Abstract
Molecular studies of speciation in birds over the last three decades have been dominated by a focus on the geography, ecology, and timing of speciation, a tradition traceable to Mayr's Systematics and the Origin of Species. However, in the recent years, interest in the behavioral and molecular mechanisms of speciation in birds has increased, building in part on the older traditions and observations from domesticated species. The result is that many of the same mechanisms proffered for model lineages such as Drosophila—mechanisms such as genetic incompatibilities, reinforcement, and sexual selection—are now being seriously entertained for birds, albeit with much lower resolution. The recent completion of a draft sequence of the chicken genome, and an abundance of single-nucleotide polymorphisms on the autosomes and sex chromosomes, will dramatically accelerate research on the molecular mechanisms of avian speciation over the next few years. The challenge for ornithologists is now to inform well studied examples of speciation in nature with increased molecular resolution—to clone speciation genes if they exist—and thereby evaluate the relative roles of extrinsic, intrinsic, deterministic, and stochastic causes for avian diversification.
Presumably, it was only a distant dream of mid-century participants in the Modern Synthesis that one day genetic analysis would come to dominate so completely the analysis of speciation in a clade such as birds. That genetics would be a useful tool in the analysis of speciation in model organisms had been evident since the early days of Drosophila genetics. Yet Mayr's Systematics and the Origin of Species (1), published 11 years before the discovery of the structure of DNA, was a treasure-trove of speciation stories not of logistically tractable species with easily sampled and manipulated populations; rather, this book focused on speciation stories from the distant South Pacific, on what were, even for ornithologists, virtually inaccessible taxa with ranges straddling some of the most remote and challenging habitats of the planet. The allure of the exotic continues for ornithologists: Mayr and Diamond have recently undertaken a complete taxonomic and biogeographic revision of the birds of the Solomon Islands (2), and detailed molecular phylogeographic tests of several speciation stories in this assemblage are finally underway (3-5). Indeed, the role of molecular techniques introduced to ornithology with the first allozyme surveys of avian populations nearly 35 years ago has been primarily to inform the geography and timing of speciation, thereby emphasizing extrinsic aspects of the speciation process: species delimitation, allopatric speciation, ecological divergence, bottlenecks, and the role of the Pleistocene (6, 7). Those mechanisms of avian speciation described by Mayr in terms of internal factors—for example, speciation resulting from so-called “genetic revolutions” (8)—were often vague and, in the case of genetic revolutions, have been largely discredited (9).
In the last 10 years, however, there has been a renewed interest in the behavioral, cognitive, and even molecular mechanisms of speciation in birds. This renaissance, spearheaded largely by recent reviews by Price (10-13), builds in part on the ancient tradition of avian husbandry and domestication, and in part on theoretical models suggesting a role for diverse behavioral factors in bird speciation, including sexual selection, sexual imprinting, learning, reinforcement, and genetic incompatibilities. Although biogeographic analyses still largely support allopatric speciation models (14), recent years have also witnessed the first serious attempts to document sympatric speciation in birds (15, 16), and the frequency of cases of sympatric speciation and divergence due to hybrid incompatibilities or reinforcement is an open question (17). Now, a draft of the complete chicken genome and >2.8 million chicken SNPs have been determined roughly 60 years after Mayr's landmark book (18, 19). This treasure-trove of genes and genetic markers will no doubt spur rapid advances in both the geography and genetics of speciation in birds. This article reviews recent studies of extrinsic and intrinsic aspects of speciation in birds, focusing specifically on systematic and mechanistic issues that challenge the universality of the allopatric speciation paradigm.
Gene Trees, Species Delimitation, and Phylogeographic Patterns
Genetic data are serving an ever-increasingly important role in the delimitation of species, yet considerable controversy remains over which criteria to apply to this age-old problem (17, 20-24). Indeed, determining which of myriad species delimitation methods and species definitions is most appropriate for one's focal taxa remains one of the paramount challenges in systematics, with important consequences for evolutionary biologists as well as for conservation biologists (25-27). The fact that many avian sister taxa occur in allopatry—particularly in Gondwanan continents such as South America and Australia (28, 29)—makes interpretation of biogeographic histories more straightforward but can also make attempts at species delimitation particularly challenging. It has long been recognized that the biological species concept (BSC), with its emphasis on reproductive isolation, is inapplicable in many allopatric situations because there is no opportunity to test for reproductive isolation, rendering the concept arbitrary (30). Species concepts emphasizing genetic clustering of forms can be equally arbitrary (reviewed in ref. 13). Diagnosibility—the ability to delimit and identify distinguishing character states for a given collection of individuals or taxa, usually but not always in a phylogenetic context—has been proffered as a general consideration when delimiting species (20). Although diagnosibility is sometimes construed as being equivalent to “fixed” character or genetic differences between taxa, alternate fixations are not a requirement for diagnosibility. The rise in sophisticated statistical genetic algorithms and large-scale multilocus analyses of variation in birds and other taxa confirm that it is trivial to diagnose species, subspecies, or even populations, even in the presence of abundant shared polymorphisms; a recent study of native and introduced house finch populations in North America was able to readily diagnose populations that had been separated for 50-100 years (31), even in the absence of fixed genetic or morphological differences. A naïve evaluation of the genetic patterns in this study would have reasonably inferred the existence of three diagnosable “species” of native and introduced House Finches within the continental United States and Hawaii. A similar ease of diagnosibility using large-scale multilocus approaches is observed for geographic populations of humans (32). Thus, diagnosibility will become highly problematic as the resolving power of multilocus approaches increases; the specter of statistical significance without biological significance will be perennial.
The criterion of monophyly advocated by proponents of the phylogenetic species concept (PSC) also suffers from arbitrariness, particularly given the disproportionate emphasis on mtDNA. It is not surprising that a number of workers have advocated the use of mtDNA in species delimitation, given its rapid attainment of reciprocal monophyly (relative to nuclear genes) and frequent ability to diagnose populations (21, 27, 33, 34). Reciprocal monophyly of mtDNA is attractive because it is an objective criterion that can be applied to any animal system. However, use of mtDNA alone to delimit species has been criticized, because the complete organismal history is not captured and because many other loci in the genome may not exhibit reciprocal monophyly (21, 26, 35).
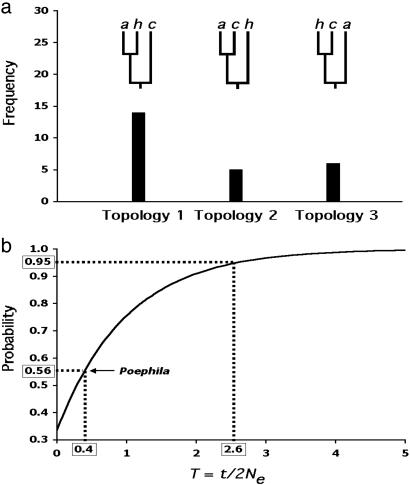
Another gene-tree-based criterion for species delimitation calls for finding reciprocal monophyly among a large majority (≈95%) of sampled nuclear genes (23, 36). Because nuclear DNA (nDNA) achieves reciprocal monophyly much slower than mtDNA, this criterion is considered conservative (26, 35). However, to date, and certainly at the time this species concept was put forward, there have been no avian data sets consisting of multiple independent gene trees with which to test the utility of this approach. A recent study of speciation in three Australian grassfinches (Poephila) using 30 independent nuclear loci provides a test of this concept. One of the taxa (Poephila cincta) examined is phenotypically and geographically very distinct from the other two, and its species status has never been questioned (37); the two allopatric western lineages of long-tailed finches (Poephila acuticauda/hecki) are distinguished by fewer characters but are nonetheless diagnosable morphologically and have been designated phylogenetic species (38). Coalescent estimates of population divergence times suggest a split of ≈0.5 million years ago for the two western lineages, and >0.7 million years ago for the basal split with cincta. Given the dynamics of nuclear genes under reasonable assumptions of ancestral population sizes for birds, it is therefore not surprising that considerable heterogeneity and conflict among the gene trees in Poephila was observed, even with regard to alleles sampled from the divergent cincta lineage (Fig. 1a) (39, 42). There is reason to believe that these conflicts are the result of incomplete lineage sorting, rather than of hybridization or gene flow (7). Application of the criterion of genealogical concordance among ≈95% of sampled gene trees would result in lumping all three taxa together despite their considerable temporal and phenotypic divergence. We suggest that concordance among multiple nuclear genes will rarely be achieved among avian lineages that are considered good species by multiple other criteria; as in Drosophila (40), if the characters leading to diagnosibility diverge by natural selection, they may outpace the well known but slow progression of the neutral nuclear genome from paraphyly to polyphyly to reciprocal monophyly (41).
Fig. 1.
Conflicts between gene and species trees in Poephila finches, and their implications. (a) Frequency distribution of all three possible gene trees encountered in a survey of nuclear DNA sequence variation among three species of Australian grassfinches (Poephila). Branch tips are labeled as follows: a, P. acuticauda; h, P. hecki; c, P. cincta. P. cincta vs. P. acuticauda/hecki is a well established divergence across the Carpentarian barrier in northern Australia and is supported by numerous species-level phenotypic differences; P. hecki vs. acuticauda are allopatric but represent a more recent taxonomic split which is regarded by some (38) as phylogenetic species. In the genetic survey, a single allele was sampled from each of the three taxa for thirty presumably unlinked loci. Twenty-five gene trees exhibiting all three possible topologies were unambiguously reconstructed. Gene trees for five loci could not be resolved. Topology 1 reflects the presumed species tree. (b) Unconditional probabilities of a gene tree being congruent with the species tree over a range of values for T (after ref. 43). The curve is based on the equation for three species [P(congruent gene tree) = 1-2/3e-T (42, 43), where T is equal to t/2Ne, t is defined as the time between speciation events (in generations), and 2Ne is twice the size of the effective population size of the basal ancestor. When T = 0, the topology of a gene tree of three sampled alleles is expected to be random with respect to the species tree (i.e., probability of 0.33). Note that T must be at least as large as 2.6 for a 0.95 probability of congruence, yet the empirically derived probability for Poephila finches of 0.56 results in a T value of only 0.4. The Poephila tree congruence probability is based on the fact that 14 of 25 independent gene trees matched the presumed species tree observed by W.B.J. and S.V.E. (unpublished work).
Although the incidence of reciprocal monophyly among genes and organisms may vary considerably, it is instructive to examine further the efficacy of this criterion in delimiting species of Poephila. The probability that a gene tree will match the species tree in a three-species scenario of divergence and isolation without gene flow has been known for some time (42, 43) and is determined by T, the ratio of time elapsed between speciation events to the ancestral effective population size (Fig. 1b). T must therefore equal or exceed 2.6 if there is to be significant concordance between the gene trees and species tree. The multilocus Poephila data suggests a value for T of 0.4, implying a substantial ancestral population size relative to divergence time, also the likely cause for the lack of concordance among gene trees. The probability of incongruence between gene and species trees for the Poephila loci are not strictly equivalent to the genealogical concordance among loci described by Avise and Ball (19), because in the Poephila study only a single allele was sampled per species; thus, some of the loci exhibiting congruence with the species tree (Topology 1, Fig. 1a) may in fact exhibit genealogical incongruence, manifested as incomplete lineage sorting, upon sampling of further alleles. Nonetheless, these data, the first substantial sampling of multiple gene trees for an avian species, suggest that species arising rapidly or having ancestors with large effective population sizes will not satisfy the concordant genealogies criterion even though they are reasonable species under the BSC or morphological PSC. For avian species, finding any nuclear gene that has achieved reciprocal monophyly, whether by neutral or selective means (akin to finding a fixed diagnostic phenotypic trait), may be a reasonable criterion for delimiting species.
It is certain that molecular approaches will continue to play an important role in species delimitation. However, the battle over species delimitation between the nuclear and mitochondrial genomes, and between the BSC and PSC, will have no victor. Nuclear gene trees will not provide enough phylogenetic resolution to satisfy avian systematists. Furthermore, the high levels of recombination detected in the first surveys of avian SNP variation appear prohibitive for standard phylogenetic analysis (19, 44). On the other hand, in our view, maternally inherited mtDNA can never capture enough of a species history to delimit species on its own. Although mtDNA will frequently deliver clean phylogeographic breaks within avian species, these breaks need not have their origin in reproductive isolation (45) and may in some cases be driven by natural selection (46). These same species showing clean mitochondrial breaks will frequently look very messy with regard to nuclear gene splits, as decades of allozyme analyses have already confirmed. We suggest that, despite its disproportionate contribution to revealing phylogeographic patterns and its ability to reflect cessation of female gene flow more rapidly than nuclear genes, mtDNA should not have priority over nuclear genes in avian species delimitation. Ours is a generation of avian systematists raised primarily on single locus analyses of avian phylogeny and divergence. But nuclear genes can and should be interrogated in questions of avian taxonomy, even if the interpretation of nuclear histories and the contrast with mtDNA histories will be challenging.
The Role of Song in Allopatric and Sympatric Speciation. As suggested by Mayr (1), divergence of populations in allopatry appears to be the dominant mode of speciation in birds. Molecular phylogenies have provided new opportunities for testing alternative geographic models, and avian sister taxa generally meet the expectation of having allopatric distributions, whether because of vicariance or dispersal (47-50). Current distributions, however, do not necessarily reflect the geographic context of speciation given the potential for dispersal and range expansion in birds (51). When expectations are derived from a model incorporating random changes in geographic range, phylogenetic data suggest allopatric speciation as the predominant mode in several avian groups, with sympatry due to range changes after speciation (52). Greater asymmetry in range size for recently evolved species also implies a role for peripatric speciation (52), as suggested in other recent avian studies (53-55). The lability of geographic ranges, however, ultimately limits the power of phylogenetic approaches to distinguish between alternative geographic models of speciation (56).
Greater insight into avian speciation is perhaps gained by focusing on the processes of population divergence and mechanisms of reproductive isolation, particularly in closely related taxa and/or diverging populations. Both ecological and sexual selection may contribute to rapid morphological and behavioral divergence in allopatric or parapatric populations (57-59). Whether these changes lead to speciation, however, depends on the evolution of reproductive isolation before or after secondary contact. Avian species typically retain hybrid viability and fertility for millions of years after speciation, reflecting a general lack of intrinsic isolating mechanisms among closely related species (11). Although ecological and/or sexual selection against hybrids may help to maintain species boundaries, reproductive isolation in birds will often depend on prezygotic mechanisms. Thus, divergence in characters involved in mate choice, such as song, plumage, and behavioral displays, likely play a central role in avian speciation. The role of song is particularly interesting given the multiple factors influencing vocal evolution and the potential for rapid change through learning and cultural evolution.
Gradual divergence of song in allopatric populations may result in reproductive isolation upon secondary contact. Although generally difficult to observe, this process is captured in present day populations of the greenish warbler (Phylloscopus trochiloides) complex. The range of this Old World species forms a narrow ring around the southern margin of the Tibetan plateau with eastern and western populations extending northward, expanding longitudinally, and meeting in Siberia. Genetic composition and songs change gradually through the nearly continuous ring of intergrading populations, but eastern and western populations in the north are reproductively isolated because of differences in song (60). Parallel sexual selection for increased song complexity in northern latitudes has apparently resulted in the stochastic divergence of songs in eastern and western populations (61). The gradual intergradation through intermediate populations in this ring species are analogous to the gradual changes that might occur over time in geographically isolated populations diverging in a similar manner (62).
Songs may diverge as a direct result of habitat-dependent selection or indirectly as a consequence of morphological adaptations, such as those related to foraging. It has long been recognized that different types of vocalizations vary in their quality as signals in different habitats, and recent studies suggest a role for habitat-dependent selection in population divergence and reproductive isolation. Two subspecies of song sparrows (Melospiza melodia) differ both in song characteristics and preferred habitat, with Melospiza melodia hermanii occupying more dense vegetation than Melospiza melodia fallax and singing a lower frequency song with more widely spaced elements, a pattern consistent with acoustic adaptation (63). Playback experiments further indicate that both males and females show greater response to homotypic songs, suggesting a role for song in reproductive isolation and the consequent development of significant genetic differentiation between the subspecies (63). Similar habitat-dependent vocal divergence accompanies morphological differentiation in the little greenbul (Andropodus virens) and may promote population differentiation across ecological gradients (64).
Ecological selection on other characters also may result in correlated vocal evolution that contributes to prezygotic reproductive isolation. In Darwin's finches, bill morphology and vocal characteristics are correlated because of physical constraints on sound production, perhaps contributing to the diversification of these species (65). In contrast, the black-bellied seedcracker (Pyrenestes ostrinus), which shows a similar pattern of divergent selection on bill morphology, shows no effect of bill size on vocal performance, contributing to the conclusion that bill size variation in this species reflects only intraspecific niche polymorphism and not incipient speciation (66, 67).
As a learned behavior in many birds, song is subject to rapid cultural evolution in which stochastic innovations or errors in copying are spread as individuals learn songs from their parents and/or neighbors (68, 69). Song learning may also increase the rate at which genetic predispositions to learn or prefer certain songs evolve in allopatry (70). Song learning, however, may sometimes inhibit reproductive isolation upon secondary contact if there has been only minimal divergence in the capacity to learn particular songs and/or the morphological structures affecting sound production (71). A well known example of reproductive character displacement involves the Ficedula flycatchers in which a sexually selected male plumage trait shows greater divergence in sympatry than in allopatry (72). Results were mixed, however, in a recent analysis of vocal divergence in this system (73). In sympatry, songs of pied flycatchers Ficedula hypoleuca have converged on those of collared flycatchers Ficedula albicollis because of heterospecific copying and singing of mixed songs. Although collared flycatcher songs in the zone of contact have diverged away from pied flycatcher songs typical of allopatric populations, the net effect of these changes is greater song similarity in sympatry.
Allopatric divergence of songs among suboscines and other birds in which differences in song are genetically determined may evolve more slowly but should also contribute to reproductive isolation. In a large comparative analysis of antbirds (Thamnophilidae), Seddon (74) found evidence of vocal divergence both as a correlated effect of morphological evolution and as a response to habitat-dependent selection on signal transmission. In addition, among trios of closely related species, sympatric forms exhibited striking vocal divergence in comparison to allopatric taxa, providing strong evidence for reproductive character displacement and the role of song in reproductive isolation. Despite divergence of song in allopatry, individuals of different species may recognize each other as potential mates upon secondary contact, leading to hybridization. If song differences are genetically determined, hybrid offspring may have intermediate songs (75).
Given divergence in vocalizations, the development of song preferences through sexual imprinting may contribute to reproductive isolation even without genetic evolution of female preferences (10, 13). In Darwin's finches, for example, cultural inheritance clearly plays a greater role than bill morphology in determining songs and song preferences (69, 76) and is critical in promoting reproductive isolation after secondary contact of populations that have diverged in feeding adaptations in allopatry (77). Ecological selection against hybrid individuals also helped maintain species boundaries, at least before changes associated with El Niño events in the 1980s (78). In the past 20 years, however, increased fitness of hybrids has resulted in substantial genetic introgression from Geospiza fortis to Geospiza scandens on Daphne Major (79). Although learned songs and song preferences are strong determinants of pair formation in these species, reproductive isolation is imperfect because of constraints on mate choice imposed by asymmetries in population size and operational sex ratios as well as infrequent cases of individuals misimprinting on heterospecific songs (69, 76, 78).
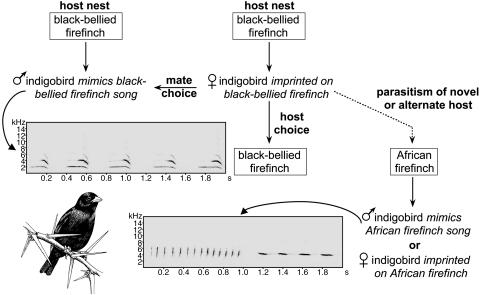
Song learning and sexual imprinting explain the recent diversification of brood parasitic indigobirds (genus Vidua), the best and perhaps only example of sympatric speciation in birds (Fig. 2). Male indigobirds include mimicry of host song in a repertoire that also includes species-specific indigobird vocalizations learned from other male indigobirds mimicking the same host (80). Likewise, female preferences for both male song and host nests result from imprinting on the host (81). Thus, speciation in indigobirds begins with reproductive isolation as a consequence of host colonization and only then proceeds to divergence in other characters, including host-specific mimicry of mouth markings and colors by indigobird chicks. Indigobird species within a region show a pattern of incomplete but significant genetic differentiation (16) but also genetic continuity across intermediate spatial scales (82), a pattern consistent with recent sympatric speciation and current reproductive isolation (83). As in Darwin's finches, song learning likely plays a role in hybridization between indigobird species. When females parasitize the host of another indigobird species (84), their offspring are likely to hybridize in the subsequent generation because they have imprinted on the songs of the alternate host (85). The frequency of misimprinting and hybridization, however, appears to be lower in indigobirds than among the Geospiza ground finches (79, 84).
Fig. 2.
Behavioral imprinting maintains host specificity and genetic cohesion of indigobird species but also provides a mechanism for rapid speciation when new hosts are colonized. Male indigobirds mimic the songs of their hosts, whereas females use song cues to choose both their mates and the nests they parasitize. Rarely, females lay in the nest of a novel or alternate host. The resulting offspring imprint on the novel host and are therefore reproductively isolated from their parent population. Indigobird drawing by Karen Klitz.
In birds generally, the importance of prezygotic isolating mechanisms may allow for rapid speciation, whereas the slow development of intrinsic postzygotic isolation will facilitate continuing hybridization. Closely related taxa may therefore be strongly differentiated at only a small number of loci influenced by divergent ecological or sexual selection. Loci “that can be shown to cause some degree of ecological, sexual or postmating isolation between young, or even nascent, species” are good candidates for speciation genes (86). Finding such genes and understanding the genetics of avian speciation are challenging but increasingly realistic objectives as genomic resources and molecular methods continue to evolve.
Sex Chromosomes and Avian Speciation. The architects of the Modern Synthesis laid the foundation for a body of work that has resulted in two “rules” of speciation that directly implicate sex chromosomes: Haldane's rule and the large X(Z)-effect (87). Haldane's rule is the preferential sterility or inviability of the heterogametic sex in hybrid crosses, when a sex-biased fitness loss in hybrids occurs. This phenomenon is found across diverse taxa, including butterflies and birds in which the female is heterogametic (ZW) (88). Although the X chromosome was experimentally implicated in Haldane's rule and hybrid male sterility in Drosophila decades ago (89), it was not until recently that this large effect of the hemizygous sex chromosome was documented for birds, using genetic data from natural hybrid zones and domesticated species (12, 90). In this section, we review empirical and theoretical work that explores these two rules of speciation in birds.
The phenomenon of Haldane's rule describes patterns of postzygotic incompatibilities in hybrids and is likely caused by negative epistatic interactions between loci derived from divergent parental genomes (17). Heterogametic hybrids are more severely affected by these interactions because, unlike the homogametic sex, they fully express recessive sex-linked genes. Interestingly, avian and Lepidopteran F1 hybrid females may suffer from an additional source of negative epistasis between parental genomes, namely that between the maternally derived mitochondria or cytoplasm and the paternally derived Z chromosomes (91). There is debate over the extent to which Haldane's rule is driven by interactions among sometimes rapidly diverging sex chromosomes per se, or whether it is the peculiar dominance patterns exhibited by the hemizygous sex chromosomes that underlie the rule. Support for Haldane's rule is excellent in birds based on experimental studies of hybrid fitness in ducks (92), pigeons and doves (93), and many other avian taxa (11).
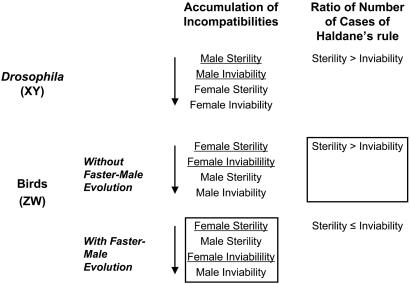
Price and Bouvier (11) characterized patterns of postzygotic incompatibilities in birds using published data from 254 hybrid crosses and found that the order in which incompatibilities accumulate with increasing species divergence differs between birds and other taxa, a pattern that informs the causes of Haldane's rule in birds (Fig. 3). In Drosophila, male sterility appears at early stages of divergence, followed in turn by male inviability, then female sterility, and finally female inviability. By contrast, avian incompatibilities accumulate in the following order: female sterility, male sterility, female inviability, and male inviability (11). Thus, in birds, homogametic (male) sterility evolves at earlier stages than does homogametic (female) sterility in Drosophila (87). The appearance of homogametic (male) sterility before heterogametic (female) inviability in birds may reflect a general trend, regardless of sex-chromosome system, of the rapid evolution of male reproductive genes via sexual selection, resulting in high divergence between species at these loci (94). However, the rapid evolution of male reproductive genes via sexual selection—so-called “faster-male evolution” or the “sexual selection model” of Wu and Davis (94)—works in opposition to Haldane's rule in birds because this particular force will negatively affect the homogametic sex (males), not the heterogametic sex (females), in ZW species. By contrast, sexual selection accentuates the pattern for sterility in XY species, such as Drosophila, perhaps explaining the preponderance of cases of hybrid male sterility in the genus (17).
Fig. 3.
Summary of Haldane's rule in birds. Predictions are based on the presence or absence of faster-male evolution (sexual selection hypothesis), which changes the order of accumulation of incompatibilities in WZ species. The predicted order of accumulation of incompatibilities for the case without sexual selection is based on the pattern in Drosophila. The data observed by Price and Bouvier (11) are boxed. The heterogametic sex is underlined.
Support for sexual selection as a modulator of Haldane's rule in birds was first detected in the relative proportions of cases of Haldane's rule for sterility and inviability (96). Specifically, the majority of cases of Haldane's rule in XY species involve heterogametic sterility, whereas Wu and Davis (96) initially found as many cases for heterogametic inviability as for sterility in birds and other ZW species. This pattern is expected when traits expressed only in males are evolving rapidly because it results in male hybrid sterility in early stages of divergence, resulting in fewer cases of Haldane's rule for sterility. However, in the larger avian data set of Price and Bouvier (11), five times as many cases of Haldane's rule involve sterility as inviability, a pattern that does not support sexual selection as a modulator of Haldane's rule. Direct evidence for a role for sexually selected genes in avian speciation, such as an examination of the molecular evolution of avian reproductive genes (see next section), is a necessary but not sufficient condition for their role Haldane's rule in birds. Additionally, the domain of such sexually selected genes needs to be determined: Do they include sexually selected traits such as plumage and song that are not directly involved in the physical production of gametes? Because males and females of many bird species exhibit correlated evolution of plumage and other sexually selected traits, the specific expression patterns of genes related to both reproduction and morphology need to be investigated to determine how such traits will influence Haldane's rule.
The rate of evolution of sex chromosomes has important implications for their role in avian speciation. In general, when rapidly evolving loci diverge between species, incompatibilities between these and other loci can arise when parental genomes come together in hybrids. In the same way that rapid evolution of sexual traits in males can produce incompatibilities in hybrids at loci involved in male sexual traits—whether autosomal or sex-linked—a faster rate of evolution of the sex chromosomes, combined with effects of dominance (17), is one hypothesis for the large X(Z)-effect in low-fitness hybrids. So-called “faster-X(Z) evolution” owing to selection on favorable mutations on the hemizygous X(Z) when they are partially recessive, manifested as more rapid substitution rates on the X chromosome than on autosomes, was demonstrated theoretically by Charlesworth et al. (95). More recently, Kirkpatrick and Hall (96) modeled the relative rates of evolution of sex chromosomes and autosomes while accounting for interactions between mode of inheritance and the intrinsically higher mutation rate in males known as “male-driven evolution.” Male-driven evolution (not to be confused with faster-male evolution) has been documented in birds, mammals, and plants, most likely because of a greater number of cell divisions in the male than in the female germ line (97); critically, such studies have documented faster neutral evolution on the avian Z but not faster adaptive, presumably nonsynonymous evolution, as predicted by Charlesworth et al. (95). Values of α, the ratio of male to female mutation rates, have been estimated from sequence data and range from ≈1.8 to ≈6.4 in both birds and mammals (reviewed in ref. 86). The avian Z chromosome is expected to have a higher neutral mutation rate than the autosomes because it passes through the male germ line twice as often as the female germ line, whereas avian autosomes spend equal time in both germ lines. In contrast to predictions of hemizygous sex chromosome evolution in mammals, where mutations must be strongly recessive for the X to evolve faster than autosomes (95), the higher mutational flux on the Z of birds is predicted to produce faster rates of Z chromosome evolution relative to autosomes even when mutations are strongly dominant (96). Perhaps not surprisingly, support for fast-X evolution is inconsistent (98, 99), whereas evidence of fast-Z evolution is more compelling (100-104). Sequence data flowing from the chicken genome project have already facilitated confirmation of faster-Z evolution in birds, both at the intra- and interspecific levels (104).
The first empirical inquiries into the role of sex chromosomes in the speciation process of natural avian groups have focused on the well studied system provided by hybridization in Old World Ficedula flycatchers. Saetre et al. (92) found that Z-linked single nucleotide polymorphism (SNP) markers in F. hypoleuca and F. albicollis showed little evidence for introgression, whereas substantial introgression was documented for autosomal SNPs. The sex chromosomes had a large effect on the fertility of hybrids: Among birds with heterozygous sex chromosomes (one from each parental species), 7 of 7 females were sterile, as opposed to 3 of 11 males; a pattern that is consistent with Haldane's rule, although whether interactions among Z loci or between the Z and W or autosomes is the cause remains unclear. The possibility of linkage of loci involved in pre- and postzygotic isolation in this system (92) motivated the development of a novel model of reinforcement (105). The model focused on the evolution of linkages among a male trait locus, a female preference locus (collectively referred to as “mating loci”), and two postzygotic incompatibility loci. Consistent with previous theory, prezygotic isolation is reinforced through tight linkage between the mating loci and incompatibility loci. As incompatibility loci diverge between the two populations, causing a decrease in the fitness of hybrids, the frequency of assortative mating increases, thereby reducing the occurrence of interspecific matings. The tight linkage between the mating loci and the incompatibility loci creates a positive feedback loop because the frequency of linked incompatibility loci increases in tandem with the loci causing population-specific mating as a result of genetic hitchhiking. The positive feedback loop is enhanced when the linkage group occurs on a sex chromosome for two reasons. First, if as assumed in the model crossing over does not occur between sex chromosomes in the heterogametic sex, hitchhiking is enhanced. (This lack of crossing over does not strictly represent the case in nature because, as in mammals, many birds possess a pseudoautosomal region of varying size in which crossing over does in fact occur.) Second, recessive mutations are more exposed to positive selection in the heterogametic sex and may rapidly sweep through the population. The model reveals a number of interesting features about the dynamics of the Ficedula hybrid zone (90). However, it is unclear how well the Ficedula hybrid zone represents the diversity of avian hybrid zones. Many avian hybrids show little evidence of fitness loss (106) and may even enjoy an ecological advantage over parental species (107), conditions that do not favor reinforcement of prezygotic isolation.
Studies in birds and other taxa indicate that sex chromosomes may disproportionately harbor genes related to sex and reproduction (108, 109). Literature on a variety of domesticated avian species suggests that 22% of traits that distinguish breeds, including likely targets of sexual selection in natural populations such as plumage and vocalizations, are sex-linked (12), an excess when one considers that the Z chromosome comprises ≈2.7% of the chicken genome, by our estimate using data from the chicken genome project (19). Avian speciation is commonly demonstrated to involve prezygotic isolation in traits such as song or display (ref. 76; see previous section), making the genes that underlie these traits promising candidate speciation genes (86). The dawn of the genomic era for ornithologists provides exciting opportunities to study the genomic composition of avian sex chromosomes and will allow a better understanding of the complex interaction between their gene content, gene expression patterns, and rate of evolution in the context of speciation.
Cryptic Mate Choice and Conflict: A Role for Reproductive Proteins in Avian Speciation?
Birds provide an abundance of examples of intense sexual selection through cryptic female choice and sperm competition (110). The dramatic evolutionary consequences of this competition have been documented at the molecular level in mammals and invertebrates, through genes collectively known as reproductive proteins. Rapid evolution of reproductive proteins has been documented in a diverse array of taxonomic groups ranging from humans to corn and is thought to be a component of the speciation process (111). However, to our knowledge there are no examples of this process from birds. One potential protein is the female-specific gene HNTW, which, although its function is unknown, shows adaptive evolution (112). With the recent draft release of the chicken genome (18) we can expect that the molecular evolution of avian reproductive genes will come under intense scrutiny, a development that is particularly exciting because birds offer unique opportunities to study the selective forces driving the rapid evolution of reproductive proteins.
Reproductive genes can be split into two classes. First, there are gamete recognition proteins on the surface of gametes that are directly involved in sperm-egg interaction. The classic example of a rapidly evolving gamete recognition protein is the abalone sperm protein lysin, perhaps the most rapidly diverging protein yet discovered (113). Lysin acts to dissolve the egg vitelline envelope, a process that demonstrates species-specificity. In mammals it has been demonstrated that sperm and egg molecules are among the most diverse found within the genome, with a minimum of 10 reproductive genes showing evidence of adaptive evolution (114). One such gene is the mammalian egg coat protein ZP3. The region within this protein undergoing adaptive evolution corresponds to experimentally determined binding sites (115), suggesting that the rapid evolution relates to fertilization.
The second class of reproductive proteins exhibiting rapid evolutionary change are not directly involved in surface recognition of the gametes. These include components of seminal fluid (116), pheromones and protamines. In Drosophila seminal fluid, an estimated 10% of the genes show the signatures of adaptive evolution (117). Many of these genes, called accessory gland proteins (ACPs) act to manipulate female reproductive behavior, thus increasing male fitness (118). In primates, seminogellin II (SEM2), a major component of seminal fluid, shows rapid adaptive evolution. This protein is involved in copulatory plug formation in rodents, and in primates its rate of evolution shows a correlation with mating system (119).
The recent comparison of predicted genes in chicken genome with the human genome supports the pattern of divergence found in reproductive proteins across taxa (18). Genes implicated in reproduction appear less conserved between chicken and human than genes involved in typical “house-keeping” functions. For example, among genes classified into 10 different tissue specificities, those expressed in the testis showed the most divergence: 65% sequence conservation compared with the mean of 75% across all genes. Many of these genes, such as ZP3, have orthologues in birds and would be important candidates for targets of natural selection. Other potential target genes in birds include seminal fluid proteins. The prediction of natural selection on such genes in other species can be inferred from reproductive observations. Adkins-Reagan (120) documented a viscous mucoprotein produced by Japanese quail (Corturnix japonica) thought to increase the probability of fertilization when a hard-shelled egg is in the uterus. The origin of such viscosity must have a basis in protein evolution, although the target loci have not yet been identified.
Cryptic female choice, sperm competition, and sexual conflict are three nonexclusive hypotheses for the forces driving the rapid evolution of these proteins (111). Cryptic female choice of reproductive proteins involves the “preference” of male proteins on the surface of the sperm or in the seminal fluid by egg coat proteins, egg proteins, or proteins in the female reproductive tract, examples of which come mostly from invertebrates (121). The ability of many birds to store sperm provides a ready mechanism for cryptic female choice. Sperm competition involves the direct competition between sperm of different males providing a potent source selection acting to improve sperm motility (110). Sexual conflict occurs when the reproductive goals of the sexes differ and is thought to drive rapid coevolutionary arms races in reproductive proteins at the molecular level (122-124). Testing the various hypotheses for the rapid evolution of reproductive proteins is particularly promising in birds. In no other taxonomic group is so much known about the diversity of mating systems and the natural history of female preferences driving trait differences (125). Combining evolutionary data on reproductive proteins with predictions of sperm competition and mate choice from behavioral studies, as has been attempted with the SEM2 locus in primates (119), is a promising avenue of research in birds. Furthermore, reproductive traits unique to birds, such as physiological polyspermy, the ability of multiple sperm to penetrate the egg without rendering it inviable, permit testing of specific hypothesis underlying rapid adaptive protein evolution. Also, variation across avian species in particular traits such as the presence and type of intromittent organs, from penises to cloacal protuberances to the absence of any intromittent organ, allows for hypothesis-testing that would be difficult in taxonomic groups without such variation. Conversely, the study of reproductive protein evolution in birds might help clarify the roles of these traits on reproductive evolution and reinforcement. Avian mating systems are thought to play an important role in the speciation process (126), and reproductive proteins provide a convenient link between these two arenas.
Conclusion
Rich natural histories, diverse biogeographies, and complex character traits and mating systems have made birds central to the formulation of many speciation theories. Now, these and other ideas are more amenable to direct testing with the increased molecular access provided by the chicken genome and by new genomic technologies and resources. These new tools will increase the resolving power of both phylogeographic analysis of speciation and of interactions among diverging genomes. Large-scale EST surveys, bacterial artificial chromosome (BAC) libraries, and other genetic resources for dimorphic species such as zebra finches, turkeys, and Japanese quail are available, and high-resolution linkage maps will soon follow. We can expect the information from these model avian species to inform the analysis of speciation within their respective clades and beyond (127). Linking of these genetic analyses with predictions from theory and application to natural populations will make for exciting times to come in avian speciation studies.
Acknowledgments
We thank K. De Quieroz, J. Hey, H. A. Orr, C. Filardi, and J. Wakeley for helpful discussion and R. Payne, G. P. Saeter, T. Price, M. Servedio, and J. Feder for very constructive comments on the manuscript. This work was supported in part by National Science Foundation grants (to S.V.E. and M.D.S.). W.J.S. was supported by National Institutes of Health Grant HD42563-01A1 and National Science Foundation Grant DEB-0410112.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Systematics and the Origin of Species: On Ernst Mayr's 100th Anniversary,” held December 16-18, 2004, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
References
- 1.Mayr, E. (1942) Systematics and the Origin of Species (Columbia Univ. Press, New York).
- 2.Mayr, E. & Diamond, J. (2001) The Birds of Northern Melanesia: Speciation, Ecology, and Biogeography (Oxford Univ. Press, New York).
- 3.Filardi, C. E. (2003) Ph.D. dissertation. (Univ. of Washington, Seattle).
- 4.Smith, C. E. (2003) Ph.D. dissertation (Univ. of Washington, Seattle).
- 5.Filardi, C. E. & Smith, C. E. (2005) Mol. Phylogenet. Evol., in press. [DOI] [PubMed]
- 6.Barrowclough, G. F. (1983) in Perspectives in Ornithology: Essays Presented for the Centennial of the American Ornithologists' Union, eds. Brush, A. H. & Clark, G. A., Jr. (Cambridge Univ. Press, New York), pp. 223-261.
- 7.Avise, J. C. (2000) Phylogeography: The History and Formation of Species (Harvard Univ. Press, Cambridge, MA).
- 8.Mayr, E. (1963) Animal Species and Evolution (Harvard Univ. Press, Cambridge, MA).
- 9.Barton, N. H. & Charlesworth, B. (1987) Annu. Rev. Ecol. Syst. 15, 133-164. [Google Scholar]
- 10.Price, T. D. (1998) Philos. Trans. R. Soc. London B 353, 251-260. [Google Scholar]
- 11.Price, T. D. & Bouvier, M. M. (2002) Evolution (Lawrence, Kans.) 56, 2083-2089. [PubMed] [Google Scholar]
- 12.Price, T. D. (2002) Genetica 116, 311-327. [PubMed] [Google Scholar]
- 13.Irwin, D. E. & Price, T. D. (1999) Heredity 82, 347-354. [DOI] [PubMed] [Google Scholar]
- 14.Coyne, J. A. & Price, T. D. (2000) Evolution (Lawrence, Kans.) 54, 2166-2171. [DOI] [PubMed] [Google Scholar]
- 15.Grant, B. R. & Grant, P. R. (1979) Proc. Natl. Acad. Sci. USA 76, 2359-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorenson, M. D., Sefc, K. M. & Payne, R. B. (2003) Nature 424, 928-931. [DOI] [PubMed] [Google Scholar]
- 17.Coyne, J. A. & Orr, H. A. (2004) Speciation (Sinauer, Sunderland, MA).
- 18.International Chicken Genome Sequencing Consortium (2004) Nature 432, 695-716. [DOI] [PubMed] [Google Scholar]
- 19.International Chicken Polymorphism Map Consortium (2004) Nature 432, 717-722.15592405 [Google Scholar]
- 20.Cracraft, J. (1983) Curr. Ornithol. 1, 159-187. [Google Scholar]
- 21.Wiens, J. J. & Penkrot, T. A. (2002) Syst. Biol. 51, 69-91. [DOI] [PubMed] [Google Scholar]
- 22.Hey, J. (2001) Genes, Categories, and Species: The Evolutionary and Cognitive Causes of the Species Problem (Oxford Univ. Press, New York).
- 23.Avise, J. C. & Ball, R. M. (1990) Oxf. Surv. Evol. Biol. 7, 45-67. [Google Scholar]
- 24.Sites, J. W. & Marshall, J. C. (2003) Trends Ecol. Evol. 18, 462-470. [Google Scholar]
- 25.Crandall, K. A., Bininda-Emonds, O. R. P., Mace, G. M. & Wayne, R. K. (2000) Trends Ecol. Evol. 15, 290-295. [DOI] [PubMed] [Google Scholar]
- 26.Sites, J. W. & Crandall, K. A. (1997) Conserv. Biol. 11, 1289-1297. [Google Scholar]
- 27.Moritz, C. (2002) Syst. Biol. 51, 238-254. [DOI] [PubMed] [Google Scholar]
- 28.Bates, J. M., Hackett, S. J. & Cracraft, J. (1998) J. Biogeogr. 25, 783-793. [Google Scholar]
- 29.Cracraft, J. (1991) Aust. Syst. Bot. 4, 211-227. [Google Scholar]
- 30.Zink, R. M. & McKitrick, M. C. (1995) Auk 112, 701-719. [Google Scholar]
- 31.Wang, Z., Hill, G. E., Baker, A. J. & Edwards, S. V. (2003) Evolution (Lawrence, Kans.) 57, 2852-2864. [DOI] [PubMed] [Google Scholar]
- 32.Tishkoff, S. A. & Kidd, K. K. (2004) Nat. Genet. 36, S21-S27. [DOI] [PubMed] [Google Scholar]
- 33.Moritz, C. (1994) Mol. Ecol. 3, 401-411. [DOI] [PubMed] [Google Scholar]
- 34.Moore, W. S. (1995) Evolution (Lawrence, Kans.) 49, 718-726. [Google Scholar]
- 35.Hudson, R. R. & Coyne, J. A. (2002) Evolution (Lawrence, Kans.) 56, 1557-1565. [DOI] [PubMed] [Google Scholar]
- 36.Baum, D. & Shaw, K. L. (1995) in Experimental and Molecular Approaches to Plant Biosystematics, eds. Hoch, P. C. & Stephenson, A. C. (Missouri Botanical Garden, St. Louis), pp. 289-303.
- 37.Schodde, R. & Mason, I. J. (1999) The Directory of Australian Birds (CSIRO, Canberra, Australia).
- 38.Cracraft, J. (1986) Evolution (Lawrence, Kans.) 40, 977-996. [DOI] [PubMed] [Google Scholar]
- 39.Wakeley, J. & Hey, J. (1997) Genetics 145, 847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ting, C. T., Tsaur, S. C. & Wu, C. I. (2000) Proc. Natl. Acad. Sci. USA 97, 5313-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neigel, J. E. & Avise, J. C. (1986) in Evolutionary Processes and Theory, eds. Karlin, S. & Nevo, E. (Academic, New York), pp. 515-534.
- 42.Nei, M. (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York).
- 43.Hudson, R. R. (1992) Genetics 131, 509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edwards, S. V. & Dillon, M. (2004) Genet. Res. 84, 175-192. [DOI] [PubMed] [Google Scholar]
- 45.Irwin, D. E. (2002) Evolution (Lawrence, Kans.) 56, 2383-2394. [DOI] [PubMed] [Google Scholar]
- 46.Hudson, R. R. & Turelli, M. (2003) Evolution (Lawrence, Kans.) 57, 182-190. [DOI] [PubMed] [Google Scholar]
- 47.Voelker, G. (1999) Evolution (Lawrence, Kans.) 53, 1536-1552. [DOI] [PubMed] [Google Scholar]
- 48.Zink, R. M., Blackwell-Rago, R. C. & Ronquist, F. (2000) Proc. R. Soc. London Ser. B 267, 497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drovetski, S. V., Zink, R. M., Rohwer, S., Fadeev, I. V., Nesterov, E. V., Karagodin, I., Koblik, E. A. & Red'kin, Y. A. (2004) Proc. R. Soc. London Ser. B 271, 545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pereira, S. L. & Baker, A. J. (2004) Auk 121, 682-694. [Google Scholar]
- 51.Chesser, R. T. & Zink, R. M. (1994) Evolution (Lawrence, Kans.) 48, 490-497. [DOI] [PubMed] [Google Scholar]
- 52.Barraclough, T. G. & Vogler, A. P. (2000) Am. Nat. 155, 419-434. [DOI] [PubMed] [Google Scholar]
- 53.Omland, K. E. (1997) Evolution (Lawrence, Kans.) 51, 1636-1646. [DOI] [PubMed] [Google Scholar]
- 54.Johnson, N. K. & Cicero, C. (2002) Mol. Ecol. 11, 2065-2081. [DOI] [PubMed] [Google Scholar]
- 55.Drovetski, S. V. & Ronquist, F. (2003) J. Biogeogr. 30, 1173-1181. [Google Scholar]
- 56.Losos, J. B. & Glor, R. E. (2003) Trends Ecol. Evol. 18, 220-227. [Google Scholar]
- 57.Uy, J. A. C. & Borgia, G. (2000) Evolution (Lawrence, Kans.) 54, 273-278. [DOI] [PubMed] [Google Scholar]
- 58.Rasner, C. A., Yeh, P., Eggert, L. S., Hunt, K. E., Woodruff, D. S. & Price, T. D. (2004) Mol. Ecol. 13, 671-681. [DOI] [PubMed] [Google Scholar]
- 59.Yeh, P. J. (2004) Evolution (Lawrence, Kans.) 58, 166-174. [Google Scholar]
- 60.Irwin, D. E., Irwin, J. H. & Price, T. D. (2001) Genetica 112, 223-243. [PubMed] [Google Scholar]
- 61.Irwin, D. E. (2000) Evolution (Lawrence, Kans.) 54, 998-1010. [DOI] [PubMed] [Google Scholar]
- 62.Irwin, D. E., Bensch, S. & Price, T. D. (2001) Nature 409, 333-337. [DOI] [PubMed] [Google Scholar]
- 63.Patten, M. A., Rotenberry, J. T. & Zuk, M. (2004) Evolution (Lawrence, Kans.) 58, 2144-2155. [DOI] [PubMed] [Google Scholar]
- 64.Slabbekoorn, H. & Smith, T. B. (2002) Evolution (Lawrence, Kans.) 56, 1849-1858. [DOI] [PubMed] [Google Scholar]
- 65.Podos, J. (2001) Nature 409, 185-188. [DOI] [PubMed] [Google Scholar]
- 66.Smith, T. B. (1993) Nature 363, 618-620. [Google Scholar]
- 67.Slabbekoorn, H. & Smith, T. B. (2000) Biol. J. Linn. Soc. 71, 737-753. [Google Scholar]
- 68.Payne, R. B. (1996) in Ecology and Evolution of Acoustic Communication in Birds, eds. Kroodsma, D. E. & Miller, E. H. (Cornell Univ. Press, Ithaca, NY), pp. 198-220.
- 69.Grant, B. R. & Grant, P. R. (1996) Evolution (Lawrence, Kans.) 50, 2471-2487. [Google Scholar]
- 70.Lachlan, R. F. & Servedio, M. R. (2004) Evolution (Lawrence, Kans.) 58, 2049-2063. [DOI] [PubMed] [Google Scholar]
- 71.Slabbekoorn, H. & Smith, T. B. (2002) Philos. Trans. R. Soc. London Ser. B 357, 493-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saetre, G. P., Moum, T., Bures, S., Kral, M., Adamjan, M. & Moreno, J. (1997) Nature 387, 589-592. [Google Scholar]
- 73.Haavie, J., Borge, T., Bures, S., Garamszegi, L. Z., Lampe, H. M., Moreno, J., Qvarnstrom, A., Torok, J. & Saetre, G. P. (2004) J. Evol. Biol. 17, 227-237. [DOI] [PubMed] [Google Scholar]
- 74.Seddon, N. (2005) Evolution (Lawrence, Kans.) 59, 97-112. [Google Scholar]
- 75.de Kort, S. R., den Hartog, P. M. & ten Cate, C. (2002) J. Avian Biol. 33, 150-158. [Google Scholar]
- 76.Grant, P. R. & Grant, B. R. (1997) Am. Nat. 149, 1-28. [Google Scholar]
- 77.Grant, B. R. & Grant, P. R. (2002) Biol. J. Linn. Soc. 76, 545-556. [Google Scholar]
- 78.Grant, B. R. & Grant, P. R. (1998) in Endless Forms: Species and Speciation, eds. Howard, D. J. & Berlocher, S. H. (Oxford Univ. Press, Oxford).
- 79.Grant, P. R., Grant, B. R., Markert, J. A., Keller, L. F. & Petren, K. (2004) Evolution (Lawrence, Kans.) 58, 1588-1599. [DOI] [PubMed] [Google Scholar]
- 80.Payne, R. B., Payne, L. L. & Woods, J. L. (1998) Anim. Behav. 55, 1537-1553. [DOI] [PubMed] [Google Scholar]
- 81.Payne, R. B., Payne, L. L., Woods, J. L. & Sorenson, M. D. (2000) Anim. Behav. 59, 69-81. [DOI] [PubMed] [Google Scholar]
- 82.Sefc, K. M., Payne, R. B. & Sorenson, M. D. (2005) Mol. Ecol., in press. [DOI] [PubMed]
- 83.Via, S. (2001) Trends Ecol. Evol. 16, 381-390. [DOI] [PubMed] [Google Scholar]
- 84.Payne, R. B., Payne, L. L., Nhlane, M. E. D. & Hustler, K. (1993) Proc. VIII Pan-Afr. Ornithol. Congr. 40-52.
- 85.Payne, R. B. & Sorenson, M. D. (2004) Auk 121, 156-161. [Google Scholar]
- 86.Wu, C. I. & Ting, C. T. (2004) Nat. Rev. Genet. 5, 114-122. [DOI] [PubMed] [Google Scholar]
- 87.Coyne, J. A. & Orr, H. A. (1989) in Speciation and Its Consequences, eds. Otte, D. & Endler, J. A. (Sinauer, Sunderland, MA), pp. 180-207.
- 88.Haldane, J. B. S. (1922) J. Genet. 12, 101-109. [Google Scholar]
- 89.Dobzansky, T. (1936) Genetics 21.
- 90.Saetre, G.-P., Borge, T., Lindroos, K., Haavie, J., Sheldon, B. C., Primmer, C. & Syvänen, A.-C. (2003) Proc. R. Soc. London Ser. B 270, 53-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Presgraves, D. C. (2002) Evolution (Lawrence, Kans.) 56, 1168-1183. [DOI] [PubMed] [Google Scholar]
- 92.Tubaro, P. L. & Lijtmaer, D. A. (2002) Biol. J. Linn. Soc. 77, 193-200. [Google Scholar]
- 93.Lijtmaer, D. A., Mahler, B. & Tubaro, P. L. (2003) Evolution (Lawrence, Kans.) 57, 1411-1418. [DOI] [PubMed] [Google Scholar]
- 94.Wu, C. I. & Davis, A. W. (1993) Am. Nat. 142, 187-212. [DOI] [PubMed] [Google Scholar]
- 95.Charlesworth, B., Coyne, J. A. & Barton, N. H. (1987) Am. Nat. 130, 113-146. [Google Scholar]
- 96.Kirkpatrick, M. & Hall, D. W. (2004) Evolution (Lawrence, Kans.) 58, 437-440. [PubMed] [Google Scholar]
- 97.Hurst, L. D. & Ellegren, H. (1998) Trends Genet. 14, 446-452. [DOI] [PubMed] [Google Scholar]
- 98.Countermann, B. A., Ortiz-Barrientos, D. & Noor, M. A. F. (2004) Evolution (Lawrence, Kans.) 656-660. [PubMed]
- 99.Betancourt, A. J., Presgraves, D. C. & Swanson, W. J. (2002) Mol. Biol. Evol. 1816-1819. [DOI] [PubMed]
- 100.Kahn, N. W. & Quinn, T. W. (1999) J. Mol. Evol. 49, 750-759. [DOI] [PubMed] [Google Scholar]
- 101.Ellegren, H. & Fridolfsson, A. K. (1997) Nat. Genet. 17, 182-184. [DOI] [PubMed] [Google Scholar]
- 102.Carmichael, A. N., Fridolfsson, A. K., Halverson, J. & Ellegren, H. (2000) J. Mol. Evol. 50, 443-447. [DOI] [PubMed] [Google Scholar]
- 103.Bartosch-Harlid, A., Berlin, S., Smith, N. G., Moller, A. P. & Ellegren, H. (2003) Evolution (Lawrence, Kans.) 57, 2398-2406. [DOI] [PubMed] [Google Scholar]
- 104.Axelsson, E., Smith, N. G., Sundstrom, H., Berlin, S. & Ellegren, H. (2004) Mol. Biol. Evol. 21, 1538-1547. [DOI] [PubMed] [Google Scholar]
- 105.Servedio, M. R. & Sætre, G. P. (2003) Proc. R. Soc. London Ser. B 270, 1473-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grant, P. R. & Grant, B. R. (1992) Science 256, 193-197. [DOI] [PubMed] [Google Scholar]
- 107.Good, T. P., Ellis, J. C., Annett, C. A. & Pierotti, R. (2000) Evolution (Lawrence, Kans.) 54, 1774-1783. [DOI] [PubMed] [Google Scholar]
- 108.Reinhold, K. (1998) Behav. Ecol. Sociobiol. 44, 1-7. [Google Scholar]
- 109.Wang, P. J., McCarrey, J. R., Yang, F. & Page, D. C. (2001) Nat. Genet. 27, 422-426. [DOI] [PubMed] [Google Scholar]
- 110.Birkhead, T. R. & Møller, A. P. (1992) Sperm Competition in Birds; Evolutionary Causes and Consequences (Academic, London).
- 111.Swanson, W. J. & Vacquier, V. D. (2002) Nat. Rev. Genet. 3, 137-144. [DOI] [PubMed] [Google Scholar]
- 112.Ceplitis, H. & Ellegren, H. (2004) Mol. Biol. Evol. 21, 249-254. [DOI] [PubMed] [Google Scholar]
- 113.Lee, Y. H. & Vacquier, V. D. (1992) Biol. Bull. (Woods Hole, MA) 182, 97-104. [DOI] [PubMed] [Google Scholar]
- 114.Swanson, W. J., Nielsen, R. & Yang, Q. (2003) Mol. Biol. Evol. 20, 18-20. [DOI] [PubMed] [Google Scholar]
- 115.Swanson, W. J., Yang, Z., Wolfner, M. F. & Aquadro, C. F. (2001) Proc. Natl. Acad. Sci. USA 98, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kingan, S. B., Tatar, M. & Rand, D. M. (2003) J. Mol. Evol. 57, 159-169. [DOI] [PubMed] [Google Scholar]
- 117.Swanson, W. J., Clark, A. G., Waldrip-Dail, H. M., Wolfner, M. F. & Aquadro, C. F. (2001) Proc. Natl. Acad. Sci. USA 98, 7375-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wolfner, M. F. (1997) Insect Biochem. Mol. Biol. 27, 179-192. [DOI] [PubMed] [Google Scholar]
- 119.Dorus, S., Evans, P. D., Wyckoff, G. J., Choi, S. S. & Lahn, B. T. (2004) Nat. Genet. 36, 1326-1329. [DOI] [PubMed] [Google Scholar]
- 120.Adkins-Reagan, E. (1999) Auk 116, 184-193. [Google Scholar]
- 121.Palumbi, S. R. (1999) Proc. Natl. Acad. Sci. USA 96, 12632-12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gowaty, P. A. (1996) in Partnerships in Birds, ed. Black, J. M. (Oxford Univ. Press, Oxford), pp. 21-52.
- 123.Rice, W. R. (1996) Nature 381, 232-234. [DOI] [PubMed] [Google Scholar]
- 124.Gavrilets, S. (2000) Nature 403, 886-889. [DOI] [PubMed] [Google Scholar]
- 125.Parker, P. G. & Burley, N. T. (1998) Avian Reproductive Tactics: Female and Male Perspectives (Am. Ornithol. Union, Washington, DC).
- 126.Jennions, M. D. & Petrie, M. (2000) Biol. Rev. 75, 21-64. [DOI] [PubMed] [Google Scholar]
- 127.Edwards, S. V., Jennings, W. B. & Shedlock, A. M. (2005) Proc. R. Soc. London Ser. B, in press.