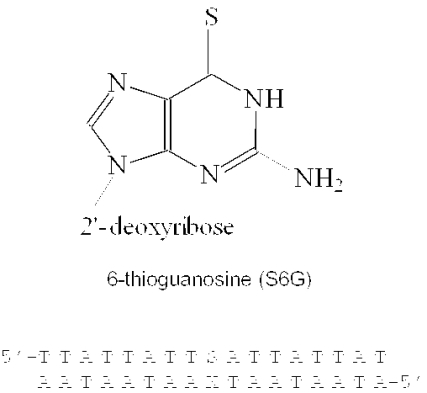
Abstract
The incorporation of 6-thioguanine (S6G) into DNA is a prerequisite for its cytotoxic action, but duplex structure is not significantly perturbed by the presence of the lesion [J. Bohon and C. R. de los Santos (2003) Nucleic Acids Res., 31, 1331–1338]. It is therefore possible that the mechanism of cytotoxicity relies on a loss of stability rather than a pathway involving direct structural recognition. The research described here focuses on the changes in thermodynamic properties of duplex DNA owing to the introduction of S6G as well as the kinetic properties of base pairs involving S6G. Replacement of a guanine in a G•C pair by S6G results in ∼1 kcal/mol less favorable Gibbs free energy of duplex formation at 37°C. S6G•T and G•T mismatch-containing duplexes have almost identical Gibbs free energy at 37°C, with values ∼3 kcal/mol less favorable than that of the control. Base pair stability is affected by S6G. The lifetime of the normal G•C base pair is ∼125 ms, whereas that of the G•T mismatch is below the detection limit. The lifetimes of S6G•C and S6G•T pairs are ∼7 and 2 ms, respectively, demonstrating that, although S6G significantly decreases the stability of the pairing with cytosine, it slightly increases that of a mismatch.
INTRODUCTION
Thioguanine (S6G) and other thiopurines have been used as therapeutic agents in the treatment of cancer and a variety of other diseases since the early 1950s (1). Although a large body of research is now available concerning thiopurines, the exact mechanism by which they operate has yet to be fully understood. It is known that all these pharmaceuticals act through the same biochemical intermediates, requiring activation to 6-thiodeoxyguanosine 5′-triphosphate through metabolic cellular pathways. This modified nucleotide is the active metabolite which is subsequently incorporated into the DNA duplex, a prerequisite for cytotoxicity (2,3). Many biochemical and molecular biological studies have examined the effects of S6G incorporation on cellular processes, increasing the overall understanding of its cytotoxic activity (4–22). However, a more basic question remains: what causes the initial recognition of S6G as abnormal?
The S6G residue itself is almost identical to an unmodified guanine nucleotide, differing only in the substitution of a sulfur atom for the oxygen at the sixth position of the purine ring (23). An initial NMR structural study had concluded that S6G substantially disrupted the duplex structure and blocked the formation of G-four tetrads (24). However, recent data confirms that S6G does not greatly perturb the normal helical form of DNA, where it can form Watson–Crick base pairs with cytosine (25,26) and adopt a wobble alignment with thymine (25). This indicates that the recognition mechanism for S6G lesions may not be structural in nature. Theoretical simulations predict a slight destabilization of the hydrogen bonding owing to the increased size and decreased electronegativity of the sulfur atom compared with the normal oxygen (27,28). These studies were later confirmed by the observation that a S6G•C pair has a greater degree of mobility and is marginally less stable than a normal G•C pair (25,26). The comparative stability of an S6G•T mismatch to that of a G•T mismatch or a S6G•C pair is difficult to predict, but, owing to the absence of a strong mutagenic effect for S6G (16), it is likely that the pairing with cytosine will be more stable.
This work describes a thorough investigation of the impact of the S6G residue upon the stability of duplex DNA through quantification of thermodynamic differences between lesioned and control duplexes as well as of the kinetic properties of the base pairs of interest. For quantification of the global thermodynamic properties of these duplexes, van't Hoff methods are used to analyze ultraviolet (UV) absorption as a function of temperature. The widespread use of this approach allows comparisons to previously determined values for many different sequence contexts (29), including mismatched and lesioned duplexes (26,30). To assess the impact of the S6G residue on the local base pair stability, NMR inversion-recovery experiments are used to quantify the kinetic properties of the lesioned base pairs. Values are also determined for a control duplex and a duplex containing a G•T mismatch in the same sequence context for comparison. In order to facilitate isolation of NMR signals, the sequences are designed to consist entirely of A•T base pairs except for a single G•C pair in the central position. The chemical structure of S6G and the sequence of the duplex used in this research are shown in Figure 1.
Figure 1.
Chemical structure of 6-thioguanine and sequence of the lesion containing duplexes studied here. S is either 6-thioguanine or guanosine and X is either cytosine or thymine.
MATERIALS AND METHODS
Synthesis of deoxyribo-oligonucleotides
A protected 6-thioguanosine precursor for solid-phase DNA synthesis was purchased from Glen Research, Inc., and was incorporated into the oligodeoxynucleotide sequence using standard phosphoramidite chemistry procedures. Then, 17mer sequences d(TTATTATTSATTATTAT), d(ATAATAATCAATAATAA) and d(ATAATAATTAATAATAA) (where S represents 6-thiodeoxyguanosine) were synthesized and purified at the SUNY Stony Brook DNA synthesis facility. Purification of modified and unmodified oligodeoxynucleotides was performed as described previously (31). Sample purity was estimated to be >96%, as revealed by analytical high-performance liquid chromatography. Oligodeoxynucleotides were subsequently desalted using a Sephadex G-25 column and then converted to the sodium salt by passing them through a Dowex cation exchange resin column.
Duplex formation and sample preparation
A 1:1 stoichiometry was obtained for each duplex by addition of the appropriate amount of each strand determined using extinction coefficients calculated using Generunner v3.00 (Hastings Software, Inc.). Samples used for the thermodynamic studies consisted of between 0.5 and 2 OD260 U of duplex dissolved in 1 ml of a water solution, pH 6.8, containing 100 mM NaCl and 0.5 mM EDTA, for DNA concentrations between 0.5 and 8 μM. Samples used for the base pair kinetic experiments consisted of between 100 and 150 OD260 U of duplex dissolved in 25 mM borate buffer, pH 8.9, containing 100 mM NaCl, 0.05 mM 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) and 0.5 mM EDTA, for DNA concentrations between 1.2 and 1.5 mM. Ammonia catalyst concentration was determined by titration analysis of a stock ammonium hydroxide and confirmed as unchanged by following, in a control sample, the NH3 peak integrals comparative to DSS over time. The loss of ammonia was negligible over the course of the experiments (usually totaling 1–2 weeks).
Thermodynamic experiments
UV-absorption thermal denaturation experiments were carried out using the ‘thermal’ program of a CARY100 Bio UV-VIS spectrophotometer equipped with a multicell block temperature regulation unit and a fluid conduction thermal regulation enhancement attachment (Varian, Inc.). Up to four duplex samples at a time, plus the blank, were placed in stoppered cuvettes and inserted into the spectrophotometer for nearly simultaneous measurement. The pre-run temperature was allowed to equilibrate for at least 10 min to either 1 or 80°C depending upon the experiment. Temperatures for each experiment generally were increased from 1 to 80°C or decreased from 80 to 1°C at a rate of 0.3°C/min for complete temperature equilibration at each point. A minimum of 40 experiments was performed for each duplex, with varied DNA concentrations. The calibration and temperature regulation was verified by utilizing a digital thermometer placed inside one of the sample chambers and comparing thermometer temperature values with those displayed by the computer software. Although it was impossible to use this method during an actual experiment, the calibration run showed that the temperature was stably controlled to within 1°C.
Thermodynamic data analysis
Absorbance versus temperature profiles were analyzed within the Meltwin (v3.5, McDowell) program to generate thermodynamic parameters. Curves were fitted using a Marquardt–Levenburg algorithm and processed via two different methods. In method I, each individual curve was analyzed separately to establish thermodynamic values (32–34). In method II, a plot of versus ln(Ct/4) yields a straight line with slope R/ΔH0 and intercept of ΔS0/ΔH0 via the following equation, allowing determination of the enthalpy and entropy simultaneously (35):
The Gibbs free energy (ΔG0) is then calculated from these values. A detailed description of the theory and error analysis is given elsewhere (36,37). Error values are stated as standard deviations of the data. Plots of the fraction of DNA in the helical conformation versus temperature, α curves, were generated for each data set in Microsoft Excel (Figure 2).
Figure 2.
Representative α curves for the control G•C duplex (filled triangles), S6G•C duplex (open triangles), G•T mismatch duplex (filled circles) and S6G•T duplex (open circles).
Kinetic experiments
In order to investigate base pair kinetics, imino proton exchange rates were measured at pH 8.9 with increasing catalyst concentrations. The imino proton chemical shifts at neutral and basic conditions were very close, indicating a similar duplex structure at those pH values. Longitudinal relaxation times (T1) were determined at 5°C using 1D frequency-selective inversion-recovery experiments. Spectra were collected on a Varian INOVA 600 MHz spectrometer, using a designed pulse program that, by means of a shaped pulse, inverted the imino proton signal of interest at the beginning of the sequence (37) (Sh2jr, available upon request). The water signal was suppressed by using a jump-return reading pulse. Low-temperature calibration was performed before each set of experiments by analyzing chemical shift differences of the methanol signals and using the macro ‘tempcal’ provided by Varian. NMR signals were referenced to external DSS at 0 p.p.m. A minimum of 64 scans was taken for each recovery time. The number of data points taken for a given ammonia concentration differed to maximize the values in the steepest portion of the sloped region of the curve. A typical inversion-recovery profile included recovery times of 0, 0.0005, 0.001, 0.002, 0.004, 0.008, 0.016, 0.032, 0.064, 0.1, 0.12, 0.14, 0.16, 0.18, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45, 0.5, 0.6, 0.7, 0.8, 0.9, 1, 2, 3 and 4 s. Recovery times between 1 and 4 s were used in each experiment to verify complete signal relaxation. The spectra were processed using Felix97 (Accelrys, San Diego, CA). Within each data set, the spectra were subjected to identical solvent suppression routine, complex Fourier transformation, baseline correction and peak integral measurement. Only the phase parameters were readjusted for each spectrum. For each duplex, the imino proton chemical shifts at neutral and basic pH were very close, indicating similar structures within this pH range.
Kinetic data analysis
In order to determine the T1 relaxation time for each catalyst concentration (ammonium), peak integral values obtained at each recovery time were fitted to the following exponential equation (38):
I(t) is the integral of the NMR peak at a given recovery time, I(0) is the integral of the peak immediately following inversion and I(∞) is the integral of the peak after full recovery. Error values for T1 were calculated as standard deviations of multiple experiments at each catalyst concentration. Imino proton exchange theory has previously been thoroughly described (39) and only the most directly utilized equations are mentioned here.
The solvent-imino proton exchange time (τex) is calculated for each catalyst concentration from T1 and values via the following equation (where is T1 in the absence of added catalyst):
The error in τex was propagated from errors in T1 as described elsewhere (37):
In this expression, τ0 is the base pair lifetime, [Cat] is the catalyst concentration and C is a constant that incorporates all parameters relating to accessibility and proton transfer probability. This constant was calculated using previously determined values (39–43) and extrapolated to 5°C. Kd represents the dissociation constant of the base pair, which is equal to the ratio of the base pair opening and closing rates (kop/kcl). Extrapolating this equation to an infinite catalyst concentration, the base pair lifetime becomes equal to the solvent exchange time and Kd can be calculated from the slope value. Thus, the measurement of exchange times at different concentrations of catalyst allows calculation of the base pair lifetime as well as opening and closing rates.
RESULTS
Thermal and thermodynamic stability
As shown in Figure S1 of the Supplementary Material, all duplexes display the characteristic hyperchromic shift in UV absorbance as the temperature is raised from 1 to 80°C. Representative α curves are shown in Figure 2, and Tm values, calculated for a 1.0 × 10−4 M concentration, are listed in Table 1. Replacement of the central G•C base pair of the duplex by a S6G•C pair causes a decrease of ∼10% in the thermal stability of duplex formation. In contrast, the Tm values of the G•T-containing and S6G•T-containing duplexes are essentially identical, indicating that within the concentration range investigated in this study, S6G does not diminish the stability of the mismatch any further.
Table 1.
Thermodynamic parameters of DNA duplexes
| Duplex | Tma (°C) | ΔH0 (kcal/mol) | ΔS0 (kcal/mol °K) | ΔG025°C (kcal/mol) | ΔG037°C (kcal/mol) |
|---|---|---|---|---|---|
| G•Cb | 42.4 | −103 ± 6 | −0.31 ± 0.02 | −12.0 ± 0.3 | −8.3 ± 0.2 |
| S6G•Cb | 39.6 | −105 ± 7 | −0.32 ± 0.02 | −11.2 ± 0.3 | −7.4 ± 0.1 |
| G•Tb | 33.2 | −91 ± 4 | −0.28 ± 0.01 | −8.7 ± 0.1 | −5.4 ± 0.1 |
| S6G•Tb | 33.6 | −97 ± 6 | −0.30 ± 0.02 | −9.0 ± 0.2 | −5.4 ± 0.2 |
| G•Cc | (39.9)d | (−171 ± 19)d | (−0.53 ± 0.06)d | (−14.8 ± 0.7)d | −8.1 ± 0.5 |
| S6G•Cc | 39.8 | −101 ± 8 | −0.30 ± 0.02 | −11.0 ± 0.2 | −7.4 ± 0.2 |
| G•Tc | 32.9 | −97 ± 9 | −0.30 ± 0.03 | −8.9 ± 0.1 | −5.2 ± 0.3 |
| S6G•Tc | 34.3 | −85 ± 5 | −0.26 ± 0.02 | −8.9 ± 0.1 | −5.8 ± 0.2 |
Reported errors are SDs.
aCalculated for a 1.0 × 10−4 M DNA concentration.
bDetermined from individual melting curves.
cDetermined from (Tm)−1 versus ln(Ct/4) plots.
dValues between brackets are included to emphasize the non-two-state process for melting of the unmodified duplex but are not valid parameters.
The thermodynamic properties computed for the duplexes studied are also listed in Table 1. The different methods of data analysis are used to verify the presence of a two-state process of duplex denaturation. Previous studies have defined that an agreement within 10% of the enthalpy values calculated by the two methods indicates a simple one-step transition. Enthalpy differences in the 10–20% range suggest a ‘marginally non-two-state’ transition, and discrepancies >20% are indicative of a non-two-state process (30). The G•C control duplex has enthalpy values that differ by almost 70%. In addition, a careful examination of the versus ln(Ct/4) plot (van't Hoff plot) reveals a curvature in the data, further supporting its departure from a one-step process (Figure 3, filled triangles). This deviation from a linear plot is postulated to be due to a non-zero change in heat capacity of the sample over different concentrations (44). Therefore, in the case of the G•C duplex, only the free energy values as computed near the melting temperature are meaningful (34). For the G•T mismatch and both S6G-containing samples enthalpy values are generally within 10% agreement (Table 1) and display linear van't Hoff plots (Figure 3), indicating the presence of a two-state dissociation process for these duplexes.
Figure 3.
Plots of versus ln(Ct/4) for the control G•C duplex (filled triangles), S6G•C duplex (open triangles), G•T mismatch duplex (filled circles) and S6G•T duplex (open circles).
In agreement with a previously calculated decrease in stability (28), the ΔG0 for the S6G•C duplex is ∼1 kcal/mol less favorable than that of the control duplex. In contrast, the G•T and S6G•T mismatch duplexes have similar ΔG0 values, which are ∼3 kcal/mol less favorable than that of the G•C control duplex, with some dependence on the temperature at which it is calculated. ΔG0 values are reported at both 25 and 37°C for all duplexes in Table 1.
Kinetic experiments
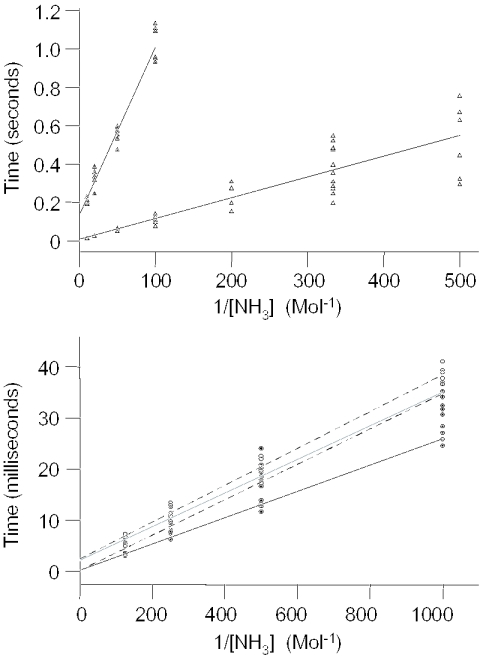
Typical inversion-recovery plots collected for the G9H1 or S6G9H1 imino protons of each duplex are shown in Figure S2 of the Supplementary Material. The T1 values derived from these curves are then used to calculate imino proton exchange times (τex) at different catalyst concentrations (see Materials and Methods). Figure 4 displays plots of τex versus (catalyst concentration)−1, which in turn are employed to determine base pair Kd, τ0 and τopen values. Table 2 lists the base pair kinetic parameters computed for each duplex.
Figure 4.
Plots of imino proton exchange times as a function of catalyst (ammonia) concentration. The top panel shows τex for the G•C G9H1 (filled triangles) and S6G•C S6G9H1 (open triangles); the lower panel depicts values for the G•T G9H1 (filled circles, solid line), G•T T26H3 (filled circles, dashed line), S6G•T S6G9H1 (open circles, solid line) and S6G•T T26H3 (open circles, dashed line).
Table 2.
Base pair kinetics parameters
| Duplex | G•C | S6G•C | G•T | S6G•T | ||
|---|---|---|---|---|---|---|
| Imino proton | G(H1) | S6G(H1) | G(H1) | T(H3) | S6G(H1) | T(H3) |
| τ0(ms) | 126 ± 8 | 7.3 ± 1.2 | 0.1 ± 0.3 | 0.1 ± 1 | 2.4 ± 0.9 | 2.3 ± 0.3 |
| Kd × 10−7 (mol−1) | 5.4 ± 0.3 | 52 ± 4 | 1760 ± 55 | 3612 ± 113 | 1534 ± 220 | 3472 ± 95 |
| τopen(ns) | 68 ± 3 | 38 ± 4 | 17 ± 12 | 36 ± 8 | 366 ± 91 | 770 ± 103 |
Kinetic parameters measured at 5°C. Reported errors are SDs τ0 is the base pair lifetime. Kd is the dissociation constant. τopen is the open pair lifetime.
The base pair lifetime, τ0, computed for the G•T mismatch should be viewed with a great degree of caution because the value is well within an error of zero. This will also greatly affect the lifetime of the open state of the base pair and the opening rate calculations, as a difference of a single millisecond in τ0 causes, at this level, an order of magnitude change in these calculated values. The dissociation constant, Kd, of the G•T mismatch is more reliable and is comparable to previously determined values on the order of 10−4 for this mismatch (41). The dissociation constants for the G•T and S6G•T mismatches must be considered equivalent, because they are within one standard deviation of each other when the same residues of the mismatches are compared. Although Kd values for the G•T mismatch are quite similar to those of the S6G•T mispair, both the open pair and closed pair lifetimes are significantly longer in the case of S6G, suggesting that, despite the similarity in lifetime ratios, the energy barrier between the open and closed states is higher for the S6G•T mismatch.
The values of the base pair lifetime in both the open and closed states for the control G•C pair are relatively large compared with most other values found in the literature (45,46). This is probably due to the lower temperature of our measurement and the positioning of the pair in the center of the 17mer, quite far from any end-fraying effect. This is not a unique result, as a very long base pair lifetime was found for the G4H1 proton in a 5′-d(CGCGAATTCGCG)-3′ context (41). The S6G•C pair is much less stable than the normal G•C pair, having a higher Kd and shorter τ0. Although the lifetime of the open state is only a little more than half that of the normal G•C pair, that of the closed state is >17 times shorter for S6G•C, indicating that its opening rate is faster and that the S6G•C pair spends more total time in the open conformation than its control counterpart. Another way to look at these differences is to compare base pair opening rates. In a second, the normal G•C pair opens ∼8 times, while the S6G•C pair opens ∼137 times and the S6G•T mismatch opens ∼400 times. Because the resolution of the method used in these experiments cannot detect lifetimes <1 ms and therefore was not high enough to measure the lifetime of the G•T mismatch, the best estimate of the opening rate is given as a lower boundary. Thus, the opening rate of the G•T mismatch can reasonably be said to be faster than 1000 times per second.
DISCUSSION
The cytotoxic events triggered by the presence of S6G in DNA are extensive and well characterized, but the precise mechanism by which this residue is perceived as ‘damage’ by the cellular machinery are not yet fully understood (4–22). Because it is so similar to normal guanine, it is important to address what differentiates S6G such that it is recognized as abnormal and thus initiates the process that eventually leads to cell death. Previous structural studies have shown the lesion to be highly conservative because it does not perturb duplex formation, or disturb Watson–Crick base pairs (25,26) or wobble mismatch alignments (25). Only the formation of G-four tetrads seems to be affected by a single substitution of S6G for guanine (24,47). Thus, a next logical step is to investigate changes in duplex and base pair stability induced by the presence of S6G.
The results indicate that the substitution of S6G for guanine on a G•C base pair moderately decreases the thermal and thermodynamic stability of DNA duplexes. This is a much weaker effect than the destabilization caused by the introduction of a single G•T mismatch in the same duplex or by the presence of a single abasic site (48). The impact of S6G on stability does not extend to the G•T mismatch, where, within the sequence context studied here, Tm and ΔG0 values of G•T-containing and S6G•T-containing duplexes are essentially identical. More significantly, the base pair kinetics are severely changed by the presence of S6G, suggesting that in genomic DNA, where its effect on overall thermal and thermodynamic stability is minimized, local dynamics of S6G-containing pairs are responsible for initiating the biologically relevant responses. The comparative differences in stability between S6G•C pairs and S6G•T mismatches are significantly smaller than the differences between normal G•C pairs and G•T mismatches. The G•T-containing and S6G•T-containing duplexes display nearly identical ΔG0 of duplex formation, having ∼3 kcal/mol less stability and melting temperatures almost 10°C lower than the G•C duplex. The S6G•C duplex is also thermodynamically less stable than the control, but by a much smaller margin. The difference is ∼3°C in melting temperature and the ΔG°37 of duplex formation is less favorable by just 1 kcal/mol.
It is interesting to note that, aside from the control duplex, the samples display apparently two-state melting behavior. This difference is perhaps not surprising given the relative base pair stabilities on the local level. The duplex sequence used in our studies contains a series of A•T base pairs, stabilized by two Watson–Crick hydrogen bonds, with a central G•C pair that has three hydrogen bonds. A typical A•T base pair has a lifetime on the order of 10 ms (49) and dissociation constant on the order of 10−5 (50). The G•C pair in the control duplex has a lifetime of 125 ms and a dissociation constant of 5.39 × 10−7. The high affinity (1/Kd) of a central G•C pair could result in an additional phase of melting where all A•T pairs have dissociated but the strands are still held together by the intact G•C pair. In this case, the separated bases may re-associate with each other or with bases on neighboring duplexes, forming transient intermediate structures that disrupt the two-state melting behavior. In contrast, this stable core is not present in the modified duplexes, for which the central pairs have lifetimes of <10 ms and Kd values close to that of A•T pairs, facilitating a two-state transition for the A•T rich duplex.
The instability of G•T, S6G•T and S6G•C pairs, indicated by higher Kd values and shorter base pair lifetimes, points to a greater degree of mobility at the S6G site compared with the control. This is particularly evident for the G•T mismatch, where the physical properties are difficult to quantify owing to a base pair lifetime within the experimental detection limit. It is true to a lesser extent for S6G•C pairs as well, given the relatively short lifetime of 7.3 ms compared with 125 ms for a G•C pair in the same sequence context. The lifetime of the S6G•C base pair computed here is almost identical to the value of 8 ms reported for the lesion in a different sequence context (26), suggesting a lack of influence of flanking base pairs on the kinetics of S6G•C base pairs. The S6G•C base pair lifetime is ∼3 times longer and the dissociation constant ∼30 times lower than the values for S6G•T. This disparity is much smaller than that between a normal G•C pair and a G•T mismatch, offering a possible explanation for the mild mutagenicity observed for S6G (16).
The destabilization of DNA as a result of the substitution of guanine for S6G is most striking on the single base pair level, where the opening rate of the S6G•C pair is 17-fold faster than that of the normal G•C pair. The decreased local stability of the modified base pairs is likely to have physiological significance relevant to the recognition of this lesion by downstream cytotoxic factors. Because the modified base pairs are relatively unstable and the bases spend a relatively large amount of time out of the helix, it is likely that an outside force, such as a DNA-binding protein, could stabilize an open conformation of the base pair. The increase in the single-stranded character of the DNA could cause a high-fidelity polymerase to sense any base incorporated opposite S6G, including dC, as a mistake and initiate a futile cycle of insertion and excision events, a process that eventually leads to apoptosis (51). Another possibility is that the instability might enhance the binding of mismatch repair proteins, the activity of which is thought to be important in the cytotoxic pathway of S6G (13). The increased mobility of S6G certainly makes it more exposed than a normal guanine to any number of cellular components that could be involved in the cytotoxic mechanism of S6G.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Ms Cecilia Torres for the synthesis and purification of oligodeoxynucleotides. NIH Grants CA93502 and CA98232 supported this research. Funding to pay the Open Access publication charges for this article was provided by NIH Grant CA98232.
Conflict of interest statement. None declared.
REFERENCES
- 1.Elion G.B. The purine path to chemotherapy. Science. 1989;244:41–47. doi: 10.1126/science.2649979. [DOI] [PubMed] [Google Scholar]
- 2.Lepage G.A. Basic biochemical effects and mechanism of action of 6-thioguanine. Cancer Res. 1963;23:1202–1206. [PubMed] [Google Scholar]
- 3.Lee S.H., Sartorelli A.C. The effects of inhibitors of DNA biosynthesis on the cytotoxicity of 6-thioguanine. Cancer Biochem. Biophys. 1981;5:189–194. [PubMed] [Google Scholar]
- 4.Maybaum J., Mandel H.G. Unilateral chromatid damage: a new basis for 6-thioguanine cytotoxicity. Cancer Res. 1983;43:3852–3856. [PubMed] [Google Scholar]
- 5.Christie N.T., Drake S., Meyn R.E., Nelson J.A. 6-thioguanine-induced DNA damage as a determinant of cytotoxicity in cultured Chinese hamster ovary cells. Cancer Res. 1984;44:3665–3671. [PubMed] [Google Scholar]
- 6.Bodell W.J., Morgan W.F., Rasmussen J., Williams M.E., Deen D.F. Potentiation of 1,3-bis(2-chloroethyl)-1-nitrosurea (BCNU)-induced cytotoxicity in 9L cells by pretreatment with 6-thioguanine. Biochem. Pharmacol. 1985;34:515–520. doi: 10.1016/0006-2952(85)90182-0. [DOI] [PubMed] [Google Scholar]
- 7.Fairchild C.R., Maybaum J., Kennedy K.A. Concurrent unilateral chromatid damage and DNA strand breakage in response to 6-thioguanine treatment. Biochem. Pharmacol. 1986;35:3533–3541. doi: 10.1016/0006-2952(86)90623-4. [DOI] [PubMed] [Google Scholar]
- 8.Covey J.M., D'Incalci M., Kohn K.W. Production of DNA–protein crosslinks (DPC) by 6-thioguanine (TG) and 2′-deoxy-6-thioguanosine (TGdR) in L1210 cells in vitro. Proc. Am. Assoc. Cancer Res. 1986;27:17. [Google Scholar]
- 9.Pan B.F., Nelson J.A. Characterization of the DNA damage in 6-thioguanine-treated cells. Biochem. Pharmacol. 1990;40:1063–1069. doi: 10.1016/0006-2952(90)90494-6. [DOI] [PubMed] [Google Scholar]
- 10.Iwaniec L.M., Kroll J.J., Roethel W.M., Maybaum J. Selective inhibition of sequence-specific protein–DNA interactions by incorporation of 6-thioguanine: cleavage by restriction endonucleases. Mol. Pharmacol. 1990;39:299–306. [PubMed] [Google Scholar]
- 11.Ling Y., Chan J.Y., Beattie K.L., Nelson J.A. Consequences of 6-thioguanine incorporation into DNA on polymerase, ligase, and endonuclease reactions. Mol. Pharmacol. 1992;42:802–807. [PubMed] [Google Scholar]
- 12.Griffin S., Branch P., Xu Y., Kurran P. DNA mismatch binding and incision at modified guanine bases by extracts of mammalian cells: implications for tolerance to DNA methylation damage. Biochemistry. 1994;33:4787–4793. doi: 10.1021/bi00182a006. [DOI] [PubMed] [Google Scholar]
- 13.Swann P.F., Waters T.R., Moulton D.C., Xu Y., Zheng Q., Edwards M., Mace R. Role of postreplicative DNA mismatch repair in the cytotoxic activity of thioguanine. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 14.Waters T.R., Swann P.F. Cytotoxic mechanism of 6-thioguanine: hMutSa, the human mismatch binding heterodimer, binds to DNA containing S6-methylthioguanine. Biochemistry. 1997;36:2501–2506. doi: 10.1021/bi9621573. [DOI] [PubMed] [Google Scholar]
- 15.Hampson R., Humbert O., Macpherson P., Aquilina G., Karran P. Mismatch repair defects and O6-methylguanine-DNA methyltransferase expression in acquired resistance to methylating agents in human hells. J. Biol. Chem. 1997;272:28596–28606. doi: 10.1074/jbc.272.45.28596. [DOI] [PubMed] [Google Scholar]
- 16.Uribe-Luna S., Quintana-Hau J.D., Maldonado-Rodriguez R., Espinosa-Lara M., Beattie K.L., Farquhar D., Nelson A.J. Mutagenic consequences of the incorporation of 6-thioguanine into DNA. Biochem. Pharmacol. 1997;54:419–424. doi: 10.1016/s0006-2952(97)00200-1. [DOI] [PubMed] [Google Scholar]
- 17.Aquilina G., Crescenzi M., Bignami M. Mismatch repair, G2/M cell cycle arrest and lethality after DNA damage. Carcinogenesis. 1999;20:2317–2325. doi: 10.1093/carcin/20.12.2317. [DOI] [PubMed] [Google Scholar]
- 18.Krynetskaia N.F., Krynetski E.Y., Evans W.E. Human RNase H-mediated RNA cleavage from DNA–RNA duplexes is inhibited by 6-deoxythioguanosine incorporation into DNA. Mol. Pharmacol. 1999;56:841–848. [PubMed] [Google Scholar]
- 19.Krynetskaia N.F., Cai X., Nitiss J.L., Krynetski E.Y., Relling M.V. Thioguanine substitution alters DNA cleavage mediated by topoisomerase II. FASEB J. 2000;14:2339–2344. doi: 10.1096/fj.00-0089com. [DOI] [PubMed] [Google Scholar]
- 20.Bignami M., O'Driscoll M., Aquilinia G., Karran P. Unmasking a killer: DNA O6-methylguanine and the cytotoxicity of methylating agents. Mutat. Res. 2000;462:71–82. doi: 10.1016/s1383-5742(00)00016-8. [DOI] [PubMed] [Google Scholar]
- 21.de las Alas M.M., Los G., Lin X., Kurdi-Haidar B., Manorek G., Howell S. identification of transdominant-negative genetic suppressor elements derived from hMSH2 that mediate resistance to 6-thioguanine. Mol. Pharmacol. 2002;62:1198–1206. doi: 10.1124/mol.62.5.1198. [DOI] [PubMed] [Google Scholar]
- 22.Yan T., Berry S.E., Desai A.B., Kinsella T.J. DNA mismatch repair (MMR) mediates 6-thioguanine genotoxicity by introducing single-strand breaks to signal a G2-M arrest in MMR-proficient RKO cells. Clin. Cancer Res. 2003;9:2327–2334. [PubMed] [Google Scholar]
- 23.Bugg C.E., Thewalt U. The crystal and molecular structure of 6-Thioguanine. J. Am. Chem. Soc. 1970;92:7441–7445. doi: 10.1021/ja00728a031. [DOI] [PubMed] [Google Scholar]
- 24.Marathias V.M., Sawicki M.J., Bolton P.H. 6-Thioguanine alters the structure and stability of duplex DNA and inhibits quadruplex DNA formation. Nucleic Acids Res. 1999;27:2860–2867. doi: 10.1093/nar/27.14.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohon J., de los Santos C. Structural effect of the anticancer agent 6-thioguanine on duplex DNA. Nucleic Acids Res. 2003;31:1331–1338. doi: 10.1093/nar/gkg203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommervile L., Krynetski E.Y., Krynetskaia N.F., Beger R.D., Zhang W., Marhefka C.A., Evans W.E., Kriwacki R.W. Structure and dynamics of thioguanine-modified duplex DNA. J. Biol. Chem. 2003;278:1005–1011. doi: 10.1074/jbc.M204243200. [DOI] [PubMed] [Google Scholar]
- 27.Singh K., Groth-Vasselli B., Farnsworth P., Rai D. Effect of thiobase incorporation into duplex DNA during the polymerization reaction. Res. Commun. Mol. Pathol. Pharmacol. 1996;94:129–140. [PubMed] [Google Scholar]
- 28.Sponer J., Leszczynski J., Hobza P. Thioguanine and thiouracil: hydrogen-bonding and stacking properties. J. Phys. Chem. 1997;101:9489–9495. [Google Scholar]
- 29.Owczarzy R., Vallone P.M., Gallo F.J., Paner T.M., Lane M.J., Benight A.S. Predicting sequence-dependent melting stability of short duplex DNA oligomers. Biopolymers. 1997;44:217–239. doi: 10.1002/(SICI)1097-0282(1997)44:3<217::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 30.Allawi H.T., SantaLucia J., Jr Thermodynamics and NMR of internal G-T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 31.Cullinan D., Johnson F., Grollman A.P., Eisenberg M., de los Santos C. Solution structure of a DNA duplex containing the exocyclic lesion 3, N4-Etheno-2′-deoxycytidine opposite 2′-deoxyguanosine. Biochemistry. 1997;36:11933–11943. doi: 10.1021/bi9705725. [DOI] [PubMed] [Google Scholar]
- 32.Gralla J., Crothers D.M. Free energy of imperfect nucleic acid helices. J. Mol. Biol. 1973;78:301–319. doi: 10.1016/0022-2836(73)90118-6. [DOI] [PubMed] [Google Scholar]
- 33.Petersheim M., Turner D.H. Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGU, and ACCGGUp. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 34.Longfellow C.E., Kierzek R., Turner D.H. Thermodynamic and spectroscopic study of bulge loops in oligoribonucleotides. Biochemistry. 1990;29:278–285. doi: 10.1021/bi00453a038. [DOI] [PubMed] [Google Scholar]
- 35.Borer P.N., Dengler B., Tinoco I., Jr, Uhlenbeck O.C. Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 1974;86:843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- 36.Marky L.A., Breslauer K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 37.Bohon J. 2004. Characterization of duplex DNA containing 6-thioguanine, PhD Dissertation, Department of Physiology and Biophysics, Stony Brook University, NY.
- 38.Folta-Stogniew E., Russu I. Base-catalysis of imino proton exchange in DNA: effects of catalyst upon DNA structure and dynamics. Biochemistry. 1996;35:8439–8449. doi: 10.1021/bi952932z. [DOI] [PubMed] [Google Scholar]
- 39.Gueron M., Leroy J.-L. Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 40.Ts'o P.O.P. Basic Principles in Nucleic Acid Chemistry. NY: Academic Press; 1974. [Google Scholar]
- 41.Moe J.G., Russu I. Kinetics and base-pair opening in 5′-d(CGCGAATTCGCG)-3′ and a substituted dodecamer containing G-T mismatches. Biochemistry. 1992;31:8421–8428. doi: 10.1021/bi00151a005. [DOI] [PubMed] [Google Scholar]
- 42.Snoussi K., Leroy J.-L. Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry. 2001;40:8898–8904. doi: 10.1021/bi010385d. [DOI] [PubMed] [Google Scholar]
- 43.Warmlander S., Sponer J.E., Sponer J., Leijon M. The influence of the thymine C5 methyl group on spontaneous base pair breathing in DNA. J. Biol. Chem. 2002;277:28491–28497. doi: 10.1074/jbc.M202989200. [DOI] [PubMed] [Google Scholar]
- 44.Chaires J.B. Possible origin of differences between van't Hoff and calorimetric enthalpy estimates. Biophys. Chem. 1997;64:15–23. doi: 10.1016/s0301-4622(96)02205-3. [DOI] [PubMed] [Google Scholar]
- 45.Dornberger U., Leijon M., Fritzsche H. High base pair opening rates in tracts of GC base pairs. J. Biol. Chem. 1999;274:6957–6962. doi: 10.1074/jbc.274.11.6957. [DOI] [PubMed] [Google Scholar]
- 46.Bhattacharya P.K., Cha J., Barton J.K. 1H NMR determination of base-pair lifetimes in oligonucleotides containing single base mismatches. Nucleic Acids Res. 2002;30:4740–4750. doi: 10.1093/nar/gkf601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spackova N., Cubero E., Sponer J., Orozco M. Theoretical study of the guanine—(6-thioguanine substitution in duplexes, triplexes, and tetraplexes. J. Am. Chem. Soc. 2004;126:14642–14650. doi: 10.1021/ja0468628. [DOI] [PubMed] [Google Scholar]
- 48.Gelfand C.A., Plum G.E., Grollman A.P., Johnson F., Breslauer K.J. Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry. 1998;37:7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- 49.Leroy J.-L., Charretier E., Kochoyan M., Gueron M. Evidence from base-pair kinetics for two types of adenine tract structures in solution: their relation to DNA curvature. Biochemistry. 1988;27:8894–8898. doi: 10.1021/bi00425a004. [DOI] [PubMed] [Google Scholar]
- 50.Gueron M., Charretier E., Hagerhorst J., Kochoyan M., Leroy J.L., Moraillon A. Applications of imino proton exchange to nucleic acid kinetics and structures. In: Sarma R.H., Sarma M.H., editors. DNA & RNA: Structure and Methods. Vol. 3. Adenine Press; 1990. pp. 113–135. [Google Scholar]
- 51.Khare V., Eckert K. The 3′-(5′ exonuclease of T4 DNA polymerase removes premutagenic alkyl mispairs and contributes to futile cycling at O6-methylguanine lesions. J. Biol. Chem. 2001;276:24286–24292. doi: 10.1074/jbc.M011025200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.