Abstract
The complete genome of Marek's disease virus serotype 1 (MDV-1) strain 584Ap80C was cloned in Escherichia coli as a bacterial artificial chromosome (BAC). BAC vector sequences were introduced into the US2 locus of the MDV-1 genome by homologous recombination. Viral DNA containing the BAC vector was used to transform Escherichia coli strain DH10B, and several colonies harboring the complete MDV-1 genome as an F plasmid (MDV-1 BACs) were identified. DNA from various MDV-1 BACs was transfected into chicken embryo fibroblasts, and from 3 days after transfection, infectious MDV-1 was obtained. Growth of MDV-1 recovered from BACs was indistinguishable from that of the parental virus, as assessed by plaque formation and determination of growth curves. In one of the MDV-1 BAC clones, sequences encoding glycoprotein B (gB) were deleted by one-step mutagenesis using a linear DNA fragment amplified by PCR. Mutant MDV-1 recovered after transfection of BAC DNA that harbored a 2.0-kbp deletion of the 2.6-kbp gB gene were able to grow and induce MDV-1-specific plaques only on cells providing MDV-1 gB in trans. The gB-negative virus reported here represents the first MDV-1 mutant with a deletion of an essential gene and demonstrates the power and usefulness of BACs to analyze genes and gene products in slowly growing and strictly cell-associated herpesviruses.
Marek's disease virus (MDV) is a member of the Alphaherpesvirinae subfamily of the Herpesviridae (8, 12, 36). Based on virulence for chickens, ability to induce T-cell lymphomas, and antigenic properties, there are three serotypes of MDV (MDV serotype 1 [MDV-1], MDV-2, and MDV-3) (24, 29). MDV-3 represents the herpesvirus of turkeys (HVT) which has been widely used for vaccination against MD. According to the most recent nomenclature, MDV-1 is classified as gallid herpesvirus 2 (GHV-2), MDV-2 is classified as GHV-3, and HVT is classified as meleagrid herpesvirus. All three viruses belong to the new genus Marek's disease-like viruses within the Alphaherpesvirinae (36).
Control of MDV-1 infection was achieved by vaccination, primarily with HVT. However, after vaccination failures and description of the so-called “very virulent” MDV-1, MDV-2 strains and later attenuated MDV-1 strains (e.g., strain Rispens CVI 988) have been used in vaccine formulations (38, 40). In recent years and first reported in the United States, even more virulent MDV-1, “very virulent plus” (vv+), MDV-1 variants appeared and caused high mortality in vaccinated flocks (39). One of these vv+ strains, 584A, was passaged serially on chicken embryo fibroblasts (CEF) and lost pathogenicity for chickens (39). The reasons for the differences or changes in the pathogenicity of 584A or other MDV-1 strains are poorly understood, because molecular analyses of MDV-1 using recombinant virus mutants are difficult to perform as no infectious virus progeny is released into the supernatants of cultured cells and only primary or secondary chicken or duck cells have allowed efficient growth of MDV-1 (29). Due to these problems, multiple rounds of purification of virus recombinants by coseeding infected and uninfected CEF are needed (1, 9, 21, 22, 23, 27, 30).
In recent years, manipulations of the large herpesvirus genomes have been facilitated by using bacterial artificial chromosome (BAC) vectors. The genomes of murine and human cytomegaloviruses (HCMV) (3, 15), herpes simplex virus type 1 (34), pseudorabies virus (PrV) (32, 33), and Epstein-Barr virus (10) have been cloned as infectious BACs using this technique. Targeted and random mutagenesis of herpesvirus genomes cloned as BACs is considerably faster and more reliable because mutagenesis is no longer dependent on growth of the viruses in eukaryotic cells but can be performed in Escherichia coli (3, 6, 15, 33, 34, 37).
The aim of this study was to provide a basis for fast and efficient production of MDV-1 recombinants by cloning of the complete 180-kbp genome in E. coli as a stable F plasmid and to apply a recently developed recE- and recT-based mutagenesis system (17, 18, 41) to cloned MDV-1 DNA. Infectious MDV-1 was readily recovered after transfection of cloned BAC DNA, and MDV-1 BACs were stable after several rounds of bacterial growth or serial propagation in CEF. Last, because one-step deletion of an essential MDV-1 gene in E. coli was possible, the system was shown to be of advantage for analysis of essential and nonessential MDV-1 genes and may serve as a tool for production of biologically safe modified live virus and/or DNA vaccines.
MATERIALS AND METHODS
Virus and cells.
Primary or secondary CEF or quail muscle cells (QM7; ATCC cell line CRL-1962) were maintained in Dulbecco's modified essential medium supplemented with 5 to 10% fetal calf serum. MDV-1 strain 584Ap80C was kindly provided by Richard L. Witter, Avian Diseases and Oncology Laboratory (ADOL), East Lansing, Mich. Strain 584Ap80C represents an avirulent, cell-culture-passaged descendant of vv+ strain 584A (39) and was grown on primary or secondary CEF cells as previously described (19). From a variety of permanent avian cells tested, QM7 cells were shown to support growth of MDV-1. Subsequently, the absence of MDV-1 sequences in QM7 cells was tested by Southern blot hybridization and PCR targeting different regions of the genome before they were used for propagation of MDV-1 (V. Zelnik, R. Riebe, and N. Osterrieder, unpublished results). Virus growth curves were determined as described previously (23) with slight modifications. Briefly, 100 PFU were used to infect 2 × 106 freshly seeded CEF cells. At various times after infection (0, 12, 24, 48, 72, 96, and 120 h), infected cells were trypsinized and titrated on fresh CEF cells. Virus growth curves were determined in two independent experiments. A QM7 cell line constitutively expressing MDV-1 glycoprotein B (gB) was obtained by transfection of 106 QM7 cells with 10 μg of plasmid pcMgB. To obtain pcMgB, which is based on pcDNA3 (Invitrogen), the MDV-1 gB gene from strain Rispens was amplified by PCR using gB-specific primers (Table 1) and cloned under the control of the HCMV immediate-early promoter. Transfected QM7 cells were grown in the presence of 1 mg of G418 per ml, and gB-expressing clones were identified using anti-gB monoclonal antibody (MAb) 2K11 (kindly provided by Jean-Francois Vautherot, Institut National de la Recherche Agronomique, Tours, France). The resulting cell line constitutively expressing MDV-1 gB was designated MgB1.
TABLE 1.
Primers used to generate plasmids pDS and pcMgB and to delete gB
| Primer | Sequencea | Fragment or plasmid generated |
|---|---|---|
| MUS21 | 5′-ACAggatccGTGTTTGAATACTGG-3′ | 2.1-kb pDS |
| MUS22 | 5′-ATAgtcgacTttaattaaCCGGTAGTCATTAGC-3′ | 2.1-kb pDS |
| MUS23 | 5′-ATCgcatgcttaattaaTTTGGCAAAACGGAATAGG-3′ | 3.0-kb pDS |
| MUS24 | 5′-CGCaagcttAATATGAATCTCTAAAACTTCTCGGC-3′ | 3.0-kb pDS |
| gB-up | 5′-GATAgaattcATGCACTATTTTAGGCGG-3′ | pcMgB |
| gB-low | 5′-ATACctcgagTTACACAGCATCATCTTCTG-3′ | pcMgB |
| gBkana | 5′-TTTTCTTTCATCAATAGATGTTCGTTCCAAATCATCTGATTCCTCGCCAT AAAGCACTAAATCGGAACCCTAAAGGGAGC-3′ | Kanr gene for gB deletion |
| gBkanb | 5′-ATATAGACAGATCACTAATCGATATACAGATAGGACGCCCGTTTCCATTG ATTGTCTCCTTCCGTGTTTCAGTTAGCCTC-3′ | Kanr gene for gB deletion |
For primers MUS21 to gB-low, bold sequences indicate the restriction enzyme sites, and sequences in italics indicate additional bases not present in the MDV-1 sequence. For primers gBkana and gBkanb, underlined sequences indicate the sequences from pEGFP-N1 used to amplify the Kanr gene, and sequences in bold italics indicate the gB sequences to allow homologous recombination for recE- and recT-mediated deletion of gB.
Construction of MDV-1 BACs.
MDV-1 DNA was purified from infected cells by sodium dodecyl sulfate-proteinase K extraction as described earlier (16). For construction of plasmid pDS-pHA1, 2.1- and 3.0-kbp fragments on either side of the MDV-1 US2 gene (Fig. 1) were amplified by PCR using primers containing appropriate restriction enzyme sites (Table 1). Both fragments were subsequently cloned into pTZ18R (Pharmacia-Amersham) to obtain plasmid pDS (Fig. 1). A BAC vector containing the Eco-gpt gene under the control of the HCMV immediate-early promoter was released as a PacI fragment from plasmid pHA1 (15; kindly provided by M. Messerle, Ludwig-Maximilians Universität, Munich, Germany) and inserted into the PacI sites of the 2.1- and 3.0-kbp fragment cloned in pDS (Fig. 1; Table 1). Primary CEF cells were cotransfected with approximately 2 μg of 584Ap80C DNA and 10 μg of pDS-pHA1. At 5 days after transfection, infected cells were plated on primary CEF cells in the presence of 250 μg of mycophenolic acid (MPA) per ml, 50 μg of xanthine per ml, and 100 μg of hypoxanthine per ml. To avoid death of actively dividing CEF, cells were seeded at a density of 1.5 × 107 cells per 75-cm2 flask and grown overnight, and selection medium was applied 1 h before addition of virus-containing cells. The MPA-xanthine-hypoxanthine selection was repeated for a total of four times. After complete cytopathic effect had developed, viral DNA was prepared from infected cells (16) and 1 μg of infected-cell DNA was electroporated into E. coli DH10B cells. Electrocompetent bacteria were prepared as described previously (17, 18), and electroporation was performed in 0.1-cm-wide cuvettes at 1,250 V, a resistance of 200 Ω, and a capacitance of 25 μF (Easyject electroporation system; Eurogenentec). Transformed bacteria were incubated in 1 ml of Luria-Bertani (LB) medium (28) supplemented with 0.4% glucose for 1 h at 37°C and then plated on LB agar containing 30 μg of chloramphenicol per ml (28). Single colonies were picked and placed in liquid LB medium, and small-scale preparations of BAC DNA were performed by alkaline lysis of E. coli (28). Large-scale preparation of BAC DNA was achieved by silica-based affinity chromatography using commercially available kits (Qiagen; Macherey & Nagel).
FIG. 1.
Schematic illustration of the cloning procedure of the transfer plasmid to introduce the BAC vector into the MDV-1 genome. The organization of the approximately 180-kbp MDV-1 genome (A) and the BamHI restriction map (B) as determined by Fukuchi et al. (11) are shown. The unique short region (US) and the ORFs located in the US are shown (C and D). A 2.1- and a 3.0-kbp fragment bordering the US2 gene (grey boxes) were amplified by PCR and cloned into plasmid pTZ18R to give rise to recombinant plasmid pDS. The 7.2-kbp BAC vector released from recombinant plasmid pHA1 (15) was inserted into pDS and resulted in plasmid pDS-pHA1 (E). Restriction enzyme sites were determined by Brunovskis and Velicer (7) and are abbreviated as follows: B, BamHI; E, EcoRI; P, PstI; Pa, PacI; S, SalI. The variable lengths of terminal DNA fragments are indicated (var).
Mutagenesis of MDV-1 BACs.
For mutagenesis of MDV-1 BAC DNA in E. coli, recE-catalyzed reactions promoting homologous recombination between linear DNA fragments, also referred to as E/T cloning, was performed (18, 41). Plasmid pGETrec (kindly provided by Panos Ioannou, Murdoch Institute, Melbourne, Australia) harboring recE, recT, and bacteriophage λ gam gene was transformed into BAC20-containing DH10B cells (18). After induction of recE, recT, and gam by addition of 0.2% arabinose, electrocompetent cells were prepared essentially as described previously (18). To delete the gB gene in BAC20, the kanamycin resistance (Kanr) gene of plasmid pEGFP-N1 (Clontech) was amplified by PCR. The designed primers contained 50-nucleotide homology arms bordering the desired deletion within gB and 20 nucleotides for amplification of the Kanr gene (Table 1). The resulting 1.6-kbp fragment was purified from an agarose gel (Qiagen) and electroporated in pGETrec-containing BAC20 cells. Colonies harboring the Camr and Kanr genes were identified on plates containing both antibiotics (18).
DNA analyses.
BAC or viral 584Ap80C DNA isolated from prokaryotic or eukaryotic cells was cleaved with EcoRI, BamHI, BglI, or StuI and separated on 0.8% agarose gels. DNA fragments were transferred to positively charged nylon membranes (Pharmacia-Amersham), and Southern blot hybridization was performed using digoxigenin-labeled BAC19 DNA or individual BamHI fragments of MDV-1 strain GA (11, 19). In addition, a gB-specific probe from plasmid pcMgB and a probe harboring the Kanr gene were prepared for analysis of gB-negative MDV-1 BAC. Chemoluminescence detection of DNA hybrids using CSPD was done according to the supplier's instructions (Roche Biochemicals).
IIF.
For indirect immunofluorescence (IIF) analysis, cells were grown on 6- or 24-well plates (Greiner) or on glass coverslips and subsequently infected where indicated. Cells were fixed with 90% acetone at various times after infection or transfection, IIF was done exactly as described, and samples were analyzed by conventional fluorescence microscopy or confocal laser scanning microscopy (14). The antibodies used were anti-gB MAb 2K11, anti-pp38 MAb H19 (9) (kindly provided by Lucy Lee, ADOL) or a convalescent serum from a chicken infected with MDV-1 (anti-MDV).
RESULTS
Construction and analysis of BACs containing complete MDV-1 genomes.
584Ap80C viral DNA was transfected into primary CEF cells together with pDS-pHA1 (Fig. 1). Five days after transfection, infected cells were seeded on fresh CEF and overlaid with selection medium. This procedure was repeated for a total of four times. Finally, DNA from recombinant MDV-1 that were able to grow in the presence of MPA-xanthine-hypoxanthine was isolated, cleaved with BamHI, and subjected to Southern blot analysis using labeled pDS as a probe. In addition to the BamHI-A fragment, two additional bands of approximately 17 and 10 kbp in size were specifically detected. This hybridization pattern indicated that approximately 10% of the viral DNA contained BAC vector sequences (data not shown). This DNA was used to transform E. coli DH10B cells. Transformed bacteria were plated on agar containing chloramphenicol, and single colonies were picked.
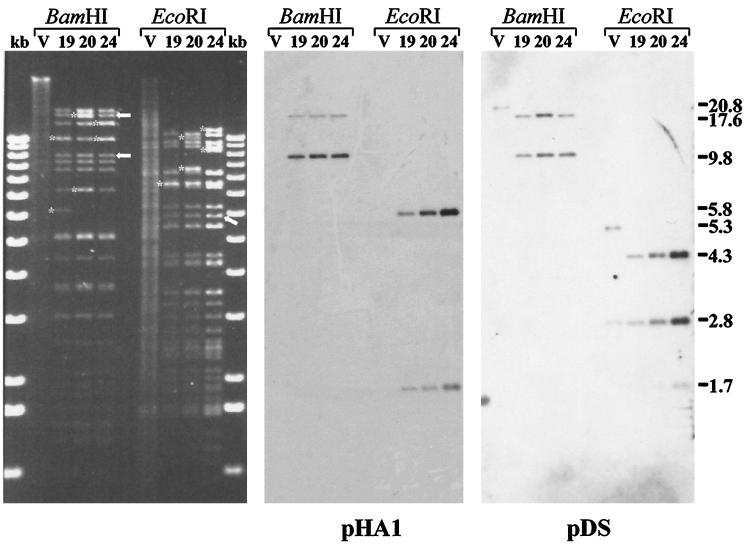
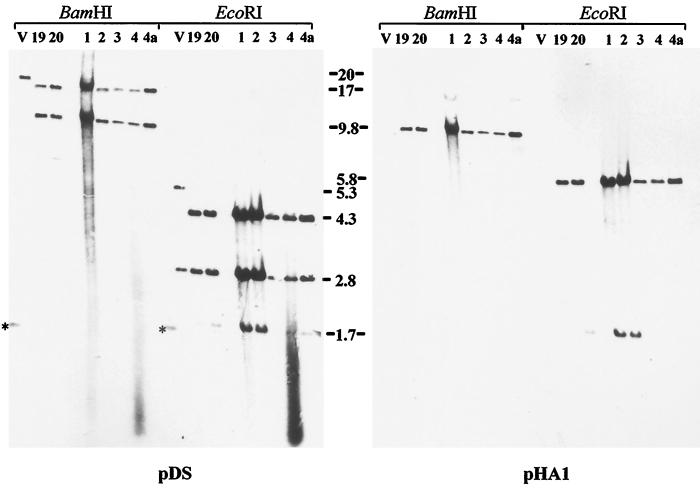
BAC DNA isolated from bacterial colonies was extracted and run on agarose gels. Several of the bacterial colonies were shown to contain high-molecular-weight extrachromosomal DNA, and three of the clones (BAC19, BAC20, and BAC24) exhibiting similar BamHI and EcoRI restriction fragment patterns were chosen for further analysis (Fig. 2). The selected BAC clones were characterized by restriction enzyme digestion and Southern blotting after cleavage with BamHI or EcoRI. It was demonstrated that compared to the parental MDV-1 strain 584Ap80C, BAC19, BAC20, and BAC24 DNA exhibited almost identical restriction enzyme fragment patterns (Fig. 2 and 3). Two notable exceptions, however, were readily recognized. The 20.8-kbp BamHI-A fragment present in 584Ap80C DNA was absent in all analyzed BAC clones. Instead, fragments of 17.4 and 9.8 kbp in size were detected in DNA from BAC19, BAC20, and BAC24 (Fig. 2). These two bands (Fig. 2) represented subfragments of BamHI-A in which an additional BamHI site was introduced by insertion of the BAC vector sequences (Fig. 1). In EcoRI-digested BAC DNA, one additional band of 5.8 kbp (BAC vector sequences) (Fig. 2) and minor alterations in sizes of fragments caused by the deletion of the US2 gene were observed (Fig. 2). The correct insertion of the BAC sequences in the various clones was proven by Southern blot hybridizations using labeled inserts of plasmid pDS or pHA1 as a probe. The expected reaction pattern in BamHI- or EcoRI-digested DNA was observed. In BamHI-digested BAC DNA, 17.4- and 9.8-kbp BamHI fragments specifically reacted with both probes, whereas in parental 584Ap80C DNA, the 20.8-kbp BamHI-A fragment hybridized only with the pDS probe (Fig. 2). In EcoRI-digested BAC19, BAC20, or BAC24 DNA, fragments of 4.3, 2.8, and 1.7 kbp specifically reacted with the pDS probe, whereas 5.8- and 1.7-kbp fragments specifically hybridized with the pHA1 probe (Fig. 2). These fragments corresponded exactly to those predicted after insertion of the pHA1 sequences (Fig. 1), and it was concluded that the BAC vector was correctly inserted instead of the US2 open reading frame (ORF) in all MDV-1 BAC clones analyzed.
FIG. 2.
Digitally scanned image of DNA of 584Ap80C (V) and DNA isolated from chloramphenicol-resistant E. coli DH10B colonies which were named BAC19, BAC20, and BAC24. Viral or BAC DNA was cleaved with BamHI or EcoRI, separated by 0.8% agarose gel electrophoresis, and stained with ethidium bromide (left panel). The restriction enzyme digests are flanked by the 1-kb ladder (Gibco-BRL). Asterisks indicate additional bands or size variations of individual fragments for the three BAC clones (in some cases the additional bands comigrate with other bands). Arrows indicate bands arising by insertion of the BAC vector sequences (left panel). Two subfragments of Bam-HI-A in which an additional BamHI site was introduced by inserting BAC vector sequences and one additional band of 5.8 kbp in EcoRI-digested BAC DNA are indicated by the arrows. After Southern transfer of DNA fragments to nylon membranes, hybridization with digoxigenin-labeled fragments released from plasmid pDS or pHA1 were performed. The sizes (in kilobase pairs) of reactive bands are given to the right.
FIG. 3.
Digitally scanned images of Southern blots to analyze size variations in BAC19, BAC20, and BAC24 DNA. Viral DNA from strain 584Ap80C and individual BACs was cleaved with BamHI or EcoRI and transferred to nylon membranes. Sheets were incubated with digoxigenin-labeled BAC19 DNA, labeled BamHI-C, or BamHI-D. The positions (in kilobase pairs) of size markers (1-kb ladder; Gibco-BRL) are given to the left. The smear-like bands 584Ap80C DNA hybridized with BamHI-D sequences are bracketed. Abbreviations: V, viral DNA from strain 584Ap80C; 19, 20, and 24, DNA from BAC19, BAC20, and BAC24, respectively.
Some variation in banding patterns of BAC19, BAC20, and BAC24 was noted in either BamHI- or EcoRI-digested DNA, e.g., an additional band of approximately 6.2 kbp in BamHI-digested BAC19 DNA or additional bands in BamHI- or EcoRI-digested DNA of BAC20 and BAC24. These additional bands, some of which were comigrating with other fragments, are indicated by asterisks in Fig. 2. To address the question of these variations of restriction enzyme patterns, hybridization with labeled BamHI-D fragment (Fig. 1) was performed because size variations in the terminal and internal repeats of the unique long region (TRL and IRL, respectively) are common in cell culture-adapted MDV-1 strains (35). It was shown by Southern blotting that the additional fragments observed in either BamHI- or EcoRI-digested DNA from BAC19, BAC20, or BAC24 resulted from variations in the TRL and IRL. Two broad smears were detected with the BamHI-D probe in viral 584Ap80C DNA digested with BamHI which ranged from approximately 9 to 15 kbp and from 4 to 8 kbp (corresponding to the BamHI-D and -H fragments of virulent MDV-1, respectively; Fig. 1). Similar observations of a smear-like bands were made after digestion of 584Ap80C DNA with EcoRI (Fig. 3). In contrast, distinct but different bands were detected with the BamHI-D probe in all BAC clones analyzed (Fig. 3). It was noted that in BAC19 only one band of ca. 8 kbp in size was apparently detected with this probe after EcoRI digestion, whereas two bands were detected in EcoRI-cleaved DNA from BAC20 and BAC24. The 8-kbp reactive band may represent a double molar band because in BamHI-digested BAC19 DNA two bands reacted with the same probe and because of its relatively high intensity compared to those of the other smaller reactive fragments and those detected in the other BACs. It must also be noted that on the original blot the 4.5-kbp EcoRI fragment present at the left terminus of the UL region (12) was visible not only in BAC24 but also in DNA from the other BACs and parental 584Ap80C. All other restriction enzyme fragments of the different BAC clones generated after cleavage of the DNA with BamHI or EcoRI appeared to be identical with those of viral 584Ap80C DNA. These observations were confirmed by Southern blot analyses using either labeled BAC19 DNA or labeled BamHI-A, -B, -C, and -I2 fragments as probes (BAC19 and BamHI-C probe shown in Fig. 3).
Reconstitution of infectious MDV-1 from cloned DNA.
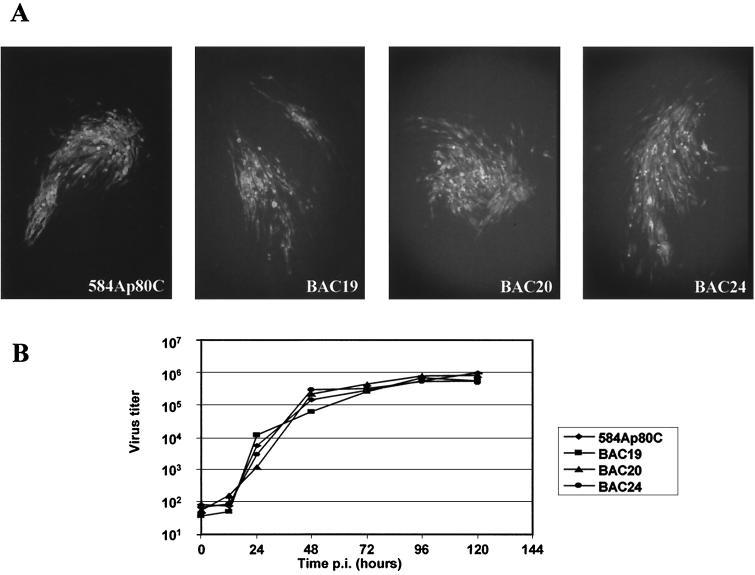
DNA from BAC19, BAC20, or BAC24 was transfected into primary CEF. From day 1 after transfection, MDV-1-specific IIF signals with several MDV-1-specific antibodies were detected. From day 3 after transfection, MDV-1-specific virus plaques clearly appeared as demonstrated by IIF using anti-MDV-1 MAbs. To determine whether growth of recombinant MDV-1 was comparable to that of parental virus, plaque sizes and virus growth curves were determined. First, MDV-1 rescued after transfection of the various BACs was coseeded with fresh CEF and sizes of plaques were compared to those induced by parental 584Ap80C. As shown for plaques stained on day 5 postinfection (p.i.), no appreciable differences in plaque sizes between MDV-1 rescued after transfection of the various MDV-1 BACs and parental viruses were detected (Fig. 4A). Second, virus growth curves of 584Ap80C and MDV-1 recovered from BACs were determined. In case of BACs, 100 PFU of virus harvested at day 5 after transfection was used to infect fresh CEF seeded on six-well plates. Similarly, 100 PFU of 584Ap80C was used to infect fresh CEF in the same way. At various times p.i., virus-infected CEF were harvested and titrated by coseeding 10-fold virus dilutions with fresh CEF. The results of these experiments are summarized in Fig. 4B. It was demonstrated that all MDV-1 reconstituted from BACs exhibited growth characteristics that were virtually identical to each other and to those of parental 584Ap80C (Fig. 4B). Virus titers steadily increased from 12 to 96 h p.i., when maximal titers were reached (Fig. 4B). From the plaque sizes and growth characteristics, we concluded that the biological properties of MDV-1 BACs in vitro were indistinguishable from those of the parental strain.
FIG. 4.
(A) IIF analysis of representative MDV-1 plaques after infection with 584Ap80C or recombinant viruses obtained after transfection of BAC19, BAC20, or BAC24 DNA. At 5 days p.i., infected cells were fixed and subjected to IIF using anti-gB MAb 2K11. Detection of bound antibodies was performed with anti-mouse Alexa 488 (Molecular Probes). Approximately 100 plaques induced by each virus were scanned under the fluorescence microscope and no significant differences between virus reconstituted from BAC clones and parental virus or between the reconstituted viruses were observed. Magnification, ×250. (B) Growth curves of MDV-1 strain 584Ap80C and viruses recovered after transfection of various BACs. After infection of CEF cells with 100 PFU of 584Ap80C or transfection progeny of BAC19, BAC20, or BAC24, virus titers were determined at the indicated times p.i. by coseeding with fresh CEF cells. Virus plaques were counted after immunofluorescent staining with MAb 2K11. Each point represents the mean of two independent experiments.
To ascertain the stability of the BAC-derived viruses, progeny of BAC transfections of BAC19 and BAC20 was passaged four times and viral DNA was prepared. Isolated DNA was cleaved with BamHI or EcoRI, and Southern blot hybridization was performed using the pDS or pHA1 probe. Identical DNA fragments as described above for BAC DNA isolated from DH10B cells were detected with the two probes (Fig. 5), and we concluded that BAC vector sequences remained stably inserted within the 584Ap80C genomes recovered from individual MDV-1 BAC clones even after serial passage in CEF. However, hybridization with the BamHI-D fragment and PCR analysis indicated that variability of the 132-bp tandem repeat sequences in the virus population was restored after the first passage in CEF (data not shown).
FIG. 5.
Digitally scanned images of Southern blots to analyze the stability of BAC vector sequences in viruses recovered after transfection of BAC19 and BAC20. Transfection progeny was passaged four times, and viral DNA was isolated after each passage. Virus DNA was cleaved with BamHI or EcoRI, separated by 0.8% agarose gel electrophoresis, and transferred to nylon membranes. Southern blot hybridization was performed using digoxigenin-labeled fragments of plasmid pDS or pHA1. Lanes: V, 584Ap80C; 19, BAC19; 20, BAC20; 1 to 4, passages 1 to 4 after transfection of BAC19 DNA, respectively; 4a, DNA isolated after passage 4 after transfection of BAC20 DNA. The sizes (in kilobases) of reactive fragments are given. Asterisks indicate the reactive 1.6-kb band of the marker (1-kb ladder; Gibco-BRL).
Mutagenesis of BAC20 and deletion of gB-encoding sequences.
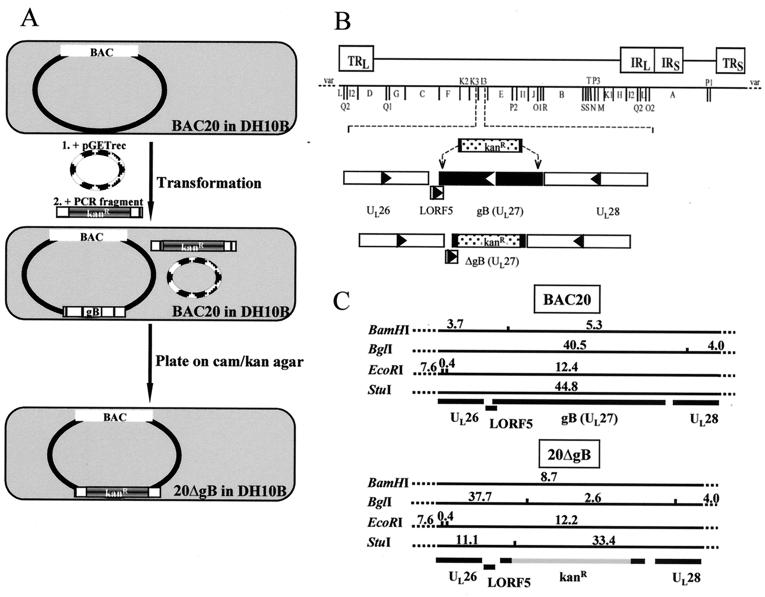
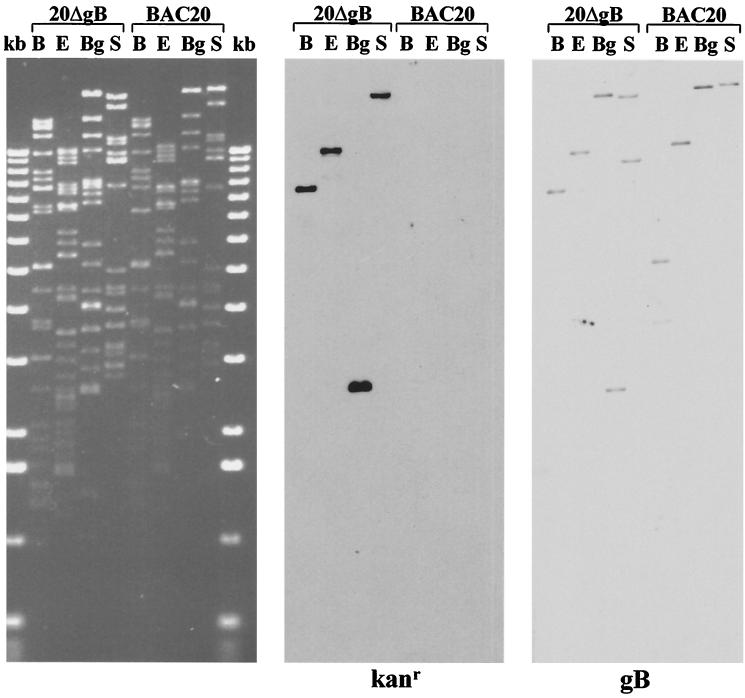
In the next experiments, a recently developed method for mutagenesis of BACs was applied to remove 2.0 kbp of the 2.6-kbp gB gene from BAC20 (Fig. 6; Table 1), i.e., nucleotides 59867 to 61881 of the MDV-1 sequence according to the numbering system of Lee et al. (12) were deleted from the gB ORF. This deletion would also affect the putative 360-nucleotide LORF5 and cause deletion of 49 nucleotides from the 3′ end of LORF5 (12). To perform one-step mutagenesis of BAC20 in E. coli, plasmid pGETrec conferring ampicillin resistance was transformed into BAC20-containing DH10B cells. Subsequently, the Kanr gene was amplified with primers of approximately 70 nucleotides in length that allowed recA-independent homologous recombination with MDV-1 gB sequences (Table 1; Fig. 6). The resulting PCR product was purified and electroporated into BAC20-pGETrec cells. Bacteria were plated on LB agar containing chloramphenicol and kanamycin, and colonies resistant to both were picked. After DNA isolation of individual colonies, Southern blot and sequence analysis of recombinant BAC20 harboring a deletion within the gB gene (20ΔgB) was performed. A Kanr- and a gB-specific probe detected fragments of mutant BAC 20ΔgB after cleavage with BamHI, EcoRI, BglI, or StuI that were in perfect agreement with those calculated after insertion of the Kanr resistance gene into gB-encoding sequences (Fig. 6C and 7). In addition, DNA cycle sequencing using primers that bind to the Kanr gene and allowed sequence determinations of the recombination sites proved the correct insertion of the Kanr gene within gB (data not shown). It was noted that pGETrec was easily lost from E. coli cells grown in the absence of ampicillin (Fig. 7). From the results of the restriction enzyme patterns, the Southern hybridizations, and the nucleotide sequencing of recombination sites, we concluded that almost the entire gB ORF was removed from mutant BAC clone 20ΔgB and that no appreciable alterations in other regions of the genome were present.
FIG. 6.
(A) Schematic illustration of mutagenesis of BAC20 to remove gB-encoding sequences. First, recombinant plasmid pGETrec encoding l-arabinose-inducible recE, recT, and bacteriophage λ gam gene was transformed into BAC20-containing DH10B cells (indicated by step 1). Subsequently, after PCR amplification of the Kanr gene from plasmid pEGFP-N1 (Clontech) with primers that also contained 50-nucleotide homology arms bordering the gB deletion, a 1.6-kbp PCR amplicon was electroporated into DH10B cells harboring both BAC20 and pGETrec (indicated by step 2). Bacterial suspensions were plated on agar containing 30 μg of kanamycin per ml and 30 μg of chloramphenicol per ml. Double-resistant colonies were picked and subjected to further analysis. (B) Schematic illustration of the location of the gB gene in the 180-kbp MDV-1 genome and the replacement of the gB ORF with the Kanr gene by homologous recombination in the recombinant BAC clone 20ΔgB. (C) Schematic representation of the BamHI, BglI, EcoRI, and StuI restriction fragment sizes of 20ΔgB generated by the insertion of the Kanr gene instead of the gB ORF. Also indicated are the restriction fragment sizes for parental BAC20. Fragment sizes are given in kilobase pairs and were calculated according to published sequences (12).
FIG. 7.
Scanned image of an ethidium bromide-stained 0.8% agarose gel containing BAC20 and 20ΔgB DNA which was cleaved with BamHI (B), EcoRI (E), BglI (Bg), or StuI (S) and separated by 0.8% agarose gel electrophoresis (left panel). DNA fragments were transferred to nylon membranes and hybridized with a digoxigenin-labeled Kanr- or gB-specific probe. The Kanr probe was prepared by labeling a 1,015-bp BlnI fragment from pEGFP-N1, and the gB probe was prepared by labeling gB sequences released from plasmid pcMgB.
Analysis of gB-negative MDV-1 reconstituted from 20ΔgB.
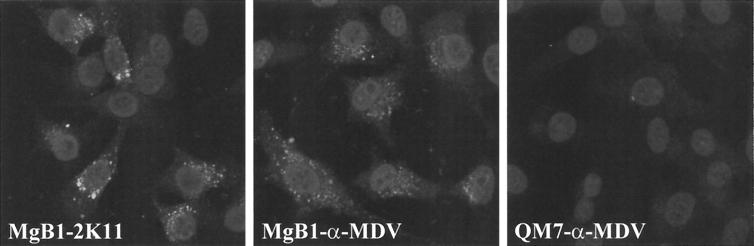
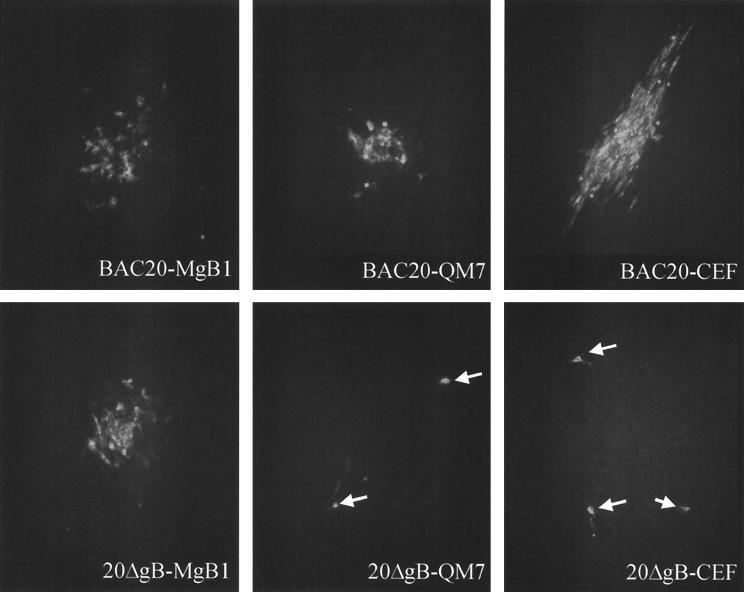
Because gB is essential for growth of all herpesviruses analyzed to date (reviewed in reference 25), a QM7 cell line which expressed MDV-1 gB under the control of the HCMV immediate-early promoter was generated (Table 1). IIF analyses demonstrated that more than 90% of the cells of the MgB1 cell line constitutively expressed MDV-1 gB as demonstrated using MAb 2K11 or a convalescent chicken serum (anti-MDV) (Fig. 8). To analyze growth of BAC20 and 20ΔgB in various cell types, DNA was prepared and used to transfect CEF, QM7, or MgB1 cells. At 3 to 5 days after transfection, virus plaques were observed in all cells transfected with BAC20 (Fig. 9). However, after transfection of 20ΔgB DNA, MDV-1-specific plaques were observed in gB-expressing MgB1 cells only (Fig. 9). In CEF and QM7 cells transfected with 20ΔgB, single cells expressed the MDV-1 pp38 protein as demonstrated by reactivity with MAb H19 (9), but plaque formation was inhibited (Fig. 9). These results of gB being required for MDV-1 cell-to-cell spread in vitro were confirmed by coseeding 20ΔgB-infected MgB1 cells with fresh CEF, QM7, or MgB1 cells. After primary transfection, plaque formation was observed only after coseeding with gB-expressing MgB1 cells (data not shown). From these results, we concluded that (i) gB is essentially required for cell-to-cell spread of MDV-1 in cultured cells and (ii) that at least the 49 nucleotides at the extreme 3′ end of the MDV-1-specific putative LORF5 are not required for growth of MDV-1 strain 584Ap80C, because the 20ΔgB virus was able to grow on cells that provided MDV-1 gB but not LORF5 in trans.
FIG. 8.
Confocal laser scan analysis of MgB1 cells constitutively expressing MDV-1 gB. MgB1 or QM7 cells were seeded on glass coverslips and incubated with anti-gB MAb 2K11 or convalescent chicken serum anti-MDV (α-MDV). Secondary antibodies were anti-mouse or anti-chicken immunoglobulin G conjugated to Alexa 488 (Molecular Probes). Nuclei were counterstained with propidium iodide. The view shown is 115 by 115 μm.
FIG. 9.
IIF analysis of MgB1, QM7, or CEF cells after transfection with BAC20 (upper panels) or 20ΔgB (lower panels). At 4 days after transfection, cells were fixed with acetone and incubated with anti-pp38 MAb H19. The secondary antibody was anti-mouse immunoglobulin G conjugated to Alexa 488 (Molecular Probes). Whereas MDV-1 plaques were observed on all cell lines after transfection of BAC20 DNA, viral plaques were observed on MgB1 cells only after transfection with 20ΔgB. Only single infected cells were observed on QM7 and CEF cells (arrowheads). Magnification, ×250.
DISCUSSION
The salient findings of this report are that (i) the complete genome of the attenuated MDV-1 strain 584Ap80C (38) was cloned as an infectious BAC and that (ii) a recently developed recE-catalyzed mutagenesis protocol was successfully applied to delete the essential gB gene from the MDV-1 genome. To our knowledge, the gB-negative virus reported here represents the first MDV-1 mutant with a deletion of an essential gene and demonstrates the power and usefulness of BACs to analyze genes and gene products in slowly growing and strictly cell-associated herpesviruses.
Although MDV is an important pathogen of chickens that causes T-cell tumors and high mortality in infected animals (24, 29), little is known about the function of individual genes and gene products in the lytic, latent, or tumor phase of the infection. Functional analyses of MDV-1 genes and gene products using mutant virus have been impaired for two main reasons. First, cultured cells infected with MDV-1 do not yield free infectious virus, and second, efficient growth of MDV-1 in cultured cells appeared to be restricted to primary or secondary chicken or duck cells (29). Hence, generation of virus recombinants by cotransfection of eukaryotic cells, i.e., by homologous recombination which is used to mutagenize other Alphaherpesvirinae has been laborious and time-consuming and has required constant supply of primary cells.
The introduction of BAC cloning into herpesvirus genomics by Messerle and coworkers (15) provided a basis for maintaining infectious herpesvirus genomes independently from eukaryotic cells and for rapid and efficient mutagenesis exploiting the recombination apparatus of prokaryotes. The BAC cloning and mutagenesis methods have since been applied to various herpesviruses comprising all three subfamilies, including the Alphaherpesvirinae herpes simplex virus and PrV (3, 6, 15, 32, 33, 34). Particularly for MDV-1, however, BAC cloning and mutagenesis could certainly be a major advantage. Once the MDV-1 genome is cloned as a BAC and can be stably maintained in E. coli, generation of mutants and analyses of essential genes should be relatively easy. We have now cloned the complete genome of MDV-1 strain 584Ap80C as an infectious BAC. Strain 584Ap80C is a descendant of the vv+ MDV-1 strain 584A after 80 serial passages on CEF (39). Analysis of the cloned MDV-1 genomes present in BAC19, BAC20, and BAC24 demonstrated that despite high similarities, variations of restriction enzyme patterns were obvious. This heterogeneity could be attributed to variations in the BamHI-D and -H fragments, i.e., the TRL and IRL region of the genome (11). It is known that different numbers of 132-bp tandem repeats are present in different MDV-1 strains and that the number of repeats increases after serial passage in cultured cells (4, 5, 11, 13, 31). Amplification of the tandem 132-bp repeats was associated with a loss of oncogenicity because a constant low number of these units was demonstrated in virulent strains (5, 6). However, recent work on the widely used Rispens vaccine strain demonstrated that there might be no direct correlation of small numbers of the 132-bp repeats and virulence (35). For MDV-1 strain 584Ap80C, hybridization of cleaved viral DNA with the BamHI-D fragment gave diffuse banding patterns, indicating a variable number of repeats present in the virus population. In contrast, only single bands were identified in each of the BAC clones with the same probe. The sizes of these bands after cleavage with BamHI or EcoRI varied in BAC19, BAC20, and BAC24, indicating that genomes containing different numbers of the 132-bp repeats had been cloned. This interpretation was substantiated by PCR analyses targeting the 132-bp repeats. Whereas the ladder-like appearance of PCR products was obtained with DNA from 584Ap80C, which is typical for attenuated MDV-1 strains (2), distinct bands were amplified from cloned viral DNA from BAC19, BAC20, or BAC24 (Zelnik et al., unpublished). It was therefore concluded that the variations of restriction enzyme patterns of the different BAC clones resulted from various numbers of tandem 132-bp repeats present in the individual clones. These variations in the TRL and IRL did not have any influence on the infectivity of the cloned DNA because infectious virus was recovered after transfection of DNA isolated from each of the different BAC clones.
After cloning of the complete MDV-1 genome and proof of infectivity of MDV-1 DNA maintained and propagated as an F plasmid in E. coli, a recently developed mutagenesis system which permits homologous recombination of linear DNA fragments with BACs (17, 18, 41) was used to delete gB-encoding sequences of BAC20. The mutagenesis is based on recE, recT, and the recB- and recC-suppressing bacteriophage λ gam gene present on plasmid pGETrec (18). The big advantages of this mutagenesis system follow. (i) Only 30- to 50-bp homology arms are needed to target a specific sequence to be deleted, i.e., deletion of any ORF can be achieved without the need to clone recombination cassettes. (ii) The method is very fast. (iii) The pGETrec vector is rapidly lost from bacterial cells in the absence of antibiotic selection. After electroporation of the gB knockout PCR product into pGETrec-containing BAC20 cells, between 10 and 30 Camr and Kanr double-resistant colonies were obtained. One of the clones was named 20ΔgB and chosen for further analyses because it had lost pGETrec immediately after it was plated on agar containing chloramphenicol and kanamycin. Southern blot analyses demonstrated successful deletion of the gB gene and the insertion of the Kanr gene in 20ΔgB. MDV-1 recovered after transfection of CEF cells with 20ΔgB was unable to spread from infected cells to neighboring cells. However, 20ΔgB growth on MgB1 cells that provide MDV-1 gB in trans was similar to that of BAC20, indicating that MDV-1 gB, like its counterparts in other herpesviruses, is essential for cell-to-cell spread of infectivity. Because MDV-1 is highly cell associated in cultured cells and does not release infectious virus to the culture medium, we were not able to investigate a possible role of MDV-1 gB in virus entry. It must be noted that by deleting the majority of the gB gene in BAC20, the putative and MDV-specific LORF5 (12) was also affected. However, because 20ΔgB was able to grow on gB trans-complementing cells which do not express LORF5, the observed phenotype of 20ΔgB virus could be attributed solely to the absence of gB expression. The generated gB mutant represents the first example of an MDV-1 with deletion of an essential gene and demonstrates the power of the BAC cloning and mutagenesis system, which is especially useful for MDV-1. MDV-1 BACs and the permanent cell line QM7, which allows MDV-1 propagation and which—unlike the quail fibroblast cell line QT35—does not harbor MDV-1 sequences (Zelnik et al., unpublished), represent an excellent combination to analyze essential MDV-1 genes. In addition, comparative analyses on gene and protein functions of various Alphaherpesvirinae can now include MDV-1 and the use of this system allows studies on very distantly related members of the viral subfamily, because for instance, PrV is able to grow on QM7 cells.
In summary, cloning of the complete MDV-1 genome as an infectious BAC, establishment of a fast and efficient mutagenesis system, and the use of the permanent QM7 cell line should greatly facilitate future analyses of MDV-1 genes. In addition, it may be possible to generate a novel generation of MDV-1 vaccines based on BACs. At present, other MDV-1 BACs containing genomes of virulent strains like RB1B are being generated. With these cloned genomes in hand, a detailed assessment of genes expressed in the lytic, latent, and tumor phases of MDV-1 infection should be possible.
ACKNOWLEDGMENTS
We gratefully acknowledge the expert technical assistance of Kerstin Wink. We are indebted to Richard L. Witter, ADOL, East Lansing, Mich., for generously providing MDV-1 strains, including 584Ap80C, and to Martin Messerle, LMU, Munich, Germany, for providing plasmid pHA1 and for help and support in BAC cloning. We thank Panos Ioannou, Murdoch Institute, Melbourne, Australia, for providing plasmid pGETrec and constant advice. Lucy Lee, ADOL, and Jean-Francois Vautherot, Institut National de la Recherche Agronomique, Tours, France, generously provided MAbs H19 and 2K11, respectively.
This work was supported by grant QLK2-CT-1999-00601 from the Commission of the European Union.
REFERENCES
- 1.Anderson A S, Parcells M S, Morgan R W. The glycoprotein D (US6) homolog is not essential for oncogenicity or horizontal transmission of Marek's disease virus. J Virol. 1998;72:2548–2553. doi: 10.1128/jvi.72.3.2548-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker Y, Tabor E, Asher Y, Davidson I, Malkinson M, Witter R L. PCR detection of amplified 132 bp repeats in Marek's disease virus type 1 (MDV-1) DNA can serve as an indicator for critical genomic rearrangement leading to the attenuation of virus virulence. Virus Genes. 1993;7:277–287. doi: 10.1007/BF01702588. [DOI] [PubMed] [Google Scholar]
- 3.Borst E M, Hahn G, Koszinowski U H, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J Virol. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley G, Hayashi M, Lancz G, Tanaka A, Nonoyama M. Structure of the Marek's disease virus BamHI-H gene family: genes of putative importance for tumor induction. J Virol. 1989;63:2534–2542. doi: 10.1128/jvi.63.6.2534-2542.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley G, Lancz G, Tanaka A, Nonoyama M. Loss of Marek's disease virus tumorigenicity is associated with truncation of RNAs transcribed within BamHI-H. J Virol. 1989;63:4129–4135. doi: 10.1128/jvi.63.10.4129-4135.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brune W, Menard C, Hobom U, Odenbreit S, Messerle M, Koszinowski U H. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat Biotechnol. 1999;17:360–364. doi: 10.1038/7914. [DOI] [PubMed] [Google Scholar]
- 7.Brunovskis P, Velicer L F. The Marek's disease virus (MDV) unique short region: alphaherpesvirus-homologous, fowlpox virus-homologous, and MDV-specific genes. Virology. 1995;206:324–338. doi: 10.1016/s0042-6822(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 8.Cantello J L, Anderson A S, Francesconi A, Morgan R W. Isolation of a Marek's disease virus (MDV) recombinant containing the lacZ gene of Escherichia coli stably inserted within the MDV US2 gene. J Virol. 1991;65:1584–1588. doi: 10.1128/jvi.65.3.1584-1588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui Z Z, Yan D, Lee L F. Marek's disease virus gene clones encoding virus-specific phosphorylated polypeptides and serological characterization of fusion proteins. Virus Genes. 1990;3:309–322. doi: 10.1007/BF00569038. [DOI] [PubMed] [Google Scholar]
- 10.Delecluse H J, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuchi K, Sudo M, Lee Y-S, Tanaka A, Nonoyama M. Structure of Marek's disease virus DNA: detailed restriction enzyme map. J Virol. 1984;51:102–109. doi: 10.1128/jvi.51.1.102-109.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee L F, Wu P, Sui D, Ren D, Kamil J, Kung H J, Witter R L. The complete unique long sequence and the overall genomic organization of the GA strain of Marek's disease virus. Proc Natl Acad Sci USA. 2000;97:6091–6096. doi: 10.1073/pnas.97.11.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maotani K, Kanamori K, Ikuta K, Ueda S, Kato S, Hirai K. Amplification of a tandem repeat within inverted repeats of Marek's disease virus DNA during serial in vitro passage. J Virol. 1986;58:657–659. doi: 10.1128/jvi.58.2.657-660.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meindl A, Osterrieder N. The equine herpesvirus type 1 US2 homolog encodes a nonessential membrane-associated virion component. J Virol. 1999;73:3430–3437. doi: 10.1128/jvi.73.4.3430-3437.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan R W, Cantello J L, McDermott C H. Transfection of chicken embryo fibroblasts with Marek's disease virus DNA. Avian Dis. 1990;34:345–351. [PubMed] [Google Scholar]
- 17.Muyrers J P, Zhang Y, Testa G, Stewart A F. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayanan K, Williamson R, Zhang Y, Stewart A F, Ioannou P A. Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther. 1999;6:442–447. doi: 10.1038/sj.gt.3300901. [DOI] [PubMed] [Google Scholar]
- 19.Osterrieder N. Sequence and initial characterization of the UL10 (glycoprotein M) and UL11 homologous genes of serotype 1 Marek's Disease Virus. Arch Virol. 1999;144:1853–1863. doi: 10.1007/s007050050710. [DOI] [PubMed] [Google Scholar]
- 20.Osterrieder N, Neubauer A, Brandmüller C, Braun B, Kaaden O-R, Baines J D. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J Virol. 1996;70:4110–4115. doi: 10.1128/jvi.70.6.4110-4115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parcells M S, Anderson A S, Cantello J L, Morgan R W. Characterization of Marek's disease virus insertion and deletion mutants that lack US1 (ICP22 homolog), US10, and/or US2 and neighboring short-component open reading frames. J Virol. 1994;68:8239–8253. doi: 10.1128/jvi.68.12.8239-8253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parcells M S, Anderson A S, Morgan R W. Retention of oncogenicity by a Marek's disease virus mutant lacking six unique short region genes. J Virol. 1994;69:7888–7898. doi: 10.1128/jvi.69.12.7888-7898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parcells M S, Anderson A S, Morgan R W. Characterization of a Marek's disease virus mutant containing a lacZ insertion in the US6 (gD) homologue gene. Virus Genes. 1994;9:5–13. doi: 10.1007/BF01703430. [DOI] [PubMed] [Google Scholar]
- 24.Payne L N. Pathology. In: Payne L N, editor. Marek's disease. Hingham, Mass: Kluwer Academic Publishers; 1985. pp. 43–76. [Google Scholar]
- 25.Pereira L. Function of glycoprotein B homologues of the family herpesviridae. Infect Agents Dis. 1994;3:9–28. [PubMed] [Google Scholar]
- 26.Ross N L J, Binns M M, Pastorek J. DNA sequence and organization of genes in a 5.5 kbp EcoRI fragment mapping in the short unique segment of Marek's disease virus (strain RB1B) J Gen Virol. 1991;72:949–954. doi: 10.1099/0022-1317-72-4-949. [DOI] [PubMed] [Google Scholar]
- 27.Sakaguchi M, Urakawa T, Hirayama Y, Miki N, Yamamoto M, Zhu G S, Hirai K. Marek's disease virus protein kinase gene identified within the short unique region of the viral genome is not essential for viral replication in cell culture and vaccine-induced immunity in chickens. Virology. 1993;195:140–148. doi: 10.1006/viro.1993.1354. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch D F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schat K A. Characteristics of the virus. In: Payne L N, editor. Marek's disease. Hingham, Mass: Kluwer Academic Publishers; 1985. pp. 77–112. [Google Scholar]
- 30.Schat K A, van Iddekinge B J, Boerrigter H, O'Connell P H, Koch G. Open reading frame L1 of Marek's disease herpesvirus is not essential for in vitro and in vivo virus replication and establishment of latency. J Gen Virol. 1998;79:841–849. doi: 10.1099/0022-1317-79-4-841. [DOI] [PubMed] [Google Scholar]
- 31.Silva R F, Witter R L. Genomic expansion of Marek's disease virus DNA is associated with serial in vitro passage. J Virol. 1985;54:690–696. doi: 10.1128/jvi.54.3.690-696.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith G A, Enquist L W. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J Virol. 1999;73:6405–6414. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith G A, Enquist L W. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc Natl Acad Sci USA. 2000;97:4873–4878. doi: 10.1073/pnas.080502497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suter M, Lew A M, Grob P, Adema G J, Ackermann M, Shortman K, Fraefel C. BAC-VAC, a novel generation of (DNA) vaccines: a bacterial artificial chromosome (BAC) containing a replication-competent, packaging-defective virus genome induces protective immunity against herpes simplex virus 1. Proc Natl Acad Sci USA. 1999;96:12697–12702. doi: 10.1073/pnas.96.22.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Iddekinge B J, Stenzler L, Schat K A, Boerrigter H, Koch G. Genome analysis of Marek's disease virus strain CVI-988: effect of cell culture passage on the inverted repeat regions. Avian Dis. 1999;43:182–188. [PubMed] [Google Scholar]
- 36.van Regenmortel M H V, Fauquet C M, Bishop D H L, Carstens E, Estes M K, Lemon S, Maniloff J, Mayo M A, McGeoch D, Pringle C R, Wickner R B, editors. Virus taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. New York, N.Y: Academic Press; 1999. [Google Scholar]
- 37.Wagner M, Jonjić S, Koszinowski U H, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Witter R L. Principles of vaccination. In: Payne L N, editor. Marek's disease. Hingham, Mass: Kluwer Academic Publishers; 1985. pp. 203–250. [Google Scholar]
- 39.Witter R L. Increased virulence of Marek's disease virus field isolates. Avian Dis. 1997;41:149–163. [PubMed] [Google Scholar]
- 40.Witter R L, Sharma J M, Fadly A M. Pathogenicity of variant Marek's disease virus isolants in vaccinated and unvaccinated chickens. Avian Dis. 1980;24:210–232. [Google Scholar]
- 41.Zhang Y, Buchholz F, Muyrers J P, Stewart A F. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]