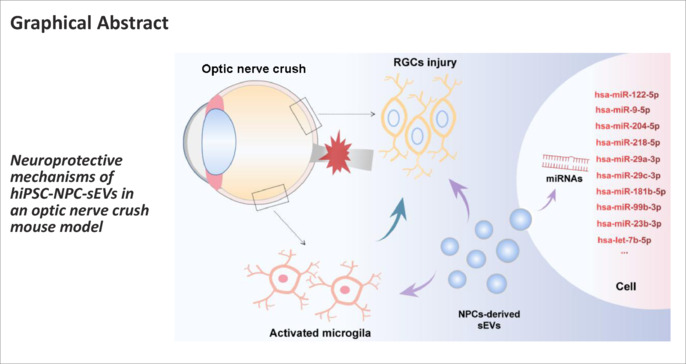
Keywords: exosome, miRNA, neural progenitor cell, neurodegeneration, neuroinflammation, neuroprotection, optic nerve crush, optic neuropathy, retinal ganglion cell, small extracellular vesicles
Abstract
Several studies have found that transplantation of neural progenitor cells (NPCs) promotes the survival of injured neurons. However, a poor integration rate and high risk of tumorigenicity after cell transplantation limits their clinical application. Small extracellular vesicles (sEVs) contain bioactive molecules for neuronal protection and regeneration. Previous studies have shown that stem/progenitor cell-derived sEVs can promote neuronal survival and recovery of neurological function in neurodegenerative eye diseases and other eye diseases. In this study, we intravitreally transplanted sEVs derived from human induced pluripotent stem cells (hiPSCs) and hiPSCs-differentiated NPCs (hiPSC-NPC) in a mouse model of optic nerve crush. Our results show that these intravitreally injected sEVs were ingested by retinal cells, especially those localized in the ganglion cell layer. Treatment with hiPSC-NPC-derived sEVs mitigated optic nerve crush-induced retinal ganglion cell degeneration, and regulated the retinal microenvironment by inhibiting excessive activation of microglia. Component analysis further revealed that hiPSC-NPC derived sEVs transported neuroprotective and anti-inflammatory miRNA cargos to target cells, which had protective effects on RGCs after optic nerve injury. These findings suggest that sEVs derived from hiPSC-NPC are a promising cell-free therapeutic strategy for optic neuropathy.
Introduction
Retinal ganglion cell (RGC) degeneration leads to irreversible visual impairment, and is a common feature of glaucoma, traumatic optic neuropathy, inherited optic neuropathy, and other types of optic atrophy. RGCs transmit visual signals to the brain via the optic nerve, and studies suggest that RGCs cannot regenerate after injury, resulting in permanent visual impairment (Guo et al., 2021; Kerschensteiner, 2022; Liu et al., 2023). The mechanisms of RGC degeneration are complex, and include genetics, oxidative stress, apoptosis, hypoxia-ischemia, mitochondrial dysfunction, gliosis, and inflammation (Ahmed et al., 2022; Fu et al., 2022; Hu et al., 2022; Ju and Tam, 2022; Rong et al., 2022; Wang et al., 2022a; Newman et al., 2023; Zhang et al., 2023). Unfortunately, there are limited effective therapeutic strategies for the treatment of optic neuropathy. Therefore, exploring novel treatment strategies is urgently needed.
Human induced pluripotent stem cells (hiPSCs) are invaluable tools with broad application prospects for the therapy of various human diseases, such as neural regeneration. hiPSCs can differentiate into various specialized cells or tissue organoids under specific conditions without ethical restrictions. Accordingly, hundreds of clinical trials using hiPSCs for human disease therapy are ongoing (Yokobayashi et al., 2021; Sugimoto et al., 2022; Wang et al., 2022b; Ito et al., 2023). Induced pluripotent stem cell (iPSC)-derived mesenchymal stem cells (MSCs) can promote the survival of neurons in mice with a mitochondrial complex I defect and in a rat model of hypoxic-ischemic encephalopathy (Jiang et al., 2019; Huang et al., 2022). In particular, neural progenitor cells (NPCs), or neural stem cells, differentiate into nervous system-related cell types, which gives them advantages for the treatment of neural degenerative diseases (Liu et al., 2019; Fischer et al., 2020; Baloh et al., 2022). iPSC-derived neural stem cells/NPCs show directional differentiation potential into neurons, and play important roles in degenerative diseases of the central nervous system (Strnadel et al., 2018; Huang and Zhang, 2019; Van Gelder et al., 2022). However, studies have suggested a poor rate of cell integration after cell transplantation, and increasing evidence indicates that transplanted cells play roles through nutrition and the microenvironment (Shi et al., 2018; Gokoffski et al., 2020; Yuan et al., 2022). Notably, stem cell transplantation has tumorigenic and immunogenic risks (Goldman, 2016; Yamanaka, 2020). Therefore, we have focused on small extracellular vesicles (sEVs), which are secreted from stem cells.
sEVs are surrounded by a phospholipid bilayer membrane of 30–150 nm diameter, and contain biologically active factors from the parent cells, including RNAs, proteins, and lipids (Gandham et al., 2020; Kalluri and LeBleu, 2020). These bioactive factors in sEVs can be delivered into recipient cells and act as modulators of pathophysiological processes (Channon et al., 2022; Chong et al., 2022). Moreover, sEVs have emerged as important paracrine regulators in local tissues, and also act as endocrine mediators of systemic regulation (Sheller-Miller et al., 2019). Numerous studies have revealed that stem/progenitor cell-derived sEVs have therapeutic effects in various diseases without immunogenicity or obvious side effects (Kalluri and LeBleu, 2020; Ding et al., 2021). In ocular neurodegenerative diseases and ocular diseases, neuron survival and nerve function are rescued by stem/progenitor cell-derived sEVs (Mead and Tomarev, 2017; Pan et al., 2019; Cui et al., 2021). Thus, sEVs have potential as a promising cell-free biological therapeutic strategy for neurodegenerative diseases.
In this study, we aimed to examine the protective effects of sEVs derived from hiPSCs and NPCs on RGC degeneration, and explore the potential mechanism. hiPSCs were directly differentiated into NPCs, and hiPSC- and NPC-derived sEVs were collected and characterized. These two types of sEVs were then intravitreally administered to verify their neuroprotective effects in a mouse model of optic nerve crush (ONC). Finally, a set of neuroprotective miRNAs were detected and analyzed by miRNA sequencing.
Methods
Animals
All animal experiments were performed following the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research (Association for Research in Vision and Ophthalmology, 2021), and were approved by the Animal Care and Use Committee of Wenzhou Medical University on November 27, 2022 (approval No. wydw2022-0735). C57BL/6JNifdc mice were purchased from Beijing Vital River Laboratory (Beijing, China; license No. SCXK (Jing) 2021-0011). Thy1-GFP and CX3CR1-GFP mice on a C57BL/6J background were obtained from Nanjing Biomedical Research Institute of Nanjing University (Nanjing, China) and Shanghai Model Organisms Center (Shanghai, China), respectively. Mice were maintained on a 12-hour dark/light cycle with good ventilation at 22°C, and provided standard laboratory chow and water, with up to five animals in every cage. Activation of microglia may be affected by the estrogen cycle of female mice, therefore male mice aged 2–3 months (20–26 g) were used in this study. For all experiments, experimenters and observers were blinded to group assignment and outcome assessment. The number distribution of mice in each experiment is shown in Additional Figure 1 (2.3MB, tif) .
Cell culture and differentiation
Cell culture and differentiation were performed as described in previous studies (Kirkeby et al., 2012). Briefly, hiPSCs were purchased from SiDanSai Biotech, Shanghai, China (Stock No. 0232-100) and verified by Guangzhou Institutes of Biomedicine and Health (Xue et al., 2013). Cells were cultured to 70–80% confluence with Essential 8 (A1517001, Gibco, Billings, MT, USA) and Matrigel (356234, Corning, Corning, NY, USA). After washing with sterilized phosphate-buffered saline (PBS), medium was replaced with neural induction medium (NIM) composed of DMEM/F12 1:1 Neurobasal A (10888022, Gibco) with 2% B27 supplement (12587010, Gibco), 1% N2 supplement (17502048, Gibco), 1% non-essential amino acids (11140050, Gibco), 1% Glutamax (35050061, Gibco), 10 µM SB431542 (S1067, Selleck, Houston, TX, USA), and 0.1 µM LDN193189 (S2618, Selleck). Cells were cultured at 37°C and 5% CO2 for 10 days. Then, NIM was changed to neural expansion medium composed of DMEM/F12 1:1 neurobasal A with 2% B27, 1% N2, 1% non-essential amino acids, 1% Glutamax, 10 ng/mL basic fibroblast growth factor (10014-HNAE, SinoBio Inc., Beijing, China), and 10 ng/mL epidermal growth factor (10605-HNAE, SinoBio Inc.) for 7 days. The medium was changed and supernatant collected every day.
sEVs isolation, characterization, and labeling
sEVs of hiPSCs and NPCs were isolated from serum-free cell culture medium as previously reported (Gurunathan et al., 2019). Briefly, the medium was collected and sequentially centrifuged at 300 × g for 10 minutes, 2000 × g for 10 minutes, and 10,000 × g for 30 minutes at 4°C to remove whole cells and debris. After filtration using a 0.22 μm pore filter, the supernatant was ultracentrifuged at 100,000 × g at 4°C for 70 minutes using a Beckman Optima XE-100 ultracentrifuge (Beckman, Bria, CA, USA), and the pellet was obtained. The sizes and concentration of purified sEVs were determined by ZetaView nanoparticle tracking analysis (PMX 110, Particle Metrix, Meerbusch, Germany). The ultrastructure of sEVs was analyzed by transmission electron microscopy (TEM; H-7650, Hitachi Ltd., Tokyo, Japan). Western blotting was used to examine protein expression of ALIX, CD9, TSG101, and calnexin in sEVs. Pellets were resuspended in 100 µL sterile PBS, and 50 µL sEVs were labeled with PKH67 Fluorescent Cell Linker Kits (PKH67GL, Sigma-Aldrich, Darmstadt, Germany).
Optic nerve crush and sEV administration
Mice were anesthetized by intraperitoneal injection of 30 mg/kg 0.3% sodium pentobarbital (P3761, Sigma-Aldrich) with 5 mg/kg xylazine (Aladdin, Shanghai, China) and 0.5% proparacaine hydrochloride (Alcon Laboratories, Fort Worth, TX, USA). ONC was performed as previously described (Cameron et al., 2020). Briefly, the optic nerve (both sides) was exposed and crushed using forceps (0203-N6-PO, Dumont, Montignez, Switzerland), 0.5 mm posterior to the lamina cribrosa. Then immediately after the crush, 200 µg (roughly 2 × 1011 particles) hiPSC- or NPC-sEVs in PBS were intravitreally injected using a 33 g Hamilton syringe (Hamilton Company, Reno, NV, America). Ofloxacin eye ointment (14200059683, Shenyang Xingqi Pharmaceutical Company, Shenyang, China) was normally administered after surgery.
Immunofluorescence staining
After washing three-times in PBS, hiPSCs or NPCs were fixed in 4% paraformaldehyde for 15 minutes and permeabilized in PBS with 0.5% Triton X-100 for 15 minutes. Mice were anesthetized as described above and sacrificed by cervical dislocation. Eyeball cryosections were prepared using a cryostat (CM1950, Leica, Nussloch Germany) at a thickness of 12 μm. Then, samples were blocked in blocking buffer (PBS with 10% bovine serum albumin and 0.1% Triton X-100) for 1 hour and incubated overnight at 4°C with primary antibody. Cells were then incubated with secondary antibody at 37°C for 2 hours. Antibody information is shown in Additional Table 1. After washing in PBS, samples were mounted in antifade mounting medium with 4′,6-diamidino-2-phenylindole (P0131, Beyotime Biotechnology, Shanghai, China).
Additional Table 1.
Antibodies used in this study.
| Name | Full name | Species | Dilution | Source | Catalog No. | RRID | Experiment |
|---|---|---|---|---|---|---|---|
| SOX-2 | SRY-box transcription factor 2 | Rabbit | 1:200 | Thermo, Waltham, MA, USA | PA1-094 | AB_2539862 | IF |
| Tra-1-60 | Tumor-related antigen-1-60 | Mouse | 1:200 | Chemicon, Moses Lake, Washington WA, USA | MAB4360 | AB_2119183 | IF |
| PAX6 | Paired box 6 | Mouse | 1:200 | Thermo Fisher | MA1-109 | AB_2536820 | IF |
| NESTIN | NESTIN | Mouse | 1:200 | Thermo Fisher | MA1-110 | AB_2536821 | IF |
| Iba1 | Ionized calcium-bin ding adapter molecule 1 | Rabbit | 1:400 | Wako Pure Chemical Industries, Tokyo, Japan | 019-19741 | AB_839504 | IF |
| RBPMS | RNA binding protein with multiple splicing | Rabbit | 1:100 0 | Proteintech Group, Chicago, IL, USA | 15187-1-AP | AB_2238431 | IF |
| GFAP | Glial fibrillary acidic protein | Rabbit | 1:1000 | Bioss Biotechnology, Beijing, China | bs-0199R-1 | IF | |
| Goat anti-rabbit IgG H&L/Alexa Fluor 488 | Goat Antianti-Rabbit rabbit | 1:500 | Bioss Biotechnology | bs-0295G-A F488 | IF | ||
| Goat anti-mouse IgG H&L/Alexa Fluor 488 | Goat Antianti-Mouse mouse | 1:500 | Bioss Biotechnology | bs-0296G-AF488 | IF | ||
| ALIX | Apoptosis-li nked gene 2-interactin g protein X | Rabbit | 1:500 | PTM BIO, Hangzhou, China | PTM-6407 | WB | |
| TSG101 | Tumor susceptibility 101 | Mouse | 1:500 | PTM BIO | PTM-5108 | WB | |
| SDCBP | Syndecan binding protein | Rabbit | 1:500 | BBI Life Sciences Corporation , Hong Kong, China | D223488-0025 | WB | |
| Calnexin | Calnexin | Rabbit | 1:500 | Proteintech Group | 10427-2-AP | AB_2069033 | WB |
| Horseradish peroxidase-la beled goat anti-rabbit | Goat Antianti-Rabbit rabbit | 1:1000 | Beyotime, Shanghai, China | A0208 | WB | ||
| IgG (H+L) Horseradish peroxidase-la beled goat anti-mouse IgG(H+L) | Goat Antianti-Mouse mouse | 1:1000 | Beyotime | A0216 | WB |
IF: Immunofluorescence; WB: western blotting.
Western blot assay
The concentration of sEVs was quantified using the Pierce BCA Protein Assay Kit (23225, Thermo Fisher Scientific, Waltham, MA, USA). Equal amounts of samples were electrophoresed in sodium dodecyl sulfate-polyacrylamide gels (10%; P0052A, Beyotime Biotechnology) and transferred to polyvinylidene fluoride membranes (FFP39, Beyotime Biotechnology). Blots were blocked in 5% milk for 1 hour and incubated overnight with primary antibodies. Anti-mouse or anti-rabbit immunoglobulin G and horseradish peroxidase-conjugated antibodies were used as secondary antibodies, and incubated for 2 hours at room temperature. Blots were developed with a chemiluminescence reagent (1863096, Thermo Fisher Scientific), and images were captured by Invitrogen iBright 1500 (iBright FL1500, Thermo Fisher Scientific). Gray values of bands were then analyzed by ImageJ software (Version 1.8.0; National Institutes of Health, Bethesda, MD, USA; Schneider et al., 2012). Antibody information is shown in Additional Table 1.
Optical coherence tomography
Optical coherence tomography (OCT) was performed on anesthetized animals at 7 and 14 days post-ONC. Pictures of the retina around the optic nerve head were taken by Micron IV Retinal Imaging Microscope (Phoenix Research Labs, Pleasanton, CA, USA). The thickness of the ganglion cell complex (GCC; consisting of a retinal nerve fiber layer, a ganglion cell layer, and an inner plexiform layer), which corresponds exactly to the anatomical distribution of RGCs in the retina (Nakano et al., 2011), was then measured by ImageJ.
Hematoxylin-eosin staining
Anesthetized mice were transcardially perfused with 4% paraformaldehyde. The eyes were collected and fixed in 4% paraformaldehyde for 48 hours at 4°C. Fixed eyes were then dehydrated and embedded in paraffin. Specimens were cut into 5-mm thick retinal cross-sections. Paraffin tissue sections were dewaxed, rehydrated, and stained with hematoxylin and eosin (C0105S, Beyotime Biotechnology). The GCC thickness was measured using ImageJ.
Retinal flat mount analysis
Eyes were enucleated after the animals were sacrificed, and fixed in 4% paraformaldehyde for 20 minutes. The anterior portion of the eyes was removed, and then the lens was removed after fixation in 4% paraformaldehyde for 40 minutes. Retinas were separated from the eyes and cut into quarters with ophthalmic scissors. Next, whole retinas were placed on slides and mounted on coverslips. Eight images of each retinal area (central, paracentral, and peripheral) were captured. The average number of Thy1-GFP-RGCs (n = 8), and average number and grid-crossed points of CX3CR1-GFP-microglia (n = 5) were counted by ImageJ software.
Flash visual evoked potentials
Retinal functional variations were evaluated using an electrophysiological diagnostic system (RETI-Port21, Roland, Munich, Germany) at 7 days post-ONC. Light stimulation intensity was 3.0 cd·s/m2, stimulation frequency was 2 Hz, and stimulation number was 100. Three silver electrodes were inserted into anesthetized mice under the skin of the occipital bone, cheek, and tail. Waves were recorded from the eyes.
sEV component analysis
Total RNA was extracted using TRIzol Reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA quality checks were performed using a Nanodrop2000 (Thermo Fisher Scientific) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA samples with concentrations > 200 ng/μL, and absorbance at 260/280 nm (A260/A280) > 1.8 and < 2.2, and A260/A230 ≥ 2.0 were used for library construction. A miRNA-seq library was constructed using TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Briefly, after adding adapters at 3 and 5 terminations, miRNAs were reverse transcribed to 1st strand complementary DNA (cDNA), which was used for PCR amplification. PCR products were purified by 6% polyacrylamide gel electrophoresis, and the library insert size determined using the Agilent 2100 Bioanalyzer. Finally, qualified libraries were sequenced using the NovaSeq 6000 platform (Illumina), and 50 bp single-end reads were obtained.
Adapter sequences and low-quality reads of miRNA sequences were removed using cutadapt (v3.2, National Bioinformatics Infrastructure Sweden [NBIS], Uppsala University, Uppsala, Sweden; Kechin et al., 2017). Clean reads were then mapped to the latest miRbase (Faculty of Life Sciences, University of Manchester, Manchester, UK; Griffiths-Jones et al., 2006) and Rfam database (v14.4, European Bioinformatics Institute, Wellcome Genome Campus, Cambridge, UK; Kalvari et al., 2021) using Bowtie2 (v2.4.5, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA; Langmead and Salzberg, 2012) to identify annotated miRNAs. Sequences that did not overlap with any annotated sequences were classified as unannotated reads. MirDeep2 (v2.0.1.2, Berlin Institute for Medical Systems Biology [BIMSB], Hannoversche, Berlin, Germany; Friedländer et al., 2012) was used to identify novel miRNAs based on secondary structure, dicer enzyme cleavage site, and minimum free energy indices. Raw read counts estimated by MirDeep2 were used to analyze differentially expressed miRNAs (DEmiRNAs) in four stages using DEseq2 (v1.34.0, Genetics Department, Biostatistics Department, UNC-Chapel Hill, NC, USA; Love et al., 2014). miRNAs with thresholds of P value < 0.05 and log2(fold-change) > 1 or < –1 were considered as DEmiRNAs.
Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using clusterProfiler (v4.4.4, School of Basic Medical Sciences, Southern Medical University, Guangzhou, China; Yu et al., 2012). Enriched GO terms or KEGG pathways with P values < 0.05 were considered significant.
Statistical analysis
All experiments were repeated at least three times and no animals were excluded from analysis. All data were analyzed with the evaluator blinded to grouping. Student’s t-test (two groups) or one-way analysis of variance (three groups and more) followed by Tukey’s post hoc test was performed by GraphPad Prism (version 8.0.1 for Windows, GraphPad Software, Boston, MA, USA, www.graphpad.com). Differences were considered to be significant when P < 0.05.
Results
Isolation and characterization of sEVs from hiPSCs and hiPSC-NPCs
To obtain sEVs from different stages of stem cells, hiPSCs were directly differentiated into NPCs in serum-free medium using an established method (Figure 1A). The morphologies of hiPSCs and NPCs, as well as immunostaining of specific biomarkers (SOX2 and TRA-1-60 for hiPSCs, and PAX6 and NESTIN for NPCs), indicated typical characteristics (Figure 1B and C). hiPSC-sEVs and NPC-sEVs were isolated from cell culture supernatants using an ultracentrifugation method (Figure 1D). Characterization of sEVs was confirmed by nanoparticle tracking analysis, TEM, and western blotting. The sEVs were shown to have a typical cup-like structure by TEM analysis (Figure 1E). Nanoparticle tracking analysis indicated the mean size of sEVs was approximately 100 nm (Figure 1F). Western blot analysis further confirmed that sEVs positively expressed representative biomarkers (SDCBP, TSG101 and ALIX), while the negative biomarker, calnexin, was undetectable (Figure 1G).
Figure 1.
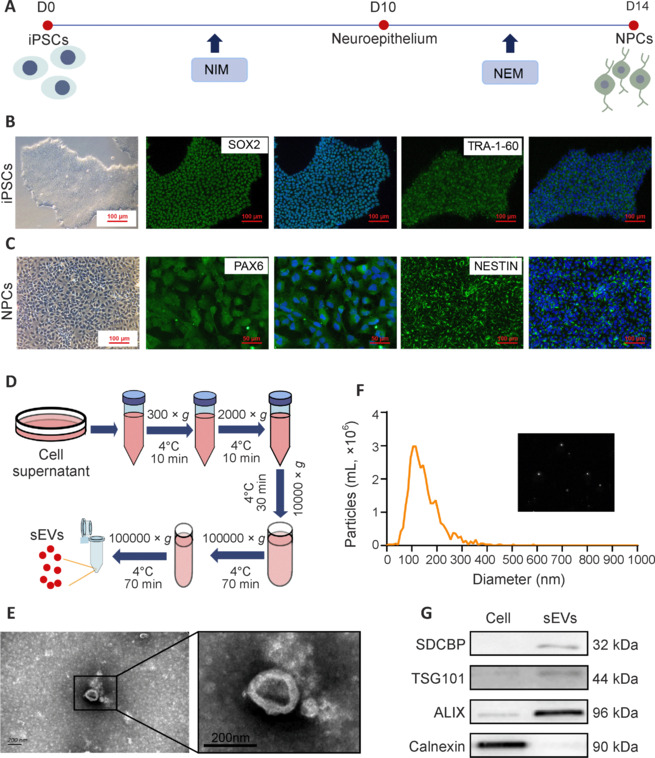
Isolation and characterization of sEVs from hiPSCs and hiPSC-NPCs.
(A) Schematic illustration of direct NPC differentiation of hiPSCs. (B, C) Representative morphology and immunostaining of hiPSCs and hiPSC-NPCs. hiPSCs showed a typical clone morphology with SOX2 and TRA-1-60 expression. hiPSC-NPCs showed a typical epithelium morphology with PAX6 and NESTIN expression. Scale bars: 100 μm. (D) Schematic of sEV isolation protocol. (E) Representative TEM images of NPC-sEVs showed a typical cup-like structure. Scale bar: 200 nm. (F) Size distribution of NPC-sEVs. The inset is a snapshot image of video tracking. (G) Western blot results for positive (SDCBP, TSG101, and ALIX) and negative (calnexin) biomarkers of NPC-sEVs and parental cells. ALIX: Apoptosis-linked gene 2-interacting protein X; hiPSC: human induced pluripotent stem cell; NEM: neural expansion medium; NIM: neural induction medium; NPC: neuronal progenitor cell; PAX6: paired box 6; SDCBP: syndecan binding protein; sEV: small extracellular vesicle; SOX2: SRY-box transcription factor 2; TRA-1-60: tumor-related antigen-1-60; TSG101: tumor susceptibility 101.
NPC-sEVs mitigate RGC degeneration after optic nerve injury
To investigate the uptake of sEVs by retinal cells, sEVs were labeled with fluorescently linked PKH67 and injected intravitreally. Flat mount analysis revealed green fluorescence in the retina 48 hours (Additional Figure 2 (8.1MB, tif) A) and 7 days (Additional Figure 2 (8.1MB, tif) B) after injection. Additionally, we noted that green fluorescent-labeled sEVs existed the vitreous cavity at 48 hours and were reduced by day 7 (Additional Figure 2 (8.1MB, tif) C). In contrast, increased green fluorescent-labeled sEVs were observed in the GCC layer on day 7 (Additional Figure 2 (8.1MB, tif) D). Immunostaining showed that NPC-sEVs were internalized by multiple cell types 48 hours after injection, including RGCs, microglia, and astrocytes (Additional Figure 2 (8.1MB, tif) E).
To evaluate the role of hiPSC-sEVs and NPC-sEVs in neuroprotection after optic nerve injury, sEVs were intravitreally injected into ONC mice, and RGC survival was analyzed on days 7 and 14 (Figure 2A). OCT analysis showed a significant increase in mean GCC layer thickness after NPC-sEV treatment, particularly from days 7 to 14 post-ONC (Figure 2B and C). Histopathological changes after optic nerve injury were also confirmed by hematoxylin-eosin staining, with NPC-sEV-treated retinas showing a thicker GCC layer on day 14 (Figure 2D and E). In addition, NPC-sEV-treated retinas contained more cells in the ganglion cell layer after ONC on both days 7 and 14 (Figure 2F).
Figure 2.
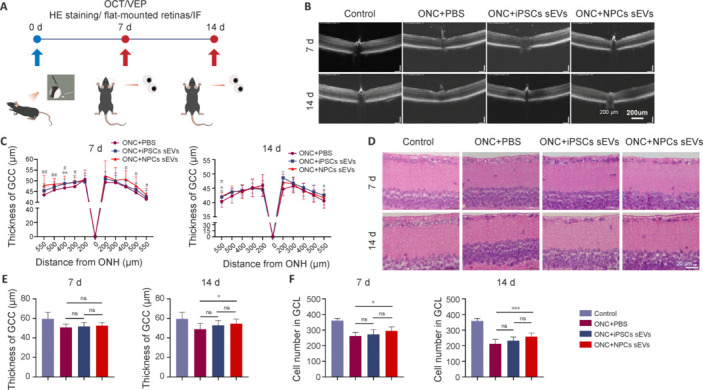
Neuroprotective effects of sEVs from hiPSCs and hiPSC-NPCs by OCT and HE staining.
(A) Schematic of animal experiments. (B) Representative OCT images of mice at 7 and 14 days post-ONC. Thickness of the retina decreased with time. Scale bar: 200 μm. (C) OCT statistical results of mean thickness of the GCC layer (n = 16). *P < 0.05, **P < 0.01, hiPSC-sEVs group vs. PBS group; #P < 0.05, ##P < 0.01, NPC-sEVs group vs. PBS group. (D) Representative image of the GCC layer from mice at 7 and 14 days post-ONC. Similar to the results obtained by OCT, thickness of the retina decreased with time. Scale bars: 20 μm. (E) Statistical results of mean thickness of the GCC layer (n = 12). (F) Quantification of mean number of cells in the GCL (n = 12). *P < 0.05, ***P < 0.001. Data are expressed as mean ± SEM (C) or mean ± SD (E and F), and were analyzed by Student’s t-test (C) or one-way analysis of variance followed by Tukey’s post hoc test (E and F). GCC: Ganglion cell complex; GCL: ganglion cell layer; HE: hematoxylin-eosin; hiPSC: human induced pluripotent stem cell; IF: immunofluorescence; iPSC: induced pluripotent stem cell; NPC: neural progenitor cell; ns: not significant; OCT: optical coherence tomography; ONC: optic nerve crush; ONH: optic nerve head; PBS: phosphate buffer saline; sEV: small extracellular vesicle; VEP: visual evoked potential.
We then conducted flat mount analysis for the whole retina in Thy1-GFP transgenic mice, which show specific GFP labeling in RGCs (Figure 3A and B). NPC-sEV treatment resulted in greater RGC survival than PBS and hiPSC-sEV treatments (Figure 3C and D). These results suggest that NPC-sEVs provide more significant RGC protection than hiPSC-sEVs after optic nerve injury.
Figure 3.
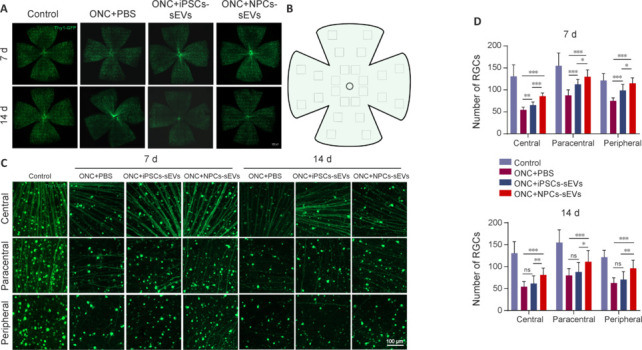
RGC survival of ONC mice with or without sEV treatment.
(A) Flat-mounted retinas of ONC mice with or without treatment with sEVs on day 7 and day 14 post-ONC. RGC degeneration was progressive after ONC. (B) Schematic diagram of different regions of flat-mounted retinas (central, paracentral, and peripheral). Created with Microsoft PowerPoint Professional 2019. (C) Representative image of central, paracentral, and peripheral retinal regions in ONC model mice with or without treatment with sEVs. Following treatment with hiPSC-sEVs, more RGCs survived in retinas on day 7 post-ONC. However, following treatment with NPC-sEVs, more RGCs survived in retinas on both days 7 and 14. Scale bars: 100 μm. (D) Quantification of mean number of RGCs in different retinal regions at 7 and 14 days (n = 8/group). Data are expressed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (one-way analysis of variance followed by Tukey’s post hoc test). hiPSC: Human induced pluripotent stem cell; iPSC: induced pluripotent stem cell; NPC: neural progenitor cell; ns: not significant; ONC: optic nerve crush; PBS: phosphate buffer saline; RGC: retinal ganglion cell; sEV: small extracellular vesicle.
NPC-sEVs suppress microglial activation and rescue visual impairment
Neuroinflammation is an important pathological process of neurodegenerative diseases. Microglia are key mediators for the progression of optic neuropathy. Suppressing the activation and proliferation of microglia can alleviate neuroinflammation and contribute to visual function restoration (Ramirez et al., 2017). To explore the potential role of NPC-sEVs in neuroinflammation, we used CX3CR1-GFP transgenic mice, which show specific GFP labeling in microglia, and performed anti-ionized calcium binding adaptor molecule 1 (IBA1) immunostaining of retinal sections, along with flat-mount analysis of the retina. Immunostaining of retinal sections showed a significant reduction in the number of migrated microglia in sEV-treated retinas on day 7 but not day 14 after ONC (Figure 4A and B). Flat-mounted retinas of CX3CR1-GFP mice further confirmed this reduced number of microglia in sEV-treated retinas on day 7, except for the paracentral region of hiPSC-sEV-treated retinas (Figure 4C–E). Moreover, analysis of grid-crossed points per microglia also revealed suppressed microglial activation after sEV treatment (Figure 4F).
Figure 4.
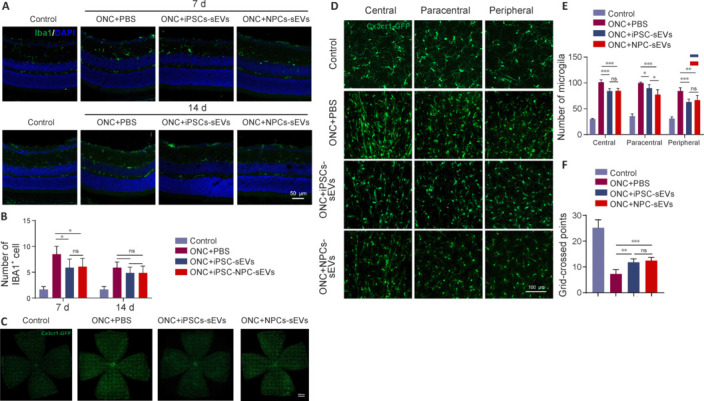
Anti-inflammatory effects of sEVs from hiPSCs and hiPSC-NPCs.
(A) Frozen sections of retinas on days 7 and 14. Migrated microglia were reduced in sEV-treated retinas on day 7 but not on day 14 after ONC. Scale bar: 50 μm. (B) Quantification of mean number of IBA1-positive cells per field (n = 5). (C) Flat-mounted retinas of ONC CX3CR1-GFP mice on day 7. The number of microglia increased in the retina after ONC, but decreased in sEV-treated retinas. (D) Representative images of central, paracentral, and peripheral retinal regions in the ONC model of CX3CR1-GFP mice with or without treatment with sEVs on day 7. The number of microglia and grid-crossed points per microglia were reduced in sEV-treated retinas on day 7. Scale bar: 100 μm. (E) Statistical results of mean number of microglia (CX3CR1-GFP positive cells) per field of different retinal regions (n = 5). (F) Quantification of mean grid-crossed points per microglia (n = 5). Data are expressed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (one-way analysis of variance followed by Tukey’s post hoc test). CX3CR1: CX3C chemokine receptor 1; DAPI: 4’,6-diamidino-2-phenylindole; GFP: green fluorescent protein; Iba1: ionized calcium-binding adapter molecule 1; hiPSC: human induced pluripotent stem cell; iPSC: induced pluripotent stem cell; NPC: neural progenitor cell; ns: not significant; ONC: optic nerve crush; PBS: phosphate buffer saline; sEVs: small extracellular vesicles.
To determine whether NPC-sEVs protect the visual function of ONC mice, we then conducted flash visual evoked potential analysis, and quantitatively measured the amplitude of the N1-P1 wave on day 7 after ONC (Figure 5A). The results showed that the amplitude of the N1-P1 wave was significantly restored by NPC-sEV treatment (Figure 5B and C). These results suggest that NPC-sEVs mitigate optic nerve injury and contribute to restoration of visual function.
Figure 5.
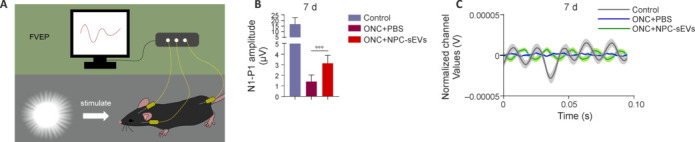
Protective effect of NPC-sEVs on visual function in ONC mice.
(A) Schematic diagram of FVEP. Created with Microsoft PowerPoint Professional 2019. (B) Mean amplitude of N1–P1 waves detected by FVEP on day 7 (n = 8). Data are expressed as mean ± SD. ***P < 0.001 (one-way analysis of variance followed by Tukey’s post hoc test). (C) Overlapping FVEP waves in ONC mice with or without treatment with NPC-sEVs on day 7. FVEP: Flash visual evoked potentials; NPC: neural progenitor cell; ONC: optic nerve crush; PBS: phosphate buffer saline; sEV: small extracellular vesicle.
NPC-sEVs play a neuroprotective role through specific miRNA delivery
sEVs play important roles in cell–cell communication by transporting bioactive molecules involved in the regulation of various biological processes into recipient cells. miRNA enrichment in sEVs play important roles in the pathophysiological processes of retinal diseases (Mead and Tomarev, 2020). Therefore, we subsequently performed component miRNA analysis by high-throughput sequencing of hiPSC-sEVs and NPC-sEVs. NPC-sEVs were found to have a total of 163 differentially expressed miRNAs compared with hiPSC-sEVs, as shown by volcano plot (Figure 6A). GO analysis of DEmiRNAs showed that these miRNAs were classified and enriched mainly in nervous system-related biological processes (Figure 6B). KEGG pathway analysis also found that these DEmiRNAs were involved in common neurodegenerative disorders as well as nervous system development- and growth-related pathways (Figure 6C). A heatmap shows the DEmiRNAs between the NPC-sEV and hiPSC-sEV groups, which were classified and enriched in the nervous system (Figure 6D). Among these miRNAs, 22 miRNAs attracted our attention because they are reported to participate in neuroprotection or anti-inflammation. We then selected the top 50 most abundant miRNAs in NPC-sEVs according to sequencing data. A total of 17 miRNAs considered neuroprotective and abundant in NPC-sEVs were identified by intersection analysis. These accounted for as many as 44.49% of the top 50 miRNAs (Figure 6E and F). Consequently, we speculated that NPC-sEVs exhibit neuroprotective effects on RGC degeneration through miRNA delivery.
Figure 6.
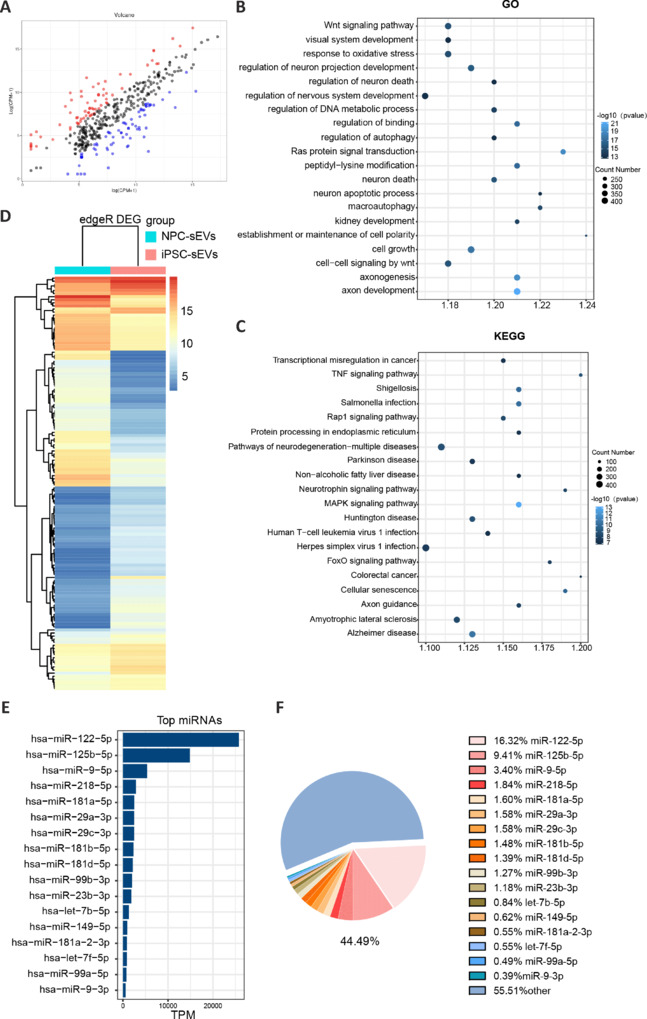
miRNA sequencing analysis of sEVs derived from hiPSCs and NPCs.
(A) Volcano plot of differentially expressed miRNAs from hiPSC-sEVs and NPC-sEVs. (B, C) KEGG and GO pathway enrichment analyses of differentially expressed miRNAs. (D) Heatmap of differentially expressed miRNAs related to neural pathways. (E) Expression rank of miRNAs in NPC-sEVs. (F) Pie chart showing the percentage of 17 neuroprotective miRNAs in the top 50 miRNAs in term of expression. DEG: Differentially expressed gene; GO: Gene Ontology; hiPSC: human induced pluripotent stem cell; KEGG: Kyoto Encyclopedia of Genes and Genomes; miRNA: microRNA; NPC: neural progenitor cell; sEV: small extracellular vesicle; TPM: tag per million.
Discussion
RGC protection and regeneration are long-term challenges for the clinical treatment of optic neuropathy. The pathogenesis of neurodegenerative diseases is complex, therefore treatment targeting a single gene may not meet clinical needs. Stem cell therapy is advantageous for neurodegenerative diseases because it can act on multiple targets. hiPSC-differentiated NPCs avoid the ethical issues of using human embryos, solve the problem of finding a source of human neural cells, and have beneficial treatment potential for nervous system disorders. Studies have shown that hiPSC-NPCs promote neuronal protection as well as RGC survival (Ju and Tam, 2022). However, few transplanted cells are integrated into tissue, and instead they are mainly involved in the secretion of neurotrophic factors and regulation of the microenvironment (Karagiannis et al., 2019). In view of this, we focused on sEVs, which are recognized as a cell-free therapeutic source to deliver biomolecules from their parent cells to recipient cells with good biocompatibility and low immunogenicity. Previous studies have shown that MSC-derived sEVs have potential for the treatment of neurodegenerative diseases (Mead et al., 2018; Hade et al., 2021). Compared with MSCs and iPSCs, which have pluripotency, NPCs can differentiate into cells of the nervous system. For this reason, we were interested in whether NPC-derived sEVs are more advantageous for neurodegenerative disease therapy.
In this study, we successfully developed a hiPSC-NPC differentiation system and obtained high-purity sEVs from cell supernatants. We found that sEVs crossed the inner limiting membrane and were internalized by ganglion cell layer cells (including RGCs, microglia, and astrocytes) as early as 48 hours after intravitreal injection, which is consistent with a previous study (Mathew et al., 2021). We found that treatment with hiPSC-NPC-sEVs significantly reduced RGC loss and partially restored visual function. Once the damage occurs in neurodegenerative diseases, the degeneration is progressive and difficult to stop. To maximize the therapeutic effect, sEV treatment was performed at the same time as injury. Even although RGCs were still observable in hiPSC-NPC-sEV-treated retina 14 days after ONC, the amplitude of flash visual evoked potentials had decreased to a very low level. More interestingly, hiPSC-NPC-sEV treatment caused significantly potent and durable neuroprotective effects compared with hiPSC-sEV treatment. It has been reported that MSCs play a major role through paracrine effects rather than cell replacement (Andrzejewska et al., 2021). Intercellular communication via sEVs may therefore play an important role in the nervous system (Budnik et al., 2016). Alternatively, the optic nerve lamina region is a NPC niche that secretes growth factors to promote neuronal extension (Bernstein et al., 2020). Thus, our novel study suggests that NPC-derived sEVs may transport bioactive molecules of parent cells and play roles in RGC neuroprotection. Considering that sEVs have good biocompatibility and low immunogenicity, even allogeneic hiPSC- or MSC-derived, NPC-sEV therapy in the clinic can effectively avoid immune rejection and other unexpected side effects.
Neuroinflammation in the central nervous system plays an essential role in protecting tissue from injury. However, uncontrolled and prolonged neuroinflammation leads to secondary damage and is a contributing factor to neurodegeneration (Ramirez et al., 2017). sEVs react to neural damage with morphological changes, proliferation, migration, and production of inflammatory cytokines, which further propagate neuroinflammation (Wei et al., 2019). Microglia are the principal modulatory cells of the nervous system (including the brain and retina) after injury, and play important roles in nutrition, protection, and repair of damaged neurons (Mundt et al., 2022). In this study, we found that hiPSC-NPC-sEVs minimize microglial activation, resulting in reconstruction of a microenvironment that benefits RGC survival. It was previously shown that sEVs derived from primary NPCs preserve photoreceptors in a mouse model of retinal degeneration by inactivating microglia (Bian et al., 2020). Interestingly, a report showed that MSCs treated with GW4869 (an inhibitor of sEV biogenesis) had no suppressive effect on glial activation, suggesting that the therapeutic role of MSCs in neural injury may occur through sEV secretion (Deng et al., 2021).
Next, we investigated the mechanisms of hiPSC-NPC-sEVs in RGC protection. Numerous studies have shown that sEVs derived from various stem cells are involved in neuroprotection by transporting miRNAs (Mead and Tomarev, 2020). Consequently, sEV cargo miRNAs were detected by high-throughput sequencing, and DEmiRNAs in hiPSC-NPC-sEVs were analyzed and compared with those in hiPSC-sEVs. We found miRNAs such as miR-181, miR-9, miR-125b-5p, and the let-7 family, which are reported to play important roles in microglial activation and neurogenesis (Bian et al., 2020; Fernández et al., 2020; Tian et al., 2021). Moreover, other miRNAs, such as miR-122-5p, miRNA-9-5p, miR-218-5p, and miRNA-29s are known to exert neuroprotective effects (Jayaram et al., 2015; Ma et al., 2023). Overall, upregulated miRNAs in hiPSC-NPC-sEVs were enriched in neuronal survival, neurogenesis-related biological processes, and TNF-, MAPK-, neurotrophin- and common neurodegenerative disorder-related signaling pathways (Ding et al., 2022). Twenty-two of the upregulated miRNAs are involved in the regulation of neuroprotection and anti-inflammation, and 17 out of 22 are the top 50 most abundant miRNAs. This analysis confirms that sEVs have a multitarget therapeutic effect on retinal cells through the transportation of cargo miRNA. We also investigated RGC protection of NPC-sEVs in ONC mice, but target delivery of sEVs to the optic nerve and the regeneration of RGC axons needs further investigation. Because the role of MSC-sEVs in neural protection is widely studied, a comparative study of MSC-sEVs and NPC-sEVs is essential for identifying the underlying effect and mechanism of neural protection. Additionally, we found a combination of 17 miRNAs in NPC-sEVs play a multitarget therapeutic effect in RGC protection. However, optimization of the miRNA complex and delivery of the optimized miRNA complex would be interesting for further study.
In summary, our novel study shows that intravitreous injection of hiPSC-NPC-sEVs were successfully taken up by retinal cells, which alleviated RGC degeneration, suppressed microglial activation, and contributed to visual function restoration after optic nerve injury. Additionally, multitarget regulation of neuroretinal protection and the inflammatory microenvironment through bioactive miRNAs is a potential therapeutic mechanism of hiPSC-NPC-sEVs. These findings suggest that hiPSC-NPC-sEVs may be a promising cell-free biological therapeutic strategy for the treatment of optic neuropathy.
Additional files:
Additional Figure 1 (2.3MB, tif) : The number distribution of mice in each experiment.
The number distribution of mice in each experiment.
CX3CR1: CX3C chemokine receptor 1; FVEP: flash visual evoked potentials; HE: hematoxylin-eosin; IF: immunofluorescence; iPSC: induced pluripotent stem cell; NPC: neural progenitor cell; RGC: retinal ganglion cell; OCT: optical coherence tomography; ONC: optic nerve crush; PBS: phosphate buffer saline; sEV: small extracellular vesicle.
Additional Figure 2 (8.1MB, tif) : Internalization of sEVs by retinal cells.
Internalization of sEVs by retinal cells.
(A, B) Flat-mounted retinas at 48 hours (A) and 7 days (B) after intravitreous injection (n = 3). Uniform distribution of the sEVs and internalization by retina cells were observed at each time point. sEVs were labeled with PKH67 (green). (C) Frozen sections of vitreous cavity (left of the white line) at 48 hours and 7 day (n = 3). More green PKH67-labeled sEVs (green) were observed in the vitreous cavity at 48 hours than day 7. (D) Frozen sections of retina at 48 hours and 7 day (n = 3). More green PKH67-labeled sEVs were observed in the retina on day 7 than 48 hours. (E) Immunostaining of frozen sections demonstrating that EVs were internalized by RGC (RBPMS, green, Alexa Fluor 488), microglia (Iba1, green, Alexa Fluor 488) and astrocyte (GFAP, green, Alexa Fluor 488) 48 hours after injection, and sEVs were labeled with PKH67 (red). The arrow indicated PKH67-labeled sEVs internalized in RBPMS positive RGC, Iba1 positive microglia and GFAP positive astrocyte. DAPI: 4',6-diamidino-2-phenylindole; GFAP: glial fibrillary acidic protein; Iba1: ionized calcium-binding adapter molecule 1; RBPMS: RNA binding protein with multiple splicing; RGC: retinal ganglion cell; sEV: small extracellular vesicle.
Additional Table 1: Antibodies used in this study.
Additional file 1: Open peer review report 1 (82.8KB, pdf) .
Funding Statement
Funding: This study was supported by the National Natural Science Foundation of China, No. 82271114; the Natural Science Foundation of Zhejiang Province of China, No. LZ22H120001 (both to ZLC).
Footnotes
Conflicts of interest: The authors declare no competing interests.
Data availability statement: All data generated or analyzed during this study are included in this published article and its Additional files.
Open peer reviewer: Alicia Mansilla, Universidad de Alcala, Spain.
P-Reviewer: Mansilla A; C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Yu J, Song LP; T-Editor: Jia Y
References
- Ahmed M, Kojima Y, Masai I. Strip1 regulates retinal ganglion cell survival by suppressing Jun-mediated apoptosis to promote retinal neural circuit formation. Elife. 2022;11:e74650. doi: 10.7554/eLife.74650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrzejewska A, Dabrowska S, Lukomska B, Janowski M. Mesenchymal stem cells for neurological disorders. Adv Sci (Weinh) 2021;8:2002944. doi: 10.1002/advs.202002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Association for Research in Vision and Ophthalmology ARVO Statement for the Use of Animals in Ophthalmic and Vision Research 2021 [Google Scholar]
- Baloh RH, et al. Transplantation of human neural progenitor cells secreting GDNF into the spinal cord of patients with ALS: a phase 1/2a trial. Nat Med. 2022;28:1813–1822. doi: 10.1038/s41591-022-01956-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein SL, Guo Y, Kerr C, Fawcett RJ, Stern JH, Temple S, Mehrabian Z. The optic nerve lamina region is a neural progenitor cell niche. Proc Natl Acad Sci U S A. 2020;117:19287–19298. doi: 10.1073/pnas.2001858117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian B, Zhao C, He X, Gong Y, Ren C, Ge L, Zeng Y, Li Q, Chen M, Weng C, He J, Fang Y, Xu H, Yin ZQ. Exosomes derived from neural progenitor cells preserve photoreceptors during retinal degeneration by inactivating microglia. J Extracell Vesicles. 2020;9:1748931. doi: 10.1080/20013078.2020.1748931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik V, Ruiz-Cañada C, Wendler F. Extracellular vesicles round off communication in the nervous system. Nat Rev Neurosci. 2016;17:160–172. doi: 10.1038/nrn.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron EG, Xia X, Galvao J, Ashouri M, Kapiloff MS, Goldberg JL. Optic nerve crush in mice to study retinal ganglion cell survival and regeneration. Bio Protoc. 2020;10:e3559. doi: 10.21769/BioProtoc.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channon LM, Tyma VM, Xu Z, Greening DW, Wilson JS, Perera CJ, Apte MV. Small extracellular vesicles (exosomes) and their cargo in pancreatic cancer: Key roles in the hallmarks of cancer. Biochim Biophys Acta Rev Cancer. 2022;1877:188728. doi: 10.1016/j.bbcan.2022.188728. [DOI] [PubMed] [Google Scholar]
- Chong MC, Silva A, James PF, Wu SSX, Howitt J. Exercise increases the release of NAMPT in extracellular vesicles and alters NAD(+) activity in recipient cells. Aging Cell. 2022;21:e13647. doi: 10.1111/acel.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Liu C, Huang L, Chen J, Xu N. Protective effects of intravitreal administration of mesenchymal stem cell-derived exosomes in an experimental model of optic nerve injury. Exp Cell Res. 2021;407:112792. doi: 10.1016/j.yexcr.2021.112792. [DOI] [PubMed] [Google Scholar]
- Deng CL, Hu CB, Ling ST, Zhao N, Bao LH, Zhou F, Xiong YC, Chen T, Sui BD, Yu XR, Hu CH. Photoreceptor protection by mesenchymal stem cell transplantation identifies exosomal MiR-21 as a therapeutic for retinal degeneration. Cell Death Differ. 2021;28:1041–1061. doi: 10.1038/s41418-020-00636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Wu Y, Dabas H, Hammarlund M. Activation of the CaMKII-Sarm1-ASK1-p38 MAP kinase pathway protects against axon degeneration caused by loss of mitochondria. Elife. 2022;11:e73557. doi: 10.7554/eLife.73557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Li Y, Sun Z, Han X, Chen Y, Ge Y, Mao Z, Wang W. Cell-derived extracellular vesicles and membranes for tissue repair. J Nanobiotechnology. 2021;19:368. doi: 10.1186/s12951-021-01113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández V, Martínez-Martínez M, Prieto-Colomina A, Cárdenas A, Soler R, Dori M, Tomasello U, Nomura Y, López-Atalaya JP, Calegari F, Borrell V. Repression of Irs2 by let-7 miRNAs is essential for homeostasis of the telencephalic neuroepithelium. EMBO J. 2020;39:e105479. doi: 10.15252/embj.2020105479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer I, Dulin JN, Lane MA. Transplanting neural progenitor cells to restore connectivity after spinal cord injury. Nat Rev Neurosci. 2020;21:366–383. doi: 10.1038/s41583-020-0314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Siggs OM, Knight LS, Staffieri SE, Ruddle JB, Birsner AE, Collantes ER, Craig JE, Wiggs JL, D’Amato RJ. Thrombospondin 1 missense alleles induce extracellular matrix protein aggregation and TM dysfunction in congenital glaucoma. J Clin Invest. 2022;132:e156967. doi: 10.1172/JCI156967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandham S, Su X, Wood J, Nocera AL, Alli SC, Milane L, Zimmerman A, Amiji M, Ivanov AR. Technologies and standardization in research on extracellular vesicles. Trends Biotechnol. 2020;38:1066–1098. doi: 10.1016/j.tibtech.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokoffski KK, Lam P, Alas BF, Peng MG, Ansorge HRR. Optic nerve regeneration: how will we get there? J Neuroophthalmol. 2020;40:234–242. doi: 10.1097/WNO.0000000000000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman SA. Stem and progenitor cell-based therapy of the central nervous system: hopes, hype, and wishful thinking. Cell Stem Cell. 2016;18:174–188. doi: 10.1016/j.stem.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhou J, Starr C, Mohns EJ, Li Y, Chen EP, Yoon Y, Kellner CP, Tanaka K, Wang H, Liu W, Pasquale LR, Demb JB, Crair MC, Chen B. Preservation of vision after CaMKII-mediated protection of retinal ganglion cells. Cell. 2021;184:4299–4314.e12. doi: 10.1016/j.cell.2021.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S, Kang MH, Jeyaraj M, Qasim M, Kim JH. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells. 2019;8:307. doi: 10.3390/cells8040307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hade MD, Suire CN, Suo Z. Mesenchymal stem cell-derived exosomes: applications in regenerative medicine. Cells. 2021;10:1959. doi: 10.3390/cells10081959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Meng S, Zhang Q, Song S, Tan C, Huang J, Chen D. Astrocyte derived TSP2 contributes to synaptic alteration and visual dysfunction in retinal ischemia/reperfusion injury. Cell Biosci. 2022;12:196. doi: 10.1186/s13578-022-00932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, U KP, Yang F, Ji Z, Lin J, Weng Z, Tsang LL, Merson TD, Ruan YC, Wan C, Li G, Jiang X. Human pluripotent stem cell-derived ectomesenchymal stromal cells promote more robust functional recovery than umbilical cord-derived mesenchymal stromal cells after hypoxic-ischaemic brain damage. Theranostics. 2022;12:143–166. doi: 10.7150/thno.57234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Zhang L. Neural stem cell therapies and hypoxic-ischemic brain injury. Prog Neurobiol. 2019;173:1–17. doi: 10.1016/j.pneurobio.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Morimoto S, Takahashi S, Okada K, Nakahara J, Okano H. Maiden voyage: induced pluripotent stem cell-based drug screening for amyotrophic lateral sclerosis. Brain. 2023;146:13–19. doi: 10.1093/brain/awac306. [DOI] [PubMed] [Google Scholar]
- Jayaram H, Cepurna WO, Johnson EC, Morrison JC. MicroRNA expression in the glaucomatous retina. Invest Ophthalmol Vis Sci. 2015;56:7971–7982. doi: 10.1167/iovs.15-18088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Xiong G, Feng H, Zhang Z, Chen P, Yan B, Chen L, Gandhervin K, Ma C, Li C, Han S, Zhang Y, Liao C, Lee TL, Tse HF, Fu QL, Chiu K, Lian Q. Donation of mitochondria by iPSC-derived mesenchymal stem cells protects retinal ganglion cells against mitochondrial complex I defect-induced degeneration. Theranostics. 2019;9:2395–2410. doi: 10.7150/thno.29422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Y, Tam KY. Pathological mechanisms and therapeutic strategies for Alzheimer’s disease. Neural Regen Res. 2022;17:543–549. doi: 10.4103/1673-5374.320970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Nawrocki EP, Ontiveros-Palacios N, Argasinska J, Lamkiewicz K, Marz M, Griffiths-Jones S, Toffano-Nioche C, Gautheret D, Weinberg Z, Rivas E, Eddy SR, Finn RD, Bateman A, Petrov AI. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021;49:D192–D200. doi: 10.1093/nar/gkaa1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis P, Takahashi K, Saito M, Yoshida Y, Okita K, Watanabe A, Inoue H, Yamashita JK, Todani M, Nakagawa M, Osawa M, Yashiro Y, Yamanaka S, Osafune K. Induced pluripotent stem cells and their use in human models of disease and development. Physiol Rev. 2019;99:79–114. doi: 10.1152/physrev.00039.2017. [DOI] [PubMed] [Google Scholar]
- Kechin A, Boyarskikh U, Kel A, Filipenko M. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol. 2017;24:1138–1143. doi: 10.1089/cmb.2017.0096. [DOI] [PubMed] [Google Scholar]
- Kerschensteiner D. Feature detection by retinal ganglion cells. Annu Rev Vis Sci. 2022;8:135–169. doi: 10.1146/annurev-vision-100419-112009. [DOI] [PubMed] [Google Scholar]
- Kirkeby A, Nelander J, Parmar M. Generating regionalized neuronal cells from pluripotency, a step-by-step protocol. Front Cell Neurosci. 2012;6:64. doi: 10.3389/fncel.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Michowski W, Kolodziejczyk A, Sicinski P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat Cell Biol. 2019;21:1060–1067. doi: 10.1038/s41556-019-0384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xue J, Liu C, Tang J, Wu S, Lin J, Han J, Zhang Q, Wu C, Huang H, Zhao L, Zhuo Y, Li Y. Selective deletion of zinc transporter 3 in amacrine cells promotes retinal ganglion cell survival and optic nerve regeneration after injury. Neural Regen Res. 2023;18:2773–2780. doi: 10.4103/1673-5374.373660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZX, Liu Z, Xiong HH, Zhou ZP, Ouyang LS, Xie FK, Tang YM, Wu ZD, Feng Y. MicroRNAs: protective regulators for neuron growth and development. Neural Regen Res. 2023;18:734–745. doi: 10.4103/1673-5374.353481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew B, Torres LA, Gamboa Acha L, Tran S, Liu A, Patel R, Chennakesavalu M, Aneesh A, Huang CC, Feinstein DL, Mehraeen S, Ravindran S, Roth S. Uptake and distribution of administered bone marrow mesenchymal stem cell extracellular vesicles in retina. Cells. 2021;10:730. doi: 10.3390/cells10040730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead B, Tomarev S. Bone marrow-derived mesenchymal stem cells-derived exosomes promote survival of retinal ganglion cells through miRNA-dependent mechanisms. Stem Cells Transl Med. 2017;6:1273–1285. doi: 10.1002/sctm.16-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead B, Tomarev S. Extracellular vesicle therapy for retinal diseases. Prog Retin Eye Res. 2020;79:100849. doi: 10.1016/j.preteyeres.2020.100849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead B, Amaral J, Tomarev S. Mesenchymal stem cell-derived small extracellular vesicles promote neuroprotection in rodent models of glaucoma. Invest Ophthalmol Vis Sci. 2018;59:702–714. doi: 10.1167/iovs.17-22855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt S, Greter M, Becher B. The CNS mononuclear phagocyte system in health and disease. Neuron. 2022;110:3497–3512. doi: 10.1016/j.neuron.2022.10.005. [DOI] [PubMed] [Google Scholar]
- Nakano N, Ikeda HO, Hangai M, Muraoka Y, Toda Y, Kakizuka A, Yoshimura N. Longitudinal and simultaneous imaging of retinal ganglion cells and inner retinal layers in a mouse model of glaucoma induced by N-methyl-D-aspartate. Invest Ophthalmol Vis Sci. 2011;52:8754–8762. doi: 10.1167/iovs.10-6654. [DOI] [PubMed] [Google Scholar]
- Newman NJ, Yu-Wai-Man P, Biousse V, Carelli V. Understanding the molecular basis and pathogenesis of hereditary optic neuropathies: towards improved diagnosis and management. Lancet Neurol. 2023;22:172–188. doi: 10.1016/S1474-4422(22)00174-0. [DOI] [PubMed] [Google Scholar]
- Pan D, Chang X, Xu M, Zhang M, Zhang S, Wang Y, Luo X, Xu J, Yang X, Sun X. UMSC-derived exosomes promote retinal ganglion cells survival in a rat model of optic nerve crush. J Chem Neuroanat. 2019;96:134–139. doi: 10.1016/j.jchemneu.2019.01.006. [DOI] [PubMed] [Google Scholar]
- Ramirez AI, de Hoz R, Salobrar-Garcia E, Salazar JJ, Rojas B, Ajoy D, López-Cuenca I, Rojas P, Triviño A, Ramírez JM. The role of microglia in retinal neurodegeneration: Alzheimer’s disease, Parkinson, and glaucoma. Front Aging Neurosci. 2017;9:214. doi: 10.3389/fnagi.2017.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong R, Zhou X, Liang G, Li H, You M, Liang Z, Zeng Z, Xiao H, Ji D, Xia X. Targeting cell membranes, depleting ROS by dithiane and thioketal-containing polymers with pendant cholesterols delivering necrostatin-1 for glaucoma treatment. ACS Nano. 2022;16:21225–21239. doi: 10.1021/acsnano.2c09202. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheller-Miller S, Trivedi J, Yellon SM, Menon R. Exosomes cause preterm birth in mice: evidence for paracrine signaling in pregnancy. Sci Rep. 2019;9:608. doi: 10.1038/s41598-018-37002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Wang Y, Li Q, Liu K, Hou J, Shao C, Wang Y. Immunoregulatory mechanisms of mesenchymal stem and stromal cells in inflammatory diseases. Nat Rev Nephrol. 2018;14:493–507. doi: 10.1038/s41581-018-0023-5. [DOI] [PubMed] [Google Scholar]
- Strnadel J, et al. Survival of syngeneic and allogeneic iPSC-derived neural precursors after spinal grafting in minipigs. Sci Transl Med. 2018;10:eaam6651. doi: 10.1126/scitranslmed.aam6651. [DOI] [PubMed] [Google Scholar]
- Sugimoto N, et al. iPLAT1: the first-in-human clinical trial of iPSC-derived platelets as a phase 1 autologous transfusion study. Blood. 2022;140:2398–2402. doi: 10.1182/blood.2022017296. [DOI] [PubMed] [Google Scholar]
- Tian T, Cao L, He C, Ye Q, Liang R, You W, Zhang H, Wu J, Ye J, Tannous BA, Gao J. Targeted delivery of neural progenitor cell-derived extracellular vesicles for anti-inflammation after cerebral ischemia. Theranostics. 2021;11:6507–6521. doi: 10.7150/thno.56367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder RN, Chiang MF, Dyer MA, Greenwell TN, Levin LA, Wong RO, Svendsen CN. Regenerative and restorative medicine for eye disease. Nat Med. 2022;28:1149–1156. doi: 10.1038/s41591-022-01862-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wiggs JL, Aung T, Khawaja AP, Khor CC. The genetic basis for adult onset glaucoma: Recent advances and future directions. Prog Retin Eye Res. 2022a;90:101066. doi: 10.1016/j.preteyeres.2022.101066. [DOI] [PubMed] [Google Scholar]
- Wang ZB, Wang ZT, Sun Y, Tan L, Yu JT. The future of stem cell therapies of Alzheimer’s disease. Ageing Res Rev. 2022b;80:101655. doi: 10.1016/j.arr.2022.101655. [DOI] [PubMed] [Google Scholar]
- Wei X, Cho KS, Thee EF, Jager MJ, Chen DF. Neuroinflammation and microglia in glaucoma: time for a paradigm shift. J Neurosci Res. 2019;97:70–76. doi: 10.1002/jnr.24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Cai X, Wang L, Liao B, Zhang H, Shan Y, Chen Q, Zhou T, Li X, Hou J, Chen S, Luo R, Qin D, Pei D, Pan G. Generating a non-integrating human induced pluripotent stem cell bank from urine-derived cells. PLoS One. 2013;8:e70573. doi: 10.1371/journal.pone.0070573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Pluripotent stem cell-based cell therapy-promise and challenges. Cell Stem Cell. 2020;27:523–531. doi: 10.1016/j.stem.2020.09.014. [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Yabuta Y, Nakagawa M, Okita K, Hu B, Murase Y, Nakamura T, Bourque G, Majewski J, Yamamoto T, Saitou M. Inherent genomic properties underlie the epigenomic heterogeneity of human induced pluripotent stem cells. Cell Rep. 2021;37:109909. doi: 10.1016/j.celrep.2021.109909. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Stewart KS, Yang Y, Abdusselamoglu MD, Parigi SM, Feinberg TY, Tumaneng K, Yang H, Levorse JM, Polak L, Ng D, Fuchs E. Ras drives malignancy through stem cell crosstalk with the microenvironment. Nature. 2022;612:555–563. doi: 10.1038/s41586-022-05475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, Liu JC, Zhou WJ, Yu YZ, Tang CC, Zou XL, Zou YP. Protective effect of stem cells on retinal ganglion cell regeneration. Zhongguo Zuzhi Gongcheng Yanjiu. 2023;27:1593–1602. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The number distribution of mice in each experiment.
CX3CR1: CX3C chemokine receptor 1; FVEP: flash visual evoked potentials; HE: hematoxylin-eosin; IF: immunofluorescence; iPSC: induced pluripotent stem cell; NPC: neural progenitor cell; RGC: retinal ganglion cell; OCT: optical coherence tomography; ONC: optic nerve crush; PBS: phosphate buffer saline; sEV: small extracellular vesicle.
Internalization of sEVs by retinal cells.
(A, B) Flat-mounted retinas at 48 hours (A) and 7 days (B) after intravitreous injection (n = 3). Uniform distribution of the sEVs and internalization by retina cells were observed at each time point. sEVs were labeled with PKH67 (green). (C) Frozen sections of vitreous cavity (left of the white line) at 48 hours and 7 day (n = 3). More green PKH67-labeled sEVs (green) were observed in the vitreous cavity at 48 hours than day 7. (D) Frozen sections of retina at 48 hours and 7 day (n = 3). More green PKH67-labeled sEVs were observed in the retina on day 7 than 48 hours. (E) Immunostaining of frozen sections demonstrating that EVs were internalized by RGC (RBPMS, green, Alexa Fluor 488), microglia (Iba1, green, Alexa Fluor 488) and astrocyte (GFAP, green, Alexa Fluor 488) 48 hours after injection, and sEVs were labeled with PKH67 (red). The arrow indicated PKH67-labeled sEVs internalized in RBPMS positive RGC, Iba1 positive microglia and GFAP positive astrocyte. DAPI: 4',6-diamidino-2-phenylindole; GFAP: glial fibrillary acidic protein; Iba1: ionized calcium-binding adapter molecule 1; RBPMS: RNA binding protein with multiple splicing; RGC: retinal ganglion cell; sEV: small extracellular vesicle.


