Abstract
Spumaretroviruses, commonly referred to as foamy viruses, are complex retroviruses belonging to the subfamily Spumaretrovirinae, family Retroviridae, which naturally infect a variety of animals including nonhuman primates (NHPs). Additionally, cross-species transmissions of simian foamy viruses (SFVs) to humans have occurred following exposure to tissues of infected NHPs. Recent research has led to the identification of previously unknown exogenous foamy viruses, and to the discovery of endogenous spumaretrovirus sequences in a variety of host genomes. Here, we describe an updated spumaretrovirus taxonomy that has been recently accepted by the International Committee on Taxonomy of Viruses (ICTV) Executive Committee, and describe a virus nomenclature that is generally consistent with that used for other retroviruses, such as lentiviruses and deltaretroviruses. This taxonomy can be applied to distinguish different, but closely related, primate (e.g., human, ape, simian) foamy viruses as well as those from other hosts. This proposal accounts for host-virus co-speciation and cross-species transmission.
Keywords: Foamy virus, ICTV, International Committee on Taxonomy of Viruses, Retrovirus, Spumaretrovirinae, Spumaretrovirus, Spumavirus, Virus classification, Virus nomenclature, Virus taxonomy, Zoonosis
1. Introduction
Spumaretroviruses belong to the subfamily Spumaretrovirinae in the family Retroviridae. They are highly prevalent in animals of diverse non-primate mammalian families [Bovidae (cloven-hooved ruminants), Felidae (cats), Equidae (horses and relatives), and Rhinolophidae (horseshoe bats)], as well as nonhuman primates (NHPs), including apes, Old World monkeys (OWM), New World monkeys (NWM), and prosimians (Rethwilm and Lindemann, 2013; Rethwilm and Bodem, 2013). Members of the subfamily Spumaretrovirinae, commonly referred to as foamy viruses, have a broad tissue and cell tropism and virus infection is generally latent, except in some tissues of the oral cavity (Falcone et al., 1999; Murray et al., 2008).
The earliest reports of foamy virus isolates were of simian origin (NWM, OWM, and apes) and were originally classified serologically using neutralization assays (Hooks and Gibbs, 1975; Meiering and Linial, 2001). The first “human” spumaretrovirus isolate was obtained from cultures of a human nasopharyngeal carcinoma in 1971 and was originally designated “human foamy virus (HFV)” (Achong et al., 1971a, 1971b), but was later found to be of chimpanzee origin based upon sequence identity to chimpanzee foamy virus and renamed as prototype foamy virus (PFV) (Meiering and Linial, 2001; Herchenroder et al., 1994). Cross-species transmissions of simian foamy viruses (SFVs) to humans have occurred by exposure to fluids or tissues from infected NHPs (Betsem et al., 2011; Calattini et al., 2007; Engel et al., 2013; Heneine et al., 2003, 1998; Mouinga-Ondémé et al., 2012; Schweizer et al., 1995; Stenbak et al., 2014; Switzer et al., 2004, 2012; Wolfe et al., 2004; Khan, 2009; Brooks et al., 2002; Callahan et al., 1999). Although foamy viruses are exogenously transmitted viruses, endogenous foamy virus sequences have been identified and characterized in genomes of many species, including aye-aye (Daubentonia madagascariensis), sloth (Choloepus hoffmanni), Cape golden mole (Chrysochloris asiatica), coelacanth (Latimeria chalumnae), platyfish (Xiphophorus maculatus), and zebrafish (Danio rerio) (Katzourakis et al., 2014, 2009; Aiewsakun and Katzourakis, 2017; Han and Worobey, 2012a, 2012b, 2014; Ruboyianes and Worobey, 2016). In fact, molecular evolution studies have indicated that spumaretroviruses have co-speciated with their hosts for millions of years (Katzourakis et al., 2014, 2009; Ghersi et al., 2015; Switzer et al., 2005a). Recently, phylogenetic analyses using amphibian and fish genomes have estimated the date of retrovirus emergence at > 450 million years ago (Aiewsakun and Katzourakis, 2017) with foamy viruses inferred to be the most ancient retrovirus. A recent increase in the number of foamy virus isolates and sequences using a variety of novel molecular and genomic techniques highlights the need for updating and expanding spumaretrovirus taxonomy (Mouinga-Ondémé et al., 2012; Ghersi et al., 2015; Ayouba et al., 2013; Liu et al., 2008; Wu et al., 2012; Muniz et al., 2013; Troncoso et al., 2015; Blasse et al., 2013; Calattini et al., 2006, 2004; Goldberg et al., 2009; Huang et al., 2013; Jones-Engel et al., 2007; Mouinga-Ondémé et al., 2010; Richard et al., 2015; Yoshikawa et al., 2014; Kehl et al., 2013; Hechler et al., 2012).
The taxonomic history of Spumaretrovirinae has been based only on the few early virus isolates. Although “foamy virus” was discovered in rhesus monkey kidney cultures in 1955 (Rustigian et al., 1955), simian foamy retroviruses were not formally classified until the Second Report (1976) of the International Committee on Taxonomy of Viruses [ICTV], which assigned bovine syncytial virus (BSV), feline syncytial virus (FSV), hamster syncytial virus (HSV), human foamy virus (HFV), and simian foamy virus (SFV) to the “genus-less” subfamily Spumavirinae (not italicized at the time) (Fenner, 1976). In the Third (1979) ICTV Report, the still genus-less subfamily Spumavirinae contained only four viruses (BSV, FSV, HFV, and SFV) (Matthews, 1979). By the Fifth (1991) ICTV Report, all three subfamilies (Oncovirinae, Lentivirinae, and Spumavirinae) of the Retroviridae were abandoned and seven genera created, including Spumavirus, which contained BSV, FSV, HFV, and SFV (Coffin et al., 1991), with HFV designated as the type species. This taxonomy was upheld in the Sixth (1995) ICTV Report, but HFV was referred to as human spumavirus (Coffin et al., 1995). The Seventh (2000) ICTV Report for the first time differentiated between species and viruses. Chimpanzee foamy virus was designated the type species and chimpanzee foamy virus (formerly human spumavirus) was assigned to it (according to the ICTV rules, virus names are not italicized). The other species were Bovine foamy virus (containing bovine foamy virus; formerly bovine syncytial virus), Feline foamy virus (containing feline foamy virus, formerly feline syncytial virus), Simian foamy virus 1 (containing simian foamy virus type 1), and Simian foamy virus 3 (containing simian foamy virus type 3) (Hunter et al., 2000). In the Eighth (2005) ICTV Report, based upon the distinct replication pathway of foamy viruses, which contains features homologous to both retroviruses and hepadnaviruses (Linial, 1999), the subfamily Spumaretrovirinae was reinstated for the genus Spumavirus. The species Simian foamy virus 1 was renamed Macaque simian foamy virus and the virus assigned to this species, simian foamy virus 1, was renamed macaque simian foamy virus. Similarly, the species Simian foamy virus 3 was renamed African green monkey simian foamy virus and the virus in this species, simian foamy virus 3, was renamed African green monkey simian foamy virus; the species Chimpanzee foamy virus was renamed Simian foamy virus and the virus in this species, chimpanzee foamy virus was renamed simian foamy virus. In addition, the species Equine foamy virus was established for equine foamy virus. No changes were made to Bovine foamy virus and Feline foamy virus (Linial et al., 2005). The most recent, Ninth (2011) ICTV Report, lists six spumavirus species for six viruses (Stoye et al., 2012), and this current classification is shown in Table 1.
Table 1.
Current spumaretrovirus taxonomy according to ICTVa.
| Family | Subfamily | Genus | Species | Virus name |
|---|---|---|---|---|
| Retroviridae | Spumaretrovirinae | Spumavirus | African green monkey simian foamy virus | African green monkey simian foamy virus (SFVagm)b |
| Bovine foamy virus | bovine foamy virus (BFV) | |||
| Equine foamy virus | equine foamy virus (EFV) | |||
| Feline foamy virus | feline foamy virus (FFV) | |||
| Macaque simian foamy virus | macaque simian foamy virus (SFVmac)c | |||
| Simian foamy virus d | simian foamy virus, human isolate (SFVcpz(hu))e |
Ref. Stoye et al. (2012).
Synonym: simian foamy virus 3 (SFV-3).
Synonym: simian foamy virus 1 (SFV-1).
Type species.
Synonyms: chimpanzee foamy virus (CFV), human foamy virus (HFV) and prototype foamy virus (PFV).
The current taxonomy for spumaretroviruses is outdated as it does not accommodate new foamy viruses discovered in NHPs of various species, including OWM and NWM (Ghersi et al., 2015; Muniz et al., 2013, 2015). Furthermore, formal nomenclature for designating virus isolates is lacking, particularly for simian foamy viruses, which has led to some confusion in virus descriptions and references in publications and retrieval of sequences deposited in public databases. The critical need for updating spumaretrovirus taxonomy and for developing a consensus nomenclature for different foamy viruses was recognized by the scientific community and discussed at the International Foamy Virus Conference held in 2014 in Puławy, Poland (Materniak et al., 2015) and in 2016 in Paris, France (Buseyne et al., 2016). Here, we present an updated and expanded spumaretrovirus taxonomy based upon current knowledge that accounts for host-virus co-speciation and cross-species transmission. Furthermore, the proposed virus nomenclature can be applied to all spumaretroviruses, including non-primate and primate viruses.
2. Updated 2017 spumaretrovirus classification
The recent increase in the number of primate and non-primate spumaretroviruses warranted an update of the current ICTV classification as well as establishment of a more consistent nomenclature. This updated taxonomy, which was proposed by the authors to the Executive Committee of the ICTV on June 8, 2017, is shown in Table 2. The following changes were proposed based on analyses of sequences of spumaretrovirus genomes available in public databases:
Table 2.
Updated spumaretrovirus taxonomy as proposed in June 2017a.
| Family | Subfamily | Genus | Species | Virus (name) |
|---|---|---|---|---|
| Retroviridae | Spumaretrovirinae | Simiispumavirus | Eastern chimpanzee simian foamy virus b,c | simian foamy virus Pan troglodytes schweinfurthii (SFVpsc) |
| Western chimpanzee simian foamy virus | simian foamy virus Pan troglodytes verus (SFVpve) | |||
| Central chimpanzee simian foamy virus | simian foamy virus Pan troglodytes troglodytes (SFVptr) | |||
| Western lowland gorilla simian foamy virus | simian foamy virus Gorilla gorilla gorilla (SFVggo) | |||
| Bornean orangutan simian foamy virus | simian foamy virus Pongo pygmaeus pygmaeus (SFVppy) | |||
| Taiwanese macaque simian foamy virus d | simian foamy virus Macaca cyclopis (SFVmcy) | |||
| Rhesus macaque simian foamy virus | simian foamy virus Macaca mulatta (SFVmmu) | |||
| Japanese macaque simian foamy virus | simian foamy virus Macaca fuscata (SFVmfu) | |||
| Grivet simian foamy virus e | simian foamy virus Chlorocebus aethiops (SFVcae) | |||
| Guenon simian foamy virus | simian foamy virus Cercopithecus nictitans (SFVcni) | |||
| Spider monkey simian foamy virus | simian foamy virus Ateles species (SFVaxx) | |||
| White-tufted-ear marmoset simian foamy virus | simian foamy virus Callithrix jacchus (SFVcja) | |||
| Squirrel monkey simian foamy virus | simian foamy virus Saimiri sciureus (SFVssc) | |||
| Yellow-breasted capuchin simian foamy virus | simian foamy virus Sapajus xanthosternos (SFVsxa) | |||
| Prosimiispumavirus | Brown greater galago prosimian foamy virus c | simian foamy virus Otolemur crassicaudatus (SFVocr) | ||
| Bovispumavirus | Bovine foamy virus c | bovine foamy virus Bos taurus (BFVbta) | ||
| Felispumavirus | Feline foamy virus c | feline foamy virus Felis catus (FFVfca) | ||
| Puma feline foamy virus | feline foamy virus Puma concolor (FFVpco) | |||
| Equispumavirus | Equine foamy virus c | equine foamy virus Equus caballus (EFVeca) |
Based upon available complete virus genome sequences.
The former species Simian spumavirus.
Type species.
The former species Macaque simian foamy virus.
The former species African green monkey simian foamy virus.
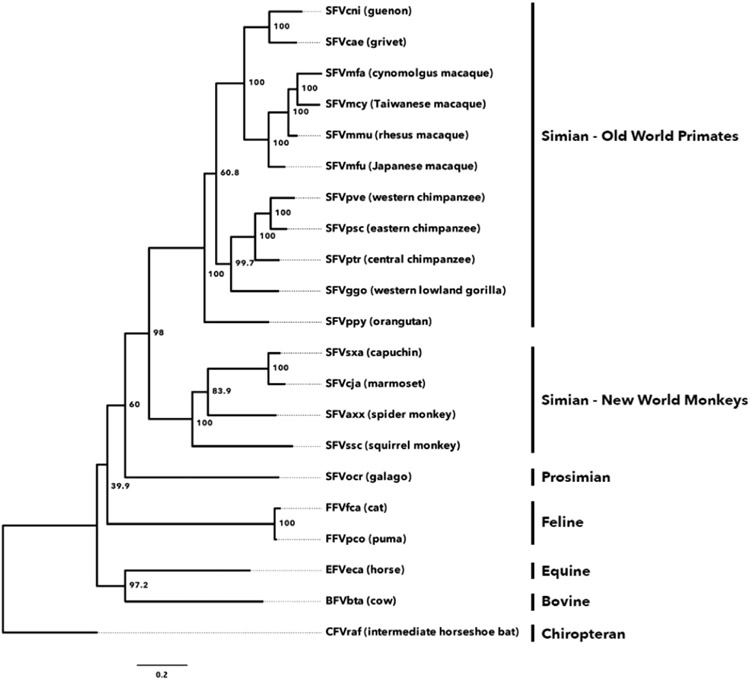
the existing genus Spumavirus was replaced by five genera named Bovispumavirus, Equispumavirus, Felispumavirus, Prosimiispumavirus, and Simiispumavirus because the spumaretrovirus phylogeny largely reflects the host phylogeny (Fig. 1), supporting the hypothesis of spumaretrovirus-host co-evolution (Switzer et al., 2004, 2005a; Katzourakis et al., 2014, 2009; Ghersi et al., 2015);
the established species Bovine foamy virus, Feline foamy virus, and Equine foamy virus were included in the new genera Bovispumavirus, Felispumavirus, and Equispumavirus, respectively. Additionally, the species Puma feline foamy virus was included in genus Felispumavirus;
a new species, Brown greater galago simian foamy virus, was included in a new genus Prosimiispumavirus;
the previously established species were included in the new genera, and new species were established for previously unclassified spumaretroviruses. Species names were chosen based upon the viral host animal names. The previous species simian foamy virus was renamed Eastern chimpanzee simian foamy virus. The species Macaque simian foamy virus and African green monkey simian foamy virus were renamed Taiwanese macaque simian foamy virus and Grivet simian foamy virus, respectively, and together with Eastern chimpanzee simian foamy virus were included in genus Simiispumavirus. Eleven additional NHP spumaretrovirus species were added to this genus. Each proposed species is represented by at least one virus for which a complete genome sequence has been deposited in public databases.
Fig. 1.
Foamy virus phylogeny reflects long-term virus – host co-speciation. Evolutionary relationships were inferred using maximum likelihood analysis of an alignment comprising the polymerase gene (pol) and envelope gene (env) nucleotide sequences of 21 foamy viruses from diverse mammalian hosts. Branch tips are labeled with viral species names and the host common names; brackets indicate common names for subfamily/group within the class Mammalia. Nodes are labeled with bootstrap support values (based on 1000 replicates). An alignment (including insertions and deletions) beginning approximately in the middle of the pol gene and extending to approximately the middle of the env gene was generated using the MUSCLE algorithm as implemented in Geneious 10.1.3. For reference, alignment corresponded to nucleotide position 5089 – 7927 of SFVpsc (NCBI accession number KX08159). Alignment of all of the sequences are submitted in the Supplement. Unrooted tree was generated using PhyML with the HKY85 substitution model and the NNI search option. Tree was visualized using FigTree 1.4.2 (note that unrooted tree is shown as rooted for ease of visualization). Topology is similar to those published by others (Rethwilm and Bodem, 2013; Katzourakis et al., 2014; Switzer et al., 2005b). Input taxa and accession numbers were: SFVcni (JQ867466); SFVcae (NC_010820); SFVmfa (LC094267); SFVmcy (NC_010819); SFVmmu (MF280817); SFVmfu (AB923 518); SFVpve (NC_001364); SFVpsc (KX08159); SFVptr (JQ867463); SFVggo (HM245790); SFVppy (AJ5445 79); SFVsxa (KP143760); SFVcja (GU356395); SFVaxx (EU010385); SFVssc (GU356394); SFVocr (KM233 624); FFVfca (Y08851); FFVpco (KC292054); EFVeca (AF201902); BFVbta (NC_001831); CFVraf (JQ8148 55).
In addition to the viruses included in the updated taxonomy submitted to the ICTV in 2017, there are other foamy virus isolates that are phylogenetically distinct from the currently included ones and were not included in the proposal because of the lack of a complete coding genome sequence. These are shown in Table 3 and are envisioned to be included in future taxonomic updates, along with other available sequences at that time. It should be noted that simian foamy virus Macaca fascicularis (SFVmfa) is also included in this table (and in Fig. 1) since it is expected to be classified in the next update based upon a published full-genome sequence (Sakai et al., 2016). Additionally, based upon recent discussions related to viruses identified by metagenomics, sequences that are distinct phylogenetically from those of the currently classified viruses and have been completely sequenced in their coding regions may be classified officially by the ICTV even in the absence of virus isolates (Simmonds et al., 2017).
Table 3.
Projected updates for spumaretrovirus taxonomya.
| Family | Subfamily | Genus | Species | Virus names and designations |
|---|---|---|---|---|
| Retroviridae | Spumaretrovirinae | Simiispumavirus | Cynomolgus macaque simian foamy virus | simian foamy virus Macaca fascicularis (SFVmfa)b |
| Drill simian foamy virus | simian foamy virus Mandrillus leucophaeus (SFVmle)c | |||
| Mandrill simian foamy virus | simian foamy virus Mandrillus sphinx (SFVmsp)c | |||
| Yellow baboon simian foamy virus | simian foamy virus Papio cynocephalus (SFVpcy)c | |||
| Chispumavirus | Intermediate horseshoe bat foamy virus d | chiropteran foamy virus Rhinolophus affinis (CFVraf)c |
Potential candidates for new species based upon phylogenetic distance from current ones.
To be included in next update since full-genome sequence has been published.
To be included in update when sequence of at least full genome open reading frames is publicly available.
Potential candidate for type species.
3. Updated 2017 spumaretrovirus nomenclature
Previously, different spumaretroviruses were named by adding three-letter lower-case abbreviations to “SFV” (simian foamy virus) derived from the common names of the hosts from which the viruses were isolated. For instance, SFVmac was used for simian spumaretroviruses infecting macaques and SFVagm was used for simian spumaretroviruses infecting grivets (African green monkeys, agm) (Meiering and Linial, 2001). However, this general designation of viruses does not distinguish strains infecting hosts of different species or subspecies. The genus Macaca (macaques) includes more than 20 species the members of which are distributed throughout Asia, with one also in Africa, and the genus Chlorocebus (African green monkeys) includes six species, the members of which are distributed across sub-Saharan Africa. The most confusing virus name was “simian foamy virus”, which referred to foamy viruses from all NHPs except macaques and African green monkeys.
Here we propose a spumaretrovirus nomenclature similar to that used for simian immunodeficiency virus and simian T-lymphotropic virus isolates. Accordingly, all spumaretrovirus names will contain the words “foamy virus” preceded by an adjectival classifier indicating the host (e.g., bovine, feline, simian) followed by the name of the species, or in some cases subspecies, of the particular host. The virus host name and “foamy virus” are abbreviated in capital letters (e.g., feline foamy virus is abbreviated FFV, simian foamy virus is abbreviated SFV) followed by lower-case three-letter abbreviations for the host species name. These abbreviations are comprised of the first letter of the host genus and the first two letters of the species or subspecies (e.g., simian foamy virus Macaca cyclopis is abbreviated SFVmcy) (Tables 2 and 3), or the first letter of the species or subspecies and the next unique letter, when the letters are the same (e.g. simian foamy viruses isolated from New World primate species such as Leontopithecus chrysomelas and Leonthopithecus chrysopygus would be abbreviated as SFVlcm and SFVlcp, respectively). In the case of an unknown host species, the first letter of the genus name is followed by “xx” (italics letters). Virus isolate names are designated by using the virus abbreviation followed by an underscore that appends additional, isolate-identifying information such as the host from which it was isolated (for instance, “hu” for human), isolate designation, and other identifying information. For example, simian foamy virus Pan troglodytes schweinfurthii, human isolate HSRV, clone 13 will be designated as SFVpsc_huHRSV.13, and feline foamy virus Felis catus strain FUV7 will be designated as FFVfca_FUV7 (Table 4). Similarly, in case of inter-species transmission involving nonhuman primates, SFV from western red colobus monkey (Philiocolobus badius badius) isolated from a wild chimpanzee subspecies Pan troglodyes verus (Leendertz et al., 2008) will be designated as SFVpba_pveLeo. Furthermore, for recombinant viruses (including naturally-occurring and those genetically-engineered), the designation “[RF]” is included at the end (e.g., SFVmcy_FV34[RF] (Galvin et al., 2013)).
Table 4.
Proposed spumaretrovirus isolate nomenclature.
| Host | Spumaretrovirus name and designation | Isolate description: type, strain, isolate [Previous designation(s)] |
New isolate designation | GenBank accession number |
Reference(s) a |
|---|---|---|---|---|---|
| Bovine | bovine foamy virus Bos taurus (BFVbta) | BSV-11 [BSV, BFV] | BFVbta_BSV11b | U94514 c | (Renshaw and Casey, 1994) |
| BFV-Riems | BFVbta_Riems | JX307862 c | (Hechler et al., 2012) | ||
| BFV100 | BFVbta_100 | JX307861 c | (Hechler et al., 2012) | ||
| Feline | feline foamy virus Felis catus (FFVfca) | type FUV7 [FSV, FFV] | FFVfca_FUV7b | Y08851 c | (Bodem et al., 1996; Winkler et al., 1997) |
| type F17/951 | FFVfca_F17/951 | U85043 c | (Helps and Harbour, 1997) | ||
| feline foamy virus Puma concolor (FFVpco) | X102 | FFVpco_X102 | KC292054 c | (Kehl et al., 2013) | |
| Equine | equine foamy virus Equus caballus (EFVeca) | isolate clone 1 [EFV] | EFVeca_1b | AF201902 c | (Tobaly-Tapiero et al., 2000) |
| Chiropteran | chiropteran foamy virus Rhinolophus affinis (CFVraf) | isolate 1 [RaFV-1] | CFVraf_1 | JQ814855 d | (Wu et al., 2012) |
| Prosimian | simian foamy virus Otolemur crassicaudatus (SFVocr) | type 5, strain 1557 [SFV type 5; SFVgal] | SFVocr_1557b | KM233624 c | (Katzourakis et al., 2014) |
| Simian—New World monkeys | simian foamy virus Ateles species (SFVaxx) | serotype 8, strain S-140, Hooks40 [SFV type 8; SFVspm] | SFVaxx_Hooks40 | EU010385 c | (Thumer et al., 2007) |
| simian foamy virus Callithrix jacchus (SFVcja) | isolate FXV [SFVmar] | SFVcja_FXV | GU356395 c | (Pacheco et al., 2010) | |
| simian foamy virus Saimiri sciureus (SFVssc) | serotype 4, strain 1224 [SFV type 4; SFVsqu] | SFVssc_1224 | GU356394 c | (Pacheco et al., 2010) | |
| simian foamy virus Sapajus xanthosternos (SFVsxa) | isolate Z17 | SFVsxa_Z17 | KP143760 c | (Troncoso et al., 2015) | |
| Simian—apes | simian foamy virus Pan troglodytes schweinfurthii (SFVpsc) | human isolate HSRV clone 13 [PFV; CFV; HSRV; SFVcpz (hu); HFV] | SFVpsc_huHSRV.13b | KX08159 c | (Wagner and Bodem, 2017) |
| simian foamy virus Pan troglodytes troglodytes (SFVptr) | human isolate BAD327 | SFVptr_huBAD327 | JQ867463 c | (Rua et al., 2012) | |
| human isolate AG15 | SFVptr_huAG15 | JQ867462 c | (Rua et al., 2012) | ||
| simian foamy virus Pan troglodytes verus (SFVpve) | [SFVcpz] | SFVpve | NC_001364 c | (Herchenroder et al., 1994) | |
| serotype 6, strain CV-1, Pan-1 [SFV type 6; SFVcpz] | SFVpve_Pan1 | ||||
| serotype 7, strain CV-11, Pan-2 [ SFV type 7; SFVcpz] | SFVpve_Pan2 | ||||
| simian foamy virus Gorilla gorilla gorilla (SFVggo) | [SFV-Gg; SFVgor] | SFVggo_Gg | HM245790 c | (Schulze et al., 2011) | |
| human isolate BAK74 | SFVggo_huBAK74 | JQ867464 c | (Rua et al., 2012) | ||
| human isolate BAD468 | SFVggo_huBAD468 | JQ867465 c | (Rua et al., 2012) | ||
| simian foamy virus Pongo pygmaeus pygmaeus (SFVppy) | serotype 11, strain bella [SFV type 11; SFVora] | SFVppy_bella | AJ544579 c | (Verschoor et al., 2003) | |
| Simian—Old World monkeys | simian foamy virus Chlorocebus aethiops (SFVcae) | strain LK3 [African green monkey SFV; SFVagm; SFVagm-3] | SFVcae_LK3 | M74895 c | (Renne et al., 1992) |
| serotype 3, strain FV2014 [SFV 3; SFV type 3; SFVagm] | SFVcae_FV2014 | MF582544 c | (Nandakumar et al., 2018) | ||
| simian foamy virus Cercopithecus nictitans (SFVcni) | human isolate AG16 | SFVcni_huAG16 | JQ867466 c | (Rua et al., 2012) | |
| simian foamy virus Macaca cyclopis (SFVmcy) | serotype 1, strain FV21 [SFV-1; macaque SFV; SFV type 1; SFVmac; SFVmcy-1] | SFVmcy_FV21 | NC_010819 c | (Kupiec et al., 1991) | |
| serotype 2, strain FV34 [SFV type 2; SFVmac; SFVmcy-2] | SFVmcy_FV34[RF]d | KF026286.1 c | (Galvin et al., 2013) | ||
| simian foamy virus Macaca fascicularis (SFVmfa) | isolate Cy5061 | SFVmfa_Cy5061 | LC094267 c | (Sakai et al., 2016) | |
| simian foamy virus Macaca fuscata (SFVmfu) | Isolate WK1, clone pJM356 [Japanese macaque SFV; SFVjm] | SFVmfu_WK1.pJM356 | AB923518 c | (Yoshikawa et al., 2014) | |
| simian foamy virus Macaca mulatta (SFVmmu) | isolate K3T | SFVmmu_K3T | MF280817 c | (Nandakumar et al., 2017) | |
| simian foamy virus Mandrillus leucophaeus (SFVmle) | isolate Mnd205 | SFVmle_Mnd205 | AY583777 d | (Calattini et al., 2004) | |
| simian foamy virus Mandrillus sphinx (SFVmsp) | isolate Mnd301 | SFVmsp_Mnd301 | AY583775 d | (Calattini et al., 2004) | |
| simian foamy virus Papio cynocephalus (SFVpcy) | serotype 10 [SFV type 10; SFVbab] | SFVpcy | AF049083 d |
reference provided for published sequences
Type species.
full-length genome.
partial sequence.
4. Conclusions
The updated and expanded spumaretrovirus taxonomy presented here was initially submitted to the ICTV on June 8, 2017, and is now approved by the ICTV Executive Committee. The proposal awaits ratification by vote of the International Union of Microbiological Societies (IUMS) Virology Division, which will occur in the first quarter of 2018. The new taxonomy format is easily expandable to accommodate new genera and species for classification of newly discovered foamy viruses. Additionally, we have established a spumaretrovirus nomenclature that considers host-virus co-speciation and cross-species transmission, and incorporates host animal species information and strain and/or animal identification codes in spumaretrovirus names and their abbreviations. This collaborative effort is aimed toward unifying classification and designations of spumaretroviruses to facilitate easy comparisons of reported foamy virus isolates.
Supplementary Material
Acknowledgements
We appreciate the expertise provided by Prof. Dr. Gerrit Kloss for deriving the Latin names of the genera in Spumaretrovirinae.
Footnotes
Disclaimer
The views and conclusions that are contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of Health and Human Services, the U.S. Centers for Disease Control and Prevention, or of the institutions and companies affiliated with the authors. The work by J.H.K. was supported through Battelle Memorial Institute's prime contract with the U.S. National Institute of Allergy and Infectious Diseases (NIAID) under contract number HHSN272200700016I.
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2017.12.035.
References
- Achong BG, Mansell PW, Epstein MA, 1971a. A new human virus in cultures from a nasopharyngeal carcinoma. J. Pathol 103, P18. [PubMed] [Google Scholar]
- Achong BG, Mansell PW, Epstein MA, Clifford P, 1971b. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst 46, 299–307. [PubMed] [Google Scholar]
- Aiewsakun P, Katzourakis A, 2017. Marine origin of retroviruses in the early Palaeozoic Era. Nat. Commun 8, 13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayouba A, Duval L, Liegeois F, Ngin S, Ahuka-Mundeke S, Switzer WM, Delaporte E, Ariey F, Peeters M, Nerrienet E, 2013. Nonhuman primate retroviruses from Cambodia: high simian foamy virus prevalence, identification of divergent STLV-1 strains and no evidence of SIV infection. Infect. Genet. Evol 18, 325–334. [DOI] [PubMed] [Google Scholar]
- Betsem E, Rua R, Tortevoye P, Froment A, Gessain A, 2011. Frequent and recent human acquisition of simian foamy viruses through apes' bites in central Africa. PLoS Pathog. 7, e1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasse A, Calvignac-Spencer S, Merkel K, Goffe AS, Boesch C, Mundry R, Leendertz FH, 2013. Mother-offspring transmission and age-dependent accumulation of simian foamy virus in wild chimpanzees. J. Virol 87, 5193–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodem J, Lochelt M, Winkler I, Flower RP, Delius H, Flugel RM, 1996. Characterization of the spliced pol transcript of feline foamy virus: the splice acceptor site of the pol transcript is located in gag of foamy viruses. J. Virol 70, 9024–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JI, Rud EW, Pilon RG, Smith JM, Switzer WM, Sandstrom PA, 2002. Cross-species retroviral transmission from macaques to human beings. Lancet 360, 387–388. [DOI] [PubMed] [Google Scholar]
- Buseyne F, Gessain A, Soares MA, Santos AF, Materniak-Kornas M, Lesage P, Zamborlini A, Löchelt M, Qiao W, Lindemann D, Wöhrl BM, Stoye JP, Taylor IA, Khan AS, 2016. Eleventh International Foamy Virus Conference-Meeting Report. Viruses 8:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calattini S, Nerrienet E, Mauclère P, Georges-Courbot M-C, Saïb A, Gessain A, 2004. Natural simian foamy virus infection in wild-caught gorillas, mandrills and drills from Cameroon and Gabon. J. Gen. Virol 85, 3313–3317. [DOI] [PubMed] [Google Scholar]
- Calattini S, Nerrienet E, Mauclère P, Georges-Courbot M-C, Saïb A, Gessain A, 2006. Detection and molecular characterization of foamy viruses in Central African chimpanzees of the Pan troglodytes troglodytes and Pan troglodytes vellerosus subspecies. J. Med. Primatol 35, 59–66. [DOI] [PubMed] [Google Scholar]
- Calattini S, Betsem EBA, Froment A, Mauclère P, Tortevoye P, Schmitt C, Njouom R, Saib A, Gessain A, 2007. Simian foamy virus transmission from apes to humans, rural Cameroon. Emerg. Infect. Dis 13, 1314–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan ME, Switzer WM, Matthews AL, Roberts BD, Heneine W, Folks TM, Sandstrom PA, 1999. Persistent zoonotic infection of a human with simian foamy virus in the absence of an intact orf-2 accessory gene. J. Virol 73, 9619–9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM, Essex M, Gallo R, Graf TM, Hinuma Y, Hunter E, Jaenisch R, Nusse R, Oroszlan S, Svoboda J, Teich N, Toyoshima K, Varmus H, 1991. Family Retroviridae. In: Classification and Nomenclature of viruses—Fifth Report of the International Committee on Taxonomy of Viruses. Archives of Virology Supplement, vol 2. Springer-Verlag, Vienna, Austria, pp. 193–204. [Google Scholar]
- Coffin JM, Essex M, Gallo R, Graf TM, Hinuma Y, Hunter E, Jaenisch R, Nusse R, Oroszlan S, Svoboda J, Teich N, Toyoshima K, Varmus H, 1995. Family Retroviridae. In: Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Matelli GP, Mayo MA, Summers MD (Eds.), Virus Taxonomy - Sixth Report of the International Committee of Viruses 10. Springer-Verlag, Vienna, Austria, pp. 193–204. [Google Scholar]
- Engel GA, Small CT, Soliven K, Feeroz MM, Wang X, Kamrul Hasan M, Oh G, Rabiul Alam SM, Craig KL, Jackson DL, Matsen Iv FA, Linial ML, Jones-Engel L, 2013. Zoonotic simian foamy virus in Bangladesh reflects diverse patterns of transmission and co-infection. Emerg. Microbes Infect 2, e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone V, Leupold J, Clotten J, Urbanyi E, Herchenroder O, Spatz W, Volk B, Böhm N, Toniolo A, Neumann-Haefelin D, Schweizer M, 1999. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 257, 7–14. [DOI] [PubMed] [Google Scholar]
- Fenner F, 1976. Classification and nomenclature of viruses. Second report of the International Committee on Taxonomy of Viruses. Intervirology 7, 1–115. [DOI] [PubMed] [Google Scholar]
- Galvin TA, Ahmed IA, Shahabuddin M, Bryan T, Khan AS, 2013. Identification of recombination in the envelope gene of simian foamy virus serotype 2 isolated from Macaca cyclopis. J. Virol 87, 8792–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi BM, Jia H, Aiewsakun P, Katzourakis A, Mendoza P, Bausch DG, Kasper MR, Montgomery JM, Switzer WM, 2015. Wide distribution and ancient evolutionary history of simian foamy viruses in New World primates. Retrovirology 12, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TL, Sintasath DM, Chapman CA, Cameron KM, Karesh WB, Tang S, Wolfe ND, Rwego IB, Ting N, Switzer WM, 2009. Coinfection of Ugandan red colobus (Procolobus [Piliocolobus] rufomitratus tephrosceles) with novel, divergent delta-, lenti-, and spumaretroviruses. J. Virol 83, 11318–11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GZ, Worobey M, 2012b. An endogenous foamy-like viral element in the coelacanth genome. PLoS Pathog. 8, e1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GZ, Worobey M, 2014. Endogenous viral sequences from the Cape golden mole (Chrysochloris asiatica) reveal the presence of foamy viruses in all major placental mammal clades. PLoS One 9, e97931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G-Z, Worobey M, 2012a. An endogenous foamy virus in the aye-aye (Daubentonia madagascariensis). J. Virol 86, 7696–7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hechler T, Materniak M, Kehl T, Kuzmak J, Löchelt M, 2012. Complete genome sequences of two novel European clade bovine foamy viruses from Germany and Poland. J. Virol 86, 10905–10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helps CR, Harbour DA, 1997. Comparison of the complete sequence of feline spumavirus with those of the primate spumaviruses reveals a shorter gag gene. J. Gen. Virol 78 (Pt 10), 2549–2564. [DOI] [PubMed] [Google Scholar]
- Heneine W, Switzer WM, Sandstrom P, Brown J, Vedapuri S, Schable CA, Khan AS, Lerche NW, Schweizer M, Neumann-Haefelin D, Chapman LE, Folks TM, 1998. Identification of a human population infected with simian foamy viruses. Nat. Med 4, 403–407. [DOI] [PubMed] [Google Scholar]
- Heneine W, Schweizer M, Sandstrom P, Folks T, 2003. Human infection with foamy viruses. Curr. Top. Microbiol. Immunol 277, 181–196. [DOI] [PubMed] [Google Scholar]
- Herchenroder O, Renne R, Loncar D, Cobb EK, Murthy KK, Schneider J, Mergia A , Luciw PA, 1994. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV). Virology 201, 187–199. [DOI] [PubMed] [Google Scholar]
- Hooks JJ, Gibbs CJ Jr., 1975. The foamy viruses. Bacteriol. Rev 39, 169–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Yu W, He Z, 2013. Foamy virus in the tree shrew Tupaia belangeri is highly related to simian foamy virus in Macaca mulatta. AIDS Res. Hum. Retroviruses 29, 1177–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter E, Casey J, Hahn B, Hayami M, Korber B, Kurth R, Neil J, Rethwilm A, Sonigo P, Stoye J, 2000. Family Retroviridae. In: van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB (Eds.), Virus Taxonomy—Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, U.S.A, pp. 369–387. [Google Scholar]
- Jones-Engel L, Steinkraus KA, Murray SM, Engel GA, Grant R, Aggimarangsee N, Lee BPY-H, May C, Schillaci MA, Somgird C, Sutthipat T, Vojtech L, Zhao J, Linial ML, 2007. Sensitive assays for simian foamy viruses reveal a high prevalence of infection in commensal, free-ranging Asian monkeys. J. Virol 81, 7330–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Gifford RJ, Tristem M, Gilbert MT, Pybus OG, 2009. Macroevolution of complex retroviruses. Science 325, 1512. [DOI] [PubMed] [Google Scholar]
- Katzourakis A, Aiewsakun P, Jia H, Wolfe ND, LeBreton M, Yoder AD, Switzer WM, 2014. Discovery of prosimian and afrotherian foamy viruses and potential cross species transmissions amidst stable and ancient mammalian co-evolution. Retrovirology 11, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl T, Bleiholder A, Roßmann F, Rupp S, Lei J, Lee J, Boyce W, Vickers W, Crooks K, VandeWoude S, Löchelt M, 2013. Complete genome sequences of two novel Puma concolor foamy viruses from California. Genome Announc. 1, e0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AS, 2009. Simian foamy virus infection in humans: prevalence and management. Expert Rev. Anti Infect. Ther 7, 569–580. [DOI] [PubMed] [Google Scholar]
- Kupiec J-J, Kay A, Hayat M, Ravier R, Périés J, Galibert F, 1991. Sequence analysis of the simian foamy virus type 1 genome. Gene 101, 185–194. [DOI] [PubMed] [Google Scholar]
- Leendertz FH, Zirkel F, Couacy-Hymann E, Ellerbrok H, Morozov VA, Pauli G, Hedemann C, Formenty P, Jensen SA, Boesch C, Junglen S, 2008. Interspecies transmission of simian foamy virus in a natural predator-prey system. J. Virol 82, 7741–7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial ML, 1999. Foamy viruses are unconventional retroviruses. J. Virol 73, 1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial ML, Fan H, Hahn B, Lwer R, Neil J, Quackenbush S, Rethwilm A, Sonigo P, Stoye J, Tristem M, 2005. Family Retroviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (Eds.), Virus Taxonomy—Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, San Diego, U.S.A, pp. 421–440. [Google Scholar]
- Liu W, Worobey M, Li Y, Keele BF, Bibollet-Ruche F, Guo Y, Goepfert PA, Santiago ML, Ndjango J-B, Neel C, Clifford SL, Sanz C, Kamenya S, Wilson ML, Pusey AE, Gross-Camp N, Boesch C, Smith V, Zamma K, Huffman MA, Mitani JC, Watts DP, Peeters M, Shaw GM, Switzer WM, Sharp PM, Hahn BH, 2008. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 4, e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materniak M, Kubiś P, Rola-Luszcza M, Khan AS, Buseyne F, Lindemann D, Löchelt M, Kuźmak J, 2015. Proceedings of the Tenth International Foamy Virus Conference 2014–achievements and perspectives. Viruses 7:1651–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews REF, 1979. Classification and nomenclature of viruses. third report of the International Committee on Taxonomy of Viruses. Intervirology 12, 131–296. [DOI] [PubMed] [Google Scholar]
- Meiering CD, Linial ML, 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev 14, 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouinga-Ondémé A, Betsem E, Caron M, Makuwa M, Salle B, Renault N, Saib A, Telfer P, Marx P, Gessain A, Kazanji M, 2010. Two distinct variants of simian foamy virus in naturally infected mandrills (Mandrillus sphinx) and cross-species transmission to humans. Retrovirology 7, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouinga-Ondémé A, Caron M, Nkoghé D, Telfer P, Marx P, Saïb A, Leroy E, Gonzalez J-P, Gessain A, Kazanji M, 2012. Cross-species transmission of simian foamy virus to humans in rural Gabon, Central Africa. J. Virol 86, 1255–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz CP, Troncoso LL, Moreira MA, Soares EA, Pissinatti A, Bonvicino CR, Seuánez HN, Sharma B, Jia H, Shankar A, Switzer WM, Santos AF, Soares MA, 2013. Identification and characterization of highly divergent simian foamy viruses in a wide range of New World primates from Brazil. PLoS One 8, e67568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz CP, Jia H, Shankar A, Troncoso LL, Augusto AM, Farias E, Pissinatti A, Fedullo LP, Santos AF, Soares MA, Switzer WM, 2015. An expanded search for simian foamy viruses (SFV) in Brazilian New World primates identifies novel SFV lineages and host age-related infections. Retrovirology 12, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SM, Picker LJ, Axthelm MK, Hudkins K, Alpers CE, Linial ML, 2008. Replication in a superficial epithelial cell niche explains the lack of pathogenicity of primate foamy virus infections. J. Virol 82, 5981–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar S, Bae EH, Khan AS, 2017. Complete genome sequence of a naturally occurring simian foamy virus isolate from rhesus macaque (SFVmmu_K3T). Genome Announc. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar S, Bae EH, Khan AS, 2018. Complete genome sequence of the African green monkey simian foamy virus serotype 3 strain FV2014. Genome Announc. 6 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco B, Finzi A, McGee-Estrada K, Sodroski J, 2010. Species-specific inhibition of foamy viruses from South American monkeys by New World Monkey TRIM5a proteins. J. Virol 84, 4095–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne R, Friedl E, Schweizer M, Fleps U, Turek R, Neumann-Haefelin D, 1992. Genomic organization and expression of simian foamy virus type 3 (SFV-3). Virology 186, 597–608. [DOI] [PubMed] [Google Scholar]
- Renshaw RW, Casey JW, 1994. Transcriptional mapping of the 3′ end of the bovine syncytial virus genome. J. Virol 68, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethwilm A, Bodem J, 2013. Evolution of foamy viruses: the most ancient of all retroviruses. Viruses 5, 2349–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethwilm A, Lindemann D, 2013. Foamy viruses. In: Knipe DM, Howley P (Eds.), 6 ed. Field's Virology 2. Lippincott, Willilams &Wilkins, Philadelphia, PA, pp. 1613–1632. [Google Scholar]
- Richard L, Rua R, Betsem E, Mouinga-Ondeme A, Kazanji M, Leroy E, Njouom R, Buseyne F, Afonso PV, Gessain A, 2015. Cocirculation of two env molecular variants, of possible recombinant origin, in gorilla and chimpanzee simian foamy virus strains from Central Africa. J. Virol 89, 12480–12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua R, Betsem E, Calattini S, Saïb A, Gessain A, 2012. Genetic characterization of simian foamy viruses infecting humans. J. Virol 86, 13350–13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruboyianes R, Worobey M, 2016. Foamy-like endogenous retroviruses are extensive and abundant in teleosts. Virus Evol. 2, vew032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustigian R, Johnston P, Reihart H, 1955. Infection of monkey kidney tissue cultures with virus-like agents. Proc. Soc. Exp. Biol. Med 88, 8–16. [DOI] [PubMed] [Google Scholar]
- Sakai K, Ami Y, Suzaki Y, Matano T, 2016. First complete genome sequence of a simian foamy virus isolate from a cynomolgus macaque. Genome Announc. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Lemey P, Schubert J, McClure MO, Rethwilm A, Bodem J, 2011. Complete nucleotide sequence and evolutionary analysis of a gorilla foamy virus. J. Gen. Virol 92, 582–586. [DOI] [PubMed] [Google Scholar]
- Schweizer M, Turek R, Hahn H, Schliephake A, Netzer K-O, Eder G, Reinhardt M, Rethwilm A, Neumann-Haefelin D, 1995. Markers of foamy virus infections in monkeys, apes, and accidentally infected humans: appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res. Hum. Retroviruses 11, 161–170. [DOI] [PubMed] [Google Scholar]
- Simmonds P, Adams MJ, Benko M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R, King AM, Koonin EV, Krupovic M, Kuhn JH, Lefkowitz EJ, Nibert ML, Orton R, Roossinck MJ, Sabanadzovic S, Sullivan MB, Suttle CA, Tesh RB, van der Vlugt RA, Varsani A, Zerbini FM, 2017. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol 15, 161–168. [DOI] [PubMed] [Google Scholar]
- Stenbak CR, Craig KL, Ivanov SB, Wang X, Soliven KC, Jackson DL, Gutierrez GA, Engel G, Jones-Engel L, Linial ML, 2014. New World simian foamy virus infections in vivo and in vitro. J. Virol 88, 982–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye JP, Blomberg J, Coffin JM, Fan H, Hahn BH, Neil J, Quackenbush S, Rethwilm A, Tristem M, 2012. Family Retroviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (Eds.), Virus Taxonomy—Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, U.K, pp. 477–494. [Google Scholar]
- Switzer WM, Bhullar V, Shanmugam V, Cong M-E, Parekh B, Lerche NW, Yee JL, Ely JJ, Boneva R, Chapman LE, Folks TM, Heneine W, 2004. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J. Virol 78, 2780–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer WM, Salemi M, Shanmugam V, Gao F, Cong M-E, Kuiken C, Bhullar V, Beer BE, Vallet D, Gautier-Hion A, Tooze Z, Villinger F, Holmes EC, Heneine W, 2005a. Ancient co-speciation of simian foamy viruses and primates. Nature 434, 376–380. [DOI] [PubMed] [Google Scholar]
- Switzer WM, Salemi M, Shanmugam V, Gao F, Cong ME, Kuiken C, Bhullar V, Beer BE, Vallet D, Gautier-Hion A, Tooze Z, Villinger F, Holmes EC, Heneine W, 2005b. Ancient co-speciation of simian foamy viruses and primates. Nature 434, 376–380. [DOI] [PubMed] [Google Scholar]
- Switzer WM, Tang S, Ahuka-Mundeke S, Shankar A, Hanson DL, Zheng H, Ayouba A, Wolfe ND, LeBreton M, Djoko CF, Tamoufe U, Esteban A, Heneine W, Peeters M, Wright LL, Muyembe-Tamfum JJ, Wemakoy EO, Mulembakani P, Hoff NA, Rimoin AW, 2012. Novel simian foamy virus infections from multiple monkey species in women from the Democratic Republic of Congo. Retrovirology 9, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumer L, Rethwilm A, Holmes EC, Bodem J, 2007. The complete nucleotide sequence of a New World simian foamy virus. Virology 369, 191–197. [DOI] [PubMed] [Google Scholar]
- Tobaly-Tapiero J, Bittoun P, Neves M, Guillemin M-C, Lecellier C-H, Puvion-Dutilleul F, Gicquel B, Zientara S, Giron M-L, de Thé H, Saïb A, 2000. Isolation and characterization of an equine foamy virus. J. Virol 74, 4064–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso LL, Muniz CP, Siqueira JD, Curty G, Schrago CG, Augusto A, Fedullo L , Soares MA, Santos AF, 2015. Characterization and comparative analysis of a simian foamy virus complete genome isolated from Brazilian capuchin monkeys. Virus Res. 208, 1–6. [DOI] [PubMed] [Google Scholar]
- Verschoor EJ, Langenhuijzen S, van den Engel S, Niphuis H, Warren KS, Heeney JL, 2003. Structural and evolutionary analysis of an orangutan foamy virus. J. Virol 77, 8584–8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TC, Bodem J, 2017. Sequence errors in foamy virus sequences in the GenBank database: resequencing of the prototypic foamy virus proviral plasmids. Arch. Virol 162, 1141–1144. [DOI] [PubMed] [Google Scholar]
- Winkler I, Bodem J, Haas L, Zemba M, Delius H, Flower R, Flügel RM, Löchelt M , 1997. Characterization of the genome of feline foamy virus and its proteins shows distinct features different from those of primate spumaviruses. J. Virol 71, 6727–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, Prosser AT, Torimiro JN, Wright A, Mpoudi-Ngole E, McCutchan FE, Birx DL, Folks TM, Burke DS, Heneine W, 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363, 932–937. [DOI] [PubMed] [Google Scholar]
- Wu Z, Ren X, Yang L, Hu Y, Yang J, He G, Zhang J, Dong J, Sun L, Du J, Liu L, Xue Y, Wang J, Yang F, Zhang S, Jin Q, 2012. Virome analysis for identification of novel Mammalian viruses in bat species from Chinese provinces. J. Virol 86, 10999–11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa R, Nakagawa S, Okamoto M, Miyazawa T, 2014. Construction of an infectious clone of simian foamy virus of Japanese macaque (SFVjm) and phylogenetic analyses of SFVjm isolates. Gene 548, 149–154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.