Abstract
Panax notoginseng is a famous perennial herb widely used as material for medicine and health-care food. Due to its various therapeutic effects, research work on P. notoginseng has rapidly increased in recent years, urging a comprehensive review of research progress on this important medicinal plant. Here, we summarize the latest studies on the representative bioactive constituents of P. notoginseng and their multiple pharmacological effects, like cardiovascular protection, anti-tumor, and immunomodulatory activities. More importantly, we emphasize the biosynthesis and regulation of ginsenosides, which are the main bioactive ingredients of P. notoginseng. Key enzymes and transcription factors (TFs) involved in the biosynthesis of ginsenosides are reviewed, including diverse CYP450s, UGTs, bHLH, and ERF TFs. We also construct a transcriptional regulatory network based on multi-omics data and predicted candidate TFs mediating the biosynthesis of ginsenosides. Finally, the current three major biotechnological approaches for ginsenoside production are highlighted. This review covers advances in the past decades, providing insights into quality evaluation and perspectives for the rational utilization and development of P. notoginseng resources. Modern omics technologies facilitate the exploration of the molecular mechanisms of ginsenoside biosynthesis, which is crucial to the breeding of novel P. notoginseng varieties. The identification of functional enzymes for biosynthesizing ginsenosides will lead to the formulation of potential strategies for the efficient and large-scale production of specific ginsenosides.
Introduction
Panax notoginseng (Burk.) F.H. Chen, also known as Sanchi, is a perennial herb belonging to the Araliaceae family, extensively detailed in the classical Chinese pharmacopeia, Ben-Cao-Gang-Mu [1]. Originating from East Asia and North America around 25 million years ago, it predominantly thrives in China’s southwestern Yunnan and Guangxi regions, with the Yunnan variant highly regarded for its medicinal properties [2]. As both a medicinal and edible plant, P. notoginseng has garnered considerable interest for its applications in pharmaceuticals, functional foods, and health-care products [3]. Its notable effectiveness in enhancing blood circulation and alleviating pain renders it crucial in cardiovascular disease treatments, featuring prominently in traditional Chinese medicine prescriptions such as Yunnan Baiyao, Pientzehuang Compound, Danshen Dripping Pills, and Xuesaitong Injection [4]. Beyond its medical uses, P. notoginseng is prized for its health benefits and culinary applications. Its flowers are brewed into herbal teas that help cool the body and lower blood pressure, while its stems and leaves are utilized in cough-relieving teas. The roots, known for their anti-inflammatory and cardioprotective attributes, are often added to soups [5]. With its widespread popularity in various commercial sectors, including health and beauty, and dietary supplements, P. notoginseng enjoys broad recognition across Asia and in the West [6]. Responding to increasing demand, the industry has seen significant growth, with production reaching 28 000 tons and generating annual revenues of 16.2 billion Chinese yuan in 2017 [7].
The metabolites of P. notoginseng play a crucial role in its pharmacological efficacy. This plant harbors a diverse array of secondary metabolites, including ginsenosides, organic acids, esters, polysaccharides, amino acids, sterols, and flavonoids. These ingredients have significant therapeutic effects on the nervous system and immune response [8]. Ginsenosides stand out as the foremost bioactive elements, driving the medicinal effectiveness of this herb [9]. As the demand for P. notoginseng escalates, so does the interest in its agricultural production and the enhancement of biosynthetic methods for these active compounds. Such advancements are vital for the progression of herbal medicine and resource conservation. Plant metabolites are synthesized, transported, and metabolized through complex metabolic networks. Despite this, there remains a notable gap in the comprehensive analysis of the biosynthesis of these crucial active ingredients and their transcriptional regulation networks.
This review focuses on four principal topics: (i) bioactive constituents of P. notoginseng, (ii) its pharmacological benefits, (iii) the biosynthesis and regulation of ginsenosides, and (iv) biotechnological approaches to ginsenoside production. An in-depth exploration of the structures, distributions, biosynthetic routes, transcriptional regulatory frameworks, and biotechnological production techniques of ginsenosides is indispensable for the qualitative assessment and optimal exploitation of P. notoginseng.
Bioactive ingredients
The therapeutic effects of P. notoginseng are mainly attributed to their bioactive components, such as saponins, amino acids, polysaccharides, and flavonoids [10]. Saponins are mainly divided into three dammarane-type tetracyclic triterpenoids, oleanane-type pentacyclic triterpenoids, and ocotilloltype pentacyclic triterpenoids, including ginsenosides Rb1, Rc, Rb2, Rg1, Re, Ro, and Rd, notoginsenoside R1, and majonoside R2. Amino acids include a non-protein amino acid (dencichine) and protein amino acids (such as arginine, aspartic acid, glutamic acid, and leucine). Polysaccharides include polysaccharide I, II (IIa, IIb), and III categories, and are mainly made up of glucose, galactose, and arabinose. The basic parent nucleus of flavonoids is 2-phenylketonuria and the basic skeleton of flavonoids is C6-C3-C6, and they mainly consist of quercetin, quercetin-7-glucoside, flavonoside quercetin-3-O sophoroside, and kaempferic acid. The types and contents of these bioactive components can be influenced by region, tissue, and development age, as discussed below.
Saponins
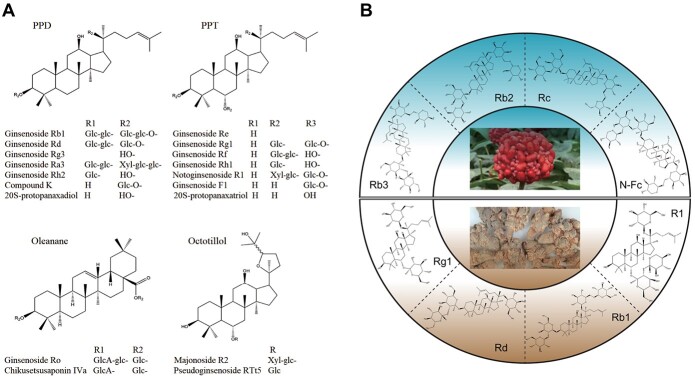
Saponins are the foremost active compounds in P. notoginseng, with >200 types identified by 2023 as noted in Supplementary Data Table S1 [4, 11–13]. These ginsenosides are classified into three main types: dammarane-type tetracyclic triterpenoids; oleanane-type pentacyclic triterpenoids; and ocotillol-type pentacyclic triterpenoids, as depicted in Fig. 1A [14]. Among them, dammarane-type ginsenosides are split into two subtypes: protopanaxadiol-type (PPD) and protopanaxatriol-type (PPT). PPD-type ginsenosides are typically bonded with sugar moieties at C-3, C-20, or both, whereas PPT-type ginsenosides feature sugars at C-3, C-6, and C-20. The ocotillol-type ginsenosides are distinguished by a five-membered epoxy ring on the C-20 side chain, and the oleanane-type by a modified C-20 side chain. Notably, PPT-type ginsenosides such as Rg1, R1, Re, and notoginsenoside R2 are found in concentrations ranging from 2.0 to 40.0 mg/g [15–17]. The most prevalent PPD-type ginsenosides, Rb1 and Rd, are found in concentrations of 26.7–30.6 and 5.7–8.4 mg/g, respectively [18]. Collectively, R1, Rb1, Rg1, Rd, and Re make up 90% of all ginsenosides in P. notoginseng [19].
Figure 1.
Specialized ginsenosides isolated from P. notoginseng. A Structures of major ginsenoside types. B Relative ginsenoside content distribution in the aerial and underground parts.
The distribution and concentration of individual ginsenosides vary across different plant parts and developmental stages, as shown in Fig. 1B [20]. The total ginsenoside content in 3-year-old plants is 1.4 times higher than that in 2-year-old plants, with PPD-type ginsenosides predominantly distributed in aboveground parts and PPT-type ginsenosides mainly distributed in underground parts [21]. Leaves are particularly rich in PPD-type ginsenosides, including Rb3, Rc, and Rb2, while the main roots harbor significant levels of PPT-type ginsenosides such as Rg1, Re, and N-R1 [22, 23].
Amino acids
Dencichine (β-N-oxalo-L-α-β-diaminopropionic acid, β-ODAP, C5H8N2O5), a non-protein amino acid with strong hemostatic activity and strong neuroexcitatory toxicity, is a characteristic component of P. notoginseng [24] (Supplementary Data Fig. S1 and Supplementary Data Table S1). The concentration of dencichine varies depending on the age and part of the plant, with the highest levels found in the flowers and leaves [24]. Specifically, dencichine content in 3-year-old flowers is 2.98%, which is 1.48 times greater than in 2-year-old flowers at 2.01%. Additionally, the method of drying significantly influences dencichine levels, the highest yield being observed with a drying treatment at 50°C and the lowest with freeze-drying at 40°C [25].
Furthermore, P. notoginseng is rich in protein amino acids, including arginine, aspartic acid, glutamic acid, and leucine (Supplementary Data Table S1). The concentration and composition of these amino acids vary across different plant parts and environments [26, 27]. Notably, the flowers possess the highest total amino acid content but lack arginine, whereas the roots are devoid of both arginine and methionine. To date, researchers have identified 19 amino acids in the roots of P. notoginseng [26].
Polysaccharides
Polysaccharides are another crucial group of pharmacodynamically active components in P. notoginseng, as catalogued in Supplementary Data Table S1. Polysaccharides mainly include polysaccharide I, II (IIa, IIb), and III categories, and are predominantly made up of glucose, galactose, arabinose, lactose, rhamnose, xylose, and arabose, with the overall content of polysaccharides around 9.45% [28]. Notably, a starch-like polysaccharide (PNPN) along with six pectin variants, namely PNPA-1A, PNPA-1B, PNPA-2A, PNPA-2B, PNPA-3A, and PNPA-3B, have been identified [29]. Moreover, the crude polysaccharide extract of P. notoginseng consists of arabinose, galacturonic acid, d-mannose, d-glucose, and galactose, in a molar ratio of 2:1:2:83:7 [30]. As of 2022, researchers have documented a total of 41 polysaccharides from P. notoginseng [31, 32].
The polysaccharide levels in P. notoginseng are significantly affected by factors such as geographical location, time of harvest, and specific plant parts. The highest levels of polysaccharides are typically found in April and the lowest in July. Geographically, Yanshan shows the highest levels of polysaccharides, in contrast to the lower levels found in Guangxi [33]. Within the plant, the polysaccharide content varies, with ribs exhibiting the highest levels and stems and leaves the lowest [34].
Flavonoids
The basic parent nucleus of flavonoids is 2-phenylketonuria and the basic skeleton of flavonoids is C6-C3-C6. A total of 13 varieties have been isolated from P. notoginseng [35, 36]. These primarily consist of quercetin, quercetin-7-glucoside, flavonoside quercetin-3-O sophoroside, kaempferic acid, quercetin-3-O-sophroside, kaempferol, kaempferol-7-O-α-l-rhamnoside, kaempferol-3-O-β-d-galactoside, kaempferol-3-O-β-d-galactose-(2 → 1)-glucoside, and quercetin-3-O-β-d-galactose-(2 → 1)-glucoside, as detailed in Supplementary Data Table S1. The concentration of these flavonoids varies by region, with Wenshan showing the highest levels at 0.7% and Honghe the lowest at 0.29%. The contents of flavonoids in different parts are significantly different: 1.77% in stem and leaf, 1.43% in flower, 0.50% in rhizome, 0.34% in fibril, and 0.19% in taproot [37].
Pharmacological activities
An increasing number of studies demonstrate that P. notoginseng has been widely used in the treatment of some chronic diseases, such as atherosclerosis, diabetes, cancer, and cardiovascular and cerebrovascular diseases [38]. Panax notoginseng saponins (PNS) are the major active ingredient of P. notoginseng and plays an important role in the medical and health fields. All kinds of PNS medicinal preparations, such as Sanqi-Tongshu capsules, Xuesaitong injection and Xuesaitong capsules, have been extensively used clinically. Modern research reveals that PNS possess a range of biological properties, including cardioprotective, cerebrovascular protective, neuroprotective, anti-tumor, anti-inflammatory, hemostatic, and anticoagulant effects. The following is an overview of these therapeutic effects and their relevant mechanisms (Table 1 and Fig. 2).
Table 1.
Pharmacological activities of P. notoginseng.
| Effect type | Extract/compound | Material/model | Results | Mechanism | Reference |
|---|---|---|---|---|---|
| Cardioprotective effect | Notoginsenoside R1 | Mouse femoral artery endothelium denudation model | Inhibiting restenosis following percutaneous transluminal angioplasty | Hindering activation of the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) signaling pathway | [39] |
| Notoginsenoside Ft1 | Human umbilical vein endothelial cells | Stimulating angiogenesis via HIF-1α-mediated VEGF expression | Inducing activation of the PI3K/AKT and Raf/MEK/ERK signaling pathways | [40] | |
| PNS | Left anterior descending ligation-operated mice | Enhancing endothelial cell migration and angiogenesis following myocardial infarction | Via phosphorylation of AMPK Thr 172 and CaMKII Thr 287 | [41] | |
| Cerebrovascular protective effect | PNS | Middle cerebral artery occlusion and oxygen–glucose deprivation/reperfusion model | Repairing the function of impaired nerves | Decreasing expression of Nogo-A and NgR | [42] |
| PNS | Rat model of cerebral ischemia and SH-SY5Y cells | Providing neuroprotective effects | Inhibiting expression of NgR1, RhoA, and ROCK2 | [43] | |
| Notoginsenoside R1 | Hypoxic–ischemic brain damage in neonates | Inhibiting neuronal apoptosis and promoting cell survival | Via the PI3K-Akt–mTOR/JNK signaling pathways by targeting estrogen receptors | [44] | |
| Ginsenoside Rd | Sprague–Dawley rats with focal cerebral ischemic injury | Attenuating the pathogenesis of cerebral ischemia-induced blood–brain barrier damage | Inhibiting proteasome activity and suppressing the NF-κB/MMP-9 pathway | [45] | |
| Ginsenoside Rb1 | Rats with induced photothrombotic stroke | Alleviating the morphological lesion and cognitive and sensorimotor deficits | Via the modulation of the Akt/mTOR/PTEN signaling pathway | [46] | |
| Ginsenoside Rg1 | Cerebral ischemia/reperfusion injury model | Preventing IR-induced neurological injuries | Blocking NF-κB nuclear translocation and phosphorylation of IκBα | [47] | |
| Neuroprotective effect | PNS | β-Mediated neurotoxicity in Caenorhabditis elegans | Inhibiting Alzheimer’s disease-like symptoms | Involving the SKN-1 signaling pathway and expression of Alzheimer’s disease-related circRNAs | [48] |
| Ginsenosides Rg1 and Rg2 | Rats with Alzheimer’s disease | Enhancing cognitive function and reducing hippocampal amyloid-β deposition | Via modulation of associated metabolic pathways | [49] | |
| Ginsenoside Rg1 | Rats exposed to isoflurane anesthesia | Combating cognitive decline | Via antioxidant, anti-inflammatory and anti-apoptotic effects, mediated by the PI3K/AKT/GSK-3β pathway | [50] | |
| Anti-tumor effect | PNS | Mice with AOM/DSS-induced colorectal cancer | Reducing the immunosuppress | Inhibiting the expression of IDO1 | [51] |
| Ginsenoside Rb1 | H9c2 and liver carcinoma HepG-2 cells | Curtailing production of tumor necrosis factor-α induced MMP-9 | Suppressing the RNA-dependent protein kinase and NF-κB signaling pathways | [52] | |
| Notoginsenoside R1 | Hepatocellular carcinoma cells | Boosting antihepatoma efficacy | Inactivation of the PI3K/Akt pathway by suppressing miR-21 | [53] | |
| Ginsenoside Rg3 | Hepatocellular carcinoma cells | Decreasing NHE1 expression | Blocking the epidermal growth factor receptor pathway, including phosphorylated extracellular signal-regulated kinase and hypoxia-inducible factor 1 alpha | [54] | |
| Ginsenoside Rg5 | Breast cancer in a mouse model | Inducing apoptosis and autophagy | Disrupting the PI3K/Akt signaling pathway | [55] | |
| Anti-inflammatory effect | Ginsenoside Re | BV2 microglial cells | Neuroprotective events in neuroinflammation occurred | Via the phospho-p38, iNOS, and COX2 signaling pathways | [56] |
| Ginsenoside Rg1 | Rodent model mimicking alcoholic hepatic injury | Alleviating symptoms of alcoholic hepatitis and TNBS-induced colitis | Inhibiting the NF-κB pathway | [57] |
Figure 2.
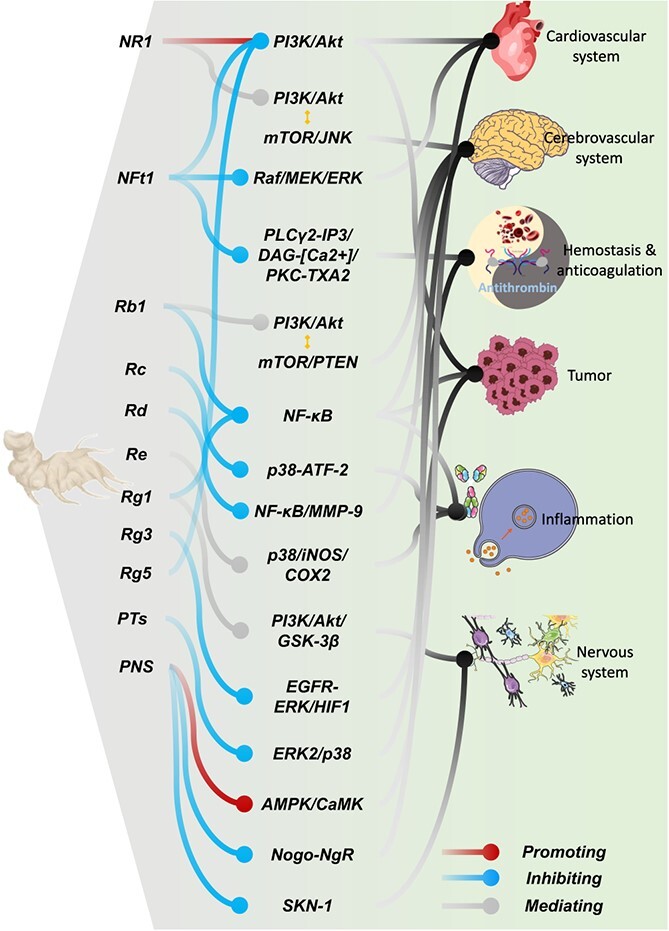
Summary of pharmacological effects and relevant mechanisms in P. notoginseng.
Table 1.
Continued.
| Effect type | Extract/compound | Material/model | Results | Mechanism | Reference |
|---|---|---|---|---|---|
| Ginsenoside Rc | HEK293 cells transfected with various inducers of inflammation | Exerting anti-inflammatory actions | Suppressing TANK-binding kinase 1/IκB kinase ε/interferon regulatory factor-3 and p38/ATF-2 signaling pathway | [58] | |
| Hemostasis and anticoagulation | Panaxatriol saponins (Rg1, Re, and R1) | Rabbit or human platelets | Inhibiting platelet aggregation | Reducing intracellular calcium mobilization and deactivating the ERK2/p38 pathway | [59] |
| Notoginsenoside Ft1 | Rats | Influencing coagulation processes | Enhancing the PLCγ2-IP3/DAG-[Ca2+]/PKC-TXA2 signaling pathway | [60, 61] |
Cardioprotective effect
Cardiovascular diseases are the primary causes of mortality in developed nations, accounting for half of all deaths [62]. Research has demonstrated that P. notoginseng has potent protective effects on myocardial cells and combats cardiovascular ailments through a variety of complex signaling pathways. For instance, notoginsenoside R1 is acknowledged as a promising therapeutic to inhibit restenosis following percutaneous transluminal angioplasty, by hindering the activation of the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) signaling pathway [39]. Moreover, notoginsenoside Ft1 boosts the expression of vascular endothelial growth factor (VEGF), stimulated by HIF-1α, linked to the PI3K/Akt and Raf/MEK/ERK signaling pathways [40]. Additionally, PNS and its main constituents, ginsenosides Rg1 and R1, are known to significantly enhance endothelial cell migration and angiogenesis following myocardial infarction through the phosphorylation of AMPK Thr 172 and CaMKII Thr 287 [41].
Cerebrovascular protective effect
Cerebrovascular diseases rank as significant causes of disability and the third leading cause of mortality in developed nations [63]. PNS have shown efficacy in mitigating neurological deficits, cerebral infarctions, edema, and neuronal death [42, 43]. For instance, ginsenoside Rd is noted for potentially reducing tau phosphorylation induced by cerebral ischemia through the PI3K/Akt/GSK-3β pathway, and for lessening damage to the blood–brain barrier via the NF-κB/MMP-9 pathway [44, 45]. Additionally, ginsenoside Rb1 influences the Akt/mTOR/PTEN signaling pathway, offering therapeutic benefits for various neurological conditions [46]. Ginsenoside Rg1 also plays a role in preventing IR-induced neurological injuries by blocking NF-κB nuclear translocation and the phosphorylation of IκBα, and by lowering glutamate and aspartate levels [47].
Neuroprotective effect
The incidence of neurodegenerative disorders such as Alzheimer’s and Parkinson’s is on the rise in developed nations, a trend attributed to longer life expectancies [64, 65]. Research into the therapeutic benefits of ginsenosides for these conditions has identified potential mechanisms involving the SKN-1 signaling pathway and the expression of Alzheimer’s disease-related circRNAs [48]. Ginsenosides Rg1 and Rg2 are noted for enhancing cognitive function and reducing hippocampal amyloid-β (Aβ) deposition through modulation of associated metabolic pathways [49]. Additionally, ginsenoside Rg1 combats cognitive decline through its antioxidant and anti-inflammatory actions mediated by the PI3K/Akt/GSK-3β pathway [50]. Ginsenoside Rb1 is also used in managing Huntington’s disease, traumatic brain injuries, and ischemia [66], while ginsenoside Rd is known to mitigate excitatory toxicity, modulate nerve growth factors, and promote nerve regeneration [67].
Anti-tumor effect
Research has demonstrated that P. notoginseng has significant anti-tumor effects against several types of cancer, including liver cancer [8]. PNS influence immune responses and curb tumor proliferation through various mechanisms. For example, they reduce the immunosuppression of Treg cells within the colorectal cancer environment by inhibiting the expression of IDO1, which is controlled by signal transducer and activator of transcription 1 (STAT1) [51]. Rb1 curtails the production of tumor necrosis factor-α (TNF-α)-induced MMP-9 by modulating the RNA-dependent protein kinase and the NF-κB signaling pathways [52]. R1 boosts antihepatoma efficacy by activating the PI3K/Akt pathway and suppressing miR-21, clarifying its role in combating hepatocellular carcinoma [53]. Rg3 decreases NHE1 expression by blocking the epidermal growth factor receptor pathway, including phosphorylated extracellular signal-regulated kinase and hypoxia-inducible factor 1α in hepatocellular carcinoma [54]. Additionally, ginsenoside Rg5 promotes apoptosis and autophagy by disrupting the PI3K/Akt signaling pathway [55].
Anti-inflammatory effect
Inflammation plays a crucial role in the body’s immune response and is linked to various health conditions. Ginsenosides are effective in reducing pro-inflammatory cytokines, such as IL-1β and TNF-α, thus mitigating cerebral ischemic injuries in the brain [68, 69]. Rb1, along with its metabolite compound K, blocks the activation of NF-κB, MAP kinases (ERK, JNK, and p-38), IKK-β, and interleukin-1 receptor-associated kinase-1 in lipopolysaccharide (LPS)-stimulated murine peritoneal macrophages [70]. The anti-inflammatory properties of ginsenoside Rd stem from its ability to downregulate NF-κB, leading to decreased expression of iNOS and COX-2 [71]. Ginsenoside Re is neuroprotective against 1 μg/ml LPS-treated microglial cells, and the neuroprotective events in neuroinflammation occurred via the phospho-p38, iNOS, and COX2 signaling pathways [56]. Furthermore, ginsenoside Rg1 has been shown to alleviate symptoms of alcoholic hepatitis and TNBS-induced colitis by inhibiting the NF-κB pathway [57]. Additionally, ginsenoside Rc targets the TANK-binding kinase 1/IκB kinase ε/interferon regulatory factor-3 and p38/ATF-2 signaling pathways, exerting its anti-inflammatory effects [58].
Hemostasis and anticoagulation
Atherosclerosis, a major contributor to cardiovascular and cerebrovascular diseases, can be mitigated by P. notoginseng [72]. Research demonstrated that panaxatriol saponins (Rg1, Re, and R1) effectively inhibit platelet aggregation by reducing intracellular calcium mobilization and deactivating the ERK2/p38 pathway [59]. Further studies indicate that Rg1 and Rg2 significantly extend blood clotting times, with Rg2 exhibiting a more potent anticoagulant effect than Rg1 [73]. Additionally, Rg2, Rg3, and protopanaxatriol are identified as potential natural inhibitors of clotting, contributing to their anti-platelet aggregation properties [74]. Rh1 and ginsenoside F1 also show capabilities in preventing platelet aggregation [75]. Moreover, notoginsenoside Ft1 influences coagulation processes by enhancing the PLCγ2-IP3/DAG-[Ca2+]/PKC-TXA2 signaling pathway [60, 61].
Biosynthesis and regulation of saponins
The most important active ingredients of P. notoginseng that exert pharmacological effects are ginsenosides, a series of triterpenoid saponins. However, the resources of P. notoginseng are currently limited. In order to better solve the source problem of ginsenosides, we need to comprehend the biosynthesis and regulation of ginsenosides in P. notoginseng, providing more clues for the breeding of novel P. notoginseng varieties with high ginsenoside contents and potential biotechnological methods for efficient and large-scale production of ginsenosides. The genes and transcription factors (TFs) involved in the ginsenoside biosynthesis pathways of P. notoginseng primarily govern the enzymes that facilitate the structurally diverse biosynthesis of ginsenosides. Additionally, environmental factors significantly influence ginsenoside production through interactions within hormonal signal-transcription regulatory networks. The biosynthesis and regulation of ginsenosides of P. notoginseng are reviewed below.
Biosynthesis of ginsenosides
Ginsenosides in the Panax genus and their biosynthesis have been reported previously [76]. The biosynthesis of ginsenosides primarily encompasses three stages: formation of the ginsenoside skeleton, glycosome synthesis, and modification of the skeleton [76]. The synthesis of ginsenosides predominantly occurs through the mevalonic acid (MVA) pathway in the cytoplasm and the methylerythritol phosphate (MEP) pathway in plastids, as illustrated in Fig. 3.
Figure 3.
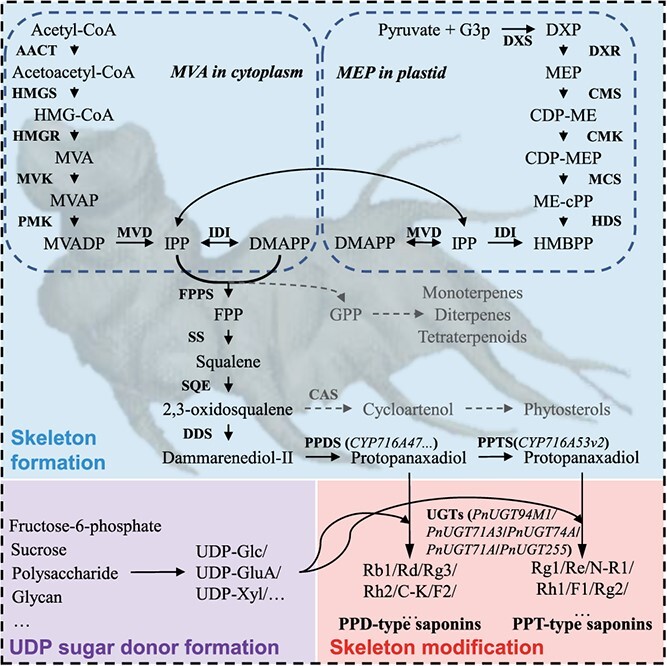
Possible biosynthesis pathway of ginsenosides in P. notoginseng.
Skeleton formation
The biosynthesis of ginsenosides involves upstream MVA and MEP pathways, which are basically similar in Panax [77]. Initially, the terpenoid precursors isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP) are synthesized. These are then converted into geranyl pyrophosphate and farnesyl pyrophosphate (FPP). Two molecules of FPP are combined by squalene synthase (SS) to form squalene [78]. This squalene is transformed into 2,3-oxidosqualene by squalene epoxidase (SE) and dammarenediol synthase (DS). Finally, 2,3-oxidosqualene undergoes cyclization, hydroxylation, and glycosylation to become ginsenosides [79].
3-Hydroxy-3-methylglutaryl CoA reductase (HMGR) is a crucial rate-limiting enzyme in ginsenoside biosynthesis [80]. The enhancement of PnHMGR expression leads to an increase in ginsenoside production in P. notoginseng [81]. Key enzymes such as PnFPS, PnSS, PnSE1, PnSE2, and PnDS are involved in this process and exhibit organ-specific expression patterns, showing particularly high levels in the flowers of 4-year-old P. notoginseng [82]. PnSE1 is present in all organs but predominantly in flowers, while PnSE2 shows relatively low expression [83]. The overall ginsenoside content is largely dependent on the activity of farnesyl diphosphate synthase (FPS), followed by SS and DS, with the concentration of Rb1 specifically linked to the activities of HMGR and FPS [84]. Research has demonstrated that the genes PnFPS, PnSS, PnSE1, PnSE2, and PnDS have tissue-specific expression patterns and show significantly higher levels in the flowers and leaves compared with the roots and fibrils, being 5.2 times greater [21, 85].
Cytochrome P450
After the initial formation of the basic ginsenoside skeleton, it is further transformed into ginsenosides with great diversity in structure and function in the Panax genus via hydroxylation by cytochrome P450 enzymes (CYPs) and glycosylation by uridine diphosphate (UDP)-dependent glycosyltransferases (UGTs). CYP enzymes are part of large and functionally diverse gene families [79]. The subfamily CYP716, belonging to the CYP85 clade, is a major contributor to the diversification of triterpenoid biosynthesis [86]. Specifically, three CYP716s – CYP716A47 (protopanaxadiol synthase, PPDS), CYP716A53v2 (protopanaxatriol synthase, PPTS), and CYP716A52v2 (oleanolic acid synthase, OAS) – have been identified [87–89]. Dammarenediol-II undergoes hydroxylation by CYP716A47 and CYP716A53v2 to form protopanaxadiol and protopanaxatriol in four common Panax species (P. notoginseng, P. ginseng, P. quinquefolius, and P. japonicus) [90]. Distinct from CYP716A47 and CYP716A53v2, CYP716A52v2 serves as a multifunctional oxygenase involved in the biosynthesis of oleanolic acid in P. ginseng, P. quinquefolius, and P. japonicus, but is not expressed in P. notoginseng. So CYP716 plays a pivotal role in the diversification of ginsenosides in Panax species. Thus, the expression of some differential genes in downstream synthetic pathways in the Panax genus will result in the unique monomer ginsenosides of Panax species.
The CYP716 subgroup is not only hydroxylated at different carbon sites in different members of the Panax genus, but also has different expression levels in different plant tissues (leaf and root tissues). Thus, dammarane-type [synthesized by Y subgroup (CYP716A47) and S subgroup (CYP716A53v2)] and oleanane-type [synthesized by A subgroup (CYP716A52v2)] ginsenosides presented different concentrations in leaf and root tissues [91]. The CYP716 genes, including PN006374, PN008424, PN011429, and PN029913, show high expression levels in the roots and are likely crucial for the biosynthesis of PPT-type ginsenosides [92, 93].
UDP-dependent glycosyltransferases
The UGTs are instrumental in forming various monomeric ginsenosides by attaching monosaccharides to aglycones, primarily at position C-3 or C-20 for PPD-type ginsenosides, and at C-6 and/or C-20 for PPT-type ginsenosides [94]. Propanaxanediol and propanaxatriol are glycosylated by various UGT enzymes to form PPD-type and PPT-type ginsenosides, which exist universally in Panax plants. For example PnUGT71A3 is capable of catalyzing both the C6 hydroxyl glycosylation of PPT and F1 into Rh1 and Rg1, and the C20 hydroxyl glycosylation of Rg3 into Rd [95]. PnUGT94M1 facilitates the C2 hydroxyl rhamnosylation of Rg1 and Rh1 into Re and Rg2, respectively. PnUGT74A is able to convert CK into F2, PnUGT94C transforms Rh2 into Rg3 and F2 into Rd, and PnUGT71A modifies PPT into F1 and PPD into CK [96]. The presence of PnUGT255 is associated with R1 levels, and PnUGT190 significantly influences Rd concentrations [97]. Elevated expression of three UGT71 genes (PN000453, PN025151, and PN025152) and three UGT74 genes (PN000316, PN024572, and PN033259) in the aerial parts is linked to the biosynthesis of PPD-type ginsenosides [98].
Transcriptional regulation
The biosynthesis and accumulation of ginsenosides are characterized by distinct spatio-temporal patterns. The expression of genes involved in biosynthesis is regulated by a variety of TFs and non-coding RNAs (ncRNAs) through intricate regulatory networks. These TFs and ncRNAs can respond to both biotic and abiotic signals, playing crucial roles in the regulation of ginsenoside production.
Transcription factors
TFs regulate gene transcription through complex networks, thereby influencing ginsenoside concentrations. Key TF families, such as WRKY, basic helix–loop–helix (bHLH), myeloblastosis (MYB), and AP2/ERF (ERF1), are crucial in ginsenoside biosynthesis. A comprehensive identification has revealed 2150 TFs across 57 families in P. notoginseng [99].
Overexpression of PnMYB1 in P. notoginseng upregulates the expression of PnFPS, PnSS, and PnDS genes and subsequently increases the levels of ginsenosides R1, Rg1, Re, Rb1, and Rd [100]. PnMYB2 enhances the expression of PnSS and PnSE1 by interacting with their promoters [101]. Conversely, inhibition of PnMYB4 can increase expression of ginsenoside biosynthetic genes, including PnSS, PnSE, and PnDS, and thus boosts ginsenoside content. Both PnMYB4 and PnMYB1 collaborate with PnbHLH to fine-tune ginsenoside biosynthesis [102]. In transgenic cell lines expressing PnbHLH1, the transcription of key biosynthetic genes – PnDS, PnSS, PnSE, and PnFPS – is elevated, doubling the total ginsenoside content compared with wild-type cell lines [103]. The PnbHLH TF indirectly augments ginsenoside levels by regulating key biosynthetic genes at multiple points [104]. With PnWRKY1 overexpression in P. notoginseng, the expression of PnDS, PnSS, and PnSE escalates by 3.1, 4.0, and 4.5 times, respectively, influencing ginsenoside concentrations [105]. In cell lines overexpressing PnERF1, transcription levels of PnDS and PnSS are increased by 1.6 and 1.9 times, respectively, enhancing the content of six ginsenosides: Rg3, Rh1, Rd, Rg1, F1, and Re [106]. Additionally, PnERF2 and PnERF3 show significant correlation with DS and SE expression in various tissues of methyl jasmonate (MeJA)-induced P. notoginseng [107].
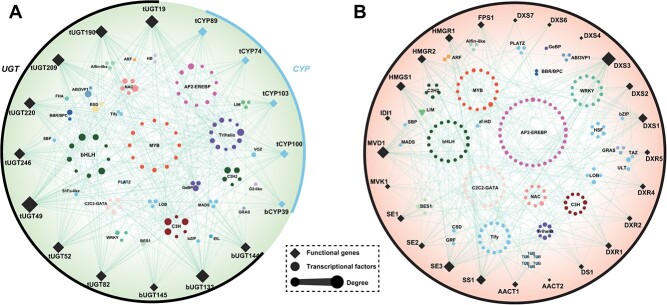
Data from the Panax genome and transcriptome database (NCBI accession number PRJNA488357) significantly enhance gene research, although reports on the transcriptional regulatory networks for ginsenoside biosynthesis are scarce. Utilizing multi-omics data and patterns of ginsenoside accumulation, co-expression networks within P. notoginseng have been developed to hypothesize the regulatory interactions between TFs and genes involved in ginsenoside biosynthesis (Fig. 4 and Supplementary Data File 2). TFs were categorized using the Plant Transcription Factor Database PlantTFDB (http://planttfdb.gao-lab.org/), and Pearson’s correlation coefficients were calculated to determine the relationships between TFs and ginsenoside biosynthetic genes using tools from OmicStudio (https://www.omicstudio.cn/tool) [108]. Analysis of the P. notoginseng transcriptome database identified 103 TFs that are predicted to have significant positive or negative correlations with the regulation of ginsenoside biosynthetic genes (Pearson’s |r| > 0.8, adjusted P < 0.05). This correlation underscores the complexity of the transcriptional regulatory network that mediates ginsenoside biosynthesis. Each biosynthetic gene is influenced by multiple TFs, which can either enhance or suppress its activity. Further research is necessary to understand the regulatory functions and interactions among TFs in P. notoginseng to clarify how they control the synthesis of specialized ginsenosides. Additionally, many TFs are capable of simultaneously regulating multiple biosynthetic genes; thus, uncovering the molecular mechanisms that influence these transcriptional regulations is crucial for developing innovative breeding strategies.
Figure 4.
Regulation relationship between TFs and genes involved in ginsenoside synthesis from P. notoginseng. A Regulatory network of CYPs and UGTs with TFs. B Regulatory network of genes from ginsenoside skeleton formation and TFs. A total of 103 TFs were identified, the transcript abundances of which were significantly correlated with functional genes (Pearson’s |r| > 0.8, adjusted P < 0.05). Edges represent significant correlations, and node size is mapped to degree.
Non-coding RNAs
In the roots of P. notoginseng, miR156 and miR166 are recognized as the predominant miRNA families, with miR156i and miR156g being the most abundant within these groups [109]. The miR156 family, alongside one of its target genes from the squamosa promoter-binding protein-like (SPL) category, exhibits inversely related expression levels that are closely associated with the increase in root biomass content [110].
Environmental regulation
Environmental factors collaboratively influence the synthesis of active ingredients through hormone signal-transcription regulatory networks in medicinal plants [111]. This environmental regulation encompasses responses to heavy metals, various environmental conditions, and fertilizer applications, as illustrated in Fig. 5.
Figure 5.
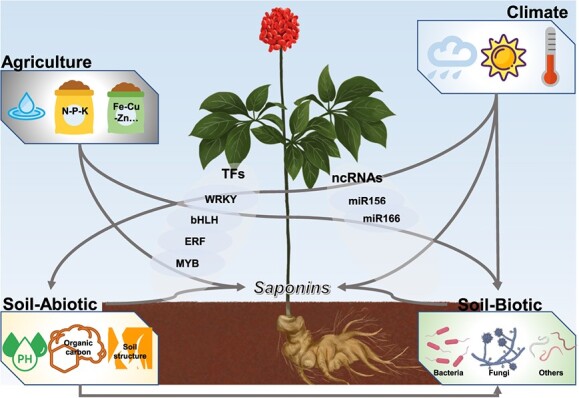
Environmental regulation (climatic factors, edaphic factors, and agricultural practice) of ginsenoside biosynthesis.
Heavy metal regulation
The relationship between heavy metals and plant growth, alongside their impact on metabolite synthesis, can be described as ‘low promotion and high inhibition’ [112]. At low concentrations, cadmium (Cd) increases the expression of DS, whereas high concentrations inhibit it; additionally, the presence of Cd significantly reduces the expression of cytochrome P450 enzymes [113]. However, the expression levels of DS and P450 genes do not show a strong correlation with the contents of R1, Rb1, Rg1, and total ginsenoside (P > 0.5). Endogenous nitric oxide (NO) boosts the accumulation of Rb1 but lowers the levels of Re and Rg1. Furthermore, endogenous NO increases the transcription of β-amyrin synthase (β-AS), cycloartenol synthase (CAS), and squalene epoxidase (SE), yet decreases the transcription of DS under Cd stress [114].
Environmental factor regulation
Recent studies have demonstrated that environmental factors such as light, temperature, water, and salinity influence the synthesis and accumulation of secondary metabolites in medicinal plants [115]. Both the rhizosphere and endophytic microflora of P. notoginseng play a role in enhancing plant health, biomass production, and the synthesis or biotransformation of ginsenosides, either directly or indirectly [116]. Climate, soil, and microbial interactions synergistically affect the contents of ginsenosides in P. notoginseng [117]. The concentrations of total and individual ginsenosides in the taproot of P. notoginseng show regional variations, with higher ginsenoside levels achievable by increasing temperature in January, atmospheric humidity, and soil calcium content, and by reducing latitude and average July temperatures [118]. Additionally, sunshine duration is a significant factor in ginsenoside production, with content increasing alongside longer exposure to sunlight [119].
Fertilizer regulation
Optimal fertilizer application during the cultivation of P. notoginseng enhances both biomass and ginsenoside content [120]. The expression of PnbZIP TFs in roots varies under different nitrogen fertilizer conditions; notably, PnbZIP46 is significantly upregulated under ammonium nitrogen fertilizer stress, which may contribute to nitrogen stress resistance [121]. Adjusting the nitrogen/potassium application ratio to 1:2 can improve the synergistic effect on both biomass and ginsenoside content in P. notoginseng [122]. A lower nitrogen/potassium ratio enhances photosynthesis, sugar accumulation, and the expression of genes involved in ginsenoside biosynthesis. Additionally, the use of ammonium and nitrate fertilizers stimulates the tricarboxylic acid (TCA) cycle, thereby increasing both biomass and ginsenoside content in P. notoginseng roots [123]. Conversely, high nitrogen concentrations can inhibit biomass and ginsenoside content; excessive nitrogen reduces root biomass and ginsenoside accumulation, likely due to decreased nitrogen efficiency and reduced photosynthetic capacity [124].
Biotechnological methods for saponin production
Panax notoginseng has a slow growth rate, typically requiring 3–4 years from seed germination to root harvest in field cultivation. Additionally, challenges such as plant pathogens and pests must be managed during the cultivation process. Metabolic engineering offers a viable alternative for enhancing the production of natural products. Consequently, biotechnological approaches, including tissue culture, adventitious roots, transgenic plants, and microbial cell factories, are recommended to boost ginsenoside production (Table 2).
Table 2.
Biosynthesis of ginsenosides by metabolic engineering
| Types | Cultured material | Operational method | Products | Reference |
|---|---|---|---|---|
| Plants | Suspension cultures | 2-Hydroxyethyl jasmonate elicitation | Increasing P6H and Rb1 and Rg content | [125] |
| Suspension cultures | 2-Hydroxyethyl jasmonate elicitation | The total ginsenoside content and Rb/Rg ratio increased about 60 and 30% | [126] | |
| Suspension cultures | Phenobarbital treatment | Enhancing the production of protopanaxatriol-type (Rg1 + Re) ginsenosides | [127] | |
| Suspension cultures | MeJA treatment | About 9-fold PPD-type and 2-fold PPT-type ginsenosides | [128] | |
| Suspension cultures | 2-Hydroxyethyl jasmonate and jasmonic acid treatment | Enhancing the synthesis of ginsenosides | [129] | |
| Adventitious root | Multi-branched root induced | 17.92 mg/g ginsenosides | [130] | |
| Adventitious root | Jasmonate treatment | 71.94 mg/g ginsenosides | [131] | |
| Transgenic cell lines | Overexpression of FPS and RNAi of CAS | 2.46-Fold total ginsenosides | [132] | |
| Transgenic cell lines | Overexpression of PnFPS | 2.66 times Rh1, 1.76 times Rg1, 4.35 times Re, and 2.90 times Rd | [133] | |
| Transgenic cell lines | Overexpression of β-AS | Chikusetsusaponin IV and chikusetsusaponin IVa were obtained | [134] | |
| Transgenic cell lines | Overexpression of PnbHLH1 | 2.27 Times total saponin contents | [103] | |
| Transgenic cell lines | Overexpression of PnERF1 | Total saponins increased ~2-fold from 40 to 80 mg/g | [106] | |
| Transgenic cell lines | Overexpression of PnbHLH | Total saponins increased ~2.6-fold | [104] | |
| Yeast | S. cerevisiae | Co-expression of HMGR, DDS and β-AS, OAS, PPDS, PPTS and cytochrome P450 | 17.2 mg/l PPD, 15.9 mg/l PPT, and 21.4 mg/l oleanolic acid | [135] |
| S. cerevisiae | PPDSeATR1 protein fusion and ROS tolerance enhancement | 4.25 g/l PPD | [136] | |
| S. cerevisiae | Overexpression of UGTPg45 | 529.0 mg/l PPD | [137] | |
| S. cerevisiae | Overexpression of PgUGT74AE2 and UGTPg1 | 2.4 g/l 3b-O-Glc-DM and 5.6 g/l 20S-O-Glc-DM | [138] | |
| S. cerevisiae | Overexpression of UGTPg1 | 1.4 mg/l compound K | [139] | |
| S. cerevisiae | Overexpression of Pn3-32-i5 | >1 g/l compound K | [140] | |
| S. cerevisiae | Overexpression of tHMG1 | 9.05 mg/l 3β,12β-Di-O-Glc-PPD | [141] | |
| S. cerevisiae | Overexpression of PgUGT71A53 and PgUGT71A54 | 1.95 g/L Rg1 | [142] | |
| Bacteria and fungi | Dictyoglomus turgidum and Pyrococcus furiosus | Conversion of main ginsenosides to minor ginsenosides | 210 mg/l/h aglycon protopanaxatriol | [143] |
| Cladosporium xylophilum | Transformation of main ginsenosides to minor ginsenosides | 0.99 mg/ml F2, 0.67 mg/ml Rd2, 0.24 mg/ml Fd, and 0.24 mg/ml Fe | [144] | |
| β-Glucosidase (Bgp1) | Ginsenosides Re and Rg1 to ginsenosides Rg2 and Rh1 | 0.83 mg/ml Rg2 and 0.6 mg/ml Rh1 | [145] | |
| Enterobacter chengduensis; Trichoderma koningii; Penicillium chermesinum | Biotransformation of main ginsenosides to minor ginsenosides | Rg1 to F1 at a rate of 13.24%; Rb1 to Rd (40.00%); Rb1 to Rg3 (32.31%); Rb1 to Rd (74.24%) | [146] | |
| Coniochaeta sp. | Conversion of ginsenoside Rb1 to C-K | Conversion rate of 11. 62% | [147] | |
| Pestalotiopsis biciliata | Conversion of ginsenoside Rb1 into rare ginsenoside F2, C-K, and ginsenoside Rd | Conversion rates of 53, 11 and 10%, respectively | [148] | |
| Bifidobacterium lactis Bi-07 | Transformation of ginsenosides Rd to F2 | Conversion rate of 25% | [149] | |
| Aspergillus niger | Ginsenoside Rb1 to generate Rg3 | Conversion rate of 48.5% | [150] |
Tissue culture
Suspension culture is an effective method for producing ginsenosides in P. notoginseng. The components of the medium, such as sugar types, significantly impact the ginsenoside concentration in cultured cells. Specifically, 2-hydroxyethyl jasmonate (HEJA) enhances the activity of protopanaxadiol 6-hydroxylase, thereby increasing the contents of Rb1 and Rg1 in cell cultures [125]. HEJA is more effective than MeJA in stimulating ginsenoside biosynthesis and altering its composition. Treatment with HEJA results in a 60% increase in total ginsenoside content and a 30% increase in the Rb/Rg ratio [126]. Additionally, adding 1 mM phenobarbital to the cultures boosts the levels of PPT-type ginsenosides (Rg1 + Re) [127]. Introducing 200 μM MeJA to the cultures can raise the levels of both PPD-type and PPT-type ginsenosides 9- and 2-fold, respectively [128]. Plant hormones such as HEJA, endogenous jasmonic acid (JA), and MeJA can induce the upregulation of squalene epoxidase (SE) and the suppression of cycloartenol synthase (CAS), promoting ginsenoside synthesis in P. notoginseng cells [129].
Hairy root and adventitious root cultures have been effectively used to produce stable biomass and high production of ginsenosides in Panax plants for over 15 years [151]. Research has revealed that an adventitious root line derived from wild-type roots of P. notoginseng has a high total ginsenoside yield of 17.92 mg/g [130]. In P. notoginseng adventitious roots, the highest total ginsenoside content reaches 71.94 mg/g after treatment with 5 mg/l JA, marking an 8.45-fold increase compared with the control [131]. Both JA and methyl dihydrojasmonate significantly boost Rd and Rg contents in these roots [131].
Transgenic plants
Despite considerable efforts to produce ginsenosides through tissue and cell culture, the yield remains relatively low. Employing genetic engineering to manipulate gene expression has proven to be an effective strategy for enhancing ginsenoside content. Transgenic P. notoginseng cell lines show higher expression of farnesyl pyrophosphate synthase (FPS) and lower expression of cycloartenol synthase (CAS) compared with wild-type cells, resulting in increased total ginsenoside production and decreased phytosterol levels [132]. In transgenic FPS-positive P. notoginseng cell lines, both relative PnFPS expression and ginsenoside contents are significantly higher, with increases of 2.66 times for Rh1, 1.76 times for Rg1, 4.35 times for Re, and 2.90 times for Rd, than in non-transgenic controls [133]. Moreover, enhancing β-amyrin synthase (β-AS) expression in transgenic P. notoginseng cells elevates the expression of key enzymes involved in ginsenoside biosynthesis and increases the levels of specific ginsenosides such as chikusetsusaponin IV and IVa [134]. Additionally, when three enzyme genes (PnDDS, CYP12H, and UGTPn3) were introduced into tobacco the three exogenous genes were expressed in the roots, stems, and leaves of the transgenic plants, and thus ginsenoside Rh2 and its precursors were successfully synthesized [152].
TFs such as PnbHLH1 and PnERF1 are known to enhance ginsenoside biosynthesis and accumulation. In P. notoginseng cell lines engineered to express PnbHLH1, the total ginsenoside content is more than double that of the control, specifically 2.27 times greater [103]. Similarly, in cell lines with PnERF1 transgenic modifications, total ginsenoside content approximately doubles from 40 in control lines to 80 mg/g [106]. Additionally, both RNAi targeting PnCAS and overexpression of PnbHLH in P. notoginseng cell lines result in increased levels of total and individual ginsenosides (Rd, Rb1, Re, Rg1, and R1) compared with wild-type and PnCAS RNAi cells [104].
Microbial cell factories
Yeast cell factories
Yeast cell factories, particularly those based on synthetic biology and heterogeneous expression systems, are proving effective for synthesizing various natural products [153]. For instance, co-expression of HMGR, DS, β-AS, OAS, PPDS, PPTS, and cytochrome P450 genes in Saccharomyces cerevisiae can enable the simultaneous production of PPD (17.2 mg/l), PPT (15.9 mg/l), and oleanolic acid (21.4 mg/l) [135]. Enhanced fed-batch fermentation techniques incorporating PPDSeATR1 protein fusion and ROS tolerance in S. cerevisiae boost PPD levels up to 4.25 g/l [136]. A newly developed yeast chassis can achieve 529.0 mg/l PPD in shake flasks and 11.02 g/l in 10-l fed-batch fermentations [137]. Additionally, 3b-O-Glc-DM and 20S-O-Glc-DM ginsenosides can reach concentrations of 2.4 and 5.6 g/l, respectively, via fed-batch fermentation [138]. Engineering yeast strains with UGTPg1 led to the production of ginsenoside compound K at a concentration of 1.4 mg/l [139]. A novel UDP-xylose-dependent glycosyltransferase enzyme, Pn3-32-i5, identified for R1 biosynthesis, allows a yeast cell factory to yield over 1 g/l of ginsenoside compound K [140]. Furthermore, overexpression of tHMG1 in yeast increases the production of 3β,12β-Di-O-Glc-PPD from 6.17 to 9.05 mg/l [141]. Finally, Rg1-producing yeast strains, incorporating PgUGT71A53 and PgUGT71A54, can produce Rg1 at 1.95 g/l [142].
Bacterial and fungal cell factories
Rare ginsenosides, which are more readily absorbed into the bloodstream and act as active substances, have garnered increasing attention. Microbial fermentation is utilized for producing these rare ginsenosides through the hydrolysis of substrate ginsenosides by microbial enzymes. For instance, enzymes from Dictyoglomus turgidum and Pyrococcus furiosus achieve a complete conversion of PPT-type ginsenosides (such as R1, R2, Re, Rg1, Rg2, Rh1, Rf, F1, F3, and F5) into 210 mg/l of aglycon PPT [143]. Additionally, Cladosporium xylophilum converts main ginsenosides Rb1, Rb2, Rb3, Rc, and notoginsenoside Fa into minor ginsenosides F2 (0.99 mg/ml), Rd2 (0.67 mg/ml), Fd (0.24 mg/ml), and Fe (0.24 mg/ml) [144]. Recombinant β-glucosidase has been used to transform Re and Rg1 into Rg2 (0.83 mg/ml) and Rh1 (0.6 mg/ml), respectively [145]. Enterobacter chengduensis converts Rg1 into F1 with a conversion rate of 13.24%; Trichoderma koningii converts Rb1 into Rd (40.00%) and Rg3 (32.31%); and Penicillium chermesinum transforms Rb1 into Rd at a high rate of 74.24% [146]. The endophyte Coniochaeta sp. can convert Rb1 into the rare ginsenoside CK with an 11.62% conversion rate [147]. Pestalotiopsis biciliata converts Rb1 into rare ginsenosides F2, CK, and Rd, with conversion rates of 53, 11, and 10% respectively [148]. Bifidobacterium lactis transforms Rb1, Rc, and Rb2 into Rd, and further into the rare ginsenoside F2, achieving a conversion rate of 25% [149]. β-Glucosidase from Aspergillus niger hydrolyzes Rb1 to produce Rg3 at a conversion rate of 48.5% [150].
Conclusions and future perspectives
Panax notoginseng and its secondary metabolites are invaluable for human therapy and healthcare, leading to an increased use of this plant as a primary ingredient in numerous products. Consequently, demand for P. notoginseng continues to rise. However, limited varieties, challenges with continuous cropping, and other issues pose significant obstacles to the sustainable development of the P. notoginseng industry. Cultivating new and high-quality varieties is crucial for the ongoing sustainability and growth of this sector.
Modern omics technology accelerates breeding of P. notoginseng
Currently, the affordability and accuracy of high-throughput sequencing have enhanced the feasibility of accessing gene resources. Utilizing high-throughput sequencing technologies and bioinformatics to deeply explore genomic, transcriptomic, and proteomic data of plants aids in the discovery of novel genes. Advanced assays such as quantitative mass spectrometry-based, fluorescence-based, and other high-throughput methods enable rapid detection of enzyme activity and efficient screening of genes. Modern omics technologies are pivotal in accelerating the breeding of new P. notoginseng varieties and providing valuable germplasms for the sustainable development of the P. notoginseng industry.
Transcriptional regulatory networks reveal synthesis mechanisms
TFs play crucial roles in regulating both biotic and abiotic stresses during the growth of P. notoginseng plants, with ginsenoside accumulation closely tied to their regulatory functions and environmental adaptability. Efforts to construct a co-expression network of TFs and gene expression patterns in P. notoginseng have been made; however, the intricate regulatory mechanisms of these TFs require further exploration. Discovering novel genes and TFs will significantly enhance molecular plant breeding. Many TFs involved in ginsenoside production also control multiple genes simultaneously, highlighting the need for a comprehensive understanding of the molecular mechanisms that influence transcriptional regulation of ginsenosides to develop innovative breeding strategies.
Synthetic biology increases ginsenoside production
Given that the cultivation of P. notoginseng is both time-consuming and labor-intensive, the development of bioengineering approaches, such as tissue culture, adventitious roots, transgenic plants, and microbial cell factories, has been pursued to enhance ginsenoside production. Thoroughly understanding the biosynthesis and regulatory mechanisms of ginsenosides will greatly benefit their biotechnological production. These advancements provide an affordable and efficient industrial platform for producing ginsenosides. Identifying new ginsenoside synthesis is expected to pave the way for novel methods that facilitate efficient and large-scale production of ginsenosides.
Supplementary Material
Acknowledgements
This study was supported by grants from the National Key R&D Plan (2022YFC3501801, 2022YFC3501802, and 2022YFC3501804), Fundamental Research Funds for the Central Public Welfare Research Institutes (ZXKT21037, ZZ15-YQ-044, ZXKT22050, and ZXKT22001), and the Scientific Research Project of the Hainan Academician Innovation Platform (SQ2021PTZ0052).
Author contributions
G.W., G.Z., and M.L. performed this work. Y.Z. and W.Z.analysed data and wrote the manuscript. G.Z., B.W., Z.Z., and X.Z helped to draw the figure. Z.H., T.W., and L.S. made the chart data in the manuscript. L.D. and S.C. reviewed the manuscript and supervised this work.
Contributor Information
Guangfei Wei, State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, No.16 Nanxiaojie, Dongzhimennei Ave., Beijing, 100700, China.
Guozhuang Zhang, State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, No.16 Nanxiaojie, Dongzhimennei Ave., Beijing, 100700, China.
Mengzhi Li, Nanyang Institute of Technology, Nanyang, No.80, Changjiang Road, Wulibao Street, Wancheng District, 473000, China.
Yuqing Zheng, Zhangzhou Pien Tze Huang Pharmaceutical Co., Ltd, No. 1 Amber Road, Xiangcheng District, Zhangzhou, Fujian, 363099, China.
Wenke Zheng, Tianjin University of Traditional Chinese Medicine, No. 312, Anshan West Road, Nankai District, Tianjin, 301617, China.
Bo Wang, Hubei Institute for Drug Control, No.54, Dingziqiao Road, Zhongnan Road, Wuchang District, Wuhan, 430012, China.
Zhaoyu Zhang, State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, No.16 Nanxiaojie, Dongzhimennei Ave., Beijing, 100700, China.
Xiao Zhang, Zhangzhou Pien Tze Huang Pharmaceutical Co., Ltd, No. 1 Amber Road, Xiangcheng District, Zhangzhou, Fujian, 363099, China.
Ziying Huang, State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, No.16 Nanxiaojie, Dongzhimennei Ave., Beijing, 100700, China.
Tengyun Wei, Zhangzhou Pien Tze Huang Pharmaceutical Co., Ltd, No. 1 Amber Road, Xiangcheng District, Zhangzhou, Fujian, 363099, China.
Liping Shi, State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, No.16 Nanxiaojie, Dongzhimennei Ave., Beijing, 100700, China.
Shilin Chen, State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, No.16 Nanxiaojie, Dongzhimennei Ave., Beijing, 100700, China; Institute of Herbgenomics, Chengdu University of Traditional Chinese Medicine, No. 37, 12 Qiao Road, Jinniu District, Chengdu, 611137, China.
Linlin Dong, State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, No.16 Nanxiaojie, Dongzhimennei Ave., Beijing, 100700, China.
Data availability
The accession number of the RNA sequencing data is PRJNA488357.
Conflict of interest
The authors declare no conflicts of interest.
Supplementary data
Supplementary data are available at Horticulture Research online.
References
- 1. Ng TB. Pharmacological activity of sanchi ginseng (Panax notoginseng). J Pharm Pharmacol. 2006;58:1007–19 [DOI] [PubMed] [Google Scholar]
- 2. Meng XX, Huang LF, Dong LL. et al. Analysis of global ecology of Panax notoginseng in suitability and quality. Acta Pharm Sin. 2016;51:1483–93 [PubMed] [Google Scholar]
- 3. Yang WZ, Hu Y, Wu WY. et al. Saponins in the genus Panax L. (Araliaceae): a systematic review of their chemical diversity. Phytochemistry. 2014;106:7–24 [DOI] [PubMed] [Google Scholar]
- 4. Wang T, Guo RX, Zhou GH. et al. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) F.H. Chen: a review. J Ethnopharmacol. 2016;188:234–58 [DOI] [PubMed] [Google Scholar]
- 5. Ji C, Zhang Q, Shi R. et al. Determination of the authenticity and origin of Panax notoginseng: a review. J AOAC Int. 2022;105:1708–18 [DOI] [PubMed] [Google Scholar]
- 6. Ding LW. Ginseng production and marketing history, current situation and market outlook. Spec Econ Anim Plant. 2014;17:15–20 [Google Scholar]
- 7. People.cn . Polishing the "Golden Sign" Wenshan Notoginseng Output Value Will Reach 100 Billion in the Future. 2018.
- 8. Liu HB, Lu XY, Hu Y. et al. Chemical constituents of Panax ginseng and Panax notoginseng explain why they differ in therapeutic efficacy. Pharmacol Res. 2020;161:105263 [DOI] [PubMed] [Google Scholar]
- 9. Ma LJ, Ma N, Cao JL. et al. Characterizing the influence of different drying methods on chemical components of Panax notoginseng leaves by heart-cutting two-dimensional liquid chromatography coupled to orbitrap high-resolution mass spectrometry. Food Chem. 2022;369:130965 [DOI] [PubMed] [Google Scholar]
- 10. Liu YC, Zhang TJ, Guo HB. et al. Research progress on Notoginseng radix et Rhizoma and predictive analysis on its Q-marker. Chin Tradit Herb Drug. 2021;52:2733–45 [Google Scholar]
- 11. Zheng YJ, Xia PG, Zhao HG. et al. Suitable soil moisture contents for water use efficiency and saponins accumulation in Panax notoginseng. Chin Herb Med. 2021;13:267–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang YD, Cheng JX, Shi Y. et al. Panax notoginseng: a review on chemical components, chromatographic analysis, P. notoginseng extracts, and pharmacology in recent five years. China J Chin Mater Med. 2022;47:2584–96 [DOI] [PubMed] [Google Scholar]
- 13. Sun XJ, Deng HB, Shu TY. et al. Study on chemical constituents of Panax notoginseng leaves. Molecules. 2023;28:2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim DH. Chemical diversity of Panax ginseng, Panax quinquifolium, and Panax notoginseng. J Ginseng Res. 2012;36:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan CS, Huo YQ, An XJ. et al. Panax notoginseng and its components decreased hypertension via stimulation of endothelial-dependent vessel dilatation. Vasc Pharmacol. 2012;56:150–8 [DOI] [PubMed] [Google Scholar]
- 16. Dou L, Lu YG, Shen T. et al. Panax notoginseng saponins suppress RAGE/MAPK signaling and NF-kappaB activation in apolipoprotein-E-deficient atherosclerosis-prone mice. Cell Physiol Biochem. 2012;29:875–82 [DOI] [PubMed] [Google Scholar]
- 17. Xiong Y, Chen LJ, Man JH. et al. Chemical and bioactive comparison of Panax notoginseng root and rhizome in raw and steamed forms. J Ginseng Res. 2019;43:385–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu HS, Zhang LJ, Song XB. et al. Chemical constituents from processed rhizomes of Panax notoginseng. China J Chin Mater Med. 2013;38:3910–7 [PubMed] [Google Scholar]
- 19. Liu JJ, Wang YT, Qiu L. et al. Saponins of Panax notoginseng: chemistry, cellular targets and therapeutic opportunities in cardiovascular diseases. Expert Opin Investig Drugs. 2014;23:523–39 [DOI] [PubMed] [Google Scholar]
- 20. Zhang SQ, Chen C, Lu WX. et al. Phytochemistry, pharmacology, and clinical use of Panax notoginseng flowers buds. Phytother Res. 2018;32:2155–63 [DOI] [PubMed] [Google Scholar]
- 21. Wei GF, Dong LL, Yang J. et al. Integrated metabolomic and transcriptomic analyses revealed the distribution of saponins in Panax notoginseng. Acta Pharm Sin B. 2018;8:458–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jia XH, Wang CQ, Liu JH. et al. Comparative studies of saponins in 1-3-year-old main roots, fibrous roots, and rhizomes of Panax notoginseng, and identification of different parts and growth-year samples. J Nat Med. 2013;67:339–49 [DOI] [PubMed] [Google Scholar]
- 23. Liu F, Ma N, He CW. et al. Qualitative and quantitative analysis of the saponins in Panax notoginseng leaves using ultra-performance liquid chromatography coupled with time-of-flight tandem mass spectrometry and high performance liquid chromatography coupled with UV detector. J Ginseng Res. 2018;42:149–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duan SF, Chen G, Yan J. et al. Optimization of extraction process and detection method of dencichine. Chin J Tropic Crops. 2019;40:2255–60 [Google Scholar]
- 25. Song JP, Lou J, Zhang ZX. et al. Determination of dencichine in Panax notoginseng medium under different drying conditions. J Wenshan Univ. 2019;32:6–812. [Google Scholar]
- 26. Chen ZJ, Sun YQ, Dong TX. et al. Comparison of amino acid contents in Panax notoginseng from different habitats. J Chin Med Mater. 2003;26:86–8 [PubMed] [Google Scholar]
- 27. Li C, Zhang H, Ma H. et al. Analysis of amino acids in Panax notoginseng. Amino Acids J. 1992;4:46 [Google Scholar]
- 28. Xiong YH, Li J, Huang S. et al. Content determination of total polysaccharides in Panax notoginseng. Asia-Pacific Tradit Med. 2011;7:7–9 [Google Scholar]
- 29. Chan MK, Yu Y, Wulamu S. et al. Structural analysis of water-soluble polysaccharides isolated from Panax notoginseng. Int J Biol Macromol. 2020;155:376–85 [DOI] [PubMed] [Google Scholar]
- 30. Xia YG, Li X, Yu LS. et al. Structural-fingerprinting of polysaccharides to discern Panax species by means of gas-liquid chromatography and mass spectrometry. Int J Biol Macromol. 2020;151:932–43 [DOI] [PubMed] [Google Scholar]
- 31. Qi HY, Zhang ZP, Liu JQ. et al. Comparisons of isolation methods, structural features, and bioactivities of the polysaccharides from three common Panax species: a review of recent progress. Molecules. 2021;26:4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu YH, Li S, Pu MD. et al. Structural characterization of polysaccharides isolated from Panax notoginseng medicinal residue and its protective effect on myelosuppression induced by cyclophosphamide. Chem Biodivers. 2022;19:e202100681 [DOI] [PubMed] [Google Scholar]
- 33. Cui XM, Dong X, Chen ZJ. et al. Studies on the change of polysaccharides in Panax notoginseng collected at different times and from regions. Chin Pharm J. 2002;11:18–20 [Google Scholar]
- 34. Liu Y, Fan K, Li LJ. et al. Studies on content determination the polysaccharides in Panax notoginseng and change of polysaccharides in Panax notoginseng collected at different parts. Chin J Exp Tradit Med Formulae. 2012;18:118–20 [Google Scholar]
- 35. Xia PG, Zhang SC, Liang ZS. et al. Research history and overview of chemical constituents of Panax notoginseng. Chin Tradit Herb Drug. 2014;45:2564–70 [Google Scholar]
- 36. Huang ZY, Liu WJ, Chen Y. et al. Research progress of Panax notoginseng flavonoids. J Liaoning Univ Tradit Chin Med. 2020;22:81–4 [Google Scholar]
- 37. Liu Y, Qu Y, Wang CX. et al. Determination of the flavonoids in the different parts of Panax notoginseng from different areas. J Anhui Agric Sci. 2015;43:54–5 [Google Scholar]
- 38. Xu CC, Wang WW, Wang B. et al. Analytical methods and biological activities of Panax notoginseng saponins: recent trends. J Ethnopharmacol. 2019;236:443–65 [DOI] [PubMed] [Google Scholar]
- 39. Fang HH, Yang SL, Luo YY. et al. Notoginsenoside R1 inhibits vascular smooth muscle cell proliferation, migration and neointimal hyperplasia through PI3K/Akt signaling. Sci Rep. 2018;8:7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen K, Ji L, Gong CY. et al. Notoginsenoside Ft1 promotes angiogenesis via HIF-1a mediated VEGF secretion and the regulation of PI3K/AKT and Raf/MEK/ERK signaling pathways. Biochem Pharmacol. 2012;84:784–92 [DOI] [PubMed] [Google Scholar]
- 41. Wang DD, Lv LY, Xu Y. et al. Cardioprotection of Panax notoginseng saponins against acute myocardial infarction and heart failure through inducing autophagy. Biomed Pharmacother. 2021;136:111287 [DOI] [PubMed] [Google Scholar]
- 42. Liu LX, Zhu LQ, Zou YH. et al. Panax notoginseng saponins promotes stroke recovery by influencing expression of Nogo-a, NgR and p75NGF, in vitro and in vivo. Biol Pharm Bull. 2014;37:560–8 [DOI] [PubMed] [Google Scholar]
- 43. Shi XW, Yu WJ, Yang TT. et al. Panax notoginseng saponins provide neuroprotection by regulating NgR1/RhoA/ROCK2 pathway expression, in vitro and in vivo. J Ethnopharmacol. 2016;190:301–12 [DOI] [PubMed] [Google Scholar]
- 44. Tu L, Wang Y, Chen D. et al. Protective effects of notoginsenoside R1 via regulation of the PI3K-Akt-mTOR/JNK pathway in neonatal cerebral hypoxic-ischemic brain injury. Neurochem Res. 2018;43:1210–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang X, Liu XD, Hu GY. et al. Ginsenoside Rd attenuates blood-brain barrier damage by suppressing proteasome-mediated signaling after transient forebrain ischemia. Neuroreport. 2020;31:466–72 [DOI] [PubMed] [Google Scholar]
- 46. Liu Y, Fan K, Li LJ. et al. Studies on content determination the polysaccharides in Panax notoginseng and change of polysaccharides in Panax notoginseng collected at different parts. Behav Brain Res. 2018;345:83–9229501622 [Google Scholar]
- 47. Zheng TY, Jiang H, Jin RH. et al. Ginsenoside Rg1 attenuates protein aggregation and inflammatory response following cerebral ischemia and reperfusion injury. Eur J Pharmacol. 2019;853:65–73 [DOI] [PubMed] [Google Scholar]
- 48. Zhou L, Huang PP, Chen LL. et al. Panax notoginseng saponins ameliorate a β-mediated neurotoxicity in C. elegans through antioxidant activities. Evid Based Complement Alternat Med. 2019;4:7621043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li NJ, Liu Y, Li W. et al. A UPLC/MS-based metabolomics investigation of the protective effect of ginsenosides Rg1 and Rg2 in mice with Alzheimer's disease. J Ginseng Res. 2016;40:9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang YN, Zhang Z, Wang HT. et al. Neuroprotective effect of ginsenoside Rg1 prevents cognitive impairment induced by isoflurane anesthesia in aged rats via antioxidant, antiinflammatory and anti-apoptotic effects mediated by the PI3K/AKT/GSK-3β pathway. Mol Med Rep. 2016;14:2778–84 [DOI] [PubMed] [Google Scholar]
- 51. Li X-M, Yuan D-Y, Liu Y-H. et al. Panax notoginseng saponins prevent colitis-associated colorectal cancer via inhibition IDO1 mediated immune regulation. Chin J Nat Med. 2022;20:258–69 [DOI] [PubMed] [Google Scholar]
- 52. Sun WT, Yang CLH, Or TCT. et al. Ginsenoside Rb1 from Panax notoginseng suppressed TNF-α-induced matrix metalloproteinase-9 via the suppression of double-strand RNA-dependent protein kinase (PKR)/NF-κB pathway. Molecules. 2022;27:8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Y, Li Z, Jia YH. et al. In vitro anti-hepatoma activities of notoginsenoside R1 through downregulation of tumor promoter miR-21. Dig Dis Sci. 2022;65:1364–75 [DOI] [PubMed] [Google Scholar]
- 54. Li X, Tsauo J, Geng C. et al. Ginsenoside Rg3 decreases NHE1 expression via inhibiting EGF-EGFR-ERK1/2-HIF-1 α pathway in hepatocellular carcinoma: a novel antitumor mechanism. Am J Chin Med. 2018;46:1915–31 [DOI] [PubMed] [Google Scholar]
- 55. Liu YN, Fan DD. Ginsenoside Rg5 induces apoptosis and autophagy via the inhibition of the PI3K/Akt pathway against breast cancer in a mouse model. Food Funct. 2018;9:5513–27 [DOI] [PubMed] [Google Scholar]
- 56. Lee KW, Jung SY, Choi SM. et al. Effects of ginsenoside Re on LPS-induced inflammatory mediators in BV2 microglial cells. BMC Complement Altern Med. 2012;12:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gao Y, Chu S, Li J. et al. Anti-inflammatory function of ginsenoside Rg1 on alcoholic hepatitis through glucocorticoid receptor related nuclear factor-kappa B pathway. J Ethnopharmacol. 2015;173:231–40 [DOI] [PubMed] [Google Scholar]
- 58. Yu T, Yang Y, Kwak YS. et al. Ginsenoside Rc from Panax ginseng exerts anti-inflammatory activity by targeting TANK-binding kinase 1/interferon regulatory factor-3 and p38/ATF-2. J Ginseng Res. 2017;41:127–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Qi HY, Huang YL, Yang Y. et al. Anti-platelet activity of panaxatriol saponins is mediated by suppression of intracellular calcium mobilization and ERK2/p38 activation. BMC Complement Altern Med. 2016;16:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu YQ, Liu TY, Zhao J. et al. Phospholipase Cγ2 signaling contributes to the haemostatic effect of notoginsenoside Ft1. J Pharm Pharmacol. 2019;71:878–86 [DOI] [PubMed] [Google Scholar]
- 61. Li WN, Zhou Z, Li XL. et al. Biosynthesis of plant hemostatic dencichine in Escherichia coli. Nat Commun. 2022;13:5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jokinen E. Obesity and cardiovascular disease. Minerva Pediatr. 2015;67:25–32 [PubMed] [Google Scholar]
- 63. López M, Dávalos A. Advances in cerebrovascular disease research in the last year. J Neurol. 2011;258:168–72 [DOI] [PubMed] [Google Scholar]
- 64. Liu FF, Zhang Z, Chen W. et al. Regulatory mechanism of microRNA-377 on CDH13 expression in the cell model of Alzheimer’s disease. Eur Rev Med Pharmacol Sci. 2018;22:2801–8 [DOI] [PubMed] [Google Scholar]
- 65. Zhong LL, Song YQ, Cao H. et al. The non-motor symptoms of Parkinson’s disease of different motor types in early stage. Eur Rev Med Pharmacol Sci. 2017;21:5745–50 [DOI] [PubMed] [Google Scholar]
- 66. Ahmed T, Raza SH, Maryam A. et al. Ginsenoside Rb1 as neuroprotective agent: a review. Brain Res Bull. 2016;125:30–43 [DOI] [PubMed] [Google Scholar]
- 67. Chen YY, Liu QP, An P. et al. Ginsenoside Rd: a promising natural neuroprotective agent. Phytomedicine. 2022;95:153883 [DOI] [PubMed] [Google Scholar]
- 68. Liu RZ, Qin S, Li WJ. Phycocyanin: anti-inflammatory effect and mechanism. Biomed Pharmacother. 2022;153:113362 [DOI] [PubMed] [Google Scholar]
- 69. Shi XW, Yu WJ, Liu LX. et al. Panax notoginseng saponins administration modulates pro-/anti-inflammatory factor expression and improves neurologic outcome following permanent MCAO in rats. Metab Brain Dis. 2017;32:221–33 [DOI] [PubMed] [Google Scholar]
- 70. Joh EH, Lee IA, Jung IH. et al. Ginsenoside Rb1 and its metabolite compound K inhibit IRAK-1 activation—the key step of inflammation. Biochem Pharmacol. 2011;82:278–86 [DOI] [PubMed] [Google Scholar]
- 71. Kim DH, Chung JH, Yoon JS. et al. Ginsenoside Rd inhibits the expressions of iNOS and COX-2 by suppressing NF-kB in LPS-stimulated RAW264.7 cells and mouse liver. J Ginseng Res. 2013;37:54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lau AJ, Toh DF, Chua TK. et al. Antiplatelet and anticoagulant effects of Panax notoginseng: comparison of raw and steamed Panax notoginseng with Panax ginseng and Panax quinquefolium. J Ethnopharmacol. 2009;125:380–6 [DOI] [PubMed] [Google Scholar]
- 73. Li CT, Wang HB, Xu BJ. A comparative study on anticoagulant activities of three Chinese herbal medicines from the genus Panax and anticoagulant activities of ginsenosides Rg1 and Rg2. Pharm Biol. 2013;51:1077–80 [DOI] [PubMed] [Google Scholar]
- 74. Xiong LX, Qi Z, Zheng BZ. et al. Inhibitory effect of triterpenoids from Panax ginseng on coagulation factor X. Molecules. 2017;22:649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang J, Huang ZG, Cao H. et al. Screening of anti-platelet aggregation agents from Panax notoginseng using human platelet extraction and HPLC-DAD-ESI-MS/MS. J Sep Sci. 2008;31:1173–80 [DOI] [PubMed] [Google Scholar]
- 76. Hou MQ, Wang RF, Zhao SJ. et al. Ginsenosides in Panax genus and their biosynthesis. Acta Pharm Sin B. 2021;11:1813–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhao SJ, Wang L, Liu L. et al. Both the mevalonate and the non-mevalonate pathways are involved in ginsenoside biosynthesis. Plant Cell Rep. 2014;33:393–400 [DOI] [PubMed] [Google Scholar]
- 78. Luo HM, Sun C, Sun YZ. et al. Analysis of the transcriptome of Panax notoginseng root uncovers putative saponin-biosynthesis genes and genetic markers. BMC Genomics. 2011;12:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Augustin JM, Kuzina V, Andersen SB. et al. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry. 2011;72:435–57 [DOI] [PubMed] [Google Scholar]
- 80. Kim YK, Lee OR, Oh JY. et al. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme A reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol. 2014;165:373–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Deng B, Zhang P, Ge F. et al. Enhancement of triterpenoid saponins biosynthesis in Panax notoginseng cells by co-overexpressions of 3-hydroxy-3-methylglutaryl CoA reductase and squalene synthase genes. Biochem Eng J. 2017;122:38–46 [Google Scholar]
- 82. Niu YY, Luo HM, Sun C. et al. Expression profiling of the triterpene saponin biosynthesis genes FPS, SS, SE, and DS in the medicinal plant Panax notoginseng. Gene. 2014;533:295–303 [DOI] [PubMed] [Google Scholar]
- 83. Niu YY, Zhu XX, Luo HM. et al. Development of the devices for synthetic biology of triterpene saponins at an early stage: cloning and expression profiling of squalene epoxidase genes in Panax notoginseng. Acta Pharm Sin. 2013;48:211–8 [PubMed] [Google Scholar]
- 84. Xu S, Zhao C, Wen G. et al. Longitudinal expression patterns of HMGR, FPS, SS, SE and DS and their correlations with saponin contents in green-purple transitional aerial stems of Panax notoginseng. Ind Crop Prod. 2018;119:132–43 [Google Scholar]
- 85. Xia PG, Zheng YJ, Liang ZS. Structure and location studies on key enzymes in saponins biosynthesis of Panax notoginseng. Int J Mol Sci. 2019;20:6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Miettinen K, Pollier J, Buyst D. et al. The ancient CYP716 family is a major contributor to the diversification of eudicot triterpenoid biosynthesis. Nat Commun. 2017;8:14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Han JY, Kim HJ, Kwon YS. et al. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011;52:2062–73 [DOI] [PubMed] [Google Scholar]
- 88. Han JY, Hwang HS, Choi SW. et al. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2012;53:1535–45 [DOI] [PubMed] [Google Scholar]
- 89. Han JY, Kim MJ, Ban YW. et al. The involvement of β-amyrin 28-oxidase (CYP716A52v2) in oleanane-type ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2013;54:2034–46 [DOI] [PubMed] [Google Scholar]
- 90. Wei GF, Wei FG, Yuan C. et al. Integrated chemical and transcriptomic analysis reveals the distribution of protopanaxadiol- and protopanaxatriol-type saponins in Panax notoginseng. Molecules. 2018;23:1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang ZH, Wang XF, Lu TY. et al. Reshuffling of the ancestral core-eudicot genome shaped chromatin topology and epigenetic modification in Panax. Nat Commun. 1902;13:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Luo HM, Sun C, Sun YZ. et al. Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthetic genes and genetic markers. BMC Genomics. 2011;12:S5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fan GY, Liu XC, Sun S. et al. The chromosome level genome and genome-wide association study for the agronomic traits of Panax notoginseng. iScience. 2020;23:101538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Khorolragchaa A, Kim YJ, Rahimi S. et al. Grouping and characterization of putative glycosyltransferase genes from Panax ginseng Meyer. Gene. 2014;536:186–92 [DOI] [PubMed] [Google Scholar]
- 95. Hou MQ, Nie F, Zhao JN. et al. New glycosyltransferases in Panax notoginseng perfect main ginsenosides biosynthetic pathways. J Agric Food Chem. 2013;71:963–73 [DOI] [PubMed] [Google Scholar]
- 96. Li CW. Identification of Glycosyltransferases Related to Panax Notoginseng Saponins Biosynthesis. Harbin: Northeast Forestry University, 2021: [Google Scholar]
- 97. Wei GF, Yang F, Wei FG. et al. Metabolomes and transcriptomes revealed the saponin distribution in root tissues of Panax quinquefolius and Panax notoginseng. J Ginseng Res. 2020;44:757–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fan GY, Liu XC, Sun S. et al. The chromosome level genome and genome-wide association study for the agronomic traits of Panax notoginseng. iScience. 2020;23:101538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jiang ZQ, Tu LC, Yang WF. et al. The chromosome-level reference genome assembly for Panax notoginseng and insights into ginsenoside biosynthesis. Plant Commun. 2020;2:100113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lei J, Chen Q, Deng B. et al. Biosynthesis of Panax notoginseng saponins regulated by R2R3-MYB transcription factor PnMYB1. Biotechnol Bull. 2022;38:74–83 [Google Scholar]
- 101. Xia PG, Hu WY, Zheng YJ. et al. Structural and interactions analysis of a transcription factor PnMYB2 in Panax notoginseng. J Plant Physiol. 2022;275:153756 [DOI] [PubMed] [Google Scholar]
- 102. Man JH, Shi Y, Huang YY. et al. PnMYB4 negatively modulates saponin biosynthesis in Panax notoginseng through interplay with PnMYB1. Hortic Res. 2023;10:uhad134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang X, Ge F, Deng B. et al. Molecular cloning and characterization of PnbHLH1 transcription factor in Panax notoginseng. Molecules. 2017;22:1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jiang L, Yu YL, Cui XM. et al. Synergistic effect on the biosynthesis of Panax notoginseng saponins by overexpressing a transcription factor PnbHLH and RNA interference of cycloartenol synthase gene. China J Chin Mater Med. 2021;46:94–102 [DOI] [PubMed] [Google Scholar]
- 105. Jiang S, Yu YL, Jiang L. et al. Effect of transcription factor PnWRKY1 on the biosynthesis of Panax notoginseng saponins. Acta Bot Boreal Occident Sin. 2019;39:0430–8 [Google Scholar]
- 106. Deng B, Huang Z, Ge F. et al. An AP2/ERF family transcription factor PnERF1 raised the biosynthesis of saponins in Panax notoginseng. J Plant Growth Regul. 2017;36:691–701 [Google Scholar]
- 107. Lin T, Du J, Zheng X. et al. Comparative transcriptome analysis of MeJA-responsive AP2/ERF transcription factors involved in notoginsenosides biosynthesis. 3 Biotech. 2020;10:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Shannon P, Markiel A, Ozier O. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wei RC, Qiu DY, Wilson IW. et al. Identification of novel and conserved microRNAs in Panax notoginseng roots by high-throughput sequencing. BMC Genomics. 2015;16:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zheng Y, Chen K, Xu ZN. et al. Small RNA profiles from Panax notoginseng roots differing in sizes reveal correlation between miR156 abundances and root biomass levels. Sci Rep. 2017;7:9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zheng H, Fu XQ, Shao J. et al. Transcriptional regulatory network of high-value active ingredients in medicinal plants. Trends Plant Sci. 2023;28:429–46 [DOI] [PubMed] [Google Scholar]
- 112. Wang F, Xu T, Guo Q. et al. Studies on response of Alternanthere philoxeroides to stress of Pb and Cd. Environ Sci Technol. 2013;36:8–16 [Google Scholar]
- 113. Zhu ML, Zeng XC, Jiang YX. et al. Effect on the expression of key enzymes DS and P450 genes in the medicinal composition synthesis of Panax notoginseng under cadmium stress. Chin Tradit Herb Drugs. 2016;47:4428–32 [Google Scholar]
- 114. Kan Q. The Mechanisms of NO in Cd Accumulation and the Saponins Synthesis in Panax Notoginseng Roots. Kunming: Kunming University of Science and Technology, 2016: [Google Scholar]
- 115. Li YQ, Kong DX, Fu Y. et al. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol Biochem. 2020;148:80–9 [DOI] [PubMed] [Google Scholar]
- 116. Xu Y, Zhu MJ, Feng YB. et al. Panax notoginseng-microbiota interactions: from plant cultivation to medicinal application. Phytomedicine. 2023;119:154978 [DOI] [PubMed] [Google Scholar]
- 117. Wei GF. Study on the Quality Ecology of Panax Notoginseng. Jinan: Shandong University of Traditional Chinese Medicine, 2019 [Google Scholar]
- 118. Ran YY, Liang SW, Weng J. et al. Relationships between saponin component contents in Panax notoginseng taproot and ecological factors. J Agro-Environ Sci. 2022;41:1908–16 [Google Scholar]
- 119. Jin H, Cui XM, Zhu Y. et al. Effects of meteorological conditions on the quality of radix notoginseng. Southwest China J Agric Sci. 2005;18:825–8 [Google Scholar]
- 120. Xia PG, Guo HB, Zhao HG. et al. Optimal fertilizer application for Panax notoginseng and effect of soil water on root rot disease and saponin contents. J Ginseng Res. 2016;40:38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mao YC, Chen X, Yan KJ. et al. Multi-algorithm cooperation comprehensive research of bZIP genes under nitrogen stress in Panax notoginseng. Gene. 2022;841:146768 [DOI] [PubMed] [Google Scholar]
- 122. Ou XH, Cui XM, Zhu DW. et al. Lowering nitrogen and increasing potassium application level can improve the yield and quality of Panax notoginseng. Front. Plant Sci. 2020;21:595095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ou XH, Li SP, Liao PR. et al. The transcriptome variations of Panax notoginseng roots treated with different forms of nitrogen fertilizers. BMC Genomics. 2019;20:965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Cun Z, Wu HM, Zhang JY. et al. High nitrogen inhibits biomass and saponins accumulation in a medicinal plant Panax notoginseng. Peer J. 2023;11:e14933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hu FX, Zhong JJ. Role of jasmonic acid in alteration of ginsenoside heterogeneity in elicited cell cultures of Panax notoginseng. J Biosci Bioeng. 2007;104:513–6 [DOI] [PubMed] [Google Scholar]
- 126. Wang W, Zhao ZJ, Xu YF. et al. Efficient induction of ginsenoside biosynthesis and alteration of ginsenoside heterogeneity in cell cultures of Panax notoginseng by using chemically synthesized 2-hydroxyethyl jasmonate. Appl Microbiol Biotechnol. 2006;70:298–307 [DOI] [PubMed] [Google Scholar]
- 127. Yue CJ, Zhong JJ. Manipulation of ginsenoside heterogeneity of Panax notoginseng cells in flask and bioreactor cultivations with addition of phenobarbital. Bioprocess Biosyst Eng. 2008;31:95–100 [DOI] [PubMed] [Google Scholar]
- 128. Wang W, Zhang ZY, Zhong JJ. Enhancement of ginsenoside biosynthesis in high-density cultivation of Panax notoginseng cells by various strategies of methyl jasmonate elicitation. Appl Microbiol Biotechnol. 2005;67:752–8 [DOI] [PubMed] [Google Scholar]
- 129. Hu FX, Zhong JJ. Jasmonic acid mediates gene transcription of ginsenoside biosynthesis in cell cultures of Panax notoginseng treated with chemically synthesized 2-hydroxyethyl jasmonate. Process Biochem. 2008;43:113–8 [Google Scholar]
- 130. Yao L, Wang SH, Liang WX. et al. Screening and evaluation of adventitious root lines of Panax notoginseng by morphology, gene expression, and metabolite profiles. Appl Microbiol Biotechnol. 2019;103:4405–15 [DOI] [PubMed] [Google Scholar]
- 131. Li JX, Wang J, Wu XL. et al. Jasmonic acid and methyl dihydrojasmonate enhance saponin biosynthesis as well as expression of functional genes in adventitious root of Panax notoginseng F.H. Chen. Biotechnol Appl Biochem. 2017;64:225–38 [DOI] [PubMed] [Google Scholar]
- 132. Yang Y, Ge F, Sun Y. et al. Strengthening triterpene saponins biosynthesis by over-expression of farnesyl pyrophosphate synthase gene and RNA interference of cycloartenol synthase gene in Panax notoginseng cells. Molecules. 2017;22:581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Yang Y, Liu DQ, Ge F. et al. Effect of over-expressing farnesyl pyrophosphate synthase (FPS) gene of Panax notoginseng cell on saponin synthesis. Mod Food Sci Technol. 2015;31:59–64 [Google Scholar]
- 134. Zhang X, Yu YL, Jiang S. et al. Oleanane-type saponins biosynthesis in Panax notoginseng via transformation of β-amyrin synthase gene from Panax japonicus. J Agric Food Chem. 2019;67:1982–9 [DOI] [PubMed] [Google Scholar]
- 135. Dai ZB, Wang BB, Liu Y. et al. Producing aglycons of ginsenoside in bakers’ yeast. Sci Rep. 2014;4:3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Zhao FL, Du YH, Bai P. et al. Enhancing Saccharomyces cerevisiae reactive oxygen species and ethanol stress tolerance for high-level production of protopanoxadiol. Bioresour Technol. 2017;227:308–16 [DOI] [PubMed] [Google Scholar]
- 137. Wang PP, Wei W, Ye W. et al. Synthesizing ginsenoside Rh2 in Saccharomyces cerevisiae cell factory at high-efficiency. Cell Discov. 2019;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hu ZF, Gu AD, Liang L. et al. Construction and optimization of microbial cell factories for sustainable production of bioactive dammarenediol-II glucosides. Green Chem. 2019;21:3286–99 [Google Scholar]
- 139. Yan X, Fan Y, Wei W. et al. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wang D, Wang JH, Shi YS. et al. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab Eng. 2020;61:131–40 [DOI] [PubMed] [Google Scholar]
- 141. Liang HC, Hu ZF, Zhang TT. et al. Production of a bioactive unnatural ginsenoside by metabolically engineered yeasts based on a new UDP-glycosyltransferase from Bacillus subtilis. Metab Eng. 2017;44:60–9 [DOI] [PubMed] [Google Scholar]
- 142. Li XD, Wang YM, Fan ZJ. et al. High-level sustainable production of the characteristic protopanaxatriol-type saponins from Panax species in engineered Saccharomyces cerevisiae. Metab Eng. 2021;66:87–97 [DOI] [PubMed] [Google Scholar]
- 143. Yang EJ, Kim TH, Shin KC. et al. Complete conversion of all typical glycosylated protopanaxatriol ginsenosides to aglycon protopanaxatriol by combined bacterial β-glycosidases. AMB Express. 2018;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Li YF, Liang YZ, Cui XM. et al. Production of minor ginsenosides from Panax notoginseng flowers by Cladosporium xylophilum. Molecules. 2022;27:6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Quan LH, Min JW, Sathiyamoorthy S. et al. Biotransformation of ginsenosides Re and Rg1 into ginsenosides Rg2 and Rh1 by recombinant β-glucosidase. Biotechnol Lett. 2012;34:913–7 [DOI] [PubMed] [Google Scholar]
- 146. Wei GF, Chen ZJ, Wang B. et al. Endophytes isolated from Panax notoginseng converted ginsenosides. Microb Biotechnol. 2021;14:1730–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Guo CL, Yang XY, Chen ZM. et al. The content determination of biotransformation of Rb1 in the total saponins of Panax notoginseng by a plant endophyte Coniochaeta sp. Chin Med Mater. 2016;39:1075–8 [PubMed] [Google Scholar]
- 148. Fei LY. Screening of Stains for Main Saponins Transformation Form Flower and Root of Panax Notoginseng and Identification of its Transformation Products. Kunming University of Science and Technology; 2023: [Google Scholar]
- 149. Tan JS, Yeo CR, Popovich DG. Fermentation of protopanaxadiol type ginsenosides (PD) with probiotic Bifidobacterium lactis and Lactobacillus rhamnosus. Appl Microbiol Biotechnol. 2017;101:5427–37 [DOI] [PubMed] [Google Scholar]
- 150. Chang KH, Jo MN, Kim KT. et al. Evaluation of glucosidases of Aspergillus niger strain comparing with other glucosidases in transformation of ginsenoside Rb1 to ginsenosides Rg3. J Ginseng Res. 2014;38:47–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Murthy HN, Georgiev MI, Kim YS. et al. Ginsenosides: prospective for sustainable biotechnological production. Appl Microbiol Biotechnol. 2014;98:6243–54 [DOI] [PubMed] [Google Scholar]
- 152. Chen Q, Liu DQ, Qu Y. et al. Construction of plant cell factory for biosynthesis of ginsenoside Rh2 in tobacco. Ind Crop. Prod. 2023;192:116057 [Google Scholar]
- 153. Luo XZ, Reiter MA, d’Espaux L. et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature. 2019;567:123–6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number of the RNA sequencing data is PRJNA488357.