Abstract
Increased HIV transmission in persons who inject drugs (PWIDs) has led to subepidemics and outbreaks in several countries in Europe, including Bulgaria. In this study in Bulgaria, we investigate the origin and spatiotemporal evolutionary history of HIV-1 infections in PWIDs and the distribution of antiretroviral resistance mutations and hepatitis co-infections in these populations. We analyzed HIV-1 polymerase sequences available from 117 of 359 PWIDs diagnosed with HIV/AIDS from 1999 to 2011. Of these, 50 (42.7%) were classified as CRF02_AG, 41 (35.0%) CRF01_AE, 12 (10.3%) URFs, ten (8.5%) subtype B, two (1.7%) subtype F1 and two (1.7%) CRF14_BG. Most recent common ancestor dating suggests that CRF01_AE was likely first introduced from Southeast Asia into persons reporting heterosexual infection in Bulgaria in 1992 and spread subsequently to PWIDs in the capital city of Sofia around 2003. Conversely, CRF02_AG in Bulgaria was likely first introduced into PWID from Germany in 2000 and later entered heterosexual populations around 2009. The overall prevalence of resistance mutations was 6.8% (8/117), of which 5.1% (5/117) was observed in patients on antiretroviral therapy and 1.7% (2/117) was from transmitted drug resistance mutations in drug-naïve individuals. 189/204 (92.6%) PWIDs were also co-infected with hepatitis C (HCV) and 31/183 (16.9%) were co-infected with hepatitis B (HBV). Our study provides valuable molecular epidemiological information on the introduction and distribution of the main HIV-1 subtypes, resistance mutations and hepatitis co-infections among PWIDs with HIV-1 in Bulgaria which can be used to target prevention efforts.
Keywords: HIV, Molecular epidemiology, Injection drug use, Subtype, Drug resistance, Hepatitis
1. Introduction
In Europe the main HIV-1 transmission routes vary by geographic region with transmission by heterosexual (HET) contact and in men who have sex with men (MSM) predominating in Eastern and West-Central Europe, respectively (Bozicevic et al. 2013). While this trend has remained relatively stable over time, transmission from and within PWIDs is rapidly expanding in some East European countries, including Ukraine and Russia, and a number of outbreaks among PWIDs have been reported in Greece and Romania, both of which border Bulgaria (Paraskevis et al. 2015).
Since the early 1990s, drug abuse and drug trafficking have emerged as visible and serious socioeconomic and public health problems in Bulgaria (EMCDDA, 2015a,b). Injecting drug use or the regular use of opioids, cocaine and/or amphetamines was estimated to affect 6 per 1000 citizens (EMCDDA, 2015a,b). Thus, between 2003 and 2011 a significant spike in HIV-1 incidence among PWIDs was observed (Alexiev et al. 2013). As of 2013, 359/1446 (24.8%) registered HIV/AIDS cases in Bulgaria were PWIDs (Hedrich et al. 2013). Concomitantly, a high prevalence of other blood-borne infections, including HCV and to a lesser extent HBV, were also seen in this population (Vassilev et al. 2006). This finding contrasts with the early epidemic in Bulgaria that predominated in heterosexual groups (Alexiev et al. 2013). Nonetheless, little is known about how and where HIV-1 originated in PWIDs or if those groups will be bridges for resurgence of the epidemic in other risk groups as has been reported in Eastern Europe (Bozicevic et al. 2013). We performed molecular epidemiological and phylogeographic analyses using polymerase (pol) sequences from PWIDs and other risk groups to infer the spatiotemporal evolutionary history of the HIV-1 epidemic in Bulgaria. In addition, we investigated the prevalence of HIV-1 resistance mutations as well as concomitant hepatitis B virus (HBV) and hepatitis C virus (HCV) co-infections to better understand the epidemiology of HIV-1 in PWIDs in Bulgaria.
2. Materials and methods
2.1. Ethics statement
All patients provided written informed consent to participate in this study approved by the Ethical Committee at the National Centre of Infectious and Parasitic Diseases, Sofia, Bulgaria (NCIPD IRB IORG0005305). The CDC IRB determined participant consent was not required for HIV sequence analyses in this study.
2.2. Study population and specimen preparation
The majority of the HIV/AIDS cases in Bulgaria (853/1446, 59%) have been attributed to HET transmission followed by PWIDs (359/1446, 24.8%), MSM (201/1446, 13.9%), blood transfusion which occurred in the early years of the epidemic (19/1446, 1.3%), and mother-to-child transmission (14/1446, 1.0%) (Alexiev et al. 2013; Alexiev et al. 2015).
Our study included data from 359 HIV-1-infected PWIDs diagnosed with HIV/AIDS at the National HIV Reference Laboratory between 1999 and 2011. HIV-1 diversity was studied in 117 patients from this group for whom plasma samples were available, some of whom have been described in earlier studies (Alexiev et al. 2013; Alexiev et al. 2015). Of these, 99 PWIDs (84.6%) were naive to antiretroviral therapy. Blood samples were collected at the National HIV Reference Laboratory and/or in the five clinics responsible for the management of patients with HIV in the cities of Sofia, Plovdiv, Varna, Pleven and Stara Zagora. Patients were from 11 regions, including Sofia and Plovdiv where most of the PWIDs were found and 9 other cities from various regions of the country (Fig. 1). Specimens and sequences were linked to demographic and clinical data through an anonymous numerical code in accordance with the ethical standards of the National Centre of Infectious and Parasitic Diseases, Sofia, Bulgaria.
Fig. 1.
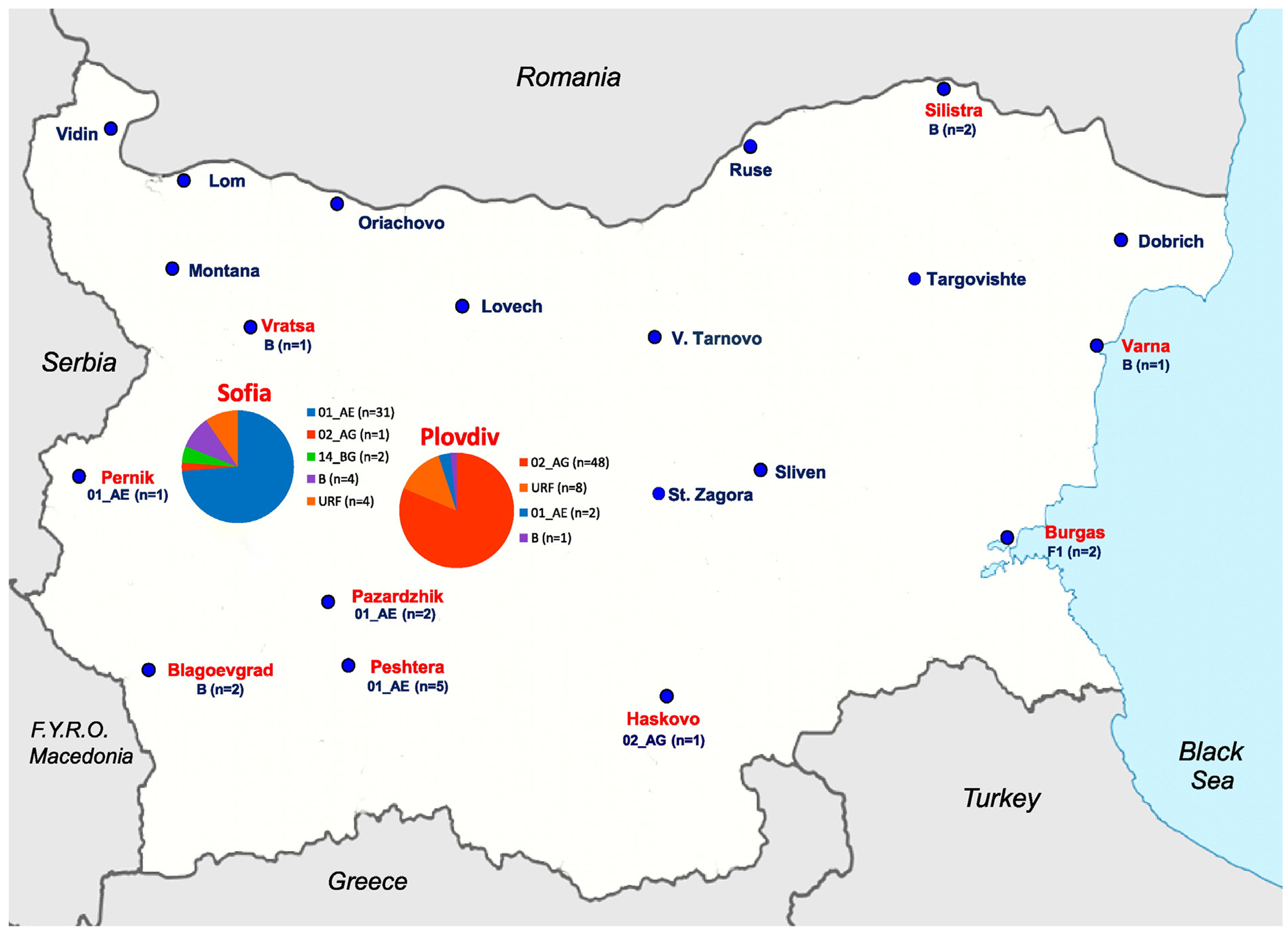
Geographic distribution of HIV-1 subtypes among persons who inject drugs (PWIDs) in Bulgaria. Pie charts show subtype distribution within Sofia and Plovdiv. Numbers of each subtype are in parentheses for the smaller towns. Cities where HIV-1-infected PWIDs reported living and that were included in our study are shown in red text.
2.3. Sequence analysis
Viral RNA was extracted from plasma samples using the QIAamp® Ultra Sens™ Virus Kit 50 QIAGEN, Cat. No.53704). The HIV-1 polymerase (pol) gene (protease and reverse transcriptase regions) was sequenced using the Viroseq HIV-1 Genotyping Test (Abbott) and/or TruGene DNA Sequencing System (Siemens Healthcare) and either the Applied Biosystems 3130xl genetic analyzer or an OpenGene DNA sequencing system following the manufacturer’s protocol (Alexiev et al. 2013). HIV-1 subtypes were determined using the automated tool COMET (https://comet.lih.lu/) and phylogenetic analysis as previously described (Alexiev et al. 2013).
HIV-1 drug resistance mutations were determined according to the WHO 2009 SDRM list (Bennett et al. 2009) using the Calibrated Population Resistance tool v5.0 of the Stanford University HIV Drug Resistance Database (http://cpr.stanford.edu/cpr.cgi).
Nucleotide substitution model selection and alignments for phylogenetic analyses were performed using MEGA6 (Tamura et al. 2013) and contained the Bulgarian pol sequences along with reference sequences from the Los Alamos HIV database (LANL 2014). All 23-drug resistance mutation codons were manually removed from the alignment to exclude the possibility of convergent evolution at these sites.
The best-fitting nucleotide substitution model for the phylogenetic alignments was inferred to be the Hasegawa-Kishino-Yano (HKY) with discrete gamma (Γ) and invariant (I) among-site rate variation. Phylogenetic relationships and HIV-1 subtypes were co-inferred using Bayesian analysis (BEAST v1.7.5) (Drummond et al. 2012). Statistical support for the inferred Bayesian trees was assessed by posterior probabilities. For the Bayesian phylogenetic analyses, an uncorrelated lognormal relaxed molecular clock model was used and each run consisted of two independent 100 million Markov chain Monte Carlo (MCMC) generations with sampling every 10,000th generation and a constant coalescent tree prior. Convergence of the MCMC was assessed by calculating the effective sampling size (ESS) of the runs using the program Tracer v1.6 (Rambaut et al. 2014). All parameter estimates showed significant ESSs >300 indicating sufficient MCMC mixing. The tree with the maximum product of the posterior clade probabilities (maximum clade credibility (MCC) tree) was chosen from the posterior distribution of 9001 sampled trees after burning in the first 1000 sampled trees with the program TreeAnnotator version 1.7.5 (Drummond et al. 2012).
Potential epidemiologic clusters were defined using a stringent set of criteria and included those sequences grouping together with posterior probabilities ≥0.97 and sharing >90% nucleotide identity per total sampling period between related sequences analyzed.
Global phylogenetic relationships of the CRF01_AE and CRF02_AG subtypes in Bulgarian PWIDs and HETs were analyzed by comparison with all available sequences of those two subtypes in GenBank using the program FastTree v2.1 (Price et al. 2010) which performs an approximate maximum likelihood (ML) analysis. Confidence values of the tree topology were tested by using the Shimodaira-Hasegawa test (SH) implemented in FastTree. The inferred tree was edited and displayed using FigTree v1.4.2.
Phylodynamics of the CRF01_AE and CRF02_AG sub-epidemics in PWIDs and HETs in Bulgaria were inferred using two methods. First, changes in effective population sizes (Ne) over time in PWIDs and HETs combined for each subtype were inferred using Bayesian skyline plots implemented in BEAST v1.7.5 and the date of specimen collection for each HIV-1 pol sequence, a Bayesian skyline model with 10 groups, and the HKY + Γ + I nucleotide substitution model. The skyline analysis was run with 50–200 million MCMC chains with sampling every 10,000th generation until convergence was reached in Tracer. The second analysis inferred the potential geographic origin and time to the most recent common ancestors (TMRCAs) using a Gaussian Markov random field (GMRF) Bayesian skyride model and included subsets of sequences with reported heterochronous collection dates identified as nearest neighbors in the ML trees of each Bulgarian CRF01_AE or CRF02 AG subtype as described above to facilitate effective computational analysis (Drummond et al. 2012). A relaxed molecular clock and time aware smoothing of the GMRF coalescent and a Bayesian stochastic search variable selection diffusion model with a symmetric substitution of transition rates between locations were used to simultaneously infer the discrete phylogeography and TMRCAs. Two MCMC chains of 500 million iterations each were run for each subtype dataset until convergence was reached as assessed in Tracer and both log and tree files were then combined using LogCombiner. MCC trees were obtained using TreeAnnotator using a 10% burn-in and viewed in FigTree. Spatial reconstruction of the inferred evolutionary dynamics were visualized, and support for diffusion rates and locations were inferred using Bayes factors (BF) in the program SPREAD v1.0.2 (Bielejec et al. 2011).
2.4. HBV and HCV co-infection analysis
Samples from PWIDs were also tested for HBV and HVC co-infection using separate ELISAs (HBsAg; DIA.PRO, Milano, Italy and Monolisa HCV Ag-Ab Ultra Bio-Rad, Marnes La Coquette, France) following the manufacturer’s instructions.
2.5. Statistical analysis
Fisher’s exact test was used to compare the demographic and epidemiologic characteristics of HIV-1-infected PWIDs and other risk groups, and to evaluate the geographic distribution of HIV-1 diversity in the 117 PWIDs with pol sequences (Campbell, 2007). Two-tailed tests were used to determine the significance of the comparisons using the software Graph Pad Prism v5.0 (GraphPad, 2015).
2.6. GenBank accession numbers of Bulgarian pol sequences
New pol sequences generated in this study have the accession numbers KT805893-KT805918. Bulgarian pol sequences from previous studies include EF517439, KJ765564, JQ259094, EF517472, KJ765424, KJ765391, KJ765425, JQ259115, JQ259116, KJ765573, KJ765608, JQ259122, KJ765609, KJ765576, KJ765578, KJ765580, KJ765585, KJ765586, KJ765429, KJ765588, JQ259139, JQ259141, JQ259142, JQ259143, KJ765589, JQ259147, KJ765590, JQ259149, KJ765592, KJ765392, KJ765431, KJ765432, KJ765593, KJ765433, KJ765594, KJ765596, KJ765597, JQ259163, KJ765598, JQ259168, JQ259171, JQ259172, JQ259173, KJ765600, JQ259178, KJ765439, KJ765390, KJ765395, KJ765443, KJ765444, KJ765398, KJ765445, KJ765403, KJ765405, KJ765407, KJ765453, KJ765409, KJ765454, KJ765601, KJ765603, KJ765414, KJ765603, KJ765416, KJ765468, KJ765422, KJ765469, KJ765470, KJ765472, KJ765474, KJ765480, KJ765481, KJ765482, KJ765482, KJ765486, KJ765487, KJ765492, KJ765493, KJ765500, KJ765501, KJ765503, KJ765510, KJ765512, KJ765513, KJ765516, KJ765517, KJ765525, KJ765537, KJ765543, KJ765550, KJ765552, KJ765553.
3. Results
3.1. Study population demographics and HIV-1 prevalence by risk group
The demographic and epidemiological characteristics of PWIDs with HIV-1 were compared to those from the remaining HIV-1-positive population registered in Bulgaria through 2011. As shown in Table 1, 88.9% of HIV-1-infected PWIDs were male, with a significantly higher prevalence of MSMs among PWIDs compared to other risk groups (88.9% vs. 70.4%, p < 0.001). Individual ages of PWIDs varied between 17 and 43 years, with a mean of 26.4 years. The proportion of young patients (below 19 years old) was significantly higher among PWIDs as compared to other risk groups (11.1% vs. 5.1%, p < 0.0001). PWIDs were concentrated in the major cities of Plovdiv and Sofia, with a higher prevalence in Plovdiv, as compared to other risk groups (46.2% vs. 8.5%, p < 0.0001). The vast majority of PWIDs (96.4%) indicated Bulgaria as the most likely place of infection, and only 3.6% reported likely acquiring infection abroad. The proportion of Roma (Gypsy) persons among PWIDs with HIV-1 was significantly higher as compared to other risk groups (39.8% vs. 2.1%, p < 0.0001). We observed a similar trend in incarcerated persons (33.1% PWIDs vs. 2.8% in other risk groups, p < 0.0001).
Table 1.
Demographic and epidemiological characteristics of the Bulgarian study population.
| Characteristics | Genotyped HIV-1-infected PWIDs | Registered HIV-1-infected PWIDs | Registered HIV-1-infected non-PWIDs | p-Valuea | p-Valueb | |
|---|---|---|---|---|---|---|
| Number of patients | 117 | 359 | 1087 | – | – | |
| Male | 107 (91.5) | 319 (88.9) | 766 (70.4) | ns | p < 0.0001 | |
| Female | 10 (8.5) | 40 (11.1) | 321 (29.5) | ns | p < 0.0001 | |
| Age (years) ≤ 19 | 15 (12.8) | 40 (11.1) | 56 (5.1) | ns | p < 0.001 | |
| Age (years) ≥ 20 | 102 (87.2) | 319 (87.5) | 1032 (94.9) | ns | p < 0.001 | |
| HCV co-infection | 103/112 (92.0) | 189/204 (92.6) | 56/406 (13.8) | ns | p < 0.0001 | |
| HBV co-infection | 15/81 (18.5) | 31/183 (16.9) | 37/399 (9.7) | ns | ns | |
| Most likely infected in Bulgaria | 113 (96.6) | 346 (96.4) | 875 (80.5) | ns | p < 0.0001 | |
| Most likely infected abroad | Immigrants | 0 | 2 (0.6) | ns | ns | ns |
| Bulgarian nationals | 4 (3.4) | 11 (3.1) | 179 (16.5) | ns | p < 0.0001 | |
| Sofia | 42 (35.9) | 119 (33.1) | 430 (39.5) | ns | ns | |
| Plovdiv | 59 (50.4) | 166 (46.2) | 93 (8.5) | ns | p < 0.0001 | |
| Other cities | 16 (13.7) | 74 (20.6) | 555 (51.1) | ns | p < 0.0001 | |
| Roma (Gypsy) | 41 (35.0) | 143 (39.8) | 34 (2.1) | ns | p < 0.0001 | |
| Patients who reported imprisonment | 43 (36.8) | 119 (33.1) | 30 (2.8) | ns | p < 0.0001 | |
| Bulgarian citizens infected in Bulgaria (not Roma, not prisoner) | 43 (36.8) | 129 (35.9) | 821 (75.7) | ns | p < 0.0001 | |
Comparison between the registered HIV-1-infected persons who inject drugs (PWIDs) (n = 359) and the subgroup of genotyped HIV-1-infected PWIDs (n = 117), Fisher’s exact test. ns, not found to be statistically significant.
Comparison between HIV-1-infected PWIDs (n = 359) and HIV-1-infected non-PWIDs (n = 1087), Fisher’s exact test.
3.2. Drug resistance prevalence in PWIDs
The overall prevalence of antiretroviral resistance in the genotyped subgroup of PWIDs with HIV-1 was 6.8% (8/117), including six (5.1%) treatment-experienced patients and two (1.7%) cases of transmitted drug resistance mutations (TDRM) in drug-naïve individuals that we previously reported (Table 2) (Alexiev et al. 2015). 3/117 (2.6%) PWIDs had resistance to nucleoside reverse transcriptase inhibitors (NRTIs; M41L, D67N, K70R, M184V, T215F, K219E) and 6/117 (5.1%) to non-nucleoside reverse transcriptase inhibitors (NNRTIs; K103N, Y181C, Y188L and G190E). Resistance mutations to protease inhibitors (PIs) were not found in this group. The only female with HIV-1 resistance mutations was a carrier of six resistance mutations to NRTIs, one patient had resistance to both NRTI and NNRTI, and the other six individuals had single resistance mutations to either NRTI or NNRTI (Table 2).
Table 2.
Drug resistance mutations present in HIV-1-infected persons who inject drugs in Bulgaria.
| Patient Codea | Gender | Region | Subtype | Naïve/HAARTb | Year of diagnosis | Year of specimen collection | Antiretroviral mutationsc |
||
|---|---|---|---|---|---|---|---|---|---|
| NRTI | NNRTI | PI | |||||||
| 06BG481 | F | Silistra | B | HAART | 2004 | 2006 | M41L, D67N, K70R, M184V, T215F, K219E | None | None |
| 11BG606 | M | Sofia | B | HAART | 2006 | 2011 | M184V | None | None |
| 10BG704 | M | Plovdiv | CRF02_AG | HAART | 2007 | 2010 | None | K103N | None |
| 11BG769 | M | Plovdiv | CRF02_AG | HAART | 2007 | 2011 | M184V | Y188L | None |
| 11BG892 | M | Peshtera | CRF01_AE | Naïve | 2008 | 2011 | None | G190E | None |
| 11BG1012 | M | Plovdiv | CRF02_AG | HAART | 2009 | 2011 | None | Y181C | None |
| 11BG1023 | M | Sofia | CRF01_AE | HAART | 2009 | 2011 | None | K103N | None |
| 11BG 1362 | M | Plovdiv | CRF02_AG | Naïve | 2011 | 2011 | None | K103N | None |
Drug resitance results for patients with codes in bold italics were reported previously (Alexiev et al., 2015).
HAART, highly active antiretroviral therapy.
NRTI, nucleotide reverse transcriptase inhibitors; NNRTI, non-nucleotide reverse transcriptase inhibitors; PI, protease inhibitors.
3.3. HIV-1 diversity and geographic distribution of subtypes in PWIDs
Phylogenetic analysis showed a broad distribution of subtypes in the sub-group of PWIDs (n = 117) with a predominance of CRFs, significant numbers of URFs and <9% subtype B infections. Of the 117 pol gene sequences, 50 (42.7%) were classified as CRF02_AG, 41 (35.0%) CRF01_AE, 12 (10.3%) URFs, 10 (8.5%) subtype B, two (1.7%) subtype F1 and two (1.7%) CRF14_BG. (Fig. 1 and Table 3).
Table 3.
HIV-1 subtype diversity in persons who inject drugs in Bulgaria.
| Subtype | Number (%) | Sofia | Plovdiv | Other cities | Pa | Prisoner | Not prisoner | P | Roma (Gypsy) | Not Roma | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRF02_AG | 50 (42.7) | 1 (2.0) | 48 (96.0) | 1 (2.0) | <0.0001 | 16 (32.0) | 34 (68.0) | <0.0001 | 19 (38.0) | 31 (62.0) | ns |
| CRF01_AE | 41 (35.0) | 31 (75.6) | 2 (4.9) | 8 (19.5) | <0.0001 | 27 (65.9) | 14 (34.1) | <0.01 | 17 (41.5) | 24 (58.5) | ns |
| URFsb | 12 (10.3) | 4 (33.3) | 8 (66.7) | 0 | nsc | 11 (91.7) | 1 (8.3) | <0.01 | 4 (33.3) | 8 (66.7) | ns |
| B | 10 (8.5) | 4 (40.0) | 1 (10.0) | 5 (50.0) | 0.01 | 1 (10.0) | 9 (90.0) | <0.05 | 1 (10.0) | 9 (90.0) | ns |
| F1 | 2 (1.7) | 0 | 0 | 2 (100) | ns | 1 (50.0) | 1 (50.0) | ns | 0 | 2 (100) | ns |
| CRF14_BG | 2 (1.7) | 2 (100) | 0 | 0 | ns | 0 | 2 (100) | ns | 0 | 2 (100) | ns |
| Total | 117 | 42 | 59 | 16 | 56 | 61 | 41 | 76 |
P, probability determined using Fisher’s exact test.
URFs, unique recombinant forms.
ns, not statistically significant.
Analysis of the geographic distribution of viral genotypes revealed a significantly higher prevalence of CRF01_AE in PWIDs in Sofia 73.8% (31/42) vs. 13.3% (10/75) in other regions (p < 0.0001). in contrast, the vast majority of PWIDs from Plovdiv had CRF02_AG infections 48/59 (81.3%) vs. 2/58(3.4%) in other regions (p < 0.0001). in contrast, subtype B was significantly less prevalent in PWIDs from Plovdiv compared to the rest of the country (1/59 or 1.7% vs. 9/58 or 15.5%, p < 0.01) (Fig. 1 and Table 3).
43/117 (36.8%) of genotyped PWIDs with HIV-1 reported that they had been imprisoned and the majority of these were found to be URFs (11/12 or 91.7% vs. 1/12 or 8.3% among non-prisoners, p < 0.001). Comparison of the HIV-1 genetic diversity among imprisoned participants and the rest of the study population showed significant differences for CRF01_AE (p < 0.01), URFs (p < 0.01) and subtype B (p < 0.05) that predominated among prisoners, unlike CRF02_AG (p < 0.0001) that was typically found among non-prisoners. Comparison of Roma and non-Roma patients showed a relatively uniform distribution of the main subtypes with no significant differences observed (Table 3).
3.4. Expanded phylogenetic analysis of predominant subtypes in PWIDs
To further investigate the potential origin of HIV-1 in PWIDs in Bulgaria we created independent alignments of pol sequences from the two major HIV-1 clades (CRF01_AE and CRF02_AG) found in PWIDs and combined those with the respective pol subtype sequences from Bulgarian HET individuals and all available CRF01AE (n = 4680) and CRF02_AG (n = 2933) pol sequences from GenBank at the time of the search, thus representing the possible global dissemination of these respective CRFs. We then performed phylogenetic analysis using maximum likelihood (ML) to determine their genetic relationships and to identify closely related global pol sequences for further analysis.
In both ML trees (Figs. 2 and 3), Bulgarian PWIDs sequences form distinct clusters with high statistical support (SH values of 0.89 and 0.98 for CRF01_AE and 02_AG, respectively) indicating strongly that these are local subepidemics. In contrast, most Bulgarian HET sequences were independently dispersed in the trees or were grouped in small and distant clusters. Exceptions to this finding were 14 CRF01_AE and six CRF02_AG HET sequences that formed part of the respective PWIDs clusters, indicating possible spillover between PWIDs and HET populations in Bulgaria. Furthermore, some CRF01_AE sequences from Bulgarians reporting HET transmission clustered with sequences from Asia, predominantly from Thailand, as expected for this subtype (data not shown). In contrast, Bulgarian HET CRF02_AG sequences were closest phylogenetically to other CRF02_AG sequences from either Spain, Nigeria, Senegal, or Cameroon (data not shown).
Fig. 2.
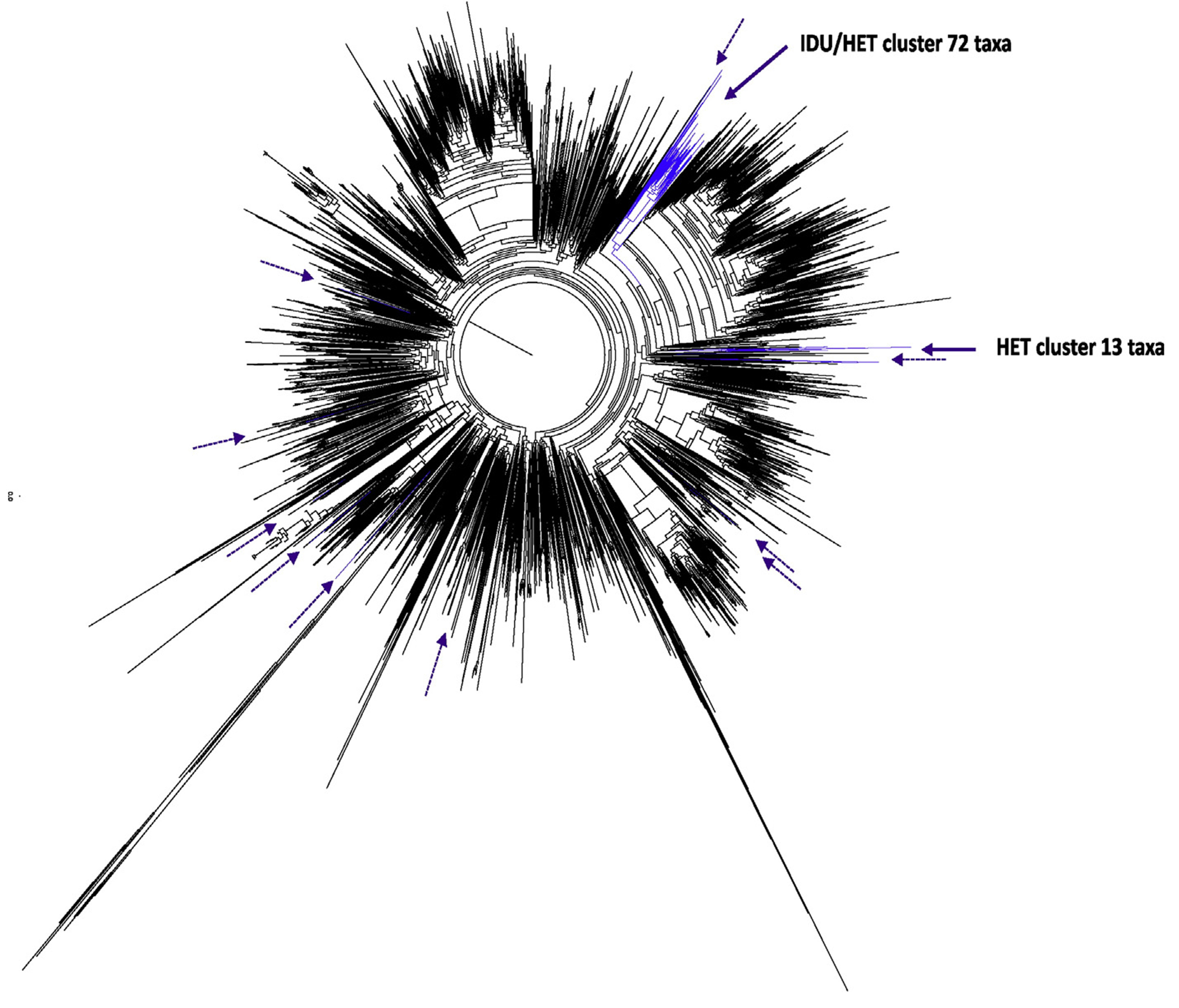
Global maximum likelihood phylogenetic tree of CRF01_AE. The phylogenetic analysis is based on an alignment of 721 HIV-1 CRF01_AE polymerase (pol) nucleotides inferred with 97 pol sequences from Bulgaria, 41 from persons who inject drugs (PWIDs) and 56 heterosexual (HET) individuals and 4680 pol sequences of the respective CRF available in GenBank using FastTree. Confidence values of tree branches and clusters were assessed by using the Shimodaira-Hasegawa test and are given as probabilities. Black and blue branches indicate globally distributed and Bulgarian pol sequences, respectively. Solid and dashed purple arrows indicate positions of Bulgarian PWID and HET only pol sequences in the tree, respectively.
Fig 3.

Global maximum likelihood phylogenetic tree of CRF02_AG. The phylogenetic analysis is based on an alignment of 774 HIV-1 CRF02_AG polymerase (pol) nucleotidesmferred with 69 pol sequences from Bulgaria, 50 from persons who inject drugs (PWIDs) and 19 heterosexual (HET) individuals and 2933 pol sequences of the respective CRF available in GenBank using FastTree. Confidence values of tree branches and clusters were assessed by using the Shimodaira-Hasegawa test and are given as probabilities. Black and blue branches indicate globally distributed and Bulgarian pol sequences, respectively. Solid and dashed purple arrows indicate positions of Bulgarian PWID and HET only pol sequences in the tree, respectively.
Interestingly, two HIV-1 sequences from Cyprus were sister taxa of one CRF02_AG cluster containing six Bulgarian HET sequences (SH = 1.0, data not shown). Similarly, two CRF01_AE sequences from the Czech Republic clustered with a group of CRF01_AE sequences from 11 Bulgarians reporting heterosexual transmission (SH = 0.88, data not shown).
3.5. Spatiotemporal evolutionary dynamics of CRF01_AE and CRF02_AG subepidemics in PWIDs and HETs in Bulgaria
Timing of the introduction and the population dynamics of HIV-1 CRF01_AE and CRF02_AG subtypes among PWIDs and heterosexual individuals in Bulgaria was inferred using Bayesian coalescent methods implemented in BEAST. Bayesian skyline analysis inferred a biphasic growth of CRF01_AE in Bulgaria with one log increase in effective population growth before 1990 which then plateaued between 1990 and 1996 (Fig. 4A). A second exponential growth phase for CRF01_AE occurred over a short period around 2000 and then plateaued shortly thereafter. For CRF02_AG, Bayesian skyline analysis inferred a stable growth period from 1985 until 1998 when about a half log increase was observed for approximately one year followed by another plateau period (Fig. 5A).
Fig. 4.
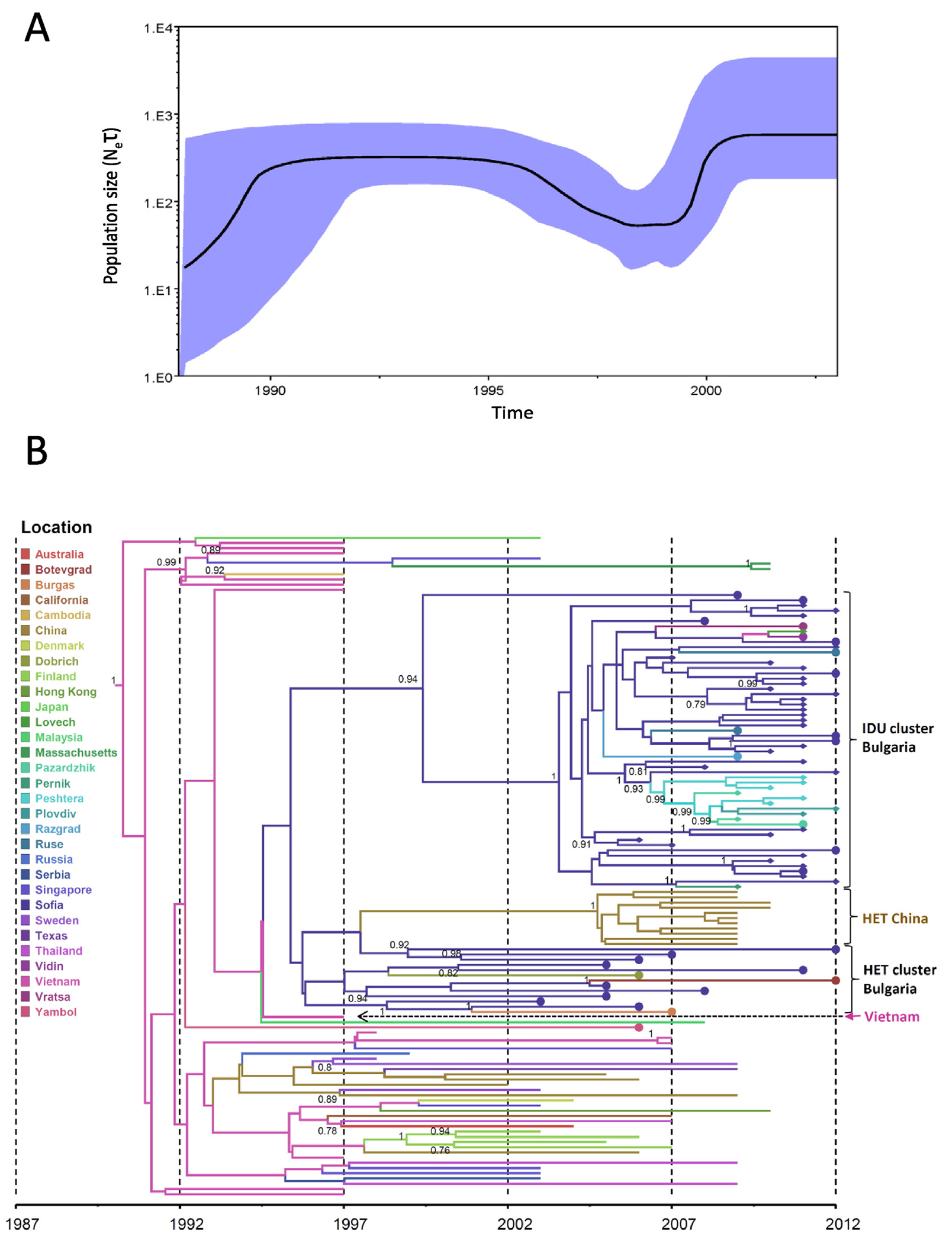
Phylodynamics and phylogeography of HIV-1 CRF01_AE in Bulgaria inferred by Bayesian analysis. (A) Bayesian skyride plot showing the inferred growth of the CRF01_AE epidemic in Bulgaria. Inferred effective population sizes (Ne) over time in years are on the y- and x-axes, respectively. (B) Phylogenetic tree showing the most recent common ancestor and geographic spread of CRF01_AE. Analysis included 70 polymerase (pol) sequences from Bulgaria, 41 sequences from persons who inject drugs (PWIDs) and 29 heterosexual (HET) individuals and 56 pol sequences from other countries. Filled circles and diamonds at branch tips indicate Bulgarian HETs and PWIDs, respectively. Time scale in years provided on x-axis and country or city origin of the sequences provided in color and defined in key.
Fig. 5.
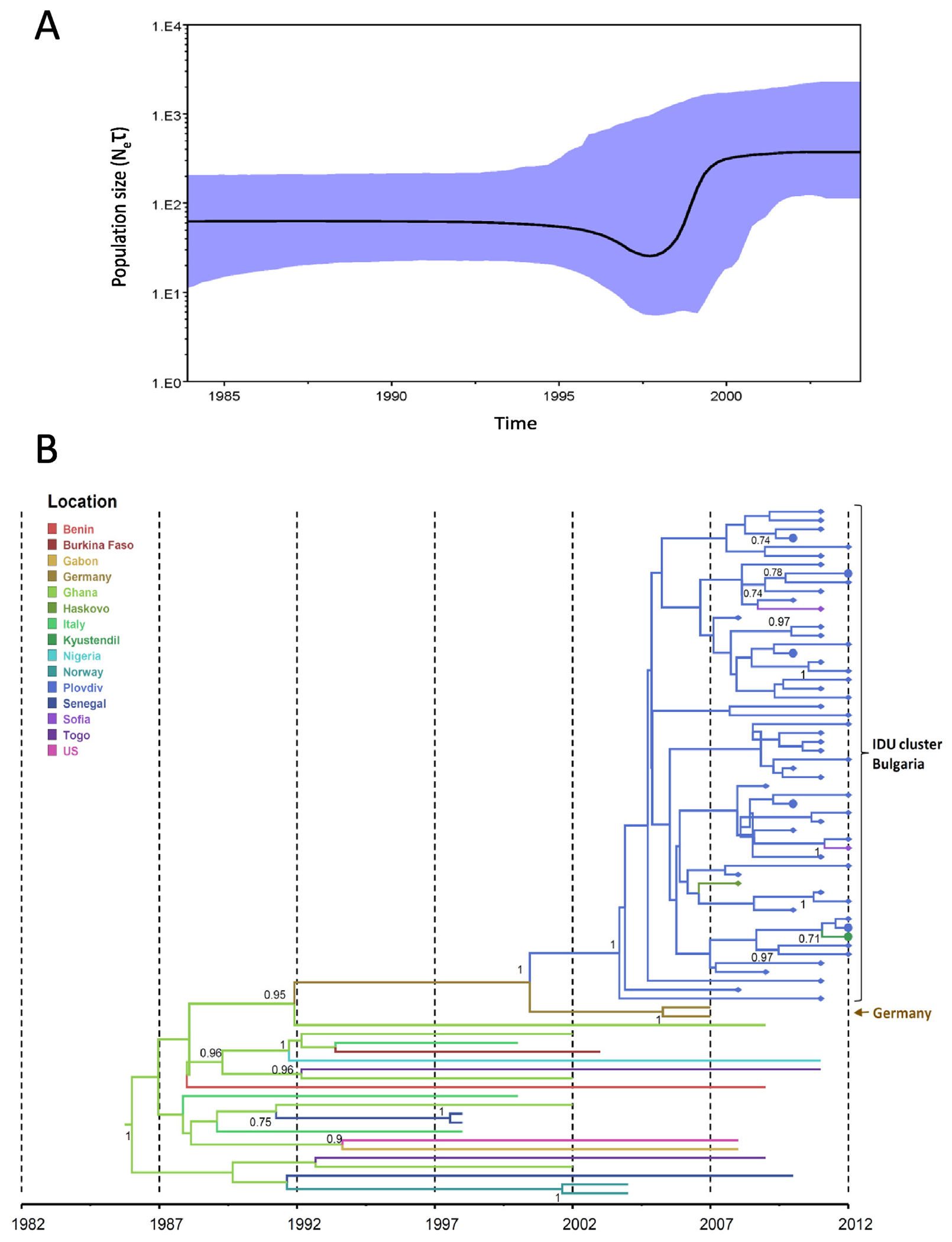
Phylodynamics and phylogeography of HIV-1 CRF02_AG in Bulgaria inferred by Bayesian analysis. (A) Bayesian skyride plot showing the inferred growth of the CRF02_AG epidemic in Bulgaria. Inferred effective population sizes (Ne) over time in years are on the y- and x-axes, respectively. (B) Phylogenetic tree showing the most recent common ancestor and geographic spread of CRF02_AG. Analysis included 69 polymerase (pol) sequences from Bulgaria, 50 sequences from persons who inject drugs (PWIDs), 19 heterosexual (HET) individuals and 22 pol sequences from other countries. Filled circles and diamonds at branch tips indicate Bulgarian HET and PWIDs, respectively. Time scale in years provided on x-axis and country or city origin of the sequences provided in color and defined in key.
The TMRCA date for CRF01_AE in Bulgaria was inferred to occur in 1992, represented by a single HET sequence from Yambol followed by expansion through 1995 (95% HPD: 1993–1996) mostly in Sofia and predominantly in heterosexuals (Fig. 4B). Around 2003 (95% HPD: 2000–2004) CRF01_AE spread to PWIDs in Sofia from the HET population and this subtype is still found in recent heterosexual infections. Around 2006 (95% HPD: 2004–2008), CRF01_AE spread from Sofia to other PWID populations in Peshtera, Plovdiv, and Pazardzhik. Interestingly, around 1998 the HET Bulgarian sequences shared a common ancestor with a cluster of CRF01_AE sequences from a Chinese heterosexual population. In addition, one sequence from Vietnam was located between the initial Bulgarian HET sequences in this cluster and the large HET/PWIDs cluster, but without strong support. The majority of the ancestral CRF01_AE sequences in this analysis were from Asia where this subtype originated (Hemelaar 2012). Phylogeographic analysis using a discrete traits approach in the programs BEAST and SPREAD inferred Vietnam as the likely origin of the strain that was introduced into the capital city of Sofia in 1995. Within Bulgaria, CRF01_AE then spread from Sofia to Dobrich around 2006 (PP = 0.99, BF = 210.917), to Razgard and Peshtera in 2007 (PP = 0.99, BFs = 1977 and 18,672, respectively), to Vidin during 2008 (PP = 0.99, BF 3736), and to Ruse in 2010 (PP = 0.99, BF = 1415).
In contrast, the introduction of CRF02_AG in Bulgaria occurred later around the year 2000 (95% HPD: 1996–2004) and shared ancestry with a pair of sequences from newly diagnosed infections from Germany with strong support (SH = 1.0) (Fig. 5B). The dispersal of CRF02_AG from Germany to Plovdiv was also supported using the program SPREAD with a discrete BF of 9.59. Risk factor information was not reported at GenBank for these two sequences to allow inference of behavioral or epidemiological linkage to the Bulgarian PWIDs. The CRF02_AG cluster contains predominantly PWIDs sequences from Plovdiv with more recent spillover into HETs between 2009 and 2010.
3.6. HIV-1, HBV, and HCV co-infection analysis in PWIDs
We determined the prevalence of HCV and HBV co-infections among HIV-1-infected PWIDs in Bulgaria to investigate the spread of other blood-borne infections in this risk group. Serological analysis showed that 189/204 (92.6%) HIV-1-infected PWIDs were also HCV-positive and 31/183 (16.9%) were HBV-positive (Table 1). Importantly, 11/81 (13.6%) HIV-1-positive patients were co-infected with both HBV and HCV. Similar to our previous study in Bulgaria (Vassilev et al. 2006), the frequency of HCV and HBV co-infection among PWIDs in our current study was significantly higher compared to the other risk groups as a whole (92.6% vs.13.8%; p < 0.0001% and 16.9% vs. 9.7%, p < 0.05, respectively).
We also found similar results for the subgroup of genotyped HIV-1-infected PWIDs. Of these PWIDs, 103/112 (92.0%) tested HCV-positive (p < 0.0001) and 15/81 (18.5%) were HBV-positive as compared to 56/406 (13.8%) HCV-positive, and 37/399 (9.3%) HBV-positive persons in other risk groups (p < 0.05, Table 1).
We then compared HCV and HBV co-infection results for all registered HIV-1-positive PWIDs (n = 359) with those from the subgroup of patients we genotyped (n = 117) and found no significant differences, indicating that selection bias likely did not affect our results. HIV subtype was not associated with the presence of HCV or HBV co-infection (p > 0.05 for all comparisons between the groups of co-infected and mono-infected patients, data not shown).
4. Discussion
In this nationwide study, we analyzed the origin, spread and epidemiological characteristics of the HIV-1 sub-epidemic among PWIDs in Bulgaria. As previously reported, the early HIV-1 epidemic in Bulgaria was driven mostly by heterosexual transmission with PWIDs composing only a small fraction of infections (Salemi et al. 2008). During that time, other blood-borne pathogens, including HCV, and to a lesser extent HBV, were widely prevalent among PWIDs (Vassilev et al. 2006; Kevorkyan et al. 2015). However, since the initial introduction of HIV-1 into Bulgaria, there has been a significant increase in cases among PWIDs (Stanojevic et al. 2012; Alexiev et al. 2013; Hedrich et al. 2013). As a population at increased risk to blood-borne infections, PWIDs can potentially form a large social and transmission network allowing rapid spread of infectious agents to other PWIDs, as well as to their sexual partners. Recent reports of HIV-1 outbreaks among PWIDs in the neighboring countries of Greece and Romania (Paraskevis et al. 2015), and the increasing incidence of HIV-1 cases among PWIDs in Bulgaria alerted public health officials of the importance of conducting a detailed study of infection in PWIDs and their role in the HIV-1 epidemic in Bulgaria.
We found that the vast majority of HIV-1-infected PWIDs were men and the proportion of patients under the age of nineteen was significantly higher in PWIDs compared to the remaining HIV-1-positive population in Bulgaria. In addition, we found that a significant number of PWIDs were Roma individuals or were persons with a history of incarceration. These characteristics of the PWID populations in Bulgaria confirm those reported earlier that also had a higher proportion of vulnerable groups, including youths and minority groups with high-risk behaviors. (Kelly et al. 2006; Kabakchieva et al. 2002; Kabakchieva et al. 2006). In contrast, similar characteristics weres not found in HIV-1-infected PWIDs in the neighboring countries of Romania and Greece (Paraskevis et al. 2015).
Importantly, we found low levels of TDRM among PWIDs, which agrees with our previous analysis in which we reported TDRM prevalence in the general HIV-1-infected population to be about 5% (Santoro et al. 2008; Alexiev et al. 2015), and is reassuring from a public health perspective. However, the presence of NRTI and NNRTI resistance mutations in some individuals receiving highly active antiretroviral therapy (HAART) was expected because PWIDs are not very likely to adhere to treatment, and which continues to be a major cause of HIV-1 drug resistance (Spire et al. 2007). Additional concerns arise from the fact that resistance mutations found in most at-risk populations can result in accelerated dissemination of resistant strains within the risk group, as it was recently reported for the role of transmission clusters of MSM with HIV-1 in Croatia (Grgic et al. 2013).
As in our previous reports (Salemi et al. 2008; Alexiev et al. 2013; Alexiev et al. 2015), we found very high levels of HIV-1 diversity in Bulgaria. However, unlike other groups at risk for HIV-1 acquisition, the PWID sub-epidemic in Bulgaria was characterized by several significant features, including the extremely low prevalence of subtype B and the dominance of two recombinant subtypes, CRF01_AE and CRF02_AG. It is interesting to note that both CRFs were divided between the two major cities, Sofia and Plovdiv, demonstrating the existence of independent PWID sub-epidemics in two distinct geographical areas within Bulgaria. In addition, almost all URFs found in PWIDs were found predominantly in a subgroup of incarcerated individuals suggesting possible superinfections of these persons with different HIV-1 strains which will require further study. Our study showed that there were no specific viral strains predominant among Roma individuals who were equally infected with CRF01_AE and CRF02_AG.
Phylogenetic analysis of Bulgarian CRF01_AE and CRF02_AG sequences together with all respective subtype sequences available at the time of our sampling GenBank were conducted to investigate their global relationships. This analysis showed that most of the sequences isolated from HET patients of both CRF01_AE and CRF02_AG were dispersed across the inferred phylogenetic trees showing that these CRFs found in Bulgaria originated from different areas of the world and thus represent multiple independent viral introductions in Bulgaria. Moreover, so far most Bulgarian HET sequences showed only limited transmission to a single partner following the primary infection from a PWID. In contrast, our phylogenetic analyses suggest that both CRF01_AE and CRF02_AG began to circulate in the respective PWIDs sub-epidemics possibly following a likely single viral introduction from a HET for CRF01_AE and a separate introduction from PWIDs for CRF02_AG. The rapid spread of HIV within PWIDs likely then led to limited local geographic outbreaks of these two subtypes. This specific feature of the HIV-1 epidemic among PWIDs in Bulgaria significantly differs from the PWIDs outbreak in neighboring Romania where about 40% of PWIDs were reported to have been infected abroad (Niculescu et al. 2014).
Our spatiotemporal evolutionary dynamics analysis supported these findings and inferred that subtype CRF01_AE was first introduced and spread among the heterosexual population around 1992 and only after ten years did this subtype reach PWIDs in the capital city Sofia. Interestingly, the most recent common ancestors of Bulgarian CRF01_AE were from Vietnam where this subtype was highly prevalent in both heterosexuals and PWIDs in the 1980s and 1990s, though we cannot exclude the possibility that these infections were acquired outside of Bulgaria (Liao et al. 2009). While this contrasts with another study showing that CRF01_AE infection in Europe originated mostly from Thailand, Japan, and Africa (Angelis et al. 2015), there was a significant number of Vietnamese workers in Sofia in the 1980s and 1990s who could have transmitted their infections within Bulgaria during that period of time. It is also interesting to note that CRF01_AE sequences isolated in China from a heterosexually driven epidemic, that also likely originated from Vietnam, clustered with HIV-1 sequences from heterosexual infections in Bulgaria (Lin et al. 2013). This could indicate an introduction of CRF01_AE from China into a subset of Bulgarians infected via heterosexual contact or it could be an artifact of the subsampling for this analysis. Interestingly, a previous study investigating the global dispersal of CRF01_AE showed that heterosexual sex was the main HIV transmission risk in Europe, which appears to no longer be the case in Bulgaria (Angelis et al. 2015). We also found that HIV-1 CRF01_AE from two Czech Republic patients were phylogenetically related to those from Bulgarian patients with reported heterosexual transmission. This potential transmission link is consistent with other global studies of CRF01_AE dissemination suggesting that Bulgaria may have exported CRF01_AE viruses to the Czech Republic (Angelis et al. 2015) and are consistent with the limited spread of HIV-1 across borders in Europe (Frentz et al. 2013).
Unlike CRF01_AE, CRF02_AG was inferred to have a much shorter evolutionary history in Bulgaria. Our analysis suggested that this subtype in PWIDs may have originated in Germany, where the majority of infections are in MSM, though the risk group for these two German HIV-1 sequences basal to those from Bulgaria was not provided in that report (Frentz et al. 2013). CRF02_AG was first identified in Plovdiv among PWIDs in 2000 and later spread to HET individuals in 2009–2010. PWIDs are known bridges for HIV-1 infection with their sex partners who are either PWIDs or non-PWIDs (Jenness et al. 2010). We found that transmission to HET from PWIDs is also prevalent in Bulgaria; CRF01_AE originated in heterosexuals, moved into PWIDs, and has returned to heterosexuals via PWIDs. Like for CRF01_AE, we found evidence of spillover of CRF02_AG outside of Bulgaria. Two sequences isolated from individuals in Cyprus were nested within a cluster of CRF02_AG sequences from six Bulgarians who reported heterosexual HIV-1 transmission further demonstrating the global dispersal of this subtype in that particular risk group (Kousiappa et al. 2011).
PWIDs are at increased risk for HCV and to a lesser extent to HBV infections (Aceijas and Rhodes, 2007; ECDC, 2015b, Hepatitis C surveillance in Europe; ECDC, 2015a, Hepatitis B surveillance in Europe). In our study we found a high prevalence of HCV, HBV and dual HCV/HBV co-infections among HIV-1-infected PWIDs compared to the general population of Bulgaria (Kevorkyan et al. 2015). The identification of hepatitis and HIV coinfections is important and will help in the concerted public health response of care and treatment management of PWIDs. Furthermore, screening for HCV could be utilized to identify populations at increased risk for HIV-1 infection to provide targeted interventions to prevent these dual infections and rapid spread of HIV-1.
Given the additional risks associated with the management of HIV-1 infection as well as the high potential for transmission of these pathogens in PWIDs, our findings highlight the need for additional public health strategies for preventing the spread of these pathogens. Increasing access to care and treatment and needle exchange programs have been successful in this regard (Frieden et al. 2015). Indeed, our molecular analyses initiated a concerted public health response using specific prevention interventions targeting PWIDs and their heterosexual partners and has been successful in reducing HIV infection in PWIDs and their contacts. For example, PWIDs have been offered free counseling, testing and needles, which has resulted in a decline in the number of PWIDs diagnosed with HIV since 2009 (EMCDDA, 2015b). Our epidemiological data also revealed that PWIDs with HIV-1 in Bulgaria are often observed in vulnerable populations, including Roma and those reporting incarceration. These vulnerable groups may require specialized public health intervention strategies.
While we highlight important outcomes of our study, it is not without limitations. The cross-sectional design of our study, which included all available specimens collected from patients who were diagnosed or came into clinics for care, may not be fully representative of the other 62% of reported PWIDs in Bulgaria. We were also unable to test all persons in the study for hepatitis viruses as sample volumes for some persons were not sufficient for this testing. The effects of these possible sampling biases in our analyses is unknown but may influence genotype introduction and distribution over time as well as patterns of resistance mutations and prevalences of hepatitis co-infections. In addition, any potential association of infection with demographic and epidemiological characteristics is based on self-reporting and may be affected by recall or other biases. Further, our phylogeographic studies are limited to those sequences deposited in public databases and thus the global spatiotemporal spread of HIV-1 could change if additional sequences become available. Nonetheless, our phylogeographic results for CRF01_AE and CRF02_AG are in general agreement with the global dispreal of these subtypes reported by others (Angelis et al. 2015; Frentz et al. 2013).
5. Conclusions
Detailed phylogenetic analysis revealed the origin and introduction of the two major HIV-1 clades among PWIDs in Bulgaria. While it is reassuring that drug resistance mutations are low in both treatment experienced and naïve individuals, additional public health interventions are needed for the increasing numbers of persons with HIV and hepatitis co-infections to limit the spread of these pathogens and to prevent the diseases associated with these viruses. Our study provides valuable molecular epidemiological information on HIV-1-infected PWIDs in Bulgaria and highlight the importance of sustained molecular surveillance of high-risk groups to better understand and control the changing epidemic in Bulgaria.
Acknowledgments
Use of trade names is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services, the Public Health Service, or the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention, or any of the authors’ affiliated institutions.
Footnotes
Transparency declaration
The authors declare no conflicts of interest.
References
- Aceijas C, Rhodes T, 2007. Global estimates of prevalence of HCV infection among injecting drug users. Int. J. Drug Policy 18, 352–358. [DOI] [PubMed] [Google Scholar]
- Alexiev I, Beshkov D, Shankar A, Hanson D, Paraskevis D, Georgieva V, Karamacheva L , Taskov H, Varleva T, Elenkov I, Stoicheva M, Nikolova D, Switzer W, 2013. Detailed molecular epidemiologic characterization of HIV-1 infection in Bulgaria reveals broad diversity and evolving phylodynamics. PloS one 8, e59666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiev I, Shankar A, Wensing A, Beshkov D, Elenkov I, Stoycheva M, Nikolova D, Nikolova M, Switzer W, 2015. Low HIV-1 transmitted drug resistance in Bulgaria against a background of high clade diversity. J. Antimicrob. Chemother 70, 1874–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelis K, Albert J, Mamais I, Magiorkinis G, Hatzakis A, Hamouda O, Struck D, Vercauteren J, Wensing AM, Alexiev I, Asjo B, Balotta C, Camacho RJ, Coughlan S, Griskevicius A, Z. G, Horban A, Kostrikis LG, Lepej S, Liitsola K, Linka M, Nielsen C, Otelea D, Paredes R, Poljak M, Puchhammer-Stockl E, Schmit JC, Sonnerborg A, Stanekova D, Stanojevic M, Boucher CA, Kaplan L, Vandamme AM, Paraskevis D, 2015. Global dispersal pattern of HIV type 1 subtype CRF01_AE: a genetic trace of human mobility related to heterosexual sexual activities centralized in Southeast Asia. J. Infect. Dis 211, 1735–1744. [DOI] [PubMed] [Google Scholar]
- Bennett E, Camacho J, Otelea D, Kuritzkes R, Fleury H, 2009. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance. PloS one 4, e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielejec F, Rambaut A, Suchard M, Lemey P, 2011. SPREAD: Spatial Phylogenetic Reconstruction of Evolutionary Dynamics Bioinformatics. 20 pp. 2910–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozicevic I, Handanagic S, Lepej S, Begovac J, 2013. The emerging and re-emerging human immunodeficiency virus epidemics in Europe. Clin. Microbiol. Infect 10, 917–929. [DOI] [PubMed] [Google Scholar]
- Campbell I, 2007. Chi-squared and Fisher–Irwin tests of two-by-two tables with small sample recommendations. Stat Med 19, 3661–3675. [DOI] [PubMed] [Google Scholar]
- Drummond J, Suchard A, Xie D, Rambaut A, 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol 29, 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC, 2015a. Hepatitis B Surveillance in Europe. http://ecdc.europa.eu/en/publications/Publications/hepatitis-b-surveillance-in-europe-2013.pdf. [Google Scholar]
- ECDC, 2015b. Hepatitis C Surveillance in Europe. http://ecdc.europa.eu/en/publications/Publications/hepatitis-c-surveillance-in-europe-2013.pdf. [Google Scholar]
- EMCDDA, 2015a. European Monitoring Centre for Drugs and Drug Addiction European Drug Report. http://www.emcdda.europa.eu/edr2015.
- EMCDDA, 2015b. Bulgaria Country Overview. http://www.emcdda.europa.eu/countries/bulgaria.
- Frentz D, Wensing M, Albert J, Paraskevis D, Abecasis B, Hamouda O, Jorgensen B, Kucherer C, Struck D, Schmit C, Asjo B, Balotta C, Beshkov D, Camacho J, Clotet B, Coughlan S, De Wit S, Griskevicius A, Grossman Z, Horban A, Kolupajeva T, Korn K, Kostrikis G, Liitsola K, Linka M, Nielsen C, Otelea D, Paredes R, Poljak M, Puchhammer-Stockl E, Sonnerborg A, Stanekova D, Stanojevic M, Vandamme M, Boucher A, Van de Vijver A, 2013. Limited cross-border infections in patients newly diagnosed with HIV in Europe. Retrovirology 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden T, Foti K, Mermin J, 2015. Applying public health principles to the HIV epidemic — how are we doing? New Engl. J. Med 23, 2281–2287. [DOI] [PubMed] [Google Scholar]
- GraphPad (2015). Graph Pad Prism v5.0 www.graphpad.com.
- Grgic I, Lepej S, Lunar M, Poljak M, Vince A, Vrakela I, Planinic A, Seme K, Begovac J, 2013. The prevalence of transmitted drug resistance in newly diagnosed HIV-infected individuals in Croatia: the role of transmission clusters of men who have sex with men carrying the T215S surveillance drug resistance mutation. AIDS Res. Hum. Retrovir 29, 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich D, Kalamara E, Sfetcu O, Pharris A, Noor A, Wiessing L, Hope V, Van de Laar M , 2013. Human immunodeficiency virus among people who inject drugs: is risk increasing in Europe. Euro Surveill. 18, 20648. [DOI] [PubMed] [Google Scholar]
- Jenness S, Neaigus A, Hagan H, Murrill C, Wendel T, 2010. Heterosexual HIV and sexual partnerships between injection drug users and noninjection drug users. AIDS Patient Care and STDs 3, 175–181. [DOI] [PubMed] [Google Scholar]
- Kabakchieva E, Amirkhanian Y, Kelly J, McAuliffe T, Vassileva S, 2002. High levels of sexual HIV/STD risk behaviour among Roma (Gypsy) men in Bulgaria: patterns and predictors of risk in a representative community sample. Int. J. STD AIDS 13, 184–191. [DOI] [PubMed] [Google Scholar]
- Kabakchieva E, Vassileva S, Kelly J, Amirkhanian Y, DiFranceisco W, McAuliffe T, Antonova R, Mihaylova M, Vassilev B, Khoursine R, Petrova E, 2006. HIV risk behavior patterns, predictors, and sexually transmitted disease prevalence in the social networks of young Roma (Gypsy) men in Sofia, Bulgaria. Sex. Transm. Dis 33, 485–490. [DOI] [PubMed] [Google Scholar]
- Kelly J, Amirkhanian Y, Kabakchieva E, Vassileva S, Vassilev B, McAuliffe T, DiFranceisco W, Antonova R, Petrova E, Khoursine R, Dimitrov B, 2006. Prevention of HIV and sexually transmitted diseases in high risk social networks of young Roma (Gypsy) men in Bulgaria: randomised controlled trial. BMJ 333, 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevorkyan A, Teoharov P, Lernout T, Petrova N, Raycheva R, Ivanov I, van Damme P, Kojouharova M, 2015. Prevalence of HBV and HCV among outpatients in the Plovdiv region of Bulgaria, 2010–2011. J. Med. Virol 87, 401–406. [DOI] [PubMed] [Google Scholar]
- Kousiappa I, Achilleos C, Hezka J, Lazarou Y, Othonos K, Demetriades I, Kostrikis G, 2011. Molecular characterization of HIV type 1 strains from newly diagnosed patients in Cyprus (2007–2009) recovers multiple clades including unique recombinant strains and lack of transmitted drug resistance. AIDS Res. Hum. Retrovir 27, 1183–1199. [DOI] [PubMed] [Google Scholar]
- LANL, 2014. Los Alamos HIV Databases. http://www.HIV.lanl.gov.
- Liao H, Tee K, Hase S, Uenishi R, Li J, Kusagawa S, Thang H, Hien T, Pybus G, Takebe Y, 2009. Phylodynamic analysis of the dissemination of HIV-1 CRF01_AE in Vietnam. Virology 391, 51–56. [DOI] [PubMed] [Google Scholar]
- Lin L, Chen L, Liang S, Liu W, Li T, Liu Y, Li H, Bao Z, Wang X, Li J, 2013. Subtype CRF01_AE dominate the sexually transmitted human immunodeficiency virus type 1 epidemic in Guangxi, China. J. Med. Virol 85, 388–395. [DOI] [PubMed] [Google Scholar]
- Niculescu I, Paraschiv S, Paraskevis D, Abagiu A, Batan I, Banica L, Otelea D, 2014. Recent HIV-1 outbreak among intravenous drug users in Romania: evidence for cocirculation of CRF14_BG and subtype F1 strains. AIDS Res. Hum. Retrovir 31, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D, Paraschiv S, Sypsa V, Nikolopoulos G, Tsiara C, Magiorkinis G, Psichogiou M, Flampouris A, Mardarescu M, Niculescu I, Batan I, Malliori M, Otelea D, Hatzakis A, 2015. Enhanced HIV-1 surveillance using molecular epidemiology to study and monitor HIV-1 outbreaks among intravenous drug users (IDUs) in Athens and Bucharest. Infect, Genet Evol 35, 109–121. [DOI] [PubMed] [Google Scholar]
- Price N, Dehal S, Arkin P, 2010. FastTree 2 — approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Suchard A, Xie D, Drummond J, 2014. Tracer v1.6 http://beast.bio.ed.ac.uk/Tracer. [Google Scholar]
- Salemi M, Goodenow M, Montieri S, de Oliveira T, Santoro M, Beshkov D, Alexiev I, Elenkov I, Elenkov I, Yakimova T, Varleva T, Rezza G, Ciccozzi M, 2008. The HIV type 1 epidemic in Bulgaria involves multiple subtypes and is sustained by continuous viral inflow from West and East European countries. AIDS Res. Hum. Retrovir 24, 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M, Ciccozzi M, Alteri C, Montieri S, Alexiev I, Dimova I, Ceccherini-Silberstein F, Beshkov D, Rezza G, Perno C, 2008. Characterization of drug-resistance mutations in HIV type 1 isolates from drug-naive and ARV-treated patients in Bulgaria. AIDS Res. Hum. Retrovir 24, 1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spire B, Lucas G, Carrieri P, 2007. Adherence to HIV treatment among IDUs and the role of opioid substitution treatment (OST). Int. J. Drug Policy 18, 262–270. [DOI] [PubMed] [Google Scholar]
- Stanojevic M, Alexiev I, Beshkov D, Gokengin D, Mezei M, Minarovits J, Otelea D, Paraschiv S, Poljak M, Zidovec-Lepej S, Paraskeveis D, 2012. HIV1 molecular epidemiology in the Balkans — a melting pot for high genetic diversity. AIDS Rev 14, 28–36. [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev P, Hagan H, Lyubenova A, Tomov N, Vasilev G, Krasteva D, Des Jarlais C, 2006. Needle exchange use, sexual risk behaviour, and the prevalence of HIV, hepatitis B virus, and hepatitis C virus infections among Bulgarian injection drug users. Int. J. STD AIDS 17, 621–626. [DOI] [PubMed] [Google Scholar]


