Abstract
Most plant viruses rely on the production of subgenomic RNAs (sgRNAs) for the expression of their genes and survival in the plant. Although this is a widely adopted strategy among viruses, the mechanism(s) whereby sgRNA production occurs remains poorly defined. Turnip yellow mosaic tymovirus (TYMV) is a positive-stranded RNA virus that produces an sgRNA for the expression of its coat protein. Here we report that the subgenomic promoter sequence of TYMV is located on a 494-nucleotide fragment, containing previously identified highly conserved sequence elements, which are shown here to be essential for promoter function. After duplication, the subgenomic promoter can be inserted into the coat protein open reading frame, giving rise to the in vivo production of a second sgRNA. It is suggested that this promoter can function when contained on a different molecule than viral genomic RNA. This interesting trait may be of general use for plant and plant virus research.
The genomes of the vast majority of positive-stranded RNA viruses contain more than one gene. Synthesis of internal genes is achieved by one or several strategies, including the formation of subgenomic RNAs (sgRNAs), internal translation initiation, leaky scanning, frameshift, and readthrough. Of these, the production of sgRNAs is by far the most common strategy used by viruses. Indeed, among positive-stranded RNA viruses of plants, only viruses of the Potyviridae and Comoviridae families and the sequiviruses of the Sequiviridae family do not resort to this strategy (reviewed in reference 29).
Synthesis of sgRNA by internal initiation of transcription on promoters located on the complementary minus-strand RNA of genomic length is well documented for plant viruses such as brome mosaic bromovirus (BMV) (18), turnip yellow mosaic tymovirus (TYMV) (10), alfalfa mosaic ilarvirus (A1MV) (26), cucumber mosaic cucumovirus (CMV) (4), beet necrotic yellow vein benyvirus (BNYVV) (2), and turnip crinkle carmovirus (TCV) (27, 28). For certain viruses such as potato potexvirus X (15) and tomato bushy stunt tombusvirus (31), internal initiation has been shown to also depend on interaction between distantly located cis regions and the promoter region in the genome. On the other hand, in red clover necrotic mosaic dianthovirus (24), sgRNA synthesis appears to occur by premature termination during minus-strand synthesis, with subsequent sgRNA production from the truncated nascent RNA. An interesting feature of this termination process is that it depends on trans activation between the two RNA components that make up the viral genome.
The promoters for the synthesis of several sgRNAs (sg promoters) have been examined. Defining such regions has relied on several methods including site-specific mutations, duplication of the hypothetical promoter region followed by deletion mutations, RNA sequence and/or structure predictions, phylogenetic sequence comparisons, and the use of chemical and enzymatic probes. Frequently, sg promoters are composed of more than one region: a core promoter and one or more enhancer elements, which most frequently overlap the transcription start site. The boundaries of the core promoter and enhancer regions appear to vary depending on the viral genome and on whether the promoter is examined in vitro or in vivo.
The sg promoters of A1MV and BMV have been examined in detail, and those of CMV and BNYVV RNA 3 have been delineated (reviewed in reference 17). In the latter case, the sg promoter is located mainly downstream of the transcription start site. The sg promoter of cucumber necrosis necrovirus is unique in so far as a core region of 26 nucleotides (nt) spanning the transcription start site suffices for the production of wild-type (wt) levels of sgRNA 2 in vivo (14). More recently, the sg promoters of TCV have been defined, and the structural requirements for the production of the larger sgRNA have been established (27, 28).
TYMV, the type member of the tymoviruses, contains a monopartite single-stranded positive sense RNA of 6,318 nt. Its genome bears a cap structure at its 5′ end and a tRNA-like structure that can be valylated in vitro and in vivo at its 3′ end. It contains three open reading frames (ORFs). The genomic RNA codes for a 206-kDa polyprotein and for a 69-kDa movement protein whose ORF nearly totally overlaps that of the polyprotein. The polyprotein possesses a methyltransferase, a proteinase, a nucleoside triphosphatase/helicase, and an RNA-dependent RNA polymerase (RdRp) domain. It undergoes autocatalytic processing yielding a 140-kDa and a 66-kDa product, the latter containing the RdRp domain (22). The coat protein (CP), whose ORF is 3′ coterminal on the genomic RNA, is synthesized via an sgRNA of 694 nt in which the CP ORF is 5′ proximal. The sgRNA which is capped is contained within the virus particles (16, 21). The transcription initiation site for the sgRNA has been mapped to position 5625 on the genomic RNA (11). It thus overlaps the end of the RdRp ORF, whose termination triplet occupies positions 5627 to 5629.
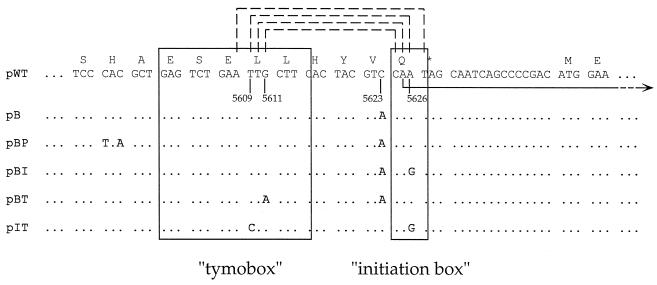
A close examination of the region surrounding the initiation site for sgRNA synthesis in TYMV RNA and in 14 other tymovirus genomes clearly demonstrated the presence of two highly conserved sequence blocks in a region of about 40 nt designated the tymobox region (8). The 5′-terminal block, the tymobox itself, is 16 nt long and is identical in 11 of the 15 tymovirus sequences compared. It is followed, 7 or 8 nt downstream, by the 4-nt initiation box (Fig. 1). In the three tymovirus genomes for which such information is available, initiation of sgRNA synthesis commences in the initiation box at the triplet AAU and is preceded by a C residue. The sequence CAAU is common to 13 of the 15 tymovirus RNA sequences examined. It has thus been proposed that the two conserved sequence motifs may function as an sg promoter (8). This would imply that transcriptional control regions overlap coding regions.
FIG. 1.
Construction of plasmids with mutations in the tymobox region of the TYMV cDNA. The relevant sequence of the plasmid carrying the wt cDNA (pWT) is displayed. Numbers below the sequence refer to the TYMV genome. Above the sequence, the encoded amino acids are indicated in the one-letter code. The sgRNA is represented by a bent arrow starting at nt 5625. The two regions of high conservation, the tymobox and the initiation box, are boxed. Potential base pairing of the nucleotides of the initiation box with 4 nt of the tymobox is indicated by dashed lines above the sequence. For mutant plasmids, identical nucleotides are represented by dots, and mutated nucleotides are indicated.
In this report we define the elements that compose the TYMV sg promoter. Two main approaches were adopted. First, mutations in the conserved sequences were introduced into the full-length infectious TYMV transcript. Second, a large segment as well as truncated versions of the viral RNA encompassing the conserved sequences were introduced within the CP ORF in the infectious transcript. The effects of these modifications were examined in vivo in Arabidopsis thaliana protoplasts by Western and Northern blot analyses. Thereby, we established that the highly conserved tymobox and initiation box are indeed part of the sg promoter and that they are essential for promoter function. Moreover, the promoter can operate ectopically and in trans. These features make the sg promoter of TYMV an attractive candidate for the development of a vector for the model plant A. thaliana.
MATERIALS AND METHODS
Plasmid constructions and probes.
All plasmids (except pFF19G and pCP) were derivatives of pTYFL84 (renamed here pWT [Fig. 1]), a TYMV cDNA clone from which infectious genome-length transcripts can be obtained with T7 RNA polymerase (5). All oligonucleotides used are listed in Table 1. To introduce point mutations into the tymobox region of the TYMV cDNA, subclones of pWT were constructed in pBluescript II SK(+) or pUC18. Subclone pSC1 contained a 1,980-bp SmaI/SalI fragment of pWT, while subclone pSC2 contained a 664-bp PstI/SmaI fragment of pSC1. Point mutations were introduced into pSC2 using a Clontech Transformer site-directed mutagenesis kit or a Pharmacia Biotech U.S.E. mutagenesis kit and appropriate oligonucleotides. The point mutations were then introduced into pSC1 by replacing the 664-bp PstI/XmaI fragment of pSC1 by those of the pSC2 derivatives before being introduced into pWT by replacing the 1,980-bp XmaI/SalI fragment of pWT by those of the pSC1 derivatives.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequencea (5′-3′) | Locationb | Constructc |
|---|---|---|---|
| o1 | CAGGAAAGAAGATCTGAGCAAAAG | pB, pBI, pBT | |
| o2 | TGCTTCACTACGTACAATAGC | 5610–5630 | pB |
| o3 | CACTACGTACAGTAGCAATC | 5615–5634 | pBI |
| o4 | GAATTACTTCACTACGTACAATAGC | 5606–5630 | pBT |
| o5 | GCTTTGACTTCTTCTGCAG | 5388–5406 | pBP |
| o6 | CTATTGTACGTAGTGAAGCAATTCAGACTCAGCTTAGGAAACCTCG | 5584–5629 | pBP |
| o7 | CTGTGACTGGTGACGCGTCAACCAAGTC | pIT | |
| o8 | CGCTGAGTCTGAACTGCTTCACTACGTCCAGTAGCAATCAGCC | 5596–5638 | pIT |
| o9 | CCTGAAACCCGGGTTCAGTATCAGTCAGCCTGC | 5357–5389 | pLs, pLa |
| o10 | GTGACCCGGGGTGATTCCAGAGATGCATGACG | 5842–5873 | pLs, pLa |
| o11 | GACTCCACACCCGGGCTCAGC | 5441–5461 | pMs, pMa |
| o12 | GTGATGGCCCGGGGACAGCTG | 5705–5725 | pMs, pMa |
| o13 | CTTCCCGGGATCCATCGAGGTTTC | 5569–5592 | pSs, pSa |
| o14 | GGTCTTCCCGGGCGAGTTCTTTG | 5655–5677 | pSs, pSa |
| o15 | CAGCGGATCCATGGAAATCGACAAAGAACTCGC | 5650–5666 | pCP |
| o16 | GCAAGATCCTGCAGTTAGGTGGAAGTGTCCGTG | 6195–6213 | pCP |
| o17 | GCTTCACGAGCTCCAATAGCAATCAGCC | 5611–5638 | pProbe |
| o18 | GGCGGGTACCCAGCAGACACC | 5917–5937 | pProbe |
Restriction sites are underlined, mutated nucleotides are in bold, and vector nucleotides are in italics.
Position on the TYMV genome.
Name code: p, plasmid; B, contains SnaBI site; I, mutation in initiation box; T, mutation in tymobox; P, contains premature stop codon in RdRp ORF; L, large insert; s, sense orientation; a, antisense orientation; M, middle-sized insert; S, small insert.
To produce pB, the selection primer o1 and the mutagenic oligonucleotide o2, which introduced a C5623A mutation to generate a SnaBI site, were used. For the construction of pBI, we used o3, which introduced the C5623A and A5626G mutations. Likewise, to generate pBT, we used o4, which introduced the C5623A and G5611A mutations. For the construction of pBP, with two mutations upstream of the tymobox introducing a stop codon into the RdRp ORF, the selection primer o5 was used together with o6, which introduced a total of three point mutations into the wt viral cDNA, one of which is a stop codon in the viral RdRp ORF. To construct pIT, the selection primer o7 was used together with o8, which introduced the A5626G and T5609C mutations (Fig. 1).
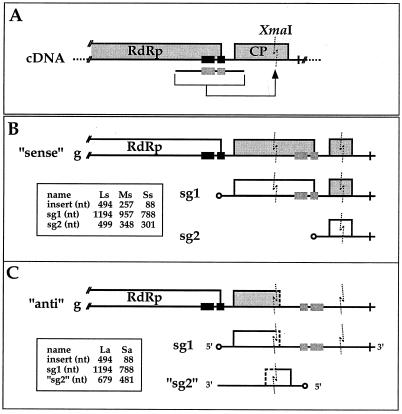
To construct mutants with a duplicated sg promoter, the sg promoter region was amplified by PCR and inserted into the XmaI restriction site of pWT (see Fig. 4). For the construction of pLs and pLa, a large region corresponding to nt 5357 to 5873 of the TYMV cDNA was amplified by PCR using pB along with o9 and o10, which engineered XmaI sites in the PCR product. The PCR fragment was inserted into the XmaI site of pWT in either the sense (pLs) or antisense (pLa) orientation. Likewise, pMs contained the middle-sized PCR fragment corresponding to nt 5441 to 5725 of the TYMV cDNA obtained with o11 and o12 in the sense orientation, while a small PCR product corresponding to nt 5569 to 5677 of the TYMV cDNA and obtained with o13 and o14 was introduced in the sense (pSs) or antisense (pSa) orientation.
FIG. 4.
Construction of mutants carrying a duplicated sg promoter region. (A) Mutants were constructed by amplification of a fragment containing the tymobox region (black or grey stippled rectangles) as the central element and insertion of the fragment at the single XmaI site (6059 to 6064; dashed zigzag line) within the CP ORF of the TYMV cDNA. Dashed bar, plasmid DNA. (B) Schematic representation of the 3′ end of the genomic RNA of mutants tLs, tMs, and tSs, with fragments of 494, 257, and 88 nt, respectively, containing the tymobox region inserted in the sense orientation into the CP ORF. The native sg promoter (black rectangles) recognized on the genomic minus strand leads to the production of sgRNA 1 (sg1). The duplicated sg promoter (grey stippled rectangles), also recognized on the genomic minus strand, leads to the production of a second sgRNA (sg2). (C) Schematic representation of the 3′ end of the genomic RNA of mutants tLa and tSa, with fragments of 494 and 88 nt, respectively, containing the tymobox region inserted in the antisense orientation. The sgRNA 1 (sg1) is produced from the native sg promoter located on the genomic minus strand, while a second small RNA (“sg2”) is produced from the duplicated promoter on sgRNA 1. Expressed (open boxes) and unexpressed (grey boxes) ORFs are shown. In chimeric ORFs, amino acid sequences with no viral equivalent (C) are indicated by partially dashed boxes. (B) Coding capacity of the RNAs of mutant tLs, the only mutant for which the interrupted ORF and the introduced ORF happen to be in frame. The proteins expressed from sgRNAs 1 and 2 of tLs are an N-terminal CP–C-terminal RdRp and an N-terminal CP–C-terminal CP fusion protein (Table 2). For mutants tMs and tSs, the CP ORFs are not in frame with other viral ORFs. (C) Coding capacity of the RNAs of mutant tLa. Open circle, cap structure; cross, tRNA-like structure; insets, sizes of the fragments introduced and of the resulting sgRNAs. Diagrams are not to scale.
For the construction of pCP containing the CP gene downstream of the cauliflower mosaic caulimovirus (CaMV) 35S promoter, pFF19G was cut with PstI and religated after dilution to yield pFF19 (25). The gene for the CP ORF was amplified by PCR from pWT using o15 and o16, which introduced a BamHI site upstream of the ATG codon and a PstI site downstream of the termination codon of the CP ORF. The PCR fragment was ligated into the BamHI and PstI sites of pFF19 to yield pCP.
For the production of digoxygenin-labeled probes used for Northern analyses, plasmid pProbe was constructed by inserting a 327-bp PCR fragment, obtained with o17 and o18 from pUCSX-WT, into the SacI and KpnI sites of pBluescript II SK(+). The probe to detect viral plus strand RNA was prepared by in vitro transcription of a 387-bp BssHII fragment of pProbe with T7 RNA polymerase in the presence of digoxigenin-UTP as described previously (23). It hybridized to nt 5622 to 5934 of the viral genomic RNA and was thus suitable for detection of the viral genomic plus-strand RNA and the sgRNA. The probe to detect viral minus strand corresponded to nt 5622 to 5934 of the viral genome and was similarly produced, except that a 353-bp KpnI/BssHII fragment of pProbe was used as template for transcription by T3 RNA polymerase. This probe did not hybridize to viral plus-strand RNA but showed extensive hybridization to 18S and 25S rRNAs. All final plasmid constructs were verified by sequencing using an ABI Prism 310 automatic sequencer (Perkin-Elmer).
In vitro transcription.
Plasmid constructs containing the full-length TYMV cDNA were linearized by AgeI and used for in vitro transcription with T7 RNA polymerase (New England Biolabs) under standard conditions in the presence of 2 mM each ATP, CTP, and UTP, 0.1 mM GTP, and 1 mM m7GpppG. After 25 min at 37°C, GTP was added to 2 mM and incubation continued for 35 min. Transcripts were not further purified if used for transfection experiments. To determine the RNA concentration, the samples were treated with RQ1 DNase (Promega) and purified using a Qiagen RNeasy Mini kit; the concentration was estimated by absorption at 260 nm or by comparison to a standard on an ethidium bromide-stained agarose gel.
Protoplast and plant inoculations.
Protoplasts of A. thaliana ecotype Columbia were prepared from a cell suspension culture (1) and transfected using polyethylene glycol as a mediating agent and 2 to 4 μg of the in vitro transcripts or 100 ng of viral RNA per 106 protoplasts as described elsewhere (23). For coinfection experiments, 10 μg of circular pCP was added to the protoplasts prior to addition of the transcripts. For plant inoculations, 2-μg aliquots of transcripts were rubbed on two young half leaves each of 2- to 3-week-old Chinese cabbage (Brassica pekinensis cv. Granaat) plants, using carborundum as an abrasive.
Tissue extraction; Western and Northern blotting.
Transfected protoplasts were harvested after 42 h and analyzed as described elsewhere (23), using 2 × 104 or 1 × 106 protoplasts for Western or Northern blotting, respectively. The equivalent of 2 × 103 protoplasts was loaded onto sodium dodecyl sulfate (SDS)-polyacrylamide (12%) gels for Western blot analyses, and 1 μg of total RNA was loaded onto agarose-formaldehyde gels for Northern blot analyses (23).
Plant tissues (50 to 100 mg) were harvested 3 weeks after inoculation, when wt symptoms were well developed, from young expanding leaves above the inoculated leaf or from the inoculated leaf of the half-side that had been inoculated or the half-side that had not been inoculated. Plant tissues were ground in liquid nitrogen and further ground in 200 μl of tissue extraction buffer (6), vortexed, and centrifuged in a tabletop centrifuge. For Western blot analyses, 95-μl aliquots of the tissue samples were heated to 95°C for 5 min. Aliquots of 20 μl were loaded onto SDS-polyacrylamide gels for mutants that did not show systemic infection, whereas 2 to 10 μl of a 1:100 dilution was loaded for mutants producing systemic symptoms. Another 95-μl aliquot of the tissue samples was subjected to two phenol (pH 4.5)-chloroform-isoamyl alcohol (25:24:1) extractions, and the nucleic acids were recovered by ethanol precipitation. For Northern blot analyses, 1 μg of total RNA was loaded onto agarose-formaldehyde gels.
Secondary structure prediction.
The secondary structure of the sg promoter region was predicted by analyzing a sequence of 500 nt (corresponding to nt 5367 to 5866 of the TYMV genome) of the minus-strand RNA sequence with the program mfold version 3.0 (32, 33) without constraint and using default values.
RESULTS
Replication of mutants with altered sequences in the tymobox region.
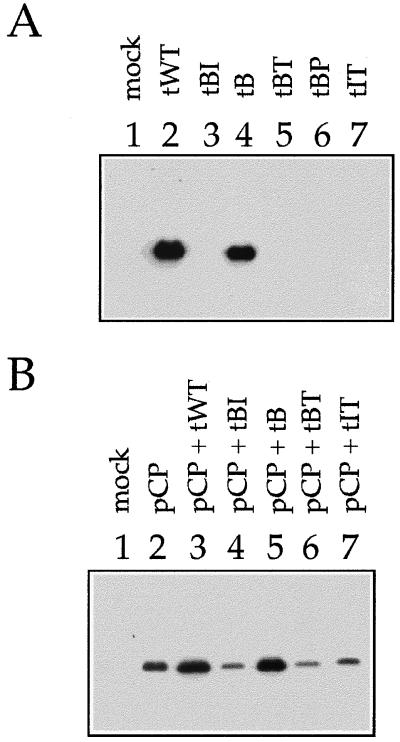
To examine the possible double role of the tymobox region as a coding region for the RdRp and as a regulatory control element for the production of the sgRNA, pBP (Table 1) was constructed by site-directed mutagenesis of wt TYMV cDNA (pWT). This mutant contains a termination codon upstream of the conserved tymobox (Fig. 1). This mutation would presumably not interfere with sgRNA regulation, since it modifies none of the highly conserved nt (nt 5600 to 5615 and 5624 to 5627) but results in an RdRp lacking the 11 terminal amino acids. Western blot analysis of A. thaliana protoplasts transfected with capped full-length transcripts of pBP (tBP) showed that this mutant did not produce CP (Fig. 2A). To determine whether lack of CP production was due to a defect in replication or in sgRNA production, total RNA was extracted from transfected protoplasts and subjected to Northern blot analyses. Accumulation of viral RNA products (subgenomic, genomic plus strand, or genomic minus strand) was not detected for this mutant (Fig. 3), presumably because of the production of an inactive RdRp, establishing the importance of the coding potential of the tymobox region.
FIG. 2.
Western blot analysis of total protein extracts of transfected protoplasts using polyclonal CP antibodies. (A) Protoplasts were treated with water (mock) or with mutant transcripts as indicated. (B) Protoplasts were treated with water, with pCP alone, or with pCP in combination with mutant transcripts as indicated. Samples were harvested 42 h after transfection. Protoplasts (4 × 103) were loaded on each lane of an SDS-polyacrylamide (12%) gel.
FIG. 3.
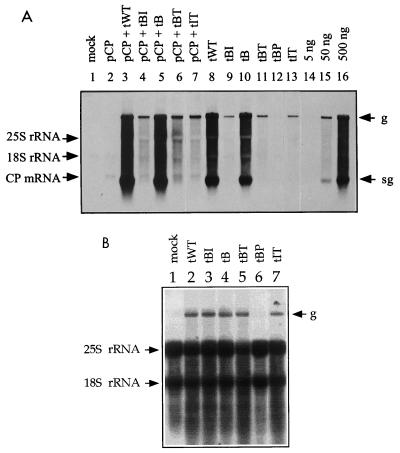
Northern blot analyses of total RNA isolated from transfected protoplasts, using a probe to detect viral plus-strand (A) or minus-strand (B) RNAs. (A) Protoplasts were either mock inoculated (lane 1), transfected with pCP (lane 2), cotransfected with pCP and mutant transcripts (lanes 3 to 7), or transfected with mutant transcripts (lanes 8 to 13) as indicated above the lanes. In lanes 14 to 16, increasing amounts of TYMV RNA were loaded on the gel as indicated. The position of the CP mRNA derived from transcription from pCP is indicated on the left; positions of the TYMV genomic (g; 6,318 nt) and subgenomic (sg; 694 nt) RNAs are indicated on the right. The detection limit of viral RNA is about 5 ng. (B) Protoplasts were either mock inoculated (lane 1) or transfected with mutant transcripts as indicated. The position of the genomic minus strand is indicated on the right. The probe designed to detect viral minus-strand RNAs shows extensive hybridization to rRNAs but does not hybridize to sgRNA. In both panels, positions of the A. thaliana 25S rRNA (3,375 nt) and 18S rRNA (1,804 nt) are indicated on the left.
To examine the possible role of the tymobox region for the control of sgRNA production, we constructed mutants that contained single nucleotide exchanges in one of the conserved boxes and/or in the less well conserved region between the boxes, such that the nature of the encoded amino acid was not changed (Fig. 1). Plasmid pB contained a C5623A change located between the tymobox and the initiation box. This mutation introduced a SnaBI site that was used for screening purposes and was also present in pBP, pBI, and pBT. Additionally, pBI contained a mutation in the initiation box (A5626G), and pBT had a mutation in the tymobox (G5611A) (Fig. 1). The mutant transcripts (tB, tBI, and tBT) were tested for the ability to replicate in A. thaliana protoplasts. Western blot analyses of protoplast extracts showed that tB supported CP accumulation to wt (tWT) levels, while tBI and tBT did not lead to detectable amounts of CP (Fig. 2A). Northern blot analyses confirmed that tB accumulated both genomic RNA and sgRNA to wt levels (Fig. 3A, compare lanes 10 and 8). In contrast, tBI and tBT did not lead to the accumulation of sgRNA, and the amount of genomic plus strand was reduced (Fig. 3A). Nevertheless, tBI and tBT, as well as tB, produced genomic minus strand to wt levels (Fig. 3B).
To determine whether the deleterious effect of the mutation in pBI was caused by disruption of possibly essential hybridization between the 4 nt of the initiation box and 4 nt (5608 to 5611) of the tymobox (Fig. 1), pIT was constructed. This mutant contains the A5626G mutation of pBI and a T5609C mutation in the tymobox that could restore possible hybridization between these conserved regions (Fig. 1). Accumulation of CP could not be detected in protoplasts transfected with tIT (Fig. 2A). RNA analysis showed that for tIT, as for tBI, genomic plus strand was produced, but at lower than wt levels (Fig. 3A), while virtually wt quantities of genomic minus strand were detected (Fig. 3B). Transcript tIT did not support the production of sgRNA (Fig. 3A), indicating that the T5609C mutation did not complement the defect of tBI caused by the A5626G mutation.
We tested wt and mutant transcripts for replication in Chinese cabbage plants. Only tWT and tB induced local and systemic symptoms; tBI, tBT, and tIT failed to produce symptoms on infected plant leaves or young leaves located above the infected leaf (not shown). The presence of CP and viral plus-strand RNAs were searched for in different parts of the infected plants. For tWT and tB, viral CP as well as genomic and subgenomic RNAs were detected in all samples; for tBI, tBT, and tIT, none of these viral products were found (not shown).
Complementation of CP-deficient mutants.
The level of viral genomic plus strand in transfected protoplasts was reduced for those mutants (tBI, tBT, and tIT) that did not produce sgRNA and hence did not produce CP. This phenomenon has been described by Bransom et al. (6), whose full-length TYMV mutant transcript no longer coded for the CP. This mutant produced genomic minus strand to wt levels in turnip protoplasts along with reduced levels of genomic plus strand and sgRNA. The authors concluded that the CP did not play an essential role in viral replication but appeared to influence the accumulation of plus-strand genomic and subgenomic RNAs.
To test whether this hypothesis was applicable to our mutants, the effect of the presence of CP on the level of genomic plus strand for mutants tBI, tBT, and tIT was investigated. The plasmid used, pCP, contained the TYMV CP ORF downstream of the CaMV 35S promoter and upstream of the 35S polyadenylation signal and, when introduced into plant cells, leads to expression of TYMV CP. A. thaliana protoplasts were cotransfected with pCP and mutant transcripts, harvested 42 h after infection, and analyzed by Western blotting (Fig. 2B). In all samples transfected with pCP, the presence of TYMV CP could be detected. Its level was increased for those samples that were cotransfected with tWT and tB, transcripts that supported CP production even in the absence of pCP (Fig. 2A), but was reduced for samples cotransfected with tBI, tBT, or tIT (Fig. 2B). Northern blot analyses confirmed the presence of the CP mRNA derived from pCP in all samples transfected with pCP (Fig. 3A). Production of both genomic and subgenomic RNAs seemed to be increased for tWT and tB in the presence of pCP (Fig. 3A, compare lanes 8 and 10 with lanes 3 and 5). Transcripts tBI, tBT, and tIT did not support the production of sgRNA, whether in the presence or in the absence of pCP. In contrast, the level of genomic RNA in protoplasts transfected with tBI, tBT, and tIT was clearly increased in the presence of pCP but not to wt levels (Fig. 3A).
Replication of mutants with a duplicated sg promoter.
To identify the sg promoter on the genomic sequence and test whether it remained functional outside its native environment, a fragment containing the tymobox region as the central element was amplified by PCR and introduced into the single XmaI site of pWT, located within the CP ORF (Fig. 4A).
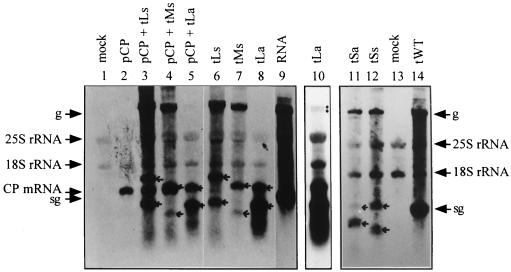
Plasmid pLs contained the potential sg promoter region of 494 nt (nt 5370 to 5863 of the TYMV genome) introduced into the XmaI site in the sense orientation (Fig. 4B). Transfection of A. thaliana protoplasts with the mutant transcript tLs resulted in the production of viral genomic RNA of plus sense (Fig. 5) and minus sense (not shown) and two small RNA species, which corresponded in migration position to the expected sizes of 1,194 and 499 nt for sgRNAs 1 and 2, respectively (Fig. 4B and 5). The two sgRNAs were produced in similar amounts, indicating that the introduced 494-nt region contains all elements required for the production of sgRNA and that the sg promoter of TYMV can be functional when not in its natural environment.
FIG. 5.
Northern blot analysis of total RNAs isolated from transfected protoplasts using a probe to detect viral plus-strand RNAs. Notations are as in Fig. 3. Arrows indicate positions of the mutant sgRNAs. Lanes 1 to 10 are from a different gel than lanes 11 to 14. In lane 9, protoplasts were transfected with TYMV RNA. Lane 10 shows the blot of lane 8 after longer exposure of the film. The position of the doublet band is indicated by two dots on the right side of the lane. In lanes 11 and 12, the sgRNAs show unusually strong signals.
To determine the minimal sequence required for sgRNA synthesis, two other mutants (pMs and pSs) were constructed so as to contain shorter regions inserted in the CP ORF (Fig. 4B). Plasmid pMs contained a fragment of 257 nt (nt 5456 to 5712 of the TYMV genome), while plasmid pSs contained a fragment of 88 nt (nt 5578 to 5665). Both fragments contained the tymobox region and were inserted into the CP ORF of TYMV in the sense orientation. Northern blot analyses of total RNA extracts of transfected protoplasts revealed that tMs and tSs supported genomic plus-strand RNA and sgRNA synthesis (Fig. 5) as well as genomic minus-strand RNA synthesis (not shown). Infection with tMs resulted in the production of two small RNA species of the expected sizes of 957 and 348 nt (Fig. 5), which represent the sgRNAs produced from the native sg promoter (sgRNA 1) and from the sg promoter introduced into the CP ORF (sgRNA 2), respectively. The level of sgRNA 2 was much lower than that of sgRNA 1, indicating that the 257-nt insert of pMs still retained the information for sgRNA synthesis but either lacked essential enhancing elements or contained elements (e.g., structures) inhibitory for efficient sgRNA production. Infection with tSs also resulted in the production of small RNA species corresponding to sgRNAs of the expected sizes of 788 and 301 nt for sgRNAs 1 and 2, respectively (Fig. 5). Apparently, the 88-nt fragment inserted into the CP ORF in pSs still contained the information necessary for the production of sgRNA but seemed to be somewhat less efficient than the 494-nt fragment of pLS.
Two of the mutants constructed contained the tymobox region on a fragment of 494 nt (pLa) or of 88 nt (pSa) inserted into the CP ORF in the antisense orientation (Fig. 4C). Replication of these mutants in protoplasts showed that for tLa, genomic RNA production could not be detected. Surprisingly, however, it supported the production of two small RNA species, which could be detected with both viral plus-strand (Fig. 5) and viral minus-strand (not shown) probes. The larger of these two RNA species (sgRNA 1) presumably derives from transcription from the native sg promoter and should therefore be 1,194 nt long. However, it migrates as a smaller RNA (Fig. 5, lanes 8 and 6; compare the 1,194-nt sgRNA 1 bands of tLa and tLs), possibly because of stable undenatured secondary structures (3). Since the 494-nt fragment was inserted in the antisense orientation, sgRNA 1 of tLa should be able to fold into a stable structure with a stem of 239 perfect base pairs by virtue of base complementarity. The second small RNA that is produced in protoplasts transfected with tLa and hybridizes to plus- and minus-strand probes (Fig. 5 and data not shown) could have resulted from transcription using the introduced sg promoter on sgRNA 1 as the template (Fig. 4C). This RNA (designated “sg2” in Fig. 4C) should be 679 nt long and thus almost as long as the wt sgRNA (694 nt). However, like sgRNA 1, it migrates as a smaller RNA (Fig. 5, compare lanes 8 and 9) presumably for the same reasons, since also in sg2 the 239-bp stem can form and might influence migration. The sg2 of tLa was produced in higher quantities than sgRNA 1, which also suggests that sg2 is a transcription product from sgRNA 1 rather than from genomic RNA. The sgRNAs 2 of tLs, tMs, and tSs, which are transcription products from genomic RNA, are produced in the same or lower amounts than the sgRNAs 1 of these mutants. However, in the absence of detailed product characterization, it cannot be excluded that the second small RNA could be a specific degradation product of the first.
Infection of protoplasts with tSa containing the 88-nt sg promoter sequence inserted into the CP ORF in the antisense orientation also led to the accumulation of two small RNA species (Fig. 5). The larger one, produced to barely detectable levels, was derived by transcription from the native sg promoter and had the expected size of 788 nt, as does sgRNA 1 produced by tSs (Fig. 5, compare sgRNA 1 bands of lanes 11 and 12). The smaller RNA species (sg2) had the expected size of 481 nt and was produced by transcription from the sg promoter situated in the inserted fragment on sgRNA 1 (Fig. 4C). As in tLa, the amount of sg2 was higher than that of sgRNA 1 (Fig. 5). However, detection of these small RNA species with the probe to reveal viral minus-strand RNA was inconclusive (not shown).
Infection of protoplasts with tLa initially did not seem to lead to the production of genomic RNA (Fig. 5, lane 8). This deficiency in genome replication of tLa could have been caused by extensive secondary structure formation due to the antisense insert. Upon longer exposure of the Northern blot, a faint doublet band was visible at the migration position of the genomic RNA (Fig. 5, lane 10), which could also be detected with the probe against the minus strand (not shown). The more slowly migrating RNA possibly represented viral genomic plus strand of 6,818 nt produced by tLa, which should also hybridize to both probes since it contained the 494-nt duplication in the antisense orientation. The slightly faster migrating RNA species of the doublet might have arisen by transcription from the second sg promoter, located on the genomic plus strand in the antisense orientation. This would lead to a mainly minus-sense RNA of 6,303 nt that should hybridize to both probes.
Likewise, infection of protoplasts with tSa led to the production of full-length genomic RNA of 6,412 nt and also to a slightly smaller RNA species that hybridized to both probes (Fig. 5 and data not shown). Like for tLa, this faster-migrating RNA species of genomic length might have been the result of transcription from the second sg promoter, located on the 88-nt fragment inserted in the antisense orientation. This would lead to an RNA of 6,095 nt of largely minus sense. In contrast to the situation in tLa, where both large RNA species are produced to comparable amounts, in tSa the 6,095-nt RNA species is produced to lower levels than the genomic RNA (Fig. 5). This is in accordance with the observation that the 88-nt sg promoter is functional, but not to wt levels.
Influence of the CP on genome replication of mutants with a duplicated sg promoter.
The level of genomic plus-strand RNA produced by most mutants with a duplicated sg promoter introduced into the CP ORF was lower than that of the wt (Fig. 5). Cotransfection of protoplasts with the viral transcripts and pCP increased the level of genomic plus-strand RNA for transcripts tLs and tMs (Fig. 5, compare lanes 6 and 7 with lanes 3 and 4). This is in accordance with the results published by Bransom et al. (6), from which it was concluded that the absence of the CP had a negative influence on the accumulation of plus-sense genomic and subgenomic RNAs. Transcript tLa supported the accumulation of genomic RNA poorly, whether in the absence or in the presence of pCP (Fig. 5).
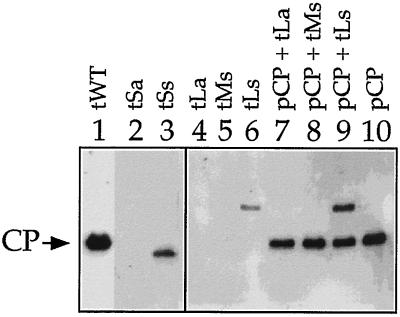
We investigated the possible presence of TYMV CP-related proteins in extracts of transfected protoplasts. Theoretically, since in these mutants the duplicated sequence was introduced near the C-terminal end of the CP ORF, the production of C-terminally truncated versions of the CP or of CP fusion proteins should be possible (Fig. 4B and C; Table 2). Western blot analyses confirmed the presence of a low amount of a 19-kDa C-terminally truncated CP derived from translation of sgRNA 1 of tSs (Fig. 6). A 25-kDa CP fusion protein could be detected for tLs (Fig. 6), which should be an N-terminal CP–C-terminal RdRp fusion protein (see the legend to Fig. 4 and Table 2). No CP or CP fusion proteins could be detected in protoplast extracts transfected with tMs, tSa, or tLa (Fig. 6). They were either not produced in sufficient quantities or rapidly degraded.
TABLE 2.
Calculated sizes of CP fusion proteins derived from translation of sgRNAs 1 and 2 from different mutant transcripts
| Mutant | Calculated size (kDa) of CP fusion protein derived from:
|
|
|---|---|---|
| sgRNA 1 | sgRNA 2b | |
| tWT | 20.1 (189a) | NA |
| tLs | 24.9 (140) | 13.2 (73 + 51) |
| tMs | 20.3 (140) | 5.8 (24) |
| tSs | 19.3 (140) | 6.6 (8) |
| tLa | 15.5 (140) | 11.2 (73) |
| tSa | 19.1 (140) | 4.2 (8) |
Number of amino acids corresponding to those of the CP.
For tLa and tSa. NA, not applicable.
FIG. 6.
Western blot analysis of total protein extracts of transfected protoplasts using polyclonal CP antibodies. Protoplasts were transfected with mutant transcripts, with pCP, or with transcripts and pCP as indicated. Lanes 1 to 3 are from a different gel than lanes 4 to 10.
When protoplasts were transfected with transcript and pCP, the presence of CP could be detected for all protoplast extracts (Fig. 6). In the presence of the CP, higher amounts of the CP fusion protein of tLs accumulated (Fig. 6, compare lanes 9 and 6). The positive influence of the wt CP on the accumulation of the mutant CP of tLs, which derived from transcription from sgRNA 1, might be indirect, improving either the stability or the production of this sgRNA. None of the possible protein products of sgRNAs 2 (Table 2) could be detected with antibodies against the TYMV CP, probably because of lack of recognizable epitopes.
Mutant transcripts tLs, tMs, and tSs were tested for replication in Chinese cabbage plants. None of the transcripts led to local or systemic symptoms on infected plant leaves or young leaves above the infected leaf (not shown). Possibly, the mutant CP proteins produced from the sgRNAs 1 did not lead to effective encapsidation, were not produced to sufficient levels, or were not functional in supporting replication or spread in plants.
DISCUSSION
We demonstrate here that the tymobox region of TYMV RNA operates at least at two levels, as a coding region for the RdRp and as a regulatory control element for the production of sgRNA. Mutant pBP, which contained a termination codon upstream of the tymobox region resulting in an RdRp lacking the 11 terminal amino acids, did not replicate in A. thaliana protoplasts, thus establishing the importance of the coding potential of this region. Whether the mutations introduced in pBP additionally altered the regulatory function of the tymobox region could not be ascertained.
Mutants were constructed that contained silent mutations in the highly conserved tymobox or initiation box (pBI, pBT, and pIT) or in the less well conserved region between the boxes (pB). None of these mutations altered the amino acids of the overlapping ORF (Fig. 1), and all of the mutants were able to replicate, although to different extents, in A. thaliana protoplasts. Introducing a single mutation in a region of low conservation (pB) resulted in RNA replication indistinguishable from that of the wt. Introducing an additional mutation in the initiation box (pBI) or in the tymobox (pBT) abolished sgRNA production, though genomic minus strand was produced to wt levels. Thus, while nt 5623 can be either C or A, nt G5611 of the tymobox and nt A5626 of the initiation box are essential for the function of the sg promoter. This result demonstrates the importance of the tymobox region as a control element for the production of the sgRNA.
In addition to the presence of specific sequences, an sg promoter might be defined by a conserved secondary structure. The formation of hairpin structures upstream of the initiation site for sgRNA synthesis has been described or proposed for sg promoters of other viruses, e.g., the Bromoviridae (13), red clover necrotic mosaic dianthovirus (30), and TCV (28). Comparative secondary structure analyses in silico of a stretch of 90 nt of the minus-strand sequence containing the tymobox region of six different tymoviruses (TYMV [19]; Kennedya yellow mosaic virus, Jervis Bay isolate [9]; Ononis yellow mosaic virus [7]; eggplant mosaic virus [20]; Physalis mottle virus [12]; and Clitoria yellow vein virus [8]) revealed that the tymobox region could be involved in hairpin formation (not shown). To test whether the 4 nt of the initiation box are involved in important base pairing with nt 5608 to 5611 of the tymobox (Fig. 1), we constructed mutant pIT, which should reestablish base pairing if A5626 indeed hybridized to T5609 in the wt virus. The T5609C mutation did not complement the defect caused by the A5625G mutation, indicating that possible base pairing of nt 5626 to nt 5609 is not important for promoter function. Secondary structure predictions of a stretch of 500 nt surrounding the tymobox region of minus-strand TYMV RNA suggested that a hairpin could be formed in which the tymobox and the initiation box are involved in base pairing, albeit not to each other (not shown). Secondary structure predictions of the corresponding stretch of minus-strand RNA of mutants with an altered sequence in the tymobox region revealed no obvious structural differences from the wt situation, supporting the view that for the production of sgRNA, the conserved sequence is important. However, in the absence of detailed structure probing analyses, the presence of important structural elements in the sg promoter of TYMV cannot be excluded.
The complete sg promoter sequence of TYMV is located within a stretch of 494 nt containing the tymobox region as the central element, and it is functional ectopically when introduced into the CP ORF. This represents the first identification of a tymovirus sg promoter. Attempts to reduce the size of the sgRNA promoter to 257 or 88 nt still led to sgRNA synthesis, indicating that these fragments contain the basic information necessary for sgRNA production.
The results suggest that the sg promoter of TYMV, located on a 494-nt fragment, can function when present on a different molecule than TYMV genomic RNA. Protoplasts infected with tLa led to the production of a second small RNA (sg2), a possible transcription product from sgRNA 1 (Fig. 4C and 5). Thus, it is likely that the sg promoter of TYMV can function in trans. This capacity of the TYMV promoter might be exploited for the identification of plant proteins that interact with a protein of interest (e.g., expressing a His-tagged protein from the TYMV promoter introduced into a host plant, infecting with TYMV, and purifying the His-tagged protein as well as other proteins to which it may bind), are necessary for viral replication (e.g., setting up an in vitro replication system) or spread (e.g., following the spread in the plant of a marker protein expressed from the TYMV sg promoter), or for the expression of foreign genes in host plants or the construction of a vector for the model plant A. thaliana. The latter possibility is of major interest, since though the complete A. thaliana genome sequence will soon be known and its genetic manipulation is possible, convenient vector systems for the introduction of foreign genes into this plant are scant. However, until the identity of sg2 is confirmed, this promising scenario remains prospective.
ACKNOWLEDGMENTS
This study was supported by a grant to J.S. from the Gemeinsames Hochschulsonderprogramm III von Bund und Ländern über den DAAD, Germany. The Institut Jacques Monod is an Institut Mixte, CNRS—Universités Paris 6 & 7.
We thank Olivier Voinnet, Ludovic Caussin, Yann Hennekin, Kodetham Gopinath, and H. S. Savithri for participation in different aspects of this work, Michèle Axelos (Castanet-Tolosan, France) for the A. thaliana cell suspension culture, J. M. Bové (Bordeaux, France) for anti-TYMV serum, Jim Haseloff (Cambridge, United Kingdom) for pFF19G, Andreij Palucha (Warsaw, Poland) for useful discussions, and Bernard Billoud (Paris, France) for help with RNA secondary structure analysis.
REFERENCES
- 1.Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem. 1992;30:123–128. [Google Scholar]
- 2.Balmori E, Gilmer D, Richards K, Guilley H, Jonard G. Mapping the promoter for subgenomic RNA synthesis on beet necrotic yellow vein virus RNA 3. Biochimie. 1993;75:517–521. doi: 10.1016/0300-9084(93)90056-x. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya A, Murchie A I H, Lilley D M J. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990;343:484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- 4.Boccard F, Baulcombe D. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology. 1993;193:563–578. doi: 10.1006/viro.1993.1165. [DOI] [PubMed] [Google Scholar]
- 5.Boyer J-C, Drugeon G, Séron K, Morch-Devignes M-D, Agnès F, Haenni A-L. In vitro transcripts of turnip yellow mosaic virus encompassing a long 3′ extension or produced from a full-length cDNA clone harbouring a 2 kb-long PCR-amplified segment are infectious. Res Virol. 1993;144:339–348. doi: 10.1016/s0923-2516(06)80049-x. [DOI] [PubMed] [Google Scholar]
- 6.Bransom K L, Weiland J J, Tsai C-H, Dreher T W. Coding density of the turnip yellow mosaic virus genome: roles of the overlapping coat protein and the p206-readthrough coding regions. Virology. 1995;206:403–412. doi: 10.1016/s0042-6822(95)80056-5. [DOI] [PubMed] [Google Scholar]
- 7.Ding S, Keese P, Gibbs A. Nucleotide sequence of the ononis yellow mosaic tymovirus genome. Virology. 1989;172:555–563. doi: 10.1016/0042-6822(89)90198-0. [DOI] [PubMed] [Google Scholar]
- 8.Ding S, Howe J, Keese P, Mackenzie A, Meek D, Osorio-Keese M, Skotnicki M, Srifah P, Torronen M, Gibbs A. The tymobox, a sequence shared by most tymoviruses: its use in molecular studies of tymoviruses. Nucleic Acids Res. 1990;18:1181–1187. doi: 10.1093/nar/18.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding S, Keese P, Gibbs A. The nucleotide sequence of the genomic RNA of kennedya yellow mosaic tymovirus-Jervis Bay isolate: relationships with potex- and carlaviruses. J Gen Virol. 1990;71:925–931. doi: 10.1099/0022-1317-71-4-925. [DOI] [PubMed] [Google Scholar]
- 10.Gargouri R, Joshi R L, Bol J F, Astier-Manifacier S, Haenni A-L. Mechanism of synthesis of turnip yellow mosaic virus coat protein subgenomic RNA in vivo. Virology. 1989;171:386–393. doi: 10.1016/0042-6822(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 11.Guilley H, Briand J P. Nucleotide sequence of turnip yellow mosaic virus coat protein mRNA. Cell. 1978;15:113–122. doi: 10.1016/0092-8674(78)90087-9. [DOI] [PubMed] [Google Scholar]
- 12.Jacob A N, Murthy M R, Savithri H S. Nucleotide sequence of the 3′ terminal region of belladonna mottle virus-Iowa (renamed Physalis mottle virus) RNA and an analysis of the relationships of tymoviral coat proteins. Arch Virol. 1992;123:367–377. doi: 10.1007/BF01317270. [DOI] [PubMed] [Google Scholar]
- 13.Jaspars E M. Genome activation in alfamo- and ilarviruses. Arch Virol. 1999;144:843–863. doi: 10.1007/s007050050551. [DOI] [PubMed] [Google Scholar]
- 14.Johnston J C, Rochon D M. Deletion analysis of the promoter for the cucumber necrosis virus 0.9-kb subgenomic RNA. Virology. 1995;214:100–109. doi: 10.1006/viro.1995.9950. [DOI] [PubMed] [Google Scholar]
- 15.Kim K H, Hemenway C. Mutations that alter a conserved element upstream of the potato virus X triple gene block and coat protein genes affect subgenomic RNA accumulation. Virology. 1997;232:187–197. doi: 10.1006/viro.1997.8565. [DOI] [PubMed] [Google Scholar]
- 16.Klein C, Fritsch C, Briand J P, Richards K E, Jonard G, Hirth L. Physical and functional heterogeneity in TYMV RNA: evidence for the existence of an independent messenger coding for coat protein. Nucleic Acids Res. 1976;3:3043–3061. doi: 10.1093/nar/3.11.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maia I G, Séron K, Haenni A-L, Bernardi F. Gene expression from viral RNA genomes. Plant Mol Biol. 1996;32:367–391. doi: 10.1007/BF00039391. [DOI] [PubMed] [Google Scholar]
- 18.Miller W A, Dreher T W, Hall T C. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−) sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 19.Morch M-D, Boyer J-C, Haenni A-L. Overlapping open reading frames revealed by complete nucleotide sequencing of turnip yellow mosaic virus genomic RNA. Nucleic Acids Res. 1988;16:6157–6173. doi: 10.1093/nar/16.13.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osorio-Keese M E, Keese P, Gibbs A. Nucleotide sequence of the genome of eggplant mosaic tymovirus. Virology. 1989;172:547–554. doi: 10.1016/0042-6822(89)90197-9. [DOI] [PubMed] [Google Scholar]
- 21.Pleij C W A, Neeleman A, Van Vloten-Doting L, Bosch L. Translation of turnip yellow mosaic virus RNA in vitro: a closed and an open coat protein cistron. Proc Natl Acad Sci USA. 1976;73:4437–4441. doi: 10.1073/pnas.73.12.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozanov M N, Drugeon G, Haenni A-L. Papain-like proteinase of turnip yellow mosaic virus: a prototype of a new viral proteinase group. Arch Virol. 1995;140:273–288. doi: 10.1007/BF01309862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schirawski J, Planchais S, Haenni A-L. An improved protocol for the preparation of protoplasts from an established Arabidopsis thaliana cell suspension culture and infection with RNA of turnip yellow mosaic tymovirus: a simple and reliable method. J Virol Methods. 2000;86:85–94. doi: 10.1016/s0166-0934(99)00173-1. [DOI] [PubMed] [Google Scholar]
- 24.Sit T L, Vaewhongs A A, Lommel S A. RNA-mediated trans-activation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- 25.Timmermans M C P, Malgia P, Vieira J, Messing J. The pFF plasmids: cassettes utilizing CaMV sequences for expression of foreign genes in plants. J Biotechnol. 1990;14:333–344. doi: 10.1016/0168-1656(90)90117-t. [DOI] [PubMed] [Google Scholar]
- 26.van der Kuyl A C, Langereis K, Houwing C J, Jaspars E M J, Bol J F. Cis-acting elements involved in replication of alfalfa mosaic virus RNAs in vitro. Virology. 1990;176:346–354. doi: 10.1016/0042-6822(90)90004-b. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Carpenter C D, Simon A E. Minimal sequence and structural requirements of a subgenomic RNA promoter for turnip crinkle virus. Virology. 1999;253:327–336. doi: 10.1006/viro.1998.9538. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Simon A E. Analysis of the two subgenomic RNA promoters for turnip crinkle virus in vivo and in vitro. Virology. 1997;232:174–186. doi: 10.1006/viro.1997.8550. [DOI] [PubMed] [Google Scholar]
- 29.Zaccomer B, Haenni A-L, Macaya G. The remarkable variety of plant RNA virus genomes. J Gen Virol. 1995;76:231–247. doi: 10.1099/0022-1317-76-2-231. [DOI] [PubMed] [Google Scholar]
- 30.Zavriev S K, Hickey C M, Lommel S A. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology. 1996;216:407–410. doi: 10.1006/viro.1996.0076. [DOI] [PubMed] [Google Scholar]
- 31.Zhang G, Slowinski V, White K A. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA. 1999;5:550–561. doi: 10.1017/s1355838299982080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 33.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. NATO ASI Series. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. [Google Scholar]