Abstract
Even in lieu of a dependable species concept for asexual organisms, the classification of bacteria into discrete taxonomic units is considered to be obstructed by the potential for lateral gene transfer (LGT) among lineages at virtually all phylogenetic levels. In most bacterial genomes, large proportions of genes are introduced by LGT, as indicated by their compositional features and/or phylogenetic distributions, and there is also clear evidence of LGT between very distantly related organisms. By adopting a whole-genome approach, which examined the history of every gene in numerous bacterial genomes, we show that LGT does not hamper phylogenetic reconstruction at many of the shallower taxonomic levels. Despite the high levels of gene acquisition, the only taxonomic group for which appreciable amounts of homologous recombination were detected was within bacterial species. Taken as a whole, the results derived from the analysis of complete gene inventories support several of the current means to recognize and define bacterial species.
Keywords: genome evolution, recombination, Gammaproteobacteria, gene repertoires, speciation
Real species are typically defined by the ability of their constituents to exchange genes. This activity (i.e., sexual reproduction) goes a long way toward explaining the maintenance of species as cohesive units whose members are closely related and are of similar genetic architecture. As such, conspecifics share numerous characteristics by which they can be grouped, even when evidence of interbreeding is limited or unknown.
By lacking a mechanism that regularly homogenizes the features of different organisms, strictly asexual organisms continuously diverge from one another as independent lineages. And although the classification of these lineages is undoubtedly useful, it could be argued that any criteria for delineating asexual species, e.g., possessing of a particular suite of phenotypic traits or attaining a prescribe degree of DNA similarity, are arbitrary, inconsistent across taxa, and biologically meaningless.
Acknowledging the problems associated with classifying groups of asexual organisms into discrete species, the situation with bacteria is even worse. Bacteria reproduce asexually, yet they are also capable of obtaining genes from other organisms, even those of different kingdoms. Moreover, the amounts, types, and sources of imported genes can vary among lineages, allowing gene transfer to blur the boundaries of bacterial groups at every taxonomic level and in ways that are impossible to predict. And if patterns of vertical descent are obscured in varied and unknown ways, then the systematic classification of bacteria might not be possible (see refs. 1-4 for current reviews on the concept of bacterial species).
There is a clear advantage to examining the process of diversification in bacteria, which is the availability of complete sequences from hundreds of genomes whose relationships range in type from members of the same nominal species to representatives of groups that diverged billions of years ago. These new data allow us to follow the origin and ancestry of every gene in a genome to resolve the degree to which gene transfer has shaped the contents of bacterial genomes and has obscured the history of bacterial groups at different phylogenetic depths.
The Scope of Gene Transfer in Bacteria
There are several means by which bacteria can acquire genes: by conjugal transfer, by phage-mediated insertions and by the update of native DNA from the outside sources (5, 6). But given the diversity of mechanisms that are capable of planting virtually any gene in virtually any organism, bacterial genomes remain small (on the order of 500-10,000 kb) and are not simply arbitrary assortments of genes of mixed heritage. Although bacteria might be bombarded constantly with foreign genes, only evolutionarily relevant events of transfer, i.e., those resulting in genes that persist, are evident from the contents of extent genomes.
With the completion of each bacterial genome sequence, there is a search for horizontally acquired genes. This research most commonly proceeds by scanning the genome sequence for regions of atypical base composition, a surprisingly accurate method for identifying one class of recently acquired genes. The rationale for this approach has its foundations in research performed nearly 50 years ago, when the initial goals were to characterize the nature of nucleic acids within cellular organisms (7-9). By the early 1960s, base composition [usually expressed as the relative proportion of guanine and cytosine (G+C) residues, % G+C] had been determined for hundreds of bacterial genomes, leading to the general observations: (i) that the diversity in base composition among bacterial genomes, which ranges from 20% to 80% G+C, is much greater than that in eukaryotes, (ii) that despite this variation, the base composition within an organism is fairly consistent over the entire chromosome, and (iii) that closely related organisms have similar G+C contents (10-12). The observed heterogeneity among genomes, coupled with the compositional homogeneity within genomes, implicates gene transfer between organisms of different G+C contents as a source of intragenomic variation in base composition and codon usage patterns (13-17).
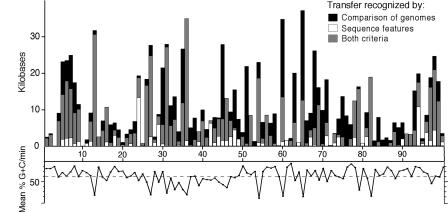
Naturally, other factors, such as the amino acid contents of a protein, might influence its overall nucleotide composition; however, phylogenetic information supports the use of G+C content as a way to identify acquired genes. Because acquired regions often manifest multiple features that denote their ancestry, it is thus not perhaps surprising that many genes with sporadic distributions, as might occur from a history of lateral transfer, have anomalous base compositions. To illustrate the utility and accuracy of these methods for recognizing acquired genes, Fig. 1 shows the amount of protein-coding DNA within each minute (≈45 kb encompassing ≈40 genes) along the E. coli MG1655 chromosome having atypical sequence features and/or a phylogenetic distribution indicative lateral transfer (18).
Fig. 1.
Linear representation of the E. coli MG1655 chromosome showing the distribution of horizontally acquired DNA. At each minute (1-100), vertical bars depict the amount of horizontally acquired, protein-coding DNA, as inferred by two methods: (i) atypical sequence features (i.e., base composition) (white) and (ii) the unique occurrence of a gene in E. coli after aligning and comparing the genome sequences of E. coli, Salmonella enterica, and Klebsiella pneumonia (black). Gray portions of vertical bars denote the overlap between the methods and the amount of protein-coding DNA in E. coli inferred to be horizontally acquired based both on its sequence features and on its phylogenetic distribution. Along the bottom of the figure is shown the base composition (% G+C) computed in discrete windows for each minute of the chromosome. The dashed horizontal line shows the overall average base composition for all protein-coding genes in this genome (51.0% G+C). Figure modified from Lawrence and Ochman (18).
These procedures rely on very different sorts of information and might be expected to identify somewhat different sets of acquired genes, the degree of overlap (gray portion of each bar in Fig. 1) is quite good: among the 755 genes originally identified as being horizontally acquired based on sequence characteristics, nearly 80% display a phylogenetic distribution compatible with lateral gene transfer (LGT). As expected, the base compositional approach does not recognize some number of acquired genes, such as those obtained from organisms of similar genomic G+C contents (black portion of bars). Taken as a whole, nearly 25% of the 4,280 protein-coding genes in this Escherichia coli lineage were introduced by LGT because it split from Salmonella enterica an estimated 100 million years ago. And the amount of acquired DNA detected in E. coli, as inferred from the base compositional features of genes, seems to be about average for a genome of its size (5, 19).
Gauging the proportion of acquired genes within a bacterial genome by evaluating its compositional features has some distinct advantages: it is computationally simple and does not rely on the availability of any other genomes. But this method divulges predominantly one class of acquired sequences, i.e., unique genes obtained from very divergent sources, and might vastly underestimate the full extent of LGT-affecting bacterial genomes. Gene exchange can also occur between close relatives and/or between genes that are conserved among organisms. Such events result in an exceptionally high degree of similarity between genes from different taxa and are usually uncovered by some type of comparative method. For example, genomes are surveyed regularly for genes whose best match (as detected by blast) lie outside their closest sequenced relatives, and in the case of certain sequenced bacteria (e.g., Thermotoga maritima and Aquifex aeolicus), substantial fractions of their genes were found to be most similar to genes present in Archaea (20-22).
Because LGT will result in different phylogenies for different portions of the genome, the most common and robust way to identify cases of transfer and exchange is by searching for evidence of discordance among gene trees. This approach has been applied from the deepest to the shallowest phylogenetic levels, and thousands of transfer events have been recognized (23, 24). The availability of full genome sequences has allowed the evaluation of the history of the genes distributed among all life forms, which might be thought to be highly constrained and less susceptible to replacement by LGT. Many of these genes, even ribosomal RNA, long touted and applied as the benchmark for determining organismal relationships, show evidence of LGT over some portion of their evolutionary history (25).
The Cohesion of Bacterial Genomes
So far, these studies confirm that LGT is pervasive and is an ongoing process within bacterial genomes. Genes with sporadic distributions and atypical sequence features arise by LGT, and there are clear cases of gene transfer occurring at all taxonomic levels, even among the genes common to all life forms. With the potential for the LGT of any gene and among all organisms, bacterial species and other taxonomic groupings might not be definable entities. Thus, there is a need to establish whether LGT is resorting the genes in bacterial genomes, eradicating the vestiges of bacterial species, and confounding attempts at phylogenetic classification.
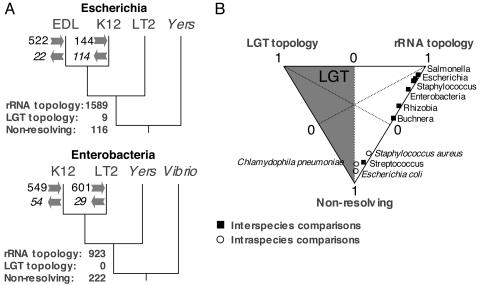
To assess the extent to which LGT is linked phylogenetic disruption, we considered the relationship between DNA acquisition and phylogenetic incongruence in fully sequenced bacteria at several taxonomic levels, including that occurring within species (E. coli, Chlamydophila pneumoniae, and Staphylococcus aureus), within genera (Escherichia, Salmonella, Buchnera, and Streptococcus), and within families (Enterobacteriaceae and Rhizobiaceae) (26). We focused on groups at these phylogenetic depths both to assure substantial overlaps in genome contents and to minimize the risk of reconstruction artifacts due to hidden paralogy or long-branch attraction. And for each group of four genomes, we inferred both the number of recently acquired and lost genes (based on their phylogenetic distributions) and the proportion of ortholog phylogenies supporting lateral transfers (by asking whether an alignment significantly supports the rRNA reference topology, either of the two alternate topologies, or no topology).
For all groups and at all taxonomic levels, the proportion of ortholog phylogenies supporting a hypothesis of LGT is always small and often zero (Fig. 2). Only among members of the same species (E. coli or C. pneumoniae) were there increased levels of LGT (i.e., where >5% of alignments support a tree other than the rRNA reference topology). In contrast to the rarity of exchange among orthologs, levels of gene acquisition (as determined from the phylogenetic occurrence of genes) remain high, as previously inferred from the compositional features of individual genomes.
Fig. 2.
Phylogenetic inference of cohesion within bacterial genomes. (A) Relationship between gene acquisition and loss and the amount of phylogenetic incongruence observed in fully sequenced bacterial genomes. For each quartet of genomes, we inferred the number of recently acquired and lost genes (shown at arrows on the corresponding branches). In addition, for each quartet of genomes, orthologous genes were inferred, aligned, and evaluated at the nucleic sequence level based on the Shimodaira-Hasegawa test implemented in Puzzle 5.1 (27, 28). Shown are the numbers of orthologs supporting each category of alignment: (i) those supporting the reference phylogeny (rRNA topology), (ii) those supporting either alternate phylogeny (LGT topology), and (iii) those with no statistical support for any phylogeny (nonresolving). (B) Relative frequencies of the three categories of alignments in diverse bacterial groups at several taxonomic levels. The shaded zone of this plot represents the area where LGT predominates. Only for intraspecies comparisons within E. coli and C. pneumoniae are the frequencies of LGT >5%. Data and figure from Daubin et al. (26).
Thus, gene acquisition is frequent but gene replacement is relatively rare, resulting in fundamentally two classes of protein-coding sequences within bacterial genomes: first are the orthologs that are conserved among taxa and not prone to gene transfer and exchange among species. Next are the acquired genes, which are generally unique to a genome and, unlike orthologs, encode proteins of uncharacterized functions. So despite high levels of LGT, bacteria seem to form coherent groups at the shallower taxonomic levels because LGT is concentrated in a class of genes that are not suitable candidates for phylogenetic analysis (26).
Determinants of Gene Exchange in Bacterial Species
Despite the massive influx of new genes into bacterial genomes, the only taxonomic group for which appreciable amounts of homologous recombination were detected was within bacterial species. This finding is remarkably similar to the concept of species that is applied to sexually reproducing eukaryotes, i.e., groups of organisms that exchange genes. But assuming that there is the potential for any sequence to be transferred among bacteria, what factors abide the integration and exchange of homologs from some sources and prevent those from others?
The extent of homologous exchange, as indexed by multilocus enzyme electrophoresis and by multilocus sequence typing, has been shown to vary enormously across bacterial species (29, 30), making it unlikely that a single mechanism regulates recombination efficiency in all bacteria. However, the process has been analyzed in some detail in E. coli and Salmonella typhimurium (Salmonella enterica serovar Typhimurium), in which gene exchange depends on the degree of similarity between donor and recipient sequences. Homologous genes from E. coli and Salmonella typhimurium differ by ≈15% in sequence, and recombination rates, as assayed in conjugal matings, are ≈105 lower for intergeneric than for intraspecies crosses (31, 32). This barrier to gene exchange is effected, in part, by mismatch repair enzymes, which inhibit recombination between divergent sequences, thereby allowing gene exchange among close relatives and preventing it among more distant strains (33-35).
If similar mechanisms that limit homologous recombination are operating in other taxa, then bacterial species can be viewed as assemblages of lineages that are sufficiently closely related to potentially exchange shared genes. Then, depending on the actual rates of recombination, population structure, and patterns of lineage extinction, these assemblages will eventually assort into distinct species that have diverged sufficiently at the DNA level to form a genetic barrier to gene exchange. In this case, the practice of delineating bacterial species on the basis of some prescribed level of sequence divergence seems to be well justified.
Why Species?
There is still an overarching issue, which stems from the fact that all of the genetic and genomic properties discussed so far were characterized in groups of lineages that were already designated as distinct bacterial species. E. coli and Salmonella typhimurium were each discovered more than a century ago, and their classification is founded on schemes devised before there was any knowledge of genes or genetics.
Bacterial species are typically recognized according to their cellular properties and metabolic capabilities; for example, E. coli, a mammalian commensal, ferments lactose but not citrate, whereas Salmonella enterica, a mammalian pathogen, is lactose negative and citrate positive. Examining the genetic basis of these traits, the lac operon is a G+C-rich region unique to E. coli, whereas the citrate utilization (as well as many Salmonella virulence determinants) is conferred by low G+C genes present only in Salmonella. Therefore, assignment of isolates to each of these bacterial species seems to have been largely on traits that were introduced by LGT (but see ref. 36 for a contrasting view of lac operon evolution). And consequently, the species so defined have turned out to be discrete biological entities that, because of genetic and mechanistic reasons, rarely exchange homologs.
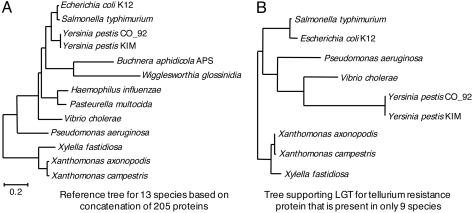
To determine how horizontally acquired genes are able to accurately define bacterial species, we need to trace the phylogenetic history of genes that occur sporadically among multiple taxa. To accomplish this, it is necessary to step back from E. coli and Salmonella enterica (where genes are either confined to one, or present in both, species) and consider families of genes within the broader taxonomic framework that subsumes these lineages. We examined the full protein-coding gene repertoires within 13 sequenced Gammaproteobacteria, including one strain each of E. coli and Salmonella enterica (37). Previously it was shown that >99% of the 205 single-copy genes that are shared by all genomes supported the same relationships for the 13 species examined (38), thereby providing a robust organismal phylogeny against which the trees based on less conserved genes can be tested (Fig. 3A).
Fig. 3.
Example of a lateral gene transfer detected by phylogenetic discordance. (A) Neighbor-joining tree based on the concatenation of 205 single-copy genes common to all 13 Gammaproteobacterial species. Note that 203 of the 205 genes individually supported the same topology. Figure adapted from Lerat et al. (38). (B) Example of a tree that conflicts with the reference topology. Among the small proportion of proteins showing statistical support for an alternate topology was tellurium resistance protein. Homologs of the gene encoding this protein have been detected in only 9 of the 13 species. Note that the topology of this tree departs from that of the reference tree (depicted in A) because of a single LGT event that occurred in the ancestor to E. coli and Salmonella enterica. Data in B are from Lerat et al. (37).
Considering single-copy genes that are absent from one of more of these genomes (i.e., those whose distributions may result from LGT, gene loss, or some combination of these processes), we found that very few display statistically supported incongruence with the organismal phylogeny (Table 1). For genes present in a single copy in only a subset of the 13 genomes, the incidence of LGT is very low and not significantly different from that observed for the 205 single-copy genes present in all species. And furthermore, those few cases of LGT can usually be accounted for by a single event, an example of which is shown in Fig. 3B.
Table 1. Incidence of lateral gene transfer among single-copy genes with different phylogenetic distributions.
| No. of species
|
||||||||
|---|---|---|---|---|---|---|---|---|
| 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| No. of single-copy genes | 167 | 188 | 137 | 78 | 69 | 109 | 98 | 205 |
| No. rejecting reference tree | 5 | 2 | 3 | 3 | 1 | 3 | 5 | 2 |
| Trees supporting LGT, % | 2.99 | 1.06 | 2.19 | 3.85 | 1.45 | 2.80 | 5.10 | 0.98 |
Only those genes whose homologs are present in 6-13 of the species are shown. Orthologs were aligned and evaluated to recover topologies that differed significantly from the reference tree.
Although LGT has been the major source of new genes in these bacterial lineages, as reflected by the large number of gene families restricted to one or two genomes (i.e., 10,728 of 14,158 total families), the lack of phylogenetic inconsistencies among the sporadically distributed genes reveals that (i) acquired genes come from sources outside of this group, and (ii) subsequent to their initial acquisition, genes are by and large transmitted vertically. These findings explain why properties introduced by LGT can serve as stable markers of bacterial species and of phylogenetics. First, genes acquired from distant sources are more likely to supply a novel trait that would set the recipient apart from its relatives. Next, those acquired genes that confer a useful (and defining) trait will persist within the descendant clade and only rarely be transferred laterally to related species.
A Bacterial Saga (Speciation Attributable to Gene Acquisition)
Taken as a whole, the effects of gene transfer and exchange on bacterial evolution and classification are opposite, or at least orthogonal, to what one might anticipate. High levels of gene transfer should, in the words of Gogarten et al. (39) “obliterate the patterns of vertical descent” and erase the boundaries between species or any other taxonomic units. But despite massive amounts of LGT, bacteria seem to form more or less cohesive groups at many taxonomic levels (26, 40). These groupings are the result of a nonarbitrary process of gene acquisition in which divergent organisms serve as a persistent source of novel genes in a genome, and the levels of recombinational exchange among homologs shared by related species are low.
As such, LGT can sometimes be viewed as an agent that promotes and maintains bacterial species (15, 41). Acquired genes play a major role in bacterial diversification by supplying previously unavailable traits, which can allow the rapid exploitation of new environments. Such capabilities, which are strictly vertically transmitted once they are acquired, can serve to subdivide the population, allowing the phenotypically distinct lineages to diverge at the sequence level to the point where there is a recombinational barrier to gene exchange. Although our saga has been based largely on the analyses of a single taxon, these results show that it is still possible to make inferences about the origin and nature of bacterial species in light of substantial lateral gene transfer.
Acknowledgments
We thank Nancy Moran and two anonymous reviewers for comments on the manuscript and Becky Nankivell for preparing the figures. Department of Energy Grant DEFG0301ER6314 and National Institutes of Health Grant GM56120 supported much of the work presented in this article.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Systematics and the Origin of Species: On Ernst Mayr's 100th Anniversary,” held December 16-18, 2004, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: G+C, guanine plus cytosine; LGT, lateral gene transfer.
References
- 1.Rossello-Mora, R. & Amann, R. (2001) FEMS Microbiol. Rev. 25, 39-67. [DOI] [PubMed] [Google Scholar]
- 2.Lan, R. & Reeves, P. R. (2001) Trends Microbiol. 9, 419-424. [DOI] [PubMed] [Google Scholar]
- 3.Young, J. M. (2001) Int. J. Syst. Evol. Microbiol. 51, 945-953. [DOI] [PubMed] [Google Scholar]
- 4.Cohan, F. M. (2002) Annu. Rev. Microbiol. 56, 457-487. [DOI] [PubMed] [Google Scholar]
- 5.Ochman, H., Lawrence, J. G. & Groisman, E. A. (2000) Nature 405, 299-304. [DOI] [PubMed] [Google Scholar]
- 6.Redfield, R. J. (2001) Nat. Rev. Genet. 2, 634-639. [DOI] [PubMed] [Google Scholar]
- 7.Rolfe, R. & Meselson, M. (1959) Proc. Natl. Acad. Sci. USA 45, 1039-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sueoka, N., Marmur, J. & Doty, P. (1959) Nature 183, 1429-1431. [DOI] [PubMed] [Google Scholar]
- 9.Sueoka, N. (1961) J. Mol. Biol. 3, 31-40. [Google Scholar]
- 10.Sueoka, N. (1962) Proc. Natl. Acad. Sci. USA 48, 582-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sueoka, N. (1988) Proc. Natl. Acad. Sci. USA 85, 2653-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muto, A. & Osawa, S. (1987) Proc. Natl. Acad. Sci. USA 84, 166-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medigue, C., Rouxel, T., Vigier, P., Henaut, A. & Danchin, A. (1991) J. Mol. Biol. 222, 851-856. [DOI] [PubMed] [Google Scholar]
- 14.Guerdoux-Jamet, P., Henaut, A., Nitschke, P., Risler, J. L. & Danchin, A. (1997) DNA Res. 4, 257-265. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence, J. G. & Ochman, H. (1998) Proc. Natl. Acad. Sci. USA 95, 9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragan, M. A. (2001) FEMS Microbiol. Lett. 201, 187-191. [DOI] [PubMed] [Google Scholar]
- 17.Daubin, V., Lerat, E. & Perriere, G. (2003) Genome Biol. 4, R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence, J. G. & Ochman, H. (2002) Trends Microbiol. 10, 1-4. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Vallve, S., Romeu, A. & Palau, J. (2000) Genome Res. 10, 1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deckert, G., Warren, P. V., Gaasterland, T., Young, W. G., Lenox, A. L., Graham, D. E., Overbeek, R., Snead, M. A., Keller, M., Aujay, M., et al. (1998) Nature 392, 353-358. [DOI] [PubMed] [Google Scholar]
- 21.Nelson, K. E., Clayton, R. A., Gill, S. R., Gwinn, M. L., Dodson, R. J., Haft, D. H., Hickey, E. K., Peterson J. D., Nelson W. C., Ketchum K. A., et al. (1999) Nature 399, 323-329. [DOI] [PubMed] [Google Scholar]
- 22.Logsdon, J. M. & Faguy, D. M. (1999) Curr. Biol. 9, R747-R751. [DOI] [PubMed] [Google Scholar]
- 23.Koonin, E. V., Makarova, K. S. & Aravind, L. (2001) Annu. Rev. Microbiol. 55, 709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boucher, Y., Douady, C. J., Papke, R. T., Walsh, D. A., Boudreau, M. E., Nesbo, C. L., Case, R. J. & Doolittle, W. F. (2003) Annu. Rev. Genet. 37, 283-328. [DOI] [PubMed] [Google Scholar]
- 25.Yap, W. H., Zhang, Z. & Wang, Y. (1999) J. Bacteriol. 181, 5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daubin, V., Moran, N. A. & Ochman, H. (2003) Science 301, 829-832. [DOI] [PubMed] [Google Scholar]
- 27.Shimodaira, H. & Hasegawa, M. (1999) Mol. Biol. Evol. 16, 1114-1116. [Google Scholar]
- 28.Strimmer, K. & von Haeseler, A. (1996) Mol. Biol. Evol. 13, 964-969. [Google Scholar]
- 29.Selander, R. K. & Musser, J. M. (1990) in The Evolution of Bacterial Pathogens (Academic, New York) Vol. XI, pp. 11-36. [Google Scholar]
- 30.Feil, E. J. & Spratt, B. G. (2001) Annu. Rev. Microbiol. 55, 561-590. [DOI] [PubMed] [Google Scholar]
- 31.Baron, L. S., Gemski, P., Johnson, E. M. & Wohlhieter, J. A. (1968) Bacteriol. Rev. 32, 362-369. [PMC free article] [PubMed] [Google Scholar]
- 32.Matic, I., Rayssiguier, C. & Radman, M. (1995) Cell 80, 507-515. [DOI] [PubMed] [Google Scholar]
- 33.Rayssiguier, C., Thaler, D. S. & Radman, M. (1989) Nature 342, 396-401. [DOI] [PubMed] [Google Scholar]
- 34.Matic, I., Taddei, F. & Radman, M. (1996) Trends Microbiol. 4, 69-72. [DOI] [PubMed] [Google Scholar]
- 35.Vulic, M., Dionisio, F., Taddei, F. & Radman, M. (1997) Proc. Natl. Acad. Sci. USA 94, 9763-9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoebel, D. M. (2005) Mol. Biol. Evol. 22, 683-690. [DOI] [PubMed] [Google Scholar]
- 37.Lerat, E., Daubin V., Ochman, H. & Moran, N. A. (2005) PLoS Biol., in press. [DOI] [PMC free article] [PubMed]
- 38.Lerat, E., Daubin V. & Moran, N. A. (2003) PLoS Biol. 1, E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gogarten, J. P., Doolittle, W. F. & Lawrence, J. G. (2002) Mol. Biol. Evol. 19, 2226-2238. [DOI] [PubMed] [Google Scholar]
- 40.Kurland, C. G., Canback, B. & Berg, O. G. (2003) Proc. Natl. Acad. Sci. USA 100, 9658-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence, J. G. (2002) Theor. Popul. Biol. 61, 449-460. [DOI] [PubMed] [Google Scholar]