Abstract
The pathways for differentiation of human CD4+ T cells into functionally distinct subsets of memory cells in vivo are unknown. The identification of these subsets and pathways has clear implications for the design of vaccines and immune-targeted therapies. Here, we show that populations of apparently naïve CD4+ T cells express the chemokine receptors CXCR3 or CCR4 and demonstrate patterns of gene expression and functional responses characteristic of memory cells. The proliferation history and T cell receptor repertoire of these chemokine-receptor+ cells suggest that they are very early memory CD4+ T cells that have “rested down” before acquiring the phenotypes described for “central” or “effector” memory T cells. In addition, the chemokine-receptor+ “naïve” populations contain Th1 and Th2 cells, respectively, demonstrating that Th1/Th2 differentiation can occur very early in vivo in the absence of markers conventionally associated with memory cells. We localized ligands for CXCR3 and CCR4 to separate foci in T cell zones of tonsil, suggesting that the chemokine-receptor+ subsets may be recruited and contribute to segregated, polarized microenvironments within lymphoid organs. Importantly, our data suggest that CD4+ T cells do not differentiate according to a simple schema from naïve → CD45RO+ noneffector/central memory → effector/effector memory cells. Rather, developmental pathways branch early on to yield effector/memory populations that are highly heterogeneous and multifunctional and have the potential to become stable resting cells.
Keywords: chemokines, immunologic memory, Th1/Th2 cells
Understanding how immunological memory develops and is sustained is a focus of ongoing research, in part because successful vaccination requires the production of long-lived memory (1, 2). Such an understanding depends on identifying subsets of memory cells, characterizing their functions, and mapping the pathways through which they are generated. As the molecular determinants of lymphocyte trafficking have been defined, the association between T cells' migratory capacities and their functions has been used to characterize T cell subsets. It was reported, for example, that expression of the chemokine receptor CCR7 identifies a subset of memory cells, so-called central memory (TCM) cells, that coexpress l-selectin (CD62L) and, in addition to being able to enter lymphoid organs, are functionally distinct from the CCR7– subset, so-called effector memory (TEM) cells (3). The key findings were that TEM but not TCM cells could produce effector cytokines and that TCM cells could serve, in vitro, as precursors for effector cells. Sallusto et al. (4) have hypothesized a linear pathway of T cell differentiation of naïve → CD45RO+/CD45RA– noneffector/TCM → effector and TEM cells, driven by the strength of the activating signals. In this model, TCM cells provide “reactive memory” and serve as a long-lived pool of precursors for additional memory and effector cells.
Our studies of how patterns of chemokine-receptor expression correlate with the function of T cell subsets have focused on CD4+ T cells, and, among the receptors we analyzed, were CXCR3 and CCR4, which are expressed on effector/memory cells. CXCR3 is the receptor for the IFN-γ-inducible chemokines CXCL9–11 and is expressed preferentially on Th1 peripheral blood lymphocytes (5). CCR4 is the receptor for CCL17 and CCL22 and is expressed preferentially on Th2 lymphocytes (5). We were surprised, therefore, that we could detect CXCR3 and CCR4 on subsets of CD4+ T cells whose other surface markers were indistinguishable from naïve cells. Our data indicate that these are very early memory cells, some of which are, nonetheless, polarized and can express immediate effector activity. An important implication of our data is that human CD4+ memory T cells are not produced along an obligatory, simple linear pathway from naïve → CD45RO+ noneffector → effector cells. Our data suggest that acquiring effector/TEM functions in vivo, including the capacity to produce cytokines and respond to inflammatory chemokines, is not necessarily a late event and is not linked causally to prior, sequential changes in a cell's phenotype. These noncausal relationships among effector/memory functions result in branched pathways of differentiation, producing highly heterogeneous memory populations, among which are the previously uncharacterized subsets described in this report.
Materials and Methods
Cells and Tissue. Whole blood and elutriated lymphocytes were obtained from donors by the Department of Transfusion Medicine at the National Institutes of Health. Cord blood was collected from term placentas at Shady Grove Adventist Hospital (Gaithersburg, MD). Tonsils were obtained as discarded tissue from the National Naval Medical Center (Bethesda). The acquisition of blood and tonsils was approved according to the policies of these institutions.
Flow Cytometry and Cell Sorting. For staining without cell sorting, cells were isolated from blood by using Ficoll/Hypaque (Amersham Pharmacia). For cell sorting, CD4+ T cells were purified from elutriated lymphocytes by using RosetteSep (StemCell Technologies, Vancouver). The following antibodies were purchased as fluorescent conjugates: anti-CXCR3-FITC (R & D Systems), anti-CD62L-FITC (Caltag, South San Francisco, CA), anti-CD45RO-phycoerythrin (PE)-Texas red (Coulter), anti-CD4-Cy5-PE, anti-CCR4-PE, anti-CXCR3-PE, anti-IL-4-allophycocyanin(APC), and anti-IFN-γ-APC (BD Pharmingen). The remaining antibodies as listed in the figure legends were obtained from BD Pharmingen and conjugated in our laboratory by using standard protocols (http://drmr.com/abcon).
Staining with anti-CXCR3 and/or anti-CCR4 was done for 15 min at room temperature before adding other antibodies for 15 min at 4°C. Cell sorting was done on a FACSVantage DiVa flow cytometer, and other analyses, except as noted, were done on an LSR II cytometer (BD Biosciences). For setting gates for defining positive and negative cells in multicolor staining, samples were stained with a mixture of all antibodies save one. Flow cytometry data were analyzed by using flowjo software (Tree Star, Ashland, OR).
T Cell Receptor (TCR) Rearrangement Excision Circles (TREC) Analysis. Cells were analyzed for TREC, as described in ref. 6, by using real-time quantitative PCR, with normalization based on copies of the albumin gene.
TCR Repertoire Analysis. Lengths of the TCR CDR3 in T cells purified by FACS were analyzed by using a modification of the protocol described by Yassai et al. (7). Numbers of cells in the purified populations ranged from 0.4 × 105 to 36 × 105. cDNA was synthesized from ≈200 ng of total RNA from each population and used in 21 PCR reactions, each containing one Vβ-specific 5′ primer plus a FAM-labeled Cβ 3′ primer, and the products were analyzed by using a 3100 Genetic Analyzer and genescan software (Applied Biosystems). Primers used were as described in ref. 8, with sequences available upon request. Each electrophoretogram was scored as Gaussian-like or non-Gaussian-like. A pattern was called non-Gaussian-like if there were one or more peak(s) that reversed the expected trend of diminishing heights in moving away from the central peak(s). Patterns where this reversal did not occur were scored as Gaussian-like. Only peaks of appropriate mobilities, differing by approximately three base pairs from neighboring peaks, were used to call patterns non-Gaussian-like. Twenty-five of the 315 electrophoretograms were of poor quality and were excluded from analysis.
Measuring CD3-Mediated Calcium Signals. Peripheral blood mononuclear cells (PBMCs) were loaded in 5 μM indo-1 AM ester (Molecular Probes) at 32°C, stained at room temperature with antibodies as noted in the legend to Fig. 3A, and incubated with 20 μg/ml biotin-conjugated anti-CD3 (SK7, BD Biosciences). For each stimulation, cells were warmed to 37°C and analyzed for 45 s before the addition of avidin (Sigma) to 10 μg/ml. The percentage of cells responding was calculated with flowjo software, and thresholds for determining responding cells were set as described in ref. 9.
Fig. 3.
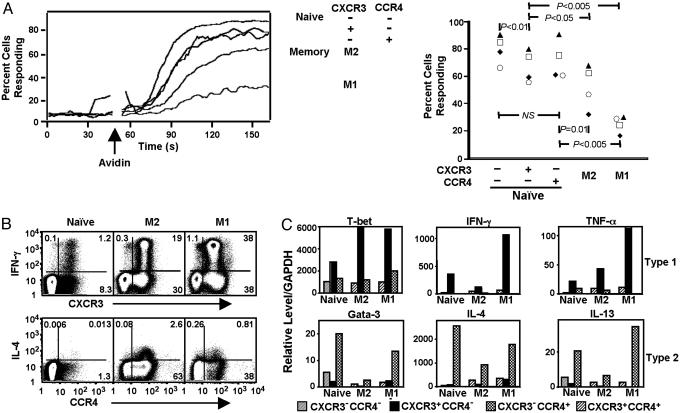
NR+ cells show responses that are not naïve. (A) PBMCs isolated from whole blood were loaded with indo-1 AM ester, stained with anti-CD4-Cy7APC, anti-CD45RO-APC, anti-CD45RA-cascade blue, anti-CD62L-Cy7PE, anti-CXCR3-FITC, and anti-CCR4-PE and anti-CD8-Cy5PE, anti-CD14-Cy5PE, and anti-CD20-Cy5PE (for the dump channel), and incubated with biotin-conjugated anti-CD3. The intracellular calcium-concentration-sensitive ratio of 405/20-nm:525/20-nm emissions was measured on the flow cytometer before and after avidin-induced CD3 cross-linking and analyzed on subsets of cells defined by gating. (Left) Within each subset for a single collection, the running means of percentages of cells vs. time having a ratio of violet/blue above a threshold that was set at the 95th percentile for the cells collected before the addition of avidin is shown. (Right) The averages of the running means obtained over the 25 s beginning with the time of peak response for the subsets from four donors in a single experiment are shown. Subsets from an individual collection/donor are identified with a unique symbol. Ps were obtained from paired t tests between groups. Similar results were obtained in two additional experiments. (B) Purified CD4+ T cells were stained with anti-CD62L-FITC, anti-CD4-Cy5PE, and anti-CD45RO-Cy7APC, separated into CD4+ naïve, M2, and M1 subsets by FACS, stained with anti-CXCR3-PE or anti-CCR4-PE, activated with PMA and ionomycin in the presence of monensin, and stained with anti-IL-4-APC or anti-IFN-γ-APC. These data are representative of two donors. Similar results were obtained in six additional experiments by using a variety of staining protocols but without purifying cells by FACS. (C) Cells were stained as for Fig. 2 A. FACS-purified populations were activated with PMA and ionomycin, and relative levels of mRNAs were determined by using real-time RT-PCR. The small subsets of naïve CXCR3+CCR4+ and M2/M1 CXCR3–CCR4– cells were not analyzed. Values are means of duplicate measurements of single samples. Values were assigned by reference to samples from cell lines and cannot be compared between different species of mRNA. Similar results were obtained with cells from three donors.
Staining for Intracellular Cytokines. Cells were stained with anti-CXCR3-PE or anti-CCR4-PE for 30 min at room temperature, stimulated with 20 ng/ml phorbol myristate acetate (PMA) and 1 μM ionomycin in the presence of 2 μM monensin for 5 h at 37°C before being stained for cytokines by using the Cytofix/CytoPerm Plus kit (BD Pharmingen), and analyzed on a FACSCalibur cytometer.
Semiquantitative Real-Time RT-PCR. Cells were stimulated with PMA and ionomycin at the doses described above for 2–3 h at 37°C. Aliquots of cDNA from ≈3–30 × 103 cells were analyzed in duplicate by using an ABI 7700 sequencer system (Applied Biosystems). cDNAs made from Th1 and Th2 cell lines were used to produce standard curves for assigning values for the experimental samples. For each standard curve, the signal obtained from cDNA synthesized from 200 ng of total RNA from the cell lines was assigned a value of 5,000. For each sample, values for the cDNAs of interest were normalized based on values for GAPDH. Probes contained the FAM reporter and the QSY7 quencher. Primer and probe sequences previously published include those for IFN-γ, TNF-α, IL-13 (10), and IL-4 (11). Primer and probe sequences for T-bet and GATA-3 are available upon request.
In Situ Hybridization. Probes for CXCL9 were synthesized as described in ref. 12. Probes for CCL22 were synthesized similarly from fragments made by RT-PCR by using primers whose sequences are available upon request. Hybridizations were done for CXCL9 and CCL22 mRNAs on serial sections of tonsil tissue by using 35S-labeled antisense and sense riboprobes by Molecular Histology (Gaithersburg, MD), as described in ref. 12.
Results
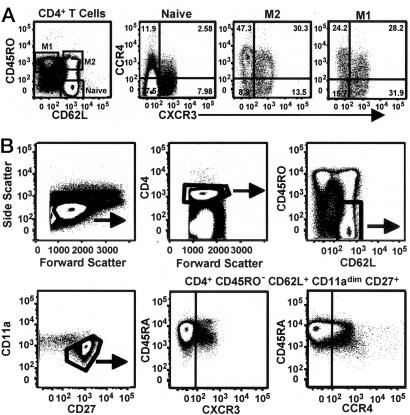
CCR4 and CXCR3 Are Expressed on Subsets of Naïve CD4+ T Cells. We analyzed the expression of the chemokine receptors CXCR3 and CCR4 on naïve and memory CD4+ T cells, as defined by CD45RO and CD62L, from the blood of healthy adults. The CD45RO–CD62L+ phenotype has been used to identify naïve cells (13, 14), and we designated the CD45RO+CD62L+ and CD45RO+CD62L– populations memory (M) 2 and M1, respectively, according to previous convention (15). As shown in Fig. 1A, the analysis of surface expression of CCR4 and CXCR3 revealed, as expected, significant percentages of CCR4+ and CXCR3+ cells in the M2 and M1 subsets. However, in analyzing >30 donors, we also uniformly found receptor+ cells in the naïve subset, principally as CXCR3+CCR4– and CXCR3–CCR4+ cells. Quantifying these cells from 10 donors by using gating, as in Fig. 1 A, yielded means of 11.5% (SEM 1.3) and 12% (SEM 0.9) for the CXCR3+CCR4– and CXCR3–CCR4+ cells, respectively, as proportions of the naïve population. To determine whether the receptor+ cells could be identified as nonnaïve by using additional markers (14), we analyzed CD4+CD45RO–CD62L+CD11adimCD27+ T cells for expression of CXCR3 and CCR4 vs. CD45RA. Both CXCR3 and CCR4 were found on naïve cells, as defined by these markers, with CXCR3 (but not CCR4) expression, showing a negative correlation with levels of CD45RA (although remaining CD45RA+). These CXCR3+ and CCR4+ subsets had the scatter profile of resting cells and did not show significant expression of the proliferation antigen Ki67 or the activation markers CD25, CD69, or HLA-DR (data not shown). We will refer to the CD4+ T cell subsets based on their expression of the conventional naïve/memory markers, using the abbreviation NR+(naïve receptor+) for the cells expressing CCR4 or CXCR3 but having an otherwise naïve surface phenotype.
Fig. 1.
CCR4 and CXCR3 are expressed on naïve CD4+ T cell subsets from adult blood. (A) PBMCs were stained with anti-CD4-Cy7APC, anti-CD45RA-Qdot 655 (data not shown), anti-CD45RO-cascade blue, anti-CD62L-Cy5.5APC, anti-CXCR3-FITC, and anti-CCR4-PE. Only CD4+ cells with the scatter profile of lymphocytes are shown. Cells within the naïve, M2, and M1 subsets (see text for details) in the left-most dot plot are shown to the right with staining for CXCR3 and CCR4. Quadrant boundaries for CXCR3 and CCR4 were set based on samples stained with all antibodies except either anti-CXCR3 or anti-CCR4. Similar results were obtained by using a variety of staining protocols and cells from >30 donors. (B) PBMCs were stained with anti-CD4-Alexa Fluor 680, anti-CD45RO-Texas red-PE, anti-CD62L-cascade blue, anti-CD11a-Cy7APC, anti-CD27-APC, anti-CD45RA-Qdot 655, anti-CXCR3-FITC, and anti-CCR4-PE. The dot plots show sequential gating to eliminate nonnaïve cells. As shown, all of the CD4+CD45RO–CD62L+CD11adimCD27+T cells are CD45RA+, although levels of CD45RA are variable. Similar results were obtained with cells from five donors.
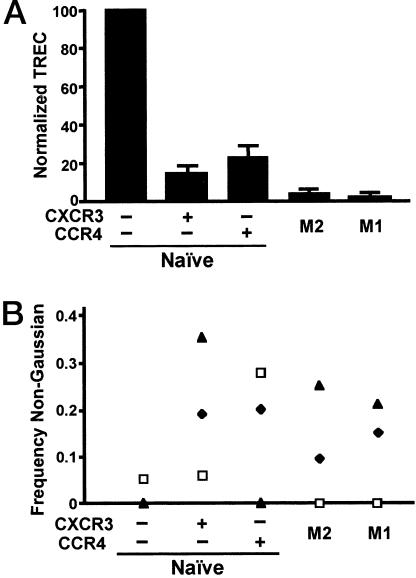
CCR4 and CXCR3 Behave Like Memory Markers. We next sought to determine whether the NR+ cells were naïve using additional criteria. We found very few NR+ cells in cord blood, and for adults, the numbers of NR+ cells showed a positive correlation with donor age, consistent with being memory cells (data not shown). If the NR+ cells are, in fact, not naïve, they would be expected to have undergone, on average, additional cell divisions as compared with naïve cells. The TREC data in Fig. 2A show that NR+ cells have undergone approximately two to three more divisions after T cell receptor gene rearrangement than have the receptor– naïve cells. As expected, TREC in the memory subsets were significantly lower. Consistent with published data on telomere lengths and the postulated precursor–product relationship between TCM and TEM (3, 16), the number of TREC was higher in the M2 as compared with M1 cells, although the difference was small. These data indicate that the NR+ cells are not derived from cells in the general memory pool that have reacquired a naïve surface phenotype. Simple marker “reversion” is also made less likely by the multiplicity of antigens showing a naïve pattern on the NR+ cells.
Fig. 2.
NR+ cells have intermediate numbers of TREC and a memory-population-like TCR repertoire. (A) Purified CD4+ T cells were stained with anti-CD4-cascade blue, anti-CD3-Cy7PE, anti-CD45RA-Ax594, anti-CD45RO-APC, anti-CD62L-Cy7APC, anti-CXCR3-FITC, and anti-CCR4-PE and anti-CD8-Cy5PE, anti-CD14-Cy5PE, anti-CD16-Cy5PE, anti-CD20-Cy5PE, and anti-CD56-Cy5PE (for the “dump” channel) and sorted into five populations. Naïve, M2, and M1 cells are as defined for Fig. 1 A. TREC numbers were calculated per 105 cells, and the values for receptor– naïve cells were normalized to 100 to allow data from six donors to be combined. Error bars show SEM. Paired t tests done on the normalized numbers showed P < 0.05 for comparisons between each subset of NR+ cells vs. receptor– naïve and vs. M2, and for M2 vs. M1. (B) Purified CD4+ T cells were stained with anti-CD4-Cy5PE, anti-CD45RO-APC, anti-CD62L-Cy5.5APC, anti-CXCR3-FITC, and anti-CCR4-PE and sorted into five populations. Electrophoretograms were scored as Gaussian-like or non-Gaussian-like, as described in Materials and Methods. Frequencies of non-Gaussian-like electrophoretograms in subsets from an individual donor are identified with a unique symbol. χ2 analysis of the frequencies obtained by summing the data for each subset for all donors gave P < 0.01 for each subset vs. receptor– naïve (significant by using the Bonferroni correction) but P > 0.05 for other pairwise comparisons. These frequencies correspond to 1 of 61, 11 of 55, 9 of 59, 4 of 47, and 6 of 57 informative electrophoretograms, respectively, for the populations, from left to right.
The above data suggested that the NR+ cells were, like other memory populations, relatively stable subsets that had accumulated during the lifetime of an individual as the result of exposures to activating antigens. If so, the NR+ TCR repertoire would, on the one hand, be expected to be broad, similar to naïve cells and to the major memory subsets, while, on the other hand, to show evidence of clonal selection. We investigated the repertoires of the NR+ and other subsets by analyzing size distributions across the CDR3 region of 21 Vβ chain mRNAs in cells from three individuals. Each of the 315 electrophoretograms was scored as Gaussian-like or non-Gaussian-like, although some were excluded for technical reasons (see Materials and Methods). Examples of electrophoretograms are shown in Fig. 6, which is published as supporting information on the PNAS web site. The data from the three donors are displayed in Fig. 2B and show significantly higher frequencies of non-Gaussian-like electrophoretograms for the NR+, M2, and M1 subsets vs. receptor– naïve cells (P < 0.01 by χ2 analysis) whose electrophoretograms were, as anticipated, virtually all Gaussian-like. These results both reveal evidence of clonal selection in the NR+ subsets and support the above data that these subsets are stable populations, similar to the M2 and M1 subsets.
NR+ Cells Show Functional Characteristics of Memory Cells. In addition to differences in surface proteins, memory and naïve cells can be distinguished by functional responses, including altered signaling through CD3 and the ability to produce effector cytokines. Previous work has demonstrated differences in the magnitude of the increase in intracellular calcium among CD4+ T cell subsets after CD3 cross-linking naïve > M2 > M1 (17). As shown in Fig. 3A, the NR+ cells demonstrated an “intermediate” phenotype. The percentages of the NR+ cells responding to CD3 cross-linking by raising their cytoplasmic calcium concentrations were lower than for the receptor– naïve cells but higher than for the M2 and M1 populations.
Given the association of CXCR3 and CCR4 with Th1 and Th2 differentiation, respectively, we tested whether the NR+ cells might produce effector cytokines. As shown in Fig. 3B, the naïve CXCR3+CCR4– population contained cells able to produce IFN-γ, and, moreover, virtually all of the IFN-γ-producing cells in the naïve population were found within the CXCR3+CCR4– subset. As expected, this same relationship between CXCR3 expression and IFN-γ staining held true for the two memory populations. Only low numbers of IL-4-staining cells were detectable within the naïve population, but these were found preferentially among the CXCR3–CCR4+ cells, as was true for the memory populations. We also determined relative levels of mRNAs for cytokines and for T-bet and GATA-3, key transcriptional regulators of Th1 and Th2 differentiation, respectively. For the naïve subsets, as shown in Fig. 3C and analogous to what was seen in the memor y populations, the CXCR3+CCR4– cells had the highest levels of the type 1 mRNAs and levels of the type 2 mRNAs that were equal to or lower than those in the naïve receptor– subset. The CXCR3–CCR4+ cells showed the inverse pattern. The CXCR3+CCR4+ memory cells showed a nonpolarized pattern, with relatively little of the selected mRNAs. In other experiments, we also measured the concentrations of cytokines in the culture medium of subsets of cells activated for 24 h with PMA and anti-CD3 (data not shown), and the results were consistent with the RT-PCR data and intracellular staining.
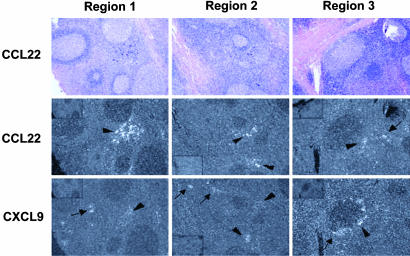
CXCL9 and CCL22 Are Expressed in Separate Foci in the T Cell Areas of Tonsil. If CXCR3 and CCR4 could be found on resting “naïve” peripheral blood lymphocytes that had undergone only a few divisions after activation, it would be expected that these receptors would be up-regulated early during the activation of naïve CD4+ T cells within lymphoid organs. Such cells can be identified in human tonsils by their simultaneous expression of CD45RA and CD45RO (13). We have already reported that these cells can express CXCR3 (12), and we found that the majority of the cells also express CCR4 (data not shown). Expression of CXCR3 and CCR4 on memory CD4+ T cells with an early central phenotype and on cells that have been recently activated within the tonsil raised the possibility that the NR+ cells might function within lymphoid organs. To investigate this possibility, we analyzed the expression in tonsils of CCL22, one of the ligands for CCR4, by in situ hybridization and compared it with the expression of CXCL9, a ligand for CXCR3, which we had previously found within interfollicular T cell areas and ringing some germinal centers (12). As shown in Fig. 4, we found that CCL22 was also expressed in foci within the T cell zones. In some cases, the CCL22 and CXCL9 mRNAs were detected in separate areas. Often, they were found in the same areas in adjacent but distinguishable foci.
Fig. 4.
CCL22 and CXCL9 are expressed in adjacent foci in tonsil. 35S-labeled RNA probes for CCL22 and CXCL9 were hybridized to serial sections of paraffin-embedded tonsil tissue. Three regions of the tissue sample, stained with hematoxylin and eosin, are shown at ×50 magnification. Bright-field illumination is shown in the first row and dark-field illumination in the second and third rows. For each region, the images in the first and second rows show results with the antisense CCL22 probe, and the image in the last row shows results from an adjacent section with the antisense CXCL9 probe. Insets in the corners of the dark-field views for regions 1 and 3 (Top Left) and region 2 (Bottom Left) show controls for each section with the CCL22 or CXCL9 sense probes. Developed grains appear black on bright-field images and white on dark-field images. The bright-field views show multiple germinal centers separated by T cell zones. In the dark-field views for each region, the arrowheads indicate foci where signals for CCL22 and CXCL9 are adjacent, and the arrows indicate foci of signal for one mRNA species without adjacent foci for the other. Different tissue sections from this tonsil hybridized to CXCL9 probes are published in ref. 12.
Discussion
Our data demonstrate that, notwithstanding their surface phenotype, the NR+ subsets contain memory cells, and therefore, the surface markers commonly used to distinguish and purify naïve and memory human CD4+ T cells, particularly in adults, are inadequate. But how do the NR+ subsets arise from naïve cells? Our analysis of CDR3 lengths suggests that the composition of the NR+ subsets has been shaped, at least in part, by TCR-mediated selection, consistent with activation by exogenous antigens. TCR-mediated selection might not, however, exclude “nonstandard” forms of TCR-driven processes, possibly including proliferation in a lymphopenic environment or homeostatic proliferation of naïve cells associated with aging. Physiological lymphopenia occurs in the neonate, where in mice it has been shown that the first thymic emigrants display polyclonal expansion and acquire a memory phenotype (18). If this were the primary source of the NR+ cells, we would not have observed, as we did, that their abundance increased with donor age. The homeostatic proliferation of naïve cells associated with diminished thymic output is another possibility. The loss of CD31 expression has been proposed as an identifier of naïve cells that have undergone homeostatic divisions. Distinguishing between homeostatic and antigen-driven proliferation was based in part on the findings that the CD31– cells were unable to produce effector cytokines (19). We found that NR+ cells were CD31– and that the NR+ cells had approximately one-half the TREC as compared with the receptor–CD45RO–CD31– subset (data not shown), suggesting that additional factors and increased proliferation are involved in producing the populations of NR+ cells. In addition, our findings that NR+ cells, unlike the larger population of receptor–CD45RO–CD31– cells, have the capacity to produce effector cytokines suggest that the NR+ cells are not the product of homeostatic proliferation. In support of this hypothesis, some naïve CD4+ T cells placed in Rag2–/– mice undergo only a limited number of IL-7-dependent homeostatic divisions. Importantly for our purposes, these cells are unable to produce IFN-γ after stimulation (B. Min and W. E. Paul, personal communication), akin to the receptor–CD45RO–CD31– human cells. In considering the data altogether, although it is impossible to rule out roles for “atypical” stimuli in producing the NR+ populations, there are no reasons to believe that such pathways have contributed to these populations any more than to the memory populations overall.
We believe that our data on the NR+ cells have implications for understanding the generation of CD4+ effector and memory T cells in vivo, providing evidence that a simple, nonbranched pathway of activation/differentiation from naïve → CD45RO+ noneffector/TCM → effector and TEM cells, as described by others (3, 20), is not obligatory. On the contrary, our data suggest that polarized subsets of effector and memory CD4+ T cells can be produced at many points along a pathway of activation, resulting in a highly heterogeneous memory population. Our data suggest that the pathway of CD4+ memory T cell differentiation in humans differs fundamentally from what has been described for CD8+ T cells in mice, where CD62L+ TCM cells were shown to have arisen by “reversion” of CD62L– TEM cells (21). Based on our analyses of multiple surface markers, signaling, and, particularly, TREC, our data strongly suggest that the NR+ populations arise from cells that have proceeded only a short way along the pathway of CD4+ effector/memory T cell differentiation before coming to rest, so that, although the pathway for CD4+ T cell differentiation is branched, it is nonetheless unidirectional. Moreover, the CDR3 analysis indicates that the NR+ cells constitute diverse, relatively stable memory populations that have accumulated with time. If the NR+ subsets consisted principally of recently activated cells that were expressing their surface phenotype transiently, we would have expected the repertoire of these subsets to be highly skewed. By contrast, the large majority of the Vβ electrophoretograms for the NR+ cells were Gaussian-like, just as for the M2 and M1 major subsets.
Current models of helper T cell differentiation include instructing and selecting components (22). Although helper T cell polarization is associated with the cells' proliferation, the causal relationship between proliferation and polarized patterns of gene expression is controversial. Our findings for the NR+ cells suggest that, at most, only limited proliferation is necessary for human CD4+ T cells to be programmed in vivo for polarized patterns of cytokine expression. Nonetheless, as shown in Fig. 3B, the percentage of cytokine-staining cells increases in the CD45RO+ compared with the NR+ populations. Our data suggest that polarization does not occur as part of a fixed temporal program in a hierarchy of differentiation steps. Rather, polarization may occur after a variable number of activation/proliferation events (presumably depending on the cells' environments and histories), which would lead to the higher frequencies of polarized cells observed in populations that have had the opportunity to undergo more and/or prolonged stimuli. Our data suggest a relationship among numbers of cell divisions, memory-associated surface markers, and polarization in T cells that is probabilistic and not causal.
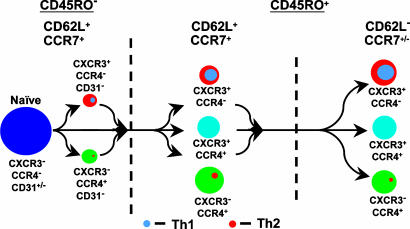
The relationships among resting CD4+ T cell subsets as defined by the expression of CD45RO, CD62L, CXCR3, and CCR4 are summarized in Fig. 5. In addition to the points made above, these data, together with results of in vitro studies (12, 23), suggest that the expression of CXCR3 and CCR4 on CD4+ T cells is driven by the two general and interrelated processes of activation and polarization. The pattern suggests that, although CXCR3 and CCR4 can be induced by T cell activation and in the absence of effector function, the conditions required for expression of CXCR3 or CCR4 at rest are necessary steps along the pathway to Th1 or Th2 differentiation, respectively, and, conversely, by the end of the pathway to Th1 or Th2 differentiation, expression of CCR4 or CXCR3, respectively, has been suppressed. The pattern of early promiscuous induction followed by selective silencing is analogous to what has been described for the principal Th1- and Th2-specific genes (22, 24). Recent data demonstrate that nonpolarized CXCR3+CCR4– and CCR4+CXCR3– TCM can serve, respectively, as precursors for Th1 and Th2 TEM, supporting a scheme in which chemokine-receptor expression occurs early along the pathway of polarization (25). For the NR+ cells, the signaling data in Fig. 3A suggest that they may have retained a “naïve-like” ability to proliferate upon TCR stimulation, a characteristic that would make the NR+ cells particularly well suited to serve as precursors for effector and additional memory cells.
Fig. 5.
CXCR3 and CCR4 in CD4+ T cell differentiation. Populations of resting CD4+ T cells are depicted based on the expression of CD45RO, CD62L, CXCR3, and CCR4. Expression of CCR7 and CD31 are also noted, based on our work (data not shown). All CD45RO+ cells are gCD31– (not depicted). Disk sizes were drawn to approximate the relative proportions of the populations in adult blood. The gray and orange discs represent Th1 and Th2 subsets, which are found principally in the CXCR3+CCR4– and CXCR3–CCR4+ populations, respectively. We hypothesize that the CD4+ T cell subsets consist of cells that have come to rest at different points along the pathway of effector cell activation and progressive differentiation from CD45RO–CD62L+ to CD45RO+CD62L+ to CD45RO+CD62L–, as indicated by the arrows. The small subsets of CD45RO–CD62L+CXCR3+CCR4+ and CD45RO+CXCR3–CCR4– cells, which we did not analyze, are omitted. Additional arrows are not drawn between subsets both for the sake of clarity and because the precise precursor–product relationships among the subsets are not known.
The fact that the earliest central memory cells are distinguished by their chemokine receptors suggests that the receptor expression itself may be important for the organization of responses and the further differentiation of these T cells within the lymphoid organs. CXCL9, like other CXCR3 ligands, is induced by IFN-γ and is expressed by macrophages and dendritic cells in human tonsils (12). CCL22 is induced by IL-4 (and inhibited by IFN-γ) and is expressed by dendritic cells in T cell regions of mouse lymph nodes (26). Here, we have shown that CXCL9 and CCL22 are expressed by separated although often adjacent clusters of cells within the T cell zones of the tonsil. Early CXCL9/CXCR3-mediated recruitment of Th1 cells or CCL22/CCR4-mediated recruitment of Th2 cells to sites of production of IFN-γ or IL-4, respectively, would be expected to help in establishing polarized microenvironments that could be used to enhance the formation of the appropriate effector (and memory) cells. Many immune responses are mixed type 1 and type 2, a finding that would seem problematic, given the mutual negative crossregulation of Th1 and Th2 cell differentiation. The use of chemokines that are selectively induced by the major regulatory cytokines to create segregated type 1 and type 2 microdomains for the aggregation, activation, and differentiation of CD4+ T cells represents one possible solution.
Supplementary Material
Acknowledgments
We thank Joanne Yu for conjugating antibodies, Calman Prussin and Barbara Foster for advice on intracellular staining, Barbara Knollmann-Ritschel for help in obtaining tonsil tissue, Carol Henry for cell sorting, Booki Min and William E. Paul for allowing us to refer to their unpublished results, and Nancy M. Hardy and Robert A. Seder for critical reading of the manuscript. The work was supported by intramural funds from the National Institute of Allergy and Infectious Diseases.
Author contributions: K.S., R.L.R., D.C.D., M.R., and J.M.F. designed research; K.S., R.L.R., B.J.H., S.C.D.R., S.P.P., H.H.Z., J.F.F., J.S.R., J.L., and J.J.M. performed research; K.S., R.L.R., D.C.D., M.R., and J.M.F. analyzed data; and J.M.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: APC, allophycocyanin; NR+, naïve receptor+ T cells expressing CCR4 or CXCR3 but having an otherwise naïve surface phenotype; PBMCs, peripheral blood mononuclear cells; PE, phycoerythrin; PMA, phorbol myristate acetate; TCM, central memory T cell; TEM, effector memory T cell; TCR, T cell receptor; TREC, TCR rearrangement excision circle.
References
- 1.Kaech, S. M., Wherry, E. J. & Ahmed, R. (2002) Nat. Rev. Immunol. 2, 251–262. [DOI] [PubMed] [Google Scholar]
- 2.Seder, R. A. & Ahmed, R. (2003) Nat. Immunol. 4, 835–842. [DOI] [PubMed] [Google Scholar]
- 3.Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. (1999) Nature 401, 708–712. [DOI] [PubMed] [Google Scholar]
- 4.Sallusto, F., Geginat, J. & Lanzavecchia, A. (2004) Annu. Rev. Immunol. 22, 745–763. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto, J., Adachi, Y., Onoue, Y., Adachi, Y. S., Okabe, Y., Itazawa, T., Toyoda, M., Seki, T., Morohashi, M., Matsushima, K. & Miyawaki, T. (2000) J. Leukocyte Biol. 68, 568–574. [PubMed] [Google Scholar]
- 6.Douek, D. C., Brenchley, J. M., Betts, M. R., Ambrozak, D. R., Hill, B. J., Okamoto, Y., Casazza, J. P., Kuruppu, J., Kunstman, K., Wolinsky, S., et al. (2002) Nature 417, 95–98. [DOI] [PubMed] [Google Scholar]
- 7.Yassai, M., Naumova, E. & Gorski, J. (1997) in The Antigen T Cell Receptor: Selected Protocols and Applications, ed. Oksenberg, J. R. (R. G. Landes, Austin, TX), pp. 326–372.
- 8.Douek, D. C., Vescio, R. A., Betts, M. R., Brenchley, J. M., Hill, B. J., Zhang, L., Berenson, J. R., Collins, R. H. & Koup, R. A. (2000) Lancet 355, 1875–1881. [DOI] [PubMed] [Google Scholar]
- 9.Rabin, R. L., Park, M. K., Liao, F., Swofford, R., Stephany, D. & Farber, J. M. (1999) J. Immunol. 162, 3840–3850. [PubMed] [Google Scholar]
- 10.Stordeur, P., Poulin, L. F., Craciun, L., Zhou, L., Schandene, L., de Lavareille, A., Goriely, S. & Goldman, M. (2002) J. Immunol. Methods 259, 55–64. [DOI] [PubMed] [Google Scholar]
- 11.Hartel, C., Bein, G., Kirchner, H. & Kluter, H. (1999) Scand. J. Immunol. 49, 649–654. [DOI] [PubMed] [Google Scholar]
- 12.Rabin, R. L., Alston, M. A., Sircus, J. C., Knollmann-Ritschel, B., Moratz, C., Ngo, D. & Farber, J. M. (2003) J. Immunol. 171, 2812–2824. [DOI] [PubMed] [Google Scholar]
- 13.Picker, L. J., Treer, J. R., Ferguson-Darnell, B., Collins, P. A., Buck, D. & Terstappen, L. W. (1993) J. Immunol. 150, 1105–1121. [PubMed] [Google Scholar]
- 14.De Rosa, S. C., Herzenberg, L. A. & Roederer, M. (2001) Nat. Med. 7, 245–248. [DOI] [PubMed] [Google Scholar]
- 15.Mitra, D. K., De Rosa, S. C., Luke, A., Balamurugan, A., Khaitan, B. K., Tung, J., Mehra, N. K., Terr, A. I., O'Garra, A., Herzenberg, L. A. & Roederer, M. (1999) Int. Immunol. 11, 1801–1810. [DOI] [PubMed] [Google Scholar]
- 16.Hengel, R. L., Thaker, V., Pavlick, M. V., Metcalf, J. A., Dennis, G., Jr., Yang, J., Lempicki, R. A., Sereti, I. & Lane, H. C. (2003) J. Immunol. 170, 28–32. [DOI] [PubMed] [Google Scholar]
- 17.Roederer, M., Bigos, M., Nozaki, T., Stovel, R. T., Parks, D. R. & Herzenberg, L. A. (1995) Cytometry 21, 187–196. [DOI] [PubMed] [Google Scholar]
- 18.Min, B., Foucras, G., Meier-Schellersheim, M. & Paul, W. E. (2004) Proc. Natl. Acad. Sci. USA 101, 3874–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimmig, S., Przybylski, G. K., Schmidt, C. A., Laurisch, K., Mowes, B., Radbruch, A. & Thiel, A. (2002) J. Exp. Med. 195, 789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iezzi, G., Scheidegger, D. & Lanzavecchia, A. (2001) J. Exp. Med. 193, 987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wherry, E. J., Teichgraber, V., Becker, T. C., Masopust, D., Kaech, S. M., Antia, R., von Andrian, U. H. & Ahmed, R. (2003) Nat. Immunol. 4, 225–234. [DOI] [PubMed] [Google Scholar]
- 22.Murphy, K. M. & Reiner, S. L. (2002) Nat. Rev. Immunol. 2, 933–944. [DOI] [PubMed] [Google Scholar]
- 23.Colantonio, L., Recalde, H., Sinigaglia, F. & D'Ambrosio, D. (2002) Eur. J. Immunol. 32, 1264–1273. [DOI] [PubMed] [Google Scholar]
- 24.Grogan, J. L., Mohrs, M., Harmon, B., Lacy, D. A., Sedat, J. W. & Locksley, R. M. (2001) Immunity 14, 205–215. [DOI] [PubMed] [Google Scholar]
- 25.Rivino, L., Messi, M., Jarrossay, D., Lanzavecchia, A., Sallusto, F. & Geginat, J. (2004) J. Exp. Med. 200, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang, H. L. & Cyster, J. G. (1999) Science 284, 819–822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.