Abstract
The biodiversity of Madagascar is extraordinarily distinctive, diverse, and endangered. It is therefore urgent that steps be taken to document, describe, interpret, and protect this exceptional biota. As a collaborative group of field and laboratory biologists, we employ a suite of methodological and analytical tools to investigate the vertebrate portion of Madagascar's fauna. Given that species are the fundamental unit of evolution, where micro- and macroevolutionary forces converge to generate biological diversity, a thorough understanding of species distribution and abundance is critical for understanding the evolutionary, ecological, and biogeographic forces that have shaped Malagasy vertebrate diversity. We illustrate the means by which we apply Mayr's “three basic tasks” of the systematist [Mayr, E. (1942) Systematics and the Origin of Species from the Viewpoint of a Zoologist (Harvard Univ. Press, Cambridge, MA)] to identify, classify, and study the organisms that together constitute Madagascar's vertebrate community. Using field inventory methods, specimen-based studies, and morphological and molecular analyses, we formulate hypotheses of species identity that then serve as the foundation for subsequent studies of biology and history. Our experience, as well as that of other investigators, has shown that much of the vertebrate species diversity in Madagascar is “cryptic” for both biological and practical reasons. Beyond issues of cryptic biological diversity, the resolution of species identity in Madagascar has been hampered because of a lack of vouchered comparative material at the population level. Through our activities, we are attempting to remedy these limitations while simultaneously enhancing research capacity in Madagascar.
Keywords: Madagascar, lemurs, biogeography, biodiversity, convervation
The actual demarcation of species taxa uses morphological, geographical, ecological, behavioral, and molecular information to infer the rank of isolated populations.
Ernst Mayr (ref. 1, p.276)
Biologists disagree, often vehemently, over the question of what constitutes a species. Virtually all agree, however, that species are a fundamental unit of evolution where micro- and macroevolutionary forces converge to generate biological diversity. Thus, the theoretical and practical issues relating to species identification are essential for the purposes of documenting, describing, and preserving biodiversity. Mayr's fundamental contribution, with the formulation of the biological species concept (BSC), was to express the question as a biological rather than a typological problem: “The most important aspect of the biological species definition is that it uses no artificial criteria, but decides each case on the basis of whether certain organisms behave as if they were conspecific or not” (ref. 2, pp. 119-120). He makes clear the importance that morphological and genetic information serve as clues to biological distinctiveness, especially in those cases where reproductive isolation cannot be determined for reasons of geographic separation. Thus, although the BSC by no means provides a universally applicable recipe whereby the working biologist can diagnose a species, and thus define its evolutionary significance, the concept provides a practicable toolkit for embarking upon the enterprise.
As a collaborative group of field and lab biologists, we are motivated by an interest in documenting, describing, understanding, and preserving the endangered vertebrate biota of Madagascar. The exceptional floral and faunal diversity of this island are well known (3). Madagascar lies ≈300 miles to the east of Africa at the narrowest point of the Mozambique Channel, where it has been isolated from the African continent for nearly 160 million years and from all other significant landmasses for the past 88 million years (4, 5). Its status as one of the world's top 12 “megadiversity” countries is without question due to the remarkable levels of taxonomic endemism found there (6). For example, 95% of the reptile species, 99% of amphibian species, and 100% of the island's primate species occur nowhere else on Earth. Certain faunas are either poorly represented (e.g., only four orders of terrestrial eutherians are currently represented) or are completely absent (e.g., there are no salamanders, vipers, or varanid lizards), whereas other groups show unrivaled diversity (e.g., chameleons). Madagascar thus generates intense interest among evolutionary biologists who wish to understand the extent to which geographic and environmental constraints influence organismal evolution. The investigation of Madagascar's biodiversity has the potential to offer insight into key concepts of ecosystem formation and function, biogeographic mechanisms such as vicariance and dispersal, and the sustainability of biodiversity (7).
Fundamental to the investigation of these key concepts is basic knowledge concerning the presence, distribution, and diversity of Madagascar's remarkable biota. In turn, this basic knowledge depends upon the identification and geographic delineation of biological species. In our efforts to understand the evolutionary forces that govern the distribution and abundance of Malagasy vertebrates, we apply a suite of empirical and analytical tools that permit us first to formulate hypotheses of species distinction, and then progress by means of genetic and morphological analysis to questions of the geographic and temporal context of the species' history. Our methods closely mirror what Mayr describes as the three basic tasks of the systematist: (i) identification (analytical stage), (ii) classification (synthetic stage), and (iii) the study of species formation and of factors of evolution (ref. 2, pp. 9-10).
New vertebrate species have been described from Madagascar at a vigorous pace over the past few years (e.g., see refs. 8-15), and the rate of discovery continues unabated. The distinct majority of these discoveries have been based on fieldwork and the collection of new material, rather than on reassessment of specimens already held in museum collections, although this latter material is paramount for points of taxonomic reference. Virtually any surveyed region of Madagascar with remaining natural habitat has been found to harbor new species of vertebrates, even for well studied groups such as mammals. Moreover, there are considerable areas of Madagascar that have received little to no inventory activity within the past century, particularly in the West, leading us to conclude that there are untold numbers of species, across the phylogenetic spectrum, that await discovery.
The process required to identify and document these species is far from an academic exercise, and indeed, is urgent. Madagascar has been designated as one of the most critical geographic priorities for conservation action (6, 16, 17), retaining <10% of the natural habitats that existed before human colonization (18-20). The coming few years offer an unprecedented opportunity for working with the Malagasy Government to establish conservation priorities, and may possibly represent the last chance to make large-scale progress in the designation of protected areas. There is keen interest among Malagasy officials to prioritize regions of the country in need of protection, and these priorities will be largely based upon basic biological knowledge relating to species diversity and distribution. Given this urgency, we as biologists do not have the luxury to contemplate and deliberate the meaning of species, without simultaneously taking the necessary action required to formulate biological hypotheses of species distribution and abundance. In other words, the urgency of the problem does not coincide well with leisurely reflection. Numerous investigators have chosen to mitigate the uncertainties of species identification by relying upon the concept of the evolutionary significant unit (ESU) (21-25). In this conceptual framework, the investigator is concerned with recognizing the evolutionary heritage and potential of a given population, typically by using genetic tools (24), with a subsequent focus on the long-term conservation of that population. This approach has significant appeal for conservation biologists, although it does not entirely avoid the theoretical and operational issues inherent to the species problem (23).
In this article, we describe a series of empirical and analytical steps that we undertake to accomplish the three goals of identification, classification, and evolutionary study that Mayr set out for the systematist bent upon species discovery. We do so by presenting four case studies, of different vertebrate groups, that are currently in various stages of development. The methodological steps that we employ in such investigations are taken in no predefined order (beyond the fact that field inventory is primary) and are certainly not unique to our enterprise. They are, however, mutually illuminating, and, when conducted as a coordinated effort among biologists of complementary basic, analytic, and organismal expertise, can be both efficient and powerful for identifying species units and for analyzing their evolutionary context.
A General Approach for Recognizing, Describing, and Understanding Species Diversity in an Underexplored Environment: Case Studies from Madagascar
The accumulated experience of our group, as well as that of other investigators (26-30), has shown that much of the vertebrate species diversity in Madagascar is “cryptic.” The proximal causes of obscure species diversity relate to a variety of issues, both biological and practical. It is often true that there are few if any externally visible diagnostic features associated with species identity, or, if they are present, the variation is often so subtle as to be detectable only by a highly trained specialist in that particular vertebrate group. In such cases, species can be said to be cryptic in the definitive sense of the word. Mayr originally described such cryptic variation among closely related species as “sibling species” (2). As he noted, cryptic variation can be especially problematic for poorly analyzed groups, with the assumption that subtle diagnostic characters exist for those species, but have not yet been discovered. Mayr also states in more recent work (1) that there are also a great number of “good biological species” that do not differ phenotypically at all, or only so slightly as to be easily confounded with intraspecific variation. All of the biological and practical variables raised by Mayr apply in the case of species discovery in Madagascar. To confound the issues of cryptic biological diversity, the resolution of species identity has been hampered due to a lack of specimen material and genetic sampling at the population level necessary for understanding patterns of variation. Thus, it is our task to first assemble the necessary specimen data within and among populations, and across their geographic distributions, before we can even attempt to penetrate the biological complexities of true cryptic variation.
As stated above, the essential first step in formulating our hypotheses of species diversity begins with field inventory activities. A field team of researchers based at WWF Madagascar, in the context of a project known as the Ecology Training Program (ETP), has a field inventory program associated with documenting the biota of the island. This team has developed a field methodology that allows for rapid yet thorough biological inventory. The information gained from these surveys is the critical first step toward establishing the sound biological data needed to support the designation of future protected areas. In determining the suitability and need for protection, it is essential first to determine the density and diversity of species contained within that area. This information provides the essential groundwork for understanding the evolution and ecosystem dynamics that uniquely define a given habitat. The data gathered are vital for understanding patterns of species turnover along different types of ecological and geographic gradients, and for understanding their relationship to a series of biotic and abiotic parameters. After years of working with a variety of terrestrial vertebrates, the survey team members have accumulated a largely unsurpassed knowledge of the relevant groups. Most team members have an exceptional ability to detect subtle differences in coloration, morphology, and other systematic characters associated with their respective study groups. It is this subtle power of detection of novel biological diversity that usually triggers a more detailed study of the morphology, genetics, phylogenetics, and patterns of geographic distribution of newly discovered organisms.
Upon completion of field collection, our first task typically involves reconciling the observed diversity with existing taxonomy, using both specimen data and molecular phylogenetic methods. Current classifications are often complex and misleading, either because certain organismal specialists have overemphasized slight morphological differences in erecting their taxonomy (i.e., over-splitting), or the converse, where others may have overlooked important biological clues, at least in part due to insufficient number of specimens across the pertinent geographical range. Thus, our first exercise in the lab is typically to generate a molecular phylogeny as a first approximation of the fit of current taxonomy to patterns of historical diversification among species. From there, we generally focus our attention on morphological and genetic covariation. In certain cases, it is the observation of subtle morphological variation that instigates the analysis of genetic patterns (31), whereas in other cases it has been the opposite situation (11). In all cases, we find that the proper estimation of species identity is essential for understanding the historical underpinnings of the species distributions and interactions (refs. 11 and 31; K.L.H., C.H., R.R., S.M.G., and A.D.Y., unpublished work; and A.L.R., J.R., E. Palkovacs, S.M.G., and A.D.Y., unpublished work).
Here, we present four case studies, progressively ordered by their degree of development, to illustrate our approach. Each study focuses on a species complex endemic to Madagascar, and each, we hope, illustrates the importance of comprehensive biological inventory and the careful examination of associated specimens for analyzing vertebrate species in the context of historical, geographic, phenotypic, and genotypic context. It is this general synthetic approach that allows us to formulate our hypotheses of species identity, which then serve as the fundamental framework to be offered to other biologists for further testing. As Mayr has so forcefully yet eloquently argued, a veritable symphony of isolating mechanisms can differentially be at play in any given species complex. Thus, the biologist is well served to investigate any and all ecological, behavioral, and life history factors that may subtly define the margins of reproductive isolation among species.
Case 1: Sorting Out Taxonomy and Generating Species Hypotheses for Malagasy Plated Lizards
The plated lizards (family Cordylidae, subfamily Zonosaurinae) of Madagascar are an ecologically and taxonomically diverse group that consists of >18 species. These animals occupy a nearly comprehensive range of habitat types in Madagascar and can be found in virtually all regions of the island. Current systematic treatments recognize two genera, Zonosaurus, which is quite speciose, containing at least 16 species, and Tracheloptychus, which contains only two species. Although field inventory has been extensive for this group, our molecular phylogenetic investigations are in their earliest stages. Accordingly, the present focus is simply on mapping the current taxonomy onto the phylogeny to identify problematic areas in need of further investigation. At the same time, we are beginning the process of examining species history in the context of species ranges and habitat preferences. It is such consideration that will ultimately allow us to formulate hypotheses of speciation mechanisms in this group, as well as to identify areas of high diversity and endemism.
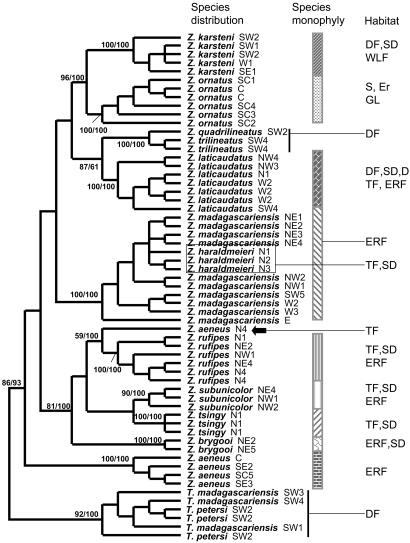
Thus far, data from the mitochondrial cytochrome b gene (Fig. 1) support the reciprocal monophyly of the two named genera. Additionally, these data support the monophyly of most of the species contained in the genus Zonosaurus. In this genus, the five most widely distributed species (Zonosaurus aeneus, Zonosaurus karsteni, Zonosaurus laticaudatus, Zonosaurus madagascariensis, and Zonosaurus ornatus) exhibit very different phylogenetic structures. Whereas Z. karsteni, Z. laticaudatus, and Z. ornatus exhibit species monophyly, the widely distributed Z. madagascariensis is paraphyletic with respect to Zonosaurus haraldmeieri, which. in turn, has a very restricted distribution. Given that Z. haraldmeieri is morphologically very similar to and genetically nested within Z. madagascariensis, it will probably be advisable to sink Z. haraldmeieri into Z. madagascariensis if no fixed, diagnostic characters are found to support the continued recognition of the former. In fact, previous distinctions between these two species have been based predominantly on their geographic distributions and habitat preferences. Z. madagascariensis is an evergreen rainforest species that is widely distributed across the island, except in the extreme southeast and in the extreme north, whereas Z. haraldmeieri is a semideciduous species found in a small area in the extreme north.
Fig. 1.
Molecular phylogeny of plated lizards. Shown is a parsimony tree based on full-length cytochrome b sequences. Numbers indicate bootstrap support and posterior probability scores for clades representing various species. Sampling localities are as follows: Northern Madagascar (N) 1, Ankarana; N2, Analamera; N3, Ambre; N4, Daraina. Northeastern Madagascar (NE) 1, Anjanaharibe-Sud; NE2, Antalaha; NE3, Betaolana; NE4, Marojejy; NE5, Tampolo. Northwestern Madagascar (NW) 1, Lokobe; NW2, Manongarivo; NW3, Ambanja; NW4, Ankarafantsika. Eastern Madagascar (E), Mantadia. Central Madagascar (C), Andranomay. South central Madagascar (SC) 1, Itremo; SC2, Vinanintelo; SC3, Manambolo; SC4 Vohipaha; SC5, Ivohibe; Western Madagascar (W) 1, Ambatomainty; W2, Bemaraha; W3, Ambohijanahary. Southeastern Madagascar (SE) 1, Petriky; SE2, Andohahela; SE3, Midongy-Sud. Southwestern Madagascar (SW) 1, Kirindy Mitea; SW2, Mike-Abrahama/Andalandomo/Ankotapike; SW3, Andotabo; SW4, Tsimanampetsotsa; SW5, Analavelona. Habitat abbreviations are: DF (dry forest); ERF (evergreen rainforest); WLS (woodland savannah); S (scrub and heath land), TF (transitional forest); SD (semideciduous forest); D (deciduous forest); GL (grass land); Er (ericoid forest). Sequences are deposited in the GenBank database under accession nos. DQ004403-DQ004461. (Adapted from A.P.R., K.P.K., and A.D.Y., unpublished work.)
In the case of Z. aeneus, an individual from Daraina (indicated with an arrow in Fig. 1) branches with Zonosaurus rufipes instead of with the rest of Z. aeneus populations. In addition, the Daraina population of Z. aeneus is geographically closer to Z. rufipes than to the rest of Z. aeneus. These findings, if upheld with additional sampling of the Daraina population, suggest the presence of a cryptic species among Z. rufipes, with the overall morphological similarity between true Z. aeneus and “Z. aeneus” from Daraina due to convergence. Alternately, we must hold open the possibility that this is simply a case of specimen misidentification until additional specimens can be collected. It is essential that we return to this locality to more fully sample this potentially crucial population, an issue that probably would not have emerged in the absence of this phylogenetic analysis. Because Daraina is ecologically different from the other areas where Z. rufipes is distributed (Daraina is drier transitional forest instead of humid evergreen rain forest), the situation is quite intriguing in that it may potentially point to an example of parapatric speciation.
The cytochrome b phylogeny also confirms the species status of the closely related species pair Zonosaurus subunicolor and Z. rufipes, which have overlapping distributions. Z. subunicolor has long been considered a subspecies of Z. rufipes and was recently elevated to species level, although without strong support (29). Two years later, in 1996, Vences et al. (32) provided more detailed information based on coloration and habitat preference to justify the resurrection of Z. subunicolor. Our phylogenetic analysis of cytochrome b supports the reciprocal monophyly of these two species, thus supporting the species designations based on morphological studies.
Continued investigation will also focus on Z. rufipes and the Zonosaurus quadrilineatus-Zonosaurus trilineatus complex for which we presently have limited geographic sampling. Z. rufipes presents geographically variable color patterns, and populations from some localities show fragile body scales, a physiological character used to distinguish some plated lizard species (33). Z. quadrilineatus and Z. trilineatus are presently recognized as separate species although they are morphologically quite similar. The number of light stripes on the back used to distinguish them show intermediary forms within the same population, and these two species are never found sympatrically. Their respective distributions are separated by the Onilahy River. Thus, it is presently unclear whether they are perhaps conspecific, although geographically isolated. It will be important to sample more individuals from the two putative species across their geographic ranges to more fully address their hypothesized species status.
Case 2: Defining Geographic Boundaries Among Species and Reconstructing the History of Trident Bats in Madagascar
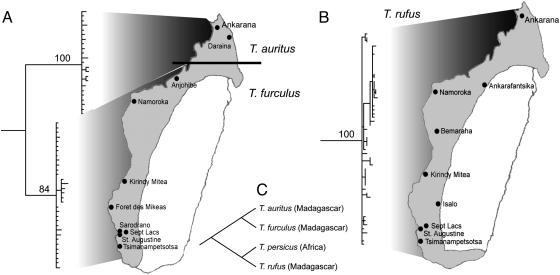
An ongoing study of trident bats (genus Triaenops, family Hipposideridae) demonstrates the ways in which extensive sampling within Madagascar yields biogeographic insights both within and beyond the island's physical limits (A.L.R., J.R., E. Palkovacs, S.M.G., and A.D.Y., unpublished work). On the basis of a recent morphological study, three species are currently recognized: Triaenops rufus, Triaenops furculus, and Triaenops auritus (J.R. and S.M.G., unpublished work). As illustrated in Fig. 2, this result is supported by molecular phylogenetic analysis. Moreover, when the Malagasy species are analyzed with their African congener (Triaenops persicus), the phylogeny reveals that the Malagasy members of this genus are paraphyletic with respect to the African species. Thus, two dispersal events between Africa and Madagascar must be invoked to explain this distribution. The unanswered question at present is whether Africa served as the center of origin, with two dispersal events to Madagascar, or whether Madagascar served as the center of diversification, with (presumably) a back migration to Africa.
Fig. 2.
Comparison of phylogenetic and biogeographic structure in Malagasy trident bats (genus Triaenops). (A) T. auritus (resurrected from synonom) (J.R. and S.M.G., unpublished work) and T. furculus show very distinct segregation into northern and southern distributions (indicated by solid line). (B) T. rufus shows diffuse distribution with no apparent biogeographic structure. (C) Comprehensive Triaenops phylogeny reveals that Malagasy taxa are paraphyletic with respect to African species, T. persicus. Sequences are deposited in the GenBank database under accession nos. DQ005718-DQ005850. (Adapted from A.L.R., E. Palkovacs, J.R., S.M.G., and A.D.Y., unpublished work.)
We are presently employing population genetic methods to address these competing dispersal hypotheses, as well as to test the hypothesis that one of the northern rivers in western Madagascar may act as a biogeographic barrier separating T. auritus and T. furculus. Neutrality tests, FS (34) and R2 (35), and mismatch distributions (36) support a history of population expansion in both T. rufus and T. furculus, with the strong indication that expansion was much more recent in T. rufus. Results from T. auritus are consistent with a history of constant population size through time, and may represent an older lineage that is at mutation-drift equilibrium. These results therefore seem to support two allochronic dispersals from Africa to Madagascar. The more northern populations of T. furculus (Namoroka and Anjohibe) are significantly differentiated from those in the south, but genetic variation within the two regions, respectively, is considerably lower, lending support to the north/south biogeographic structuring observed in some other Malagasy mammals (37). Conversely, analyses of genetic structure within T. rufus show a complete lack of geographic structure. Pastorini et al. (38) found that the Betsiboka River formed a major barrier separating populations and species in several different lemur groups. The Triaenops data, however, are not consistent with that pattern. Given the vastly different life history and dispersal characteristics in lemurs and bats, it should not be surprising that rivers might present significant barriers to dispersal for one group (lemurs), but not for another (bats).
Case 3: Revealing Unexpected Geographic and Evolutionary Patterns in Long-Tailed Shrew Tenrecs
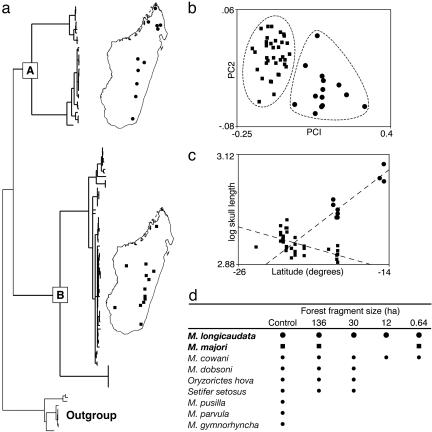
Recently, Olson et al. (11) used an integrative approach for clarifying species boundaries in one of the more broadly distributed terrestrial mammals on Madagascar (summarized in Fig. 3). The lesser long-tailed shrew tenrec (Microgale longicaudata), like many members of this most speciose genus of Malagasy mammal, has a complicated taxonomic history. Tenrecs, and shrew tenrecs in particular, exhibit numerous ontogenetic peculiarities that have stymied taxonomists for the better part of a century (see refs. 39 and 40). The number of recognized shrew tenrec species has jumped from 10 to 20 in the past two decades alone (11, 40-42), largely due to a notable increase in museum specimens from previously unsurveyed portions of the island, and to a better understanding of patterns of intra- and inter-population variation. Although traditional comparative morphology continues to advance our knowledge of shrew tenrec diversity (14, 42), molecular methods offer the advantage of rapidly uncovering cryptic genetic lineages. This proved to be the case with M. longicaudata, a forest-dwelling species with purportedly substantial levels of intraspecific morphometric variation (40). Some authors had suggested that a second described species (Microgale majori), subsequently synonymized with M. longicaudata, may warrant resurrection. The distribution of a putative M. majori was suspected to be limited to a small handful of localities. Moreover, the surveys of Ecology Training Program team members found that, at certain sites, intraspecific variation did not seem to be continuous, suggesting that two separate species might be existing in sympatry. Phylogeographic evidence from the mitochondrial ND2 gene recovered two deeply divergent and reciprocally monophyletic haplotype lineages with broad distributional overlap (indeed, members of the two respective clades have been collected in syntopy). The area of sympatric overlap spans the latitudinal extent of Madagascar's remaining forested areas (see Fig. 3). Morphometric analyses conducted on the same specimens corroborated the molecular findings, resulting in the resurrection of M. majori from synonymy. Several lines of evidence suggest that at least one additional cryptic species exists within this species complex, but current sample sizes are insufficient to rigorously test this hypothesis at present.
Fig. 3.
Overview of the approach used to clarify species limits in long-tailed shrew tenrecs and the subsequent insights into geographic variation and community structure. (a) Phylogeographic analysis of mtDNA recovers two cryptic, highly divergent, yet broadly sympatric (and in many cases syntopic), haplotype clades (clades A and B) within the single nominal species of long-tailed shrew tenrec, M. longicaudata. (b) Despite their striking morphological similarity, members of each haplotype clade are readily distinguished by both a priori and a posteriori morphometric analyses, supporting the recognition of two cryptic species, M. longicaudata (clade A, round symbols) and M. majori (clade B, square symbols). (c) The revised species-level taxonomy provides insights into biogeography and geographic variation. For example, contrasting patterns of clinal variation in body size were previously obscured. (d) Reevaluation of a published study of tenrec community assembly in fragmented forest patches (79) in light of the revised taxonomy shows that both species coexist in remarkably small habitat patches. Sequences are deposited in the GenBank database under accession nos. AY193297-AY193416. (Adapted from figures and text in ref. 11.)
Recently proposed methods that integrate molecular and morphological data to identify cryptic species (e.g., ref. 43) do not take potential sympatry among reproductively isolated lineages into account. As was shown with M. longicaudata and M. majori, widespread sympatry among cryptic species can and does occur (and, indeed, may have contributed to the continued recognition of one rather than two or more species in this case). Second, the revised taxonomy of long-tailed shrew tenrecs revealed contrasting patterns of geographic morphological variation. The failure to recognize two distinct species would have obscured the statistically significant trend toward larger body size at higher latitudes in M. majori (Fig. 3). Emerging evidence suggests that such latitudinal clinal variation may be much more widespread among Madagascar's tenrecs than has been previously appreciated (L.E.O., unpublished work). Additional insight into elevational segregation and habitat partitioning was also revealed by the phylogeographic analysis (11). Finally, the resurrection of M. majori from M. longicaudata uncovered surprising evidence that these remarkably similar species are able to coexist not only in syntopy, but within the same tiny, isolated forest fragments (Fig. 4). Whether this ability to coexist is due to subtle differences in body size or to some other mechanism, perhaps relating to differential resource utilization, remains unknown. The clarification of potential habitat partitioning is thus a question to be resolved with future field investigations. Collectively, these results challenge the assertion that, within shrew tenrecs, “no further fine adjustments to taxonomic boundaries are likely to uncover convincing examples of heretofore unknown adaptive types” (ref. 40, p. 33). Rather, continued taxonomic refinements offer the best promise for understanding adaptation and diversification in these diminutive yet unequivocally successful mammals.
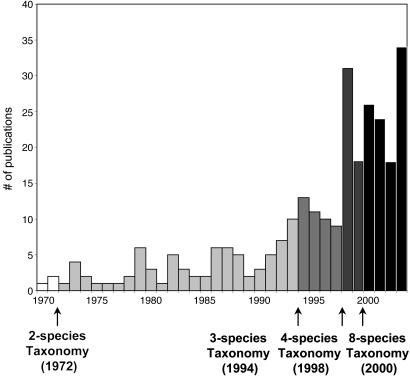
Fig. 4.
Graph of the number of publications focusing on genus Microcebus from 1970 through 2003. Increased publication activity seems to correlate with increased number of recognized species. Publication numbers were determined by means of a survey of ISI Web of Science.
Case 4: Adding a Temporal Dimension to Species Diversification in Mouse Lemurs, Moving from the Field to the Laboratory and Back Again
The genus Microcebus was considered monotypic by most authorities, containing the single species murinus (44), from the time of its original description in 1795 until the 1970s. After increased research activity and broader geographic review of mouse lemur populations, several investigators reached the conclusion that there were actually at least two distinct forms (45-47): murinus, a long-eared gray animal from the western regions of Madagascar and rufus, a short-eared reddish animal from the east. Martin (45), in particular, made note of the differing habitats and ecological constraints defining the two species, with murinus inhabiting dry deciduous and spiny desert forest and specializing on insectivory, and rufus inhabiting humid rain forest and showing dietary tendencies toward omnivory. Thus, the idea that both ecological and biogeographic mechanisms maintain species separation was an implicit assumption of the two-species taxonomy. The two-species classification remained stable until the early 1990s, after which time studies on mouse lemur biology increased sharply (48-58). The discovery that multiple species co-occurred at several localities in the west was one result of this enhanced research activity, thereby yielding an increase in the number of recognized species from two to four (59-61). Most recently, Rasoloarison et al. (31) described three new species from the western regions of Madagascar, and resurrected another two from synonymy, bringing the total count of recognized species to eight. The Rasoloarison et al. species designations were based on a combination of natural history observations, distributional data, and detailed morphometric analysis, with Yoder et al. (37) testing the species designations with mtDNA data.
Combined analysis of three mitochondrial data partitions (HV1 of the control region, cytochrome b, and cytochrome oxidase II) consistently yielded reciprocally monophyletic clades congruent with the various species recognized in the Rasoloarison et al. study (37). One of the more surprising results from the mtDNA study, however, concerns the phylogenetic placement of two population samples from eastern localities. Although it had been assumed that these two populations would belong to a single clade, congruent with their species designation of Microcebus rufus, the two populations were instead found to be paraphyletic with respect to the western species. This result strongly suggests that, contrary to the current recognition of a single species Microcebus rufus, there are at least two species of mouse lemurs in the eastern regions of Madagascar, and potentially many more (37).
The quadrupling of recognized mouse lemur species within a period of 6 years (1994-2000) undoubtedly relates (as both cause and consequence) to the enormous amount of investigative energy that has been directed toward these animals during this time interval. As Fig. 4 illustrates, the number of empirical studies focused on mouse lemurs has increased in step with the number of recognized species. One might therefore ask whether the increased scrutiny has driven the numerical expansion of recognized species, or the converse, that the appreciation of unexpected species diversity has inspired a renewed interest in these primates. Undoubtedly, both are true, although it is virtually certain that the rather uniform mouse lemur phenotype has until recently retarded our appreciation of their biological diversity. Indeed, recent studies have indicated that the process of mouse lemur diversification began at least 5 million years ago (62), making their morphological uniformity all the more intriguing.
In surveying lemur diversity at the species level, we can now appreciate that mouse lemurs are probably the most speciose of all of the Lemuriformes (with the possible exception of genus Lepilemur). Genus Eulemur ranks a close second, with at least five recognized species. The species count for Eulemur has remained nearly stable for at least the past 20 years (47) whereas that for Microcebus has changed dramatically in the past several years (31, 60, 61). Ostensibly, the differences in taxonomic stability relate to the fact that the various Eulemur species are readily identifiable according to their variety of coloration patterns and other morphological features, whereas Microcebus species are not. But why is this so? Given that we now suspect that the temporal origins of the two groups are nearly contemporaneous (63), why should rates of apparent morphological evolution have been markedly more rapid in one genus than in the other? The answer probably relates to the fact that Eulemur is primarily diurnal whereas Microcebus is strictly nocturnal.
For mammals, visual signals will be most efficiently transmitted and received by day, and other signals, such as acoustic or olfactory, will be required for nocturnal signaling. Thus, mate choice criteria will tend to mirror the signal transmission favored in a given environment (64). As discussed above, the species diversity within the genus Microcebus had been underestimated for many years, and, although we can now identify subtle patterns of coloration and morphometric variation as distinguishing among species, it is not a stretch to refer to them as a cryptic species radiation (sensu of ref. 1). Although the various species contained within the diurnal genus Eulemur show a notable array of sexually dichromic pelage variation, with males in particular showing species-specific head ornamentation, mouse lemurs are uniformly drab, showing no sexual dichromatism. These patterns perfectly fit with the prediction that diurnal animals will emphasize visual cues for mate selection whereas nocturnal animals will emphasize olfactory and auditory signals (65).
This prediction as applied to mouse lemurs seems to be born out by studies demonstrating that olfactory and hormonal signals conveyed by means of urine exposure can have powerful effects on both behavior and on basic physiological and reproductive functions in these mammals. For example, exposure to female urine can significantly increase testosterone levels in males, just as exposure to the urine of dominant males can suppress testosterone production in other males (66). Acoustic studies in particular have revealed subtleties in signaling, with two results noteworthy in their implications for potential mate-choice mechanisms. First, acoustic signals in mouse lemurs seem to evolve extremely rapidly, and second, the greatest levels of acoustic separation occur in the sexual advertisement calls of males. Relevant to the issue of rapid rates, studies of captive mouse lemur colonies in Europe reveal that colonies that have been separated for only a few generations have already begun to develop distinct dialects in their acoustic signals (67). Similarly, a detailed field study conducted in Madagascar revealed that the sexual advertisement calls of males occurring in demes separated by only 1.5 km or so showed distinct differences, even though there were no apparent biogeographic barriers separating the demes (27). Moreover, when sexual advertisement calls were compared with predator advertisement calls in two species from widely separated habitats, it was found that, although there was a great deal of overlapping interspecific variation in the predator calls, the sexual advertisement calls were entirely and profoundly distinct (54).
Thus, auditory and chemosensory data lend support to the morphological and mitochondrial hypotheses of mouse lemur species diversity. Moreover, numerous field studies are reporting that sympatric mouse lemur species practice microhabitat partitioning, either in their choice of nesting sites (68, 69) and/or with regard to competitive exclusion relating to as-yet-undetermined resources (58). It should be noted, however, that, up to the present, all published studies that have examined sympatric overlap between mouse lemur species have demonstrated sympatry only for Microcebus murinus plus another species (58, 60, 61, 68-70). Sympatric overlap of species not including Microcebus murinus has yet to be reported.
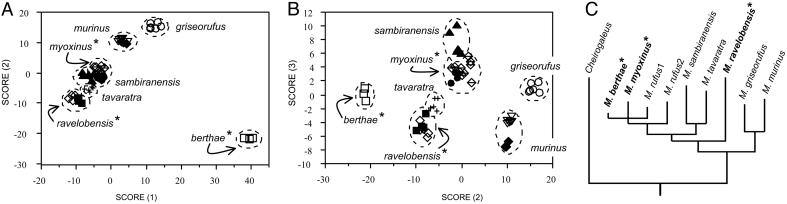
Even in light of the accumulating morphological, genetic, and behavioral evidence supporting the species level status of the eight mouse lemur groups (31, 37), more can be done to investigate the evolutionary barriers among these putative species. For example, careful scrutiny of morphological variation and genetic divergence in this group indicates that the patterns of covariation are not uniform. It seems that high levels of genetic divergence do not necessarily predict clear-cut morphological divergence, just as clear-cut levels of morphological divergence do not necessarily indicate high levels of genetic distance (Fig. 5). For these and other reasons, we are further exploring mouse lemur species patterns with nuclear loci, as advocated by many investigators (71-73). Here, the results are proving interesting, although far from decisive with relation to the question of species identity. As exemplified in Fig. 6, the nuclear markers that we have investigated thus far tend to show patterns of either incomplete lineage sorting, or perhaps persistent hybridization among several species. Only with continued genetic sampling and analysis will we be able to differentiate between these (and potentially other) explanations.
Fig. 5.
Fig. 5 illustrates the lack of precise correlation between morphometric distinctiveness and genetic divergence in mouse lemur species. The results of discriminant function analysis of 34 cranial, dental, and external morphometric characters are redrawn from ref. 37. Functions 1 and 2 (A) show conspicuous discrimination of Microcebus berthae from other species, but otherwise do not discriminate well among species. Functions 2 and 3 (B) show discrimination of all species, with Microcebus berthae remaining as highly distinct. (C) A maximum likelihood phylogram of species-specific haplotypes derived from the fibrinogen α intron 4 locus illustrates that Microcebus berthae and Microcebus myoxinus are genetically very similar. Conversely, Microcebus ravelobensis is genetically and phylogenetically divergent.
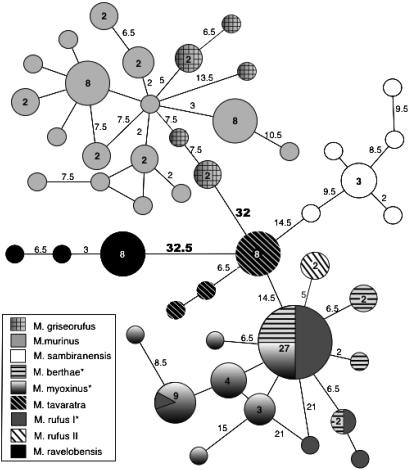
Fig. 6.
Minimum spanning network of the fibrinogen α intron 4 (609 bp) in genus Microcebus. This network was calculated in arlequin 2.000 (80) using pairwise differences between haplotypes. Each shading represents an individual species. Numbers inside circles and squares are the number of individuals sharing a haplotype; empty circles equal one individual. Numbers on connecting lines are the number of nucleotide changes separating each haplotype; empty lines equal one change. Note that all alleles for Microcebus ravelobensis are species-specific and are highly diverged from alleles sampled from other species. Conversely, the allele in greatest frequency within the myoxinus-berthae-rufus1 clade is identical among the three species. Sequences are deposited in the GenBank database under accession nos. DQ003345-DQ003479. (Adapted from K.L.H., R.R., S.M.G., and A.D.Y., unpublished work.)
Finally, among the most intriguing of the genetic results is the observation that Microcebus ravelobensis is genetically quite diverged from other mouse lemur species (Fig. 6). This result becomes particularly interesting when considered in the light of emerging physiological data. It has long been known that mouse lemurs demonstrate a physiological specialization for torpor (51, 57, 74-76). Numerous studies of naturally occurring mouse lemur populations have shown a strong seasonal pattern of daily torpor in response both to changes in ambient temperature and to photoperiodic variations, with this behavior presumably adapting them for extreme resource limitation during Madagascar's dry season (51, 75, 77). Mouse lemurs progressively accumulate fat stores during the wet season (77), after which time they spontaneously enter a period of daily torpor during the dry season. Presently, however, our information is limited to a few localities in western Madagascar, and predominantly, to a single species, Microcebus murinus. It is therefore of evolutionary consequence to ask whether this unusual system is characteristic of all mouse lemur species and populations, or only to a subset of species and habitats. At present, there is preliminary indication that Microcebus ravelobensis is perhaps unique among mouse lemurs in that it does not enter torpor (57). Given our new-found appreciation for the genetically divergent position of this species, this physiological anomaly becomes all the more meaningful. It will be a fascinating exercise to return to the field, giving refined scrutiny to the ecological and other biological characteristics of this highly derived mouse lemur species.
Summary
With the case studies above, we hope to have illustrated both the complexity and the importance of resolving species boundaries in nature. As a process of discovery, the identification of species requires a multidimensional approach that employs tools spanning everything from human intuition to molecular phylogenetics. In the cases highlighted above, we have attempted to illustrate the roles that basic biological inventory, the collection of specimen data, and the careful analysis of morphological and genetic variation can play in delimiting species boundaries, or at least for hypothesizing their existence. As we have shown, the impacts of this exercise can be far-reaching, spanning disciplines of behavioral ecology to biogeography to conservation biology. By providing biologists with a hypothesis of species identity, distribution, and abundance, a cascade of investigation is prompted that will ultimately reflect back upon and refine the initial hypothesis (78). As described above, systematic clarification, coupled with the revelation of cryptic biological diversity, is yielding insight into the myriad of ecological, evolutionary, and biogeographic forces that have shaped Madagascar's vertebrate diversity. The need for species discovery and documentation in Madagascar and other biodiversity hotspots is urgent. There remains a vast wealth of species yet to be identified, and their future is uncertain at best. To address these problems, we advocate a collaborative structure among biologists of differing organismal, methodological, and analytical skills, as well as of differing cultural backgrounds, so that the process can move rapidly and expertly from field, to lab, and back again. Furthermore, as we are discovering in our work, Mayr's “three basic tasks” of the systematist provide an ideal framework for collecting, synthesizing, and implementing the acquired data for both analytical and applied goals.
Acknowledgments
A.D.Y. thanks Francisco Ayala, Walter Fitch, and Jody Hey for their invitation to participate in this colloquium celebrating the life and works of Ernst Mayr. This work has been supported by a National Science Foundation-CAREER Award and a Biodiversity Leadership Award from the Bay and Paul Foundations (to A.D.Y.). S.M.G. has been supported by awards from the MacArthur Foundation, the Volkswagen Foundation, and the Critical Ecosystem Partnership Fund of Conservation International.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Systematics and the Origin of Species: On Ernst Mayr's 100th Anniversary,” held December 16-18, 2004, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ003345-DQ003479, DQ004403-DQ004461, and DQ005718-DQ005850).
References
- 1.Mayr, E. (1995) Philos. Sci. 63, 262-277. [Google Scholar]
- 2.Mayr, E. (1942) Systematics and the Origin of Species from the Viewpoint of a Zoologist (Harvard Univ. Press, Cambridge, MA).
- 3.Goodman, S. M. & Benstead, J. P., eds. (2003) The Natural History of Madagascar (Univ. of Chicago Press, Chicago).
- 4.Coffin, M. F. & Rabinowitz, P. D. (1992) in Geology and Geophysics of Continental Margins, eds. Watkins, J. S., Zhiqiang, F. & McMillen, K. J. (American Association of Petroleum Geologists Memoir), Vol. 53, pp. 207-246. [Google Scholar]
- 5.Storey, M., Mahoney, J. J., Saunders, A. D., Duncan, R. A., Kelley, S. P. & Coffin, M. F. (1995) Science 267, 852-855. [DOI] [PubMed] [Google Scholar]
- 6.Myers, N., Mittermeier, R. A., Mittermeier, C. G., da Fonseca, G. A. B. & Kent, J. (2000) Nature 403, 853-858. [DOI] [PubMed] [Google Scholar]
- 7.de Wit, M. (2003) Annu. Rev. Earth Planet. Sci. 31, 213-248. [Google Scholar]
- 8.Andreone, F. & Greer, A. E. (2002) J. Zool. 258, 139-181. [Google Scholar]
- 9.Goodman, S. M. & Soarimalala, V. (2004) Proc. Biol. Soc. Washington 117, 265-279. [Google Scholar]
- 10.Goodman, S. M. & Cardiff, S. G. (2004) Acta Chiropt. 69, 75-81. [Google Scholar]
- 11.Olson, L. E., Goodman, S. M. & Yoder, A. D. (2004) Biol. J. Linn. Soc. 83, 1-22. [Google Scholar]
- 12.Vences, M., Andreone, F., Glaw, F. & Mattioli, F. (2002) Copeia 2002, 1057-1062. [Google Scholar]
- 13.Vences, M. & Glaw, F. (2004) J. Nat. Hist. 38, 77-118. [Google Scholar]
- 14.Jenkins, P. D. & Goodman, S. M. (1996) Bull. Nat. Hist. Mus. London 65, 155-164. [Google Scholar]
- 15.Goodman, S. M., Hawkins, A. F. A. & Domergue, C. A. (1997) Bull. Br. Ornithol. Club 117, 4-10. [Google Scholar]
- 16.Sechrest, W., Brooks, T. M., da Fonseca, G. A., Konstant, W. R., Mittermeier, R. A., Purvis, A., Rylands, A. B. & Gittleman, J. L. (2002) Proc. Natl. Acad. Sci. USA 99, 2067-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreone, F. & Luiselli, L. M. (2003) Ital. J. Zool. 70, 53-63. [Google Scholar]
- 18.Dufils, J.-M. (2003) in The Natural History of Madagascar, eds. Goodman, S. M. & Benstead, J. P. (Univ. of Chicago Press, Chicago), pp. 88-96.
- 19.Horning, N. (2000) Lemur News 5, 28-30. [Google Scholar]
- 20.Green, G. M. & Sussman, R. W. (1990) Science 248, 212-214. [DOI] [PubMed] [Google Scholar]
- 21.Crandall, K. A., Bininda-Emonds, O. R. P., Mace, G. M. & Wayne, R. K. (2000) Trends Ecol. Evol. 15, 290-295. [DOI] [PubMed] [Google Scholar]
- 22.Karl, S. A. & Bowen, B. W. (1999) Conserv. Biol. 13, 990-999. [Google Scholar]
- 23.Moritz, C. (1994) Trends Ecol. Evol. 9, 373-375. [DOI] [PubMed] [Google Scholar]
- 24.Moritz, C. (1995) Philos. Trans. R. Soc. London B 349, 113-118. [Google Scholar]
- 25.Ryder, O. A. (1986) Trends Ecol. Evol. 1, 9-10. [Google Scholar]
- 26.Jenkins, P. D. (1992) Bull. Br. Mus. Nat. Hist. (Zool). 58, 53-59. [Google Scholar]
- 27.Hafen, T., Neveu, H., Rumpler, Y., Wilden, I. & Zimmermann, E. (1998) Folia Primatol. 69, Suppl. 1, 342-356. [DOI] [PubMed] [Google Scholar]
- 28.Carleton, M. D. & Goodman, S. M. (1996) Fieldiana (Zool.) 85, 231-256. [Google Scholar]
- 29.Glaw, F. & Vences, M. (1994) A Fieldguide to the Amphibians and Reptiles of Madagascar (Moos Druck, Leverkussen, Germany).
- 30.Vences, M. & Glaw, F. (2002) Trop. Zool. 15, 141-163. [Google Scholar]
- 31.Rasoloarison, R. M., Goodman, S. M. & Ganzhorn, J. U. (2000) Int. J. Primatol. 21, 963-1019. [Google Scholar]
- 32.Vences, M., Müller-Jung, J., Glaw, F. & Böhme, W. (1996) Senckenb. Biol. 76, 47-59. [Google Scholar]
- 33.Raselimanana, A. P., Raxworthy, C. J. & Nussbaum, R. A. (2000) Sci. Pap. Univ. Kansas Nat. Hist. Mus. 18, 1-16. [Google Scholar]
- 34.Fu, Y.-X. & Li, W.-H. (1997) Mol. Biol. Evol. 14, 195-199. [DOI] [PubMed] [Google Scholar]
- 35.Ramos-Onsins, S. E. & Rozas, J. (2002) Mol. Biol. Evol. 19, 2092-2100. [DOI] [PubMed] [Google Scholar]
- 36.Slatkin, M. & Hudson, R. R. (1991) Genetics 129, 555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoder, A. D., Rasoloarison, R. M., Goodman, S. M., Irwin, J. A., Atsalis, S., Ravosa, M. J. & Ganzhorn, J. U. (2000) Proc. Natl. Acad. Sci. USA 97, 11325-11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastorini, J., Thalmann, U. & Martin, R. D. (2003) Proc. Natl. Acad. Sci. USA 100, 5879-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenkins, P. D., Raxworthy, C. J. & Nussbaum, R. A. (1997) Bull. Nat. Hist. Mus. London (Zool.) 63, 1-12. [Google Scholar]
- 40.MacPhee, R. D. E. (1987) Am. Mus. Novit. 2889, 1-45. [Google Scholar]
- 41.Jenkins, P. D. (2003) in The Natural History of Madagascar, eds. Goodman, S. M. & Benstead, J. P. (Univ. of Chicago Press, Chicago), pp. 1273-1278.
- 42.Goodman, S. M. & Soarimalala, V. (2004) Proc. Biol. Soc. Washington 117, 250-264. [Google Scholar]
- 43.Wiens, J. J. & Penkrot, T. A. (2000) Syst. Biol. 51, 69-91. [DOI] [PubMed] [Google Scholar]
- 44.Schwarz, E. (1931) Proc. Zool. Soc. London 1931, 399-428. [Google Scholar]
- 45.Martin, R. D. (1972) Z. Tierpsychol. 9, Suppl., 43-89. [Google Scholar]
- 46.Petter, J.-J., Albignac, R. & Rumpler, Y. (1977) in Faune de Madagascar (ORSTOM and CNRS, Paris), Vol. 44, pp. 1-513. [Google Scholar]
- 47.Tattersall, I. (1982) The Primates of Madagascar (Columbia Univ. Press, New York).
- 48.Wright, P. C. & Martin, L. B. (1995) in Creatures of the Dark, eds. Alterman, L., Doyle, G. A. & Izard, M. K. (Plenum, New York).
- 49.Ortmann, S., Heldmaier, G., Schmid, J. & Ganzhorn, J. U. (1997) Naturwissenschaften 84, 28-32. [DOI] [PubMed] [Google Scholar]
- 50.Radespiel, U., Cepok, S., Zietemann, V. & Zimmermann, E. (1998) Am. J. Primatol. 46, 77-84. [DOI] [PubMed] [Google Scholar]
- 51.Ganzhorn, J. U. & Schmid, J. (1998) Int. J. Primatol. 19, 785-796. [Google Scholar]
- 52.Fietz, J. (1999) Am. J. Primatol. 48, 127-133. [DOI] [PubMed] [Google Scholar]
- 53.Atsalis, S. (2000) Am. J. Primatol. 51, 61-78. [DOI] [PubMed] [Google Scholar]
- 54.Zimmermann, E., Vorobieva, E., Wrogemann, D. & Hafen, T. G. (2000) Int. J. Primatol. 21, 837-852. [Google Scholar]
- 55.Perret, M. & Aujard, F. (2001) Am. J. Physiol. 281, R1925-R1933. [DOI] [PubMed] [Google Scholar]
- 56.Genin, F., Nibbelink, M., Galand, M., Perret, M. & Ambid, L. (2003) Am. J. Physiol. 284, R811-R818. [DOI] [PubMed] [Google Scholar]
- 57.Randrianambinina, B., Rakotondravony, D., Radespiel, U. & Zimmermann, E. (2003) Primates 44, 321-331. [DOI] [PubMed] [Google Scholar]
- 58.Schwab, D. & Ganzhorn, J. U. (2004) Int. J. Primatol. 25, 307-330. [Google Scholar]
- 59.Thalmann, U. & Rakotoarison, N. (1994) Folia Primatol. 63, 156-161. [DOI] [PubMed] [Google Scholar]
- 60.Schmid, J. & Kappeler, P. M. (1994) Folia Primatol. 63, 162-170. [DOI] [PubMed] [Google Scholar]
- 61.Zimmermann, E., Cepok, S., Rakotoarison, N., Zietemann, V. & Radespiel, U. (1998) Folia Primatol. 69, 106-114. [DOI] [PubMed] [Google Scholar]
- 62.Yang, Z. & Yoder, A. D. (2003) Syst. Biol. 52, 705-716. [DOI] [PubMed] [Google Scholar]
- 63.Yoder, A. D. & Yang, Z. (2004) Mol. Ecol. 13, 757-773. [DOI] [PubMed] [Google Scholar]
- 64.Endler, J. A. (1992) Am. Nat. 139, S125-S153. [Google Scholar]
- 65.Jones, G. (1997) Adv. Study Behav. 26, 317-354. [Google Scholar]
- 66.Perret, M. & Schilling, A. (1995) Physiol. Behav. 58, 633-639. [DOI] [PubMed] [Google Scholar]
- 67.Zimmermann, E. & Hafen, T. G. (2001) Am. J. Primatol. 54, 129-141. [DOI] [PubMed] [Google Scholar]
- 68.Weidt, A., Hagenah, N., Randrianambinina, B., Radespiel, U. & Zimmermann, E. (2004) Am. J. Phys. Anthropol. 123, 40-51. [DOI] [PubMed] [Google Scholar]
- 69.Radespiel, U., Ehresmann, P. & Zimmermann, E. (2003) Am. J. Primatol. 59, 139-151. [DOI] [PubMed] [Google Scholar]
- 70.Yoder, A. D., Burns, M. M. & Genin, F. (2002) Int. J. Primatol. 23, 1335-1343. [Google Scholar]
- 71.Zhang, D. X. & Hewitt, G. M. (2003) Mol. Ecol. 12, 563-584. [DOI] [PubMed] [Google Scholar]
- 72.Brumfield, R. T., Beerli, P., Nickerson, D. A. & Edwards, S. V. (2003) Trends Ecol. Evol. 18, 249-256. [Google Scholar]
- 73.Edwards, S. V. & Beerli, P. (2000) Evolution Int. J. Org. Evolution 54, 1839-1854. [DOI] [PubMed] [Google Scholar]
- 74.Martin, R. D. (1973) in Comparative Ecology and Behaviour of Primates, eds. Michael, R. P. & Crook, J. H. (Academic, London), pp. 1-68.
- 75.Aujard, F., Perret, M. & Vannier, G. (1998) J. Comp. Physiol. B 168, 540-548. [DOI] [PubMed] [Google Scholar]
- 76.Genin, F. & Perret, M. (2003) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136, 71-81. [DOI] [PubMed] [Google Scholar]
- 77.Genin, F. & Perret, M. (2000) Physiol. Behav. 71, 315-321. [DOI] [PubMed] [Google Scholar]
- 78.Hey, J., Waples, R. S., Arnold, M. L., Butlin, R. K. & Harrison, R. G. (2003) Trends Ecol. Evol. 18, 597-603. [Google Scholar]
- 79.Goodman, S. M. & Rakotondravony, D. (2000) J. Zool. 250, 193-200. [Google Scholar]
- 80.Schneider, S., Roessli, D. & Excoffier, L. (2000) arlequin: A Software for Population Genetics Data Analysis (Genetics and Biometry Lab., Dept. of Anthropology, Univ. of Geneva, Geneva), Version 2.000.