Abstract
Background
Organoids are approved by the US FDA as an alternative to animal experiments to guide drug development and for sensitivity screening. Stable organoids models of gastric cancer are desirable for personalized medicine and drug screening.
Methods
Tumor tissues from a primary cancer of the stomach and metastatic cancer of the lymph node were collected for 3D culture. By long-term culture for over 50 generations in vitro, we obtained stably growing organoid lines. We analyzed short tandem repeats (STRs) and karyotypes of cancer cells, and tumorigenesis of the organoids in nude mice, as well as multi-omics profiles of the organoids. A CCK8 method was used to determine the drugs sensitivity to fluorouracil (5-Fu), platinum and paclitaxel.
Results
Paired organoid lines from primary cancer (SPDO1P) and metastatic lymph node (SPDO1LM) were established with unique STRs and karyotypes. The organoid lines resulted in tumorigenesis in vivo and had clear genetic profiles. Compared to SPDO1P from primary cancer, upregulated genes of SPDO1LM from the metastatic lymph node were enriched in pathways of epithelial-mesenchymal transition and angiogenesis with stronger abilities of cell migration, invasion, and pro-angiogenesis. Based on drug sensitivity analysis, the SOX regimen (5-Fu plus oxaliplatin) was used for chemotherapy with an optimal clinical outcome.
Conclusions
The organoid lines recapitulate the drug sensitivity of the parental tissues. The paired organoid lines present a step-change toward living biobanks for further translational usage.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05512-0.
Keywords: Gastric cancer, Living biobank, Organoid models, Drug sensitivity, Translational usage
Introduction
Gastric cancer is malignancy with high incidence and mortality in East Asia [1, 2], and the poor prognosis is attributed to gastric cancer metastasis [3–5]. Lymphatic metastasis is the most common metastatic site in advanced gastric cancer [6]. The regimen of platinum plus fluorouracil (S-1 or capecitabine) is often used in gastric cancer treatment. Regimens of two or multi-drugs are generally accepted in different medical centers, such as the SOX (S-1 plus oxaliplatin), DCF (docetaxel, cisplatin and 5-fluorouracil), AS (albumin-bound paclitaxel and S-1), ECF (epirubicin, cisplatin, and 5-fluorouracil), and FLOT (5-fluorouracil, leucovorin, oxaliplatin, and docetaxel) regimens [7–10]. However, there is no conclusion about which regimen is the best balance between efficacy and toxicity. The therapeutic response may relate to phenotypic features of histological types and the degree of differentiation [10, 11]. A prospective study demonstrated that there is only a 35% of complete responsive rate to FLOT or FOLFOX (5-fluorouracil plus leucovorin and oxaliplatin) regimens [12]. If the sensitivity of cancer cells to drugs is known in advance, personalized treatments can be used to kill cancer cells to the maximum extent.
Since the successful cultivation of intestinal organoids from mouse intestinal mucosa in 2009 [13], organoid technology has developed rapidly. Organoid is a 3D cellular model in vitro, which can simulate the characteristics of parental organs in terms of tissue structure, cell function, and genomic profile. Organoid has been widely used in biomedical research, especially in cancer research, such as gastric cancer [14], lung cancer [15–17], pancreatic cancer [18], colorectal cancer [19–21], bladder cancer [22], and neuroendocrine tumors [23]. Recently, the FDA Modernization Act 2.0 was approved by the US government, which allows organoids to be used as models for drug screening and sensitivity assay as an alternative to animals [24]. Li and colleagues constructed a signet-ring cell organoid line from colon cancer, which recapitulated the histology of the original cancer tissue [25]. Hoshi and colleagues constructed an organoid line of acinar cell carcinoma from the pancreas, which could tolerate cryopreservation and recovery and showed functions similar to the original tissue [26]. However, there have been no reports on the construction of stable organoid lines from gastric cancer.
Recently, we constructed an organoid living biobank for gastric cancers [27]. In this study, a pair of organoid lines derived from a poorly-differentiated adenocarcinoma and its metastatic lymph node were successfully cultured for over fifty generations. We characterized parameters including cell doubling time, culture medium requirements, tumorigenesis in vivo, STRs, karyotypes, genetic profiles, and drug sensitivity. Importantly, based on the drug sensitivity analyses, the patient received a better therapeutic regimen and achieved a sustained response.
Materials and methods
Ethics statement
Protocols for fresh organoid culture of gastric cancer were approved by the Ethics Committee for Scientific Research of Ruijin Hospital, and informed consents were obtained from the patients. An additional number was used to register the samples in the database with no link to patient names or personal information.
Establishment of organoid lines
The primary cancer and lymphatic metastatic cancer of the station six were collected after radical gastrectomy within 30 min ex vivo and stored in advanced DMEM/F12 medium (12,634,010, Thermo Fisher Scientific). The tissues were washed with PBS containing 1×penicillin-streptomycin (C0222, Beyotime) and 1×puromycin (A1113802, Thermo Fisher Scientific) at least four times, and then minced to 1–2 mm3 for further digestive incubation in buffer including 1 mg/ml collagenase(40507ES60, Yeasen), 1 mg/ml collagenase IV (40510ES60, Yeasen), and 1 U/ml dispase II (40104ES60, Yeasen) for 1.5 h at 37℃ with shaking. The incompletely digested tissue pieces were discarded and then cells were centrifuged at 1000 rpm for 5 min. The cell precipitates were mixed with DMEM and matrigel (356,231, Corning) at a 1:1 ratio and seeded in a 24-well plate (50 µl/well). Once the matrigel was polymerized after 30 min incubation at 37 °C, complete medium (Table S1) was added (500 µl/well). The organoid culture was performed at 37 °C, 5% CO2, and the medium was replaced every 5 days. The well-grown organoids were obtained after 7 to 10 days of cultivation. The organoids were digested with TrypLE Express enzyme (12,605,010, Thermo Fisher Scientific) for 0.5–1 h. The cell suspensions were collected and centrifuged at 1000 rpm for 5 min. The cell precipitates were mixed with matrigel at a 1:1 ratio and seeded in a 24-well plate (50 µl/well, 5 × 104 cells) again for passaging or subsequent study.
Hematoxylin and eosin (H&E) staining
Well-grown organoids (diameter > 100 μm) were blown thoroughly at least 10 times and centrifuged at 1500 rpm for 5 min. The organoid spheres were collected and fixed in 4% paraformaldehyde for 30 min, put in 10% agarose, and embedded in paraffin. The paraffin block was cut into 8-µm sections. The sections were treated in xylene twice for 10 min, and immersed in pure ethanol for 5 min, 95% ethanol for 5 min, and 75% ethanol for 5 min. The sections were heated with 1×antigen repair reagent at 100℃ for 20 min, and washed with clean water. After washing, the sections were stained in hematoxylin for 10 min, immersed in hydrochloric acid and ammonium hydroxide for several seconds, and washed in running water for 1 h. The sections were stained in alcohol eosin for 2 min (G1005, Servicebio), and dehydrated in 70% and 90% alcohol for 10 min, and further immersed in 95% ethanol and pure ethanol for 5 min, respectively. The sections were transparent in xylene for 10 min twice, and sealed with neutral resin.
Immunohistochemistry (IHC) and western blotting
In immunohistochemical assays, the sections were prepared in the same way as in H&E stain. The staining methods were performed as previously described [28]. The primary antibodies included E-cadherin (GB11082, Servicebio, 1:200), N-cadherin, GB111273, Servicebio, 1:200), and Ki67 (GB111141, Servicebio, 1:200). The western blot assay was performed as previously described [29]. The primary antibodies included p-mTOR (5536, CST, 1:1000), mTOR (2972, CST, 1:1000), p-AKT (4060, CST, 1:200), AKT(4691, CST, 1:1000), HRP-conjugated goat anti-rabbit IgG (H + L) (SA00001-2, Proteintech, 1:3000), and HRP-conjugated anti-GAPDH (HRP-60,004, Proteintech, 1:3000).
Cell growth analysis
The well-grown organoids were digested with TrypLE Express enzyme for 0.5–1 h. The single-cell suspension was centrifuged at 1000 rpm for 5 min. The cell precipitate was mixed with matrigel at a 1:1 ratio and seeded in a 96-well plate (6 µl /well, 6000 cells, n = 3). After polymerization of the matrigel for 30 min at 37 °C, the complete medium (60 µl/well) was added. Cellular vitality of the organoids was measured by a CCK8 assay (CK04, DOJINDO) every 48 h for 8 days. The growth curves were plotted using time (hour) as the x-axis and cell vitality from the 450-nm OD value as the y-axis.
Short tandem repeats (STRs) analysis
The well-grown organoids were digested with TrypLE Express enzyme for 15 min and centrifuged at 1500 rpm for 5 min to collect the organoid spheres. DNA was extracted with a genome extraction kit (Ap-mn-p-500, Axygen), amplified by the 21-STR amplification protocol, and detected on an ABI 3730XL. We focused on eight core STR loci (D5S818, vWA, D7S820, D16S539, TH01, D13S317, TPOX and CSF1PO) and the sex chromosome Amelogenin. The STR results were matched with the STR data library in DSMZ tools, which covers STR data of 2455 cell lines from the ATCC, DSMZ, JCRB and RIKEN databases. When the STR matching rate was above the cut-off value (≥ 75%), the two matched lines were judged to be homologous, and otherwise, unique [30].
Karyotypes analysis
Colchicine solution (0.2 µg/ml) was added to well-grown organoids and incubated for 1.5 h at 37℃. Organoids were then digested with TrypLE Express enzyme for 0.5–1 h. The single-cell suspensions were centrifuged at 1500 rpm for 5 min. The cell precipitates were preheated with 6 ml of KCL hypotonic solution (0.075 mol/L) at 37℃ in a water bath for 30 min with gentle agitation and were then thoroughly dried. The fixative solution (1 ml of methanol and glacial acetic acid at a ratio of 3:1) was slowly added for 30 min, and then the cells were centrifuged at 1500 rpm for 5 min. The cell precipitates were further fixed by the above fixative solution (5 ml) for 30 min and then centrifuged at 1500 rpm for 5 min. The cells were resuspended in 0.5 ml fixative solution. One or two drops of the cell suspension were added to a cooled slide and fixed by an alcohol flame. After drying at room temperature, the cells were stained with Giemsa for 10 min, the dye was washed and the slide was dried at room temperature. The cells in the division phase were observed and captured using the ZEISS karyotyping system (Ikaros, ZEISS).
Whole-exome sequencing (WES)
The paired organoid lines (at passage 18) and cryopreserved primary cancer tissues were used for DNA extraction (DP304, Tiangen). The DNA was fragmented by enzyme digestion and adenine deoxynucleotide and a sequencing adaptor were added. By PCR amplification, the enriched library was hybridized by Agilent SureSelect Human All Exon V6 to capture exonic regions. Genomic sequencing was performed using an Illumina PE150.The FASTQ format of sequencing data was mapped to the human reference genome (hg38) by the BWA Tool. Single nucleotide polymorphisms (SNPs; Sentieon DNAseq 201808.05), copy number variation (CNV; CNVkit, 0.9.6), somatic single nucleotide variants (SNVs; Sentieon DNAseq 201808.05), and insertions and deletions (InDel; Sentieon DNAseq 201808.05) were analyzed. The driver gene mutations were screened based on the CGC513, Bert Vogelstein125, SMG127 and Comprehensive435 databases. The raw data (PRJNA1015608) were uploaded to SRA database (PRJNA1015608).
Single-cell transcriptome sequencing (scRNA-seq)
Cell dissociation and single-cell capture
The well-grown organoids were digested with TrypLE Express enzyme for 0.5–1 h. The single-cell suspensions with cell vitality greater than 70% were collected into nuclease-free water.
Library preparation and sequencing
The Chromium Single Cell 3ʹ Gel Beads-in-emulsion (GEM) Library &Gel Bead Kit v3 was used for library construction, and sequencing was performed on a Novaseq 60,000 (Illumina). The raw data (GSE242893) were updated to GEO database.
Quality control and data preprocessing
ScRNA-seq data analysis was performed using the 10×Genomics software package (Cell Ranger version 2.1.1) for data quality control and alignment. SCfastp was used for sequence filtering and sequence quality assessment. CellrangerCounts was used to count the number of cells. A total of 7662 cells and 6257 cells were collected from SPDO1LM and SPDO1P organoid lines, respectively. Gene expression matrixes were generated using unique molecular identifiers (UMIs). The alignment of transcriptome data to the GRCh38 reference genome (GRCh38_Ensembl_Ensembl104) was performed by CellrangerAggr. The UMIs were mapped to genes. Low-quality cells that matched the following were removed: (1)expressed gene number of cells are < 200 or > 7500.2 expressed mitochondrial genes or ribosomal genes of cells were > 25%. A total of 10,578 cells from SPDO1P and SPDO1LM were used for analysis.
Data normalization
The Seurat package (4.1.1) was used for data normalization, including “NormalizeData” and “LogNormalize”. The log-transformed UMI counts were computed to obtain the normalized gene expression value.
Dimension reduction
A subset of 2500 highly variable genes (HVGs) was selected for analysis by “FindVariableFeatures” and “ScaleData”. The “RunPCA” of the Seurat package was used for principal components analysis (PCA). The “JackStraw” was used to determine 20 PCA. The “ScoreJackStraw” was used to obtain principal components scores.
Cell clustering and gene set enrichment analysis (GSEA)
The “FindClusters” was used for cell clustering analysis. The linear dimensionality reduction and plotting of tSNE was performed by “RunTSNE” with a resolution of 0.2. The “FindAllMarkers” (min.pct = 0.25, logfc.threshold = 0.25) was used to obtain differential genes of the subclusters. The GSEA was performed to obtain enriched pathways.
Transcriptome sequencing (RNA-seq)
Tissues (5 × 5 mm) and organoid cells (1.5 × 106) were collected for RNA-seq assays. Total RNA was extracted with TRIzol Reagent. The RNA-seq libraries were prepared using the Fast RNA-seq Lib Prep Kit V2(RK20306, ABclonal, China) according to the manufacturerˊs instructions. The sequencing was performed by the NovaSeq xplus. Gene counts were normalized to transcripts per million (TPM) for data analysis.
3D invasion assay
The well-grown organoids were digested with TrypLE Express enzyme for 15 min and then centrifuged at 1000 rpm for 5 min. The precipitates were mixed with matrigel (354,231, Corning) at a ratio of 1:1 and seeded in a 24-well plate (50 µl/well, 1000 organoid spheres). After matrigel polymerization for 30 min at 37℃, complete medium (500 µl/well) was added. Images were captured at 0 and 12 h using an inverted microscope.
2D migration assay
The well-grown organoids were digested with TrypLE Express enzyme for 15 min and centrifuged at 1000 rpm for 5 min. Cells (300 µl) were resuspended in serum-free DMEM (2 × 105 cells) and added to the upper chamber of the transwell chamber. Complete medium (600 µl) was added to the lower chamber. The cells were incubated at 37℃ and 5% CO2 for 72 h, then stained with 1% crystal violet for 30 min. The cells in the upper chamber were wiped off and dried at room temperature. The cells were counted, and images were captured under a microscope.
2D invasion assay
The well-grown organoids were digested with TrypLE Express enzyme for 15 min, and centrifuged at 1000 rpm for 5 min. Matrigel (10% 100 µl) was added to the upper chamber of the transwell chamber and polymerized for 30 min at 37℃. Cells (2 × 105) were resuspended in 200 µl of serum-free DMEM (cells) and added to the upper chamber of the transwell chamber. Complete medium (600 µl) was added to the lower chamber. The cells were incubated at 37℃ and 5% CO2 for 96 h, then stained with 1% crystal violet for 30 min. The cells in the upper chamber were wiped off and dried at room temperature. The cells were counted and images were captured under a microscope.
Tubule formation assay
Human umbilical vein endothelial cells (EA.HY926) were evenly seeded in a 96-well plate coated with matrigel. The supernatants of SPDO1LM and SPDO1P organoid lines were added for incubation for 48 h. The tubular length and branch nodes were counted under a microscope.
Gene knockdown by siRNA
The siRNAs for FLNA were used for gene knockdown (Sangon Biotech, China). The knockdown was detected using western blotting. The antibodies included anti-FLNA (67,133, Proteintech, 1:1000) and HRP-conjugated goat anti-mouse IgG (H + L) (SA00001-1, Proteintech, 1:3000).
Drug sensitivity assays using CCK8 method
The well-grown organoids were digested with TrypLE Express enzyme for 15 min and centrifuged at 1000 rpm for 5 min. After cell counting, the cell suspension was mixed with matrigel at a ratio of 1:1, and seeded in a 96-well plate (6 µl/well, 6000 cells). After matrigel polymerization for 30 min at 37℃, complete medium (60 µl/well) containing various concentrations of drugs was added (Table S2). Traditional gastric cancer cell lines (AGS and NCI-N87) were used as controls. The cells were digested by TrypLE Express Enzyme into single cell suspension and 60 µl/well cells were seeded in 96-well plates for CCK8 analysis. Concentration gradients of drugs (5-Fu, 0, 0.5, 0.75, 1, 2, 5, and 10 μm), oxaliplatin (OXA, 0, 2, 5, 10, 20, 30, and 40 μm), and paclitaxel (PTX, 0, 0.1, 0.5, 1, 2, 5, and 10 nm) were added. After 5 days incubation at 37℃, a 10% CCK8 solution (CK04, DOJINDO) was added, and the matrigel was blown off with tips to make the organoids fully exposed to the reagent. After incubation at 37℃ for 2 h, the OD450 values were measured using a microplate reader to calculate cell vitality and IC50 of drugs.
Hypoxia and normoxia culture
The well-grown organoids were digested with TrypLE Express enzyme for 0.5–1 h. The cell precipitate was mixed with matrigel at a 1:1 ratio and then seeded in a 96-well plate (6 µl /well, 6000 cells, n = 3). After the matrigel polymerization, complete medium (60 µl/well) was added and cells were cultured for 72 h in hypoxic (37℃, 94% N2, 5% CO2, and 1% O2) and normoxic (37℃, 20% O2, and 5% CO2) conditions. The cellular vitality of organoids was measured using a CCK8 assay.
Lactate assay upon inhibition of glycolysis
The well-growth organoids were digested with TrypLE Express enzyme for 0.5–1 h. Organoid cells were seeded in a 96-well plate (6 µl /well, 6000 cells, n = 3). The chemical 2-deoxy-D-glucose (1 mM, M5140, AbMole, China) was added for incubation at 37℃, 20% O2 and 5% CO2 for 72 h. The culture medium was collected and tested by the lactate assay using the Cedex Bio Analyzer (06343759001, Roche, Switzerland) following the manufacturer’s instructions.
Tumorigenesis of organoid lines in nude mice
Organoid cells of SPDO1P or SPDO1LM (100 µl, 5 × 105 cells) were injected into the armpit of male BALB/c-nu mice (4 weeks of age, 4 mice/group). After 4 weeks, the mice were sacrificed and the tumors were removed. The tumor volume was calculated by: V = π/6 × L (long diameter) × W (short diameter). The xenograft tumors (PDO-X) were fixed in 4% paraformaldehyde overnight, embedded in paraffin, and stained with H&E. This study was approved by the Institutional Animal Care & Use Committee of Shanghai Jiao Tong University School of Medicine (B-2022-010).
Statistics
GraphPad Prism 6.0 (GraphPad Prism, RRID: SCR_002798) was used for statistical analysis. The Shapro-Wilk test was used for normal test. The student’s t-test (homogeneity of variance) or unpaired t-test with Welch’s correction (heterogeneity of variance) was used for analysis of two group data, respectively. ANOVA or Brown-Forsythe ANOVA was used for data analysis for the above two groups. Each experiment was repeated at least three times independently. P < 0.05 was considered statistically significant.
Results
Clinicopathological characteristics of organoid cell lines
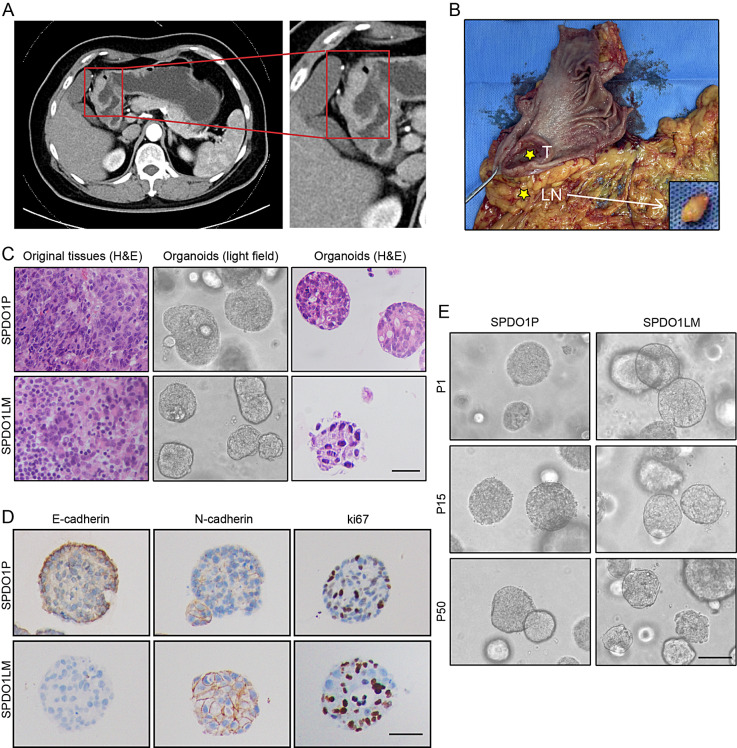
The cancer tissues were collected from a 68-year-old female patient who presented with upper abdominal pain for several months. Under endoscopy, an ulcerated mass of 3.5 × 3.5 cm was found at the greater curvature of the antrum. By enhanced computed tomography (CT) examination, localized thickening of the gastric antrum was found, resulting in an unclear three-layer structure of the gastric wall with abnormal enhancement in the arterial phase. An enlarged lymph node near the right gastroepiploic artery was identified (Fig. 1A). After radical distal gastrectomy, an ulcerative mass (Borrmann II) and an enlarged lymph node were observed (Fig. 1B). The pathological reports showed a poorly-differentiated adenocarcinoma (intestinal type by the Lauren classification) of stage A (T3N2M0) without H. pylori infection.
Fig. 1.
Clinicopathological characteristics of organoid lines (A) A thickened gastric antrum wall is shown by CT scanning. The red box marks the enlarged image. (B) An ulcerated mass is observed at the antrum in the gastrectomy sample with the enlarged lymph node near the right gastroepiploic artery (T, primary tumor; LN, lymph node). (C) From left to right, histology of poorly-differentiated adenocarcinoma of original tissues by H&E staining, organoid morphology by light- field observation, and organoid morphology by H&E staining. The histology is compatible with poorly-differentiated adenocarcinoma with irregular lumens. The upper panel indicates SPDO1P and the lower panel indicates SPDO1LM. Scale bar, 100 μm. (D) Immunohistochemical staining of E-cadherin, N-cadherin, and Ki67 for SPDO1P and SPDO1LM. Scale bar, 80 μm. (E) SPDO1P and SPDO1LM maintain cell vitality in continuous culture in vitro. Both organoid lines expanded for over 50 generations. Note: P1 = passage 1, P15 = passage 15, and P50 = passage 50. Scale bar, 100 μm
Organoids were successfully established from the primary cancer (SPDO1P) and the metastatic lymph node (SPDO1LM). By H&E staining, both organoid lines showed poorly-differentiated adenocarcinoma with cell disarrangement, and matched well with their original tissues (Fig. 1C). Organoid lines grew in nests with lost polarity. Irregular lumens were occasionally observed. By immunohistochemistry, we examined the expression of E-cadherin, N-cadherin and Ki67 biomarkers for SPDO1P and SPDO1LM (Fig. 1D). E-cadherin was expressed on SPDO1P organoids and significantly downregulated on SPDO1LM organoids. N-cadherin was weakly expressed on SPDO1P organoids and significantly upregulated on SPDO1LM organoids. The cell proliferation marker Ki67 was upregulated in SPDO1LM organoids compared to the SPDO1P organoids. The cell vitality of SPDO1P and SPDO1LM was maintained in vitro for the long-term culture (over 600 days and 50 generations) and tolerated repeated freezing and recovery (Fig. 1E).
Culture conditions
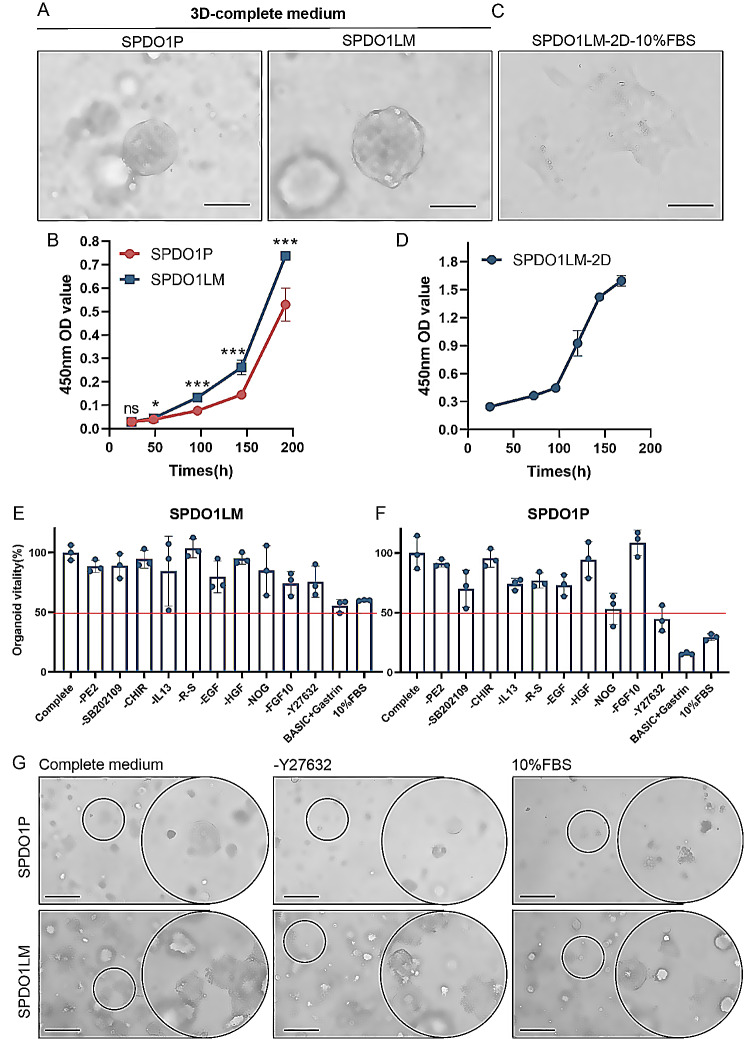
Both SPDO1P and SPDO1LM organoid lines proliferated in 3D culture with complete medium (Fig. 2A). The doubling times of SPDO1P and SPDO1LM were 120 h and 72 h, respectively (Fig. 2B). In addition, the SPDO1LM organoid line could proliferate in traditional 2D culture. During the first 24 h, a portion of cells adhered to the wall in a semi-floating state. At 48 h, SPDO1LM cells were completely adhered to the wall and could be passaged continuously (Fig. 2C). The doubling time of SPDO1LM in 2D culture was 96 h (Fig. 2D).
Fig. 2.
Culture conditions of SPDO1P and SPDO1LM. (A) The SPDO1P and SPDO1LM lines grow well in complete medium in 3D culture (light field). Scale bar, 100 μm. (B) The growth curves of SPDO1P (red) and SPDO1LM (blue) in 3D culture show that the doubling time of SPDO1P (120 h) is longer than that of SPDO1LM (72 h). (C) The SPDO1LM cells can adhere to the wall in 2D culture with 10% FBS-DMEM medium (light field). Scale bar, 100 μm. (D) The growth curve of SPDO1LM in 2D culture with a doubling time of 96 h. (E) Comparison of cell vitality of the SPDO1LM line based on the stepwise elimination of growth factors or chemicals. (F) Comparison of cell vitality of the SPDO1P line based on stepwise elimination of growth factors or chemicals. (G) The growth of SPDO1P (upper panel) and SPDO1LM (lower panel) cultured in complete medium (left), without Y27632 (middle), and 10% FBS-DMEM (right) under light-field observation. The SPDO1LM line grows well in complete medium, without Y27632, and 10% FBS-DMEM, while the SPDO1P line shows lower cell vitality without Y27632 or 10% FBS-DMEM. Scale bar, 200 μm, nsP > 0.05, *P < 0.05, ***P < 0.001. Abbreviation: DMEM, Dulbecco’s modified Eagle medium; FBS, fetal bovine serum
To clarify the requirement for growth factors or cytokines of SPDO1P and SPDO1LM lines, we examined the growth status by a stepwise elimination method. SPDO1LM could grow well even in DMEM medium supplemented only with 10% FBS and no growth factors or chemicals. The cell vitality of SPDO1LM in DMEM medium supplemented with 10% FBS was about 60% of that in complete medium (Fig. 2E, Table S1, Figure S1) However, after the removal of growth factors or chemicals, the cell vitality of SPDO1P line was only 30% of that in complete medium and was difficult to passage (Fig. 2F, Figure S2). These data indicate that the SPDO1P line was highly dependent on growth factors and chemicals such as R-spondin 1, noggin, EGF, SB202109, and Y27632. Among them, Y27632, which is a selective ROCK1 and ROCK2 inhibitor, was crucial to SPDO1P growth (Fig. 2G). Therefore, the medium requirement of the SPDO1P line was as follows: advanced DMEM/F12, HEPES, B27, N2 additive, GlutaMAX, nicotinamide, N-acetyl-L-cysteine, gastrin, noggin, Y27632, SB202109, EGF, and R-spondin 1 (Table S1). We note that both SPDO1P and SPDO1LM lines could grow well without WNT/β-catenin signaling pathway activator WNT3a or CHIR-99,021.
STRs and karyotypes analysis
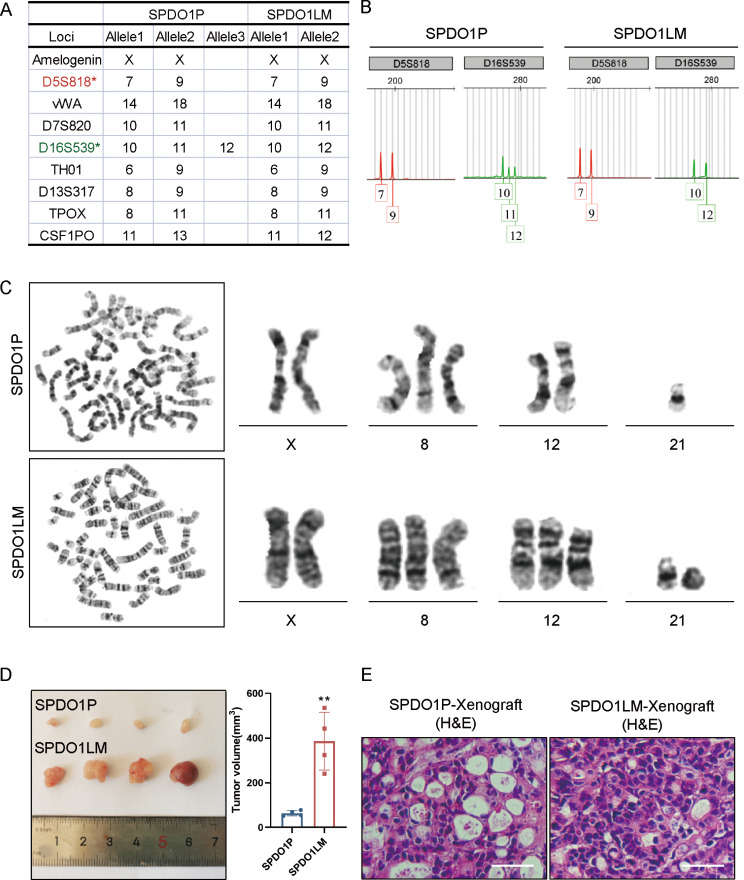
By STRs analysis, both SPDO1P and SPDO1LM lines did not match the STRs of 2455 cell lines in the public database, which suggests that SPDO1P and SPDO1LM lines were newly created organoid cell lines. By comparison of eight core STRs loci and the sex gene amelogenin between SPDO1P and SPDO1LM lines, the above loci were consistent (Fig. 3A). Allele drift at the D16S539 and CSF1PO loci was observed between the two lines (Fig. 3B, Figure S3). Overall, the matching rate between SPDO1P and SPDO1LM lines was 88.9%, which means that SPDO1P and SPDO1LM were homologous derivatives.
Fig. 3.
STRs and karyotypes analysis, as well as tumorigenesis of SPDO1P and SPDO1LM organoid lines. (A) The crucial STRs loci are compared between SPDO1P and SPDO1LM lines. Both organoid lines show XX sex alleles. STRs loci are matched well at D5S818, vWA, D7S820, TH01, D13S317, and TPOXSTR. There is allele drift at the loci of D16S539 and CSF1PO. (B) The chromatograms of the D5S818 locus (red) and D16S539 locus (green) from SPDO1P and SPDO1LM lines. The D5S818 locus is matched well, and one allele is shifted at the D16S539 locus. (C) The karyotype of SPDO1P line is 46, XX, + 8, -21 (upper), while that of the SPDO1LM line is 48, XX, + 8, +12, r(21)(p13q22.3) (lower). (D) The tumor volume of the SPDO1LM xenograft is larger than that of the SPDO1P xenograft (385.76 ± 112.16 vs. 63.46 ± 10.55 mm3). (E) By H&E staining, both xenografts of SPDO1P and SPDO1LM are poorly-differentiated adenocarcinoma. Scale bar, 100 μm.;**P < 0.01
The karyotypes of SPDO1P and SPDO1LM lines were 46, XX, + 8, -21 and 48, XX, + 8, +12, r(21)(p13q22.3), respectively (Fig. 3C). The number of chromosomes from SPDO1P was 46, including an extra chromosome 8 and a loss of chromosome 21(Fig. 3C, upper; Figure S4). The number of chromosomes from SPDO1LM was 48, with extra chromosomes 8 and 12, and a circular chromosome 21(Fig. 3E, lower; Figure S4).
Tumorigenesis of SPDO1P and SPDO1LM lines in vivo
After inoculation of SPDO1P and SPDO1LM cells into the armpit of nude mice, tumorigenesis was observed in 100% of the experimental animals (n = 4 per group). The tumor volumes of the SPDO1LM xenograft were larger than that of the SPDO1P xenograft (SPDO1LM-X vs. SPDO1P-X, 385.76 ± 112.16 vs. 63.46 ± 10.55 mm3; Fig. 3D). By H&E staining, the histology of the SPDO1P and SPDO1LM xenografts were consistent with their original poorly-differentiated adenocarcinoma (Fig. 3E).
Genetic features by whole-exome sequencing
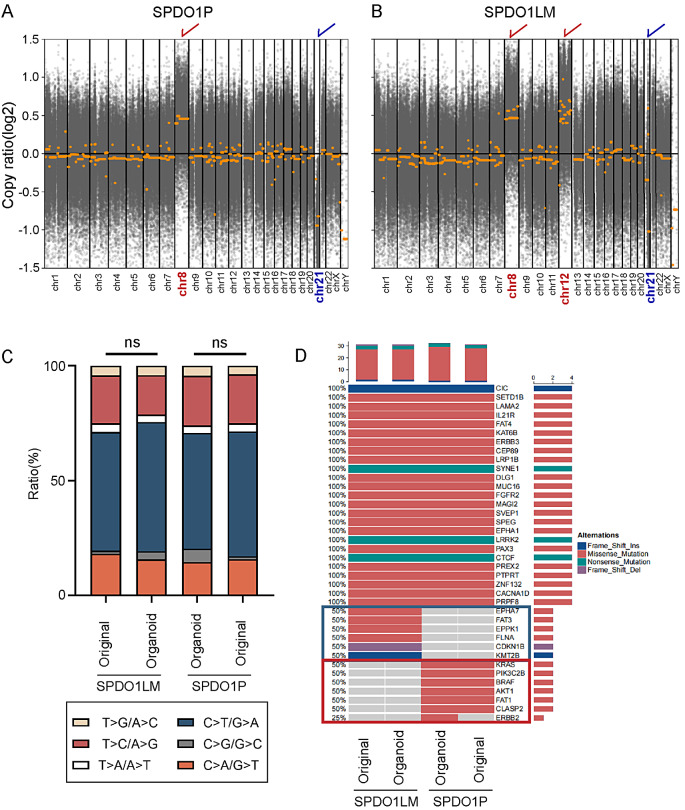
To identify genomic variation and driver mutations of SPDO1P and SPDO1LM, WES was performed for organoid lines and matched original tissues. Both SPDO1P and SPDO1LM showed genomic instability with copy number variation in some chromosomes. Among them, SPDO1P showed a gain of chromosome 8 and a loss of chromosome 21 (Fig. 4A), consistent with the results found in the karyotypes analysis. The SPDO1LM showed a gain of chromosome 8 and 12 and a loss of chromosome 21 (Fig. 4B), consistent with the result found in the karyotypes analysis. Both SPDO1P and SPDO1LM lines and their matched tissues showed similar proportions of SNPs (Fig. 4C).
Fig. 4.
Genetic features of SPDO1P and SPDO1LM by whole-exome sequencing. (A) By CNV analysis, the SPDO1P shows a chromosome 8 gain (marked by red) and chromosome 21 loss (marked by blue). (B) By CNV analysis, the SPDO1LM line shows a chromosome 8 gain (marked by red), chromosome 12 gain (marked by red), and chromosome 21 loss (marked by blue). Copy ratio > 0 indicats a gain; copy ratio < 0 indicates a loss. (C) The bar plot of SNP ratios in SPDO1P, SPDO1LM, and their matched original tissues. There is no significant difference in the ratio of SNPs between SPDO1P and its original tissue (nsP = 0.8832) or SPDO1LM and its original cancer tissue (nsP = 0.7003) by the Chi-square test. (D) By mutation analysis, 25 somatic driver mutations are shared in both SPDO1P and SPDO1LM lines, as well as their original cancer tissues. There are specific driver mutations in the SPDO1P line and the matched cancer tissue (red box) and SPDO1LM line and matched cancer tissue (blue box)
By gene mutation analysis, both SPDO1P and SPDO1LM lines, as well as matched original tissues showed wild-type for TP53. A total of 25 somatic driver mutations were shared in both organoid lines and their matched original tissues, including frameshift insertion mutations of CIC, missense mutations of SETD1B, LAMA2, IL21R, FAT4, KAT6B, ERBB3, CEP89, LRP1B, DLG1, MUC16, FGFR2, MAGI2, SVEP1, SPEG, EPHA1, PAX3, PREX2, PTPRT, ZNF132, CACNA1D, and PRPF8, and nonsense mutations of SYNE1, LRRK2, and CTCF (Fig. 4D). In addition, there were specific driver mutations in SPDO1P or SPDO1LM (Fig. 4D). Among the 63 somatic driver mutation genes in the SPDO1LM line, 34 mutated genes (53.97%) were the same as that found in the original cancer tissue, covering missense mutations of EPHA7, FAT3, EPPK1, and FLNA, a frameshift deletion mutation of CDKN1B, and a frameshift insertion mutation of KMT2B. Among the 62 somatic driver mutation genes in the SPDO1P line, 32 mutated genes (51.61%) were the same as those found in the original cancer tissue, including missense mutations of KRAS, PIK3C2B, BRAF, AKT1, FAT1, and CLASP2. A missense mutation of ERBB2 was identified in the SPDO1P line, but not in its original cancer tissue (Table S3). In addition, we compared the gene expression correlation of transcriptomic data between SPDO1P or SPDO1LM and the original tissues and found the gene expression profiles of SPDO1P and SPDO1LM correlated well with their original tissue (r = 0.9079 and 0.7992, respectively).
ScRNA-Seq and cell functions
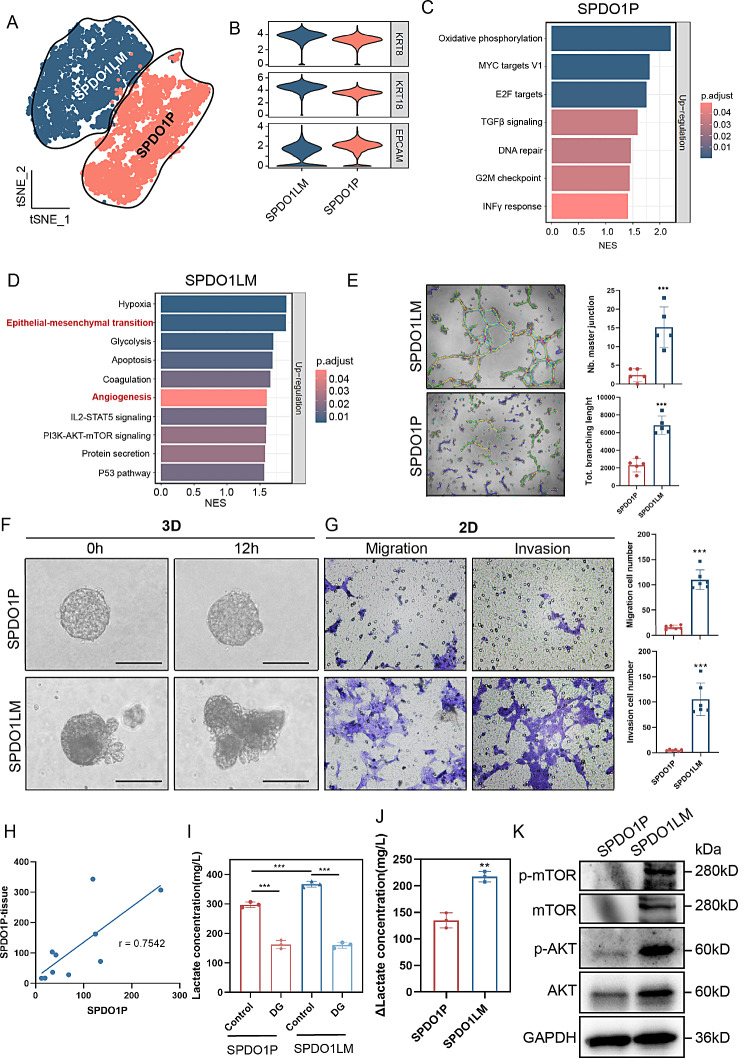
In tSNE mapping, the gene expression patterns of SPDO1P and SPDO1LM were different, suggesting significant functional differences between the two organoid lines (Fig. 5A). Both organoid lines expressed epithelial markers KRT8, KRT18 and EPCAM, indicating their epithelial origin (Fig. 5B). By GSEA pathway enrichment analysis, the highly expressed genes of SPDO1P were enriched in the oxidative phosphorylation and MYC targets V1 pathways (Fig. 5C,Table S4), and the highly expressed genes of SPDO1LM were enriched in hypoxia, epithelial-mesenchymal transition (EMT), glycolysis, apoptosis, coagulation, angiogenesis, IL2-STAT5 signaling, and PI3K-AKT-mTOR signaling pathways (Fig. 5D, Table S4). We compared the gene expression consistency between the scRNA-seq and RNA-Seq of the two organoid lines and found that 95% of the top 20 upregulated genes (19/20) in the scRNA-seq of SPDO1P were highly expressed in the RNA-seq analysis. The 90% of the top 20 upregulated genes (18/20) in the scRNA-seq of SPDO1LM were also highly expressed by RNA-seq (Figure S5).
Fig. 5.
ScRNA-Seq and biological functions assays. (A) The tSNE plot of 10,578 cells from SPDO1P and SPDO1LM. The highly expressed genes of the SPDO1P and SPDO1LM organoid lines are separated into two clusters. (B) The violin plots show high expression of KRT8, KRT18, and EPCAM in SPDO1P and SPDO1LM. (C) The highly expressed gene set of SPDO1P is enriched in oxidative phosphorylation, MYC targets V1, and E2F target pathways. (D) The highly expressed gene set of SPDO1LM is enriched in hypoxia, EMT, glycolysis, apoptosis, coagulation, angiogenesis, IL2-STAT5 signaling, PI3K-AKT-mTOR signaling, protein secretion, and P53 pathways. (E) Images of the tubule formation analysis (light field). The supernatant of SPDO1LM shows stronger proangiogenic potential than that of SPDO1P with longer branch lengths and increased tubular junctions, n = 5. (F) Images of the organoid spheres in the 3D invasion assay of SPDO1P and SPDO1LM (light field). The SPDO1P line grows stably in a sphere structure, but the SPDO1LM line extends pseudopodia around the spheres. (G) Images of migration and invasion cells. The SPDO1LM line shows more migrated cells than the SPDO1P line after 72 h of incubation and more invading cells than SPDO1P after line for 96 h. n = 6. (H) The correlation analysis shows that the gene expression levels of the top 10 genes in the oxidative phosphorylation pathway of SPDO1P and the original tissue are consistent (r = 0.7542). (I) The lactate concentrations of both organoids significantly declined upon 2-deoxy-D-glucose (DG, 1 mM, 72 h) treatment. (J) The Δlactate concentration (lactate concentration of control group - lactate concentration of DG- treated group) is different between the two organoids. (K) According to western blot analysis, the protein expression levels of p-mTOR, mTOR, p-AKT, and AKT in the SPDO1LM line are higher than those of the SPDO1P line. *P < 0.05, **P < 0.01, ***P < 0.001
To verify the functional changes of upregulated pathways of SPDO1LM line, we examined proangiogenic ability, migration and invasion of the two organoid lines. We collected the conditioned media of the SPDO1LM organoids and SPDO1P and incubated the human endothelial cell line EA.HY926 in the media for 48 h. The conditioned media of SPDO1LM promoted angiogenesis with longer branch lengths (6831 ± 927 vs. 2304 ± 671 μm, ***P = 0.001) and increased the number of tubular junctions (15.20 ± 4.83 vs. 2.40 ± 1.50, ***P = 0.001), compared to that of SPDO1P (Fig. 5E). In the 3D invasion assay, the SPDO1P organoids grew in a spherical structure, while SPDO1LM organoids extended pseudopodia around spheres, indicating stronger invasion characteristics (Fig. 5F). The migration and invasion abilities of the SPDO1P and SPDO1LM lines were identified in 2D cultivation. The migrated cells of SPDO1LM were more than that of SPDO1P for 72 h (110 ± 18 vs. 16 ± 4, ***P < 0.0001). The invaded cells of SPDO1LM were more than that of SPDO1P at 96 h (105 ± 30 vs. 5 ± 2, ***P < 0.0001)(Fig. 5G).
To verify the difference in the metabolic pathways of SPDO1P and SPDO1LM, we cultured the SPDO1P and SPDO1LM in normoxic and hypoxic conditions. The expression correlation of the top 10 oxidative phosphorylation pathway genes of SPDO1P and the original tissue was consistent (Fig. 5H, Figure S6A). The hypoxia- and EMT-related genes MXI1 and ENO2 were enriched in SPDO1LM, compared with SPDO1P (Figure S6B). SPDO1P was more oxygen-dependent than SPDO1LM (Figure S6C). The cell proliferation rate in SPDO1P decreased in hypoxia, compared with that of normoxic, while the SPDO1LM was not (Δorganoid cell vitality SPDO1P vs. SPDO1LM: 102.27 ± 24.26 vs. 63.53 ± 24.98, *P = 0.0107) (Figure S6D). Hexokinase 2 (HK2) is a glycolysis-related gene and was highly expressed in SPDO1LM (Figure S6E). We examined the lactate contents of the two organoid lines upon treatment with 2-deoxy-D-glucose (DG), a hexokinase inhibitor. The lactate content of SPDO1LM was significantly higher than that of SPDO1P, but significantly decreased upon DG treatment (SPDO1LM: 366.62 ± 7.93 vs. 160.21 ± 8.05 mg/L; SPDO1P: 297.23 ± 7.76 vs. 162.32 ± 11.57 mg/L; Fig. 5I). Upon inhibition of hexokinase by DG treatment, the reduction of lactate content in SPDO1LM was considerably different, compared to SPDO1P (Fig. 5J). These data indicate that the glycolysis of SPDO1LM was greatly inhibited by the hexokinase inhibitor.
Regarding to the highly expressed genes of PI3K-AKT-mTOR signaling pathways in SPDO1LM by scRNA-Seq, we verified the protein expression of p-mTOR, mTOR, p-AKT, and AKT of SPDO1LM by western blot (Fig. 5K). In addition, we found that the top 100 upregulated genes intersected with EMT pathway genes, including ENO1, LGALS1, and FLNA (Figure S7A). Then, the FLNA gene was knocked down by siRNA in SPDO1LM (Figure S7B). In 2D migration assay, the FLNA knockdown significantly inhibited SPDO1LM migration (Figure S7C).
Drug sensitivity and clinical association
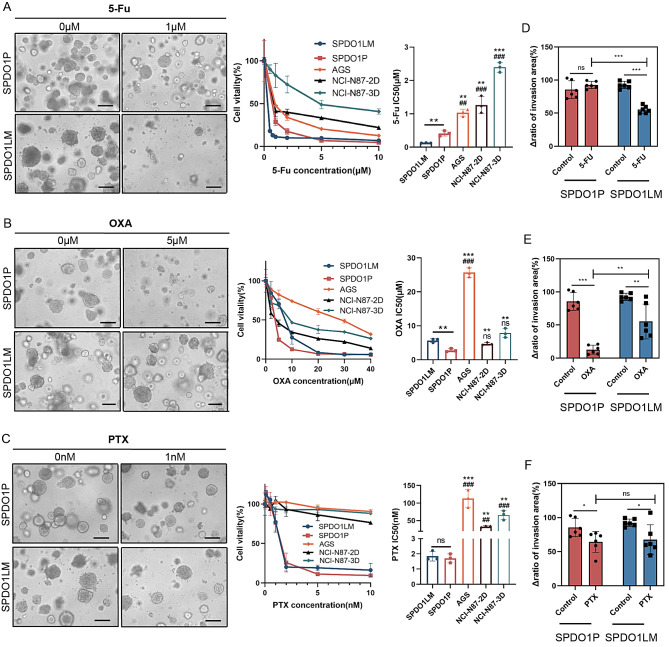
We compared the CCK8 and GTC 3D assays for drug sensitivity testing in 3D cultured organoids and found that there was no significant difference between the two methods (Figure S8). The improved CCK8 method showed smaller batch-to-batch variation than the GTC 3D method. According to CCK8 assay, the SPDO1LM line was more sensitive than SPDO1P to 5-Fu (IC50: 0.1267 ± 0.0053 vs. 0.4125 ± 0.0632 µM, **P = 0.0031), while the SPDO1P line was more sensitive to OXA than SPDO1LM (IC50: 2.7940 ± 0.3993 vs. 5.6873 ± 0.4514 µM, **P = 0.0025). In addition, both SPDO1P and SPDO1LM lines were sensitive to PTX (IC50: 1.7040 ± 0.2390 vs. 1.8463 ± 0.2614 nM, nsP = 0.6002) (Fig. 6A and B, and 6C). Since the SPDO1LM line can proliferate in both 2D and 3D culture conditions, we examined the drug sensitivity of the SPDO1LM line in 2D and 3D conditions and found that the IC 50 of SPDO1LM-2D and SPDO1LM-3D was 119.70 ± 5.61 nM and 126.67 ± 5.33 nM, respectively (P < 0.05, Figure S9). The drug sensitivity of 5-Fu was consistent between 2D and 3D culture settings. We also examined the drug sensitivity of SPDO1P or SPDO1LM and traditional cancer cells in 2D and 3D culture conditions. The IC50 of 5-Fu in AGS, NCI-N87-2D, and NCI-N87-3D spheroids was 1.0336 ± 0.0858, 1.2673 ± 0.2088, and 2.3930 ± 0.1200 µM, respectively. The IC50 of OXA in AGS, NCI-N87-2D, and NCI-N87-3D spheroids was 25.7033 ± 1.1197, 4.6780 ± 0.3169, and 7.8373 ± 1.1069 µM, respectively. The IC50 of PTX in AGS, NCI-N87-2D, and NCI-N87-3D spheroids was 113.1967 ± 20.3906, 33.0667 ± 1.8236, and 65.9033 ± 10.8147 nM, respectively. Both SPDO1P and SPDO1LM were sensitive to 5-Fu and PTX than traditional cancer cell lines of AGS, NCI-N87-2D, and NCI-N87-3D spheroids.
Fig. 6.
Drug sensitivity of SPDO1P and SPDO1LM lines and its effect on 3D cell invasion. (A) Left: The morphological changes of SPDO1P (upper) and SPDO1LM (lower) upon 5-Fu treatment. Middle: Cell vitality analysis of organoids compared to cancer cell lines AGS, NCI-N87-2D, and NCI-N87-3D spheroids upon 5-Fu treatment. Right: The bar chart shows IC50 values of organoids and cancer cell lines AGS, NCI-N87-2D, and NCI-N87-3D spheroids. (B) Left: The morphological changes of SPDO1P (upper) and SPDO1LM (lower) upon OXA treatment. Middle: Cell vitality analysis of organoids compared to cancer cell lines AGS, NCI-N87-2D, and NCI-N87-3D spheroids upon OXA treatment. Right: The bar chart shows IC50 values of organoids and cancer cell lines AGS, NCI-N87-2D, and NCI-N87-3D spheroids upon OXA treatment. (C) Left: The morphological changes of SPDO1P (upper) and SPDO1LM (lower) upon PTX treatment. Middle: Cell vitality analysis of organoids compared to cancer cell lines AGS, NCI-N87-2D, and NCI-N87-3D spheroids upon PTX treatment. Right: The bar chart shows IC50 values of organoids and cancer cell lines AGS, NCI-N87-2D, and NCI-N87-3D spheroids upon PTX treatment. n = 3. Scale bar, 100 μm. (D) In 3D cell invasion assays, 5-Fu more significantly inhibits SPDO1LM invasion in SPDO1LM compared to SPDO1P. (E) OXA significantly inhibits SPDO1P invasion compared to SPDO1LM. (F) PTX inhibits both SPDO1P and SPDO1LM invasion compared to controls. nsP > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001
We further examined the effect of drug 5-Fu, OXA, and PTX (1µM) on 3D invasion in SPDO1P and SPDO1LM lines. By the ratio of the invasion area, 5-Fu showed a significant inhibitory effect on 3D cell invasion in SPDO1LM (SPDO1P vs. SPDO1LM: 92.55 ± 4.89% vs. 55.38 ± 5.10%, ***P < 0.001) after 24 h of drug treatment (Fig. 6D). Drug OXA showed a significant inhibitory effect on 3D cell invasion of SPDO1P (SPDO1P vs. SPDO1LM: 12.72 ± 6.26% vs. 55.29 ± 24.03%,**P = 0.003; Fig. 6E). Drug PTX showed an obvious inhibitory effect on 3D cell invasion in both SPDO1P and SPDO1LM lines (SPDO1P vs. SPDO1LM: 64.41 ± 14.07% vs. 67.64 ± 19.98%) compared to controls (P < 0.05; Fig. 6F). The drug sensitivity analysis of organoids provided an important reference for chemotherapy. The patient received systemic chemotherapy of S-1 plus oxaliplatin (SOX regimen: oral S-1, 40 mg/m2, twice a day on days 1–14, and oxaliplatin 130 mg/m2 intravenously on day 1; each treatment cycle was 21 days) for totally six cycles after radical surgery. The patient has been stable for 24 months without recurrence.
Discussion
Paired organoid lines from primary gastric cancer (SPDO1P) and corresponding metastatic cancer of the lymph node (SPDO1LM) were established. The “S” in the name has two meanings: “Shanghai” and “Stomach”. PDO means patient-derived organoid. The “1” represents number one. The letter “P” means primary tumor, while “LM” represents lymphatic metastasis. The two organoid lines have been deposited in the China Center for Type Culture Collection (CCTCC) with collection number C202387 for SPDO1P and C2023149 for SPDO1LM. Although there are several gastric cancer cell lines, such as HGC-27 [31] from lymphatic metastasis of poorly-differentiated adenocarcinoma and NCI-N87 [32] and MKN-45 [33] from liver metastases of gastric cancer, there are no reports about paired cell lines from the primary cancer and the lymph node metastasis. Thus, this is the first time that such matched organoid cell lines have been established. These organoid lines will provide valuable experimental models for exploring metastatic mechanisms and therapeutic drugs for gastric cancer. More importantly, these organoid lines have translational value for drug screening. The two organoid lines showed high sensitivity to first-line therapeutic drugs and guided the choice of chemotherapy, i.e., the SOX regimen [34]. Our bench results reflected the drug response at the bedside and provided support for the FDA Modernization Act 2.0, that is, organoids can be applied as an alternative to animal experiments for drug sensitivity studies [24, 35, 36].
Although SPDO1P and SPDO1LM are derived from the same patient, they showed different biological characteristics in some aspects (Table 1). The SPDO1LM line is derived from metastatic cancer of the lymph node and showed stronger growth, invasion, and metastatic abilities than the SPDO1P line from the primary cancer. The protein expression levels of N-cadherin and Ki67 were higher in SPDO1LM than in SPDO1P. Their difference in biological characteristics can be explained by scRNA-seq analysis. The differentially expressed genes of SPDO1LM were mainly enriched in EMT and angiogenesis pathways. Activation of EMT and angiogenesis pathways plays important roles in promoting metastasis [37, 38]. In our study, the SPDO1LM organoid line revealed a stronger ability to induce neovascularization, migration, and invasion than that of SPDO1P. In the metabolic dependence analysis, the highly expressed gene set of the SPDO1P line was enriched in the oxidative phosphorylation pathway, which may be related to its dependence on oxygen for cell growth. In contrast, the highly expressed gene set of SPDO1LM was enriched in hypoxia- and glycolysis-related pathways, which likely explains the hypoxia-resistant cell growth. Lactate, a glycolytic product, was significantly higher in SPDO1LM than in SPDO1P. Recently, Zong and coworkers reported that lactate produced by cancer cells can lactylate important tumor suppressor, resulting in functional inactivation of proteins and promoting tumor progression [39]. The chemical 2-deoxy-D-glucose can inhibit glycolysis by targeting HK2 [40], which may be a new direction to inhibit glycolysis and prevent tumor progression.
Table 1.
Biological characteristics of SPDO1P and SPDO1LM organoid lines
| Parameters | Characteristics | SPDO1P (Primary tumor) |
SPDO1LM (Lymph node) |
|---|---|---|---|
| Growth | Culture time | 570 days | 570 days |
| Doubling time | 120 h (3D) | 72(3D) or 96 (2D) hours | |
| 3D culture | Yes | Yes | |
| 2D culture | No | Yes | |
| Culture medium |
Essential cytokine (Noggin, Y27632, SB202109, EGF, R-Spondin1) WNT3a-free |
Essential cytokine or 10% FBS DMEM WNT3a-free |
|
| STR | Unique | Unique | |
| Karyotype | 46, XX, + 8, -21 | 48, XX, + 8, +12, r(21)(p13q22.3) | |
| Xenograft | 100% | 100% | |
| Genomics | CNV | 8 gain, 21 loss | 8 gain, 12 gain, 21 loss |
| TP53 | Wild-type | Wild-type | |
|
Mutation (shared) |
CIC, SETD1B, LAMA2, IL21R, FAT4, KAT6B, ERBB3, CEP89, LRP1B, DLG1, MUC16, FGFR2, MAGI2, SVEP1, SPEG, EPHA1, PAX3, PREX2, PTPRT, ZNF132, CACNA1D, PRPF8, SYNE1, LRRK2, CTCF | CIC, SETD1B, LAMA2, IL21R, FAT4, KAT6B, ERBB3, CEP89, LRP1B, DLG1, MUC16, FGFR2, MAGI2, SVEP1, SPEG, EPHA1, PAX3, PREX2, PTPRT, ZNF132, CACNA1D, PRPF8, SYNE1, LRRK2, CTCF | |
|
Mutation (special) |
KRAS, PIK3C2B, BRAF, AKT1, FAT1, CLASP2 | EPHA7, FAT3, EPPK1, FLNA, CDKN1B, KMT2B | |
| ScRNA-Seq | Up-regulated pathway | Oxidative phosphorylation, MYC targets V1 | Angiogenesis, epithelial-mesenchymal transition, glycolysis, hypoxia |
| Behaviors | Migration ability | Weak | Strong |
| Invasion ability | Weak | Strong | |
| Angiogenesis | Weak | Strong | |
| Drug sensitivity | 5-Fu (IC50) | 0.4125 μm | 0.1267 μm |
| OXA (IC50) | 2.7940 μm | 5.6873 μm | |
| PTX (IC50) | 1.7040 nm | 1.8463 nm |
Histologically, both SPDO1P and SPDO1LM retained the morphology of poorly-differentiated adenocarcinoma and were consistent with those described for the adenocarcinoma organoids established by Seidlitz [41]. Cancer organoids often recapitulate the genetic profiles of the original tissues [23, 35, 41]. To date, given the high cost of genetic analysis, organoid-related studies have not profiled the genetic mutations for each generation of organoids. Therefore, the data on genetic changes between organoids and original tissues are mainly based on the detection of a specific generation. For example, Weeber et al. have examined genetic mutations in metastatic colorectal cancer biopsies and early passage organoids (within 2–3 months) and found that 90% of mutations were shared between the organoids and the original tissues [42]. Yan et al. analyzed differences in the mutation spectrum between early and late organoids (more than 6 months) from colorectal cancer and the original tissues and found that the drift of genetic mutation of short-term culture of organoids was less than 20%, while the drift of genetic mutation in long-term cultured organoids could reach 50% [43]. Our team has examined the differences in gene variants between short-term culture organoids (less than 3 months) and their original tissues and found that the shared mutation rate was over 90% (data unpublished). In the current study, we analyzed the mutations in organoids of the eighteenth passage and found that the shared mutations between organoids and original tissues account for 50–60%. Additional somatic mutations may happen during repeated cell passaging in vitro. For example, we identified an ERBB2 mutation in the SPDO1P line but did not identify it in the original cancer tissue. Although we detected 25 shared mutations in both organoid lines and the original cancer tissue, we also found some mutations that were in the organoids and the matched cancer tissues. The biological significance of the somatic mutations and whether they are related to driving cancer metastasis will be studied in the future. In addition, the newly established SPDO1P and SPDO1LM lines are wild-type for the TP53 gene, and thus they provide an important model for molecular biology and drug development for TP53 wild-type gastric tumors in the future.
Regarding the requirements for culture medium, both SPDO1P and SPDO1LM lines were independent of WNT signaling activators, such as WNT3a, indicating that cancer cells are auto-activated for the WNT pathway. SPDO1P is highly dependent on growth factors and chemicals, especially Y27632, while SPDO1LM can grow well in DMEM medium containing 10% FBS only. This may be attributed to cell dissociation during the passaging procedure, which disrupts the cell-cell adhesion and induces cell apoptosis. SPDO1P is dependent on cell-cell adhesion, while SPDO1LM is independent of cell-cell adhesion. The chemical Y27632 inhibits apoptosis caused by cell dissociation and maintains the continued growth of SPDO1P.
STRs are the main indicator for discriminating cell homology or contamination of human cell lines [30]. The concordance rate of 75% is generally used as the cut-off value [30, 44]. In STRs analysis, the STRs loci of SPDO1P and SPDO1LM lines do not match with 2455 human cell lines in public cell databases, indicating their uniqueness as cell lines. The concordance rate of STRs between SPDO1P and SPDO1LM lines reached 88.9%, implying that SPDO1P and SPDO1LM lines are homologous. The appearance of allele drift in some loci could be explained by the variation of short tandem repetitive sequences along with cancer progression [30]. Since SPDO1LM was constructed from the metastatic tumor of the lymph node, the chromosomal instability may continue along with the evolution of cancer metastasis [45]. In addition, the spatial heterogeneity in cancer progression might result in a non-uniform distribution of genetic variants of different subpopulations of cancer, such as primary tumors and metastases [46, 47]. Therefore, we recommend establishing organoids from primary cancer lesion, as well as metastatic cancer lesions, if possible, to get the real drug sensitivity for metastatic cancer. Karyotypes analysis of chromosomes is another indicator for cell line construction. We found that both SPDO1P and SPDO1LM are aneuploid, which is supported by the copy number analysis. The genomic instability of SPDO1P and SPDO1LM is in accordance with that of intestinal-type gastric cancer [41, 48].
Regarding organoid technology as part of routine healthcare, it is feasible, because the time of culture is short (10–15 days). If the drug screening is subsequently carried out, it will take a total of 30 days. The cost of organoid culture is cheaper than that of traditional PDX models [49]. Clinically, chemotherapy is usually started one month after surgery. The results of drug sensitivity screening of patient-derived organoids may be very helpful. To decrease the cost of organoid cultivation and drug screening, artificial assistant evaluation could be used, as in our previous work [27].
Conclusion
Paired organoid cell lines from primary and metastatic cancer of the lymph node were established for the first time from gastric tumors. The two organoid lines were deposited in the Chinese Typical Culture Collection Center, and form part of the living biobank at the institution. The cell biology, genetics, and drug sensitivity of the paired organoid lines were characterized, which has the potential to inform clinical decision-making and guide the adjuvant postoperative chemotherapy. These organoid lines constitute valuable models for the further investigation of metastatic mechanisms and new anti-cancer drugs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
R, Y, Y. Qi, and W. K: study conception and design. Y. D, R.Y, L. Z, X.Y, C. L, Z. Z, X. Z, H. G: data acquisition, and data analysis and interpretation. I. H. C, Z. K, and Y.Y: manuscript writing and final review.
Funding
This work was supported by grants from the Shanghai Science and Technology Committee (Grant No. 20DZ2201900), the National Natural Science Foundation of China (Grant No. 82072602, 82372933 and 82173222), the Innovation Foundation of Translational Medicine of Shanghai Jiao Tong University School of Medicine (Grant No.TM202001), and the Collaborative Innovation Center for Clinical and Translational Science by Chinese Ministry of Education & Shanghai Municipal Government (Grant No.CCTS-2022202 and CCTS-202302). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank LetPub (www.letpub.com.cn) for its linguistic assistance during the preparation of this manuscript.
Data availability
The raw data of this study (PRJNA1015608) was uploaded into SRA database
Declarations
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer/WHO, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/WHO.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ruixin Yang, Yao Qi and Wingyan Kwan contributed equally to this work.
Contributor Information
Zisis Kozlakidis, Email: kozlakidisz@iarc.fr.
Yingyan Yu, Email: yingan3y@sjtu.edu.cn.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Wong MCS, Huang J, Chan PSF, Choi P, Lao XQ, Chan SM, et al. Global incidence and mortality of gastric Cancer, 1980–2018. JAMA Netw Open. 2021;4:e2118457. 10.1001/jamanetworkopen.2021.18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L, Liang Y, Zhang C, Wang X, Ding X, Huang C, et al. Prognostic significance of lymphovascular infiltration in overall survival of gastric cancer patients after surgery with curative intent. Chin J Cancer Res. 2019;31:785–96. 10.21147/j.issn.1000-9604.2019.05.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou H, Lei PJ, Padera TP. Progression of Metastasis through Lymphatic System. Cells 2021; 10. [DOI] [PMC free article] [PubMed]
- 5.Wang Y, Li X, Zhang T, Li F, Shen Y, He Y, et al. Neutrophils promote tumor invasion via FAM3C-mediated epithelial-to-mesenchymal transition in gastric cancer. Int J Biol Sci. 2023;19:1352–68. 10.7150/ijbs.79022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JE, Kim KT, Shin SJ, Cheong JH, Choi YY. Genomic and evolutionary characteristics of metastatic gastric cancer by routes. Br J Cancer 2023. [DOI] [PMC free article] [PubMed]
- 7.Feng ZL, Chen LB, Liu ZY, Chen XJ, Ren XC, Liu YE, et al. DCF intraperitoneal and intravenous dual chemotherapy regimen for advanced gastric cancer: a feasibility study. Oncol Lett. 2015;9:491–7. 10.3892/ol.2014.2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghorbani M, Dehghani M, Fahimfar N, Namazi S, Dehshahri A. FLOT (a chemotherapy regimen for gastric/esophagogastric junction cancer): to be treated as a highly emetogenic regimen or a moderately emetogenic one? Comparison of the emetogenic potential of FLOT versus FOLFOX and TAC regimens. Support Care Cancer. 2022;30:3865–73. 10.1007/s00520-022-06832-x [DOI] [PubMed] [Google Scholar]
- 9.Farrokhi P, Sadeghi A, Sharifi M, Riechelmann R, Moghaddas A. Efficacy and safety of FLOT regimen vs DCF, FOLFOX, and ECF regimens as perioperative chemotherapy treatments for resectable gastric cancer patients; a report from the Middle East. Res Pharm Sci. 2022;17:621–34. 10.4103/1735-5362.359430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Zhang J, Wang Y, Li W, Yu S, Li Q, et al. Efficacy of AS versus SOX regimen as first-line chemotherapy for gastric cancer patients with peritoneal metastasis: a real-world study. BMC Gastroenterol. 2022;22:296. 10.1186/s12876-022-02369-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng L, Shao L, Sun S, Zhang C, Cai B. Analysis of the efficacy and influencing factors of preoperative P-SOX neoadjuvant chemotherapy regimen for progressive gastric cancer-construction of a clinical prediction model. Cancer Med. 2023;12:13031–40. 10.1002/cam4.5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arefpour AM, Hosseini S, Basi A, Novin K, Foroughi A, Garousi M. Evaluation of pathological response rate and complications of FOLFOX versus FLOT Regimen in Perioperative Chemotherapy for Resectable Gastric Cancer: a prospective study. Asian Pac J Cancer Prev. 2023;24:2791–7. 10.31557/APJCP.2023.24.8.2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- 14.Nanki K, Toshimitsu K, Takano A, Fujii M, Shimokawa M, Ohta Y, et al. Divergent routes toward wnt and R-spondin Niche Independency during Human gastric carcinogenesis. Cell. 2018;174:856–e869817. 10.1016/j.cell.2018.07.027 [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Li X, Chen H. Organoid models in lung regeneration and cancer. Cancer Lett. 2020;475:129–35. 10.1016/j.canlet.2020.01.030 [DOI] [PubMed] [Google Scholar]
- 16.Kim M, Mun H, Sung CO, Cho EJ, Jeon HJ, Chun SM, et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat Commun. 2019;10:3991. 10.1038/s41467-019-11867-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu J, Kalyani FS, Liu L, Cheng T, Chen L. Tumor organoids: synergistic applications, current challenges, and future prospects in cancer therapy. Cancer Commun (Lond). 2021;41:1331–53. 10.1002/cac2.12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TDD, et al. Organoid Profiling identifies common responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018;8:1112–29. 10.1158/2159-8290.CD-18-0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–45. 10.1016/j.cell.2015.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganesh K, Wu C, O’Rourke KP, Szeglin BC, Zheng Y, Sauvé CG, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med. 2019;25:1607–14. 10.1038/s41591-019-0584-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, van Werkhoven E et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med 2019; 11. [DOI] [PubMed]
- 22.Abugomaa A, Elbadawy M, Yamanaka M, Goto Y, Hayashi K, Mori T, et al. Establishment of 2.5D organoid culture model using 3D bladder cancer organoid culture. Sci Rep. 2020;10:9393. 10.1038/s41598-020-66229-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawasaki K, Toshimitsu K, Matano M, Fujita M, Fujii M, Togasaki K, et al. An Organoid Biobank of Neuroendocrine Neoplasms enables genotype-phenotype mapping. Cell. 2020;183:1420–e14351421. 10.1016/j.cell.2020.10.023 [DOI] [PubMed] [Google Scholar]
- 24.Wadman M. FDA no longer has to require animal testing for new drugs. Science. 2023;379:127–8. 10.1126/science.adg6276 [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Wang R, Huang D, Ma X, Mo S, Guo Q, et al. A novel human colon signet-ring cell carcinoma organoid line: establishment, characterization and application. Carcinogenesis. 2020;41:993–1004. 10.1093/carcin/bgz178 [DOI] [PubMed] [Google Scholar]
- 26.Hoshi D, Kita E, Maru Y, Kogashi H, Nakamura Y, Tatsumi Y, et al. Derivation of pancreatic acinar cell carcinoma cell line HS-1 as a patient-derived tumor organoid. Cancer Sci. 2023;114:1165–79. 10.1111/cas.15656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang R, Du Y, Kwan W, Yan R, Shi Q, Zang L et al. A quick and reliable image-based AI algorithm for evaluating cellular senescence of gastric organoids. Cancer Biol Med 2023. [DOI] [PMC free article] [PubMed]
- 28.Xiang Z, Zhou Z, Song S, Li J, Ji J, Yan R, et al. Dexamethasone suppresses immune evasion by inducing GR/STAT3 mediated downregulation of PD-L1 and IDO1 pathways. Oncogene. 2021;40:5002–12. 10.1038/s41388-021-01897-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiang Z, Li J, Song S, Wang J, Cai W, Hu W, et al. A positive feedback between IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric cancer metastasis. J Exp Clin Cancer Res. 2019;38:314. 10.1186/s13046-019-1318-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cell line misidentification. : the beginning of the end. Nat Rev Cancer. 2010;10:441–8. 10.1038/nrc2852 [DOI] [PubMed] [Google Scholar]
- 31.Tian S, Peng P, Li J, Deng H, Zhan N, Zeng Z, et al. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/β-catenin signaling pathway. Aging. 2020;12:3574–93. 10.18632/aging.102831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang L, Li JF, Su L, Zang M, Fan Z, Yu B, et al. ALEX1, a novel tumor suppressor gene, inhibits gastric cancer metastasis via the PAR-1/Rho GTPase signaling pathway. J Gastroenterol. 2018;53:71–83. 10.1007/s00535-017-1329-y [DOI] [PubMed] [Google Scholar]
- 33.Naranjo S, Cabana CM, LaFave LM, Romero R, Shanahan SL, Bhutkar A, et al. Modeling diverse genetic subtypes of lung adenocarcinoma with a next-generation alveolar type 2 organoid platform. Genes Dev. 2022;36:936–49. 10.1101/gad.349659.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sah BK, Zhang B, Zhang H, Li J, Yuan F, Ma T, et al. Neoadjuvant FLOT versus SOX phase II randomized clinical trial for patients with locally advanced gastric cancer. Nat Commun. 2020;11:6093. 10.1038/s41467-020-19965-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang R, Yu Y. Patient-derived organoids in translational oncology and drug screening. Cancer Lett. 2023;562:216180. 10.1016/j.canlet.2023.216180 [DOI] [PubMed] [Google Scholar]
- 36.Zu M, Hao X, Ning J, Zhou X, Gong Y, Lang Y, et al. Patient-derived organoid culture of gastric cancer for disease modeling and drug sensitivity testing. Biomed Pharmacother. 2023;163:114751. 10.1016/j.biopha.2023.114751 [DOI] [PubMed] [Google Scholar]
- 37.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 38.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 39.Zong Z, Xie F, Wang S, Wu X, Zhang Z, Yang B, et al. Alanyl-tRNA synthetase, AARS1, is a lactate sensor and lactyltransferase that lactylates p53 and contributes to tumorigenesis. Cell. 2024;187:2375–e23922333. 10.1016/j.cell.2024.04.002 [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z, Tan X, Luo J, Yao H, Si Z, Tong JS. The miR-30a-5p/CLCF1 axis regulates sorafenib resistance and aerobic glycolysis in hepatocellular carcinoma. Cell Death Dis. 2020;11:902. 10.1038/s41419-020-03123-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seidlitz T, Merker SR, Rothe A, Zakrzewski F, von Neubeck C, Grützmann K, et al. Human gastric cancer modelling using organoids. Gut. 2019;68:207–17. 10.1136/gutjnl-2017-314549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A. 2015;112:13308–11. 10.1073/pnas.1516689112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan HHN, Siu HC, Ho SL, Yue SSK, Gao Y, Tsui WY, et al. Organoid cultures of early-onset colorectal cancers reveal distinct and rare genetic profiles. Gut. 2020;69:2165–79. 10.1136/gutjnl-2019-320019 [DOI] [PubMed] [Google Scholar]
- 44.Almeida JL, Korch CT. Authentication of human and Mouse Cell Lines by Short Tandem Repeat (STR) DNA genotype analysis. In: Markossian S, Grossman A, Brimacombe K, Arkin M, Auld D, Austin C, et al. editors. Assay Guidance Manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda (MD),; 2004. [PubMed]
- 45.Watkins TBK, Lim EL, Petkovic M, Elizalde S, Birkbak NJ, Wilson GA, et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature. 2020;587:126–32. 10.1038/s41586-020-2698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang KK, Ma H, Chong RHH, Uchihara T, Lian BSX, Zhu F et al. Spatiotemporal genomic profiling of intestinal metaplasia reveals clonal dynamics of gastric cancer progression. Cancer Cell 2023; 41: 2019–2037.e2018. [DOI] [PMC free article] [PubMed]
- 47.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94. 10.1038/nrclinonc.2017.166 [DOI] [PubMed] [Google Scholar]
- 48.Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Liu H, Chen K. Living biobank-based cancer organoids: prospects and challenges in cancer research. Cancer Biol Med. 2022;19:965–82. 10.20892/j.issn.2095-3941.2021.0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of this study (PRJNA1015608) was uploaded into SRA database