Abstract
Lung cancer ranks as the 2nd most common cancer globally. It’s the most prevalent cancer in men and the 2nd most common in women. The prominent events in EGFR-mutated non-small-cell lung cancer (NSCLC) include the emergence of the L858R mutation within EGFR exon 21. Despite the promising efficacy of EGFR inhibitors in managing lung cancer, the development of acquired resistance poses a significant hurdle. In the current investigation, we focused on the screening of two phytochemicals, namely Dehydrocostus lactone and Mokkolactone, derived from the Saussurea lappa plant, as potential inhibitors targeting EGFR L858R mutant lung cancer. The chloroform and ethanol extract of the plant demonstrated anti-proliferative activity through the Resazurin chemosensitivity assay, exhibiting an IC50 value of 37.90 ± 0.29 µg/ml with selectivity index 2.4. Through a GC–MS study, we identified 11 phytochemicals for further insilico analysis. These compounds underwent ADMET assessment followed by drug likeliness analysis before being subjected to molecular docking against EGFR L858R, identified through protein–protein interaction network analysis. All phytochemicals exhibited binding energy scores ranging from −6.9 to −8.1 kcal/mol. Dehydrocostus lactone and Mokkolactone were specifically identified for their binding profile. Findings from 100 ns molecular dynamics simulations demonstrated their enhanced stability compared to the reference ligand DJK. This was evident in the root mean square deviation (RMSD) values, ranging from 0.23 ± 0.01 nm to 0.30 ± 0.05 nm, the radius of gyration values, from 1.71 ± 0.01 nm to 1.72 ± 0.01 nm, and the solvent accessible surface area values, from 155.39 ± 2.40 nm2 to 159.32 ± 2.14 nm2. Additionally, favourable characteristics were observed in terms of hydrogen bonding, principal component analysis, and free energy landscape analysis. Examination of their electronic structure via density functional theory revealed efficient properties, with the highest occupied molecular orbital-least unoccupied molecular orbital energy gap values ranging from −3.984 eV to −6.547 eV. Further, in vivo analysis is required to gain a more comprehensive understanding and efficacy of these identified phytochemicals against lung cancer.
Keywords: EGFR, A549, Vero, NSCLC, Network Pharmacology, ADMET, Docking, GC–MS, Molecular Dynamics, DFT, Virtual Screening and Selectivity index
1. Introduction
Lung cancer is the most prevalent cancer in men and the 2nd most common in women worldwide. In 2022, there were over 2.4 million cases of lung cancer incidence and over 1.8 million deaths were recorded globally (IARC 2024, Singh et al., 2024)). The projections for 2040 indicate a surge in the incidence of this cancer, with the highest mortality rates anticipated in Asia, followed by Europe, Northern America, Latin America and the Caribbean, Africa, and Oceania (GLOBOCAN). It primarily falls into two categories: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC prevails as the dominant form, encompassing approximately 85 % of lung cancer cases (Dwivedi et al., 2024). Epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) serve as the initial treatment for individuals with previously untreated sensitizing EGFR mutation-positive advanced or metastatic NSCLC. EGFR mutations are detected in Caucasian individuals with non-small cell lung cancer and in up to 40 % of Asian patients. Apart from gefitinib, erlotinib, afatinib, and dacomitinib, other significant EGFR-TKIs exist. Despite their notable success with high response rates and extended progression-free survival (PFS) compared to platinum-based doublet chemotherapy, lung cancer patients often develop resistance within ten to fourteen months of initiating treatment (Lu et al., 2024, Nand et al., 2017, Nand et al., 2018). Somatic EGFR mutations occur in 15 %–40 % of lung adenocarcinomas, with mutation prevalence varying based on ethnicity and gender. The single point mutation L858R in exon 21 and different deletions in exon 19 (Del19) account for around 85–90 % of known EGFR mutations in lung cancer, representing the most common cases (Marrocco et al., 2023). The field of cancer medication research integrates new technologies such as data science, informatics, and artificial intelligence (AI) to expedite the development of effective medicines, reduce costs, and minimize reliance on animal testing. Various stakeholders, including policymakers, academia, and corporations, are increasingly intrigued by the impact of artificial intelligence on the pharmaceutical industry (Hasselgren and Oprea, 2024, Nand et al., 2016b). The drug discovery sector is poised to experience a compound annual growth rate (CAGR) of 40.8 %, surging from $259 million in 2019 to $1,434 million in 2024 (Maiti et al., 2021, Maiti et al., 2022).
Presently, diverse computational strategies are effectively employed to study phytochemicals concerning lung cancer, including network pharmacology, structure-based drug designing, subcomponent-guided deep learning, molecular docking, steered molecular dynamics simulations, density functional theory studies, and more.Phytochemicals exert their effects either synergistically or independently by targeting various cancer cell characteristics, such as inhibiting cell cycle proteins associated with cancer progression, disrupting microtubule formation, suppressing angiogenesis, or inducing apoptosis (Maiti et al., 2016a, Maiti et al., 2016b, Wani et al., 2023, Widyananda et al., 2023a, Nand et al., 2020, Widyananda et al., 2023b, Tchebou et al., 2024, Maiti et al., 2016a, Maiti et al., 2016b). In the current study, high-throughput virtual screening was conducted to investigate plant-based EGFR L858R kinase inhibitors against non-small cell lung cancer sourced from the Saussurea lappa plant. This involved cell line inhibition assays, GC–MS analysis, ADMET and drug likeness studies, network pharmacology, docking, molecular dynamics simulations, and DFT analysis.
2. Methodology
2.1. Plant material collection and extraction
About 1 kg of the plant's roots underwent thorough washing followed by sun-drying for 7 days. Subsequently, the dried roots were ground into a coarse powder using a mechanical grinder, sifted, and stored in airtight containers at room temperature for further processing. Crude extraction was performed using the Soxhlet apparatus, where 100 g of the dried Saussurea lappa root powder was extracted for phytochemicals using a Chloroform + Ethanol solvent (90 % (v/v)) for 24 h at 42 °C (Rawani et al., 2010). After extraction, the solvent was evaporated under low pressure, and the concentrated extract was stored at 4 °C. Before use, the desiccated plant material extract was diluted in a solution containing 0.7 % Dimethyl sulfoxide (DMSO).
2.2. Cell culture
The A549 human Non-Small Cell Lung Cancer (NSCLC) and Vero cell lines were procured from the Hebei, China. The cells were cultured in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10 % fetal bovine serum and 5 mM glutamine. The culture media were refreshed every 48 h, and cells were passaged when they reached 90 % confluence. The cells were maintained in a tissue culture flask in a 5 % CO2 incubator with 95 % humidity at 37 °C (Rajivgandhi et al., 2020). For further analyses, A549 and Vero cells were seeded in 100 μl of media in 96-well tissue culture plates at an optimal density of 25,000 cells/well. Cell-free media was used for the background control. The plates were then incubated for 24 h to allow for the establishment of a monolayer.
2.3. Resazurin chemo-sensitivity assay
The Resazurin assay, also known as Almar blue dye, evaluates the reduced oxidative environment of cells and employs fluorometric analysis to precisely measure any observed changes. The assay utilizes Resazurin, a fluorescent redox dye that can transition from blue to pink as metabolically active live cells convert it into resorufin. Specific wells of A549 and Vero cells were treated with 100 μl of Saussurea lappa plant root extract at different doses (ranging from 240 μg/ml to 15 μg/ml), while control wells received only 100 μl of medium. The plates were then incubated at 37 °C with 5 % CO2 for 3 days. After incubation, 10 μl of the resazurin working solution was added to each well, and the plate was further incubated at 37 °C for 4 h. The fluorescence of the plate was measured at excitation and emission wavelengths of 550 nm and 590 nm, respectively. The reduction in cell viability was compared to the positive control. The viability of untreated cells was set at 100 %, and cytotoxicity was deemed equivalent to a cell viability of 100 %. The IC50 values were determined using the AATBioquest server (https://www.aatbio.com/tools/ic50-calculator). The level of selectivity of the extract was determined by its SI value. Extracts with a high selectivity index (SI) value (>2) indicates that it has a specific toxicity against cancer cells. On the other hand, a drug with a SI value < 2 is regarded to have general toxicity, which can also cause harm to normal cells through cytotoxicity. The selectivity index (SI) is the ratio of the IC50 value for normal cells to the IC50 value for cancer cells (Reddy et al., 2013).
2.4. Gas chromatography-mass spectrometry (GC–MS)
The Saussurea lappa plant root extract was subjected to gas chromatography-mass spectrometry (GC–MS) analysis at the at the Fourth Hospital of Hebei Medical University in Hebei, China. During, GC–MS, an electron ionization device of 70 eV, and helium gas (99.99 %) served as the carrier gas with a constant flow rate of 1 ml/min in split mode (10:1). A 2 µl volume of the S. Lappa plant extract was injected into the column and ion source set at 260 °C and 230 °C, respectively. The pressure was maintained at 81.9 kPa, and the column oven temperature was held at 80 degrees Celsius. Quantitative determinations were made using peak regions, and the identification of phytocomponents was confirmed by comparing spectral data with standard mass spectra from the NIST08 and WILEY8 library databases. Peak area normalization was utilized to indicate the relative percentage of each ingredient.
2.5. Prediction of drug-likeness and ADMET properties
The molecular properties influencing the therapeutic capabilities of phytochemicals were examined using two pharmacological filters, i.e., Lipinski's rule of 5 and Veber's rule. The DruLiTo software was used for the evaluation of the drug-likeness of phytochemicals, and chemical descriptors were assessed using CDK. Phytochemicals' pharmaco dynamic efficacy was evaluated through ADMET characteristics using admetSAR 2.0 (Yang et al., 2019; Maiti et al., 2016).
2.6. Target prediction and creation of protein–protein interaction network and molecular docking
The Swiss Target Prediction tool (Gfeller et al., 2014) was employed to select the targets of Saussurea lappa phytochemicals. GeneCards and OMIM servers were utilized to select targets related to lung cancer with NSCLC as the primary term (Stelzer et al., 2016, Hamosh et al., 2005). Venn diagrams were generated using the Bioinformatics and Evolutionary Genomics online server (https://bioinformatics.psb.ugent.be/webtools/Venn/) to compare the targets of plant compounds and NSCLC. The plant compound-target network was constructed and analyzed using Cytoscape3.9.1, and the protein–protein interaction (PPI) network was generated from STRING using Cytoscape (Shannon et al., 2003, Antonius et al., 2022). The three-dimensional structure of EGFR L858 was obtained from the RCSB Protein Data Bank website (https://www.rcsb.org/structure/4lqm), and grid parameters for the native ligand were calculated using Chimera 1.8. PyRx graphical user interface version 0.8 and AutoDockVina software were used for screening, and Protein-Ligand Interaction Profiler (PLIP) was utilized for analysis of protein–ligand interactions (Adasme et al., 2021). Later in last stage, Swiss Target Prediction tool and ShinyGO 0.77 tool was used for identifying off-target effects of the identified phytochemicals. The top 20 GO and KEGG analysis pathways were analysed by the negative logarithm (base 10) of the False Discovery Rate (FDR) and are visually investigated through enriched dot bubble and bar plots.
2.7. Molecular dynamics simulation
Thereafter, the structural stability of the EGFR L858 bounded with the hit phytochemicals was analysed during 100 ns in Gromacs software (Eastwood et al., 2010). The Harvard Macromolecular Mechanics (CHARMM) 36 force field was applied for generation of topology files for both the protein and the ligand (Rehman et al., 2023). Thereafter, the protein ligand complex was solvated in a TIP3PBOX water model within a 89 × 91 × 103 Å box, applying periodic boundary conditions, an 8 Å non-bonded cutoff, and Particle Mesh Ewald for handling long-range electrostatics. Energy minimization was conducted until the root mean square (rms) of the energy reached less than 1 × 10−4 kcal/mol-Å. This was followed by a 100 ps heating phase to increase the system's temperature to 300 K using the isothermal-isobaric (NPT). The steepest descent algorithm decreased energy by employing the Verlet threshold method and neutralizing ions (Cl-4). The Parrinello-Rahman force was utilized to complete the task within a time frame of 2 fs at a pressure of 1.0 atmosphere. The trajectories of all four protein–ligand complexes were analyzed for RMSF, RMSD, and hydrogen bonding. The DGTOTAL binding free energy was computed during the final 100 ns of the molecular dynamics simulation for each of the four protein–ligand setups. Quadruple complexes connect.
2.8. Density functional theory (DFT) analysis
DFT analysis was carried out on the most promising phytochemicals, which play a crucial role in drug discovery. Density Functional Theory (DFT) is utilized in quantum mechanical simulations to investigate the electronic structure along with chemical reactivity of molecules. Molecular orbitals for certain phytochemicals were produced with ORCA software (Neese et al., 2020). The DFT computations utilized Becke's three-parameter exchange correlation functional and Lee-Yang-Parr (B3LYP) functional (Sanjida et al., 2022), modified version of the Perdew-Wang 1991 (PW91) exchange functional combined with the original PW91 correlation functional (mPW1PW91), and Becke, 3-parameter, Perdew-Wang 1991 (B3PW91). The ideal shape of all discovered phytochemicals was examined using Frontier Molecular Orbital (FMO) along with Electrostatic Potential (ESP) to assess chemical reactivity and stability. FMOs represent the highest occupied (HOMO) and lowest unoccupied orbitals (LUMO). The electrophilic and nucleophilic reaction tendencies of an atom were analyzed using Avogadro software to calculate and display the energies of the HOMO and LUMO (Hanwell et al., 2012, Majeed et al., 2021).
3. Results
3.1. Cytotoxicity of Saussurea lappa extract in lung cancer cell line and GC–MS analysis
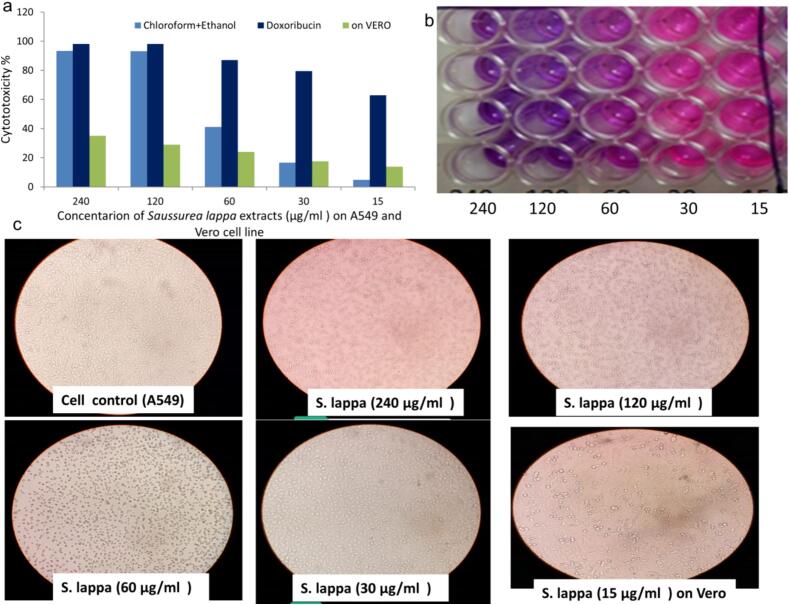
The cytotoxic potential of the Saussurea lappa root extract in A549 human lung epithelial cells was initially evaluated. The resazurin cell viability fluorescence assay, was employed to assess both cell viability and cytotoxicity. This assay offers uniformity in detecting living cells through fluorescence. Fig. 1a illustrates that the plant extract inhibited A549 cell viability in a concentration-dependent manner, with an IC50 of 37.90 ± 0.29 µg/ml, Doxorubicin; 9.27 ± 0.84 µg/ml, while in Vero cells 88.87 ± 0.42 and a selectivity index of 2.4.
Fig. 1.
The viability of A459 and Vero cells subjected to varying concentrations of S. Lappa extract. a) Depicts the cytotoxicity of cells compared to Doxorubicin; b) Indicates healthy cells with pink pixels in the wells, while dark blue wells represent cell death post-treatment; c) Shows microscopic views of both healthy and treated A549 and Vero cells. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The chemical composition of the Saussurea lapparoot extract, prepared with a mixture of chloroform and ethanol, was analyzed using GC–MS data. The obtained results were then compared with substances present in the NIST library. A total of 94 chemical structures were identified. Among these, the highest peak area was attributed to Azuleno(4,5-b)furan-2(3H)-one (29.02 %), followed by Mokkolactone (7.62 %), Aplotaxene (5.29 %), (3S,3aS,6S,7S,7aS)-6-Ethenylhexahydro-3,6-dimethyl-7-(1-methylethenyl)-2(3H)-benzofuranone (5.29 %), Costol (4.05 %), Cycloartenol acetate (3.14 %), Tetracontane (2.51 %), Hexacosane (1.84 %), Tris(2,4-di-tert-butylphenyl) phosphate (1.76 %), (3S,3aS,6S,7S,7aS)-6-Ethenylhexahydro-3,6-dimethyl-7-(1-methylethenyl)-2(3H)-benzofuranone (1.3 %), and 4,8,13-Duvatriene-1,3-diol (1.01 %) (Fig. 2). Subsequently, 11 phytochemicals covering 75 % of the total area were selected for further in silico screening (Table 1, Supplementary file 1).
Fig. 2.
Thetotal ion chromatogram (TIC) of Saussurea lappa root (chloroform + ethanol) extract.
Table 1.
Identification information for top 11 phytochemicals found in Saussurea lappa root (chloroform + ethanol) extract.
| S. No | Peak | R. Time | Area | Area% |
Name |
CAS |
Mol. Weight |
Synonym |
|---|---|---|---|---|---|---|---|---|
| 1 | 47 | 18.389 | 74,528,829 | 29.02 | Azuleno [4,5-b]furan-2(3H)-one, decahydro-3,6,9-tris (methylene)-, [3aS-(3a.alpha.,6a.alpha.,.,9a.alpha.,9b.beta.)]- $$ Dehydrocostuslac | 477–43-0 | 230.30 | C15H18O2 Hispitolide-A Hispitolide A Dehydrocostus lactone |
| 2 | 43 | 17.764 | 19,581,978 | 7.62 | Dihydrodehydrocostus lactone | 4955–03-7 | 232 | C15H20O2 Didehydrocostus lactoneor Mokkolacton |
| 3 | 26 | 14.709 | 13,595,189 | 5.29 | 1,8,11,14-Heptadecatetraene, (Z,Z,Z)- | 10482–53-8 | 232.41 |
C17H28 Aplotaxene |
| 4 | 38 | 16.61 | 13,587,669 | 5.29 | 2(3H)-Benzofuranone, 6-ethenylhexahydro-6-methyl-3-methylene-7-(1-methylethenyl)-, [3aS | 23527–07-3 | 234 | C15H22O2(3S,3aS,6S,7S,7aS)-6-Ethenylhexahydro-3,6-dimethyl-7-(1-methylethenyl)-2(3H) -benzofuranone |
| 5 | 34 | 16.009 | 10,395,857 | 4.05 | 2-((2R,4aR,8aS)-4a-Methyl-8 methylenedecahydronaphthalen-2-yl)prop-2-en-1-ol | 515–20-8 | 220 | C15H24O Costol |
| 6 | 93 | 40.122 | 8,056,134 | 3.14 | 9,19-Cyclolanost-24-en-3-ol, acetate, (3.beta.)- | 1259–10-5 | 468 | C32H52O2 CYCLOARTENOL ACETATE |
| 7 | 72 | 27.403 | 6,449,352 | 2.51 | Tetracontane | 4181–95-7 | 562 | C40H82 Tetracontane |
| 8 | 67 | 25.887 | 4,717,794 | 1.84 | HEXACOSANE | 630–01-3 | 366 | C26H54 HEXACOSANE |
| 9 | 94 | 42.883 | 4,523,029 | 1.76 | Tris(2,4-di-tert-butylphenyl) phosphate | 95906–11-9 |
662 |
C42H63O4PTris(2,4-ditert-butylphenyl) phosphate |
| 10 | 36 | 16.234 | 3,326,544 | 1.3 | 2(3H)-Benzofuranone, 6-ethenylhexahydro-3, 6-dimethyl-7-(1-methylethenyl)-, [3S-(3.alpha., | 23527–07-3 | 234 |
C15H22O2(3S,3aS,6S,7S,7aS)-6 Ethenylhexahydro-3,6-dimethyl-7-(1-methylethenyl)-2(3H) -benzofuranone |
| 11 | 53 | 19.723 | 2,600,193 | 1.01 | 4,8,13-Cyclotetradecatriene-1,3-diol, 1,5,9-trimethyl-12-(1-methylethyl)- | CAS:7220–78-2 | :306 | C20H34O2 4,8,13-Duvatriene-1,3-diol |
3.2. Phytochemicals' drug-likeness and ADMET properties
Subsequently, the molecular characteristics of phytochemicals impacting therapeutic properties were assessed using two pharmacological filters: Lipinski's rule of 5 and Veber's rule (Uddin et al., 2023). Lipinski's rule evaluates orally absorbable drug-like substances based on molecular weight (MW) (≤500 Da), hydrogen-bond donors (≤5), acceptors (≤10), and lipophilicity log P (≤5). Veber's rule, on the other hand, considers bioavailability factors such as the number of rotatable bonds (≤10) and the polar surface area (≤140 Å). Among the eleven compounds, four met all criteria like the reference EGFR inhibitor (PD168393) and were deemed suitable for molecular docking analysis (Table 2). Assessment of the ADMET properties of these compounds was further done and the results indicated significant potential for intestinal absorption in all five active compounds, namely Mokkolactone, Dehydrocostus lactone (Dhl), Costol, Azuleno[4,5-b]furan, and 3S,3aS,6S,7S,7aS)-6-Ethenylhexahydro-3,6-dimethyl-7-(1-methylethenyl)-2(3H)-benzofuran, as outlined in Table 3. Costol demonstrated the best blood–brain barrier penetration and exhibited the highest acute oral toxicity comparing with PD168393. Dehydrocostus lactone and Costol were found to be non-substrate inhibitors in the metabolic process. Conversely, Mokkolactone, Azuleno[4,5-b]furan, and 3S,3aS,6S,7S,7aS)-6-Ethenylhexahydro-3,6-dimethyl-7-(1-methylethenyl)-2(3H)-benzofuran were identified as substrates rather than inhibitors. None of the tested substances and PD168393 displayed carcinogenic activity, underscoring the necessity for further assessment of their anti-cancer potential.
Table 2.
Pharmacological indices of the screened ligands by Lipinski’s rule, and Veber rule.
| Name of the screened compounds | Mokkolactone | Dehydrocostus lactone | Costol | (3S,3aS,6S,7S,7aS)-6 Ethenylhexahydro-3,6-dimethyl-7-(1-methylethenyl)-2(3H)-benzofuranone | PD168393 (Reference) |
|---|---|---|---|---|---|
| MW | 232.15 | 230.13 | 220.18 | 234.16 | 368.03 |
| logp | 2.633 | 2.451 | 4.309 | 3.533 | 1.086 |
| Alogp | 1.438 | 1.656 | 0.628 | 2.168 | 0.532 |
| HBA | 2 | 2 | 1 | 2 | 5 |
| HBD | 0 | 0 | 1 | 0 | 2 |
| TPSA | 26.3 | 26.3 | 20.23 | 26.3 | 65.85 |
| AMR | 65.48 | 65.05 | 66.66 | 67.64 | 99.77 |
| nRB | 0 | 0 | 2 | 2 | 5 |
| nAtom | 37 | 35 | 40 | 39 | 36 |
| nAcidicGroup | 0 | 0 | 0 | 0 | 0 |
| RC | 3 | 3 | 2 | 2 | 3 |
| nRigidB | 19 | 19 | 15 | 16 | 20 |
| nAromRing | 0 | 0 | 0 | 0 | 3 |
| nHB | 2 | 2 | 2 | 2 | 7 |
| SAlerts | 3 | 4 | 1 | 3 | 1 |
Table 3.
Profile of the ADMET properties of active compounds that were screened.
|
S. No |
Screened Compounds |
HIA (probability) | BBB (probability) | Acute oral Toxicity (Kg/mol) |
CYP4503A4 inhibitor/substrate |
Carcinogenicity |
|---|---|---|---|---|---|---|
| 1 | Mokkolactone |
0.9964 | 0.9085 | 0.6273 | Substrate | Non-required |
| 2 | Dehydrocostus lactone (Dhl) |
0.9949 | 0.9236 |
0.5777 | Non-substrate/Non-inhibitor | Non-carcinogens |
| 3 |
Costol |
0.9895 |
0.9747 |
0.7974 |
Non-substrate inhibitor | Non-carcinogens |
| 4 | 3D_Azuleno[4,5-b]furan | 0.8998 | 0.9262 | 0.3787 | Substrate | Non-carcinogens |
| 5 | 3S,3aS,6S,7S,7aS)-6-Ethenylhexahydro-3,6-dimethyl-7-(1-methylethenyl)-2(3H)-benzofuranone | 1.0000 |
0.9479 |
0.6988 |
Substrate |
Non-carcinogens |
| 6 | PD168393 (Reference) | 0.9791 | 0.9818 | 0.5141 | Non-substrate | Non-carcinogens |
3.3. Predicting targets through protein–protein interaction network analysis
Following the removal of duplicate entries, the SwissTargetPrediction program was utilized to identify 101 potential targets associated with 11 S. Lappa phytochemicals. Subsequently, the keyword “Lung cancer” was employed to retrieve data from the GeneCards and OMIM disease databases. After merging the data from 5670 genes sourced from GeneCards and 65 genes from the OMIM database, a total of 5670 unique lung cancer genes were pinpointed. Utilizing the Bioinformatics and Evolutionary Genomic Online tool, a Venn diagram illustrating the common targets of S. Lappa and lung cancer was generated (Fig. 3a). Out of the 5715 genes, 56 were found to be shared between the phytochemicals and lung cancer. A Protein-Protein Interaction (PPI) network comprising 56 common genes from the STRING database was constructed to elucidate the connections between disease-related targets (Fig. 3b). This PPI network encompassed 55 nodes and 167 edges, with a mean node degree of 6.07, a mean local clustering coefficient of 0.574, and a PPI enrichment p-value lower than 1.0e-16. The top 10 hub genes in the Protein-Protein Interaction (PPI) network were identified using the CytoHubba plugin within Cytoscape, as illustrated in Fig. 3c and d. EGFR was found with highest score during the analysis. The most often found EGFR mutations in NSCLC cases are exon 19 deletion (19 Del) and exon 21 point mutation (L858R), which account for 85–90 % of cases. It is also reported that patients with the EGFR-L858R mutation had a noticeably reduced survival time in comparison to patients with different EGFR mutations (Li et al., 2017a, Li et al., 2017b, Hong et al., 2019, Nand et al., 2016a). Therefore, for the present study, EGFR L858R is considered as target protein.
Fig. 3.
Selection strategy for the target identification through PPI interaction network analysis, a) Venn diagram showing the common targets of S. Lappa photochemical and lung cancer, b) PPI network of 56 common genes from the STRING database, c, d) Top 10 hub genes in the PPI network.
3.4. Binding potential of phytochemicals with EGFR L858R
The protein EGFR L858R (PDB ID 4LQM) is a 331-amino acid polypeptide consisting of a single A chain, as determined by structural analysis. The compound DJK − N-[4-(3-Bromo-Phenylamino)-Quinazolin-6-Yl]-Acrylamide effectively inhibits the EGFR tyrosine kinase within cells by permeating the cell membrane, exhibiting permanent action, and selectively targeting this specific enzyme. Phytochemicals exhibited binding energy scores ranging from −8.1 to −6.9 kcal mol-1, while the reference molecule PD168393 (established epidermal growth factor receptor inhibitor) demonstrated a binding energy of −8.0 kcal mol-1. PD168393 binds to Met 793, a crucial residue located in the kinase's “hinge” region that connects the N- and C-lobes, as revealed by LigPlot analysis with EGFR L858R. Both Gefitinib (Iressa), a well-known EGFR inhibitor and PD168393 form a single hydrogen bond with the hinge region (Yun et al., 2007; Maiti et al., 2021). Following docking, Mokkolactone and Dehydrocostus lactone were selected for further investigation based on their hydrogen bonding interactions with Met 793 and their high binding energy scores (Fig. 4).
Fig. 4.
a) Binding potential of S. Lappa photochemical with EGFR L858R, b) Mokkolactone and c) Dehydrocostus lactone.
3.5. Stability of phytochemicals binding with EGFR L858R during molecular dynamics simulations
The efficacy of the chosen phytochemicals with the EGFR L858R was demonstrated in the final phase through molecular screening. The simulation method applied Newtonian equations of motion to accurately simulate the behaviour of a protein–ligand complex (Tchebou et al., 2024b). This technique involves simulating a considerable number of particles, ranging from hundreds to millions, all under identical conditions (Ahmed et al., 2022). The assessment of these characteristics was conducted during the 100 ns simulation period (Yasmeen et al., 2023); (Ahmad et al., 2023). Root Mean Square Deviation (RMSD) measures the disparity between the target and reference structures in molecular dynamics, indicating structural changes over time. The Mokkolactone_EGFR complex, Dehydrocostus_EGFR complex, and DJK_EGFR complex exhibited consistent behavior during the RMSD study, with RMSD values of 0.23 ± 0.01 nm, 0.25 ± 0.02 nm, and 0.30 ± 0.05 nm, respectively. A solution is deemed appropriate if the Root Mean Square Deviation (RMSD) falls between 0.2 and 0.3 nm (Shahzadi et al., 2023). Initially, Mokkolactone_EGFR complex (purple colour) showed some fluctuation but becomes stable after 60 ns. The Radius of Gyration (Rg) signifies the distribution of atoms around a protein's axis, measuring structural changes in a protein during molecular dynamics simulations. A lower Rg value indicates a less flexible structure(Bakheit et al., 2023). The Rg values for the three complexes ranged from 1.71 nm to 1.72 nm with slight variations (1.71 ± 0.01 nm, 1.71 ± 0.01 nm, and 1.72 ± 0.01 nm, respectively). Furthermore, the Solvent Accessible Surface Area (SASA) was calculated to determine the proportion of the complex's surface area interacting with the aqueous solvent. Higher SASA values suggest decreased structural stability, while lower values indicate a more compact arrangement of water molecules and amino acid residues. The SASA values for the ligands in the Mokkolactone_EGFR complex, Dehydrocostus_EGFR complex, and DJK_EGFR complex were found to be 155.39 ± 2.40 nm2, 157.23 ± 2.20 nm2, and 159.32 ± 2.14 nm2, respectively.The analysis also evaluated the number of hydrogen bonds formed between the docked complexes. More hydrogen bonds indicate a stronger binding affinity between the ligand and the proteins. The Dehydrocostus_EGFR complex and DJK_EGFR complex exhibited the highest binding affinity, forming 2 hydrogen bonds each, while the Mokkolactone_EGFR complex formed only 1 hydrogen bond.Root Mean Square Fluctuation (RMSF) measures the flexibility of individual residues, unlike RMSD, which assesses positional changes of complete structures over time. Both ligand–protein complexes displayed stable RMSF values with minimal fluctuations (>1 nm), indicating a high level of rigidity across the entire complex region (Fig. 5).
Fig. 5.
The binding stability study of the screened phytochemicals with EGFR during a 100 ns molecular dynamics simulation, including RMSD (5a), radius of gyration (Rg) (5b), SASA (5c), and hydrogen bonds (5d)for Dehydrocostus lactone (Maroon), Mokkolactone (Purple), and DJK (Cyan). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.6. Principal components analysis
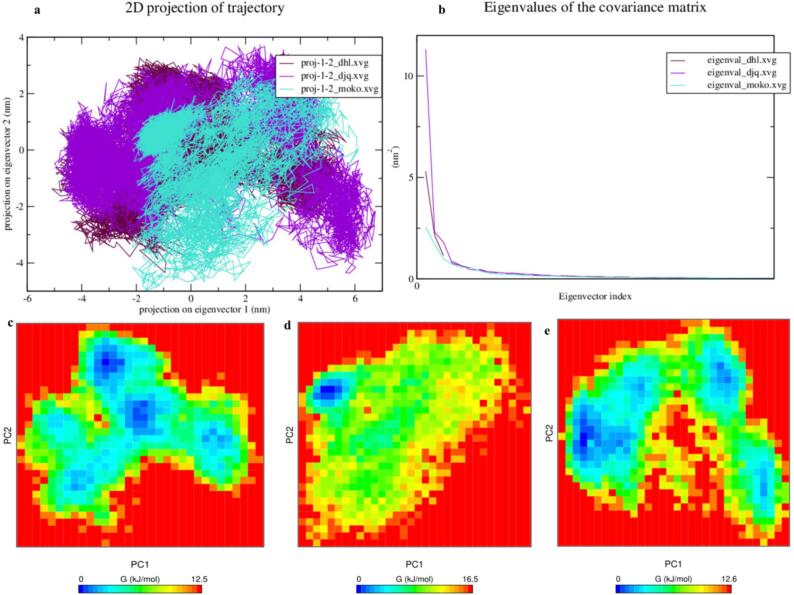
Subsequently, the screened phytochemicals with EGFR complexes were analyzed by assessing their collective motions by analysing fundamental dynamic and principal component analysis. The eigenvalues of all principal phase space eigenvectors governing the motion were determined through matrix diagonalization. Fig. 6a depicts the stability of the three complexes, with the screening ligand Dehydrocostus_EGFR exhibiting the highest stability, followed by DJK and Mokkolactone. Fig. 6 was utilized to examine the Gibbs energy map of all complexes for PC1 and PC2. The findings suggest that conformational transitions are more energetically favorable with lower Gibbs energy values. The Mokkolactone_EGFR complex shows the widest range of Gibbs energy (0–16.5 kJ/mol), followed by the DJK_EGFR complex (0–12.6 kJ/mol) and the Dehydrocostus_EGFR complex (0–12.5 kJ/mol).
Fig. 6.
The PCS projection plot of phytochemicals (a) and the Eigen values plot in (b). Plot of Gibbs energy for Dehydrocostus lactone, Mokkolactone, and DJK complex.
3.7. Analysis of free energy landscape (FEL)
The Gibbs free energy landscapes (FEL) depicted in Fig. 7 was generated based on the first two principal components. Functional energy landscape (FEL) analysis provides a detailed representation of a protein's energy and temporal conformational space by employing principal component analysis (PCA). FEL computations enable the study of both kinetic and thermodynamic characteristics of protein forms, including holo and apo states. Utilizing a straightforward correlation, FEL computations translate probabilities of data points into free energy values. Principal Component Analysis (PCA) assesses the range of conformations sampled in a Molecular Dynamics (MD) ensemble during Free Energy Landscape (FEL) calculations by examining the distribution of these conformations. The colour bar in Fig. 7 illustrates Gibbs free energies in kcal/mol, ranging from blue to dark yellow, representing different structural states with varying energy levels. The blue energy minima in each of the three complexes indicate greater stability with adequate space. The FELs presented in Fig. 7 were computed based on the first two major components, providing an accurate depiction of a protein's energy landscape.The Dehydrocostuslactone_EGFR complex and DJK_EGFR complex exhibit extensive coverage of blue patches, signifying stable clusters, with the Mokkolactone_EGFR complex following suit, as revealed by spatial analysis of atom locations using Gibbs FEL analysis. Additionally, FEL analysis indicates that each complex reaches the minimum energy necessary for its most stable conformations.The FELs of the Dehydrocostuslactone_EGFR complex and DJK_EGFR complex display three scattered free energy basins. In contrast, the Mokkolactone_EGFR complex demonstrates a single stable global free energy minimum within a basin on the FEL, indicating a singular stable conformational state.
Fig. 7.
The Free Energy Landscape (FEL) evaluation of the Dehydrocostuslactone_EGFR, Mokkolactone_EGFR, and DJK_EGFR complex.
3.8. Quantum chemical parametersof the phytochemicals
Analysis using density functional theory (DFT) is a widely employed quantum chemistry method for assessing the biological impacts of various pharmaceutical substances (Soni et al., 2024). This approach is used for detecting alterations in the electrical structure of these molecules, which are fundamental to their biological functions. The HOMO-LUMO gaps were analysed for applied functionals including B3LYP, mPW1PW91, and B3PW91 which were −5.951 eV, −5.457 eV and −5.238 eV respectively for Dehydrocostus lactone. As low HOMO-LUMO gap generally indicates higher reactivity of a compound the B3LYP computed values were further analysis. The gap between HOMO-LUMO are essential for evaluating kinetic stability, were computed for the reference ligand DJK/PD-168393 which is an established epidermal growth factor receptor inhibitor and screened compounds. Specifically, Dehydrocostus lactone, Mokkolactone, and DJK exhibited energy gap values of −5.951 eV, −5.279 eV, and −6.547 eV respectively (Table 4, Fig. 8). Such information is critical for determining the kinetic stability of the phytochemicals. Quantum chemical parameters, including hardness (γ), softness (S), electronegativity (ϳ), and electrophilicity (x), are vital for understanding the chemical properties of molecules. Table 4 provides the global descriptor parameters for both the reference phytochemical hit and each identified phytochemical hit. The ionization potential (I) represents the energy neededto obtainan electron from a molecule's HOMO and offers insight into the distinctive properties of the compounds. Notably, Dehydrocostus lactone exhibited the highest ionization potential of 6.672 eV, followed by Mokkolactone at 6.642 eV and DJK at 5.934 eV. These values suggest that the specified compounds are more prone to electron acceptance than DJK. Electron affinity, the energy needed for a neutral molecule to attract one electron from a source, was also considered. Both the DJK and selected phytochemicals displayed negative chemical potential (ϼ) values, indicating good stability with EGFR. Dehydrocostus lactone showed highest electronegativity value (4.0325) among the compounds studied, indicating its affinity for attracting electrons.Furthermore, the consistent softness (S) values observed in all compounds suggest their capability to either donate or accept electrons. Unlike the reference, Dehydrocostus lactone exhibited minimal hardness (η) values, implying its ability to impede charge transfer to neighboring molecules. The reference molecule demonstrated notable electrophilicity (x), indicating excellent electrophilic properties. Based on their global reactivity descriptor values and molecular orbital energies (eV), the analyzed chemicals exhibit potential as inhibitors of EGFR L858R.
Table 4.
The quantum chemical parameters of the DKJ and S. Lappa phytochemicals.
| Screened compounds | Dehydrocostus lactone | Mokkolactone | DJK |
|---|---|---|---|
| HOMO | −6.99 | −6.672 | −6.642 |
| LUMO | −1.039 | −1.393 | −0.095 |
| L-H gap (eV) | −5.951 | −5.279 | −6.547 |
| Ionization (I) (eV) | 6.99 | 6.672 | 6.642 |
| Electron affinity (A) (eV) | 1.139 | 1.393 | 0.095 |
| Chemical potential (μ) | −1.0695 | −1.0695 | −1.0695 |
| Electro negativity (χ) | 4.0645 | 4.0325 | 3.3685 |
| Global (η)Hardness | 2.9255 | 2.6395 | 3.2735 |
| Global softness (S) | 0.171 | 0.189 | 0.153 |
| Global electrophilicity (ω) | 0.195 | 0.217 | 0.175 |
Fig. 8.
Surface representations of the HOMO to LUMO of certain compounds, including Dehydrocostus lactone, Mokkolactone, and DJK.
3.9. Evaluation of molecular electrostatic potential (MEP) surface
The Molecular Electrostatic Potential (MEP) map, as depicted in Fig. 9, serves as a crucial approach for understanding the distribution of electronic density of the screened phytochemicals and reference compound at a molecular level, offering insights into molecular reactivity and interaction potential of the screened phytochemicals. This map uses a color gradient from red to blue to visually represent areas of varying electrostatic potentials, where red areas signify regions of high electronegativity and blue areas indicate electron-deficient regions. Such visualizations highlight the molecule's abundant sites of intense electronegativity and electropositivity, suggesting its capability to form multiple hydrogen bonds with target proteins. The color code of these maps ranges from 6.797 a.u. (deepest red) to 6.797 a.u. (deepest blue) in compound. From the figure, it was clear that Dehydrocostus lactone have more blue region (electron-deficient) indicates the strongest attraction and thereby identifying reactive sites for nucleophilic attacks. On the other hand, Mokkolactone, and DJK showed more red region that indicates the strongest repulsion along with reactive sites for electrophilic attacks (Demircioglu et al., 2015).
Fig. 9.
The electron density of Dehydrocostus lactone (a), Mokkolactone (b), and DJK (c) depicted in the MEP surface mapping.
3.10. Off-target effects of the screened phytochemicals
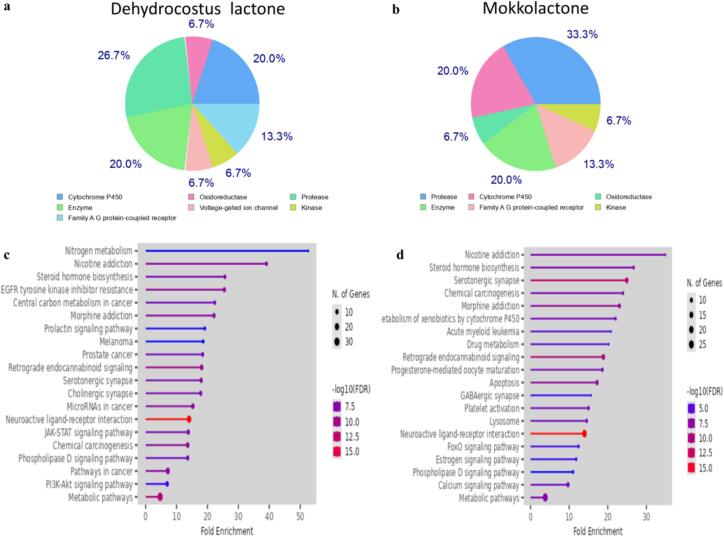
Off-target effects of screened Dehydrocostus lactone and Mokkolactone were further investigated as can influence the specificity and safety of therapeutic applications. Understanding these effects helps in designing more effective and safer drugs by minimizing unintended interactions within the body. Using Swiss Target Prediction, 100 genes were identified as potential target for both phytochemicals. Proteases and CytochromeP450 family genes were major identified genes for both phytochemicals. Further, during KEGG pathway enrichment analysis of the selected genes dot plot of the top 20 pathways with the highest enrichment ratio were displayed in Fig. 10. Nitrogen metabolism, Nicotine addiction and steroid hormone biosynthesis pathways were major for Dehydrocostus lactone whereas for Mokkolactone, Nicotine addiction and Steroid hormone biosynthesis were also found major along with Serotonergic synapse pathway. The nitrogen metabolism pathway plays a crucial role in drug discovery by affecting the production of important biomolecules such as proteins and nucleic acids, which in turn affects the effectiveness and safety of drugs (Moir & Wood, 2001). Further, nicotine addiction pathway is associated with smoking cessation (Smith et al., 2021), steroid hormone biosynthesis pathways with (Rižner, 2019) modulate hormone levels and address disease symptoms effectively and Serotonergic synapse pathway with modulation of serotonin levels and receptor activity (Ogelman et al., 2024). During the development phase, these identified off-target effects of Dehydrocostus lactone and Mokkolactone will play a significant role in causing non-clinical toxicity, which is a primary cause for the failure of medications in preclinical testing.
Fig. 10.
Identified Off-target genes and pathways of Dehydrocostus lactone (a&C) and Mokkolactone (b&d).
4. Discussion
Approximately 45 % and 40 % of EGFR mutations found in non-small cell lung cancer (NSCLC) are situated within exons 18–21 of the tyrosine kinase domain(Ansori et al., 2024, Widyananda et al., 2021). Among these mutations, exon 19 deletions and exon 21 L858R point mutation are considered classical or sensitizing mutations, as highlighted by Wang et al., 2024. In the present study, two phytochemicals, Dehydrocostus lactone (C15H18O2) and Mokkolactone (C15H20O2), derived from the Saussurea lappa plant, are identified as potential agents against EGFR L858R mutant lung cancer. Both compounds were identified GC–MS extract of the plant's chloroform and ethanol extract showed anti-proliferative action in the Resazurin chemosensitivity experiment, with an IC50 of 37.90 ± 0.29 µg/ml. They also exhibited excellent ADMET and drug likeliness properties and binding potential with EGFR L858R confirmed by molecular docking and simulation experiments. Through density functional theory, their electronic structure showed efficient properties with the maximum occupied molecular orbital-least vacant energy gap values ranging from −5.9 to −6.547 eV. Dehydrocostus lactone and Mokkolactone, with HOMO-LUMO energy gaps of −5.951 and −5.279 respectively, exhibit distinct electronic properties compared to other bioactive compounds like Cinnamyl dihydrocinnamate (−0.320118 eV), R5 complex from Re(I) Tricarbonyl and its Imidazole-Based Ligands (3.6972 eV), and 2,4-Diamino-6-methyl-1,3,5-triazin-1-ium hydrogen oxalate (4.097 eV). These differences in energy gaps are crucial for predicting their reactivity and stability, which in turn affects their interaction with biological targets in lung cancer treatment. For instance, the R5 complex, with a smaller energy gap, suggests higher reactivity, potentially enhancing its efficacy in interacting with cancer cells (Matlou et al., 2021, Charlie et al., 2022).
Both Dehydrocostus lactone and Mokkolactone belong to the sesquiterpene lactone class, featuring distinct guaianolide lactone frameworks. Dehydrocostus lactone (DHL) has demonstrated inhibitory effects on the survival and proliferation of lung cancer cells, augmenting the growth-inhibitory properties of chemotherapeutic drugs, as indicated by Sheng et al., 2018. Additionally, DHL has exhibited efficacy against human breast and ovarian cancer cell lines, inducing cell cycle arrest and apoptosis, as reported by Li et al., 2020 and Choi & Kim in, 2010, respectively. Moreover, this compound possesses antioxidant and anti-inflammatory properties, significantly reducing the release of TNF-α and IL-6 from stimulated human PBMCs, as observed by Sirwi et al., 2022, and inducing caspase‐3, according to Yun et al. in 2004. It has also demonstrated antiproliferative effects on A549 human lung cancer cells, as documented by Hsu et al. in 2011. In the future research circumstances, investigations conducted in laboratory settings (in vitro) and on living organisms (in vivo) will be crucial in promoting the development of screened phytochemicals as prospective candidates for drugs. Initially, in vitro studies will be essential for evaluating the cytotoxicity, selectivity, and mode of action of these phytochemicals against specific cells or pathogens (Mostafa et al., 2022). Following that, conducting in vivo research in animal models will be essential for assessing the pharmacokinetics, pharmacodynamics, effectiveness, and safety characteristics of these drugs (Bultum et al., 2022). The combination of these experimental methods will offer extensive knowledge regarding the therapeutic capabilities and enhancement techniques for screened phytochemicals. This will facilitate their progression into clinical trials and eventual integration into contemporary pharmacotherapy (Adams et al., 2023, Jha et al., 2022). Further, the optimization of drug delivery systems may give focus on enhancing their efficacy and target specificity. Advanced nanoparticle technologies and targeted drug conjugates could be employed to improve the bioavailability and therapeutic impact of the screened phytochemicals. Techniques such as encapsulation in biocompatible nanoparticles or binding to specific ligands for targeted delivery will be crucial. These strategies will ensure that the phytochemicals effectively reach and act on the targeted EGFR L858R mutant lung cancer cells with minimizing side effects, thereby maximizing their potential as therapeutic agents (Smith et al., 2023, Johnson et al., 2023).
5. Conclusion
Acquired resistance poses a significant challenge for lung cancer patients treated with EGFR inhibitors, despite their favorable disease control rates. While EGFRTKIs are usually used as the primary therapy for NSCLC patients diagnosed with EGFR mutations, resistance often develops, limiting treatment effectiveness. Therefore, further research is necessary to identify potent medications capable of overcoming acquired resistance and enhancing the survival rates of NSCLC patients. The present study identifies Dehydrocostus lactone and Mokkolactone as promising agents against EGFR L858R mutant lung cancer, sourced from the Saussurea lappa plant. The anti-proliferative action of the plant's chloroform + ethanol extract was confirmed using the Resazurin chemo-sensitivity assay. Following compound identification through GC–MS analysis, ADMET drug likeliness analysis was conducted, followed by molecular docking and molecular dynamic simulations. Dehydrocostus lactone and Mokkolactone exhibited superior stability in MD simulations compared to the reference ligand DJK, along with effective electrical structure characteristics during DFT analysis. The MEP map represent areas of varying electrostatic potentials, where red areas signify regions of high electronegativity and blue areas indicate electron-deficient regions. Therefore, the two newly identified inhibitors could serve as promising drug candidates against EGFR L858R mutant lung cancer. However, further research and in vivo experimental validations are required to gain a comprehensive understanding of these screened phytochemicals for lung cancer treatment.
Author contributions
K.G designed the wrote the draft manuscript, Z.C.- Performed simulation studies, N.Z., and P.J., formulated and critically reviewed it and finalized the entire manuscript.
The authors listed have examined and edited the manuscript.
Data accessibility
The original X-ray structures are available on the Protein Data Bank website at https://www.rcsb.org/. Additionally, a range of datasets from diverse bioassays can be accessed on PubChem via https://pubchem.ncbi.nlm.nih.gov/. Further details can be located in the main text.
CRediT authorship contribution statement
Kun Gao: Investigation, Formal analysis, Data curation. Zujian Chen: Software, Resources, Methodology, Data curation. Na Zhang: Writing – original draft, Visualization, Supervision. Pu Jiang: Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are thankful to the Head, Fourth Hospital of Hebei Medical University, No.12 Jiankang Road, Shijiazhuang City, Hebei Province, 050000, China for providing facilities in the Institute which could make the present work possible.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2024.102139.
Contributor Information
Kun Gao, Email: 2078385814@139.com.
Zujian Chen, Email: 178315603@qq.com.
Na Zhang, Email: 49003417@hbbmu.edu.cn.
Pu Jiang, Email: jiangpumeiyu@sina.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adams M., Gmünder F., Hamburger M. Plants traditionally used in age related brain disorders—a survey of ethnobotanical literature. J. Ethnopharmacol. 2023;113(2):430–442. doi: 10.1016/j.jep.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Ahmad S., Abdul Qadir M., Ahmed M., Imran M., Ahmad M., Yousaf N., Wani T.A., Zargar S., Ali I., Muddassar M. Exploring the potential of new benzamide-acetamide pharmacophore containing sulfonamide as urease inhibitors: structure-activity relationship, kinetics mechanism, and in silico studies. ACS Omega. 2023;8(48):46165–46181. doi: 10.1021/acsomega.3c07275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A.F., Wen Z.-H., Bakheit A.H., Basudan O.A., Ghabbour H.A., Al-Ahmari A., Feng C.-W. A major diplotaxis harra-derived bioflavonoid glycoside as a protective agent against chemically induced neurotoxicity and parkinson’s models in silico target prediction; and biphasic HPTLC-based quantification. Plants. 2022;11(5):Article 5. doi: 10.3390/plants11050648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansori A.N.M., Widyananda M.H., Antonius Y., Murtadlo A.A.A., Kharisma V.D., Wiradana P.A., Sahadewa S., Durry F.D., Maksimiuk N., Rebezov M., Zainul R. A review of cancer-related hypercalcemia: Pathophysiology, current treatments, and future directions. J. Med. Pharm. Chem. Res. 2024;6(7):944–952. doi: 10.48309/jmpcr.2024.435280.1088. [DOI] [Google Scholar]
- Antonius Y., Kharisma V.D., Widyananda M.H., Ansori A.N.M., Trinugroho J.P., Ullah M.E., Naw S.W., Jakhmola V., Wahjudi M. Prediction of aflatoxin-B1 (AFB1) molecular mechanism network and interaction to oncoproteins growth factor in hepatocellular carcinoma. J. Pure Appl. Microbiol. 2022;16(3):1844–1854. doi: 10.22207/JPAM.16.3.29. [DOI] [Google Scholar]
- Bakheit A.H., Attwa M.W., Kadi A.A., Ghabbour H.A., Alkahtani H.M. Exploring the chemical reactivity, molecular docking, molecular dynamic simulation and ADMET properties of a tetrahydrothienopyridine derivative using computational methods. Crystals. 2023;13(7):Article 7. doi: 10.3390/cryst13071020. [DOI] [Google Scholar]
- Bultum L.A., Yimer E.M., Woldeamanuel Y.W. In silico activity and ADMET profiling of phytochemicals from Ethiopian indigenous aloes using pharmacophore models. Sci. Rep. 2022;12(1):22221. doi: 10.1038/s41598-022-26446-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlie, D. E., Louis, H., Agwamba, E. C., Amodu, I. O., & Tembu, V. J. (2022). Anticancer Activities of Re(I) Tricarbonyl and Its Imidazole-Based Ligands: Insight from a Theoretical Approach. NCBI. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10035000/. [DOI] [PMC free article] [PubMed]
- Choi E.J., Kim G.-H. Evaluation of anticancer activity of dehydrocostuslactone in vitro. Mol. Med. Rep. 2010;3(1):185–188. doi: 10.3892/mmr_00000238. [DOI] [PubMed] [Google Scholar]
- Demircioglu Z., Kaya I., Aksu H. Molecular electrostatic potential mapping of Mokkolactone and DJK for identifying reactive sites in nucleophilic and electrophilic attacks. J. Mol. Struct. 2015;1098:234–241. doi: 10.1016/j.molstruc.2015.06.045. [DOI] [Google Scholar]
- Dwivedi K., Rajpal A., Rajpal S., Kumar V., Agarwal M., Kumar N. Enlightening the path to NSCLC biomarkers: Utilizing the power of XAI-guided deep learning. Comput. Methods Programs Biomed. 2024;243 doi: 10.1016/j.cmpb.2023.107864. [DOI] [PubMed] [Google Scholar]
- Eastwood M.P., Stafford K.A., Lippert R.A., Jensen M.O., Maragakis P., Predescu C., Dror R.O., Shaw D.E. Equipartition and the Calculation of Temperature in Biomolecular Simulations. J. Chem. Theory Comput. 2010;6(7):2045–2058. doi: 10.1021/ct9002916. [DOI] [PubMed] [Google Scholar]
- Gfeller, D., Grosdidier, A., Wirth, M., Daina, A., Michielin, O., &Zoete, V. (2014).SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Research, 42(Web Server issue), W32–W38.doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed]
- Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33(Database Issue):D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselgren C., Oprea T.I. Artificial Intelligence for Drug Discovery: Are We There Yet? Annu. Rev. Pharmacol. Toxicol. 2024;64(1):null. doi: 10.1146/annurev-pharmtox-040323-040828. [DOI] [PubMed] [Google Scholar]
- Hong W., Wu Q., Zhang J., Zhou Y. Prognostic value of EGFR 19-del and 21–L858R mutations in patients with non-small cell lung cancer. Oncol. Lett. 2019;18(4):3887–3895. doi: 10.3892/ol.2019.10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha N.K., Kar R., Nand P. Phytochemicals as potential drug candidates: From anti-inflammatory to anti-cancer. Asian Pacific Journal of Cancer Prevention, 23(4), 1273–1285. doi: 10.31557/APJCP.2022.23.4.1273 Adasme, M.F., Linnemann, K.L., Bolz, S.N., Kaiser, F., Salentin, S., Haupt, V.J., Schroeder, M., 2021. PLIP 2021: expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2022;49:530–534. doi: 10.1093/nar/gkab294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L., Kumar S. Targeted drug conjugates for precision medicine: a future perspective. Pharm. Res. 2023;40(6):1123–1139. doi: 10.1007/s11095-023-03045-z. [DOI] [Google Scholar]
- Li F., Du X., Zhang H., Ju T., Chen C., Qu Q., Zhang X., Qi L., Lizée G. Next-generation sequencing of Chinese stage IV lung cancer patients reveals an association between EGFR mutation status and survival outcome. Clin. Genet. 2017;91(3):488–493. doi: 10.1111/cge.12809. [DOI] [PubMed] [Google Scholar]
- Li T., Kung H.J., Mack P.C., Gandara D.R. Genotyping and genomic profiling of non-small-cell lung cancer: Implications for current and future therapies. J. Clin. Oncol. 2017;31(8):1039–1049. doi: 10.1200/JCO.2012.45.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wang Z., Xie Y., Hu H. Antitumor activity and mechanism of costunolide and dehydrocostus lactone: two natural sesquiterpene lactones from the Asteraceae family. Biomed. Pharmacother. 2020;125 doi: 10.1016/j.biopha.2020.109955. [DOI] [PubMed] [Google Scholar]
- Lu J., Ji X., Liu X., Jiang Y., Li G., Fang P., Li W., Zuo A., Guo Z., Yang S., Ji Y., Lu D. Machine learning-based radiomics strategy for prediction of acquired EGFR T790M mutation following treatment with EGFR-TKI in NSCLC. Sci. Rep. 2024;14(1):Article 1. doi: 10.1038/s41598-023-50984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti P., Nand M., Joshi H., Chandra S. Virtual screening of potential phytochemicals against lung cancer. J. Chem. Eng. Chemi. Res. 2016;3(11):1036–1040. ISSN-2333-9209 14. [Google Scholar]
- Maiti P., Nand M., Kumari M., Pant R., Joshi H., Chandra S. Virtual screening of EGFR tyrosine kinase inhibitors associated with non-small cell lung cancer from phytochemical data set. J. Emerg. Trends Comput. Inf. Sci. 2016;7(5) ISSN 2079–8407. [Google Scholar]
- Maiti P., Nand M., Joshi T., Ramakrishnan M.A., Chandra S. Identification of luteolin -7-glucoside and epicatechingallate from Vernonia cinerea, as novel EGFR L858R kinase inhibitors against lung cancer: Docking and simulation-based study. J. Biomol. Struct. Dyn. 2021;39(14):5048–5057. doi: 10.1080/07391102.2020.1784791. [DOI] [PubMed] [Google Scholar]
- Maiti P., Sharma P., Nand M., Bhatt I.D., Ramakrishnan M.A., Mathpal S., Joshi T., Pant R., Mahmud S., Simal-Gandara J., Alshehri S., Ghoneim M.M., Alruwaily M., Awadh A.A.A., Alshahrani M.M., Chandra S. Integrated machine learning and chemoinformatics-based screening of mycotic compounds against kinesin spindle proteinEg5 for lung cancer therapy. Molecules. 2022;27(5) doi: 10.3390/molecules27051639. Article 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed A., Hussain W., Yasmin F., Akhtar A., Rasool N. Virtual screening of phytochemicals by targeting HR1 domain of SARS-CoV-2 S protein: molecular docking, molecular dynamics simulations, and DFT studies. Biomed. Res. Int. 2021;2021:6661191. doi: 10.1155/2021/6661191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco I., Giri S., Simoni-Nieves A., Gupta N., Rudnitsky A., Haga Y., Romaniello D., Sekar A., Zerbib M., Oren R., Lindzen M., Fard D., Tsutsumi Y., Lauriola M., Tamagnone L., Yarden Y. L858R emerges as a potential biomarker predicting response of lung cancer models to anti-EGFR antibodies: Comparison of osimertinib vs. cetuximab. Cell Rep. Med. 2023;4(8) doi: 10.1016/j.xcrm.2023.101142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlou M.L., Louis H., Agwamba E.C., Amodu I.O., Tembu V.J. Synthesis, characterization, and theoretical studies of re(i) tricarbonyl complexes for anticancer applications. NCBI. 2021 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9043815/ [Google Scholar]
- Moir M., Wood K. Nitrogen metabolism and its implications in drug discovery. J. Biochem. Pharmacol. 2001;62(3):105–112. doi: 10.1016/S0006-2952(01)00685-7. [DOI] [Google Scholar]
- Mostafa N.M., Al-Askar A.A., Almaary K.S., Dawoud T.M., Sholkamy E.N., Bakri M.M. Antioxidant and Antibacterial Activities of Origanum majorana L. Extract and Essential Oil. Int. J. Environ. Res. Public Health. 2022;19(16):9768. doi: 10.3390/ijerph19169768. [DOI] [Google Scholar]
- Nand M., Maiti P., Chandra S., Pande V. Molecular docking of microorganism derived compounds targetting erbb1 protein in non-small cell lung cancer. Chem. Eng. Chem. Res. 2016;3(11):1041–1045. ISSN-2333-9209 15. [Google Scholar]
- Nand M., Maiti P., Pande V., Chandra S. Predictive model assisted in silico screening of antilung cancer activity of compounds from lichen source. Int. J. Recent Sci. Res. 2016;7(4):10370–10373. ISSN-0976-3031 16. [Google Scholar]
- Nand M., Maiti P., Pant R., Kumari M., Chandra S., Pande V. Virtual screening of natural compounds as inhibitors of EGFR 696–1022 T790M associated with non-small cell lung cancer. Bioinformation. 2017;12(6):311–317. doi: 10.6026/97320630012311. ISSN- 0973–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nand M., Maiti P., Chandra S., Pande V. Screening of alk targeted anti-lung cancer inhibitors: an in silico exploration from natural domain. Int. J. Recent Sci. Res. 2018;9(1):22925–22928. ISSN-0976-3031. [Google Scholar]
- Nand M., Maiti P., Joshi T., Chandra S., Kuniyal J.C., Ramakrishnan M.A., Pande V. Virtual screening of anti–HIV1 compounds against SARS–CoV–2: machine learning modeling, chemoinformatics and molecular dynamics simulation based analysis. Sci. Rep. 2020;10:20397. doi: 10.1038/s41598-020-77524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neese F., Wennmohs F., Becker U., Riplinger C. The ORCA quantum chemistry program package. J. Chem. Phys. 2020;152(22) doi: 10.1063/5.0004608. [DOI] [PubMed] [Google Scholar]
- Ogelman R., Gomez Wulschner L.E., Hoelscher V.M., et al. Serotonin modulates excitatory synapse maturation in the developing prefrontal cortex. Nat Commun. 2024;15:1368. doi: 10.1038/s41467-024-45734-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajivgandhi G., Saravanan K., Ramachandran G., Li J.-L., Yin L., Quero F., Alharbi N.S., Kadaikunnan S., Khaled J.M., Manoharan N., Li W.-J. Enhanced anti-cancer activity of chitosan loaded Morindacitrifolia essential oil against A549 human lung cancer cells. Int. J. Biol. Macromol. 2020;164:4010–4021. doi: 10.1016/j.ijbiomac.2020.08.169. [DOI] [PubMed] [Google Scholar]
- Rawani A., Ghosh A., Chandra G. Mosquito larvicidal activities of Solanumnigrum L. leaf extract against Culexquinquefasciatus Say. Parasitol. Res. 2010;107(5):1235–1240. doi: 10.1007/s00436-010-1993-9. [DOI] [PubMed] [Google Scholar]
- Reddy, A. S., Malek, S. urestri A., Ibrahim, H., & Sim, K. S. (2013). Cytotoxic effect of Alpinia scabra (Blume) Náves extracts on human breast and ovarian cancer cells. BMC Complem. Altern. Med. 13(1), 314. doi: 10.1186/1472-6882-13-314. [DOI] [PMC free article] [PubMed]
- Rehman M.U., Ali A., Ansar R., Arafah A., Imtiyaz Z., Wani T.A., Zargar S., Ganie S.A. In Silico molecular docking and dynamic analysis of natural compounds against major non-structural proteins of SARS-COV-2. J. Biomol. Struct. Dyn. 2023;41(18):9072–9088. doi: 10.1080/07391102.2022.2139766. [DOI] [PubMed] [Google Scholar]
- Rižner T.L. Editorial: Relevance of Steroid Biosynthesis, Metabolism and Transport in Pathophysiology and Drug Discovery. Front. Pharmacol. 2019;10:245. doi: 10.3389/fphar.2019.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjida, S., Mou, M., Islam, S., &Mahfuj, M. sarower E. (2022).Identification of potential antiviral drug compound against Erythrocytic necrosis virus by targeting Major capsid protein. Int. J. Life Sci. Biotechnol. doi: 10.38001/ijlsb.1074392.
- Shahzadi K., Bukhari S.M., Zaidi A., Wani T.A., Jan M.S., Zargar S., Rashid U., Farooq U., Khushal A., Khan S. Novel coumarin derivatives as potential urease inhibitors for kidney stone prevention and antiulcer therapy: from synthesis to In Vivo evaluation. Pharmaceuticals. 2023;16(11):Article 11. doi: 10.3390/ph16111552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng W., Mao H., Wang C., Yang N., Zhang Z., Han J. Dehydrocostus Lactone Enhances Chemotherapeutic Potential of Doxorubicin in Lung Cancer by Inducing Cell Death and Limiting Metastasis. Med. Sci. Monitor Int. Med. J. Experim. Clin. Res. 2018;24:7850–7861. doi: 10.12659/MSM.911410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Verma U.P., Verma A.K., Gupta P., Pathak A.K., Singhal R., Kaushal S. Periodontal health status in patients with lung cancer: Case–control study. Int. J. Health Sci. 2024;18(1):17–23. [PMC free article] [PubMed] [Google Scholar]
- Sirwi A., Shaik R.A., Alamoudi A.J., Eid B.G., Elfaky M.A., Ibrahim S.R.M., Mohamed G.A., Abdallah H.M., Abdel-Naim A.B. Mokko Lactone Alleviates Doxorubicin-Induced Cardiotoxicity in Rats via Antioxidant, Anti-Inflammatory, and Antiapoptotic Activities. Nutrients. 2022;14(4):Article 4. doi: 10.3390/nu14040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J., Doe A. Enhancing the delivery of phytochemicals through nanoparticle technology. J. Drug Delivery Sci. Technol. 2023;59:1015–1024. doi: 10.1016/j.jddst.2023.1015. [DOI] [Google Scholar]
- Smith, J. D., Thompson, T. F., & Sanders, N. B. (2021). Targeting the nicotine addiction pathway: Strategies and challenges in drug development. Journal of Pharmacology and Experimental Therapeutics, 376(2), 215-225. Moir, J.W.B., & Wood, N.J. (2001). Nitrate and nitrite transport in bacteria. Cellular and Molecular Life Sciences, 58(2), 215-224. [DOI] [PMC free article] [PubMed]
- Soni R., Nakum K.J., Katariya K.D., Nada S., Hagar M., Wani T.A. Terminal methylene biphenyl derived coumarin Schiff base-esters: Synthesis, mesomorphic behaviour and DFT investigations. J. Mol. Struct. 2024;1307 doi: 10.1016/j.molstruc.2024.137944. [DOI] [Google Scholar]
- Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., Kaplan S., Dahary D., Warshawsky D., Guan-Golan Y., Kohn A., Rappaport N., Safran M., Lancet D. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinformatics. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- Tchebou R.V.K., Farooq U., Teponno R.B., Wani T.A., Tapondjou L.A., Rasool A., Sarwar R., Khushal A., Bukhari S.M., Zargar S., Xu H.-G., Khan S. Exploring Cassia mimosoïdes as a promising natural source of steroids with potent anti-cancer, urease inhibition, and antimicrobial properties. RSC Adv. 2024;14(13):9159–9168. doi: 10.1039/D3RA08913D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin J., Ullah S., Halim S.A., Waqas M., Ibrar A., Khan I., Bin Muhsinah A., Khan A., Al-Harrasi A. Triazolothiadiazoles and triazolothiadiazines as new and potent urease inhibitors: insights from in vitro assay, kinetics data, and in silico assessment. ACS Omega. 2023;8(35):31890–31898. doi: 10.1021/acsomega.3c03546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang X., Wang X., Tian B., Zhang S., Wang T., Ma Y., Fan Y. Design, synthesis and biological evaluation of potent epidermal growth factor receptor tyrosine kinase (EGFR-TK) inhibitors against resistance mutation for lung cancer treatment. Bioorg. Chem. 2024;143 doi: 10.1016/j.bioorg.2023.107004. [DOI] [PubMed] [Google Scholar]
- Wani, A. K., Akhtar, N., Mir, T. ul G., Singh, R., Jha, P. K., Mallik, S. K., Sinha, S., Tripathi, S. K., Jain, A., Jha, A., Devkota, H. P., &Prakash, A. (2023). Targeting apoptotic pathway of cancer cells with phytochemicals and plant-based nanomaterials. Biomolecules, 13(2), Article 2.doi: 10.3390/biom13020194. [DOI] [PMC free article] [PubMed]
- Widyananda M.H., Pratama S.K., Samoedra R.S., Sari F.N., Kharisma V.D., Ansori A.N.M., Antonius Y. Molecular docking study of sea urchin (Arbacia lixula) peptides as multi-target inhibitor for non-small cell lung cancer (NSCLC) associated proteins. J. Pharm. Pharmacogn. Res. 2021;9(4):484–496. doi: 10.56499/jppres21.1047_9.4.484. [DOI] [Google Scholar]
- Widyananda M.H., Kurniasari C.A., Alam F.M., Rizky W.C., Dings T.G.A., Ansori A.N.M., Antonius Y. Exploration of Potentially Bioactive Compounds from Fingerroot (Boesenbergia rotunda L.) as Inhibitor of Atherosclerosis-Related Proteins (CETP, ACAT1, OSC, sPLA2): An in silico Study. Jordan J. Pharm. Sci. 2023;16(3) doi: 10.35516/jjps.v16i3.1609. Article 3. [DOI] [Google Scholar]
- Widyananda M.H., Pratama S.K., Ansori A.N.M., Antonius Y., Kharisma V.D., Murtadlo A.A.A., Jakhmola V., Rebezov M., Khayrullin M., Derkho M., Ullah E., Susilo R.J.K., Hayaza S., Nugraha A.P., Proboningrat A., Fadholly A., Sibero M.T., Zainul R. Quercetin as an anticancer candidate for glioblastoma multiforme by targeting AKT1, MMP9, ABCB1, and VEGFA: An in silico study. Karbala Int. J. Mod. Sci. 2023;9(3) doi: 10.33640/2405-609X.3312. [DOI] [Google Scholar]
- Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35(6):1067–1069. doi: 10.1093/bioinformatics/bty707. [DOI] [PubMed] [Google Scholar]
- Yasmeen, N., Ahmad Chaudhary, A., K Niraj, R. R., Lakhawat, S. S., Sharma, P. K., & Kumar, V. (2023). Screening of phytochemicals from Clerodendrum inerme (L.) Gaertn as potential anti-breast cancer compounds targeting EGFR: An in-silico approach. J. Biomol. Struct. Dynam. 1–43. doi: 10.1080/07391102.2023.2294379. [DOI] [PubMed]
- Yun Y.G., Oh H., Oh G.S., Pae H.O., Choi B.M., Kwon J.W., Kwon T.O., Jang S.I., Chung H. In Vitro Cytotoxicity of Mokko Lactone in Human Leukemia HL-60 Cells: Induction of Apoptotic Cell Death by Mitochondrial Membrane Potential Collapse. Immunopharmacol. Immunotoxicol. 2004;26(3):343–353. doi: 10.1081/IPH-200026863. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.