Abstract
Aim:
Valuable studies have tested the role of UCP1 on body temperature maintenance in mice, and we sought to knockout Ucp1 in rats (Ucp1−/−) to provide insight into thermogenic mechanisms in larger mammals.
Methods:
We used CRISPR/Cas9 technology to create Ucp1−/− rats. Body weight and adiposity were measured, and rats were subjected to indirect calorimetry. Rats were maintained at room temperature or exposed to 4°C for either 24 h or 14 days. Analyses of brown and white adipose tissue and skeletal muscle were conducted via histology, western blot comparison of oxidative phosphorylation proteins, and qPCR to compare mitochondrial DNA levels and mRNA expression profiles. RNA-seq was performed in skeletal muscle.
Results:
Ucp1−/− rats withstood 4°C for 14 days, but core temperature steadily declined. All rats lost body weight after 14 days at 4°C, but controls increased food intake more robustly than Ucp1−/− rats. Brown adipose tissue showed signs of decreased activity in Ucp1−/− rats, while mitochondrial lipid metabolism markers in white adipose tissue and skeletal muscle were increased. Ucp1−/− rats displayed more visible shivering and energy expenditure than controls at 4°C. Skeletal muscle transcriptomics showed more differences between genotypes at 23°C than at 4°C.
Conclusion:
Room temperature presented sufficient cold stress to rats lacking UCP1 to activate compensatory thermogenic mechanisms in skeletal muscle, which were only activated in control rats following exposure to 4°C. These results provide novel insight into thermogenic responses to UCP1 deficiency; and highlight Ucp1−/− rats as an attractive translational model for the study of thermogenesis.
Keywords: non-shivering, thermogenesis, brown adipose tissue, thermogenesis, UCP1
1 ∣. INTRODUCTION
Mammals respond to cold by increasing their heat production through both shivering and non-shivering thermogenesis (NST).1-3 Major uncoupling protein 1 (UCP1)-dependent centers of NST in adult humans are brown adipose tissue (BAT) and beige adipocytes induced in white adipose tissue (WAT).2 Although important in promoting heat production during cold exposure,6 UCP1 is expendable in mammals, as Ucp1 knockout mice (Ucp1−/−) can adapt to cold through activation of WAT oxidative capacity and alternative calcium cycling pathways in skeletal muscle.7,8 Similarly, adult humans have very little BAT, and thus rely more on skeletal muscle to generate heat, potentially independent of Ucp1.5,9
The above mentioned Ucp1−/− mouse model has proven invaluable as a tool for the study of thermogenesis.10,11 However, heat loss decreases proportionally with increased body surface area,12 making knockout models in larger mammals an attractive tool for providing additional translational application to humans. Mice and rats share a similar thermoneutral temperature (~29°C)13,14 and both expend considerable energy on thermogenesis when housed at 23°C15,16. However, mice are likely to rely more heavily on UCP1-mediated thermogenesis in BAT in response to cold than rats. In addition, due to their larger size relative to mice, as well as the high number of parallels that exist between rats and humans,17,18 we predicted Ucp1−/− rats would be an attractive translational model to provide insight into human thermogenic processes.
We used CRISPR/Cas technology to create Ucp1−/− rats on a Sprague Dawley background with the purpose of testing thermogenic adaptations in BAT, WAT, and skeletal muscle. A similar model was recently reported in a study assessing the role of UCP1 in myocardial ischemia.19 Herein we sought to characterize several aspects of Ucp1−/− rats with and without exposure to cold. While Ucp1−/− rats exhibited cold sensitivity, they appeared to rely heavily on compensatory thermogenic adaptations in inguinal white adipose tissue (iWAT) and skeletal muscle, and thereby maintained body temperature longer than mice.11,20 This phenotype provides insights beyond those from Ucp1−/− mouse models, making the Ucp1−/− rat an attractive translational model of thermoregulatory adaptations.
2 ∣. RESULTS
2.1 ∣. Phenotypic characterization of CRISPR Ucp1 knockout rats at 23°C
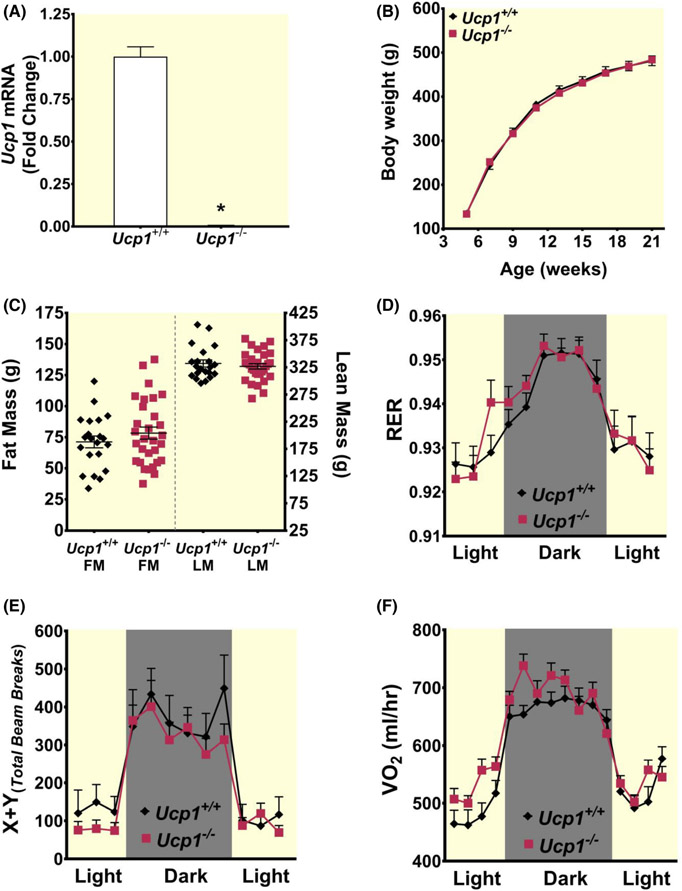
As described in Methods, Ucp1−/− rats were generated utilizing CRISPR/Cas9 technology to target exon 2 of the Ucp1 gene of Sprague Dawley rats (Figure S1A). After initial screening, three founders were chosen and deletion mutations in exon 2 in the Ucp1 gene were confirmed by DNA sequencing (Figure S1B). A 99% reduction in Ucp1 mRNA was observed in interscapular BAT of Ucp1−/− rats from all three lines compared to control rats (Figure 1A). All three founders produced viable offspring at 23°C and pups were born with the expected Mendelian inheritance ratio. Male offspring from these three founders were used in all subsequent studies.
FIGURE 1.
Few differences are immediately observed between controls and rats lacking Ucp1 at 23°C. (A) Reverse transcriptase PCR results showing ablation of Ucp1 mRNA expression in BAT from control and Ucp1−/− rats (n = 6 control; n = 9 Ucp1−/−). (B) Body weight is shown for control (black diamonds) and Ucp1−/− (red squares) rats (n = 21–30 per group). (C) 21-week fat mass (FM) and lean mass (LM) are shown for control (black diamonds) and Ucp1−/− (red squares) rats (n = 21–30 per group). (D–F) Respiratory exchange ratio (RER), total locomotor activity (X + Y total beam breaks), and volume of oxygen consumed (VO2) are plotted for control (black diamonds) and Ucp1−/− (red squares) rats (n = 7–9 per group). All data are presented as mean ± SEM, *p ≤ 0.05.
Ucp1−/− and control rats were fed a standard chow diet with 13% kcal from fat and were housed at 23°C during the 21 weeks of the study. No difference in body weight was observed between Ucp1−/− and control rats throughout the study (Figure 1B). Fat mass and lean mass at study completion were also not different (Figure 1C).
Indirect calorimetry measurements revealed few differences between Ucp1−/− males and control animals housed at 23°C in terms of respiratory exchange ratio (RER) and locomotor activity during both light and dark cycles (Figure 1D,E). Although Ucp1−/− rats show slightly elevated energy expenditure (EE) relative to controls at several time points (Figure 1F), these differences did not reach statistical significance. Collectively, these results indicate that Ucp1−/− rats did not exhibit a metabolic phenotype immediately distinct from controls when housed at room temperature.
2.2 ∣. Thermogenic response of Ucp1−/− rats to acute (24-h) and chronic (14-day) cold exposure
Ucp1−/− mice display relatively similar body composition measurements to control mice when housed at 23°C but fail to maintain core body temperature above 28°C for more than 10 h of 4°C exposure.6 We predicted that rats would not be as dependent on UCP1-mediated NST as mice. To test this prediction, two separate weight-matched cohorts of rats were placed in either an acute (24 h) or chronic (14 days) cold challenge.
Body and tissue masses from control and Ucp1−/− rats at each temperature condition are shown in Table 1. In addition to the significantly lower interscapular BAT mass observed in Ucp1−/− rats at 23°C when compared to controls, we also observed significantly lower gastrocnemius mass and significantly higher liver mass in these animals (Table 1). When rats were exposed to 24 h of cold, both genotypes experienced small, non-significant decreases in body weight along with significant decreases in interscapular BAT weight (Table 1). Interestingly, a significant (~20%) decrease in liver weight that was unique to Ucp1−/− males was observed after 24-h cold exposure; this trend persisted following the 14-day cold exposure period (Table 1).
TABLE 1.
Weight in grams (g) for body and collected tissues of control (Ucp1+/+) and Ucp1 knockout (Ucp1−/−) rats used in this study
| 23°C | 24 h at 4°C | 14 days at 4°C | ||||
|---|---|---|---|---|---|---|
| Ucp1+/+ | Ucp1−/− | Ucp1+/+ | Ucp1−/− | Ucp1+/+ | Ucp1−/− | |
| Body weight | 492.05 ± 52.45 | 502.83 ± 41.13 | 482.07 ± 42.97 | 421.014 ± 37.87 | 419.5 ± 39.27bc | 419.50 ± 39.27bc |
| Adipose tissue depots | ||||||
| BAT | 0.41 ± 0.12 | 0.28 ± 0.08a | 0.22 ± 0.10b | 0.16 ± 0.06b | 0.35 ± 0.07c | 0.23 ± 0.06a |
| eWAT | 4.57 ± 1.63 | 3.70 ± 0.64 | 5.33 ± 1.57 | 4.83 ± 1.42 | 2.24 ± 0.89c | 2.75 ± 1.06c |
| iWAT | 9.62 ± 3.86 | 10.07 ± 1.58 | 7.91 ± 1.33 | 8.78 ± 4.4 | 3.06 ± 0.77bc | 3.98 ± 1.66bc |
| rWAT | 2.52 ± 1.23 | 2.22 ± 0.59 | 3.63 ± 0.97 | 3.07 ± 0.91 | 0.78 ± 0.49bc | 0.86 ± 0.42bc |
| Skeletal muscle | ||||||
| EDL | 0.44 ± 0.16 | 0.44 ± 0.14 | 0.36 ± 0.06 | 0.37 ± 0.03 | 0.27 ± 0.07b | 0.29 ± 0.08b |
| Gastroc | 3.09 ± 0.39 | 2.65 ± 0.33 | 2.80 ± 0.353 | 2.84 ± 0.24 | 2.22 ± 0.3bc | 2.36 ± 0.27c |
| Soleus | 0.50 ± 0.13 | 0.44 ± 0.09 | 0.43 ± 0.06 | 0.47 ± 0.04 | 0.36 ± 0.04b | 0.37 ± 0.03b |
| Liver and heart | ||||||
| Heart | 1.59 ± 0.29 | 1.62 ± 0.19 | 1.64 ± 0.17 | 1.52 ± 0.13 | 1.53 ± 0.13 | 1.43 ± 0.16 |
| Liver | 18.02 ± 1.77 | 22.06 ± 1.83a | 16.95 ± 2.85 | 17.72 ± 2.92b | 18.02 ± 3.87 | 17.85 ± 2.77b |
Note: Rat tissues were either harvested without cold exposure (23°C), or following exposure to 4°C for 24 h or 14 days. Body weights were taken for all rats prior to cold exposure, and the mean 21-week body weight of all animals (n = 21 for Ucp1+/+ and 30 for Ucp1−/−) is reported. One third of each group (n = 7 for Ucp1+/+ and 10 for Ucp1−/−) was then exposed to one of the two cold challenges before tissue collection. As body temperature was recorded on day 1 of the 14-day treatment, these body weights are also included in the 24-h treatment group such that n = 14 for Ucp1+/+ and 20 for Ucp1−/− body weights at the 24-h time point. “a” indicates a statistically significant difference between genotypes within the same treatment. “b” indicates a statistically significant difference between the respective cold-challenged group and the 23°C group. “c” indicates a statistically significant difference between the 24-h cold-challenged group and the 14-day cold-challenged group. Significance is set to p ≤ 0.05.
Abbreviations: BAT, brown adipose tissue; eWAT, epididymal white adipose tissue; iWAT, inguinal white adipose tissue; rWAT, retroperitoneal white adipose tissue.
We assayed liver triglycerides to test the hypothesis that Ucp1−/− rats may be mobilizing this energy source following cold exposure. Unlike control animals, triglyceride content in Ucp1−/− rats was significantly diminished after 14 days of cold exposure relative to Ucp1−/− rats at room temperature (Figure S2). The higher liver weight in Ucp1−/− rats at room temperature cannot be entirely explained by differences in triglyceride content, as no differences were found in liver triglyceride levels between Ucp1−/− and control rats. Chronic cold exposure for 14 days induced significant weight loss in rats of both genotypes, although final body weights did not differ between the genotypes. Also similar between genotypes was an apparent recovery of interscapular BAT mass during chronic cold exposure (Table 1). Both Ucp1−/− and control animals also showed statistically significant decreases in masses of all WAT depots and skeletal muscle tissues after 14 days of cold exposure when compared to rats of the same genotype housed at 23°C or 4°C for 24 h (Table 1).
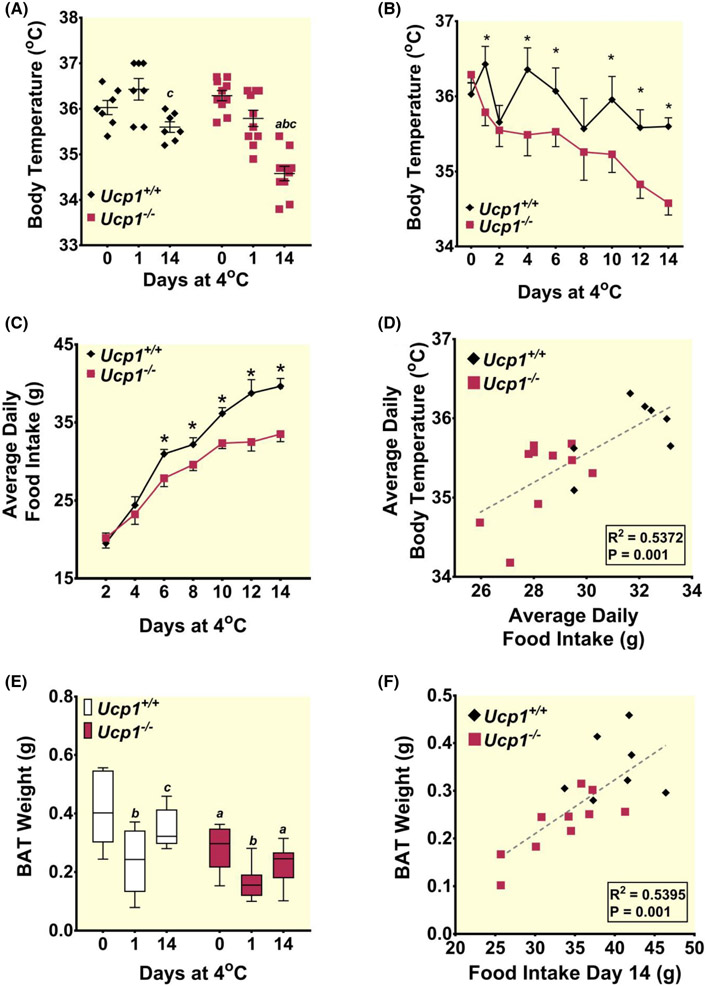
Ucp1−/− rats began to decrease body temperature upon exposure to cold (Figure 2A). Control rats maintained body temperature following the 24-h cold exposure, and this persisted throughout a 14-days chronic challenge (Figure 2B). Conversely, Ucp1−/− rats showed a steady decrease in body temperature throughout the study and had a final temperature averaging 1.71°C below baseline (Figure 2B). Importantly, while studies using Ucp1−/− mice report removal of animals within a few hours after acute cold exposure due to excess drops in core temperature,6 no rats of either genotype reached our critical removal temperature of 32°C over the 14-days cold exposure period.
FIGURE 2.
Ucp1−/− rats cannot maintain body temperature at control levels during a 14-day exposure to 4°C. (A) Body temperature is shown throughout the course of a 14-day 4°C cold exposure for control (black diamonds) and Ucp1−/− (red squares) rats. (B) Body temperature differences are shown at 0-, 1-, and 14-day time points of 4°C exposure for control (black diamonds) and Ucp1−/− (red squares) rats. (C) Average daily food intake is shown throughout the course of a 14-day 4°C cold exposure for control (black diamonds) and Ucp1−/− (red squares) rats. (D) Linear regression analysis is shown for average daily food intake versus average daily body temperature for all rats. (E) Changes in BAT weight are shown at 0-, 1-, and 14-day time points of 4°C exposure for control (black diamonds) and Ucp1−/− (red squares) rats. (F) Linear regression analysis is shown for food intake during the final day of the 14-day 4°C cold exposure versus harvested BAT weight for all rats. n = 7–10 for all experiments. Asterisks indicate statistical significance with p ≤ 0.05. “a” indicates a statistically significant difference between genotypes within the same treatment. “b” indicates a statistically significant difference between the respective cold-challenged group and the 23°C group. “c” indicates a statistically significant difference between the 24-h cold-challenged group and the 14-day cold-challenged group. All data are presented as mean ± SEM, *p ≤ 0.05.
Prolonged cold exposure correlates with increased caloric intake, and this increase supports BAT expansion.21 Food intake began to increase in both Ucp1−/− and control rats within a few days following the initiation of a cold challenge (Figure 2C). However, control rats increased their food intake more rapidly, and begin to eat significantly more than Ucp1−/− rats by day 6 of 4°C exposure. There was a direct correlation between average daily food intake and average body temperature throughout the study for all rats (Figure 2D). By the 14-day endpoint of the study, control rats increased their food intake by 103% from prior to cold exposure, whereas Ucp1−/− rats only increased food intake by 66%.
Before cold exposure, interscapular BAT was initially 32% smaller in Ucp1−/− rats than in controls (Figure 2E). Both Ucp1−/− and control rats lost interscapular BAT mass following 24-h cold exposure, with interscapular BAT weight in Ucp1−/− rats remaining significantly lower after the acute challenge. Interscapular BAT mass significantly increases in controls after 14 days of cold exposure relative to acute cold exposure; and food intake at day 14 of cold exposure shows a direct correlation with harvested interscapular BAT weight (Figure 2F). This result is consistent with hyperphagia-driven BAT expansion during prolonged cold exposure.21 Regardless of the experimental conditions, interscapular BAT mass for Ucp1−/− rats remained significantly lower than that of control, and no statistically significant increase in interscapular BAT was present for Ucp1−/− rats after 14 days of cold exposure relative to acute cold exposure.
The differences found in food intake and BAT expansion could relate to the amount of energy necessary to drive BAT thermogenesis and prompted further investigations into adaptive responses within Ucp1−/− rats. Two hormones that are secreted from BAT in response to cold challenge in mice are FGF21 and GDF15, both of which affect food intake and EE.22-24 We found no changes in the serum levels of these batokines, either between control and Ucp1−/− rats or between treatment groups (Figure S3).
2.3 ∣. White adipose tissue and skeletal muscle compensate for the loss of brown adipose tissue thermogenic capability during cold challenge
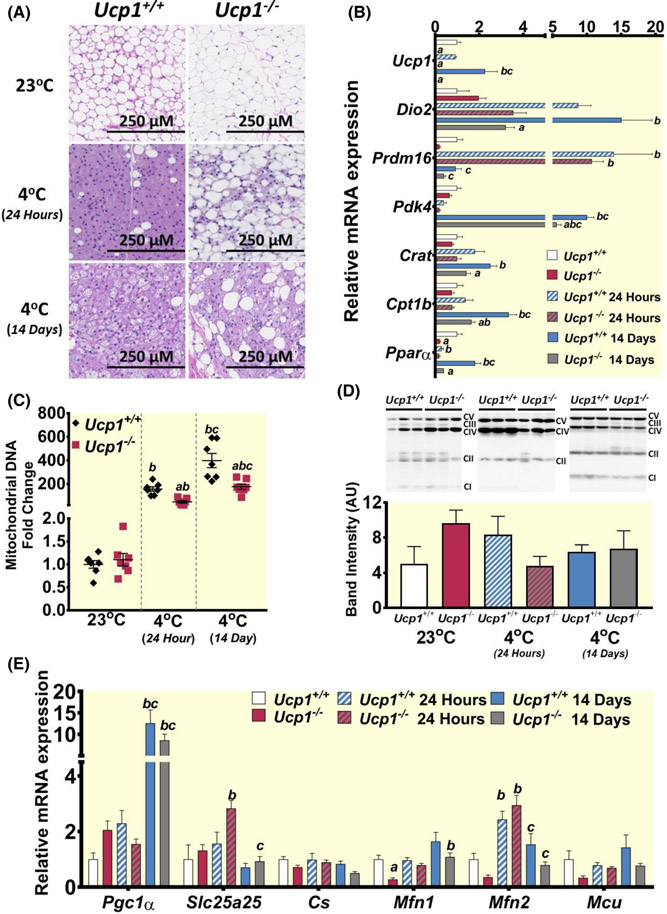
While the cold-sensitivity phenotype is much more pronounced in mice,6 our findings above showing that Ucp1−/− rats did not maintain body temperature as well as controls during cold exposure suggesting an insufficiency in activating thermogenic mechanisms. We therefore characterized the three major centers of mammalian thermogenic regulation: BAT, iWAT, and skeletal muscle.1-3,10,25 Microscopically, H&E-stained BAT from control rats at 23°C contained the multilocular adipocytes typically observed in BAT, along with unilocular adipocytes (Figure 3A, top left). Multilocular adipocytes were not apparent in Ucp1−/− rats (Figure 3A, top right). After acute cold exposure, BAT from control rats was almost entirely devoid of lipid (Figure 3A, center left), while very little lipid was mobilized from the BAT of Ucp1−/− rats (Figure 3A, center right). After 14 days of cold exposure, multilocular adipocytes are prominent in control BAT (Figure 3A, bottom left), with fewer of these cells observed in Ucp1−/− BAT (Figure 3A, bottom right). These results suggested an impairment of lipid mobilization from BAT in Ucp1−/− rats when cold-challenged.
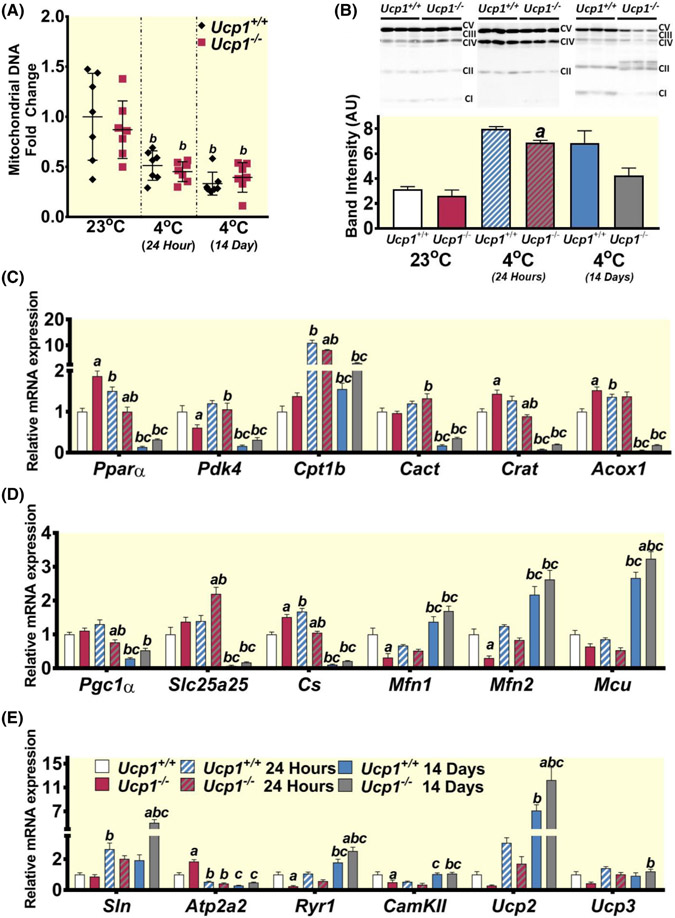
FIGURE 3.
Analysis of interscapular brown adipose tissue (BAT) shows decreased activation during cold exposure in rats lacking Ucp1. (A) H&E staining of BAT sections from control and Ucp1−/− rats at each temperature time point. (B) Gene expression of various genes involved with lipid oxidation and BAT browning. (C) Fold change relative to controls at room temperature is plotted for the ratio of mitochondrial NADH-5 DNA to genomic β-actin DNA (controls-black diamonds; Ucp1−/− – red squares). (D) Western blots and quantification are shown for all components of the mitochondrial respiratory chain. Each lane was quantified with all bands together and standardized to ponceau S. individual band quantification is further shown in Figure S4. CI-CV abbreviations represent Complex I through Complex V. (E) Gene expression of various genes involved with mitochondrial function. For gene and protein plots: White bars – controls at 23°C, red bars – Ucp1−/− at 23°C, blue and white striped bars – controls at 4°C for 24 h, red and gray striped bars – Ucp1−/− at 4°C for 24 h, blue bars – controls at 4°C for 14 days, gray bars Ucp1−/− at 4°C for 14 days. n = 6–9 for all expression experiments, n = 3 for western blots. “a” indicates a statistically significant difference between genotypes within the same treatment. “b” indicates a statistically significant difference between the respective cold-challenged group and the 23°C group. “c” indicates a statistically significant difference between the 24-h cold-challenged group and the 14-day cold-challenged group. Significance is set at p < 0.05. All data are presented as mean ± SEM.
In line with the above morphological observations, we also found that activation of the browning-associated gene Dio226 was significantly decreased in BAT from Ucp1−/− rats relative to controls after 14 days of exposure (Figure 3B). Lipid oxidation is known to extensively fuel NST,27,28 and several genes involved in lipid oxidation such as Pdk4, Cact, Crat, and Cpt1b were also less abundant in Ucp1−/− BAT than in control BAT during prolonged cold exposure (Figure 3B). Additionally, Pparα was induced after 14 days of cold exposure in control rats, while it was unchanged in Ucp1−/− rats, further supporting a decrease in the ability of Ucp1−/− rats to activate lipid oxidation to promote thermogenesis following cold exposure (Figure 3B).
To facilitate aerobic metabolism, BAT mitochondrial content increases during cold exposure through activation of Pgc1α.29 We determined levels of mitochondrial DNA as a measure of mitochondrial copy number in all conditions tested. Both Ucp1−/− and control rats show a marked increase in the content of mitochondrial DNA following acute cold exposure, which is maintained through the 14-day cold challenge (Figure 3C). However, Ucp1−/− BAT content of mitochondrial DNA remains significantly lower than that of control animals during each cold exposure treatment. Although no statistically significant differences were found, Complex IV in the mitochondrial respiratory chain trended toward showing higher protein levels in Ucp1−/− BAT at room temperature (Figure 3D and Figure S4). The marker of mitochondrial biogenesis, Pgc1α showed a similar but non-significant increase at room temperature30,31 (Figure 3E). Little difference between groups was noted for other markers of mitochondrial activity and thermogenic activity such as citrate synthase (Cs), mitochondrial fusion proteins 1 and 2 (Mfn1 and Mfn2), and mitochondrial calcium uniporter (Mcu).32 These results suggest that while Ucp1−/− BAT is responsive to exposure to 4°C, activation of BAT metabolic processing and subsequent morphological changes is less robust in Ucp1−/− rats than controls.
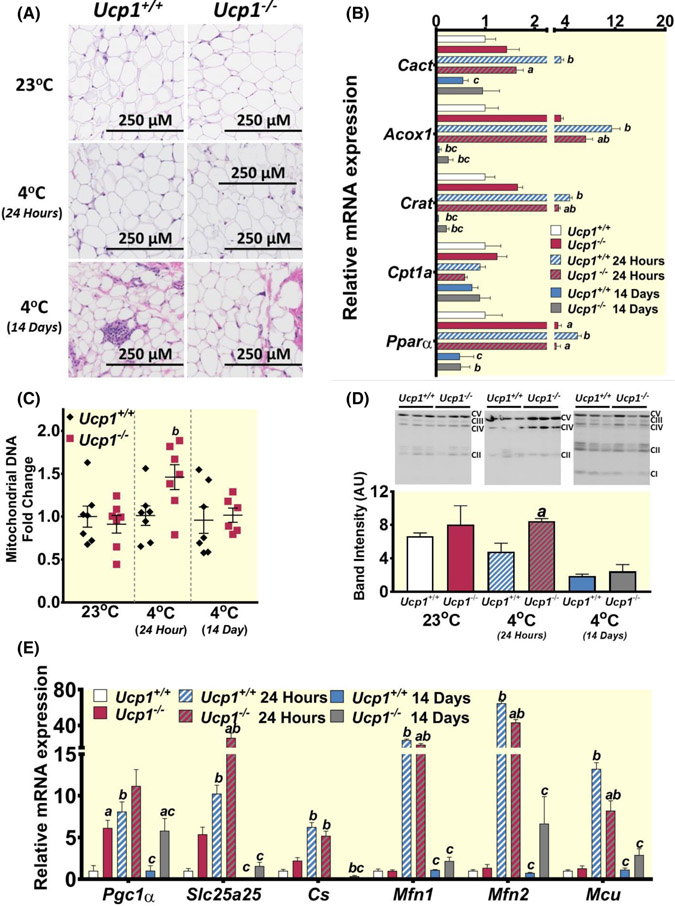
White adipose tissue depot weight significantly decreased in both control and Ucp1−/− rats following 14 days at 4°C (Table 1), and no morphological differences in iWAT were noted between the genotypes within the 23°C or 24-h 4°C treatment conditions (Figure 4A). However, following 14 days of 4°C exposure, adipocytes within control animals appeared slightly smaller on average than in Ucp1−/− rats. By assaying a similar set of genes in iWAT as those tested in BAT, we found that the lipid oxidation regulator Pparα was significantly increased in Ucp1−/− iWAT relative to controls at room temperature, and genes under its control such as Crat and Acox1 trend toward an increase, although not significantly (Figure 4B). In contrast to BAT, Pparα and its responsive are increased after 24 h of cold exposure. However, they are more robustly activated in control animals than in Ucp1−/−. Interestingly, following 14 days of cold exposure these genes returned to or below baseline levels in both genotypes. Despite higher expression of Pgc1α in Ucp1−/− iWAT at 23°C (Figure 4E), no greater mitochondrial content was seen as measured by mtDNA (Figure 4C) and respiratory chain proteins (Figure 4D). Interestingly, both contents of mitochondrial DNA and levels of various respiratory chain proteins were higher in Ucp1−/− iWAT after 24 h of cold exposure relative to room temperature (Figure 4C,D and Figure S4). However, these levels became indistinguishable from controls after 14 days at 4°C. In line with these results, markers of mitochondrial thermogenic activity such as Pgc1α, Slc25a5, Cs, Mfn1, Mfn2, and Mcu were generally activated in both genotypes after 24 h of cold exposure but returned to room temperature levels after 14 days (Figure 4E).
FIGURE 4.
Analysis of iWAT shows that UCP1-independent thermogenesis may be more active during cold exposure in the absence of Ucp1. (A) H&E staining of iWAT sections from control and Ucp1−/− rats at each temperature time point. (B) Gene expression of various genes involved with lipid oxidation. (C) Fold change relative to controls at room temperature is plotted for the ratio of mitochondrial NADH-5 DNA to genomic β-actin DNA (controls-black diamonds; Ucp1−/− – red squares). (D) Western blots and quantification are shown for all components of the mitochondrial respiratory chain. (E) Gene expression of various genes involved with mitochondrial function. For gene and protein plots: White bars – controls at 23°C, red bars – Ucp1−/− at 23°C, blue and white striped bars – controls at 4°C for 24 h, red and gray striped bars – Ucp1−/− at 4°C for 24 h, blue bars – controls at 4°C for 14 days, gray bars Ucp1−/− at 4°C for 14 days. n = 6–9 for all expression experiments, n = 3 for western blots. “a” indicates a statistically significant difference between genotypes within the same treatment. “b” indicates a statistically significant difference between the respective cold-challenged group and the 23°C group. “c” indicates a statistically significant difference between the 24-h cold-challenged group and the 14-days cold-challenged group. Significance is set at p < 0.05. All data are presented as mean ± SEM.
Both Ucp1−/− and control animals showed decreased skeletal muscle tissue mass after 14 days of cold exposure when compared to tissues from rats at earlier exposure time points; however, there was no difference between genotypes (Table 1). In gastrocnemius muscle, mitochondrial DNA levels were slightly, but significantly, lower for both genotypes following both acute and chronic cold exposure (Figure 5A). No differences were noted between genotypes in mitochondrial content (Figure 5A) or levels of respiratory chain components (Figure 5B) at room temperature. However, after being exposed to a cold environment Ucp1−/− rats had significant reductions in some components of the respiratory chain in skeletal muscle, especially at complexes I and V (Figure 5B and Figure S4). Decreased respiratory chain complexes after 14 days of cold exposure coincided with lower core body temperature in these rats (Figure 2B). It is intriguing that in several of our blots, complex II migrates as two separate bands. This appears strongly in the 14-day cold exposed Ucp1−/− muscle (Figure 5B), and different migration patterns of such complexes have been shown to be dependent on the presence of iron–sulfur clusters.33 The presence of multiple bands may thus reflect a higher percentage of complex II with decreased electron transport function due to lack of an iron–sulfur cluster, but we have not confirmed this in the present study.
FIGURE 5.
Analysis of skeletal muscle predicts higher markers of lipid oxidation and calcium cycling following cold exposure in Ucp1−/− rats. (A) Fold-change relative to controls at room temperature is plotted for the ratio of mitochondrial NADH-5 DNA to genomic β-actin DNA (controls-black diamonds; Ucp1−/− – red squares). (B) Western blots and quantification are shown for all components of the mitochondrial respiratory chain. Gene expression of various genes involved with mitochondrial function (C), lipid oxidation (D), and muscle calcium cycling/thermogenesis (E) are shown. For gene and protein plots: White bars – controls at 23°C, red bars – Ucp1−/− at 23°C, blue and white striped bars – controls at 4°C for 24 h, red and gray striped bars – Ucp1−/− at 4°C for 24 h, blue bars – controls at 4°C for 14 days, gray bars Ucp1−/− at 4°C for 14 days. n = 6–9 for all expression experiments, n = 3 for western blots. “a” indicates a statistically significant difference between genotypes within the same treatment. “b” indicates a statistically significant difference between the respective cold-challenged group and the 23°C group. “c” indicates a statistically significant difference between the 24-h cold-challenged group and the 14-days cold-challenged group. Significance is set at p < 0.05. All data are presented as mean ± SEM.
Markers of fatty acid oxidation and mitochondrial activity such as Pparα, Crat, Acox1, and cs showed significant increases in Ucp1−/− skeletal muscle relative to controls at room temperature (Figure 5C,D). Finally, several mitochondrial and thermogenic markers within the skeletal muscle were expressed at or below control levels in Ucp1−/− rats at room temperature. Following exposure to cold these markers were then expressed at control levels or higher (Figure 5D,E). Such mitochondrial markers included Slc25a25, Mfn1, Mfn2, and Mcu. The thermogenic markers which showed this trend encode such proteins as sarcolipin (Sln), SERCA2 (Atp2a2), ryanodine receptor 1 (Ryr1), Ca2+/calmodulin-dependent protein kinase II (Camkii), and uncoupling proteins 2 and 3 (Ucp2 and Ucp3); and are involved with calcium signaling and thermogenesis.8,34 Collectively, the results from these three tissues show that deficiencies in driving sufficient thermogenic adaptations in BAT of Ucp1−/− rats were at least partially compensated for by activating metabolic processes in iWAT and skeletal muscle. However, this response was not sufficient to fully maintain body temperature during prolonged cold exposure.
2.4 ∣. Shivering assessment and transcriptomics analysis in skeletal muscle predicts activation of thermogenesis pathways in Ucp1−/− rat muscle, even at 23°C
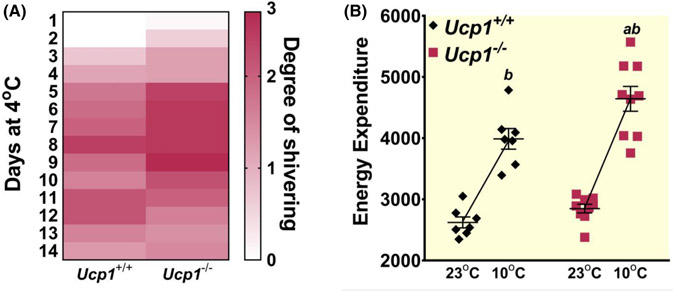
Given our evidence that skeletal muscle heat generative processes may have been compensating for the loss of Ucp1 in rats during cold exposure, we assessed indicators of bodily shivering using an adaptation of the Bedside Shivering Assessment Scale (BSAS) during the 14-day cold exposure period35; and by measuring resting energy expenditure (REE) during a mild cold challenge of 10°C for 5 h. Throughout the 14-day cold exposure study, we assigned higher BSAS scores to Ucp1−/− rats relative to controls at several time points due to visibly higher presence of body tremors (Figure 6A), which indicates increased shivering.37 Increases in REE are also commonly used as an indicator of bodily shivering,36 and both Ucp1−/− and control rats showed significant increases in REE during mild cold challenge (Figure 6B). Ucp1−/− rats showed a statistically more robust increase in REE during cold challenge relative to controls, further supporting an increased reliance upon shivering for heat generation.
FIGURE 6.
Ucp1−/− rats show indicators of increased shivering during cold exposure. (A) A modified bedside shivering assessment scale (BSAS)36 was used to visually quantify shivering of rats during a 14-day 4°C exposure from 0 (no shivering) to 3 (severe shivering). The average number for rats within each genotype was used to generate the heatmap. (B) Average energy expenditure is plotted for control (black diamonds) and Ucp1−/− (red squares) rats at room temperature for 5 h before and then at 10°C for 5 h. “a” indicates a statistically significant difference between genotypes within the same treatment. “b” indicates a statistically significant difference between the 10°C group and the 23°C group. n = 7 Control; n = 9 Ucp1−/−. Significance is set at p < 0.05. All data are presented as mean ± SEM.
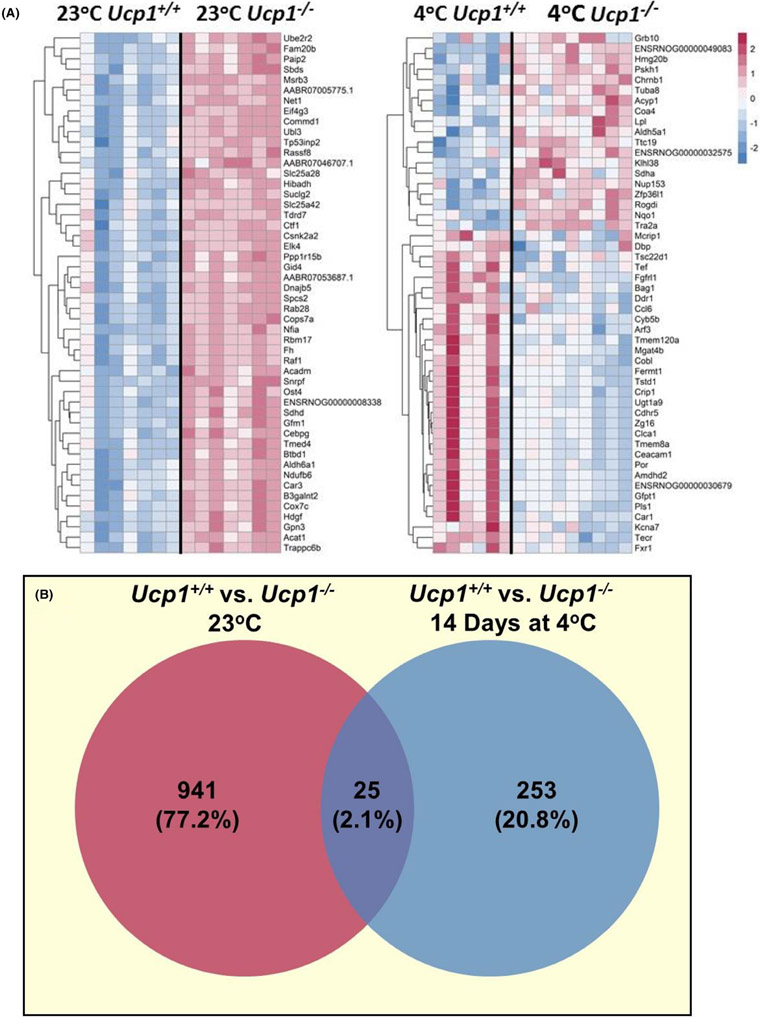
These observations, along with our findings indicating activation of thermogenic genes in Ucp1−/− rat gastrocnemius (Figure 5), led us to conduct a genome-wide transcriptomic analysis of gastrocnemius muscle from male control and Ucp1−/− rats housed at 23°C or 4°C for 14 days. Differentially expressed genes were sorted by adjusted p-value and the top 50 most significantly differentially regulated genes in Ucp1−/− rats compared to controls at 23°C are shown in Figure 7A and Table S2. Many of these genes encode for proteins involved in metabolic processing and mitochondrial function such as the coenzyme A solute carrier protein (Slc25a42), NADH/ubiquinone oxidoreductase subunit B6 (Ndufb6), 3-hydroxyisobutyrate dehydrogenase (Hibadh), succinate dehydrogenase subunit D (Sdhd), acetyl-CoA acetyltransferase 1 (Acat1), and cytochrome C oxidase subunit 7C (Cox7c). Interestingly, exposure to 4°C reduced the number of differentially expressed genes with a nominal p ≤ 0.01 from 941 at 23°C to 253 at 4°C (Figure 6B) and the top 50 differentially expressed genes at 4°C became a mixture of upregulated and downregulated genes in Ucp1−/− rats (Figure 7A and Table S3). None of these genes display an adjusted p-value below 0.1. Remarkably, these results suggest that cold exposure resulted in a more similar transcriptome between genotypes rather than driving greater differences.
FIGURE 7.
Transcriptomics analyses in skeletal muscle show more differences in gene expression between Ucp1−/− and control rats at 23°C than after 14 days at 4°C. (A) Genes were sorted by their adjusted p-values, and the top 50 genes were plotted for comparison between groups without cold exposure (left) and with 14 days of cold exposure (right). (B) Venn diagram showing the number of differentially expressed genes with p ≤ 0.01 between Control and Ucp1−/− rats without cold exposure (left) and with 14 days of cold exposure (right).
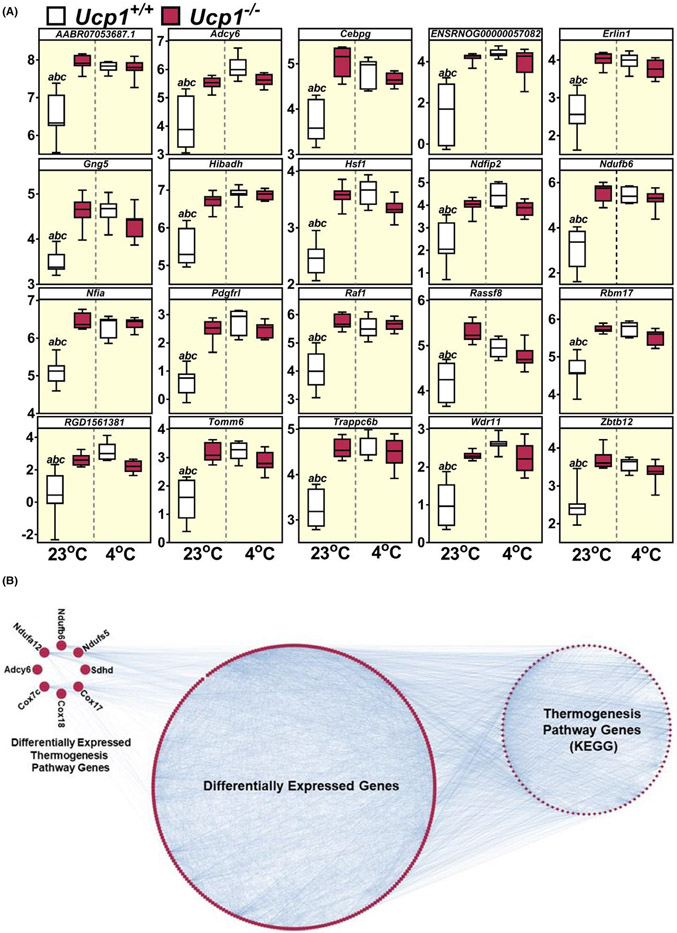
We further interrogated the top 20 genes displaying interaction effects, that is, with significant differential expression in only 1 out of the 4 tested conditions (control at 23°C vs. Ucp1−/− at 23°C vs. control at 4°C vs. Ucp1−/− at 4°C). These results showed that in all 20 genes, a significant upregulation of expression was seen in the Ucp1−/− group relative to controls only at room temperature. Two of the genes mentioned above as the most significantly differentially expressed between controls and Ucp1−/− rats were on this list (Hibadh and Ndufb6) and are involved in metabolic processing. Additionally, several mRNA-encoding proteins involved in thermogenic pathways including that of adenylate cyclase 6 (Adcy6), heat shock factor 1 (Hsf1), and nuclear factor IA (Nfia) were on this list. This is indicative of the activation of pathways consistent with exposure to cold in Ucp1−/− skeletal muscle at 23°C (Figure 8A). Additionally, Evinet analysis found many of the differentially expressed genes between control and Ucp1−/− rats at 23°C have known interactions with thermogenesis pathway genes, and that 8 of the differentially expressed genes are also members of the thermogenesis pathway (Figure 8B). Collectively, the transcriptomics analyses predicted that at 23°C skeletal muscle in Ucp1−/− rats was reprogrammed in a manner to activate thermoregulatory pathways, which are usually only activated in control rats following cold exposure. Therefore, these results suggest that even though Ucp1−/− rats maintain normal core temperature at 23°C (Figure 2A,B), they likely adapt to this mild cold stress by activating thermogenic mechanisms in skeletal muscle to compensate for the loss of Ucp1.
FIGURE 8.
Transcriptomics analyses predict activation of genes in Ucp1−/− rat gastrocnemius not activated in controls until cold exposure. (A) limma analysis showing the top 20 genes differentially expressed in only one out of the four conditions tested (control at 23°C; Ucp1−/− at 23°C; control at 4°C; Ucp1−/− at 4°C). Data are plotted as log2 of the detected signal. (B) Evinet analysis identifying the Kyoto Encyclopeida of Genes and Genomes (KEGG) thermogenesis pathway as being enriched for connectivities in analysis of Control versus Ucp1−/− rats without cold exposure. 8 genes shown at the top left are thermogenesis pathway genes, and many of the remaining differentially expressed genes show interactions with thermogenesis pathway genes. n = 7 rats per group.
3 ∣. DISCUSSION
Studies assessing the maintenance of body temperature have revealed that BAT, skeletal muscle, and WAT are important tissues in regulating thermogenic processes.1-3,10,25 However, the extent to which each of these tissues contributes to thermogenesis within mammalian systems remains unclear. It is well understood that exposure to cold can stimulate WAT lipolysis to provide fuel for thermogenic activity in BAT and skeletal muscle.11,25,27 BAT thermogenesis is known to rely heavily on futile lipid cycling for heat production via UCP1,6,38 and the removal of Ucp1 from mice results in compensatory heat production in both WAT and skeletal muscle.4,11 However, using only mice to study thermogenic adaptations as they translate to human physiology is limiting, as nearly 60% of the total EE of a mouse housed at room temperature (23°C) is due to BAT-mediated thermogenesis39 which far exceeds the energetic capacity of BAT in humans. Given the importance of the rat as a translational model,17 we used CRISPR-Cas technology to create Ucp1 knockout rats (Ucp1−/−) to study UCP1-independent thermogenic mechanisms.
Like male Ucp1−/− mice,6,11 Ucp1−/− rats do not exhibit notable differences from controls in core temperature, body weight, or food intake when housed at room temperature. Alternatively, the difference between species becomes more prominent as the temperature is decreased. Firstly, to survive a cold environment, Ucp1−/− mice must be acclimated to 4°C by a gradual decrease of ambient temperature.40 Unlike Ucp1−/− mice, Ucp1−/− rats tolerated a direct transition from 23 to 4°C and no rats approached hypothermia. Importantly, despite this proclivity of rats toward survival during cold exposure, Ucp1−/− rats do experience a gradual decrease in body temperature over 14 days when exposed to 4°C resulting in an average body temperature of 1–2°C below controls. Second, both Ucp1−/− mice11 and Ucp1−/− rats lose similar amounts of body weight as their respective controls after prolonged cold exposure; however, the dietary response to counter this negative energy balance differs between species. Specifically, increases in food intake in response to cold are similar between Ucp1−/− mice and controls,11 whereas control rats increase food intake twofold while Ucp1−/− rats only increase food intake by 66%. We did not see differences in the batokines known to regulate food intake and EE, FGF21 and GDF15 between controls and Ucp1−/− rats,22-24 and the mechanisms governing the decreased food intake of Ucp1−/− rats relative to controls during prolonged cold exposure are thus unclear. Increasing food intake as a mechanism of regulating body temperature during cold exposure is a known phenomenon in rats41; and given that we found a direct correlation between average body temperature and average daily food intake for the rats in our study, the mechanisms governing this decrease in food intake are important to investigate in future studies as a potential correlate of the inability of Ucp1−/− to maintain body temperature during prolonged cold exposure.
Another notable difference between species is that interscapular BAT mass is similar between Ucp1−/− mice and controls,6 whereas interscapular BAT mass is ~30% lower in Ucp1−/− rats than controls regardless of ambient temperature. Within the present study, a physiological and tissue-level analysis suggests that adaptive mechanisms in BAT are not activated in Ucp1−/− rats during cold exposure as potently as is seen in control rats. Firstly, BAT hypertrophy has been suggested to be driven by an increased food intake,21,42 and there was a direct correlation between daily food intake at day 14 of the cold exposure study and BAT mass of cold-exposed rats. The decreased food intake in Ucp1−/− rats relative to controls over a 14-day cold exposure may thus correlate with limited expansion of BAT mass. Collectively, results showing differences in food intake, BAT mass, and the ability to withstand cold between Ucp1−/− rats and Ucp1−/− mice suggest that (1) while rats do exhibit a need for Ucp1-mediated thermogenesis over prolonged cold exposure, their dependence on BAT to tolerate cold is far less pronounced than mice and (2) the differences between species in caloric intake in response to cold suggest that there may be unique energetic differences between the thermogenic responses that are invoked between mice and rats, which is an area of investigation that is worthy of pursuit.
Histological analysis of BAT shows a rapid depletion of lipid content in BAT of control rats after only 24 h, whereas the response was much more gradual and less robust in Ucp1−/− rats. Several additional molecular markers suggesting decreased BAT activation in Ucp1−/− rats were also observed. Specifically, mitochondrial biogenesis/content,29,43 fusion,32,44 lipid oxidation, and browning,26 are known to be upregulated in BAT during cold exposure.42,45 However, in the present study, cold-induced adaptations in these pathways were less robust in Ucp1−/− rats than in controls, suggesting a partial requirement for Ucp1 function in rats to facilitate BAT remodeling in response to a cold environment. It is important to note that several proteins within the electron transport chain have decreased expression in Ucp1−/− mice relative to controls following cold exposure.46 We find few differences between control and Ucp1−/− rats in BAT for ETC protein levels at any temperature treatment, which may reflect a greater UCP1 dependent remodeling of BAT in general for mice as compared to rats.
Although cold exposure activates most of these markers more robustly in controls than in Ucp1−/− rats, it is of note that when housed at 23°C several of these markers are significantly higher in BAT of Ucp1−/− rats relative to controls. Specifically, Ucp1−/− rats have greater expression of the mitochondrial biogenesis gene Pgc1α at room temperature. Ucp1−/− rats also display higher levels of the gene encoding the ATP-Mg2+/Pi shuttle, Slc25a25. This protein is upregulated in the absence of Ucp1 and acts as a calcium-sensitive shuttle to regulate body temperature.30 Overall, increases in these markers indicate that Ucp1−/− rats sense mild cold stress at 23°C and activate heat generating mechanisms since this is below their thermoneutral temperature of 29°C.13 A similar phenomenon was recently reported for Ucp1−/− mice housed at 20°C, which showed higher expression than controls of the metabolic markers Dgat2 and Complex V in the respiratory chain.38
As an adaptation to the inability to sufficiently activate BAT thermogenesis in the absence of Ucp1, rats in our study increase thermogenic markers in iWAT. We again observed various markers of lipid oxidation increased in Ucp1−/− rats relative to controls at 23°C, potentially reflecting the activation of thermogenic mechanisms in WAT to meet the higher energetic cost of UCP1-independent thermogenesis.30,47 Similar to our results in rats showing that markers of mitochondrial activity are higher in iWAT immediately following cold exposure, reports from Ucp1−/− mice show increased levels of Pgc1α and mitochondrial DNA relative to controls when adapted to a 4°C environment.11 That many of the markers of mitochondrial activation and lipid oxidation remain elevated in iWAT of Ucp1−/− rats relative to controls following 14 days of cold exposure is additional evidence that adaptations in WAT promote thermogenesis independent of Ucp1, which is consistent with previous reports.11,25,27
UCP1-independent thermogenesis, via both shivering and NST, has been described in skeletal muscle, which is thought to be a major center of heat production in humans.1,5,11,37,43,48,49 Muscle contraction during shivering correlates positively with increased EE, and inversely with decreasing temperature.37,50 In the present study, we observed several indications of thermogenic compensation in skeletal muscle following the loss of Ucp1 in rats, most notably in indicators of shivering. Visible tremors in Ucp1−/− rats were initially higher than in controls after exposure to cold, but this activity tapered off toward the end of the study as body temperatures dropped in Ucp1−/− rats. Furthermore, we observed a more robust increase in EE for Ucp1−/− rats following a mild 10°C cold stress relative to controls, which is an established indicator of increased shivering.36,37 Previous studies have shown that EE as calculated from oxygen consumption (VO2) is an important proxy for shivering, as it can reach 40% of VO2 max, while <1% of cold-induced increases in EE are due to BAT thermogenesis.36 This demonstrates the increased energetic cost of skeletal muscle shivering relative to BAT NST, and further advocates for the necessity to increase food intake if body temperatures are to be adequately maintained during cold exposure in the absence of Ucp1. Future studies in cold-exposed Ucp1−/− rats measuring EMG activity, which increases as ambient temperatures fall,37,51 are imperative to most efficiently quantify shivering in Ucp1−/− rats, as this may be an important mechanism affording rats the ability to persist at 4°C far longer than mice in the absence of Ucp1.
Related to the potential for NST in skeletal muscle, the ryanodine receptor (Ryr1) is an ion channel known to release Ca2+ from the endoplasmic reticulum,52 which can then be taken back up by SERCA (Atp2a2).53 This process is inhibited by sarcolipin (Sln) binding to SERCA,8 which can allow calcium ions to leak back through the ryanodine ion channel,31 thus creating a futile cycle of heat production fueled by dysregulation of the sarcoplasmic reticulum Ca2+ ion gradient. RNA levels of both Sln and Ryr1 are increased in Ucp1−/− rat muscle relative to controls after 14 days of cold exposure. Additionally, markers of similar futile cycles within mitochondria are increased, such as Ucp2 and Mcu.32,54 Ucp2 expression has been shown to increase in models lacking Ucp1,55 and calcium cycling via proteins such as MCU is thought to enhance thermogenic respiration in BAT.56 However, both these proteins are expendable for body temperature maintenance in mice,6,32 and our data suggest that muscle shivering may be a more important mechanism of heat generation in rat skeletal muscle than NST.
The impact of NST on skeletal muscle is still a subject of much debate within the thermogenesis field. A single contraction relaxation cycle has been estimated to hydrolyze up to 50-fold more ATP for the generation of heat than Ca2+ leak via the ryanodine ion channel.57 Secondly, muscle shivering is well-documented as an important mechanism of heat generation in cold-exposed rats, especially at ambient temperatures as low as −20°C, where BAT NST is not alone sufficient for temperature maintenance.58,59 Indeed, muscle contractile activity requires calcium cycling during shivering,60 and it, thus, becomes difficult to parse out whether increases in transcripts within the ryanodine receptor-SERCA Ca2+ cycling system reflect increased shivering or increased NST. Our data in Figure 6 suggest that Ucp1−/− rats may shiver earlier and more vigorously than controls during cold exposure. This increase is likely an important mechanism for allowing Ucp1−/− rats to survive longer than mice during cold exposure, as Ucp1−/− mice do not show increased shivering relative to controls at 6°C.34 Additionally, Ucp1−/− rats were unable to maintain body temperature at control levels after 14 days of 4°C exposure even with an mRNA increase in markers of muscle NST, providing more evidence of the insufficiency of this mechanism to produce enough heat to allow long term maintenance of body temperature.
Whether shivering or NST, mechanisms of heat generation in skeletal muscle are largely supported by mitochondrial lipid metabolism in rodents and humans.20,49 Skeletal muscle mitochondrial content is responsive to cold adaptation, and the directionality of the response depends on muscle fiber type.4,43 While slow-twitch, type I muscle fibers show decreased mitochondrial number during cold exposure in mice,43 type II, fast-twitch fibers harbor an increase in mitochondrial abundance driven by cold.4 In our study, EDL, which is composed primarily of type II fibers,61 shows a small and insignificant increase in the content of mitochondrial DNA in both genotypes immediately following cold exposure (Figure S5A). However, contrary to the characteristic decrease seen for mice in type I muscle, Ucp1−/− rats show an increase in mitochondrial copy number in type I fibers following 24-h cold exposure, as evidenced by our results from soleus muscle (Figure S5B).61 This again implicates the importance of muscle activity for body temperature maintenance in Ucp1−/− rats. Gastrocnemius muscle, which contains relatively equal amounts of type I and II fibers,61 showed a slight but significant decrease in mitochondrial content in both genotypes following cold exposure relative to levels at 23°C. Although markers of mitochondrial content (mtDNA and OxPHOS proteins) were inconsistent, the observed increase in Ucp1−/− muscle relative to controls in mRNA levels of Pgc1α and Cs, as well as increases in lipid oxidation markers, after 14 days of cold exposure are consistent with studies showing increased lipid-fueled aerobic metabolism in skeletal muscle in the absence of Ucp1.4,20 These data collectively suggest a role of muscle-derived thermogenesis in the attempted regulation of body temperature in Ucp1−/− rats. However, the degree to which this mechanism is activated as a consequence of muscle contractile activity remains unclear, thus making it difficult to determine whether it is tied to shivering or NST.57
Transcriptomics analyses from skeletal muscle revealed that more extensive differences exist between Ucp1−/− and control rats at 23°C than after 14 days of 4°C exposure. It is quite striking that many of the upregulated genes are related to mitochondrial function and thermogenesis. Specifically, various NADH dehydrogenase complex subunits were identified by our analyses as being upregulated in the skeletal muscle of Ucp1−/− rats at room temperature, and several of the mRNA markers of lipid oxidation tested via qPCR are also more abundant in Ucp1−/− animals relative to controls at room temperature. These results further support the assertion that Ucp1−/− rats may be oxidizing more lipid than controls for heat generation at 23°C.30,47 Such increases in markers of fatty acid oxidation and metabolic processing have been shown in hibernating rodent muscle, and may facilitate an increased shivering activity for the maintenance of body temperature.62 Furthermore, although we found that the average EE between Ucp1−/− rats and controls was not significantly different, a close examination of Figures 1F and 6B reveals that EE for Ucp1−/− rats tends to be higher than controls at room temperature. This phenomenon could reach statistical significance in future studies if sample sizes exceed 7–9, and may reveal increased oxidative metabolism in Ucp1−/− rats at 23°C. Many of the metabolic processing and thermogenic genes which we found upregulated at 23°C in Ucp1−/− gastrocnemius are not stimulated in control rats until after 14 days of 4°C exposure. Our results thus predict a modification to metabolic physiology in the absence of Ucp1 that results in compensatory mechanisms of heat generation in skeletal muscle to maintain body temperature at 23°C or below. These data may thus reflect increased reliance upon skeletal muscle and decreased reliance upon BAT for heat generation in rats relative to mice, which leads to the ability of these larger rodents to persist far longer during cold exposure in the absence of Ucp1.
4 ∣. CONCLUSION
Our results characterize for the first time the temperature sensitivity of rats lacking Ucp1. Although these rats are unable to activate BAT thermogenesis as potently as control animals in response to prolonged cold exposure, rats clearly are not as dependent as mice upon UCP1-driven thermogenesis because Ucp1−/− rats tolerate cold exposure much better than Ucp1−/− mice. Cold tolerance appears to be largely maintained in Ucp1−/− rats in part through upregulated thermogenic mechanisms in iWAT and skeletal muscle, and may be related to an increased reliance on shivering, muscle-based NST, or both. However, these mechanisms are insufficient for complete temperature maintenance during prolonged cold exposure. Overall, the results of this study support the notion that Ucp1−/− rats are a solid translational model to study thermogenic responses and further studies are necessary to more extensively characterize the relationship between UCP1-independent thermogenesis in rats and humans.
5 ∣. MATERIALS AND METHODS
5.1 ∣. Animals
Animal studies were conducted at Pennington Biomedical Research Center's American Association for the Accreditation of Laboratory Animal Care-approved facility. All rats were fed a standard chow diet, composed of 29% (wt/wt) protein, 13% (wt/wt) fat, and 58% (wt/wt) carbohydrate (Purina LabDiet no. 5001; Purina Mills) and were doubly housed at 23°C unless used for cold exposure studies as described below. Rats were 16 weeks of age at the time of sacrifice unless otherwise stated. All procedures were approved by the PBRC Institutional Animal Care and Use Committee.
5.2 ∣. Preparation of pX459 with sgRNA
CRISPR/Cas9 vector pX459 was purchased from addgene, and development of this plasmid has been described previously.63 As depicted in Figure S1A, the gRNA was targeted to the second exon of the Rattus norvegicus Ucp1 gene (5′–CAGGATTGGCCTCTACGATA–3′). The sgRNA/pX459 expression vector was constructed by ligating the annealed oligos (5′–CACCGCAGGATTGGCCTCTACGATA–3′, and 5′–AAACTATCGTAGAGGCCAATCCTGC–3′) into pX459 following BBSL digestion. The T7 promoter sequence was added to the 5′ end of the gRNA to prepare the final template using the primers (5′–TTAATACGACTCACTATAGGCAGGATTGGCCTCTACGATA–3′ and 5′–AAAAGCACCGACTCGGTGCC–3′). Following digestion with BBSL and PCR amplification, the product was transformed into Escherichia coli (DH5α) cells for amplification. Insertion of the Ucp1 gRNA coding sequence and the T7 promoter sequence was confirmed by sequencing.
5.3 ∣. Generation of Ucp1 knockout rats and genotyping
Following procedures previously described,64 the plasmid described above was microinjected into Sprague Dawley rat zygotes by the UNITECH Corporation. For validation of Ucp1 gene modification in offspring via genotyping, genomic DNA was extracted from tail tips of rat pups and PCR was run using primers (5′–CACCGCAGGATTGGCCTCTACGATA–3′, and 5′–AAACTATCGTAGAGGCCAATCCTGC–3′). Of the eight pups that were screened, three exhibited homozygous Ucp1 deletion (Figure 1B). These F0 generation rat pups were crossed with Sprague Dawley rats and germ-line transmission of Ucp1 knockout was confirmed by sequencing.
5.4 ∣. Animal procedures
Body composition was measured using a Bruker NMR Minispec (Bruker Corporation). Plasma extractions were performed following euthanasia from collected trunk blood. Indirect calorimetry was conducted in a 16-chamber indirect calorimetry chamber system (TSE Systems) as described previously.65 Briefly, rats were singly housed during metabolic chamber experiments and allowed ad libitum access to food and water. Measurements were taken every 30 min to assess VCO2 output, VO2 consumption, total activity via X and Y coordinate beam breaks, and food intake via hopper weight sensor. At day 5 of metabolic measurements, 5 h after the beginning of the light cycle, the temperature of the system was dropped to the minimum allowable temperature of 10°C for 5 h for measurement of EE during cold exposure. The Weir equation [(VO2 × 3.941)+(VCO2 × 1.11)] was used to calculate the average EE, and the average for the period of cold exposure was compared to the average for 5 h preceding cold exposure.
For cold exposure studies, rats were singly housed and placed acutely at 4°C for the indicated duration of time in the results section above. These animals were not allowed enrichment devices, which could be used to artificially maintain body temperature. Body temperature was monitored twice daily using a rectal digital thermometer during the light cycle only, and a critical body temperature of 32°C was established as the temperature at which rats were to be promptly removed from the study and euthanized. However, as discussed in the results above, this did not occur with any of the rats in our study. Tissue collection immediately followed the conclusion of cold exposure studies. All BAT used for these studies was from the interscapular region.
5.5 ∣. Quantitative RT-PCR
Total RNA was isolated from each tissue for later qRT-PCR using an RNeasy Mini Kit supplemented with DNase digestion (Qiagen). cDNA was then synthesized with an iScript cDNA synthesis kit and was used for qRT-PCR with the SYBR Green system (Bio-Rad). Analysis was conducted using the Norma-gene macro as previously described.66 Rat cyclophilin B transcript was included as an additional reference gene within the Norma-gene macro as described previously.66,68 Primer details are provided in Table S1.
5.6 ∣. RNA-sequencing (Seq)
Whole gastrocnemius muscle of male Ucp1−/− and control rats (N = 8 per group) were used for total RNA extraction as described in Section 2.5. RNA-Seq libraries were constructed using Illumina's TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero. RNA was sequenced on the Illumina NextSeq 500 using the High Output v2 Kit and paired-end sequencing forward and reverse reads (2 × 75 bp) with 75 million reads/sample. Gene-level aggregated raw counts were normalized via the TMM algorithm in edgeR69 and subsequent differential gene expression analysis was conducted via limma.70 Differentially expressed genes (absolute fold-change >1.5-fold, adjusted p < 0.05, and log average signal >2) were evaluated for topology-based biological pathway enrichment via the Evinet tool,71 using the Funcoup LE network as the background interaction network for enrichment analysis.72
5.7 ∣. Western blotting
Protein homogenates were prepared from gastrocnemius muscle, iWAT, and BAT tissue in Cell Lysis Buffer (EMD Millipore). Each sample (20 μg protein) was mixed with cell lysis buffer and Laemmli sample buffer to reach a final volume of 50 μl. Samples were loaded onto 15% polyacrylamide gels (one gel per tissue type) and subjected to electrophoresis. Proteins were then transferred to 0.2 μm polyvinylidene difluoride membranes and stained with Ponceau S to determine total protein content. After destaining, membranes were blocked with 4% non-fat dry milk for 1 h at room temperature. Membranes were quickly washed in TBST and then probed with total OXPHOS Rodent WB Cocktail (#ab110413, Abcam), at a dilution of 1:250 in 5% BSA-TBST, overnight at 4°C. After washing in TBST, membranes were incubated with anti-mouse secondary antibody (1:10 000 dilution) for 1 h at room temperature. Membranes were washed again in TBST, incubated with SuperSignal West Pico Plus Chemiluminescent Substrate (ThermoFisher) for detection, and imaged using a Bio-Rad ChemiDoc XRS+ Imaging System. Band intensities were quantified using Image J software and normalized to Ponceau S stain total protein intensity (Figure S4).
5.8 ∣. Histology
BAT and iWAT depots from rats were harvested, fixed in 10% neutral-buffered formalin, and embedded in paraffin. Samples were sectioned and stained with hematoxylin and eosin stain (H&E) by the Cell Biology and Bioimaging Core Facility at PBRC as previously described.73
5.9 ∣. Mitochondrial DNA quantification
DNA was isolated using a DNA Mini Kit (Qiagen) and used for qRT-PCR with the SYBR Green system (Bio-Rad). Analysis was conducted by determining the expression level via the formula 2−CT as described previously67 and the ratio of mitochondrial NADH-5 to genomic β-actin was used to determine mitochondrial abundance.
5.10 ∣. Serum protein and liver triglyceride quantification
ELISA kits were used for the measurement of FGF21 (Abcam, ab223589) and GDF-15 (R&D Systems, MGD150) from serum in the fed state. The manufacturer's instructions were closely followed and 50 μl of undiluted serum was used for each Kit. Liver triglycerides were also measured using a commercial kit (Abcam, ab65336). The manufacturer's instructions were again followed, and 50 μl of the sample was prepared from 100 mg of liver tissue diluted in 1 ml of 0.5% NP-40 in ddH2O.
5.11 ∣. Statistical analysis
Data are expressed as mean ± SEM. For single treatment comparisons between control and Ucp1−/− rats, Microsoft Excel software was to determine significant differences with paired two-tailed student's t tests where normality was established using GraphPad Prism software and the D'Agostino-Pearson normality test. For multiple comparisons between genotypes and treatment groups, GraphPad Prism software was used to determine significant differences with two-way anova using Tukey post-hoc analysis. For either test, p < 0.05 was considered significant. GraphPad Prism software was also used for linear regression analyses.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Tamra Mendoza, Olga Dubuisson, and Estrellita Bermudez for their assistance during animal tissue collections. J.D.W., C.M.E, R.C.N., R.L.M conceived and designed the research and wrote the manuscript; R.L.M., C.M.E., J.D.W., B.V., J.Z., D.S.B., P.O.N., A.C.S., and S.E.V. performed experiments; S.G., J.D.W., B.V., C.M.E., R.C.N., and R.L.M. analyzed data; and R.L.M., C.M.E., R.C.N, B.V., D.S.B., J.Z., A.C.S, S.E.V., S. G., and J.D.W edited the manuscript. J.D.W. was supported by NIDDK K01-DK-116194 and T32-DK-6458413. C.M.E. is supported by R03DK122121 from the National Institutes of Health. D.S.B. was supported by T32-AT-004094. R.C.N. was supported by NIH R01DK103860. R.L.M was supported by NIDDK R01-DK-089641. S.G. and R.L.M. were supported in part by a Louisiana Clinical and Translational Science Center (LaCATS) P&F grant funded by the National Institute of General Medical Sciences; NIGMS, United States GM-104940. This work used the PBRC Transgenic, Comparative Biology, and Genomics Cores; and the Clinical Research Laboratory supported in part by COBRE (NIGMS 8P20-GM-103528) and NIDDK NORC 2P30-DK-072476.
Funding information
National Center for Complementary and Integrative Health, Grant/Award Number: T32-AT-004094; National Institute of Diabetes and Digestive and Kidney Diseases, Grant/Award Number: 2P30-DK-072476, K01-DK-116194, R01-DK-089641, R01-DK-103860, R03-DK-122121 and T32-DK-6458413; National Institute of General Medical Sciences, Grant/Award Number: 8P20-GM-103528 and GM-104940; National Institutes of Health, Grant/Award Number: R01DK103860 and R03DK122121; Louisiana Clinical and Translational Science Center (LaCATS)
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DEDICATION
We dedicate this manuscript to the memory of the late Dr. Randall “Randy” L. Mynatt, the principal investigator of this study. The Ucp1−/− rat model discussed herein was the last he designed during his career as Professor and Transgenics Core Director at Pennington Biomedical Research Center. Randy and the many animal models he generated have established a lasting legacy of valuable insights into metabolic physiology, and his presence is truly missed.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. RNA-seq raw data have been submitted to GEO under accession numbers GSM5234610–GSM5234640 and are immediately available. Additionally, cryopreserved Ucp1−/− rat sperm are available from the PBRC Transgenics Core facility. Interested investigators should send all inquiries to the corresponding author at jaycob.warfel@pbrc.edu.
REFERENCES
- 1.Block BA. Thermogenesis in muscle. Annu Rev Physiol. 1994;56:535–577. [DOI] [PubMed] [Google Scholar]
- 2.Fenzl A, Kiefer FW. Brown adipose tissue and thermogenesis. Horm Mol Biol Clin Investig. 2014;19(1):25–37. [DOI] [PubMed] [Google Scholar]
- 3.Fuller-Jackson JP, Henry BA. Adipose and skeletal muscle thermogenesis: studies from large animals. J Endocrinol. 2018;237(3):R99–R115. [DOI] [PubMed] [Google Scholar]
- 4.Bal NC, Maurya SK, Singh S, Wehrens XH, Periasamy M. Increased reliance on muscle-based thermogenesis upon acute minimization of brown adipose tissue function. J Biol Chem. 2016;291(33):17247–17257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brychta RJ, Chen KY. Cold-induced thermogenesis in humans. Eur J Clin Nutr. 2017;71(3):345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enerback S, Jacobsson A, Simpson EM, et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387(6628):90–94. [DOI] [PubMed] [Google Scholar]
- 7.Koza RA, Hohmann SM, Guerra C, Rossmeisl M, Kozak LP. Synergistic gene interactions control the induction of the mitochondrial uncoupling protein (Ucp1) gene in white fat tissue. J Biol Chem. 2000;275(44):34486–34492. [DOI] [PubMed] [Google Scholar]
- 8.Rowland LA, Bal NC, Kozak LP, Periasamy M. Uncoupling protein 1 and sarcolipin are required to maintain optimal thermogenesis, and loss of both systems compromises survival of mice under cold stress. J Biol Chem. 2015;290(19):12282–12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betz MJ, Enerback S. Targeting thermogenesis in brown fat and muscle to treat obesity and metabolic disease. Nat Rev Endocrinol. 2018;14(2):77–87. [DOI] [PubMed] [Google Scholar]
- 10.Bal NC, Singh S, Reis FCG, et al. Both brown adipose tissue and skeletal muscle thermogenesis processes are activated during mild to severe cold adaptation in mice. J Biol Chem. 2017;292(40):16616–16625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ukropec J, Anunciado RP, Ravussin Y, Hulver MW, Kozak LP. UCP1-independent thermogenesis in white adipose tissue of cold-acclimated Ucp1−/− mice. J Biol Chem. 2006;281(42):31894–31908. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt-Nielsen K. Scaling, why is animal size so important? Cambridge University Press; 1984. [Google Scholar]
- 13.Gordon CJ. Thermal biology of the laboratory rat. Physiol Behav. 1990;47(5):963–991. [DOI] [PubMed] [Google Scholar]
- 14.Armitage G, Harris RB, Hervey GR, Tobin G. The relationship between energy expenditure and environmental temperature in congenitally obese and non-obese Zucker rats. J Physiol. 1984;350:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Essen G, Lindsund E, Cannon B, Nedergaard J. Adaptive facultative diet-induced thermogenesis in wild-type but not in UCP1-ablated mice. Am J Physiol Endocrinol Metab. 2017;313(5):E515–E527. [DOI] [PubMed] [Google Scholar]
- 16.Moore BJ, Stern JS, Horwitz BA. Brown fat mediates increased energy expenditure of cold-exposed overfed neonatal rats. Am J Physiol. 1986;251(3 Pt 2):R518–R524. [DOI] [PubMed] [Google Scholar]
- 17.Iannaccone PM, Jacob HJ. Rats! Dis Model Mech. 2009;2(5-6):206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenzen S. Animal models of human type 1 diabetes for evaluating combination therapies and successful translation to the patient with type 1 diabetes. Diabetes Metab Res Rev. 2017;33(7). doi: 10.1002/dmrr.2915 [DOI] [PubMed] [Google Scholar]
- 19.Hou D, Fu H, Zheng Y, et al. Uncoupling protein 1 knockout aggravates isoproterenol-induced acute myocardial ischemia via AMPK/mTOR/PPARalpha pathways in rats. Transgenic Res. 2022;31(1):107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shabalina IG, Hoeks J, Kramarova TV, Schrauwen P, Cannon B, Nedergaard J. Cold tolerance of UCP1-ablated mice: a skeletal muscle mitochondria switch toward lipid oxidation with marked UCP3 up-regulation not associated with increased basal, fatty acid- or ROS-induced uncoupling or enhanced GDP effects. Biochim Biophys Acta. 2010;1797(6-7):968–980. [DOI] [PubMed] [Google Scholar]
- 21.Johnson TS, Murray S, Young JB, Landsberg L. Restricted food intake limits brown adipose tissue hypertrophy in cold exposure. Life Sci. 1982;30(17):1423–1426. [DOI] [PubMed] [Google Scholar]
- 22.Hondares E, Iglesias R, Giralt A, et al. Thermogenic activation induces FGF21 expression and release in brown adipose tissue. J Biol Chem. 2011;286(15):12983–12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keipert S, Ost M. Stress-induced FGF21 and GDF15 in obesity and obesity resistance. Trends Endocrinol Metab. 2021;32(11):904–915. [DOI] [PubMed] [Google Scholar]
- 24.Verdeguer F, Soustek MS, Hatting M, et al. Brown adipose YY1 deficiency activates expression of secreted proteins linked to energy expenditure and prevents diet-induced obesity. Mol Cell Biol. 2015;36(1):184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartelt A, Heeren J. Adipose tissue browning and metabolic health. Nat Rev Endocrinol. 2014;10(1):24–36. [DOI] [PubMed] [Google Scholar]
- 26.Seale P, Kajimura S, Yang W, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6(1):38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blondin DP, Labbe SM, Phoenix S, et al. Contributions of white and brown adipose tissues and skeletal muscles to acute cold-induced metabolic responses in healthy men. J Physiol. 2015;593(3):701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labbe SM, Caron A, Bakan I, et al. In vivo measurement of energy substrate contribution to cold-induced brown adipose tissue thermogenesis. FASEB J. 2015;29(5):2046–2058. [DOI] [PubMed] [Google Scholar]
- 29.Barbera MJ, Schluter A, Pedraza N, Iglesias R, Villarroya F, Giralt M. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J Biol Chem. 2001;276(2):1486–1493. [DOI] [PubMed] [Google Scholar]
- 30.Anunciado-Koza RP, Zhang J, Ukropec J, et al. Inactivation of the mitochondrial carrier SLC25A25 (ATP-Mg2+/Pi transporter) reduces physical endurance and metabolic efficiency in mice. J Biol Chem. 2011;286(13):11659–11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arruda AP, Nigro M, Oliveira GM, de Meis L. Thermogenic activity of Ca2+-ATPase from skeletal muscle heavy sarcoplasmic reticulum: the role of ryanodine Ca2+ channel. Biochim Biophys Acta. 2007;1768(6):1498–1505. [DOI] [PubMed] [Google Scholar]
- 32.Flicker D, Sancak Y, Mick E, Goldberger O, Mootha VK. Exploring the In vivo role of the mitochondrial calcium uniporter in Brown fat bioenergetics. Cell Rep. 2019;27(5):1364–1375.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crooks DR, Maio N, Lane AN, et al. Acute loss of iron-sulfur clusters results in metabolic reprogramming and generation of lipid droplets in mammalian cells. J Biol Chem. 2018;293(21):8297–8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda K, Kang Q, Yoneshiro T, et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med. 2017;23(12):1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Badjatia N, Strongilis E, Gordon E, et al. Metabolic impact of shivering during therapeutic temperature modulation: the Bedside Shivering Assessment Scale. Stroke. 2008;39(12):3242–3247. [DOI] [PubMed] [Google Scholar]
- 36.Arnold JT, Hemsley Z, Hodder SG, Havenith G, Lloyd AB. Reliability and validity of methods in the assessment of cold-induced shivering thermogenesis. Eur J Appl Physiol. 2020;120(3):591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lomo T, Eken T, Bekkestad Rein E, Nja A. Body temperature control in rats by muscle tone during rest or sleep. Acta Physiol (Oxf). 2020;228(2):e13348. [DOI] [PubMed] [Google Scholar]
- 38.Oeckl J, Janovska P, Adamcova K, et al. Loss of UCP1 function augments recruitment of futile lipid cycling for thermogenesis in murine brown fat. Mol Metab. 2022;61:101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reitman ML. Of mice and men - environmental temperature, body temperature, and treatment of obesity. FEBS Lett. 2018;592(12):2098–2107. [DOI] [PubMed] [Google Scholar]
- 40.Golozoubova V, Hohtola E, Matthias A, Jacobsson A, Cannon B, Nedergaard J. Only UCP1 can mediate adaptive nonshivering thermogenesis in the cold. FASEB J. 2001;15(11):2048–2050. [DOI] [PubMed] [Google Scholar]
- 41.Brobeck JR. Food intake as a mechanism of temperature regulation. 1948. Obes Res. 1997;5(6):641–645. [DOI] [PubMed] [Google Scholar]
- 42.Labbe SM, Mouchiroud M, Caron A, et al. mTORC1 is required for brown adipose tissue recruitment and metabolic adaptation to cold. Sci Rep. 2016;6:37223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bal NC, Maurya SK, Pani S, et al. Mild cold induced thermogenesis: are BAT and skeletal muscle synergistic partners? Biosci Rep. 2017;37:BSR20171087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boutant M, Kulkarni SS, Joffraud M, et al. Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J. 2017;36(11):1543–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouellet V, Labbe SM, Blondin DP, et al. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J Clin Invest. 2012;122(2):545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazak L, Chouchani ET, Stavrovskaya IG, et al. UCP1 deficiency causes brown fat respiratory chain depletion and sensitizes mitochondria to calcium overload-induced dysfunction. Proc Natl Acad Sci USA. 2017;114(30):7981–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blondin DP, Daoud A, Taylor T, et al. Four-week cold acclimation in adult humans shifts uncoupling thermogenesis from skeletal muscles to brown adipose tissue. J Physiol. 2017;595(6):2099–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hohtola E, Gonzalez-Alonso J. Motor unit function during cold induced thermogenesis in muscle-new perspectives on old concepts. Acta Physiol. 2020;228(2):e13408. [DOI] [PubMed] [Google Scholar]
- 49.Haman F, Blondin DP. Shivering thermogenesis in humans: origin, contribution and metabolic requirement. Temperature (Austin). 2017;4(3):217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toien O, Aulie A, Steen JB. Thermoregulatory responses to egg cooling in incubating bantam hens. J Comp Physiol B. 1986;156(3):303–307. [Google Scholar]
- 51.McKay WP, Vargo M, Chilibeck PD, Daku BL. Effects of ambient temperature on mechanomyography of resting quadriceps muscle. Appl Physiol Nutr Metab. 2013;38(3):227–233. [DOI] [PubMed] [Google Scholar]
- 52.Tomiya S, Tamura Y, Kouzaki K, et al. Cast immobilization of hindlimb upregulates sarcolipin expression in atrophied skeletal muscles and increases thermogenesis in C57BL/6J mice. Am J Physiol Regul Integr Comp Physiol. 2019;317(5):R649–R661. [DOI] [PubMed] [Google Scholar]
- 53.Chemaly ER, Troncone L, Lebeche D. SERCA control of cell death and survival. Cell Calcium. 2018;69:46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouillaud F, Alves-Guerra MC, Ricquier D. UCPs, at the interface between bioenergetics and metabolism. Biochim Biophys Acta. 2016;1863(10):2443–2456. [DOI] [PubMed] [Google Scholar]
- 55.Arsenijevic D, Onuma H, Pecqueur C, et al. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet. 2000;26(4):435–439. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y, Zeng X, Huang X, Serag S, Woolf CJ, Spiegelman BM. Crosstalk between KCNK3-mediated ion current and adrenergic signaling regulates adipose thermogenesis and obesity. Cell. 2017;171(4):836–848.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campbell KL, Dicke AA. Sarcolipin makes heat, but is it adaptive thermogenesis? Front Physiol. 2018;9:714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harri M, Dannenberg T, Oksanen-Rossi R, Hohtola E, Sundin U. Related and unrelated changes in response to exercise and cold in rats: a reevaluation. J Appl Physiol Respir Environ Exerc Physiol. 1984;57(5):1489–1497. [DOI] [PubMed] [Google Scholar]
- 59.Sellers EA, Scott JW, Thomas N. Electrical activity of skeletal muscle of normal and acclimatized rats on exposure to cold. Am J Physiol. 1954;177(3):372–376. [DOI] [PubMed] [Google Scholar]
- 60.Periasamy M, Herrera JL, Reis FCG. Skeletal muscle thermogenesis and its role in whole body energy metabolism. Diabetes Metab J. 2017;41(5):327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91(4):1447–1531. [DOI] [PubMed] [Google Scholar]
- 62.Vermillion KL, Anderson KJ, Hampton M, Andrews MT. Gene expression changes controlling distinct adaptations in the heart and skeletal muscle of a hibernating mammal. Physiol Genomics. 2015;47(3):58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki H, Ito Y, Shinohara M, et al. Gene targeting of the transcription factor Mohawk in rats causes heterotopic ossification of Achilles tendon via failed tenogenesis. Proc Natl Acad Sci USA. 2016;113(28):7840–7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Albarado DC, McClaine J, Stephens JM, et al. Impaired coordination of nutrient intake and substrate oxidation in melanocortin-4 receptor knockout mice. Endocrinology. 2004;145(1):243–252. [DOI] [PubMed] [Google Scholar]
- 66.Heckmann LH, Sorensen PB, Krogh PH, Sorensen JG. NORMA-Gene: a simple and robust method for qPCR normalization based on target gene data. BMC Bioinformatics. 2011;12:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandanmagsar B, Youm YH, Ravussin A, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011;17(2):179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Connell GC, Treadway MB, Petrone AB, et al. Leukocyte dynamics influence reference gene stability in whole blood: data-driven qRT-PCR normalization is a robust alternative for measurement of transcriptional biomarkers. Lab Med. 2017;48(4):346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jeggari A, Alekseenko Z, Petrov I, Dias JM, Ericson J, Alexeyenko A. EviNet: a web platform for network enrichment analysis with flexible definition of gene sets. Nucleic Acids Res. 2018;46(W1):W163–W170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Merid SK, Goranskaya D, Alexeyenko A. Distinguishing between driver and passenger mutations in individual cancer genomes by network enrichment analysis. BMC Bioinformatics. 2014;15:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vandanmagsar B, Warfel JD, Wicks SE, et al. Impaired mitochondrial fat oxidation induces FGF21 in muscle. Cell Rep. 2016;15(8):1686–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. RNA-seq raw data have been submitted to GEO under accession numbers GSM5234610–GSM5234640 and are immediately available. Additionally, cryopreserved Ucp1−/− rat sperm are available from the PBRC Transgenics Core facility. Interested investigators should send all inquiries to the corresponding author at jaycob.warfel@pbrc.edu.