are then replaced by viruses with fewer mutations (13–15). This reemergence of the wild type is presumably due to poor replicative fitness of the mutant virus compared to the wild type in the absence of AZT (19). Even small differences in viral fitness can quickly lead to the replacement of the less fit variants in a population (10, 20, 34, 40).
In an attempt to suppress viral replication and delay or prevent the development of drug-resistant viruses, antiretroviral drugs have been used in combination. Subsequently, multidrug-resistant variants of HIV-1 have emerged and been characterized. A set of five mutations in the RT (A62V, V75I, F77L, F116Y, and Q151M) has been characterized as associated with multiple dideoxynucleoside resistance (MDR) (23, 38, 39). In contrast to most drug-resistant mutations, this MDR variant has been reported to be fitter than the wild type in the absence of drugs (27, 32). A two-amino-acid insertion between codons 69 and 70 with a Thr-to-Ser amino acid change at position 69 has been described as a novel MDR mutation (11, 31, 41, 42). More recently, several groups have reported the emergence of an amino acid deletion between codons 67 and 68 in the finger domain of the RT (22, 42). A deletion at codon 67 (Δ67 mutation) has been reported to be associated with multidrug resistance (T. Imamichi, H. Imamichi, J. C. Lopez, J. A. Metcalf, J. Fallon, and H. C. Lane, Abstr. 7th Conf. Retroviruses Opportunistic Infect., abstr. 738, 2000; E.-Y. Kim, L. Vang, B. Oberg, and T. C. Merigan, Abstr. 7th Conf. Retroviruses Opportunistic Infect., abstr. 742, 2000). This, in association with a novel change of Thr to Gly at codon 69 (T69G) and a combination of known AZT resistance mutations (K70R/T215F/K219Q) and nonnucleoside RT inhibitor (NNRTI) resistance mutations (L74I/K103N), leads to a virus with much (>1,800-fold) higher levels of AZT resistance (22).
Elucidation of the mechanism(s) by which these mutations affect the replication of resistant viruses may help us to understand and predict the nature of future mutant viruses and changes in virus population after changes in therapy. To this end, we performed the present study to determine the impact of the Δ67 and T69G mutations on the replication and resistance profile of HIV.
MATERIALS AND METHODS
Cells and viruses.
Peripheral blood mononuclear cells were obtained from a patient with HIV-1 infection who had been treated with interleukin-2 and a variety of antiretroviral drugs including RT and protease inhibitors (22) under the auspices of National Institute of Allergy and Infectious Diseases-Institution Review Board-approved protocols. The MT-2 cell line (16, 17) and the HIV-1 pNL4.3 proviral DNA clone (1) were obtained from the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases (Rockville, Md.), and were contributed by Douglas Richman and Malcolm Martin, respectively. RD (human embryonal rhabdomyosarcoma) cells were obtained from the American Type Culture Collection (Manassas, Va.). MT-2 cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (HyClone Laboratories Inc., Logan, Utah), 10 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (RPMI-10). RD cells were maintained in Eagle's minimal essential medium supplemented with 10% fetal bovine serum, 10 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (EMEM-10).
PCR amplification.
Genomic DNA was extracted from 106 peripheral blood mononuclear cells from selected time points, using a QIAamp DNA Blood Mini kit (Qiagen Inc., Valencia, Calif.). A 1,685-bp fragment of the HIV-1 genome containing the gag (p7/p1/p6) regions, the protease gene, and part of the RT gene was amplified by PCR using the Expand high-fidelity PCR system (Roche Molecular Biochemical, Indianapolis, Ind.) with forward primer (nucleotides [nt] 1881 to 1904) 5′-GAAGCAATGAGCCAAGTAACAAAT-3′ and reverse primer (nt 3543 to 3566) 5′-GATATGTCCATTGGCCTTGCCCCT-3′. A nested PCR was then carried out with forward primer (nt 1965 to 1988) 5′-TTCAATTGTGGCAAAGAAGGGCA-3′ and reverse primer (nt 3482 to 3505) 5′-ATAATACACTCCATGTACTGGTTC-3′. Reaction mixtures (50 μl) containing 1× Expand HF buffer, oligonucleotide pairs (400 nM), deoxynucleoside triphosphates (200 nM), and 1.75 U of Expand high-fidelity PCR system enzyme mix were subjected to 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 2 min, with the final extension at 72°C for 7 min. The PCR products were purified with a QIAquick spin PCR purification kit (Qiagen).
Cloning and sequencing.
The purified PCR products were ligated into the pCRII vector (Invitrogen, Carlsbad, Calif.), and the ligation products were used to transform Escherichia coli TOP10F′ competent cells (Invitrogen). Positive colonies were identified, and the presence of a 1.6-kb insert was confirmed by restriction enzyme digestion with EcoRI. Plasmid DNAs containing the PCR fragments were purified with a S.N.A.P. Miniprep kit (Invitrogen). Sequencing reactions were performed with an ABI PRISM Dye Terminator cycle sequencing kit with AmpliTaq DNA polymerase FS (PE Biosystems, Foster City, Calif.); the reaction products were resolved by electrophoresis on 6.0% polyacrylamide gels and analyzed with an Applied Biosystems 373 automated sequencing system (PE Biosystems). Nucleotide sequences of the gag (p7/p1/p6) regions, the protease gene, and the RT gene were translated and aligned with Sequence Navigator (PE Biosystems). Changes in the RT regions were compared with the HIV-1 clade B consensus sequence as a reference (33).
Construction of molecular clones.
The BalI and a PflM1 sites of pNL4.3 located at nt 4553 and 5303, respectively, were deleted using a Quickchange site mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's protocol. A new PflM1 restriction site located at nt 3492 within the RT region was introduced using sense (5′-ATAATACACTCCATGTACTGGTTC-3′) and antisense (5′-GAACCAGTACATGGAGTGTATTAT-3′) primers. The resulting plasmid (pNL4.3PFB) thus contained one BalI site at nt 2622 and one PflM1 site at nt 3492 in RT. These two restriction enzyme sites were then used to construct a series of chimeric infectious clones of HIV-1. In some clones, an aspartic acid at RT codon 67 in the chimeric clone was induced using a mutagenesis kit with a sense primer (5′-GCCATAAAGAAAAAAGACAGTTGGTAGATGGAG-3′) and corresponding antisense primer.
To construct HIV variants containing multiple mutations in the RT gene, site-directed mutagenesis reactions were carried out on plasmid pCRII containing the wild-type RT gene from pNL4.3PFB, using a Quickchange site mutagenesis kit. The mutagenesis primers were designed according to the manufacturer's protocol. The presence of the intended mutations without unexpected second-site mutations was confirmed by DNA sequencing. Fragments from BalI and PflM1 digestion were used to replace the corresponding fragment of pNL4.3PFB. DNA sequencing was used to ascertain that each of the chimeric pNLPFB and mutated pNL4.3PFB variants possessed the intended mutations.
Transfections, infections, and generation of viral stocks.
Transfections were performed with a Perfect Lipid Pfx-3 kit (Invitrogen) as previously described (43). Briefly, 3.5 × 105 RD cells in 35-mm-diameter culture dishes were used for each transfection with 2 μg of the molecular clones in 5 ml of EMEM-10. After 24 h of transfection, 106 fresh MT-2 cells were added to the dishes, which were then incubated at 37°C for further 24 h. The MT-2 cells thus infected were collected, washed, and cultured at 37°C for 3 days in 5 ml of RPMI-10. Cell-free culture supernatants were obtained and stored at −80°C until used as virus stocks. Nucleotide sequencing of the stocks was performed to confirm that each stock had the intended mutation(s) without any unexpected mutations in the RT gene. The 50% tissue culture infectious dose (TCID50) of each stock was determined as previously described (43). Cultures of MT-2 cells and serial fourfold dilutions of the appropriate recombinant HIV stock were set up in triplicate. After incubation of the tissue culture plates at 37°C for 7 days, culture supernatants were collected and p24 antigen assays were performed (p24 antigen capture kit; Beckman-Coulter, Miami, Fla.). Using a cutoff value for p24 of <50 pg/ml, TCID50s were calculated for each stock by the Spearman-Karber method (2).
Drug resistance assays.
MT-2 cells (3 × 106) were incubated with 1,250 TCID50 of HIV-1 for 2 h at 37°C, washed twice, and resuspended at a cell density of 0.2 × 106/ml in RPMI-10. A 0.2 ml aliquot of the suspension was added to each well of a 96-well flat-bottom plate in the presence or absence of various concentrations of drugs and cultured for 7 days at 37°C. p24 values in day 7 culture supernatants were measured with a p24 antigen capture kit. Each assay was performed in quadruplicate. AZT and 2′,3′-dideoxyinosine (ddI) were purchased from Sigma (St. Louis, Mo.). Sensitivities were reported as the concentrations of drugs that inhibited p24 production by 50% (IC50) (22).
HIV growth kinetics and comparative replication assays.
For growth kinetic studies, 106 infected MT-2 cells were cultured in 10 ml of RPMI-10 in T-25 culture flasks in the presence or absence of 1 μM AZT. Culture supernatants (200 μl) were collected every day and replaced with an equal volume of fresh complete medium. Drug concentrations were maintained throughout the culture period. For comparative replication assays, infected MT-2 cells were cultured in 2 ml of RPMI-10 at a density of 0.1 × 106/ml in 24-well plates. Each assay was performed in triplicate. p24 levels in day 7 culture supernatants were measured by p24 antigen assays; results were expressed as percentage of growth of the wild type, NL4.3 (Wt), in the absence of drugs.
Competitive replication assays.
Wt and mutant viruses containing the Δ67 and T69G/K70R/L74I/K103N/T215F/K219Q mutations were mixed together at Wt/mutant ratios of 1:1 and 4:1. Each culture was inoculated with 1,250 TCID50 of virus. The mixed viral samples were incubated with 3 × 106 fresh MT-2 cells for 2 h at 37°C. The infected MT-2 cells were then washed with RPMI-10 and resuspended at 0.1 × 106 cells/ml. Five milliliters of each suspension was incubated in T-25 flasks in the presence or absence of 10 μM AZT. After 4 to 6 days, 0.5 ml of cell-free supernatant was collected and used to reinfect a fresh aliquot of 3 × 106 MT-2 cells, and the entire process was repeated. HIV-1 RNA was isolated from 130 μl of cell-free supernatant at the end of each passage, using a QIAamp Viral RNA Mini kit (Qiagen). HIV-1 RNA was reverse transcribed to cDNA with a minus-strand primer (5′-TTGTTTTTACATCATTAGTGTGGGC-3′; nt 3626 to 3649 of HIV-HXB2), using the Superscript preamplification system (Life Technologies, Gaithersburg, Md.). The cDNA was PCR amplified, and then the products were subjected to direct DNA sequencing using the dRhodamine Terminator Cycle Sequencing Ready reaction with AmpliTaq DNA polymerase FS (PE Biosystems) and analyzed with an Applied Biosystems 377 automated sequencing system (PE Biosystems). The data were imported into the software package EditView (PE Biosystems) for further analysis of relative peak height (19). To assess the viral population changes in each passage, the peak heights of each nucleotide at codon 67 were compared. To determine the emergence of unexpected mutations during tissue culture passages, a single strain of virus was used as a control for infection, passage, and DNA sequencing. No spontaneous mutations were found in these cultures.
Statistical analysis.
Differences between HIV variants in comparative replicative ability were calculated by using the unpaired t test of the StarView program (Abacus Concepts, Berkeley, Calif.).
RESULTS
Impact of Δ67 on HIV replication.
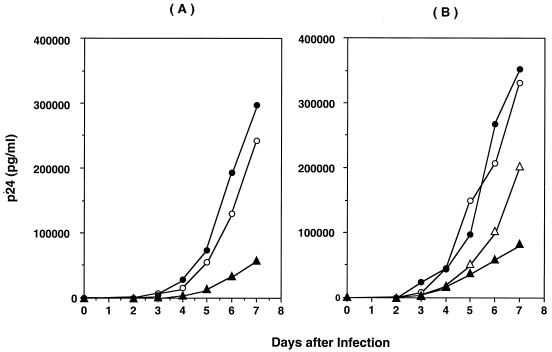
To assess the impact of the Δ67 mutation on HIV replication, a series of chimeric and site-directed-mutated recombinant viruses was created. Figure 1A compares the rates of growth of chimeric HIV containing the RT of the patient's virus predominating at 36 months following the initiation of antiviral therapy (Fig. 2, HIVRT31980), Wt, and HIVRTP31980 with an aspartic acid at codon 67, in the absence of drug (22). The chimeric HIVRT31980 possessed seven mutations associated with drug resistance: Δ67, T69G, three mutations associated with AZT resistance (K70R/T215F/K219Q), and two mutations associated with NNRTI resistance (L74I/K103N) (22). The chimera also contained 11 uncharacterized amino acid substitutions (V35I, S48T, Q102K, K122E, I135M, C162S, G196E, R277K, R284K, V291I, and E297A) which were not clearly associated with drug resistance. The chimeric HIV grew at rates comparable to that of Wt (123% of growth of Wt at day 7) in the absence of drug. However, a construct in which the wild-type amino acid aspartic acid was introduced at the codon 67 position in the chimeric HIV (HIVRT31980+D67) demonstrated a replication rate only 23% of the rate of Wt at day 7. To define the role of the 11 uncharacterized substitutions in the chimeric virus, a recombinant HIV containing only seven mutations (Δ67+T69G/K70R/L74I/K103N/T215F/K219Q) was constructed and analyzed for replicative activity. This mutant demonstrated activity comparable to that of Wt (106% of Wt [Fig. 1B]) and chimeric virus HIVRT31980. Thus, it appeared that the 11 uncharacterized mutations in the RT of the chimeric virus did not affect the rate of replication. Of note, constructs containing only Δ67 or the combination T69G/K70R/L74I/K103N/T215F/K219Q did not grow as well as Wt (60 or 25%, respectively, of Wt on day 7). Thus, the HIV mutant T69G/K70R/L74I/K103N/T215F/K219Q was an impaired virus, whereas replication was enhanced by addition of Δ67.
FIG. 1.
Growth kinetics of recombinant HIV variants. MT-2 cells were infected with chimeric (A) or mutated (B) NL4.3. Virus growth was monitored by daily quantitation of p24 antigen in culture supernatants. (A) Open circles, Wt; closed circles, HIVRT31980; triangles, HIVRT31980 with an aspartic acid at RT codon 67. (B) Open circles, Wt; open triangles, recombinant HIV with Δ67; closed triangles, HIV mutant T69G/K70R/L74I/K103N/T215F/K219Q; closed circles, HIV Δ67+T69G/K70R/L74I/K103N/T215F/K219Q mutant. Results are representative of three independent experiments.
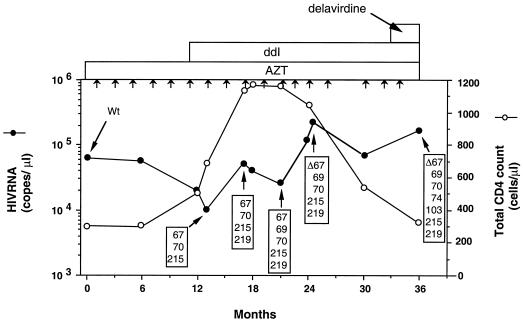
FIG. 2.
Serial total CD4+ cells count, HIV RNA copy number, and HIV-1 RT genotypes in a patient treated with AZT monotherapy followed by AZT-ddI combination therapy (27). Particle-associated HIV-1 RNA levels in plasma were determined by the branched-DNA signal amplification assay (version 1; Chiron Diagnostics Corporation, East Walpole, Mass.) (12), the detection limit of which was 10,000 copies per ml. Sequence data were obtained from PCR-amplified proviral DNA. Boxed numbers 67, 70, 74, 103, 215, and 219 refer to amino acid changes at codons 67 (Asp to Asn), 69 (Thr to Gly), 70 (Lys to Arg), 74 (Leu to Ile), 103 (Lys to Asn), 215 (Thr to Phe), and 219 (Lys to Gln), respectively. Short arrows indicate time points when interleukin-2 was administered.
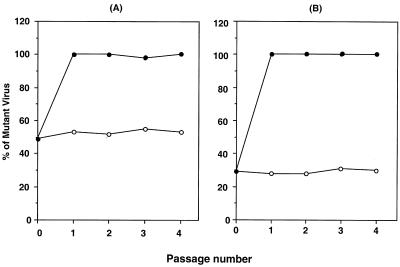
To further compare the replicative fitness of the mutant to that of Wt, a competitive fitness assay was performed. In the absence of 10 μM AZT, the ratio of the two virus populations remained the same through four tissue culture passages (Fig. 3). In the presence of 10 μM AZT, however the mutant virus rapidly became the predominant species during the first passage.
FIG. 3.
Competitive replication assays of Wt and mutant Δ67+T69G/K70R/L74I/K103N/T215F/K219Q, mixed at ratios of 1:1 (A) and 4:1 (B). Infected cells were cultured in the presence (closed circles) or absence (open circles) of 10 μM AZT. Data were generated based on relative peak heights of electropherograms produced from direct DNA sequencing of virion RNA from tissue culture supernatants at the end of each passage.
Relevance of the emergence of Δ67.
The Δ67 and T69G mutations emerged in a patient undergoing combination therapy with AZT and ddI (22). To determine the temporal relationship between the emergence of these mutations, longitudinal sequence analyses were performed at different time points. As shown in Fig. 2, the well-described AZT mutations D67N, K70R, T215F, and K219Q were the first to emerge under this combination therapy. The T69G mutation then emerged, followed by Δ67. To evaluate the effects of these amino acid changes, we constructed recombinant viruses and evaluated their replicative ability and sensitivity to AZT and ddI.
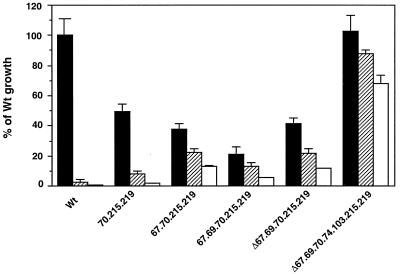
The HIV mutant K70R/T215F/K219Q replicated poorly compared to Wt (49.6% ± 4.66% of Wt, P < 0.01) (Fig. 4). Addition of the D67N mutation in this context (K70R/T215F/K219Q) led to a further decrease in replicative ability (38.0% ± 3.50% of Wt, P = 0.04). The addition of T69G to D67N/K70R/T215F/K219Q further reduced the replication potential (20.8% ± 5.30%). In contrast, deletion of amino acid 67 from D67N/T69G/K70R/T215F/K219Q (Δ67+T69G/K70R/T215F/K219Q) partially restored replicative ability (41% ± 3.72% of Wt, P = 0.024), and the addition of L74I and K103N restored it to 100% of Wt. Significant differences in replication were noted between K70R/T215F/K219Q and D67N/K70R/T215F/K219Q (P = 0.04), between D67N/K70R/T215F/K219Q and D67N/T69G/K70R/T215F/K219Q (P = 0.013), and between D67N/T69G/K70R/T215F/K219Q and Δ67+T69G/K70R/T215F/K219Q (P <0.01).
FIG. 4.
Replication of HIV variants containing mutations associated with in vivo growth. Recombinant mutants were constructed by site-directed mutagenesis. The mutations in the variants correspond to mutations existing in proviral DNA derived from a patient (Fig. 2). To generate viruses, MT-2 cells were infected with 1,250 TCID50/3 × 106 cells and cultured for 7 days. Levels of p24 antigen were measured on day 7 to determine the growth properties of each virus. Viruses were cultured in the absence (closed bars) or presence of 1 (hatched bars) or 10 (opened bars) μM AZT. Results are expressed as the percentage of growth ±/standard error of the mean compared to Wt growth in the absence of AZT. In this experiment, the p24 concentration of Wt was 644 ± 75 ng/ml. Three independent experiments were performed. Numbers 70, 215, 219, 67, 74, and 103 on the x axis indicate K70R, T215F, K219Q, D67N, L74I, and K103N, respectively.
The drug sensitivities of each variant were also analyzed (Table 1). As mutations accumulated in RT, AZT sensitivity decreased. Emergence of the T69G mutation in the D67N/K70R/T215F/K219Q backbone led to ddI resistance (Table 1). The deletion of codon 67 led to an increase in ddI sensitivity but had no effect on ddI resistance in the presence of AZT or on AZT resistance.
TABLE 1.
Drug susceptibility of recombinant HIV
| Virus | Mean IC50 (nM) ± SEMa
|
|||
|---|---|---|---|---|
| AZT
|
ddI
|
|||
| −ddI | +1 μM ddI | −AZT | +1 μM AZT | |
| Wt (NL4.3) | 14.7 ± 6.50 | NAb | 305 ± 101 | NA |
| K70R/T215F/K219Q | 170 ± 62.0 | NA | 283 ± 22 | NA |
| D67N/K70R/T215F/K219Q | 2,690 ± 556 | NA | 403 ± 55 | 281 ± 65.0 |
| D67N/T69G/K70R/T215F/K219Q | 2,260 ± 530 | 5,700 ± 1,780 | 1,780 ± 109 | 3,370 ± 633 |
| Δ67+T69G/K70R/T215F/K219Q | 3,600 ± 737 | 5,250 ± 1,320 | 327 ± 68.0 | 3,700 ± 850 |
| Δ67+T69G/K70R/L74I/K103N/T215F/K219Q | 31,700 ± 9,070 | 32,000 ± 15,300 | 381 ± 72.0 | 1,450 ± 236 |
Values were derived from at least three independent assays.
NA, not applicable (presence of 1 μM ddI or AZT prevented determination IC50 for AZT or ddI).
High-level replication profile.
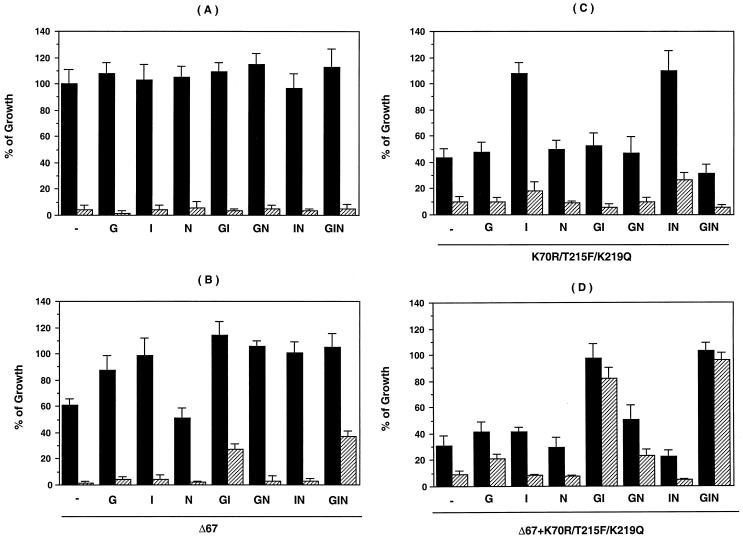
To define the interactions among the RT mutations associated with the increased replicative potential of the Δ67+T69G/K70R/L74I/K103N/T215F/K219Q mutant (Fig. 1), comparative replication assays were carried out in a series of HIV-1 mutants in the presence or absence of 1 μM AZT. Replicative activity was measured on day 7 postinfection (Fig. 5). In the absence of AZT, the T69G, L74I, K103N, T69G/L74I, T69G/K103N, L74I/K103N, and T69G/L74I/K103N mutants showed no significant difference in growth compared to Wt (P > 0.05). All were sensitive to 1 μM AZT (Fig. 5A). The deletion of codon 67 resulted in a 61% ± 5.0% decrease in the replication of Wt (P < 0.01) in the absence of AZT (Fig. 5B). The addition of T69G or L74I to the Δ67 mutant increased the replicative potential to 87% ± 10% and 99% ± 13% of Wt (P < 0.05 and P < 0.01, respectively). The addition of K103N alone to the Δ67 mutant had no effect. In the presence of 1 μM AZT, the mutants designated Δ67+GI and Δ67+GIN in Fig. 5B showed significant growth (P < 0.01) compared to Wt, indicating resistance to AZT (22).
FIG. 5.
Replication properties of HIV variants. Recombinant HIV-1 mutants were constructed by site-directed mutagenesis. MT-2 cells were infected with 1,250 TCID50/3 × 106 cells and cultured for 7 days. Levels of p24 antigen in day 7 culture supernatant were measured to determine the growth properties of each virus. Viruses were cultured in the absence (closed bars) or presence (hatched bars) of 1 μM AZT. Results are expressed as in Fig. 4. In this experiment, the p24 concentration of Wt was 468 ± 52 ng/ml. Three independent experiments were performed. G, I, N, GI, GN, IN, and GIN denote HIV variants containing T69G, L74I, K103N, T69G/L74I, T69G/K103N, L74I/K103N, and T69G/L74I/K103N mutations, respectively. (A) Recombinant viruses with Wt amino acid (aspartic acid) at codon 67; (B) recombinant viruses with Δ67; (C) viruses containing K70R, T215F, and K219Q in addition to the indicated mutations; (D) same as panel C plus Δ67.
In further studies to define the role of additional mutations in the AZT resistance backbone (K70R/T215F/K219Q), mutant K70R/T215F/K219Q demonstrated partially decreased replicative potential compared to Wt (43% ± 7.6% of Wt, P < 0.01). This impaired ability was compensated for by the addition of L74I (108% ± 8.0%, P < 0.01) or L74I/K103N (110% ± 15%, P < 0.01)) (Fig. 5C). Addition of Δ67 alone to the K70R/T215F/K219Q mutant did not increase replicative capacity (Fig. 5D). However, the constructs containing Δ67+K70R/T215F/K219Q with the GI and GIN mutations (Fig. 5D) replicated to 97% ± 12% and 103% ± 5.5% of Wt (P < 0.01), respectively.
DISCUSSION
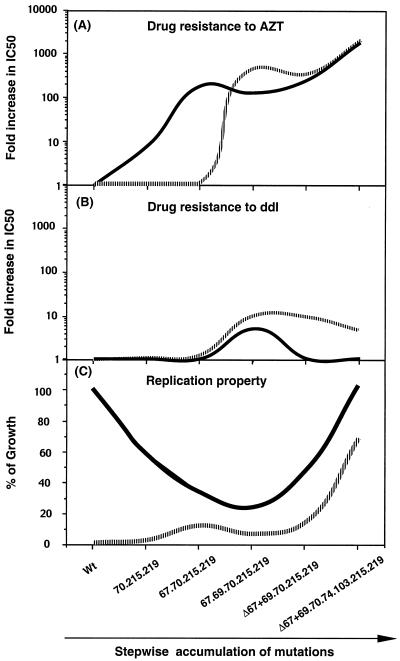
This study has characterized the Δ67 and T69G mutations that were noted to emerge in the setting of combination drug therapy. The T69G mutation appeared to be critical for the development of high-level resistance, while Δ67 restored viral fitness that was lost as a consequence of earlier mutations. A model based on our results is presented in Fig. 6.
FIG. 6.
Proposed model for the accumulation of mutations in the RT of HIV-1 during AZT-ddI combination therapy. Data points were derived from Fig. 2 and Table 1. (A) Fold increase in IC50 of AZT in the absence (solid line) and presence (dashed line) of 1 μM ddI; (B) fold increase in IC50 of ddI in the absence (solid line) and presence (dashed line) of 1 μM AZT; (C) percentage of Wt growth in the absence (solid line) and presence (dashed line) of 10 μM AZT. 70.215.219, 67.70.215.219, 67.69.70.215.219, Δ67+69.70.215.219, and Δ67+69.70.74.103.215.219 stand for K70R/T215F/K219Q, D67N/K70R/T215F/K219Q, D67N/T69G/K70R/T215F/K219Q, Δ67+T69G/K70R/T215F/K219Q, and Δ67+T69G/K70R/L74I/K103N/T215F/K219Q, respectively.
The accumulation of multiple mutations in association with the development of AZT resistance does not occur at random but follows a certain order (7, 25). Harrigan et al. performed a relative replication fitness study to address the impact of mutations conferring AZT resistance on HIV replicative fitness (19). All variants tested in that study displayed reduced replicative fitness compared to the wild type in the absence of drugs. In the presence of drugs, they observed a dose-dependent selection according to the resistance profile of each virus. Coffin has suggested that any mutations conferring resistance to drugs should be at least slightly detrimental to virus replication and thus would be present at very low levels in the absence of selective pressure by the drug (10). This proposal is consistent with the observed lower fitness of viruses resistant to AZT and/or 3TC (6, 19, 30). In our study, longitudinal nucleotide sequence analysis demonstrated that the loss of fitness that resulted as a consequence development of drug resistance could be reversed with compensatory mutations; in this case, it was a deletion at amino acid 67.
To define the relative biologic effects of the Δ67 and T69G mutations, we created a series of recombinant mutants to study virus replication capabilities. Consistent with the finding of others (19), the accumulation of a set of three mutations (K70R/T215F/K219Q) was associated with AZT resistance and impaired replicative activity (43% of Wt). Replication capacity was restored by the addition of an L74I mutation; addition of T69G to this motif, while conferring ddI resistance (data not shown), led to a new decrease in replicative capacity, which was then increased with the emergence of the Δ67 deletion (Fig. 5C and D). Thus, while the L74I change was originally reported as a mutation associated with NNRTI resistance (26), our data suggest that this change may more likely be associated with an increase in viral fitness in the setting of nucleoside resistance. In a previous study, we demonstrated that emergence of the K103N mutation following administration of delavirdine led to high-level AZT resistance in a mutant Δ67+T69G/L74I/T215F/K219Q (22). Meanwhile, addition of the K103N mutation to several variants had little effect on replication fitness (Fig. 5C and D). Taken together, these data suggest that the emergence of the K103N mutation may be associated with drug resistance but not with increased fitness. Indeed, our data suggested that the virus acquired the Δ67 and T69G mutations to achieve better fitness rather than to develop drug resistance.
Recent studies have revealed the emergence of deletions in the β3-β4 hairpin loop of the RT after AZT-ddI treatment as well as in the setting of d4T-3TC-indinavir (22, 36a). Emergence of the Δ67 mutation may be an uncommon event most likely to occur in subjects with long-term antiretroviral therapy and perhaps facilitated by prior monotherapy. Possibly current combination therapies minimize the risk of seeing these multidrug-resistant isolates.
Previous studies have shown that AZT-resistant HIV-1 isolates do not show cross-resistance to other nucleoside analogs (4, 37). However, treatment of HIV-1-infected patients with AZT plus ddI or ddC has resulted in the failure of both drugs and the emergence of resistant HIV-1 harboring the M41L and T215F mutations (9). Arts et al. have demonstrated that AZT-mediated cross-resistance to other nucleoside analogs can be induced in M41L and/or T215Y mutants (5). In our study, we were able to clearly demonstrate not only multiple nucleoside RT inhibitor resistance but also resistance to ddI that was only seen in the presence of AZT. Recently, Meyer et al. reported AZT resistance induced by an increase in nucleoside-dependent primer unblocking by AZT resistance mutations (32a). They demonstrated that HIV-1 RT containing AZT resistance mutations had a higher level of nucleotide-dependent primer unblocking activity in the presence of AZT-TTP. This mechanism may be involved in the AZT-dependent ddI resistance seen in the present study in two Δ67-containing mutants (Table 1).
It is likely that as our knowledge of different resistance profiles increase, similar types of relationships may emerge. A better understanding of these interactions should be of value in the development of rational approaches to therapy.
ACKNOWLEDGMENTS
We thank Robin Dewar for providing data on HIV RNA levels in plasma during the course of therapy and for critical reading of the manuscript, and we thank Michael Baseler, Randy Stevens, and Laurie Lambert for providing the data on total CD4 T-cell count. We also thank Siobhan Tierney for helping with quantitation of p24 antigen values in tissue culture and Ven Natarajan for critical reading of the manuscript.
The National Institute of Allergy and Infectious Diseases supported this work under contract N01-CO-56000 with SAIC-Frederick.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AIDS Clinical Trials Group. Virology manual for HIV laboratories. [Online.] Bethesda, Md: Division of AIDS, National Institute of Allergy and Infectious Diseases; 1997. http://www.niaid.nih.gov./daids/vir manual. [Google Scholar]
- 3.Albert J, Wahlberg J, Lundeberg J, Cox S, Sandstrom E, Wahren B, Uhlen M. Persistence of azidothymidine-resistant human immunodeficiency virus type 1 RNA genotypes in posttreatment sera. J Virol. 1992;66:5627–5630. doi: 10.1128/jvi.66.9.5627-5630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arts E J, Wainberg M A. Mechanisms of nucleoside analog antiviral activity and resistance during human immunodeficiency virus reverse transcription. Antimicrob Agents Chemother. 1996;40:527–540. doi: 10.1128/aac.40.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arts E J, Quinones-Mateu M E, Albright J L, Marois J P, Hough C, Gu Z, Wainberg M A. 3′-Azido-3′-deoxythymidine (AZT) mediates cross-resistance to nucleoside analogs in the case of AZT-resistant human immunodeficiency virus type 1 variants. J Virol. 1998;72:4858–4865. doi: 10.1128/jvi.72.6.4858-4865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Back N K, Nijhuis M, Keulen W, Boucher C A, Oude Essink B O, van Kuilenburg A B, van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher C A, O'Sullivan E, Mulder J W, Ramautarsing C, Kellam P, Darby G, Lange J M, Goudsmit J, Larder B A. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis. 1992;165:105–110. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- 8.Boucher C A, van Leeuwen R, Kellam P, Schipper P, Tijnagel J, Lange J M, Larder B A. Effects of discontinuation of zidovudine treatment on zidovudine sensitivity of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1525–1530. doi: 10.1128/aac.37.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brun-Vezinet F, Boucher C, Loveday C, Descamps D, Fauveau V, Izopet J, Jeffries D, Kaye S, Krzyanowski C, Nunn A, Schuurman R, Seigneurin J M, Tamalet C, Tedder R, Weber J, Weverling G J. HIV-1 viral load, phenotype, and resistance in a subset of drug-naive participants from the Delta trial. The National Virology Groups. Delta Virology Working Group and Coordinating Committee. Lancet. 1997;350:983–990. doi: 10.1016/s0140-6736(97)03380-1. [DOI] [PubMed] [Google Scholar]
- 10.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 11.De Antoni A, Foli A, Lisziewicz J, Lori F. Mutations in the pol gene of human immunodeficiency virus type 1 in infected patients receiving didanosine and hydroxyurea combination therapy. J Infect Dis. 1997;176:899–903. doi: 10.1086/516511. [DOI] [PubMed] [Google Scholar]
- 12.Dewar R L, Highbarger H C, Sarmiento M D, Todd J A, Vasudevachari M B, Davey R T, Jr, Kovacs J A, Salzman N P, Lane H C, Urdea M S. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J Infect Dis. 1994;170:1172–1179. doi: 10.1093/infdis/170.5.1172. [DOI] [PubMed] [Google Scholar]
- 13.Goudsmit J, De Ronde A, Ho D D, Perelson A S. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J Virol. 1996;70:5662–5666. doi: 10.1128/jvi.70.8.5662-5664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goudsmit J, de Ronde A, de Rooij E, de Boer R. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J Virol. 1997;71:4479–4484. doi: 10.1128/jvi.71.6.4479-4484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurusinghe A D, Land S A, Birch C, McGavin C, Hooker D J, Tachedjian G, Doherty R, Deacon N J. Reverse transcriptase mutations in sequential HIV-1 isolates in a patient with AIDS. J Med Virol. 1995;46:238–243. doi: 10.1002/jmv.1890460312. [DOI] [PubMed] [Google Scholar]
- 16.Haertle T, Carrera C J, Wasson D B, Sowers L C, Richman D D, Carson D A. Metabolism and anti-human immunodeficiency virus-1 activity of 2-halo-29, 39-dideoxyadenosine derivatives. J Biol Chem. 1988;263:5870–5875. [PubMed] [Google Scholar]
- 17.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 18.Harrigan P R, Kinghorn I, Bloor S, Kemp S D, Najera I, Kohli A, Larder B A. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J Virol. 1996;70:5930–5934. doi: 10.1128/jvi.70.9.5930-5934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrigan P R, Stuart B, Larder B A. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J Virol. 1998;72:3773–3778. doi: 10.1128/jvi.72.5.3773-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 21.Hooker D J, Tachedjian G, Solomon A E, Gurusinghe A D, Land S, Birch C, Anderson J L, Roy B M, Arnold E, Deacon N J. An in vivo mutation from leucine to tryptophan at position 210 in human immunodeficiency virus type 1 reverse transcriptase contributes to high-level resistance to 39-azido-39-deoxythymidine. J Virol. 1996;70:8010–8018. doi: 10.1128/jvi.70.11.8010-8018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamichi T, Sinha T, Imamichi H, Zhang Y-M, Metcalf J A, Falloon J, Lane H C. High-level resistance to 3′-azido-3′-deoxythymidine due to a deletion in the reverse transcriptase gene of human immunodeficiency virus type 1. J Virol. 2000;74:1023–1028. doi: 10.1128/jvi.74.2.1023-1028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iversen A K, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kellam P, Boucher C A, Larder B A. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc Natl Acad Sci USA. 1992;89:1934–1938. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellam P, Boucher C A, Tijnagel J M, Larder B A. Zidovudine treatment results in the selection of human immunodeficiency virus type 1 variants whose genotypes confer increasing levels of drug resistance. J Gen Virol. 1994;75:341–351. doi: 10.1099/0022-1317-75-2-341. [DOI] [PubMed] [Google Scholar]
- 26.Kleim J P, Rosner M, Winkler I, Paessens A, Kirsch R, Hsiou V, Arnold E, Riess G. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-743Val or Ile and Val-753Leu or Ile) HIV-1 mutants. Proc Natl Acad Sci USA. 1996;93:34–38. doi: 10.1073/pnas.93.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosalaraksa P, Kavlick M F, Maroun V, Le R, Mitsuya H. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 (HIV-1) in an in vitro competitive HIV-1 replication assay. J Virol. 1999;73:5356–5363. doi: 10.1128/jvi.73.7.5356-5363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larder B A, Darby G, Richman D D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 29.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 30.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 31.Larder B A, Bloor S, Kemp S D, Hertogs K, Desmet R L, Miller V, Sturmer M, Staszewski S, Ren J, Stammers D K, Stuart D I, Pauwels R. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 1999;43:1961–1967. doi: 10.1128/aac.43.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeda Y, Venzon D J, Mitsuya H. Altered drug sensitivity, fitness, and evolution of human immunodeficiency virus type 1 with pol gene mutations conferring multi-dideoxynucleoside resistance. J Infect Dis. 1998;177:1207–1213. doi: 10.1086/515282. [DOI] [PubMed] [Google Scholar]
- 32a.Meyer P R, Matsuura S E, Mian A M, So A G, Scott W A. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 33.Myers G, Korber B T, Wain-Hobson S, Smith R, Pavlakas G N. Human retroviruses and AIDS: a compilation and analysis of nucleicacid and amino acid sequences. [Online.] Los Alamos National La: Theoretical Biology and Biophysics Group T-10; 1995. http://hiv-web.lanl.gov boratory, Los Alamos, N.Mex. http://hiv-web.lanl.gov. . [Google Scholar]
- 34.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell lifespan, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 35.Rooke R, Tremblay M, Soudeyns H, DeStephano L, Yao X J, Fanning M, Montaner J S, O'Shaughnessy M, Gelmon K, Tsoukas C the Canadian Zidovudine Multi-Centre Study Group. Isolation of drug-resistant variants of HIV-1 from patients on long-term zidovudine therapy. AIDS. 1989;3:411–415. doi: 10.1097/00002030-198907000-00001. [DOI] [PubMed] [Google Scholar]
- 36.Ross L, Johnson M, Graham N, Shaefer M, St. Clair M. The reverse transcriptase codon 69 insertion is observed in nucleoside reverse transcriptase inhibitor-experienced HIV-1-infected individuals, including those without prior or concurrent zidovudine therapy. J Hum Virol. 1999;2:290–295. [PubMed] [Google Scholar]
- 36a.Ross L, Johnson M, Ferris R G, Short S A, Boone L R, Melby T E, Lanier R, Shaefer M, St. Clair M. Deletions in the beta3-beta4 hairpin loop of HIV-1 reverse transcriptase are observed in HIV-1 isolated from subjects during long-term antiretroviral therapy. J Hum Virol. 2000;3:144–149. [PubMed] [Google Scholar]
- 37.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance: 1999–2000 update. Int Antiviral News. 2000;7:46–69. [Google Scholar]
- 38.Schmit J-C, Cogniaux J, Hermans P, van Vaeck C, Sprecher S, van Remoortel B, Witvrouw M, Balzarini J, Desmyter J, de Clercq E, Vandamme A-M. Multiple drug resistance to nucleoside analogues and nonnucleoside reverse transcriptase inhibitors in an efficiently replicating human immunodeficiency virus type 1 patient strain. J Infect Dis. 1996;174:962–968. doi: 10.1093/infdis/174.5.962. [DOI] [PubMed] [Google Scholar]
- 39.Shirasaka T, Kavlick M F, Ueno T, Gao W-Y, Kojima E, Alcaide M L, Chockekuchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxy-nucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 41.Winters M A, Coolley K L, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merigan T C. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Investig. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yahi N, Tamalet C, Tourres C, Tivoli N, Ariasi F, Volt F, Gastaut J A, Gallais H, Moreau H J, Fantini J. Mutation patterns of the reverse transcriptase and protease genes in human immunodeficiency virus type 1-infected patients undergoing combination therapy: survey of 787 sequences. J Clin Microbiol. 1999;37:4099–4106. doi: 10.1128/jcm.37.12.4099-4106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y-M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]