Significance
In species with differentiated sex chromosomes (XY or ZW), X or Z-borne genes are present in different dosages in male and females. Gene dosage is equalized between sexes by transcriptional silencing of genes on one X in female mammals, so dosage compensation has been thought to be essential. However, imbalanced messenger RNA levels observed between sexes in several species cast doubt on the importance of dosage compensation. Here, we show that although mRNA levels between the sexes in chicken and platypus sex chromosomes were unequal, balance was restored at the protein level. We conclude that dosage compensation is essential in species with differentiated sex chromosomes but that it may be achieved by a combination of transcriptional and post-transcriptional control.
Keywords: sex chromosome, dosage compensation, post-transcriptional control, epigenetic
Abstract
Heteromorphic sex chromosomes (XY or ZW) present problems of gene dosage imbalance between sexes and with autosomes. A need for dosage compensation has long been thought to be critical in vertebrates. However, this was questioned by findings of unequal mRNA abundance measurements in monotreme mammals and birds. Here, we demonstrate unbalanced mRNA levels of X genes in platypus males and females and a correlation with differential loading of histone modifications. We also observed unbalanced transcripts of Z genes in chicken. Surprisingly, however, we found that protein abundance ratios were 1:1 between the sexes in both species, indicating a post-transcriptional layer of dosage compensation. We conclude that sex chromosome output is maintained in chicken and platypus (and perhaps many other non therian vertebrates) via a combination of transcriptional and post-transcriptional control, consistent with a critical importance of sex chromosome dosage compensation.
Despite their different sex chromosome systems, mammals and birds (and many other vertebrates and invertebrates with differentiated sex chromosomes) share the challenge that dosage of sex-linked genes is unequal between males and females, and between sex chromosomes and autosomes.
Therian (eutherian and marsupial) mammals have XY males and XX females in which approximately 859 X-borne coding genes in eutherians (see human genome assembly Ensembl GRCh38.p14) and approximately 497 X-borne coding genes in marsupials (see opossum genome assembly Ensembl A SM229v1) are unequally represented between the sexes. Birds have ZZ males and ZW females in which approximately 865 Z-borne coding genes are unequally represented between the sexes (see chicken genome assembly Ensembl bGalGal1.mat.broiler.GRCg7b). Platypus (an egg-laying monotreme mammal) has an even more acute dosage problem arising from a complex sex chromosome system, in which females have five pairs of X chromosomes and males have five X chromosomes and five degraded Y chromosomes (1). In male meiosis, these ten sex chromosomes form a chain held together by recombination within nine pseudoautosomal regions (PARs) shared between successive X and Y chromosomes. The platypus sex chromosomes have no homology with the sex chromosomes of therian mammals but share considerable homology with the chicken Z chromosome (2).
In therian mammals, parity of expression of X-borne genes between the sexes is achieved by the epigenetic silencing of one X in the somatic cells of females. X chromosome inactivation (XCI) was long assumed to be at the transcriptional level, and this was experimentally demonstrated (3) and repeatedly confirmed (4, 5). XCI is considered to be a global silencing mechanism, although many genes escape inactivation on the human X. Many of these escapers have active, though underexpressed copies on the Y chromosomes; RNA-seq data in placental mammals showed that Y gametologs have 1/3 the expression compared to ancestral levels, although combined XY gametolog expression in males surpasses the expression from both X homologs (the active X, and XCI escapers) in females (6).
However, dosage compensation between male and female was not observed in studies of platypus and birds. In platypus, male:female (M:F) transcript ratios of X-borne genes fell between complete compensation (1.0) and complete absence of dosage compensation (0.5), with a median of 0.6 (4). Similarly in birds, transcription from Z-borne genes showed M:F ratios between full compensation (1.0) and absence of compensation (2.0), with a median of 1.4 (4, 7). The Z and W chromosomes of paleognathous birds, which have large non-degenerated PARs, showed incomplete dosage compensation of transcripts from genes on the differentiated region of the Z chromosome (8). Z chromosome-wide dosage compensation was therefore considered not to occur in birds or to be restricted to dosage-sensitive genes (4, 9).
As well as dosage differences of X or Z-borne genes between the sexes, differentiated sex chromosomes must contend with differences in the expression ratio of X or Z-borne genes and the autosomal genes with which they are likely to interact. Unequal expression of X-borne and autosomal genes in XY males has been suggested to be compensated by upregulation of X-borne genes, a step considered by Ohno (10) to have driven the evolution of XCI. However, twofold transcriptional upregulation of the X chromosome in human and mouse is more complex than originally thought (4, 11–13), as upregulation appears to be a malleable process depending on location on the X and when it occurs during development (14). In eutherian mammals, X to autosome transcriptional output is less than 1 in both sexes (4), but this reduced X expression relative to the autosomes is compensated in the proteome, at least in males (15). It may be that only a subset of dosage sensitive X-borne genes have a large effect on X chromosome upregulation (11), though the process is neither global nor restricted to just these genes (16, 17). However, global upregulation of the active mammalian X to match ancestral autosome levels occurs in marsupials (4, 11).
Outside of mammals, balance between male and female expression, as well as X:autosome equivalence, is achieved by twofold upregulation of the X in XY males of the lizards Anolis carolinensis (18) and Urosaurus nigricaudus (19), as well as in the fish Poecilia parae (20). A compensatory relationship is well documented for Drosophila (21), and the small brown planthopper Laodelphax striatellus shows complete dosage compensation of the X between males and females in somatic tissues (22). Thus, dosage compensation mechanisms are widely distributed in animals with differential sex chromosomes.
However, the absence of chromosome-wide compensation at the transcriptional level in monotremes and birds provoked doubt about the universal importance of dosage compensation (11, 23, 24). The alternative possibility that dosage compensation was enacted also at the posttranscriptional level was canvassed, but there are few data on M:F dosage compensation in the proteome. Uebbing et al. (25) found that M:F expression ratios in the chicken proteome were not balanced, although 30% of Z-linked genes had a significant change in M:F ratios between the transcriptome and the proteome.
There is conflicting evidence of sex chromosome to autosome dosage compensation in the proteome. Deng et al. (21) found an X to autosome expression ratio of 1.07 in a limited mouse proteomics dataset, whereas Lin et al. (11) reported an X to autosome protein expression ratio of 0.5 in human (11). Uebbing et al. (25) found the mean Z to autosome expression ratio was close to 1 in males, but below 1 in females in the chicken proteome. Wang et al. (15) measured the translatome to circumvent the lower resolution of mass spectrometry over nucleic acid sequencing, demonstrating that ancestral (proto-sex chromosomal) expression levels are more similar to current day X expression levels in the translatome than the transcriptome in eutherians, with the most noticeable upregulation occurring in testes. However, this does not take into account any post-translational processes that could influence sex chromosome dosage compensation in the proteome.
Here, we investigate dosage compensation in both the transcriptome and proteome of representative therian mammals (mouse and opossum), a monotreme (platypus), and a bird (chicken). We demonstrate that incomplete transcriptional dosage compensation of sex chromosomes is balanced between sexes by post-transcriptional compensation in platypus and chicken.
Compensation by Transcriptional and Post-transcriptional Control
We first measured global mRNA and protein output from X-borne genes in mouse and opossum and calculated the M:F ratios of RNA and protein abundance. We found that transcriptional output was equivalent (M:F ~ 1) between the sexes in mouse and opossum (SI Appendix, Fig. S1), as previously described (4), and a 1:1 ratio was also observed in their respective proteomes. Thus, male-to-female X chromosome gene dosage equivalence is established in the transcriptome, and parity is maintained in the proteome in therian mammals.
However, this was not the case for platypus sex chromosomes. The need for dosage compensation in the platypus sex chromosomes would be expected to be acute, given that 9.23% of the genome (the non-PAR regions of the five X chromosomes) is present in a single dose in males (5). Here, we confirmed the boundaries of seven of the nine PARs demarcated by Zhou et al. (5) to 100 kbp resolution using male fibroblast HiC interaction patterns (SI Appendix, Fig. S2 and Table S1) and also defined the X3Y2 PAR to a 500 kbp resolution (SI Appendix, Table S2) but could not resolve the tiny X4Y4 PAR. Y-specific sequences were adjacent to their associated PARs and as such were clearly demarcated by genomic interactions with the Xs. The PARs were all assembled on X chromosome contigs.
We measured RNA and protein output in males and females from genes on autosomes and X chromosomes in platypus fibroblasts, heart, and liver. For RNA, M:F expression ratios were calculated. For protein abundance, MS/MS data were processed using ProteinPilot (SI Appendix), which outputs protein ratios between samples. Genes were assigned according to their location on an autosome or a sex chromosome for each species. For platypus, genes were also assigned to either a PAR or an X-specific region. Since there are protein data for only a subset of genes, RNA expression was also compared for this subset to check that they were representative of total RNA.
For all autosome genes in each tissue, 1:1 M:F ratios were observed (shown as 0 on log2 scale) for total RNA (Fig. 1A). For all PAR genes in each tissue (Fig. 1 A, Left), M:F total RNA expression ratios were also close to 1:1. The M:F ratio of protein produced by a subset of these total PAR genes was also 1:1, as was the transcription ratio of this same subset. Individual platypus PARs showed minor variation (Fig. 1 A, Right). Most had M:F expression ratios that were close to 1:1 in total RNA, subset RNA, and the proteome; however, X5-PAR1 in liver and X1-PAR1 in heart showed a male bias for the total RNA and subset RNA but balanced ratios in the proteome (Fig. 1A).
Fig. 1.
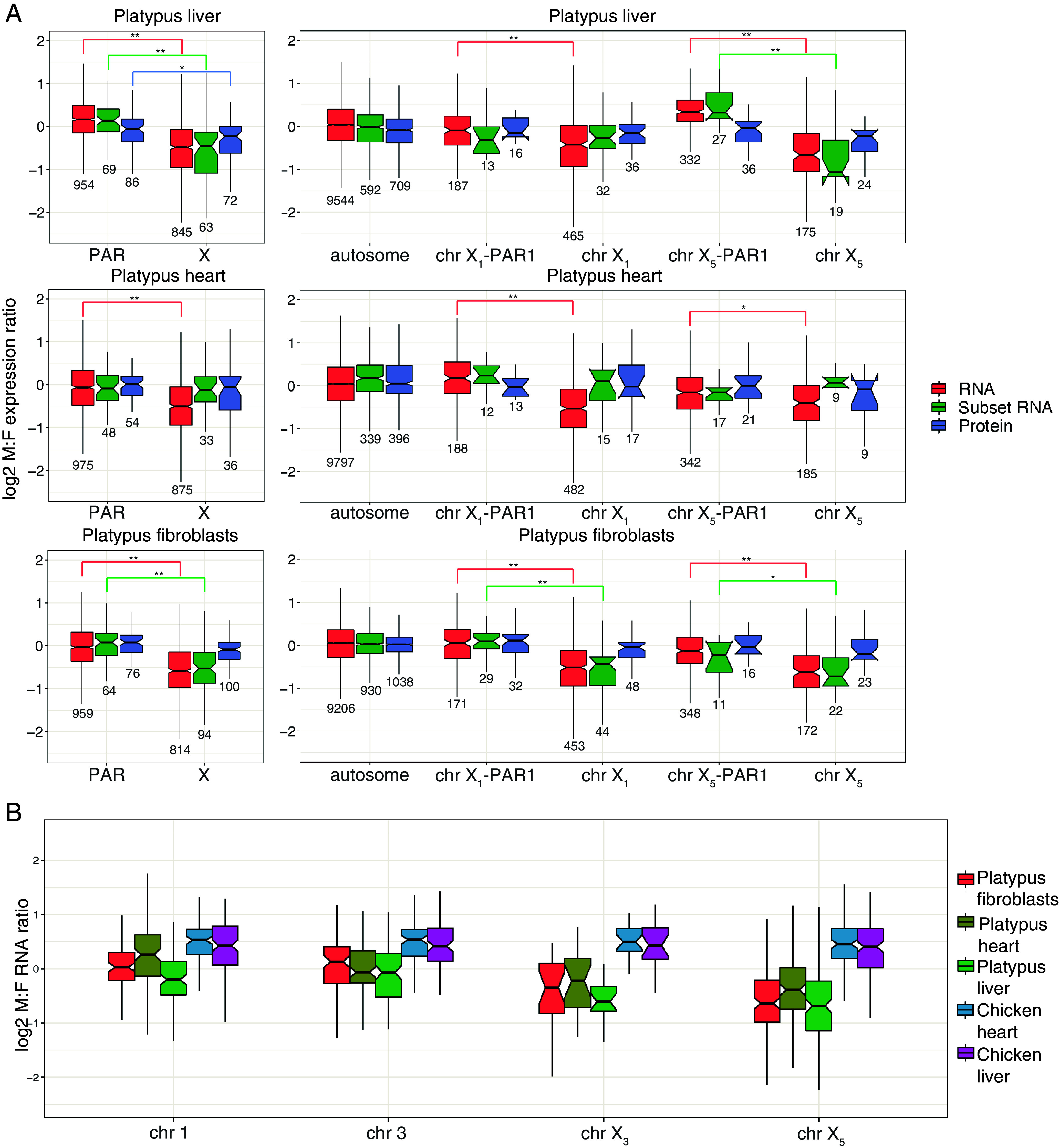
Male to female expression ratios of X-borne and autosomal genes in platypus. Median M:F transcript abundance ratios on a log2 scale of (A) autosomal and X-borne genes in platypus fibroblasts (n = 1), heart (n = 2), and liver (n = 2). On the Left, M:F ratios for total PAR genes and X-specific genes across all platypus Xs are shown. On the Right, autosomes are shown alongside chromosomes X1 and X5 PARs and X-specific regions. Ratios were calculated for all expressed mRNA (red), the subset of mRNAs sampled in the proteome (green), and proteins (blue). A ratio above zero is higher expression in males, whereas below zero is lower expression. Boxes represent the middle 50% of the data, and whiskers represent 1.5 times the interquartile range. Outliers are not plotted. Median is plotted inside the box, with the number of genes sampled below each boxplot. A Mood’s median test was used to calculate statistical difference (**P < 0.001, *P < 0.01). (B) Male-to-female ratios in the transcriptome of orthologs sampled on the chicken Z compared with two platypus autosomes (chromosomes 1 and 3) and two platypus sex chromosomes (X3 and X5). In chicken, the Z genes have higher transcriptional output in males. Genes that are autosomal in platypus have equal expression between the sexes. Genes that are on a platypus X have higher median expression in females.
Very different results were obtained for platypus X-specific genes. The total RNA of platypus X-specific genes had significantly lower transcriptional output of genes in males compared to females in all tissues (Fig. 1 A, Left). The median M:F ratio of 0.67 across all tissues implied a level of transcriptional dosage compensation between no dosage compensation (M:F = 0.5) and full dosage compensation (M:F = 1.0) for X-specific genes.
In contrast, the median protein M:F ratios were 1:1 in heart and fibroblasts, and was closer to a 1:1 ratio in liver than the total RNA (Fig. 1 A, Left). This implied that the lack of dosage compensation in the transcriptome is further compensated at the post-transcriptional level. To check that this discrepancy was not due to a sampling of genes analyzed for protein expression, ratios of mRNA expression were calculated for this subset of genes. For liver and fibroblasts, the M:F ratio reflected whole RNA, although for heart the M:F ratio was closer to 1:1.
The same pattern was observed when the X-differentiated regions were considered separately (Fig. 1A). The X-specific genes on chromosome X1 had a female bias for the total RNA for all tissues. This bias was reflected by the subset RNAs in liver and fibroblasts but not heart. For X-specific genes on X5, both fibroblasts and liver had a female bias for total RNA and a larger female bias for subset RNA. However, in heart, there was a female bias in total RNA but not subset RNA. In all three tissues, X-specific genes on X1 and X5 had near 1:1 M:F ratios in the proteome, with no significant differences between the PAR and X-specific genes (Fig. 1 A, Right).
These observations suggest that in platypus, M:F dosage compensation of X-specific genes at the protein level is accomplished by partial transcriptional regulation that is complemented by post-transcriptional regulation. The balance between transcriptional and post-transcriptional control may vary between different tissues and between different X chromosomes.
To investigate whether a similar mixture of transcriptional and post-transcriptional control occurs to compensate for Z dosage differences between ZZ male and ZW female birds, we profiled the transcriptome and proteome of chicken heart and liver. This showed that autosomal genes had M:F transcriptome ratios near 1:1 (SI Appendix, Fig. S3), whereas Z-borne genes had a strong male bias (Fig. 1B and SI Appendix, Fig. S3) as previously reported (26, 27). However, median M:F ratios of the proteins encoded by Z-borne genes were near 1:1 in both chicken tissues (SI Appendix, Fig. S3).
This trend for M:F ratios toward the homogametic sex in the platypus (females) and chicken (males) transcriptomes is demonstrated in Fig. 1B, which shows orthologous genes sampled on the chicken Z compared with two platypus autosomes (chromosomes 1 and 3) and with two platypus sex chromosomes (X3 and X5). Genes on the platypus autosomes had 1:1 M:F ratios. Genes on the platypus sex chromosomes had female-biased M:F ratios, whereas genes on the chicken Z had a male bias (Fig. 1B).
Thus, for both platypus and chicken, incomplete transcriptional compensation is complemented by post-transcriptional regulation to achieve male–female parity. Unbalanced M:F ratios in the transcriptome but balanced M:F ratios in the proteome contrasts with therian mammals, in which dosage compensation occurs largely in the transcriptome.
Molecular Basis of Partial Transcriptional Dosage Compensation in Platypus
XCI in eutherians involves DNA methylation and histone modifications (28–30). We analyzed published DNA methylation data for platypus (31), finding only small regional increases of methylation on chromosomes X1 and X2 in female platypus (SI Appendix, Fig. S4), the implications of which are unclear.
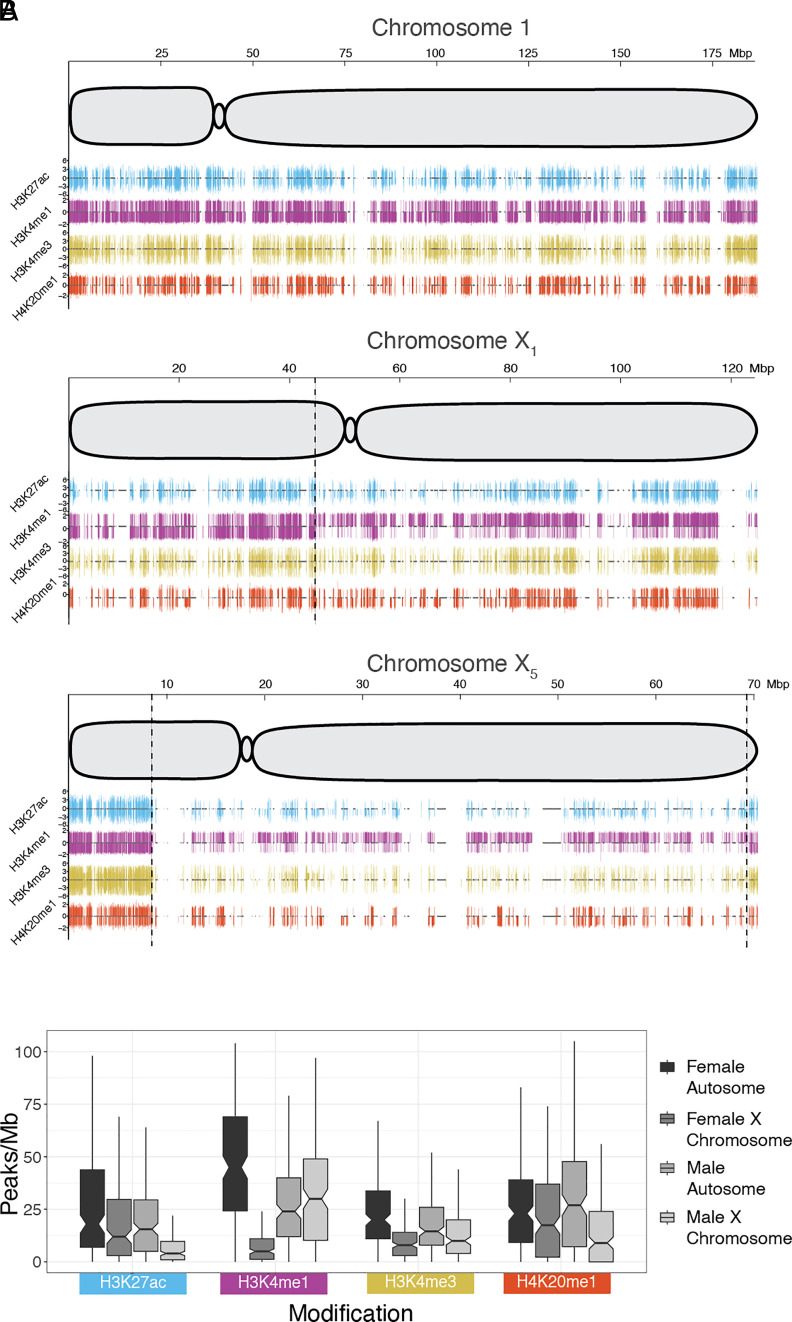
We performed Chromatin Immunoprecipitation sequencing (ChIP-seq) in platypus fibroblasts to detect histone modifications associated with active transcription: H3K27ac, H3K4me1, H3K4me3, and H4K20me1. In both sexes, fewer ChIP-peaks were detected in X-specific regions than on autosomes and PARs for all four histone modifications, except for H3K4me1 in males (Fig. 2, Table 1, and SI Appendix, Fig. S5). A random subset of 1 Mb autosome windows were taken so that the number of bins (174) was equal between the X chromosomes and autosomes (Fig. 2B and Table 1). The mean X to autosomal (X/A) ratio in both males and females were significantly different except for H4K20me1 in females (Fig. 2B and Table 1). The most striking difference between males and females was for H3K4me1, for which there was a 20% increase in peak density on the X chromosomes compared to autosomes in males, contrasting with an 83% decrease on the X chromosomes compared to autosomes in females. The higher density of this active histone mark in male platypus could up-regulate genes on the Xs.
Fig. 2.
Epigenetic profile of platypus chromosomes 1, X1, and X5. (A) Whole chromosome plots for chromosome 1 (a representative autosome), X1, and X5 in platypus. The x-axis represents the full length of the chromosome, with size (in Mbp) at the Top. Each plot shows a schematic of the chromosome with centromere position. Below the chromosomes are ChIP-seq peak tracks for four active histone marks: H3K27ac (blue), H3K4me1 (purple), H3K4me3 (yellow), and H4K20me1(red). For each histone mark, male and female peaks are shown above and below the center line, respectively. ChIP peaks were filtered using a threshold q-value of 0.05 and peak height is displayed up to a fivefold change on the y-axis. Gene locations are shown in the center of each ChIP track as grey boxes. On the X chromosome plots, vertical dashed lines indicate PAR boundaries identified with HiC interaction data (SI Appendix, Fig. S2). (B) Median peaks/Mb plots for each histone modification, with samples shown from Left to Right, respectively: female autosome, female X chromosome, male autosome, and male X chromosome. For male and female X chromosomes, only X-specific regions were analyzed. A random subset of 1 Mb autosome windows were taken so that the number of bins (174) was equal between the X chromosomes and autosomes. Boxes represent the middle 50% of the data, and whiskers represent 1.5 times the interquartile range. Outliers are not plotted. A Mood’s median test was used to calculate statistical difference, with adjusted P-values (Table 1).
Table 1.
ChIP peak density
| Peaks/Mb | ||||||||
|---|---|---|---|---|---|---|---|---|
| Modification | Male autosome mean | Male X mean | Male X/A mean ratio | Male X/A P adjust value | Female autosome mean | Female X mean | Female X/A ratio mean | Female X/A P adjust value |
| H3K27ac | 22 | 8.3 | 0.38 | 4.5E-10 | 29 | 21 | 0.71 | 3.3E-02 |
| H3K4me1 | 27 | 32 | 1.2 | 4.2E-02 | 46 | 7.8 | 0.17 | 0 |
| H3K4me3 | 20 | 16 | 0.78 | 1.1E-02 | 25 | 11.9 | 0.47 | 3.5E-12 |
| H4K20me1 | 30 | 15 | 0.49 | 3.8E-07 | 27 | 21.7 | 0.81 | 0.11 |
Thus, partial transcriptional upregulation of X chromosomes in males correlates with differential loading of the active histone modification H3K4me1 between sexes.
Full Compensation by Control at Multiple Regulatory Levels
Our most significant finding is that although X and Z-borne genes in platypus and chicken are variably and only partially compensated by differential transcription, they are fully compensated between the sexes at the protein level (Fig. 3). The median M:F ratio of mRNAs sampled in the proteome reflected the whole transcriptome, so correction in the proteome can only result from post-transcriptional dosage compensation.
Fig. 3.
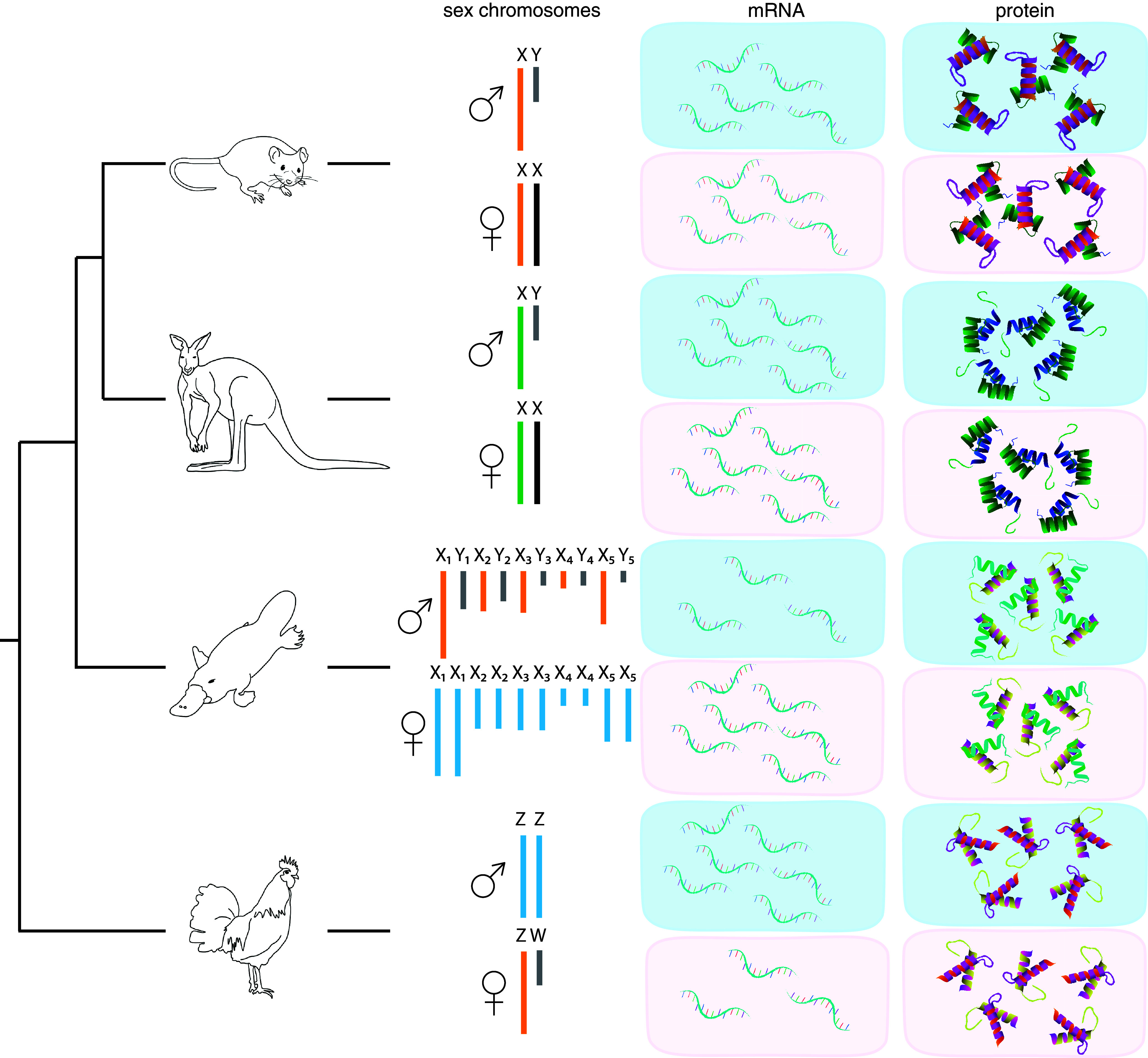
Transcriptome and proteome sex chromosome dosage compensation. Relationships of the different study species and their respective sex chromosome and dosage compensation systems. For each species, equal or unequal output between the sexes is indicated by the number of molecules shown for the transcriptome (mRNA) and proteome (protein). The green X in marsupials is up-regulated to match autosome transcriptional output. The orange X/Z in mouse, platypus, and chicken indicates partial upregulation, while blue X/Z in platypus and chicken indicates no known partial upregulation in the homogametic sex. Gray chromosomes represent the degraded sex-specific Ys or Ws. Black chromosomes indicate X/Z has undergone XCI.
There are two important implications of our findings. First, genes sampled in the proteome must be up-regulated post-transcriptionally from the X/Z in the heterogametic sex or downregulated in the homogametic sex. Regulation could be accomplished by differential translation or altered transcript decay rates of X/Z genes in males and females and relative to autosomal genes (32). Second, this post-transcriptional regulation must be gene specific because different genes are transcriptionally regulated to different extents.
How could transcripts be recognized by the translation machinery as originating from sex chromosomes? Recognition could involve epigenetic modifications of RNA (33) or sex-biased microRNAs (miRNA), which have the potential to down-regulate the expression of many genes. Sex-biased miRNAs have a high turnover rate, and there are examples in birds of a miRNA with copies on both the Z and W, and for another miRNA that preferentially down-regulates Z genes in males (34).
Thus, far from dosage compensation being optional in non-therian vertebrates, our results suggest that it is essential. However, it is accomplished by a combination of transcriptional and post-transcriptional control, rather than the transcriptional repression of one X chromosome, as in therian mammals.
Evolution of Dosage Compensation in Vertebrates
Is this combination of transcriptional and post-transcriptional control ancestral to dosage compensation in vertebrates or at least in amniotes?
Transcriptional inactivation, although common to all three mammal branches, is accomplished by mechanisms that are different at the molecular level. In eutherian mammals, XCI involves specific histone modifications as well as DNA methylation, coordinated by an inactivation center containing the gene XIST which is transcribed into a long non-coding RNA. In contrast, X inactivation in marsupials uses a different but overlapping set of histone modifications (35), a different DNA methylation profile (31), and a noncoding RNA from the unrelated RSX gene (36). In platypus, partial transcriptional upregulation was correlated to sex differences in only one of the four active histone marks, and a female-specific non-coding RNA (37) is unrelated to either the eutherian or marsupial noncoding RNAs, suggesting use of a different but overlapping set of ancient epigenetic mechanisms.
In eutherian mammals, X chromosome expression levels in the translatome are more similar to ancestral proto-sex chromosomal expression levels than in the transcriptome (15). Only in dosage compensation in non-therian vertebrates does post-transcriptional control of gene activity complement transcriptional control, but it may play a role in regulation of the autosome-sex chromosome balance in eutherians. Thus, our observations reveal different evolutionary mechanisms to compensate for sex chromosome gene dosage between the autosomes and sex chromosomes (and ultimately males and females) in different vertebrate lineages (Fig. 3). Evolutionary flexibility can be seen by comparing expression of orthologous genes on the chicken Z and platypus X chromosomes, which have higher expression in ZZ male birds but lower expression in XY male platypuses (Fig. 1B).
It is interesting to consider how these different modes of dosage compensation evolved in mammals. The balancing of incomplete transcriptional dose regulation by post-transcriptional control in birds and monotremes suggests that both mechanisms could be involved in dosage compensation in a common amniote ancestor. It will be instructive to compare dosage compensation in non-avian reptiles, amphibians, and fish. The Komodo dragon has a ZW system with no dosage compensation of Z genes at the transcriptional level (38). Conversely, the green anole has an XY system with complete dosage compensation of genes on the X at the transcriptional level (18). The complete reliance on transcriptional repression of X-borne genes, which is specific to therian mammals, would therefore appear to be a later evolutionary event, perhaps driven by regulator sequences specific to a relatively young therian X chromosome (39).
The different dosage compensation mechanisms of vertebrate groups may therefore represent different mixes of ancient epigenetic mechanisms that affect transcriptional and post-transcriptional control of gene activity.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We acknowledge that reviewer Dr. S.V.E. and author J.A.M.G. have co-published in two multiauthor articles, see: H. A. Lewin et al. (115 authors) and M. Blaxter et al. (24 authors). J.A.M.G. and P.D.W. are supported by Australian Research Council Discovery Projects (DP210103512 and DP220101429). P.D.W. is supported by National Health Medical and Research Council (NHMRC) Ideas Grants (2021172, 2027730). H.R.P. is supported by an NHMRC Ideas Grant (2021172). S.A.W. is supported by the University of New South Wales (UNSW) Scientia program and an NHMRC Ideas Grant (1188987). M.I.R is supported by Helmholz association core funding and a DFG project grant FOR2841. S.M. was supported by grant MU 880/27-1 from the Deutsche Forschungsgemeinschaft (DFG). A.R.-H. is supported by the Spanish Ministry of Science and Innovation (PID2020-112557GB-I00 founded by AEI/10.13039/501100011033) and the Agència de Gestió d’Ajuts Universitaris i de Recerca, AGAUR (2021SGR00122). F.G. and L.S.-W. is supported by an Australian Research Council Discovery Project (DP210103512).
Author contributions
P.D.W. designed research; N.C.L., A.M.L., W.B.H., L.K.W., and A.R.R. performed research; N.C.L., A.M.M., H.R.P., B.J.H., and A.R.-H. analyzed data; and N.C.L., H.R.P., S.A.W., B.J.H., K.L.M., S.M., M.I.R., L.S.-W., F.G., J.A.M.G., A.R.-H., and P.D.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: A.P.A., University of California, Los Angeles; C.M.D., University of Washington; and S.V.E., Harvard University.
Contributor Information
Jennifer A. Marshall Graves, Email: J.Graves@latrobe.edu.au.
Paul D. Waters, Email: p.waters@unsw.edu.au.
Data, Materials, and Software Availability
All sequencing data are available under BioProject PRJNA929280 (40). Mass spectrometry data for platypus, chicken, and mouse heart and liver samples, including raw files and protein summaries, are available at the ProteomeXchange Consortium via the PRIDE (41) partner repository with the dataset identifier PXD040182. The scripts for the statistical models and the datasets necessary to run analyses included in this paper, as well as protein summaries for platypus and opossum fibroblasts, have been deposited in the public depository Git Hub, and are available at: https://github.com/kango2/dcomp (42). Protein summary files are names according to sample labeling described in ProteinPilot software workflow section of methods. Previously published data were used for this work (43).
Supporting Information
References
- 1.Grützner F., et al. , In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 432, 913–917 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Veyrunes F., et al. , Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18, 965–973 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graves J. A. M., Gartler S. M., Mammalian X chromosome inactivation: Testing the hypothesis of transcriptional control. Somat. Cell Mol. Genet. 12, 275–280 (1986). [DOI] [PubMed] [Google Scholar]
- 4.Julien P., et al. , Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 10, e1001328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y., et al. , Platypus and echidna genomes reveal mammalian biology and evolution. Nature 592, 756–762 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martínez-Pacheco M., et al. , Expression evolution of ancestral XY gametologs across all major groups of placental mammals. Genome Biol. Evol. 12, 2015–2028 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh Y., et al. , Dosage compensation is less effective in birds than in mammals. J. Biol. 6, 2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L., Wa Sin S. Y., Grayson P., Edwards S. V., Sackton T. B., Evolutionary dynamics of sex chromosomes of paleognathous birds. Genome Biol. Evol. 11, 2376–2390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmer F., Harrison P. W., Dessimoz C., Mank J. E., Compensation of dosage-sensitive genes on the chicken Z chromosome. Genome Biol. Evol. 8, 1233–1242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohno S., Sex Chromosomes and Sex-Linked Genes (Springer Science & Business Media, 1967). [Google Scholar]
- 11.Lin F., Xing K., Zhang J., He X., Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc. Natl. Acad. Sci. U.S.A. 109, 11752–11757 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M., Lin F., Xing K., Liu L., Random X-chromosome inactivation dynamics in vivo by single-cell RNA sequencing. BMC Genom. 18, 90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Y., et al. , RNA sequencing shows no dosage compensation of the active X-chromosome. Nat. Genet. 42, 1043–1047 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Lentini A., et al. , Elastic dosage compensation by X-chromosome upregulation. Nat. Commun. 13, 1854 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z. Y., et al. , Transcriptome and translatome co-evolution in mammals. Nature 588, 642–647 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyu Q., et al. , A small proportion of X-linked genes contribute to X chromosome upregulation in early embryos via BRD4-mediated transcriptional activation. Curr. Biol. 32, 4397–4410.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Naik H. C., Hari K., Chandel D., Jolly M. K., Gayen S., Single-cell analysis reveals X upregulation is not global in pre-gastrulation embryos. iScience 25, 104465 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marin R., et al. , Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage. Genome Res. 27, 1974–1987 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davalos-Dehullu E., et al. , Chromosome-level genome assembly of the blacktail brush lizard, urosaurus nigricaudus, reveals dosage compensation in an endemic lizard. Genome Biol. Evol. 15, evad210 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metzger D. C. H., Sandkam B. A., Darolti I., Mank J. E., Rapid evolution of complete dosage compensation in poecilia. Genome Biol. Evol. 13, evab155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng X., et al. , Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 43, 1179–1185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Q. L., et al. , Chromosome-level assembly, dosage compensation and sex-biased gene expression in the small brown planthopper, Laodelphax striatellus. Genome Biol. Evol. 14, evac160 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graves J. A. M., Disteche C. M., Toder R., Gene dosage in the evolution and function of mammalian sex chromosomes. Cytogenet. Cell Genet. 80, 94–103 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Graves J. A. M., Disteche C. M., Does gene dosage really matter? J. Biol. 6, 1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uebbing S., et al. , Quantitative mass spectrometry reveals partial translational regulation for dosage compensation in chicken. Mol. Biol. Evol. 32, 2716–2725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellegren H., et al. , Faced with inequality: Chicken do not have a general dosage compensation of sex-linked genes. BMC Biol. 5, 40 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uebbing S., Kunstner A., Makinen H., Ellegren H., Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome Biol. Evol. 5, 1555–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heard E., Delving into the diversity of facultative heterochromatin: The epigenetics of the inactive X chromosome. Curr. Opin. Genet. Dev. 15, 482–489 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Heard E., Disteche C. M., Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes. Dev. 20, 1848–1867 (2006). [DOI] [PubMed] [Google Scholar]
- 30.J. A. M. Graves, 5-azacytidine-induced re-expression of alleles on the inactive X chromosome in a hybrid mouse cell line. Exp. Cell Res. 141, 99–105 (1982). [DOI] [PubMed] [Google Scholar]
- 31.Waters S. A., et al. , Landscape of DNA methylation on the marsupial X. Mol. Biol. Evol. 35, 431–439 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Faucillion M. L., Larsson J., Increased expression of X-linked genes in mammals is associated with a higher stability of transcripts and an increased ribosome density. Genome Biol. Evol. 7, 1039–1052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kan R. L., Chen J., Sallam T., Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 38, 182–193 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warnefors M., et al. , Sex-biased microRNA expression in mammals and birds reveals underlying regulatory mechanisms and a role in dosage compensation. Genome Res. 27, 1961–1973 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaumeil J., et al. , Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: Epigenetic modifications in distantly related mammals. PLoS One 6, e19040 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grant J., et al. , Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487, 254–258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Necsulea A., et al. , The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Rovatsos M., Rehák I., Velenský P., Kratochvíl L., Shared ancient sex chromosomes in varanids, beaded lizards, and alligator lizards. Mol. Biol. Evol. 36, 1113–1120 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Graves J. A. M., Evolution of vertebrate sex chromosomes and dosage compensation. Nat. Rev. Genet. 17, 33–46 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Lister N. C., Waters P. D., Data from “Incomplete transcriptional dosage compensation of chicken and platypus sex chromosomes is balanced by post-transcriptional compensation.” NCBI SRA. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA929280. Deposited 7 February 2023. [DOI] [PMC free article] [PubMed]
- 41.Perez-Riverol Y., et al. , The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel H. R., Lister N. C., Waters P. D., Code and protein summaries from “Incomplete transcriptional dosage compensation of chicken and platypus sex chromosomes is balanced by post-transcriptional compensation.” GitHub. https://github.com/kango2/dcomp. Deposited 15 May 2024. [DOI] [PMC free article] [PubMed]
- 43.Brawand D., et al. , The evolution of gene expression levels in mammalian organs. Nature 478, 343–348 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All sequencing data are available under BioProject PRJNA929280 (40). Mass spectrometry data for platypus, chicken, and mouse heart and liver samples, including raw files and protein summaries, are available at the ProteomeXchange Consortium via the PRIDE (41) partner repository with the dataset identifier PXD040182. The scripts for the statistical models and the datasets necessary to run analyses included in this paper, as well as protein summaries for platypus and opossum fibroblasts, have been deposited in the public depository Git Hub, and are available at: https://github.com/kango2/dcomp (42). Protein summary files are names according to sample labeling described in ProteinPilot software workflow section of methods. Previously published data were used for this work (43).