Abstract
At the end of 2022 into early 2023, the UK Health Security Agency reported unusually high levels of scarlet fever and invasive disease caused by Streptococcus pyogenes (StrepA or group A Streptococcus). During this time, we collected and genome-sequenced 341 non-invasive throat and skin S. pyogenes isolates identified during routine clinical diagnostic testing in Sheffield, a large UK city. We compared the data with that obtained from a similar collection of 165 isolates from 2016 to 2017. Numbers of throat-associated isolates collected peaked in early December 2022, reflecting the national scarlet fever upsurge, while skin infections peaked later in December. The most common emm-types in 2022–2023 were emm1 (28.7 %), emm12 (24.9 %) and emm22 (7.7 %) in throat and emm1 (22 %), emm12 (10 %), emm76 (18 %) and emm49 (7 %) in skin. While all emm1 isolates were the M1UK lineage, the comparison with 2016–2017 revealed diverse lineages in other emm-types, including emm12, and emergent lineages within other types including a new acapsular emm75 lineage, demonstrating that the upsurge was not completely driven by a single genotype. The analysis of the capsule locus predicted that only 51 % of throat isolates would produce capsule compared with 78% of skin isolates. Ninety per cent of throat isolates were also predicted to have high NADase and streptolysin O (SLO) expression, based on the promoter sequence, compared with only 56% of skin isolates. Our study has highlighted the value in analysis of non-invasive isolates to characterize tissue tropisms, as well as changing strain diversity and emerging genomic features which may have implications for spillover into invasive disease and future S. pyogenes upsurges.
Keywords: emm-type, group A Streptococcus, non-invasive, skin infection, strain diversity, Streptococcus pyogenes, throat infection, whole genome sequencing
Data Summary
All new genome sequence data is available on the NCBI short read archive under the bioproject PRJNA1062601, and individual accession numbers are listed in Tables S1 and S2, available in the online Supplementary Material (upplementary Materials 1 and 2).
Impact Statement
The human bacterial pathogen Streptococcus pyogenes, also known as group A Streptococcus or StrepA, caused a dramatic and sudden upsurge in scarlet fever in the UK at the end of 2022 into early 2023. We present molecular characterization of this upsurge, through genome sequence analysis of throat, skin and other types of non-severe infection isolates collected by the microbiology diagnostic lab at the Northern General Hospital in Sheffield, England. We found that while two strain types were the predominant cause of infections during the upsurge, other types had emerged or changed when compared with a similar collection from 2016 to 2017. We also identified differences between throat-associated isolates and skin-associated isolates and highlighted important bacterial factors that might influence infection types. Isolates from non-severe throat/skin types of infections are rarely saved, and, therefore, our knowledge of them is limited. However, here, we demonstrate that the study of such isolates may be key to understanding upsurges of more severe infections.
Introduction
The human pathogen Streptococcus pyogenes, also known as group A Streptococcus or StrepA, is a common cause of throat infections, such as pharyngitis and tonsillitis, as well as mild skin infections such as pyoderma or impetigo. More commonly in children than adults, throat infections can progress to scarlet fever, with a characteristic sandpaper-like rash and ‘strawberry tongue’. On rare occasions, S. pyogenes can also cause severe and potentially lethal invasive diseases, such as pneumonia, empyema, bacteraemia and necrotizing fasciitis (‘flesh-eating’ disease). In England, scarlet fever and invasive S. pyogenes/group A Streptococcus (iGAS) disease cases are notifiable to the UK Health Security Agency (UKHSA).
In September 2022, the UKHSA reported an unusually high level of scarlet fever notifications with ~3.7-fold more than in the same period for the previous five seasons [1]. Notifications continued to rapidly increase across England and Wales, with 8688 cases in weeks 37–48 (mid-September to end-November), compared with 333–2536 in the previous five seasons [2]. Alongside the increasing scarlet fever cases, there were also high numbers of iGAS notifications with 772 in weeks 37–48. Concerningly, during these weeks, there were more cases of iGAS in children under 15 (26.1%), compared with previous seasons (6.4–13.3%), and 14 deaths in this age group [2]. Scarlet fever notifications peaked in week 49 with 10 069 cases, and iGAS notifications peaked in week 52 with 213 cases [3]. Typing of isolates by the sequence of the emm gene, which encodes for the hypervariable M protein, identified emm1 as the most common cause of iGAS in those older than 15 (31 %) but an even higher proportion of cases in those younger than 15 (57 %). emm12 and emm4 were the second (23 %) and third (7 %) most common causes of iGAS in children, at least in the early part of the upsurge [2].
Scarlet fever demonstrates seasonality with cases typically increasing in late winter and peaking in early spring. An unexpected increase in scarlet fever cases in England was first seen in 2013–2014, peaking in early April and totalling over 13 000 notifications, compared with fewer than 3000 in previous seasons [4,5]. Notifications remained seasonally elevated, rising each year to their highest in 2017–2018 [6], until the COVID-19 (coronovirus disease 2019) pandemic when cases fell dramatically. No corresponding rise was observed for iGAS notifications until the 2015–2016 season. The link between scarlet fever cases and iGAS cases is not well understood, but it appeared that the increase in both scarlet fever and invasive disease notifications in early 2016 was due to the emergence of a new variant of a lineage carrying emm1 termed M1UK [7,8], which, prior to 2022–2023, also led to the biggest upsurge in cases in 2017–2018 [6]. The M1UK variant had steadily increased in prevalence in England since 2010 and represented 91.5 % of invasive emm1 by 2020 [8]. M1UK is characterized by 27 SNPs in comparison to a globally circulating emm1 population (M1global) and increased expression of the superantigen SpeA [7,9, 10]. The M1UK lineage has been detected in European countries, North America, Australia and New Zealand, often associated with increases in disease [7,9, 11,17].
While iGAS undergoes routine surveillance in several high-income countries, with the inclusion of scarlet fever and outbreak situations in some, these types of infections only represent a small proportion of streptococcal cases. Our knowledge of circulating non-invasive disease (from non-sterile sites) isolates is severely lacking, yet these isolates may act as an early indicator for increasing prevalence of new genotypes or lineages that could lead to more severe infections. This lack of knowledge also means we have a limited understanding of the connection between certain emm-types or lineages and preferences for causing throat infections or skin infections. As non-invasive isolates are not routinely collected, data acquisition relies on local collections.
Sheffield is a large northern city in England with a population of around 600 000. During the 2022–2023 upsurge, the region of Yorkshire and the Humber, which includes Sheffield, reported the highest rates of invasive S. pyogenes in England, at 8.7 per 100 000 population [3]. Scarlet fever notifications were also high in the region, at 132.0 per 100 000 population, although this was similar to the rate seen in the North West region and lower than the East Midlands region [3]. We began in November 2022 to routinely save all non-invasive isolates identified by the Department of Laboratory Medicine at the Northern General Hospital, Sheffield, which performs microbiological services for community care as well as surrounding hospitals. We had also previously performed a similar collection in 2016–2017 which we used as a comparative population. We undertook whole genome sequencing (WGS) analysis of both collections and characterized strain diversity and pathogenicity factors. As expected, the emm1 M1UK lineage dominated during the 2022–2023 upsurge followed by emm12, but there were some unexpected increases in other emm-types as well as differences between throat-associated isolates and skin-associated isolates.
Methods
Isolate collection
A total of 384 non-invasive isolates (from non-sterile sites), presumptively identified from culture as S. pyogenes, were collected from the Department of Laboratory Medicine, Northern General Hospital, Sheffield, UK, between November 2022 (week 45) and February 2023 (week 6). The Department of Laboratory Medicine performs microbiology diagnostics for NHS Trusts as well as primary care and community services, acting as the single regional diagnostic microbiology laboratory for a population of around 600 000 adults and children in Sheffield. Anonymized clinical data were collected for each isolate from the sample request information and electronic clinical patient records: swab source (throat/skin/ear/eye/nose), sampling date, age, sex and infection type. Cases were considered to be associated with scarlet fever where scarlet fever was queried by the clinician or a rash consistent with a scarlet fever diagnosis was described in the clinical details accompanying the request. All other throat samples, and other sample types, were deemed non-scarlet fever samples. Samples from ear, eye and nose isolates were excluded from further downstream analyses to focus the comparison on throat and skin isolates.
For comparison, a further 229 archived non-invasive S. pyogenes isolates collected in a similar manner between October 2016 and January 2017 were also included in this study, with data collected as above. The 2016–2017 season also showed a seasonal upsurge in disease compared with previous years, however with substantially fewer scarlet fever and invasive S. pyogenes cases reported compared with 2022–2023. The age distribution of cases in 2016–2017, of both scarlet fever and invasive S. pyogenes disease, was consistent with previous years, and this was therefore considered a suitably representative comparative sample.
Whole genome sequencing
Genomic DNA was extracted from all isolates using a previously described method [18]. Genomic DNA from isolates collected in 2022–2023 underwent WGS at Earlham Institute by the Genomics Pipeline group, using the LITE protocol [19] for library preparation and sequenced on a NovaSeq X plus generating 150 bp paired-end reads. Genomic DNA from isolates collected in 2016 underwent sequencing provided by MicrobesNG (https://microbesng.com) using the Nextera XT library prep kit (Illumina) and the Illumina HiSeq 2500, generating 250 bp paired-end reads.
WGS analysis
Raw sequence reads were trimmed using Trimmomatic (v0.39) with the settings LEADING:3 TRAILING:3 SLIDING WINDOW:4 : 15 MINLEN:36 [20]. For the 2022–2023 isolates, the average estimated read coverage was ~524× with some greater than 1000×. Reads were therefore randomly subsampled, using seqtk, to 1.2 million reads per isolate, to provide ~180× coverage.
Trimmed and subsampled reads were then used to perform de novo assembly using SPAdes (v.3.13.1) with k-mer sizes of 21, 33, 55 and 77 [21]. Assembly statistics were generated for each isolate using Quast [22] (Tables S1 and S2), and any draft assemblies with more than 500 contigs or a total genome size greater than 2.2 Mb were excluded from downstream analysis, as were any that were determined were not to be S. pyogenes based on their genomic sequences. MLST and emm-types were determined from the de novo assemblies using mlst (https://github.com/tseemann/mlst) with the pubmlst database [23] and the emm_typer.pl script (github.com/BenJamesMetcalf/GAS_Scripts_Reference), respectively. New MLSTs were submitted to pubmlst and new emm-types and sub-types to the CDC emm-type database (https://cdc.gov/streplab) for assignment.
De novo assemblies were annotated using Prokka (v.1.14.6) [24]. Snippy (https://github.com/tseemann/snippy) was used to determine SNP distances between sequence reads and a reference genome. RAxML v8.2.12 [25] was used to generate maximum likelihood phylogenetic trees based on the core gene alignment with a general time-reversible substitution model and 100 bootstraps. Known regions of recombination (prophage regions and other mobile genetic elements) were excluded from reference genomes prior to mapping. Where these regions were unknown in the emm22 reference genome, regions of predicted recombination were identified and removed using Gubbins [26] prior to tree construction. Phylogenetic trees were annotated using iTOL (version 6) [27].
Further comparison was made with previously published WGS data from Sheffield, consisting of 142 non-invasive skin and soft tissue isolates collected in 2019 [28], and national and international data from other published collections (Table S3).
Virulence factor, regulator and antimicrobial resistance typing
The presence of superantigen genes speA, speC, speG, speH, speI, speJ, speK, speL, speM, speK/M, speQ, speR, ssa and smeZ and DNase genes sda1, sda2, sdn, spd1, spd3 and spd4 was determined by blast (100 % coverage, 80 % identity) with representative gene sequences against assemblies and a manual check where needed. For other genes (covR/S, rocA, hasABC and the nga-ifs-slo locus with promoter region), sequences were extracted from the assembled genomes and compared with the reference genome, H293 (GenBank accession NZ_HG316453.1), as previously described [28]. Antimicrobial resistance gene carriage was determined with ABRicate (https://github .com/tseemann/abricate) using the NCBI database [29].
Statistical analysis
Statistics were performed in GraphPad Prism (v10.2.3).
Results
Collected isolates
During the unusual S. pyogenes infection upsurge period reported by the UKHSA in England in 2022–2023, we collected a total of 384 non-invasive isolates from the Department of Laboratory Medicine, Sheffield. This collection was compared with 229 isolates collected in 2016–2017, representing a time of more typical seasonal upsurge. The proportion of throat isolates was similar for both time periods (61.7% and 65.1 %), but during 2022–2023, overall isolate numbers were much higher (Table 1). Significantly more throat isolates were also associated with scarlet fever in 2022–2023 than in 2016–2017 [24.5 % vs 5.4 %, χ2(1) = 23.55, P<0.0001]. For both time periods, throat isolates were primarily from children under 9, but in 2022–2023, significantly more were from children aged 5–9 compared with 2016–2017 [42.6 % vs 19.5 %, χ2(1) = 21.96, P<0.0001]. There were fewer skin than throat isolates for both time periods, and skin isolates were predominantly from those aged 1 year and under and those aged 45 and older. There was evidence in both time periods of a bimodal age distribution with the highest number of isolates from children under 9 years of age and a second peak in individuals aged 30–39 years (Fig. S1). S. pyogenes was more frequently isolated from throat swabs in female patients [57.6 %(136/236) in 2022–2023 and 63.8 % (95/149) in 2016–2017] and from skin swabs in male patients [68.8 % (77/112) in 2022–2023 and 63.2 % (43/68) in 2016–2017] (Fig. S1).
Table 1. Clinical characteristics of isolates collected in 2022–2023 and 2016–2017.
| 2022–2023 (%) | 2016–2017 (%) | |
| Clinical presentation | ||
| Throat | 237 (61.7) | 149 (65.1) |
| Scarlet fever | 58 (24.5) | 8 (5.4) |
| No scarlet fever | 179 (75.5) | 141 (94.6) |
| Skin | 111 (28.9) | 68 (29.7) |
| Others | 36 (9.4) | 12 (5.2) |
| Ear | 28 (7.3) | 10 (4.4) |
| Eye | 7 (1.8) | 0 (0) |
| Nose | 1 (0.3) | 2 (0.9) |
| Sex | ||
| Female | 190 (49.5) | 125 (54.6) |
| Male | 194 (50.5) | 104 (45.4) |
| Age* | ||
| 1 year and under | 26 (6.8) | 10 (4.4) |
| 2–4 years | 64 (16.7) | 38 (16.6) |
| 5–9 years | 128 (33.3) | 36 (15.7) |
| 10–14 years | 33 (8.6) | 20 (8.7) |
| 15–44 years | 97 (25.3) | 103 (45.0) |
| 45–64 years | 22 (5.7) | 13 (5.7) |
| 65–74 years | 9 (2.3) | 5 (2.2) |
| 75 years and over | 5 (1.3) | 4 (1.7) |
| Total | 384 (100) | 229 (100) |
#*Isolates are grouped in accordance with UKHSA age categories.
emm-type distribution
After quality control filtering, a total of 341 whole genome sequences from the S. pyogenes isolates collected in 2022–2023 were included in study analyses: 209 (61.3 %) from throat swabs, 54 (25.8 %) of which were associated with scarlet fever, 100 (29.3 %) from skin swabs and 32 (9.4 %) from other sites. From the 2016–2017 collection, 165 isolates passed quality control filtering: 127 (77.0 %) from throat swabs and 38 (23.0 %) from skin swabs. We purposely did not sequence isolates from ‘other’ sites in this earlier collection.
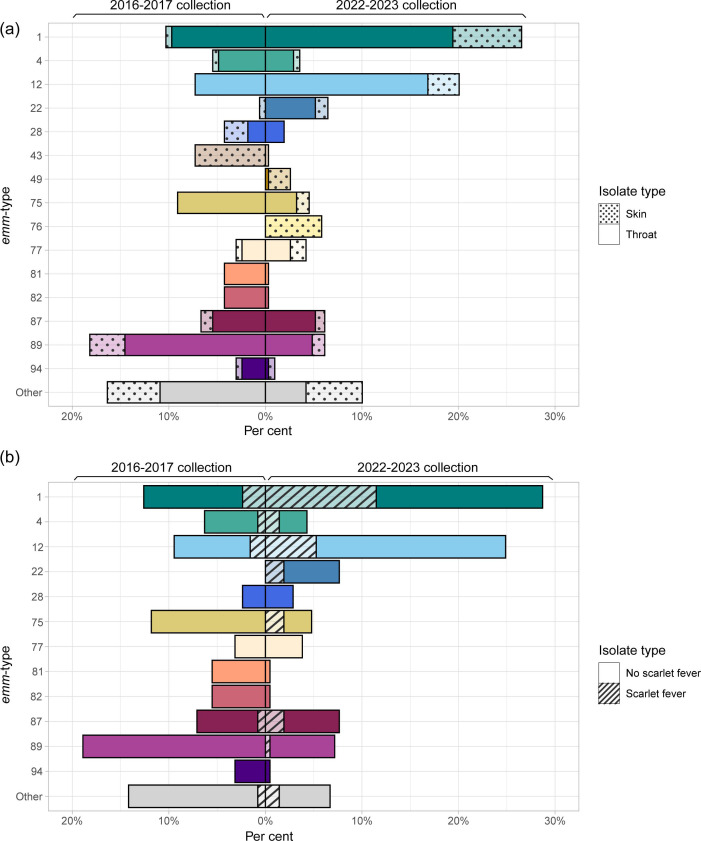
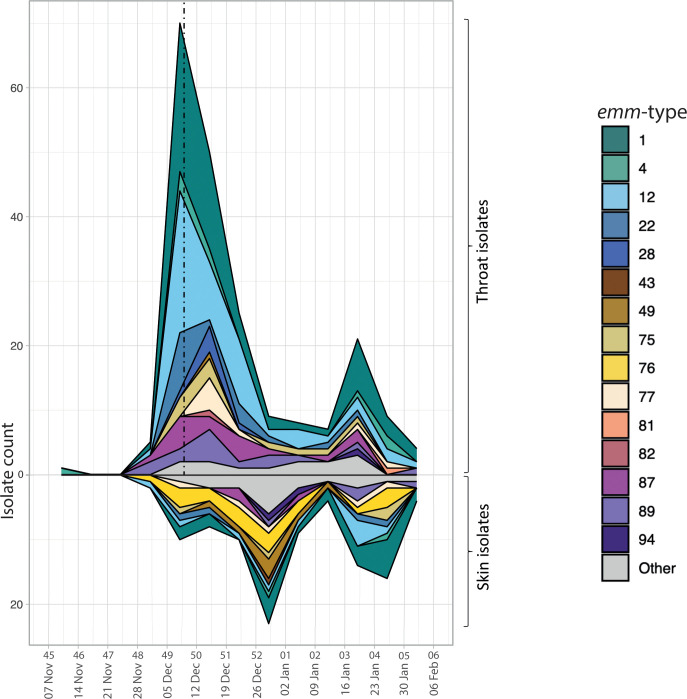
The emm-type for each isolate was extracted from the WGS data. The frequency of isolates and the distribution of emm-types varied over time in the 2022–2023 collection, with the total number peaking in week 49, in keeping with UKHSA data [3]. We observed a fall in the number of throat swabs following the issue of interim clinical guidance by NHS England on the 9th of December 2022 (week 49); this guidance advised clinicians to lower their threshold to empirically treat children with sore throats, including when their presentation may be secondary to viral respiratory illness. Across all 2022–2023 isolates, a total of 33 different emm-types were identified, with emm1 being the most common at 95/341 (27.9 %) followed by emm12, at 64/341 (18.8 %) (Fig. 1a). This high level of emm1 and emm12 cases was reflected in an increase in the number of throat samples in late November to early December 2022 (Fig. 2). Within our comparative collection from 2016–2017, 28 different emm-types were identified overall, most frequently emm89 (30/165, 18.2 %).
Fig. 1. (a) Distribution of emm-types by collection year and clinical isolate type. (b) Distribution of emm-types within throat isolates by clinical presentation. Data presented as a percentage of (a) the total number of isolates (n=341 for 2022–2023, n=165 for 2016–2017) and (b) the total number of throat isolates (n=209 for 2022–2023, n=127 for 2016–2017) in each collection period.
Fig. 2. Distribution of emm-types across time for throat and skin isolates collected in 2022–2023. Week numbers and dates are presented on the x-axis. The dashed line represents the introduction of the NHS England group A Streptococcus interim clinical guidance summary for case management, on the 9th of December 2022.
Within the 2022–2023 throat isolates, 22 different emm-types were identified, but just 6 emm-types made up 80.9 % of isolates: emm1 (28.7 %, 60/209), emm12 (24.9 %, 52/209), emm22 (7.7 %, 16/209), emm87 (7.7 %, 16/209), emm89 (7.2 %, 15/209) and emm75 (4.8 %, 10/209). A significantly higher proportion of throat isolates were emm1 and emm12 in 2022–2023 compared with 2016–2017, where 12.6 % (16/127) isolates were emm1 [χ2(1) = 11.71, P=0.0006] and 9.5 % (12/127) were emm12 [χ2(1) = 12.20, P=0.0005]. In the 2022–2023 throat isolates, emm89 made up a significantly lower proportion than the 2016–2017 collection, in which it was the most common emm-type at 18.9 % (24/127) isolates [χ2(1) = 10.58, P=0.0011]. There was also a lower proportion of emm75 within the 2022–2023 throat isolates compared with 2016–2017, where emm75 made up 11.8 % (15/127). emm87 made up a similar proportion in 2016–2017 (7.1 %, 9/127); however, no throat isolates were emm22 in 2016–2017. Throat isolates associated with scarlet fever in 2022–2023 were predominantly emm1 (44.4 %, 24/54) and emm12 (20.4 %, 11/54) (Fig. 1b). In 2016–2017, scarlet fever was associated with 8/127 throat isolates of five emm-types: emm1 (3/8), emm12 (2/8), emm4 (1/8), emm6 (1/8) and emm87 (1/8).
The 2022 upsurge in throat disease was followed by a surge in skin disease in late December 2022, with a second peak in late January 2023 (Fig. 2). This was driven by 23 different emm-types, most frequently emm1 (22/100, 22 %) and emm12 (10/100, 10 %), but also emm76 (18/100, 18 %) and emm49 (7/100,7 %), both of which were rarely found in the throat. This differed from our 2016–2017 collection, for which there were 17 different emm-types but emm43 (31.6 %, 12/38) was dominant, followed by emm89 (15.8 %, 6/38) and emm28 (10.5 %, 4/38) (Fig. 1a). Only a single skin isolate was emm1, and none were emm12 or emm76.
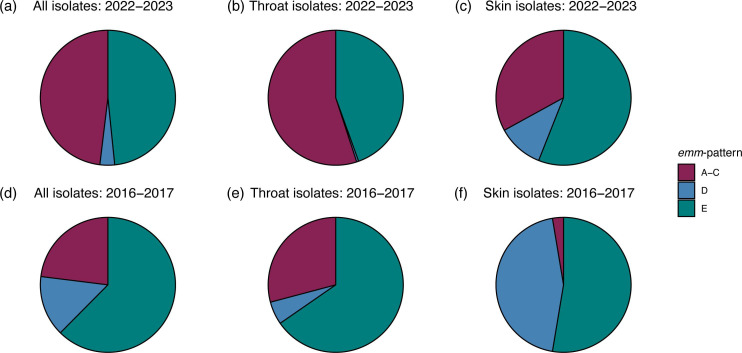
An emm-pattern was assigned to each isolate using previous emm-type assigned patterns based on the genes surrounding the emm-gene [30]. Within the 2022–2023 collection, there was a similar proportion of ‘throat-associated’ emm-pattern A–C (48.1 %, 164/341) and ‘generalist’ emm-pattern E (48.4 % 165/341), with just 3.5 %(12/341) being ‘skin-associated’ pattern D (Fig. 3). All pattern D isolates were of skin origin, except a single throat isolate. Within the 2016–2017 collection, ‘generalist’ emm-pattern E was more common at 62.4 % (103/165); 23 % (38/165) were patterns A–C. With 14.5 % (24/165) of isolates pattern D, the 2016–2017 collection had a greater proportion of pattern D than the 2022–2023 collection overall, with the most striking difference noted in the skin isolates, where pattern D made up 44.7 %(17/38) of the 2016–2017 skin isolates but just 11 % (11/100) of the 2022–2023 skin isolates. Just one skin isolate collected in 2016–2017 was of emm-pattern A–C. Despite these differences and consistent with their previously defined tissue tropism, in both time periods, emm-pattern A–C were significantly more associated with throat isolates than skin [2016–2017 χ2(1) = 11.59, P=0.0007; 2022–2023 χ2(1) = 13.15, P=0.0003], while emm-pattern D was significantly more associated with skin isolates than throat [2016–2017 χ2(1) = 36.20, P<0.0001; 2022–2023 χ2(1) = 20.06, P<0.0001], and pattern E isolates were not significantly associated with one or the other.
Fig. 3. Distribution of emm-patterns across 2022–2023 and 2016–2017 collections. (a) all 2022–2023 isolates (n=341); (b) 2022–2023 throat isolates (n=209); (c) 2022–2023 skin isolates (n=100); (d) all 2016–2017 isolates (n=165); (e) 2016–2017 throat isolates (n=127); (f) 2016–2017 skin isolates (n=38). Pie charts represent the percentage of isolates associated with each pattern.
Each isolate was also assigned an emm-cluster, a functional classification grouping closely related M proteins that share binding and structural properties (Fig. S2). Within the 2022–2023 collection, the most common was the emm1 cluster type A-C3 (27.9 %, 95/341) and the emm12 cluster type A-C4 (18.8 %, 64/341). E4 was also common (19.6 %, 67/341), representing emm22, emm77, emm89 and emm102. By comparison, emm-cluster E4 made up the largest proportion of isolates within the 2016–2017 collection (28.5 %, 47/165), reflecting the high number of emm89 isolates from both throat and skin samples. The 2016–2017 collection also had a number of samples in cluster D4 (9.7 %, 15/165), all of which were skin isolates and mostly emm43 (80 %, 12/15).
Antimicrobial resistance genes
The presence of antimicrobial resistance genes in our 2022–2023 throat and skin isolate genomes was relatively low overall (Table S1), with only 29.4 % (91/309) carrying at least one gene. By far the most common was tetM at 24.3 % (75/309) and 28.5 % of isolates carried at least one tet gene (tetL, tetM, tetO or tetT). The second most common resistance gene was ermA (4.9 %). No resistance genes were identified in emm1, and only three emm12 carried resistance genes (mefA and msrD encoding macrolide resistance). Fewer 2016–2017 throat and skin isolate genomes carried at least one resistance gene (18.2 %, 30/165) (Table S2), but all of these included a tet gene (tetM or tetO), with tetM being the most common at 15.8 % (26/165).
Hyaluronic acid capsule synthesis and the nga-ifs-slo toxin loci
We previously identified an increasing number of emm-types that had undergone recent genetic changes leading to the inability to produce the hyaluronic acid capsule, through loss or nonsense mutation of capsule synthesis genes hasABC [31]. Forty per cent of our Sheffield 2022–2023 throat and skin isolates were predicted to be unable to synthesize the hyaluronic acid capsule due to nonsense mutations in or the absence of the hasA, hasB and hasC genes. While sporadic nonsense mutations occurred in hasA or hasB in some emm-types, such as emm1 (4.9 %) and emm12 (14.5 %), 100 % of all isolates belonging to 11 different emm-types were predicted to be acapsular (Table S1). This included emm22 and emm89, for which, as expected, the entire hasABC locus was absent, emm28, emm77 and emm87 which had previously described nonsense mutations in hasA [31,32], and all emm9, emm29, emm58, emm81, emm90 and emm94 isolates. Typically, emm4 also lacked the hasABC locus although one isolate was found to carry it. Nonsense hasA mutations were also found in the majority of emm75 (78.5 %) and emm11 (66.7 %).
Only 51 % of 2022–2023 throat isolates were predicted to be able to produce capsule compared with 78 % of 2022–2023 skin isolates. Loss of capsule was predominantly associated with emm-pattern E isolates, for which only 27 % were predicted to be encapsulated compared with 91 and 92 % of patterns A–C and D, respectively. This was similar in the 2016–2017 isolates, where only 30 % of pattern E isolates were predicted to be able to make capsule (Table S2).
We previously identified convergent evolution with acapsular isolates also having undergone homologous recombination resulting in increased expression of the toxins NADase and streptolysin O (SLO) [31,33, 34]. High or low expression of these toxins can be linked to three residues in the promoter region of the nga (encoding for NADase), ifs (encoding the inhibitor of NADase) and slo locus [34]. Within the 2022–2023 throat isolates, 90 % were predicted to have high-toxin expression (as defined previously [31]), compared with 56 % of skin isolates (Table S1). Only 8 % of pattern D isolates were predicted to have high-toxin expression, compared with 99 % of A–C and 66 % of E. Additionally, only 58 % of pattern D isolates would express active NADase, based on a glycine residue at codon 330 rather than an aspartate [35], compared with 99% and 100 % of pattern A–C and E isolates, respectively. Although emm1 and emm12 isolates were predominantly encapsulated with a high-toxin expression genotype, an acapsular with a high-toxin expression genotype was found in 41 % of throat isolates compared with a significantly lower 22 % of skin isolates [χ2(1) = 10.91, P=0.001]. Only 2 % of throat isolates were predicted to be encapsulated with low-toxin expression genotype compared with 44 % of skin isolates. This was similar in 2016–2017 with 8 % of throat isolates predicted to be encapsulated with a low-toxin expression genotype but 50 % of skin isolates (Table S2). Interestingly, the association of capsule and toxin expression genotype with infection site was maintained even within pattern E isolates. A significantly higher proportion of 2022–2023 pattern E isolates from the skin were encapsulated with a low toxin expression genotype compared with the pattern E throat isolates [58.9 % vs 3.2 %, χ2(1) = 59.19, P<0.0001] and the same within 2016–2017 pattern E isolates [52.6 % vs 7.9 %, χ2(1) = 39.39, P<0.0001]. Conversely, a significantly higher proportion of 2022–2023 pattern E isolates from the throat were acapsular with a high-toxin expression genotype compared with pattern E skin isolates [78.5 % vs 37.5 %, χ2(1) = 25.22, P<0.0001], and a higher but not significantly different proportion of 2016–2017 pattern E throat isolates compared with skin isolates [47.2 % vs 31.5 %, χ2(1) = 2.918, P=0.088].
Superantigens
S. pyogenes has the potential to carry at least one of 13 different superantigen genes: speG, speJ, speQ, speR and smeZ are chromosomal, while speA, speC, speH, speI, speK, speL, speM and ssa are prophage-associated. Within 2022–2023 throat and skin isolates, 96.1 % (297/309) carried smeZ and 92.2 % (285/309) carried speG (Fig. S3A). The other chromosomal superantigens were seen less frequently, with speJ in 42.4 % (131/309) and the co-transcribed speQ and speR in 12.6 % (39/309). Of the prophage-associated superantigens, speC was the most common at 52.4 % (162/309), followed by speA at 32 % (99/309). One emm49 skin isolate carried a speK/speM fusion gene which we previously identified in an emm65 [28]. Of the 99 isolates that carried speA, 80.8 % (80/99) were emm1.
In comparison, a similar proportion of isolates from the 2016–2017 collection carried the most common superantigen genes, smeZ and speG, at 94.5 % (156/165) and 90.9 % (150/165), respectively (Fig. S3B). Prophage-associated speC was found in 64.8 % (107/165) of isolates in 2016–2017, slightly higher than in 2022–2023. However, the number of isolates carrying speJ and speA was lower in 2016–2017, where speJ was found in 22.4 % (37/165) and speA in 16.4 % (27/165); this difference reflects the 97.6 % (80/82) of 2022–2023 emm1 isolates carrying this combination of superantigens. In 2022–2023, 22 % (68/309) carried speI compared with 16.4 %(27/165) in 2016–2017, reflecting the upsurge in emm12 in 2022–2023, all of which carried speI. A lower proportion had speK in 2022–2023 (14.6 %, 45/309) than in 2016–2017 (26.1 %, 43/165), reflecting the single emm43 isolate in 2022–2023 compared with 12 emm43 skin isolates in 2016–2017. One emm25 skin isolate in 2016–2017 carried a speK/speM fusion gene.
DNases
All S. pyogenes isolates carry at least one DNase gene, out of a possible two chromosomal and six prophage-encoded genes [36]. Within the 2022–2023 throat and skin isolates, the most common prophage-associated DNase genes identified were spd3 in 60.5 % (187/309), spd1 (carried with speC) in 52.4 % (162/309) and sda2 in 43.0 % (133/309) (Fig. S4A). Most DNase combinations were seen in a range of emm-types, although the most common combinations were specific to emm1 (sda2 with spd3, 23.6 %, 73/309) and emm12 (sda2 with spd1, 13.3 %, 41/309). By comparison, in the 2016–2017 collection, sda2 was seen in fewer isolates, at 21.2 % (35/165), and spd1 in slightly more, at 64.2 % (106/165), but spd3 in similar numbers, at 56.4 % (93/165) (Fig. S4B). In both collections, sdn was less common, and spd4 was quite rare, with spd4 only being present with spd3 in a single isolate in 2022–2023 and two isolates in 2016–2017 (two emm5 and one emm87); all other isolates carrying spd3 or spd4 had just one of these two DNase genes.
CovR, CovS and RocA regulator proteins
The two-component regulator CovR/S and the regulator of Cov, RocA, negatively regulate key S. pyogenes virulence factors including capsule and toxins. For both collections, nonsense or frameshift mutations leading to premature stop codons that would alleviate virulence factor repression were rare in CovS and RocA and absent in CovR. Only one 2022–2023 isolate had a premature stop codon in CovS (an emm1) and two 2016–2017 isolates (one emm1 and one emm89). A total of 2.6 %(8/309) 2022–2023 throat or skin isolates carried premature stop codons that would truncate RocA. Consistent with previous findings, all emm3 isolates from both collections would express a truncated RocA after 416 aa. No other emm-types had nonsense mutations in rocA in 2016–2017. Other non-synonymous variations were found in covS and rocA (Tables S1 and S2), but the impact of these is difficult to determine, and many were associated with emm-type.
Although nonsense mutations in covR were absent, other non-synonymous variations in covR were found in 45/309 (14.6 %) of 2022–2023 skin and throat isolates. However, the majority were related to the emm-type. All emm22 isolates from 2022–2023 (n=20) and 2016–2017 (n=1) had the same variation in CovR (V128A) and an additional variation in RocA (V333A). Nearly all emm77 isolates from both 2022–2023 (12/13) and 2016–2017 (4/5) had the same M170I variation in CovR. 14.5 % of 2022–2023 emm12 isolates (9/62) had an A105G variation in CovR. Four other isolates of different emm-types had other CovR variations.
emm1
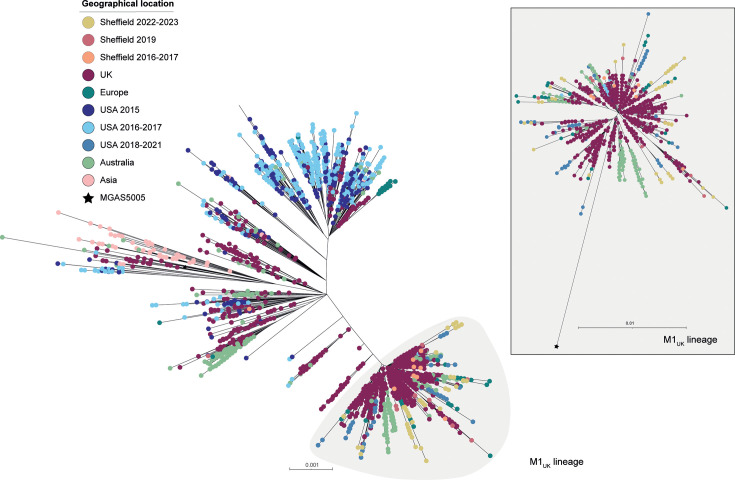
A total of 95 (27.9 %) of the 341 genomes collected in 2022–2023 were emm1. Sixty (63.2 %) of these were from throat isolates, of which 24 were associated with scarlet fever. Twenty-two (23.2 %) were skin isolates, and the remaining 13 were from other sites including 12 from ear swabs and one from an eye swab. All these emm1 isolate genomes clustered within the M1UK lineage, and all carried the 27 lineage defining SNPs (Fig. 4). Similarly, for our 2016–2017 emm1 isolates, only one out of 17 emm1 isolates was not M1UK. This is in keeping with the emergence of M1UK as the dominant emm1 strain globally. Sheffield isolates were spread throughout the M1UK phylogeny without evidence of expansion of a specific sub-clade. Two scarlet fever throat isolates lacked the speA-carrying Φ5005.1 phage. All other isolate genomes had speA; the chromosomal speG, speJ and smeZ; and a combination of other prophage-associated superantigen genes. No clear correlation was seen between clinical presentation and the presence of a particular profile of superantigens and/or DNases (Fig. S5).
Fig. 4. Phylogenetic analysis of Sheffield emm1 genomes collected in 2022–2023 and 2016–2017, within the context of global emm1 isolates. A maximum likelihood phylogenetic tree was generated with SNPs extracted from a core gene alignment (excluding prophage and SpyCI regions) to reference emm1 strain MGAS5005 [54]. Alongside our Sheffield emm1 genomes from 2022–2023 and 2016–2017, we included publicly available emm1 genome data from Sheffield, 2019 (n=14) [28]; other UK sites, 2001–2018 (n=1399) [7,31, 55,59]; Denmark, 2018–2023 (n=98) [14]; Netherlands, 2019 (n=15) [13]; Portugal, 2022–2023 (n=30) [16]; Australia, 2005–2020 (n=318) [9]; China, 2004–2016 (n=64) [60,61]; the USA, 2015 (n=316) [38], 2016–2017 (n=468) [62] and 2018–2021 (n=85) [11]. The scale bar represents the number of nucleotide substitutions per site. The grey shaded area indicates the emergent lineage M1UK defined by 27 SNPs; this lineage is shown as a separate tree within the panel.
emm12
A total of 64/341 (18.8 %) 2022–2023 isolate genomes were emm12, the majority of which were throat isolates (52/64, 81.3 %), and 11 of these were associated with scarlet fever. Ten were skin swab samples, and two were ear swabs. For the 2016–2017 collection, 12 out of 124 (9.7 %) genomes were emm12, and all were from a throat source.
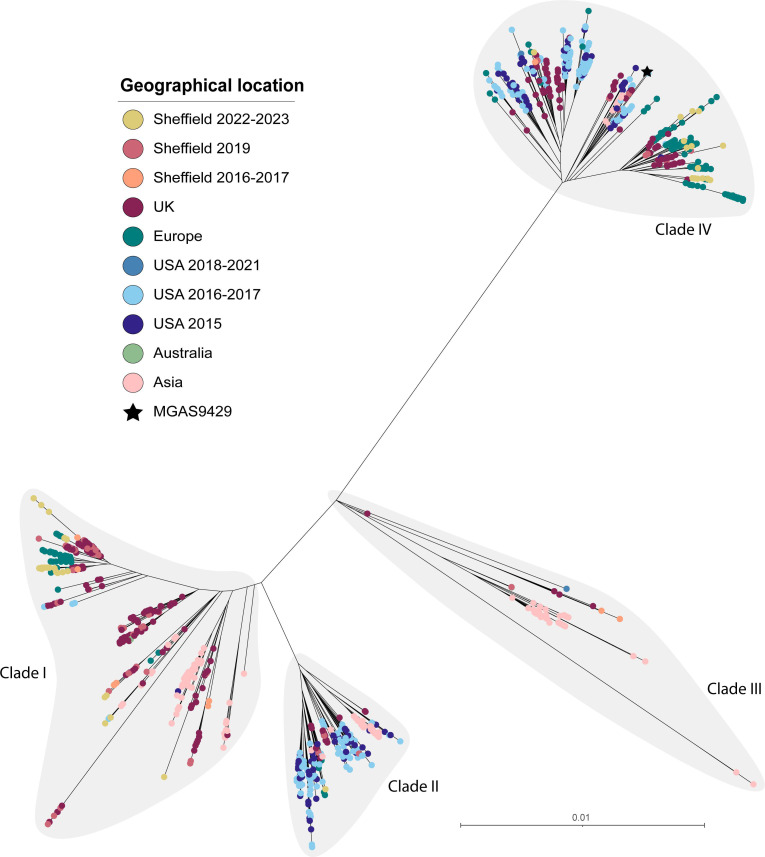
The core genome phylogeny of global emm12 isolates showed four distinct clades (Fig. 5), as described previously [37]. The majority of all Sheffield isolates were clade I or clade IV, alongside other UK and European isolates. A single 2022–2023 Sheffield isolate was clade II, and no 2022–2023 isolates were found within clade III. Our phylogeny also showed three sub-clades within clade IV, one of which was dominated by Sheffield, other UK and European isolates, while isolates from the USA were restricted to the other two sub-clades, one of which also included Asian strains. Other US isolates also dominated clade II.
Fig. 5. Phylogenetic analysis of Sheffield emm12 genomes collected in 2022–2023 and 2016–2017, within the context of global emm12 isolates. A maximum likelihood phylogenetic tree was generated with the core gene alignment (excluding prophage regions) to reference strain MGAS9429 [63]. Alongside our Sheffield emm12 genomes from 2022–2023 and 2016–2017, we included publicly available emm12 genome data from Sheffield, 2019 (n=15) [28]; the UK, 2009–2018 (n=439) [31,55, 56, 59]; Denmark, 2018–2023 (n=239) [14]; Portugal, 2022–2023 (n=12) [16]; the USA, 2015 (n=134) [38], 2016–2017 (n=228) [62] and 2018–2021 (n=2) [11]; Australia, 1995/2017 (n=2) [37,64]; Hong Kong, 2005–2011 (n=132) [37,65]; and China, 2004–2018 (n=43) [66]. Clades I–IV, previously defined by Davies et al. [37], are shaded grey. The scale bar represents the number of nucleotide substitutions per site.
All Sheffield isolates were ST36, except four 2022–2023 isolates that were ST242 and clustered within one clade IV sub-clades (Fig. S6). One other 2022–2023 isolate was a single locus (xpt) variant ST1459. The only isolates to carry the speA gene were seven 2022–2023 throat isolates (9.1 %), and they clustered together within clade I. Again, no clear correlation was seen between clinical presentation and the presence of a particular profile of superantigens and/or DNases. Three 2022–2023 throat isolates in clade I carried mefA and msrD genes, associated with acquired macrolide resistance. Interestingly, ten 2022–2023 isolates with the CovR A105G variant all clustered together within clade I; a single isolate within this cluster also carried a RocA variation (G184R). A separate cluster of four isolates in clade IV carried the same mutation in rocA leading to a premature stop codon after 427 aa.
The single emm82 isolates from 2022–2023, and all seven emm82 isolates from 2016–2017 had the same ST as emm12: ST36. These isolates were confirmed to be the lineage of emm82 recently identified to have arisen through recombination and emm-switching of emm12 to emm82 [38,39].
emm4
emm4 was associated with invasive disease in children <15 years early in the 2022–2023 upsurge [2] and was previously associated with the 2014 scarlet fever upsurge in England [4]. Of the twelve 2022–2023 emm4 isolates, nine were throat isolates, including three associated with scarlet fever; two were skin isolates, and one eye swab isolate. One skin-associated isolate had a highly divergent core genome and uniquely was ST289 and emm-subtype 4.2 rather than ST39 and emm-subtype 4.0 or 4.19. It also carried the hasABC locus which was characteristically absent in the other 11 emm4 isolates.
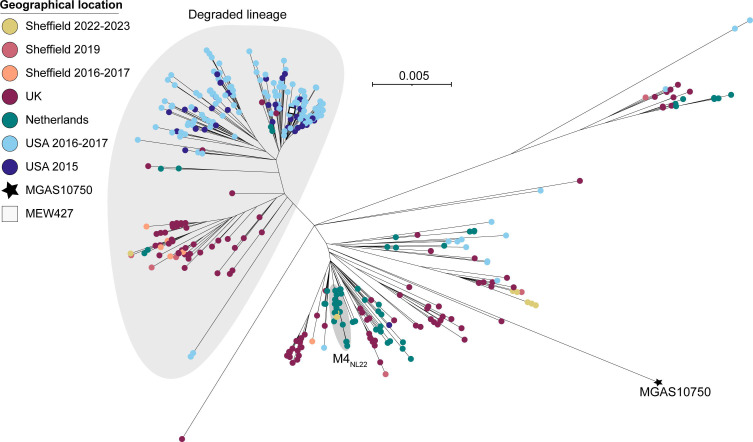
Phylogeny of the other eleven 2022–2023 isolates within a wider emm4 population showed only a single strain clustering with the previously described ‘Degraded’ lineage [40] due to substantial loss of genes within the three prophages and the integrated conjugative element associated with emm4 (Fig. 6). This lineage also has a fusion of the 5′ of emm gene with the 3′ of the downstream enn gene [41]. The five emm4.19 isolates clustered together within a ‘Complete’ sub-lineage, closely related to other UK isolates. The remaining five isolates clustered with the recently described invasive disease-associated M4NL22 lineage from the Netherlands [42]. This appears to be a recent emergence of this lineage in England as no emm4 isolates in our 2016-2017 comparative collection nor in our Sheffield 2019 isolates [28] were found clustering with M4NL22 isolates. Our seven Sheffield 2016–2017 emm4 isolates included four within the ‘Degraded’ lineage, while three were ‘Complete’. This split was similar in our 2019 Sheffield isolates with four isolates in each of these lineages. This pattern of an even divide of UK isolates between ‘Complete’ and ‘Degraded’ lineages is consistent with our phylogeny and previous findings [40], with the shift towards dominance of the ‘Complete’ lineage, at least in Sheffield, emerging during the 2022–2023 upsurge.
Fig. 6. Phylogenetic analysis of Sheffield emm4 genomes collected in 2022–2023 and 2016–2017, within the context of global emm4 isolates. A maximum likelihood phylogenetic tree was generated with the core gene alignment (excluding prophage and SpyCI regions) to reference strain MGAS10750 (GenBank CP000262.1 [63]). The position of another reference strain MEW427 (GenBank CP014138.1 [67]) is also indicated. Alongside our Sheffield emm4 genomes from 2022–2023 and 2016–2017, we included publicly available emm4 genome data from Sheffield, 2019 (n=9) [28]; the UK, 2011–2016 (n=176) [31,55, 56, 59]; the Netherlands, 2009–2022 (n=66) [42]; and the USA, 2015–2017 (n=204) [38,62]. The large grey shaded area indicates the ‘Degraded’ lineage, and the small grey shaded area indicates the M4NL22 lineage. The scale bar represents the number of nucleotide substitutions per site.
emm22
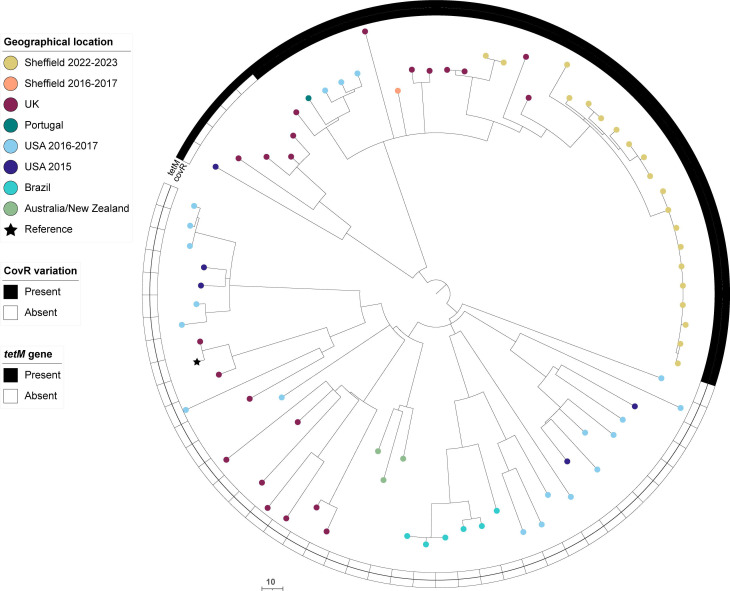
A rise in emm22 was seen within the 2022–2023 collection, overall representing 5.9 % (20/341) of isolates compared with a single, skin-associated, isolate in 2016–2017. Of the 2022–2023 isolates, four were skin isolates and the remainder from throat, with 4/16 throat isolates also associated with scarlet fever. All were ST46. Phylogenetic analysis showed the clustering of all Sheffield 2022–2023 isolates, except two, together in expansion of a single lineage (Fig. 7). All Sheffield emm22 isolates carried a CovR V128A variant, clustering together in a lineage with 12 other isolates from the UK, Europe and the USA all also carrying the same variant. These isolates also carried tetM, alongside additional strains from the same parent lineage. All emm22 isolates, Sheffield and globally, carried the same variation in RocA (V333A) compared with a reference RocA sequence.
Fig. 7. Phylogenetic analysis of Sheffield emm22 genomes collected in 2022–2023 and 2016–2017, within the context of global emm22 isolates. Alongside our Sheffield emm22 genomes from 2022–2023 and 2016–2017, we included publicly available emm22 genome data from the UK, 2001–2018 (n=22) [31,56]; Portugal, 2022–2023 (n=1) [16]; the USA, 2015 (n=5) [38] and 2016–2017 (n=20) [62]; Brazil, 2000–2013 (n=6) [68]; Australia, 1999/2004 (n=2) [68]; and New Zealand, 2010 (n=1) [68]. A maximum likelihood phylogenetic tree was generated with the core gene alignment to the de novo assembly of the oldest UK isolate, BSAC_bs1196 [31], and the removal of regions of potential recombination using Gubbins [26]. The scale bar represents the number of nucleotide substitutions per site.
emm75
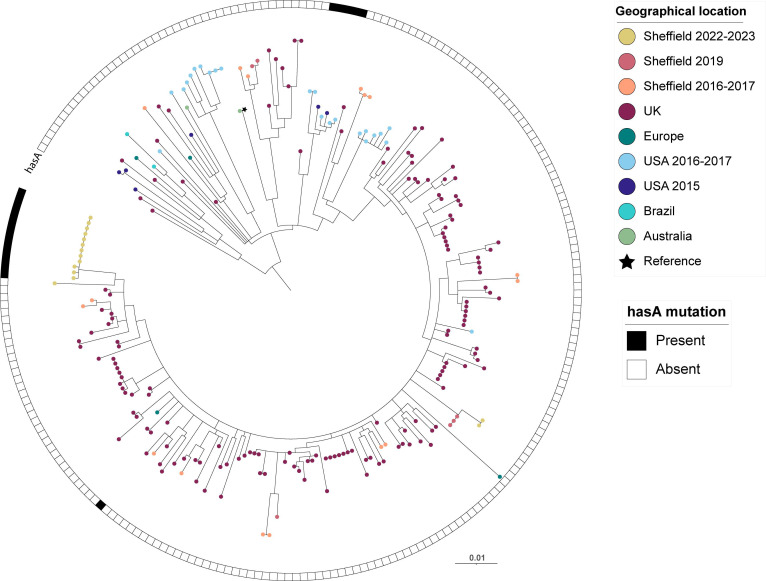
Within all 2022–2023 Sheffield isolates, 4.4 % were emm75 (15/341), of which ten were from a throat source and four of these associated with scarlet fever, four from skin and one from an ear swab. By comparison, in 2016–2017, 9.1 % of isolates were emm75 (15/165), all of which were throat isolates not associated with scarlet fever. All Sheffield emm75 isolates from both 2022–2023 and 2016–2017 were ST150, distinct from the other dominant lineage of emm75, ST49. Of the 2022–2023 isolates, 13/15 formed a new sub-lineage with identical superantigen profiles (Fig. 8). Within this sub-lineage, 12/13 had the same additional T in a 7 residue homopolymeric tract in hasA, leading to a premature stop codon after 46 amino acids, and therefore are likely to be acapsular. This is new compared with 2016–2017 Sheffield isolates, none of which carried a hasA mutation and were scattered throughout the phylogeny. Other acapsular emm75 strains with the same premature stop codon in hasA were identified in a small cluster of US isolates, and a single sporadic UK strain.
Fig. 8. Phylogenetic analysis of Sheffield emm75 genomes collected in 2022–2023 and 2016–2017, within the context of global emm75 isolates. A maximum likelihood phylogenetic tree was generated with the core gene alignment to reference strain M75 (star) (GenBank CP033621.1 [69]). Alongside our Sheffield emm75 genomes from 2022–2023 and 2016–2017, we included publicly available emm75 genome data from Sheffield, 2019 (n=6) [28]; the UK, 2011–2016 (n=158) [31,55, 56, 59]; Belgium, 2004 (n=1) [68]; the USA, 2015–2017 (n=81) [38,62]; Australia, 1991–2010 (n=7) [68]; and Brazil, 2007–2012 (n=2) [68]. The scale bar represents the number of nucleotide substitutions per site. The presence or absence of a hasA nonsense mutation is indicated for each strain. Only isolates of ST150 (and closely related STs) are shown, excluding the other major lineage of emm75 (ST49 and closely related) as these are genetically quite distant [31]. Those that were ST49 and closely related STs comprised genomes from the UK, 2014–2018 (n=3) [56]; the USA, 2015 (n=14) and 2016–2017 (n=38) [38,62]; Australia, 1991–2004 (n=6) [68]; Fiji, 2006 (n=3) [68]; India, 2007–2010 (n=2) [68]; and Kenya, 2000–2011 (n=5) [68]. No Sheffield isolates were ST49.
emm76, emm77 and emm87
Previously, we found recent expanding lineages within emm76, emm77 and emm87 that were acapsular due to mutations in hasA and had undergone recombination around the nga-ifs-slo promoter region to become high-toxin expressing variants [31]. In our 2022–2023 isolates, this was still the case for emm77 and emm87, but not for emm76.
In 2022–2023, 5.6 % (19/341) of isolates were emm76, compared with none in 2016–2017. Of these, 18 were from a skin source and one from an ear swab, and all were ST378, with no mutations in hasA or hasB, and predicted low-toxin expression (Fig. S7). All carried tetM, as described previously within this lineage [31]. This reflects a recent expansion of the encapsulated, low-toxin phenotype ST378 lineage, in contrast to the previously described ST50 lineage which was acapsular with high-toxin expression [31].
In 2022–2023, 4.1 % (14/341) of isolates were emm77, an increase from 2.4 % (4/165) in 2016–2017. Of the 2022–2023 isolates, 8 were from throat swabs and not associated with scarlet fever, 5 were from a skin source and 1 from an ear swab. The majority (11/14) were ST63, carrying resistance genes tetO and ermA, with the CovR M170I variation and belonged to the previously described acapsular (truncated HasA after 154 aa) with high-toxin expression lineage [31]. The two other emm77 isolates were ST399, quite distinct from ST63 although also predicted to be acapsular (truncated HasA after 46 aa) but with low-toxin expression.
An increase in emm87 was also observed in 2022–2023, 6.7 % (23/341), from 4.8 % in 2016–2017. Of the 2022–2023 isolates, 16 were from a throat source and 2 from ear swabs, 2 from eye swabs and 3 from skin sources. All isolates were part of the previously described lineage characterized as being acapsular, through a mutation in hasA (truncating HasA after 46 aa), and high-toxin expressing through recombination around the nga-ifs-slo locus and promoter [31] (Fig. S8). Interestingly, international phylogeny of emm87 revealed four broad clades, one of which contained a sub-lineage dominated by Sheffield and other UK strains and characterized by a premature stop codon in hasB, truncating it after 188 aa (Fig. S8). Only small clusters of USA emm87 isolates carried other truncating mutations in hasB. There was also some evidence of geographical variation across the lineages, with USA isolates dominating one lineage and UK isolates another.
Invasive S. pyogenes isolates
During the time of our 2022–2023 collection, the Department of Laboratory Medicine, Sheffield, identified 19 iGAS samples (S. pyogenes isolated from a sterile site) and these had undergone routine emm-typing by the UKHSA, but genomic data was not available. Patients ranged in age from 1 to 94 years, but were most commonly 0–9 years (36.8 %, 7/19) or over 75 years (31.6 %, 6/19), in keeping with national data from the same time period. By far, the majority were emm1 (57.9 %, 11/19), followed by emm76 (15.8 %, 3/19), emm89 (10.5 %, 2/19), emm92 (10.5 %, 2/19) and emm87 (5.3 %, 1/19). The most common sample type was blood culture, at 13/19 samples (68.4 %, 3/19), followed by empyema samples (15.8 %, 3/19), and soft tissue or joint fluid (15.8 %, 3/19). All empyema samples were emm1 and all in children aged 0–9 years.
Discussion
In recent years, the UK has experienced significant rises in morbidity and mortality in association with substantial upsurges in S. pyogenes infections, including in 2017–2018 and in 2022–2023 [3]. Despite this, study of non-invasive isolates has been limited, and the capacity for dynamic changes in non-invasive disease to drive upsurges and invasive disease is poorly understood. We sequenced 341 non-invasive isolates from Sheffield during the major 2022–2023 UK upsurge, and although we found emm1 and emm12 to be the leading causes of both throat and skin infections, they were differentially followed by emm22, emm87 and emm89 in throat infections but emm76 and emm49 in skin. A comparison to non-upsurge isolates from 2016–2017 indicated emm1 and emm12 contributed significantly to the 2022–2023 upsurge, but other emm-types had also changed over time with more emm22 in throat infections and emm76 in skin infections. All 2022–2023 emm1 isolates were the prevalent M1UK lineage, but more diverse lineages were identified in other emm-types, including emm12, and emergent lineages in others, including emm75, demonstrating that, at least local to Sheffield, the upsurge was not primarily caused by a single genotype.
For both time periods studied, non-invasive S. pyogenes infections were most common in those aged 0–9 years, but more infections were seen in 5–9-year-old children in 2022–2023 than in 2016–2017 (33.3 % vs 15.7 %). Overall, those aged 4, 5, 6 or 7 years of age had the highest rates of S. pyogenes infection of any age in 2022–2023. This may reflect accelerated exposure to infection associated with school attendance, in keeping with reduced immunity to S. pyogenes and common respiratory viruses in this age group following reduced exposure during the COVID-19 pandemic [2,43].
The dominant peak in throat isolates at week 49 of 2022 within our collection, followed by a second smaller peak in week 3 of 2023, coincides with the peaks in scarlet fever notifications reported by the UKHSA during this time period [3]. During week 49, NHS England group A Streptococcus interim clinical guidance summary for case management was issued with guidance temporarily altering the clinical scoring criteria threshold for immediate antibiotic treatment in children with a sore throat. The issue of this guidance on the 9th of December 2022 likely enhanced clinician confidence in the empirical diagnosis of S. pyogenes throat infection and therefore may have contributed to a fall in throat swabs being received by the diagnostic laboratory. However, this also coincides with the fall in scarlet fever reporting nationally [3]. In contrast, the prominent peak in skin isolates occurred in week 52, between the peaks in throat isolates. This may be due to behavioural or environmental factors, such as increased skin-to-skin transmission of S. pyogenes associated with increased social mixing or altered chronic wound care at this time of year.
Analysis of national iGAS data from this time period identified a significant association between M1UK and pleural isolates, likely the result of an early S. pyogenes upsurge coinciding with the respiratory virus season, facilitating disease progression [44]. Nationally, emm1 and emm12 were the most common emm-types among invasive isolates in all age groups during this upsurge, notably with emm4 as the next most common emm-type in children [2]. While we identified that 57.9 % of our invasive isolates were emm1, our dataset did not identify any invasive emm12 nor emm4 isolates, and the remaining 47.4 % (8/19) of invasive isolates during the 2022–2023 collection were of four other emm-types, potentially a reflection of regional variation in circulating strains within England. The skin prevalent emm76 did cause a substantial proportion of skin and iGAS infections suggesting we should not overlook this infection site. It is not known if the pronounced association of emm76 with skin infections was a local phenomenon as national data on skin infection emm-types were not collected.
Tissue tropism within S. pyogenes infections is well-recognized but bacterial molecular adaptations associated with tissue specialization remain incompletely understood [45]. An acapsular genotype was frequently seen in throat isolates in this study, with just 51 % predicted to be able to produce capsule compared with 78 % of skin isolates. In throat isolates, these acapsular strains were predominantly ‘generalist’ pattern E. Just 27 % of pattern E isolates overall were predicted to be encapsulated and made up 44.5 % of throat infections. Interestingly, although more skin than throat infections were pattern E, at 56 %, a higher proportion (62.5 %) of these were encapsulated. We also identified evidence of evolving capsule loss with the emergence of a recent acapsular sub-lineage of emm75, a pattern E type, and the increase in the prevalence of acapsular emm22.
We further observed tissue-specific differences in predicted expression of the toxins NADase and SLO based on the promoter sequence, with 90 % of throat isolates predicted to express high levels of toxins compared with 56 % of skin isolates, highlighting a key role for toxin expression in the pathogenesis of S. pyogenes throat infections. We previously provided evidence of convergent evolution with acapsular strains gaining increased toxin expression through homologous recombination [31]. Successful emergent lineages characterized by this recombination event have been identified previously in pattern E emm76, emm77 and emm87. Interestingly, while we continued to see expansion of these acapsular/high-toxin lineages in emm77 and emm87, in emm76, we observed instead an expansion of an ST378 encapsulated/low-toxin lineage to become a leading cause of skin infections. Indeed, overall, we observed significant differences between capsule/toxin-expression genotypes by infection site, with more throat isolates possessing an acapsular/high-toxin genotype and more skin isolates possessing an encapsulated/low-toxin genotype. This association was maintained even within the ‘generalist’ pattern E isolates when divided by isolate source, suggesting that differential capsule expression and NADase/SLO expression is a bacterial adaptation mechanism for tissue tropism. The emergence of lineages within pattern E emm-types 76, 77, 87 and 89 that have undergone genetic changes to become acapsular with high-toxin expression appear to be recent events [31]; the rise to dominance of the new acapsular/high-toxin variant of emm89 in the UK population occurred in 2007–2008 [33]. Here, in 2022–2023 compared with 2016–2017, we observed an increase in the prevalence of acapsular/high-toxin expression genotype emm22 causing throat infections and the emergence of an acapsular emm75 lineage. Why there has been a recent shift towards an acapsular/high toxin expression genotype and why this supports throat infections over skin infections is unclear. Although capsule has been shown to promote upper respiratory tract infections for some genotypes [46,47], long-term throat carriage isolates lose capsule expression through mutations in hasABC [48], potentially allowing for invasion into host cells that is otherwise hindered by capsule expression. Although why this would be preferential in the throat compared with the skin is also unclear. Capsule also provides protection against phagocytosis, and this has also been shown to promote both skin and throat infections [47]; however, this may be an emm-type specific phenomenon [49], and an increase in NADase/SLO expression may compensate for capsule loss [50]. An association between NADase activity and tissue tropism has also been identified previously, with NADase-inactive strains being primarily ‘skin-associated’ emm-pattern D, suggesting toxin may be less essential for S. pyogenes infection in the skin ecological niche compared with in the throat [51], although this could also be related to capsule expression in this niche, rendering NADase activity non-essential. We are limited in our study by the fact that we are only looking at genotypes and predicting phenotype, and it is possible that predicted capsule or NADase/SLO expression may be different in vivo. Work is ongoing to determine these phenotypes and the association with tissue tropism.
The most frequently identified emm-type across both throat and skin infections in 2022–2023 was emm1; all were M1UK lineage and distributed throughout the M1UK phylogeny without evidence of local expansion of a specific sub-clade. While classically ‘throat-associated’, it is likely the high number of emm1 cases in the skin reflected the high burden of throat disease and onwards transmission to the skin [52]. In contrast, emm12 isolates from our collections in both 2022–2023 and 2016–2017 were distributed predominantly across two of the four clades: I and IV. Previous work has suggested an association in clades I–III between scarlet fever and ssa (encoding streptococcal superantigen A), and an absence of scarlet fever in clade IV; however, no such clear associations were seen in our collection [37]. Geographical variation was apparent across our emm12 phylogeny, with the majority of Sheffield isolates found within a single sub-clade of clade IV and clade I, in a distribution distinct from that of USA isolates and again from Asian isolates. We observed similar geographical variation in emm87, with two of the four main clades dominated by USA strains and one clade containing the majority of Sheffield and other UK strains. These variations are in keeping with regional divergence of circulating lineages and can give rise to the emergence of geographically restricted sub-lineages, some of which have been seen to expand more widely if carrying a fitness advantage [7].
Comparison of isolates from 2022–2023 to 2016–2017 revealed differences in emm-type distribution between collections. More isolates from 2022–2023 were emm1 and emm12 compared with 2016–2017, across all infection sites. Within throat samples, 2022–2023 saw a fall in the number of emm89 and emm75 isolates compared with 2016–2017. Within skin site isolates, we saw more emm76 and emm49 in 2022–2023, and fewer emm43, emm89 and emm28 than 2016–2017. Where there is seasonality of S. pyogenes infection, as seen in Sheffield, seasonal fluctuation in emm-type distribution also frequently occurs. It is likely this is multifactorial, affected by antibiotic pressures, climate, patterns of social mixing and variations in concurrent circulating respiratory viruses. Population immunity also plays a key role in successful strain transmission; the strongest immunological protection is emm-type-specific, though infection with one emm-type can confer a degree of protection to other emm-types, thereby influencing the profile of circulating strains in subsequent seasons [53]. Genomic changes in circulating strains also have the potential to enhance strain virulence, with such changes underpinning previous upsurges in S. pyogenes disease [7]. We have demonstrated several emergent sub-lineages in our 2022–2023 isolates, including within emm22 and emm75, which is likely to further impact strain diversity between seasons.
Our study was limited by the lack of genomic data for local iGAS isolates; the addition of this data would allow a more detailed genomic comparison to better understand the interplay between non-invasive and invasive diseases. Our throat samples were deemed scarlet fever or not scarlet fever based on details provided by clinicians, which were often incomplete. Public health and media messaging may have also contributed to ascertainment bias. In weeks 49 and 50 of our study, our diagnostic laboratory received substantially more throat swabs compared with these same weeks in previous years, with high numbers of S. pyogenes identified, however, with a lower overall swab positivity.
Overall, we found that an increase in prevalence of emm1 and emm12 in non-invasive disease in 2022–2023 locally reflected the national increase of these emm-types in iGAS and scarlet fever cases. We have highlighted the need to study both throat and skin infection isolates as they can differ, and the potential for differential capsule expression and/or NADase and SLO toxin expression to drive tissue tropism. We also demonstrated that emm-types may not be represented by single lineages and these can change over time and by geographical location. Frequent monitoring with WGS is needed to determine how rapidly these changes occur, what factors influence these changes and how they might drive infection rates, including overspill from non-invasive disease into invasive and upsurges.
supplementary material
Acknowledgements
The authors are grateful to laboratory colleagues at Sheffield Teaching Hospitals NHS Foundation Trust for isolate collection. We also acknowledge the input of the genomics pipeline group at the Earlham Institute.
Abbreviations
- COVID-19
coronovirus disease 2019
- iGAS
invasive S. pyogenes/group A Streptococcus
- SLO
streptolysin O
- StrepA
Streptococcus pyogenes
- UKHSA
UK Health Security Agency
- WGS
whole genome sequencing
Footnotes
Funding: CET was a Royal Society and Wellcome Trust Sir Henry Dale Research Fellow (208765/Z/17/Z) and then a Wellcome Trust Career Development award holder (227240/Z/23/Z).
Ethical statement: The study protocol was approved by the North East – Newcastle and North Tyneside 2 Research Ethics Committee (reference 24/NE/0033; Integrated Research Application System project ID 334500).
Contributor Information
Jennifer N. Hall, Email: jennifer.hall@sheffield.ac.uk.
Saikou Y. Bah, Email: saikouybah@gmail.com.
Henna Khalid, Email: h.khalid@sheffield.ac.uk.
Alison Brailey, Email: alison.mckinven@nhs.net.
Sarah Coleman, Email: sarah.coleman24@nhs.net.
Tracey Kirk, Email: tracey.kirk2@nhs.net.
Naveed Hussain, Email: naveed.hussain3@nhs.net.
Mark Tovey, Email: mark.tovey1@nhs.net.
Roy R. Chaudhuri, Email: r.chaudhuri@sheffield.ac.uk.
Steve Davies, Email: steve@ukoms.co.uk.
Lisa Tilley, Email: lisa.tilley4@nhs.net.
Thushan de Silva, Email: t.desilva@sheffield.ac.uk.
Claire E. Turner, Email: c.e.turner@sheffield.ac.uk.
References
- 1.UK Health Security Agency Group A streptococcal infections: first update on seasonal activity in England, 2022 to 2023. 2022. https://www.gov.uk/government/publications/group-a-streptococcal-infections-activity-during-the-2022-to-2023-season/group-a-streptococcal-infections-first-update-on-seasonal-activity-in-england-2022-to-2023
- 2.Guy R, Henderson KL, Coelho J, Hughes H, Mason EL, et al. Increase in invasive group A streptococcal infection notifications, England, 2022. Euro Surveill. 2023;28:2200942. doi: 10.2807/1560-7917.ES.2023.28.1.2200942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.UK Health Security Agency . GOVUK; 2023. [15-February-2024]. Group A streptococcal infections: 15th update on seasonal activity in England.https://www.gov.uk/government/publications/group-a-streptococcal-infections-activity-during-the-2022-to-2023-season/group-a-streptococcal-infections-15th-update-on-seasonal-activity-in-england accessed. [Google Scholar]
- 4.Turner CE, Pyzio M, Song B, Lamagni T, Meltzer M, et al. Scarlet fever upsurge in England and molecular-genetic analysis in north-west London, 2014. Emerg Infect Dis. 2016;22:1075–1078. doi: 10.3201/eid2206.151726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guy R, Williams C, Irvine N, Reynolds A, Coelho J, et al. Increase in scarlet fever notifications in the United Kingdom, 2013/2014. Euro Surveill. 2014;19:20749. doi: 10.2807/1560-7917.es2014.19.12.20749. [DOI] [PubMed] [Google Scholar]
- 6.Public Health England Group A streptococcal infections: third report on seasonal activity, 2017 to 2018. 2018. https://webarchive.nationalarchives.gov.uk/ukgwa/20220202090104/https://www.gov.uk/government/publications/group-a-streptococcal-infections-activity-during-the-2017-to-2018-season
- 7.Lynskey NN, Jauneikaite E, Li HK, Zhi X, Turner CE, et al. Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study. Lancet Infect Dis. 2019;19:1209–1218. doi: 10.1016/S1473-3099(19)30446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhi X, Li HK, Li H, Loboda Z, Charles S, et al. Emerging invasive group A Streptococcus M1UK lineage detected by allele-specific PCR, England, 2020. Emerg Infect Dis. 2023;29:1007–1010. doi: 10.3201/eid2905.221887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies MR, Keller N, Brouwer S, Jespersen MG, Cork AJ, et al. Detection of Streptococcus pyogenes M1UK in Australia and characterization of the mutation driving enhanced expression of superantigen SpeA. Nat Commun. 2023;14:1051. doi: 10.1038/s41467-023-36717-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li HK, Zhi X, Vieira A, Whitwell HJ, Schricker A, et al. Characterization of emergent toxigenic M1UK Streptococcus pyogenes and associated sublineages. Microb Genom. 2023;9:000994. doi: 10.1099/mgen.0.000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Rivers J, Mathis S, Li Z, Chochua S, et al. Expansion of invasive group A Streptococcus M1UK lineage in active bacterial core surveillance, United States, 2019‒2021. Emerg Infect Dis. 2023;29:2116–2120. doi: 10.3201/eid2910.230675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolters M, Berinson B, Degel-Brossmann N, Hoffmann A, Bluszis R, et al. Population of invasive group A streptococci isolates from A German tertiary care center is dominated by the hypertoxigenic virulent M1UK genotype. Infection. 2024;52:667–671. doi: 10.1007/s15010-023-02137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rümke LW, de Gier B, Vestjens SMT, van der Ende A, van Sorge NM, et al. Dominance of M1UK clade among Dutch M1 Streptococcus pyogenes. Lancet Infect Dis. 2020;20:539–540. doi: 10.1016/S1473-3099(20)30278-4. [DOI] [PubMed] [Google Scholar]
- 14.Johannesen TB, Munkstrup C, Edslev SM, Baig S, Nielsen S, et al. Increase in invasive group A streptococcal infections and emergence of novel, rapidly expanding sub-lineage of the virulent Streptococcus pyogenes M1 clone, Denmark, 2023. Euro Surveill. 2023;28:2300291. doi: 10.2807/1560-7917.ES.2023.28.26.2300291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demczuk W, Martin I, Domingo FR, MacDonald D, Mulvey MR. Identification of Streptococcus pyogenes M1UK clone in Canada. Lancet Infect Dis. 2019;19:1284–1285. doi: 10.1016/S1473-3099(19)30622-X. [DOI] [PubMed] [Google Scholar]
- 16.Gouveia C, Bajanca-Lavado MP, Mamede R, Carvalho AA, Rodrigues F, et al. Sustained increase of paediatric invasive Streptococcus pyogenes infections dominated by M1UK and diverse emm12 isolates, Portugal, September 2022 to May 2023. Eurosurveillance. 2022;28:2300427. doi: 10.2807/1560-7917.ES.2023.28.36.2300427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lacey JA, Bennett J, James TB, Hines BS, Chen T, et al. A worldwide population of Streptococcus pyogenes strains circulating among school-aged children in Auckland, New Zealand: a genomic epidemiology analysis. Lancet Reg Health West Pac. 2024;42:100964. doi: 10.1016/j.lanwpc.2023.100964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Sepulveda BM, Heavens D, Pulford CV, Predeus AV, Low R, et al. An accessible, efficient and global approach for the large-scale sequencing of bacterial genomes. Genome Biol. 2021;22:349. doi: 10.1186/s13059-021-02536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics. 2018;34:i142–i150. doi: 10.1093/bioinformatics/bty266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolley KA, Maiden MCJ. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010;11:1–11. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 25.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bah SY, Keeley AJ, Armitage EP, Khalid H, Chaudhuri RR, et al. Genomic characterization of skin and soft tissue Streptococcus pyogenes isolates from a low-income and a high-income setting. mSphere. 2023;8:e0046922. doi: 10.1128/msphere.00469-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. 2019;63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMillan DJ, Drèze P-A, Vu T, Bessen DE, Guglielmini J, et al. Updated model of group A Streptococcus M proteins based on A comprehensive worldwide study. Clin Microbiol Infect. 2013;19:E222-9. doi: 10.1111/1469-0691.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner CE, Holden MTG, Blane B, Horner C, Peacock SJ, et al. The emergence of successful Streptococcus pyogenes lineages through convergent pathways of capsule loss and recombination directing high toxin expression. mBio. 2019;10:e02521-19. doi: 10.1128/mBio.02521-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flores AR, Chase McNeil J, Shah B, Beneden C, Shelburne SA. Capsule-negative emm types are an increasing cause of pediatric group A streptococcal infections at A large pediatric hospital in texas. J Pediatr Infect Dis Soc. 2019;8:244–250. doi: 10.1093/jpids/piy053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner CE, Abbott J, Lamagni T, Holden MTG, David S, et al. Emergence of A new highly successful acapsular group A Streptococcus clade of genotype emm89 in the united kingdom. mBio. 2015;6:e00622. doi: 10.1128/mBio.02521-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu L, Olsen RJ, Nasser W, Beres SB, Vuopio J, et al. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest. 2015;125:3545–3559. doi: 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatsuno I, Sawai J, Okamoto A, Matsumoto M, Minami M, et al. Characterization of the NAD-glycohydrolase in streptococcal strains. Microbiol Read Engl. 2007;153:4253–4260. doi: 10.1099/mic.0.2007/009555-0. [DOI] [PubMed] [Google Scholar]
- 36.Remmington A, Turner CE. The DNases of pathogenic Lancefield streptococci. Microbiology. 2018;164:242–250. doi: 10.1099/mic.0.000612. [DOI] [PubMed] [Google Scholar]
- 37.Davies MR, Holden MT, Coupland P, Chen JHK, Venturini C, et al. Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat Genet. 2015;47:84–87. doi: 10.1038/ng.3147. [DOI] [PubMed] [Google Scholar]
- 38.Chochua S, Metcalf BJ, Li Z, Rivers J, Mathis S, et al. Population and whole genome sequence based characterization of invasive group A Streptococci recovered in the United States during 2015. mBio. 2017;8:e01422-17. doi: 10.1128/mBio.01422-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unoarumhi Y, Davis ML, Rowe LA, Mathis S, Li Z, et al. A novel invasive Streptococcus pyogenes variant sublineage derived through recombinational replacement of the emm12 genomic region. Sci Rep. 2023;13:21510. doi: 10.1038/s41598-023-48035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remmington A, Haywood S, Edgar J, Green LR, de Silva T, et al. Cryptic prophages within a Streptococcus pyogenes genotype emm4 lineage. Microb Genom. 2021;7:mgen000482. doi: 10.1099/mgen.0.000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DebRoy S, Sanson M, Shah B, Regmi S, Vega LA, et al. Population genomics of emm4 group A Streptococcus reveals progressive replacement with A hypervirulent clone in North America. mSystems. 2021;6:e0049521. doi: 10.1128/mSystems.00495-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Putten BCL, Bril-Keijzers WCM, Rumke LW, Vestjens SMT, Koster LAM, et al. Novel emm4 lineage associated with an upsurge in invasive group A streptococcal disease in the Netherlands, 2022. Microb Genom. 2023;9:mgen001026. doi: 10.1099/mgen.0.001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chudasama DY, Tessier E, Flannagan J, Leeman D, Webster H, et al. Surge in SARS-CoV-2 transmission in school-aged children and household contacts, England, August to October 2021. Euro Surveill. 2021;26:2101019. doi: 10.2807/1560-7917.ES.2021.26.48.2101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vieira A, Wan Y, Ryan Y, Li HK, Guy RL, et al. Rapid expansion and international spread of M1UK in the post-pandemic UK upsurge of Streptococcus pyogenes. Nat Commun. 2024;15:3916. doi: 10.1038/s41467-024-47929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bessen DE. Tissue tropisms in group A Streptococcus: what virulence factors distinguish pharyngitis from impetigo strains? Curr Opin Infect Dis. 2016;29:295–303. doi: 10.1097/QCO.0000000000000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynskey NN, Goulding D, Gierula M, Turner CE, Dougan G, et al. RocA truncation underpins hyper-encapsulation, carriage longevity and transmissibility of serotype M18 group A streptococci. PLoS Pathog. 2013;9:e1003842. doi: 10.1371/journal.ppat.1003842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurst JR, Shannon BA, Craig HC, Rishi A, Tuffs SW, et al. The Streptococcus pyogenes hyaluronic acid capsule promotes experimental nasal and skin infection by preventing neutrophil-mediated clearance. PLoS Pathog. 2022;18:e1011013. doi: 10.1371/journal.ppat.1011013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flores AR, Jewell BE, Olsen RJ, Shelburne SA, III, Fittipaldi N, et al. Asymptomatic carriage of group A Streptococcus is associated with elimination of capsule production. Infect Immun. 2014;82:3958–3967. doi: 10.1128/IAI.01788-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vega LA, Sanson MA, Shah BJ, Flores AR. Strain-dependent effect of capsule on transmission and persistence in an infant mouse model of group A Streptococcus infection. Infect Immun. 2020;88:e00709-19. doi: 10.1128/IAI.00709-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sierig G, Cywes C, Wessels MR, Ashbaugh CD. Cytotoxic effects of streptolysin o and streptolysin s enhance the virulence of poorly encapsulated group A streptococci. Infect Immun. 2003;71:446–455. doi: 10.1128/IAI.71.1.446-455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riddle DJ, Bessen DE, Caparon MG. Variation in Streptococcus pyogenes NAD+ glycohydrolase is associated with tissue tropism. J Bacteriol. 2010;192:3735–3746. doi: 10.1128/JB.00234-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lacey JA, Marcato AJ, Chisholm RH, Campbell PT, Zachreson C, et al. Evaluating the role of asymptomatic throat carriage of Streptococcus pyogenes in impetigo transmission in remote Aboriginal communities in northern territory, Australia: a retrospective genomic analysis. Lancet Microbe. 2023;4:e524–e533. doi: 10.1016/S2666-5247(23)00068-X. [DOI] [PubMed] [Google Scholar]
- 53.Bessen DE, Smeesters PR, Beall BW. Molecular epidemiology, ecology, and evolution of group A Streptococci. Microbiol Spectr. 2018;6 doi: 10.1128/microbiolspec.CPP3-0009-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, et al. Evolutionary origin and emergence of A highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 55.Chalker V, Jironkin A, Coelho J, Al-Shahib A, Platt S, et al. Genome analysis following a national increase in Scarlet fever in England 2014. BMC Genom. 2017;18:224. doi: 10.1186/s12864-017-3603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kapatai G, Coelho J, Platt S, Chalker VJ. Whole genome sequencing of group A Streptococcus: development and evaluation of an automated pipeline for emmgene typing. PeerJ. 2017;5:e3226. doi: 10.7717/peerj.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cordery R, Purba AK, Begum L, Mills E, Mosavie M, et al. Frequency of transmission, asymptomatic shedding, and airborne spread of Streptococcus pyogenes in schoolchildren exposed to scarlet fever: a prospective, longitudinal, multicohort, molecular epidemiological, contact-tracing study in England, UK. Lancet Microbe. 2022;3:e366–e375. doi: 10.1016/S2666-5247(21)00332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma H, Ong MR, Ready D, Coelho J, Groves N, et al. Real-time whole genome sequencing to control a Streptococcus pyogenes outbreak at a national orthopaedic hospital. J Hosp Infect. 2019;103:21–26. doi: 10.1016/j.jhin.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 59.Turner CE, Bedford L, Brown NM, Judge K, Török ME, et al. Community outbreaks of group A Streptococcus revealed by genome sequencing. Sci Rep. 2017;7:8554. doi: 10.1038/s41598-017-08914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen M, Cai J, Davies MR, Li Y, Zhang C, et al. Increase of emm1 isolates among group A Streptococcus strains causing scarlet fever in Shanghai, China. Int J Infect Dis. 2020;98:305–314. doi: 10.1016/j.ijid.2020.06.053. [DOI] [PubMed] [Google Scholar]
- 61.Ben Zakour NL, Davies MR, You Y, Chen JHK, Forde BM, et al. Transfer of scarlet fever-associated elements into the group A Streptococcus M1T1 clone. Sci Rep. 2015;5:15877. doi: 10.1038/srep15877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Rivers J, Mathis S, Li Z, Velusamy S, et al. Genomic surveillance of Streptococcus pyogenes strains causing invasive disease, United States, 2016–2017. Front Microbiol. 2020;11:1547. doi: 10.3389/fmicb.2020.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beres SB, Musser JM. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One. 2007;2:e800. doi: 10.1371/journal.pone.0000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker MJ, Brouwer S, Forde BM, Worthing KA, McIntyre L, et al. Detection of epidemic Scarlet fever group A Streptococcus in Australia. Clin Infect Dis. 2019;69:1232–1234. doi: 10.1093/cid/ciz099. [DOI] [PubMed] [Google Scholar]
- 65.Tse H, Bao JYJ, Davies MR, Maamary P, Tsoi H-W, et al. Molecular characterization of the 2011 Hong Kong Scarlet fever outbreak. J Infect Dis. 2012;206:341–351. doi: 10.1093/infdis/jis362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.You Y, Davies MR, Protani M, McIntyre L, Walker MJ, et al. Scarlet fever epidemic in China caused by Streptococcus pyogenes serotype M12: epidemiologic and molecular analysis. EBioMedicine. 2018;28:128–135. doi: 10.1016/j.ebiom.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacob KM, Spilker T, LiPuma JJ, Dawid SR, Watson ME. Complete genome sequence of emm4 Streptococcus pyogenes MEW427, a throat isolate from a child meeting clinical criteria for Pediatric Autoimmune Neuropsychiatric Disorders Associated with Streptococcus (PANDAS) Genom Announc. 2016;4:e00127-16. doi: 10.1128/genomeA.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davies MR, McIntyre L, Mutreja A, Lacey JA, Lees JA, et al. Atlas of group A streptococcal vaccine candidates compiled using large-scale comparative genomics. Nat Genet. 2019;51:1035–1043. doi: 10.1038/s41588-019-0417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Osowicki J, Azzopardi KI, McIntyre L, Rivera-Hernandez T, Ong C-LY, et al. A controlled human infection Model of group A Streptococcus pharyngitis: which strain and why? mSphere. 2019;4:e00647-18. doi: 10.1128/mSphere.00647-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.