Abstract
Recent advances in DNA nanotechnology allow for the assembly of nanocomponents with nanoscale precision, leading to the emergence of DNA-based material fabrication approaches. Yet, transferring these nano- and micron-scale structural arrangements to the macroscale morphologies remains a challenge, which limits the development of materials and devices based on DNA nanotechnology. Here, we demonstrate a materials fabrication approach that combines DNA-programmable assembly with actively driven processes controlled by acoustic fields. This combination provides a prescribed nanoscale order, as dictated by equilibrium assembly through DNA-encoded interactions, and field-shaped macroscale morphology, as regulated by out-of-equilibrium materials formation through specific acoustic stimulation. Using optical and electron microscopy imaging and x-ray scattering, we further revealed the nucleation processes, domain fusion, and crystal growth under different acoustically stimulated conditions. The developed approach provides a pathway for the fabrication of complexly shaped macroscale morphologies for DNA-programmable nanomaterials by controlling spatiotemporal characteristics of the acoustic fields.
Subject terms: Molecular self-assembly, DNA nanostructures, DNA and RNA, Self-assembly
DNA nanotechnology is useful in preparing nano- and meso-components, but transfer to macroscale arrangements is challenging. Here, the authors report an assembly approach combining DNA programmable assembly with process controlled by acoustic fields to prepare macroscale morphologies.
Introduction
Nanomaterials have gained much interest due to their extraordinary mechanical, chemical, optical, and biological properties1–3, which are desirable in various emerging applications. However, translating their unique properties to bulk materials and devices requires control over macroscale morphology. Often, nanomaterial generation methods permit the creation of only small mesoscale structures, which are difficult to utilize in applications. Therefore, one of the key challenges for transferring nanomaterials toward applications is gaining control over material structure on the macroscale while maintaining a desirable nanoscale organization. This challenge has fundamental roots - thermodynamic processes are effective for material formation at small scales, but kinetic effects dominate the growth when a system size increases. Besides, the thermodynamic processes typically result in equilibrium morphologies, such as crystal habits, which limit the shape control of formed materials. This work explores how to address this challenge and shape the precisely nano-organized materials at the macroscale by combining thermodynamically controlled DNA-programmable assembly with out-of-equilibrium processes driven by acoustic fields.
DNA-guided self-assembly has emerged as a powerful method for fabricating ordered three-dimensional (3D) nanomaterials with defined nanoscale control4,5 and with ability to generate diverse types of inorganic framework architectures6,7. The tunability of DNA Watson-Crick base pair interactions allows the design of programmable materials with controlled organizations at the nanometer length scale with crystal domain sizes on the order of micrometers8–10. DNA strands can be designed to fold into a designated architecture, so-called DNA origami, which enables the construction of DNA frames with the capability to hold nanoparticles and proteins8,9,11–14. The frames can be further assembled into lattice structures using directional bonds between DNA frames8,13,15. For example, the octahedral DNA frames, which interact via ssDNA sequences (sticky ends) extended from the six vertices, form a simple cubic lattice, whose Wulff shape is a cube (Fig. 1). While recent studies show that domain, growth and morphology can be manipulated through differentiated bonds14,16, the shapes of formed crystals are determined by their type. Thus, it is challenging to shape materials formed through DNA assembly into the desired form at the macroscale.
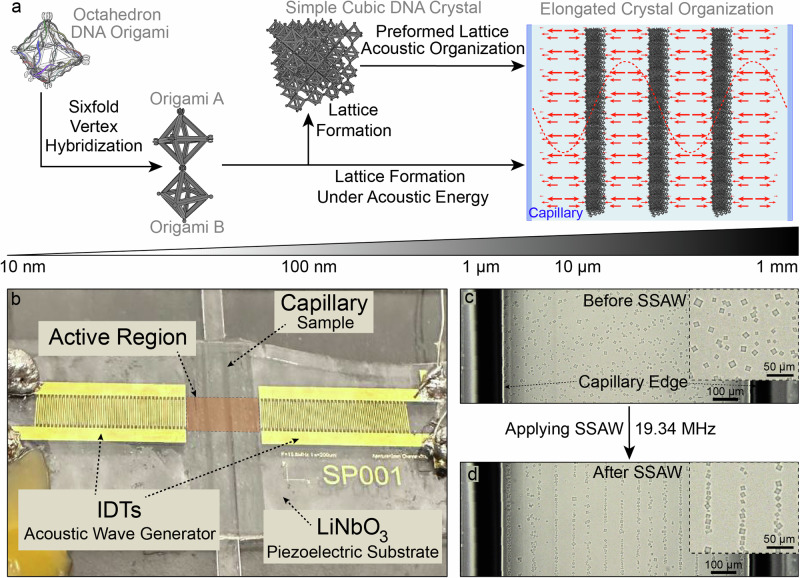
Fig. 1. Controlling DNA-assembled materials on a macroscale using acoustic waves.
a Schematic representation of the acoustic field use for directing the formation of DNA-based assemblies. DNA strands are designed to fold into octahedral frames, which assemble into simple cubic lattices. SSAW (red dashed line), either after, or during lattice formation, direct the assembly toward nodes where minimal acoustic pressure is applied within a capillary, where elongated organizations from DNA crystals are formed at the millimetric scale. Red arrows represent pressure produced by the SSAW. b Acoustic device. Two IDTs connected to a function generator produce a SSAW in the active region between the electrodes (highlighted in orange). A capillary of 1 mm inner width and 50 µm height with the sample sealed inside is placed in the active region. c Brightfield microscopy image of DNA crystals dispersed in a glass capillary. Capillary with DNA crystallites without field. Inset is a magnification of the same sample. d The same capillary shown in (c), after applying an acoustic field, crystals form chains in the nodes within the capillary. Inset is a magnification of the same sample.
The control of the macroscale nanomaterial morphology can be achieved using external factors affecting material formation, such as light, magnetic and electric fields17–21. However, these methods typically require specific material components and media characteristics, e.g., conductivity, transparency, or magnetic susceptibility, that respond to the applied field; this limits materials diversity and their applications. In this respect, the use of an acoustic field is attractive since it only requires a non-zero acoustic contrast factor (see Supplementary Information, Acoustic contrast factor) between the assembled components and medium22,23, which is determined by density or compressibility differences; this drastically reduces requirements on component material. This approach was used to manipulate micron-scale particles and their interactions with their environment24–26. More specifically, Standing Surface Acoustic Waves (SSAW)23,27,28 can be utilized to direct the assembly of nanoparticles into microstructures and manipulate chemical reactions29,30. In addition, using an acoustic field is attractive since its power density for manipulating particles can be lower than used in other methods, while the working area can be easily scaled up31,32.
In this work, we propose and investigate using an acoustic field to shape DNA-assembled material at the millimeter scale. We demonstrate (Fig. 1a) that specifically shaped acoustic pulses allow the formation of DNA-assembled materials according to the shape of the standing wave nodes (space with a minimum wave amplitude) into macroscale morphologies. Our studies reveal that specifically tailored pulses of SSAW can enhance the nucleation and crystallization of DNA lattices leading to the assembly of larger crystallites. The combination of DNA-based assembly and acoustic-directed assembly allows for bridge control over six orders of magnitude in scale (nm to mm). Due to the broad applicability of the acoustic method to diverse materials classes, our approach for sculpturing DNA-based assemblies can be extended to catalytic, biological, and inorganic materials5,33–36 metamaterials37, optical fibers and information storage devices38.
Results and discussion
Our experimental system includes two complementary DNA octahedral origami frames with about 30 nm edge length that can self-assemble into crystalline organization with a cubic lattice through vertex-driven sixfold hybridization8. The nucleation and growth of these lattices is closely dependent on the thermal annealing procedure, which starts at 50 °C to obtain a monomeric DNA-frame solution, followed by a slow ramp down of temperature to allow crystallization8,15. The acoustic setup consists of interdigital transducers (IDTs) and wave generator. IDT, fabricated by photolithography, convert electrical signals into acoustic waves through the piezoelectric effect.
Acoustic organization of pre-formed crystals
We first explored the use of acoustic waves for a macroscale organization using preassembled micron-sized DNA crystallites. A sealed glass capillary with the sample solution is placed in between the two IDTs, where the SSAW with a half wavelength of ~100 µm are generated (Fig. 1b, c). This results in the formation of a pattern of linear nodes with a width of several microns, separated by ~50 µm microns39, the length of nodes (~3 mm) is determined by the size of IDT (3.5 mm). While applying the SSAW, the acoustic forces push the DNA crystallites to the node positions within the capillary, creating a linear pattern (Fig. 1d). The entire setup is held in a temperature-controlled chamber which permits controlling DNA hybridization and pathway for crystal assembly and disassembly, while visualizing the process in real time through optical microscopy.
The standing acoustic waves are generated in pulses to stimulate crystallites in a specific spatiotemporal way. This leads to local heating at the active region between the IDTs, where the magnitude of the effect is determined by the temporal acoustic field profile (Supplementary Fig. 1). The profile consists of a wave pulse with a duration of 50 ms active SSAW and different time periods between the initiation of each pulse, ranging from 500 to 5000 ms in our experiments. This design allows us to independently control several key parameters of the system: thermal protocol (using the thermal chamber), pulse and period durations. Initially, the crystals are dispersed randomly within the capillary (Fig. 1c), yet when the acoustic field is applied, the crystals move to the nodes, forming a linear arrangement of individual crystals in the nodes (Fig. 1d and Supplementary Movies 1 and 2).
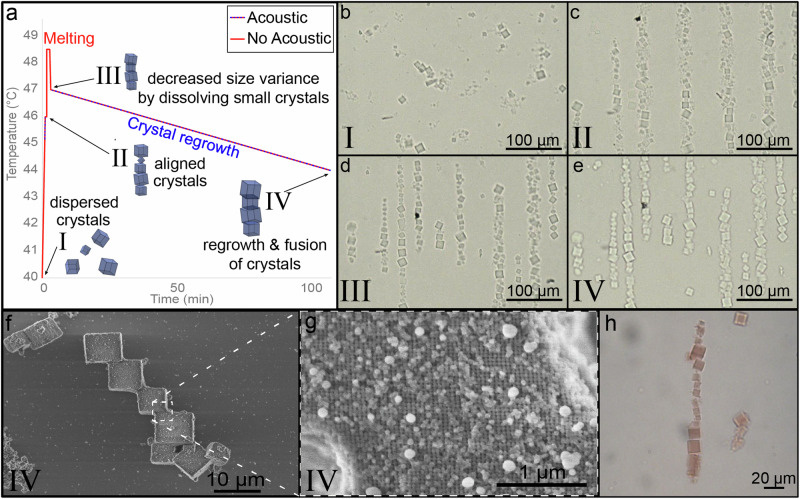
Following the successful organization of DNA crystals into linear patterns, we investigated merging the crystals into a single macroscale morphology through thermally controlled fusion. Our thermal protocol (Fig. 2a) includes the reannealing of crystals without their full melting into a single extended unit under the acoustic field. First, preassembled crystals (Fig. 2b) were aligned into the nodes (Fig. 2c). Next, the temperature was elevated to completely dissolve smaller crystals and nuclei, leaving only crystals that were initially larger, which narrowed the size distribution of crystallites (Fig. 2d). Then, the temperature was ramped down slowly to allow the remaining crystals regrowth while the acoustic field kept the growing crystals within the nodes (Fig. 2e). Due to the closely packed linear alignment and confinement to the nodes by the acoustic force, crystals have the propensity to grow one into the other. Scanning electron microscopy (SEM) images show that crystals are fused together during regrowth, creating a single continuous elongated entity formed from organized crystals. The fused crystals are coherently coordinated and form a solitary lattice that spans over several crystallites (Fig. 2f, g). We observed that the cubic crystallites within the nodes exhibit a certain degree of alignment with a preference for facets to be orientated parallel to the transducer surface (Supplementary Fig. 2), likely since such arrangement minimizes their cross-section to the SSAW. However, this orientation effect is suppressed by inter-crystallite attachments due to the hybridization between DNAs on the surfaces of different crystallites (Fig. 2b–h and Supplementary Fig. 3). Our approach for forming DNA-based morphologies by acoustic field is also suitable for lattices with nano-cargo. Such a cargo (gold nanoparticles (AuNPs), proteins, or quantum dots) allows adding a desired functionality to DNA lattices. As an example of this possibility, Fig. 2h shows the linear arrangement of lattices filled with 10 nm AuNP cargo formed under acoustic stimulation.
Fig. 2. Fusing crystallites into macroscale materials.
a Thermal profile for reannealing preformed crystals. Active acoustic waves pulsing is represented by a dashed blue line; inactive acoustic field (no pulsing waves) is represented by a solid red line. b–e Brightfield microscopy images of crystals subjected to the thermal reanneal protocol and acoustic waves at the different stages of the protocol, corresponding to the numerical notation in (a). f, g SEM images showing that the crystal reanneal protocol induces fusing of the crystals to form elongated macroscale morphology of crystals. h Crystals filled with AuNP aligned using acoustic waves. The red hue is the result of AuNP loaded into the lattice.
Crystallization under acoustic stimulation
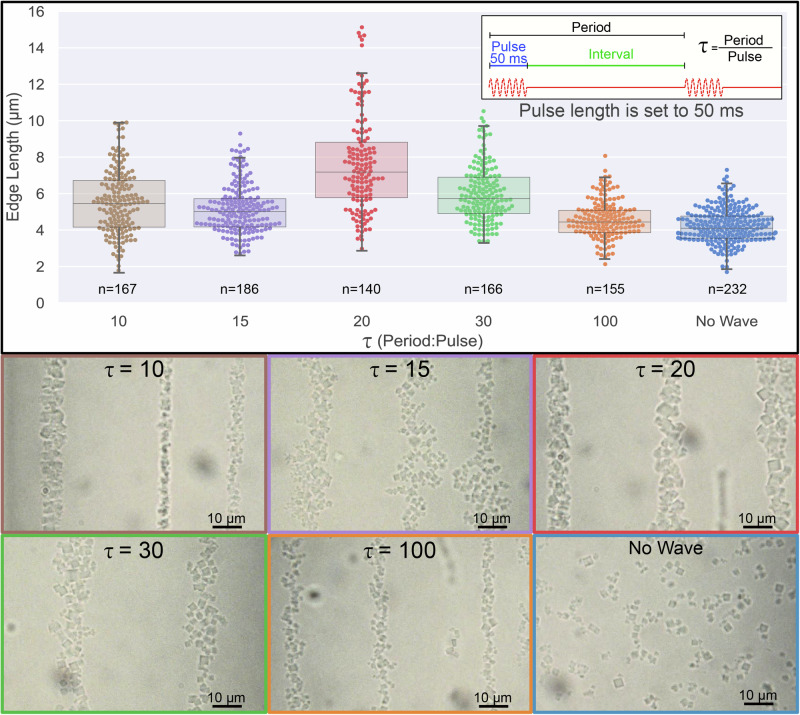
Considering the ability of SSAW to push preassembled lattices, we were interested in exploring whether lattice formation is affected by the acoustic field. We applied SSAW pulses during the initial crystal nucleation and growth and combined this with a thermal anneal process. While the wave pulse length is set to 50 ms (approximately 1 M wave cycles), the period, i.e., the time between the initiation of each pulse, is altered for different experiments; the period divided by the pulse length is denoted as τ (Fig. 3). Following this procedure, we observed a formation at the nodes of elongated morphologies of tightly packed crystals, whose appearance is similar to the morphologies formed by preformed crystals under SSAW-driven assembly. However, crystallites assembled under acoustic field pulses exhibited a noticeable increase in crystal size, particularly for certain pulse regimes, as we elaborate below (Figs. 3 and 4a).
Fig. 3. Crystal size formed with acoustic waves.
Crystal size formed with varied τ. The colors of the plotted sample correspond to the frame of the brightfield microscopy image frame. All samples (other than No Wave) were subjected to 50 ms wave pulses with varied periods (τ = period/pulse, as shown in the inset). Box-plot overlayed shows the median, Q1, Q3 and 1.5 Interquartile range whiskers. n is the number of crystals measured for each sample.
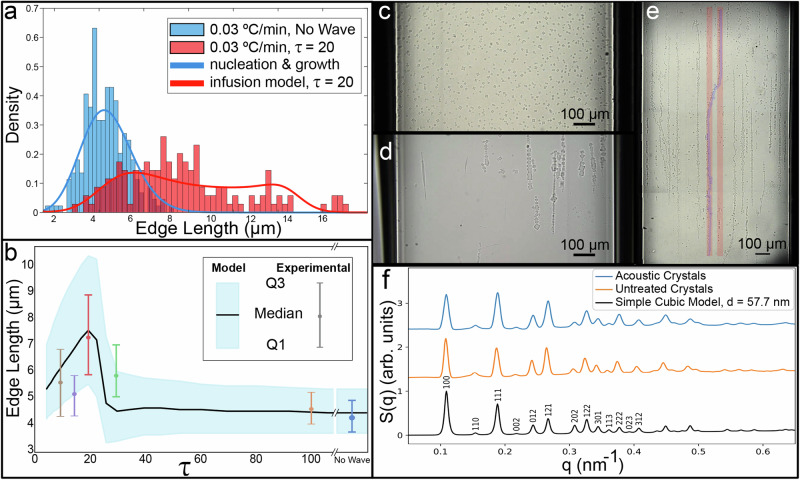
Fig. 4. Acoustic waves affect nucleation and growth.
a Histogram of crystal size distribution with no acoustic waves (blue) and with τ = 20 (red) for a fast temperature decrease rate of 0.03 °C/min (some of the data is shown in Fig. 3 with different representation). The nucleation and growth theory fit (blue line), and the infusion model (red line) account for the effect of the acoustic waves. b Crystal size distribution vs. τ, showing both discrete experimental data points (as shown in Fig. 3) and continuous model-calculated behavior. c, d Slow thermal anneal (temperature decrease rate of 0.01 °C/min) of crystals with no waves applied (c) and with τ = 20. e Fast thermal anneal (0.03 °C/min) with τ = 20, followed by a thermal reanneal to fuse crystals together, results in elongated macroscale structures at the millimetric scale. The width of the capillary (left wall to right wall) is 1 mm. f Measured SAXS structure factor (S(q)) of the crystals assembled under acoustic field and untreated crystals corresponds to modeled S(q) of simple cubic crystal structure.
To obtain a quantitative understanding of this phenomenon, we systematically studied the effect of the ratio between period and pulse, τ, on the size of formed crystals. Pulse in a narrow range of τ values yields an increase of the crystal size at a fast thermal anneal of 0.03 °C/min (Fig. 3 and Supplementary Fig. 6). Compared with thermal annealing without acoustic waves (τ → ∞), SSAW treatment also broadens crystal size distribution, which is more prominent in the narrow τ range (Figs. 3 and 4a). Specifically, at τ = 20 (1000:50 ms period to pulse ratio), crystal sizes are significantly larger (nearly doubling in edge length), even compared to the other samples formed under acoustic waves at different τ values (Fig. 3 and Supplementary Fig. 5). We also observed this effect for all explored thermal anneal rates and found that the largest crystals were formed at the same τ = 20 with a slow thermal anneal of 0.01 °C/min (Supplementary Figs. 7 and 8, for more microscopy images, see Supplementary Figs. 9–18).
We first analyzed the size distribution in the absence of acoustic waves to gain mechanistic insights into the observed phenomena of larger crystals formed under the acoustic field (Fig. 4a). This distribution arises from the interplay between nucleation dynamics and diffusion-limited crystal growth. We first ruled out the possibility that the acoustic field has an effect on the individual frames since our estimation indicates that the thermal energy exceeds an acoustic energy for ~30 nm objects (see Acoustic field considerations in Supplementary Information). Both nucleation and growth processes depend on the difference in chemical potential between the solution of free monomers frame, , and the crystal phase, denoted as:
| 1 |
Here, represents the cooling rate, and time is measured from , the point at which the crystal would be in coexistence with the solution at the original concentration . denotes the entropy associated with the unbinding of a single frame from the crystal, which corresponds to melting of 12 DNA duplexes. We introduce the quantity , that represents the thermodynamic driving force to crystallization, defined as:
| 2 |
Let be the lattice constant of the cubic crystal, with representing its surface energy, and denoting the diffusion coefficient of a free building block. The classical homogeneous nucleation rate for a cubic-shaped crystal can be expressed as follows40:
| 3 |
Once nucleated, each crystal grows at a diffusion-limited rate given by:
| 4 |
Here, represents the edge size of the cube, and is a numerical constant specific to the cubic geometry. As the number density of crystals and their average volume increase, the solution of the building blocks gets depleted:
| 5 |
By constructing the Fokker–Planck equation and solving it numerically, we obtained the final size distributions, which are in excellent agreement with the experiments conducted without acoustic waves for both fast and slow thermal anneal protocols (Fig. 4a, b). When fitting the data, we used as a single adjustable parameter (the same for all annealing rates), while the rest of the parameters have been determined based on the known geometry of the building blocks, their concentration, and the thermodynamics of DNA hybridization.
Once the system is subjected to acoustic stimulation, two new effects must be considered. Firstly, SSAW are expected to suppress the nucleation of crystallites due to local heating in the active region of the acoustic field due to their dissipation (Supplementary Fig. 1). Secondly, the forces associated with the acoustic field result in an influx of small clusters of frames into the active region from adjacent areas in the capillary (Fig. 1b), which act as nuclei for crystal growth. Thus, the number of growing crystals is dictated by both an interplay of acoustically suppressed homogeneous nucleation and the influx of nuclei from the adjacent region. As the time interval between pulses, i.e., τ decreases, both effects become more pronounced, explaining the observed non-monotonic behavior of the τ vs. crystal size relationship (Fig. 3). Namely, as τ decreases, the suppression of homogeneous nucleation is initially offset by a moderate influx of crystals from the surrounding regions. This leads to a reduction in the overall number of crystalline particles and an increase in their average size (since the total amount of material is determined by the initial concentration). Once the homogeneous nucleation is sufficiently suppressed, the further increase in influx reverses the effect, resulting in a greater number of crystalline particles and, consequently, smaller crystal sizes.
To describe this effect, we propose a simple model in which nucleation in the active region is suppressed by a factor of , while remaining unaffected in the adjacent region. The infusion process is represented by a single rate proportional to pulse frequency, . The size distribution in the region exposed to SSAW evolves according to the Fokker–Planck equation:
| 6 |
Here, the first two terms represent the nucleation and growth processes discussed above, and the last term is due to the infusion of material from the adjacent region unaffected by the acoustic waves. Thus, is the size distribution without the SSAW effect that obeys the same equation with parameters and set to 0.
Although oversimplified, this model provides a surprisingly good description of the observed phenomenon. Specifically, it captures the non-monotonic behavior and the observed broadening of the size distribution due to the acoustic field. Furthermore, this simple model gives an impressive quantitative agreement with the observed size distribution in the vicinity of the optimal conditions for larger crystal sizes (Fig. 4a, b and Supplementary Figs. 6–8). Note that the same values of adjustable parameters and , have been used for different cooling rates and inter-pulse periods. As the τ increases, homogeneous nucleation in the active region returns to its unsuppressed rate. Conversely, more frequent acoustic driving leads to an increased infusion rate κ. Both these effects cause the size distribution to revert to the no-wave case, which is described by the unmodified nucleation and growth theory.
In certain cases, the SSAW effect could produce exceedingly large crystallites compared to those assembled without a field, specifically, with a slow thermal anneal (0.01 °C/min) and an acoustic pulse of 50 ms and τ of 20 (Fig. 4c, d). Interestingly, in contrast to the formation of larger single crystals with a slow anneal, pulsing SSAW during a fast annealing (0.03 °C/min) results in assembly of long crystalline morphologies, reaching length of 2 mm (Fig. 4e). Combined with the reannealing and fusing process discussed above (Fig. 2), each of these morphologies is merged into a single unit, as indicated by the fact that such linear structures (highlighted in blue) can hop from one node to another (marked in red) without breaking. We further explored that nanoscale structure of the lattice morphologies assembled under both acoustic driving and annealing using small angle x-ray scattering (SAXS). SAXS measurements reveal that the formed crystals exhibit a simple cubic lattice, similar to crystals without acoustic stimulation (Fig. 4f). This implies that the acoustic-driven assembly does not alter the nano-scale arrangement of the DNA frames in a lattice but only affects the lattice growth at the level of crystallites and a final morphology at the scale of a hundred micrometers up to millimeters.
In conclusion, we have demonstrated that acoustic fields with specific spatiotemporal characteristics provide an effective means to drive the formation of DNA-assembled materials at the macroscale. While DNA nanotechnology enables precise spatial control at the nanoscale, achieving controlled organization at larger scales has remained challenging. By combining DNA-guided assembly with acoustically driven processes, we successfully directed the assembly and dictated the morphology of DNA-origami-based crystal lattices at scales ranging from tens of microns to millimeters, while maintaining DNA-defined nanoscale organization. This study shows that the acoustic field can drive the assembly of macroscale morphologies from preformed crystals and monomers under annealing conditions. The macroscale morphology of crystals can be potentially expanded to other geometries, beyond linear structures shown in our work. This can be achieved by changing the boundary conditions of the sample, or by changing the arrangement and geometry of the transducers. For example, complex two-dimensional patterns can be produced, much like Chladni plates41. More broadly, acoustic holography can be potentially employed to form complexly designed patterns of self-assembled materials42,43. Moreover, the acoustic field might enhance the formation of crystals within a certain pulse regime due to the combination of the two new effects caused by the acoustic waves (local heating and influx of nuclei). Our experimental observations are supported by a computational model that incorporated nucleation dynamics, diffusion-limited growth, and the effects of acoustic driving. Given the flexibility in engineering acoustic fields and a broad range of functions of DNA-based materials, our combined DNA and acoustically driven assembly approach potentially allows for the controlling structure formation over 6 orders of magnitude in scale from sub-nm to mm. Combined with an inorganic templating strategy6,15, this approach provides device-scale nanomaterials fabrication for potential applications in photonics, mechanics, electronic devices, and biomaterials.
Methods
Design, synthesis, and purification of DNA origami
The octahedron DNA origami frame was designed using the DNA origami design software caDNAano44 (http://cadnano.org/). The edges of the octahedron frame were designed to be six-helix bundles (6HB) with edge lengths of 84 base pairs. Each of the 6HBs have one sequence extending from each of the ends. This results in four ssDNA sequences extending away from each of the six vertices of the octahedron.
DNA origami frames are folded by mixing 40 nM M13mp18 scaffold purchased from Bayou Biolabs, LLC with a 5:1 staple to scaffold ratio. The staple sequences were purchased from Integrated DNA Technologies (IDT) and are presented in Supplementary Tables 4 and 5. The buffer of this solution is mixed to contain 1 mM EDTA, 40 mM Tris at a pH of approximately 8.0, and a concentration of 12.5 mM magnesium chloride. This mixture is heated to 90 °C and slowly cooled to 20 °C over a period of 20 h to allow for the correct folding of the scaffold sequence into the target 3D DNA origami frame. This protocol is adapted from previous literature8,12,45.
The folded DNA origami is washed in order to remove the excess staples used during the folding process. The origami samples are washed five times through a 100 KDa Millipore Sigma Amicon Ultra-0.5 mL centrifugal filters in an Eppendorf 5424R at a speed of 2.2k rpm for 25 min using a buffer consisting of (1X) TAE and 12.5 mM Mg2+.
To preform lattices, 2 types of origami frames with complementary sticky ends on their vertices are incubated together in a PCR tube at a concentration of 20 nM each. The mixture is heated to 50 °C and slowly cooled down to 25 °C over a period of ~72 h to allow the formation of crystalline structures.
Acoustic experiment design and setup
One end of a borosilicate hollow capillary, with dimensions 50 × 1 × 0.05 mm (VitroTubesTM, VitroCom), was immersed in the sample solution to allow capillary effects to draw the solution into the capillary. The capillary was then sealed using wax and positioned onto the acoustic device between the two IDTs. In order to ensure the proper coupling of the capillary to the piezoelectric substrate, a small amount (~1 µl) of immersion oil (Olympus) was applied between the substrate and the capillary. Subsequently, the setup was connected to a function generator (SDG 1032X, Siglent Technologies). A sine wave with a frequency of 19.34 MHz was generated at an amplitude of 20 Volts peak-to-peak. This wave was applied to both IDTs located on the sides of the sample to generate a standing wave. For the wave pulsing, a 50 ms pulse (966,980 wave cycles/19.34 MHz) was generated, followed by a variable relaxation time in the different experiments (and by that changing the τ variable). The end product can be extracted from the capillary by breaking the wax seal and pushing the liquid out using a pipette with a gel loading tip.
Crystal size measurement post acoustic-stimulated crystallization
Crystals were formed under a standing waves acoustic field with varying τ values. DNA frames were incubated inside the capillary with acoustic pulsing in effect and heated to 50 °C to dissolve any aggregates. The temperature was adjusted and regulated using a temperature-controlled Linkam LTS420 thermal stage. The temperature was ramped down at a rate of 0.03 (fast) or 0.01 (slow) °C/min. For the specific DNA frame used, crystal growth occurs at the range of approximately 47.5–45 °C. When the temperature reached 40 °C and the growth had finished, 5–10 optical microscopy images were collected with a ×50 magnification. This process was done for each τ value at least 3 times. Images were taken to cover all locations inside the active region of the acoustic device (Fig. 1b). For every set of τ values, samples were taken from the same stock of building blocks. Only crystals with clear edges were selected to measure the size of the crystals. This approach causes a bias toward larger crystals with clearer edges; however, the same bias applies to all τ values. The area of each distinct crystal was measured using Fiji46 (ImageJ; version 1.54f), and the value of crystal area was square rooted to get the edge length, assuming crystals are cubic.
Statistics and reproducibility
Each experiment, with each of the various conditions including applying acoustic waves on pre-formed crystals (Fig. 1), fusing crystals using thermal reannealing (Fig. 2), varying the value of τ (Figs. 3 and 4), varying the thermal anneal ramp (Fig. 4 and Supplementary Figs.), and measuring crystal sizes (Figs. 3 and 4) was independently repeated at least three times.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This research used resources of the Center for Functional Nanomaterials, and small angle scattering was collected at the Complex Matter Scattering (CMS) at 11-BM of the National Synchrotron Light Source II at Brookhaven National Laboratory, which are of the US DOE Office of Science Facilities, under contract no. DE-SC0012704. The DNA assembly work was supported by the US Department of Energy, Office of Basic Energy Sciences, grant DE-SC0008772. The acoustic assembly work was supported by the US Department of Defense, Army Research Office, W911NF-22-2-0111. The work of Z.A.A. was in part supported by the Human Frontier Science Program (HFSP) and the Zuckerman Israeli Postdoctoral Scholars Program (Z.A.A.). H.S. and S.P. were partially supported by the Israeli Science Foundation (grant No. 952/19).
Author contributions
Z.A.A., H.S. and O.G. conceptualized the study and designed the experiments. Z.A.A., D.C.R., and E.R. performed the experiments. Z.A.A., D.C.R., E.R. and A.V.T. analyzed the data. S.P. and H.S. designed and fabricated the acoustic devices. A.V.T. contributed to the theoretical analysis and modeling. Z.A.A., A.V.T., H.S. and O.G. wrote the manuscript with input from all other authors. O.G. supervised the project.
Peer review
Peer review information
Nature Communications thanks Jan Lipfert and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The authors declare that the data supporting the findings of this study are available from authors on request and within the paper, its Supplementary Information files.
Code availability
The code used to perform modeling for the simple cubic crystal is publicly available to download at https://github.com/CFN-softbio/scattersim.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51049-7.
References
- 1.Kadic, M., Milton, G. W., Van Hecke, M. & Wegener, M. 3D metamaterials. Nat. Rev. Phys.1, 198–210 (2019). 10.1038/s42254-018-0018-y [DOI] [Google Scholar]
- 2.Roy, K., Jaiswal, A. & Panda, P. Towards spike-based machine intelligence with neuromorphic computing. Nature575, 607–617 (2019). 10.1038/s41586-019-1677-2 [DOI] [PubMed] [Google Scholar]
- 3.Liddle, J. A. & Gallatin, G. M. Nanomanufacturing: a perspective. ACS Nano10, 2995–3014 (2016). 10.1021/acsnano.5b03299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang, D., Zhou, C., Gao, F., Wang, P. & Ke, Y. DNA‐guided assembly of molecules, materials, and cells. Adv. Intell. Syst.2, 1900101 (2020). 10.1002/aisy.201900101 [DOI] [Google Scholar]
- 5.Kahn, J. S. & Gang, O. Designer nanomaterials through programmable assembly. Angew. Chem.134, e202105678 (2022). 10.1002/ange.202105678 [DOI] [PubMed] [Google Scholar]
- 6.Michelson, A. et al. Three-dimensional nanoscale metal, metal oxide, and semiconductor frameworks through DNA-programmable assembly and templating. Sci. Adv.10, eadl0604 (2024). 10.1126/sciadv.adl0604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li, Y. et al. Open-channel metal particle superlattices. Nature611, 695–701 (2022). 10.1038/s41586-022-05291-y [DOI] [PubMed] [Google Scholar]
- 8.Tian, Y. et al. Ordered three-dimensional nanomaterials using DNA-prescribed and valence-controlled material voxels. Nat. Mater.19, 789–796 (2020). 10.1038/s41563-019-0550-x [DOI] [PubMed] [Google Scholar]
- 9.Zheng, J. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature461, 74–77 (2009). 10.1038/nature08274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stulz, E. DNA architectonics: towards the next generation of bio-inspired materials. Chem Eur. J.18, 4456–4469 (2012). 10.1002/chem.201102908 [DOI] [PubMed] [Google Scholar]
- 11.Wang, X. et al. An organic semiconductor organized into 3D DNA arrays by “Bottom‐up” rational design. Angew. Chem. Int. Ed.56, 6445–6448 (2017). 10.1002/anie.201700462 [DOI] [PubMed] [Google Scholar]
- 12.Liu, W. et al. Materials Science Diamond family of nanoparticle superlattices. Science351, 582–586 (2016). 10.1126/science.aad2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian, Y. et al. Lattice engineering through nanoparticle–DNA frameworks. Nat. Mater.15, 654–661 (2016). 10.1038/nmat4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, Z. et al. Engineering organization of DNA nano-chambers through dimensionally controlled and multi-sequence encoded differentiated bonds. J. Am. Chem. Soc.142, 17531–17542 (2020). 10.1021/jacs.0c07263 [DOI] [PubMed] [Google Scholar]
- 15.Michelson, A. et al. Three-dimensional visualization of nanoparticle lattices and multimaterial frameworks. Science376, 203–207 (2022). 10.1126/science.abk0463 [DOI] [PubMed] [Google Scholar]
- 16.Adhikari, S. et al. Controlling the self-assembly of DNA origami octahedra via manipulation of inter-vertex interactions. J. Am. Chem. Soc.145, 19578–19587 (2023). 10.1021/jacs.3c03181 [DOI] [PubMed] [Google Scholar]
- 17.Montelongo, Y. et al. Electrotunable nanoplasmonic liquid mirror. Nat. Mater.16, 1127–1135 (2017). 10.1038/nmat4969 [DOI] [PubMed] [Google Scholar]
- 18.Sim, S., Miyajima, D., Niwa, T., Taguchi, H. & Aida, T. Tailoring micrometer-long high-integrity 1D array of superparamagnetic nanoparticles in a nanotubular protein jacket and its lateral magnetic assembling behavior. J. Am. Chem. Soc.137, 4658–4661 (2015). 10.1021/jacs.5b02144 [DOI] [PubMed] [Google Scholar]
- 19.Fava, D., Winnik, M. A. & Kumacheva, E. Photothermally-triggered self-assembly of gold nanorods. Chem. Commun. 2571 10.1039/b901412h (2009). [DOI] [PubMed]
- 20.Grzelczak, M., Liz-Marzán, L. M. & Klajn, R. Stimuli-responsive self-assembly of nanoparticles. Chem. Soc. Rev.48, 1342–1361 (2019). 10.1039/C8CS00787J [DOI] [PubMed] [Google Scholar]
- 21.Harraq, A. A., Choudhury, B. D. & Bharti, B. Field-induced assembly and propulsion of colloids. Langmuir38, 3001–3016 (2022). 10.1021/acs.langmuir.1c02581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, X., Lim, K. M. & Zhai, W. A novel class of bioinspired composite via ultrasound-assisted directed self-assembly digital light 3D printing. Appl. Mater. Today26, 101388 (2022). 10.1016/j.apmt.2022.101388 [DOI] [Google Scholar]
- 23.Wood, C. D. et al. Alignment of particles in microfluidic systems using standing surface acoustic waves. Appl. Phys. Lett.92, 044104 (2008). 10.1063/1.2838748 [DOI] [Google Scholar]
- 24.Abdelaziz, M. A. et al. Ultrasonic chaining of emulsion droplets. Phys. Rev. Res.3, 043157 (2021). 10.1103/PhysRevResearch.3.043157 [DOI] [Google Scholar]
- 25.Moerman, P. G. et al. Solute-mediated interactions between active droplets. Phys. Rev. E96, 032607 (2017). 10.1103/PhysRevE.96.032607 [DOI] [PubMed] [Google Scholar]
- 26.Peng, C., Chen, M., Spicer, J. B. & Jiang, X. Acoustics at the nanoscale (nanoacoustics): a comprehensive literature review. Part I: materials, devices and selected applications. Sens. Actuators Phys.332, 112719 (2021). 10.1016/j.sna.2021.112719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, J., Huang, H., Stratton, Z., Huang, Y. & Huang, T. J. Continuous particle separation in a microfluidic channel via standing surface acoustic waves (SSAW). Lab. Chip9, 3354 (2009). 10.1039/b915113c [DOI] [PubMed] [Google Scholar]
- 28.Kim, H., Kim, T., Kim, D. & Shim, W. Wave-tunable lattice equivalents toward micro- and nanomanipulation. Nano Lett.16, 6472–6479 (2016). 10.1021/acs.nanolett.6b02973 [DOI] [PubMed] [Google Scholar]
- 29.Piperno, S., Sazan, H. & Shpaisman, H. Simultaneous polymerization and patterning: a one step acoustic directed assembly method. Polymer173, 20–26 (2019). 10.1016/j.polymer.2019.04.016 [DOI] [Google Scholar]
- 30.Sazan, H., Piperno, S., Layani, M., Magdassi, S. & Shpaisman, H. Directed assembly of nanoparticles into continuous microstructures by standing surface acoustic waves. J. Colloid Interface Sci.536, 701–709 (2019). 10.1016/j.jcis.2018.10.100 [DOI] [PubMed] [Google Scholar]
- 31.Ding, X. et al. On-chip manipulation of single microparticles, cells, and organisms using surface acoustic waves. Proc. Natl Acad. Sci.109, 11105–11109 (2012). 10.1073/pnas.1209288109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dholakia, K., Drinkwater, B. W. & Ritsch-Marte, M. Comparing acoustic and optical forces for biomedical research. Nat. Rev. Phys.2, 480–491 (2020). 10.1038/s42254-020-0215-3 [DOI] [Google Scholar]
- 33.Niemeyer, C. M., Koehler, J. & Wuerdemann, C. DNA-directed assembly of bienzymic complexes from in vivo biotinylated NAD(P)H:FMN oxidoreductase and luciferase. ChemBioChem3, 242–245 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Ellis, G. A. et al. Artificial multienzyme scaffolds: pursuing in vitro substrate channeling with an overview of current progress. ACS Catal.9, 10812–10869 (2019). 10.1021/acscatal.9b02413 [DOI] [Google Scholar]
- 35.Laramy, C. R., O’Brien, M. N. & Mirkin, C. A. Crystal engineering with DNA. Nat. Rev. Mater.4, 201–224 (2019). 10.1038/s41578-019-0087-2 [DOI] [Google Scholar]
- 36.Wang, S.-T. et al. Designed and biologically active protein lattices. Nat. Commun.12, 3702 (2021). 10.1038/s41467-021-23966-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, J. B. et al. A mechanical metamaterial made from a DNA hydrogel. Nat. Nanotechnol.7, 816–820 (2012). 10.1038/nnano.2012.211 [DOI] [PubMed] [Google Scholar]
- 38.Church, G. M., Gao, Y. & Kosuri, S. Next-generation digital information storage in DNA. Science337, 1628–1628 (2012). 10.1126/science.1226355 [DOI] [PubMed] [Google Scholar]
- 39.Maramizonouz, S. et al. Acoustofluidic patterning inside capillary tubes using standing surface acoustic waves. Int. J. Mech. Sci.214, 106893 (2022). 10.1016/j.ijmecsci.2021.106893 [DOI] [Google Scholar]
- 40.Kashchiev, D. & Van Rosmalen, G. M. Review: Nucleation in solutions revisited. Cryst. Res. Technol.38, 555–574 (2003). 10.1002/crat.200310070 [DOI] [Google Scholar]
- 41.Kopitca, A., Latifi, K. & Zhou, Q. Programmable assembly of particles on a Chladni plate. Sci. Adv.7, 7716–7738 (2021). 10.1126/sciadv.abi7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melde, K., Mark, A. G., Qiu, T. & Fischer, P. Holograms for acoustics. Nature537, 518–522 (2016). 10.1038/nature19755 [DOI] [PubMed] [Google Scholar]
- 43.Raymond, S. J. et al. A deep learning approach for designed diffraction-based acoustic patterning in microchannels. Sci. Rep.10, 8745 (2020). 10.1038/s41598-020-65453-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res.37, 5001–5006 (2009). 10.1093/nar/gkp436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian, Y. et al. Prescribed nanoparticle cluster architectures and low-dimensional arrays built using octahedral DNA origami frames. Nat. Nanotechnol.10, 637–644 (2015). 10.1038/nnano.2015.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The authors declare that the data supporting the findings of this study are available from authors on request and within the paper, its Supplementary Information files.
The code used to perform modeling for the simple cubic crystal is publicly available to download at https://github.com/CFN-softbio/scattersim.