Abstract
A minimally addressed area in Saccharomyces cerevisiae research is the mapping of transcription start sites (TSS). Mapping of TSS in S.cerevisiae has the potential to contribute to our understanding of gene regulation, transcription, mRNA stability and aspects of RNA biology. Here, we use 5′ SAGE to map 5′ TSS in S.cerevisiae. Tags identifying the first 15–17 bases of the transcripts are created, ligated to form ditags, amplified, concatemerized and ligated into a vector to create a library. Each clone sequenced from this library identifies 10–20 TSS. We have identified 13 746 unique, unambiguous sequence tags from 2231 S.cerevisiae genes. TSS identified in this study are consistent with published results, with primer extension results described here, and are consistent with expectations based on previous work on transcription initiation. We have aligned the sequence flanking 4637 TSS to identify the consensus sequence A(Arich)5NPyA(A/T)NN(Arich)6, which confirms and expands the previous reported PyA(A/T)Pu consensus pattern. The TSS data allowed the identification of a previously unrecognized gene, uncovered errors in previous annotation, and identified potential regulatory RNAs and upstream open reading frames in 5′-untranslated region.
INTRODUCTION
Comparative genomics is now possible in Saccharomyces cerevisiae with the genome sequencing of several related fungal species, including Ashbya gossypii (1), Kluyveromyces waltii (2), Saccharomyces kluyveri (3), Candida glabrata (4) and Kluyveromyces lactis (4). While comparative genomics has been an aid in identifying genes and made it possible to identify previously overlooked genes (5) and to correct errors in the yeast genome (6), prediction of promoters and starts of transcription in either yeast or higher eukaryotes is still difficult at best (7). For S.cerevisiae, only a limited number of cDNA sequences have been generated, with ∼3000 expressed sequence tags and cDNA sequences available through GenBank. Knowledge of the transcription start site (TSS) provides insights into the likely location of coding and non-coding sequence features. Recently, a high-throughput method of identifying TSS based on a combination of 5′ RACE and SAGE analysis has been reported (8–10). We have independently developed a method similar in strategy (Figure 1) and demonstrated it in S.cerevisiae. Analysis of transcript length across the S.cerevisiae genome by microarray analysis (11) has been carried out, but the resolution of this approach is insufficient to precisely identify TSS. Examination of the published literature and of the UTRdb (12) suggests that for <500 genes have the TSS been published or reported in GenBank (Table 2). In addition, unlike higher eukaryotes where the TSS is typically located 25 bp downstream of the TATA element (13), in S.cerevisiae the TSS ranges from 45 to 120 bases downstream of the TATA element (14) complicating bioinformatic prediction of the TSS. In eukaryotes, the sequence around the TSS, sometimes referred to as the Initiator sequence (Inr), has been described to play an important role in transcription initiation (14,15), particularly in the absence of a TATA element. The Inr may act as a major transcription promoter element in TATA-less genes (16,17). For mammalian genes, the Inr sequence appears to be gene specific (16) with a minimal consensus ‘CA’ (15,18) where A is the TSS. In S.cerevisiae, >80% of genes are classified as TATA-less (19). Further investigation of the nature of the TSS element depends on identifying TSS locations for a large number of genes. The small genome size, high-quality annotation (20) and ease with which high-quality RNA can be purified (21) from S.cerevisiae make high-throughput identification of TSS practical.
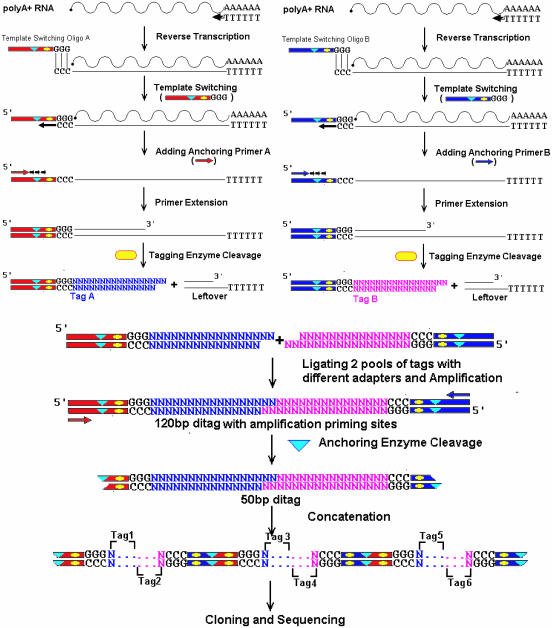
Figure 1.
Scheme of 5′ SAGE methodology. The poly(A)-rich RNA is divided into two pools. Different oligos set (blue or red) are used to carry out reverse transcription, template-switching and primer extension. After tagging enzyme (yellow oval) digestion, the two sample pools are combined together to make 120 bp ditags. The anchoring enzyme (blue triangle) is used to generate 50 bp ditag for concatenation. Specifics are discussed in Materials and Methods.
Table 2.
Validation of protein-coding gene TSS from published results
| ORF | Gene | Total occurrencea | Tags position (occurrence) | Published data | References |
|---|---|---|---|---|---|
| Gene TSS being consistent to the published results | |||||
| YAL038W | CDC19 | 4 | −30(1), 27(2), −18(1) | Around −20 | (33) |
| YBR093C | PHO5 | 4 | −35(3), −32(1) | −40, −35 | (33) |
| YCL030C | HIS4 | 7 | −63(1), −60(6) | −60 | (58) |
| YCR012W | PGK1 | 58 | −46(1), −43(4), −40(53) | Within −48 to −27 | (59) |
| YCR097W | HMRA1 | 6 | −12(4), −9(2) | Around −10 | (33) |
| YDL067C | COX9 | 14 | −46(9), −42(3), −23(1), −10(1) | −52, −42 | (60) |
| YDL081C | RPP1A | 17 | −78(2), −74(2), −73(1), −68(1), −66(8), −58(1), −44(2) | −74, −73, −67, −66, −58 | (61) |
| YDL130W | RPP1B | 7 | −115(1), −22(2), 20(2) −17(1), −13(1) | −18, −17, −16 | (61) |
| YDR035W | ARO3 | 3 | −21(2), −35(1) | −8, −9, −20 to −22, −30, −31, −37, −38, −74 to −77 | (62) |
| YDR382W | RPP2B | 11 | −122(1), −83(1), −81(7), −21(1), −10(1) | −81, −80 | (61) |
| YEL039C | CYC7 | 1 | −89(1) | −93, −89, −81 | (54) |
| YER081W | SER3b | 12 | −473(6), −271(1), −219(1), −38(1), −18(3) | −15 to −43 | (35) |
| YER055C | HIS1 | 3 | −93(1), −62(1), −33(1) | −116 to −109, −94 to −90, −72 to −70, −64 to −59, −55 to −52, −47 to −45, −39, −38, −35 to −32, −29, −30 | (37) |
| YGL030W | RPL30 | 11 | −80(1), −69(1), −64(2), −59(1), −58(4), −54(1), −43(1) | −58 | (63) |
| YHR216W | IMD2 | 14 | −240(1), −106(10), −44(3) | −106 | (64) |
| YML024W | RPS17A | 15 | −42(1), −27(1), −26(12), −17(1) | −27, −26, −19, −18, −13 | (65) |
| YML123C | PHO84 | 14 | −54(1), −45(1), −39(12) | −39 | (66) |
| YNL052W | COX5A | 9 | −34(1), −31(3), −19(4), −4(1) | −31 | (67) |
| YOL039W | RPP2A | 5 | −30(3), −28(1), −21(1) | −55, −31 to −8 | (61) |
| YOL058W | ARG1 | 5 | −72(4), −59(1) | −77, −72 | (68) |
| YOL086C | ADH1 | 16 | −61(1), −49(1), −43(2), −38(5), −33(2), −31(2), −20(1), −12(2) | −37, −38, −28, −27 | (69,70) |
| YOR065W | CYT1 | 7 | −360(1), −289(1), −282(1), −277(4) | −296, −282, −268, −182, −140 | (71) |
| YPR035W | GLN1 | 38 | −187(1), −126(2), −120(10), −117(21), −115(2), −103(1), −87(1) | −120 | (72) |
| Gene TSS having minor difference between 5′ SAGE and published result | |||||
| YBR010W | HHT1 | 26 | −29(1), −28(24), −19(1) | −30 | (73) |
| YBR118W | TEF2 | 23 | −111(1), −110(1), −87(1), −74(2), −45(1), −30(1), −22(16) | −23 | (31) |
| YBR249C | ARO4 | 2 | −235(1), −86(1) | −91 | (74) |
| YDL205C | HEM3 | 4 | −346(1), −193(1), −187(1), −143(1) | −176 | This study |
| YDR044W | HEM13 | 2 | −43(1), −38(1) | −75, −52, −48, −47 | (75) |
| YEL009C | GCN4c | 5 | −603(1), −586(2), −575(2) | −591, −574 | (45) |
| YGL187C | COX4d | 11 | −478(1), −476(1), −465(1), −406(1), −401(3), −383(1), −381(1), −374(1), −370(1) | −481, −479, −468, −464 | (76) |
| YGR192C | TDH3 | 32 | −47(2), −45(1), −39(28), −13(1) | −40 | (77) |
| YGR254W | ENO1 | 23 | −37(22), −34(1) | −39 | (78) |
| YHR018C | ARG4 | 3 | −55(3) | −57 | (79) |
| YHR174W | ENO2 | 9 | −38(3), −30(6) | −35 | (78) |
| YIL125W | KGD1 | 4 | −149(1), −137(1), −117(1), −112(1) | −254, −109 | (80) |
| YJR048W | CYC1 | 4 | −56(1), −46(2), −38(1) | −61, −47 | (37) |
| YLL041C | SDH2 | 6 | −43(3), −39(1), −16(2) | −40 | (81) |
| YLR286C | CTS1 | 2 | −428(1), −231(1) | −238 | (82) |
| YOR303W | CPA1c | 4 | −243(1), −230(2), −222(1) | −244 | (45) |
| YPL111W | CAR1 | 1 | −40(1) | −49, −48, −46, −39, −37 | (37) |
| YPR080W | TEF1 | 32 | −31(31), −26(1) | −32 | (31) |
| YPR187W | RPO26 | 2 | −44(1), −16(1) | −84, −78, −76, −75, −60, −59, −58, −51, −43, −42, −35, −34, −26, −25, −15, −1 | (83) |
| Gene TSS that are not consistent with the published results | |||||
| YBL022C | PIM1 | 1 | −133(1) | −146, −341 | (84) |
| YDR050C | TPI1 | 28 | −41(1), −38(1), −30(24), −15(2) | −20 | (32,33) |
| YDR232W | HEM1 | 1 | −260(1) | −76, −72, −68, −63 | (85) |
| YKL216W | URA1 | 5 | −62(1), −44(1), −42(2), −32(1) | −155, −151, −143, −138, −136, −133 | (86) |
| YLR420W | URA4 | 1 | −79(1) | −41, −40, −22, −18 | (87) |
| YMR186W | HSC82 | 7 | −35(3), −31(1), −30(1), −20(1), −10(1) | −98, −42 | (88) |
| YMR303C | ADH2 | 3 | −46(3) | −67, −63, −58, −55 | (70) |
For each gene, the total occurrences of tags in the 5′-UTR region are shown. Published data were collected from the literature. Each unitag position is shown relative to the translation start position followed with occurrence in parenthesis. Bold type indicates consistent position of TSS position between 5′ SAGE and previously published results.
aThe total occurrence = ∑tag occurrence.
bSER3 contains an upstream regulatory non-coding RNA SRG1, see the text for detail.
cBoth GCN4 and CPA1 contain long 5′-UTR containing uORF(s). Tags representing GCN4 TSS identified by examining all tags upstream of GCN4, as they were beyond the 500 bp 5′-UTR mapping range using in this study.
dCOX4 contains a 342 bp 5′-UTR intron.
MATERIALS AND METHODS
Strains and culturing
S.cerevisiae W303–1A (MATa ura3–1 leu2–3112 trp1–1 can1–100 ade2–1 his3–11,15 [psi+]) was the strain used in this study. Yeast was cultured in rich medium (YPD) at 30°C. Escherichia coli strain TOP10 (Invitrogen) was used in 5′ SAGE library construction.
Ditag formation
Total RNA from exponential stage (OD = 1.0) S.cerevisiae is extracted by the Qiagen RNeasy Mini kit. Poly(A)+ RNA is further isolated using NucleoTrap mRNA kit (BD Bioscience Clontech) and divided into two pools A and B. Each 300 ng poly(A)+ RNA is converted to double-stranded cDNA using a SMART cDNA library construction kit (BD Bioscience Clontech). The manufacturer's protocol is followed, except that the primer sequences are altered. First, CDS primer [5′-AAGCAGTGGTATCAACGCAGAGTAC-(T)30-VN-3′] (V = A, G or C) is added with either 5′ SAGE template switching (TS) oligos A (5′-GGGATTTGCTGGTGCAGTACAGGATCCGACggg-3′; lower case, RNA) or B (5′-GCTGCTCGAATTCAAGCTTCTGGATCCGACggg-3′; lower case, RNA) in the reaction mixture for first strand synthesis and TS at 42°C for 90 min. Each oligo final concentration is 1 μM. The newly synthesized single strand cDNA is used as template for three cycles primer extension by PCR. The anchoring primers: Anchor-A, 5′-GTGCTCGTGGGATTTGCTGGTGCAGTACAGG-3′ and Anchor-B, 5′-GAGCTCGTGCTGCTCGAATTCAAGCTTCTGG-3′ and the 5′ CDS primer are used. The primer extension products are checked by electrophoresis on 2% agarose gel (Supplementary Figure S1A).
The two primer extension products are digested by tagging enzyme MmeI (New England Biolabs) for 2 h at 37°C. The digestion mixture is extracted by phenol–chloroform once and precipitated by ammonium acetate and ethanol at −80°C, resuspended in TE (10 mM Tris–HCl, 1 mM EDTA, pH 8.0), and loaded on a 15% TBE polyacrylamide gel (BioRad) (Supplementary Figure S1B). After staining with ethidium bromide, the ∼60 bp tag band from two pools are combined and eluted from the gel. The eluted DNA is precipitated by ethanol and resuspended in 6 μl LoTE (2.5 mM Tris–HCl, 0.25 mM EDTA, pH 8.0).
The gel recovered tag pools in LoTE are ligated by T4 DNA ligase to form ∼120 bp ditag in 6 μl reaction at 16°C overnight. The ligation mixture is used to make 1:10, 1:20, 1:40, 1:80 and 1:160 dilutions for PCR optimization. Each 50 μl reaction contains: 10× reaction buffer 5 μl, dNTP (2.5 mM each) 5 μl, Anchor-A (10 μM) 5 μl, Anchor-B (10 μM) 5 μl, diluted ditag template 1 μl and ExTaq (Takara) 1 μl (5 U/μl). The PCR condition is 95°C for 2 min; 95°C for 30 s, 55°C for 1 min, 72°C for 30 s for 27 cycles; and 72°C for 5 min. The reaction mixture is loaded on a 15% TBE polyacrylamide ready gel (BioRad) to determine the dilution that was suitable according to the product size and quality (Supplementary Figure S1C). After optimization, the best PCR condition is applied in a scaled-up PCR. A total of ∼200 PCRs are pooled together and precipitated by ethanol and loaded on 12% TBE polyacrylamide gel for ∼30 reactions per lane (Supplementary Figure S1D). Similar protocols as stated previously are used to recover the 120 bp ditag from the TBE gel into 120 μl LoTE.
Ditag digestion and concatenation
The 120 bp ditag recovered from TBE gel is digested by BamHI at 37°C for 2 h. The 120 bp ditag is digested into 32, 50 and 32 bp fragments (Supplementary Figure S1E). The gel recovered 50 bp ditag sample is self-ligated at 16°C for 5–8 h. The ligation mixture is loaded on 2% agarose gel with DNA ladder (Supplementary Figure S1F). Different size ranges, 300–500 bp, 500–800 bp and 800–1000 bp are excised and recovered by gel extraction kit (Qiagen). Different concatemer size ranges are cloned into the BamHI site of pUC18. Colonies are screened by blue–white selection on ampicillin Luria–Bertani plates. The library quality assessment is similar to the standard SAGE library method provided by the Invitrogen Long SAGE kit.
Sequence analysis and tag extraction
The 5′ SAGE library is sequenced using M13 forward/reverse primers. All sequencing reads were processed by Phred (22,23) to generate quality scores for each base. Perl scripts are used to mask out low-quality sequence and get valid ditag sequence from each sequence read. All ditag sequences are further processed to get non-redundant tag pools, recording occurrence and ditag origin of each tag. Considering a possible amplification effect, we regard tags having the same ditag origin as occurring only once.
Genome-wide localization analysis
Each valid 5′ SAGE tag with occurrence information is searched against the S.cerevisiae genomic sequence to find the matching position. All tags are divided into three categories: tags with unique hit (unitag), tags with multiple hits and tags with no hit. An open reading frame (ORF) is assigned to the unitag if the matching position locates within the region from 500 bp upstream of ATG to 100 bp downstream of stop codon. The analysis results are organized using a MySQL database system.
Primer extension
The AMV Primer Extension Kit (Promega) was used. Following the manufacturer's protocol, 10 μg total RNA purified by RNeasy Mini kit (Qiagen) from S.cerevisiae was reverse transcribed by annealing with gene-specific 32P-end-labeled primer (Supplementary Table S1) at 42°C for 30 min. The extension product was separated by electrophoresis on 8% denaturing polyacrylamide gels containing 7 M urea. Bands were detected on auto-radiographic film.
Statistical analysis
The whole genome gene expression microarray data were obtained from http://web.wi.mit.edu/young/expression/ (24). Linear regression model was calculated by R (http://www.r-project.org/). All distribution results were drawn by JMP statistical software package (SAS Institute Inc.).
TSS sequence LOGO
The 20 bases of sequence flanking each TSS identified by unitag was extracted from genomic sequence and processed by WebLOGO (http://weblogo.berkeley.edu/). Since the tag is always beginning with three or more Gs from the pre-synthesized adapter, the tag extraction process eliminated all Gs at the 5′ end. Considering a possible loss of G in the tag 5′ end during tag sequence extraction, we added a G at 5′ end of tag if tag length is <17 bp and a G is actually there based on the genomic sequence.
RESULTS
Data summary and mapping to the S.cerevisiae genome
A 5′ SAGE library was constructed from S.cerevisiae strain W303–1A following the flowchart shown in Figure 1. From 2112 sequencing reads, 11 776 non-duplicated ditags were extracted. From these ditags, we obtained 21 952 tags identifying 13 746 distinct sequences. Among these 13 746 distinct tags were 10 866 tags with one or more exact matches in the S.cerevisiae genome, of which 9738 had only a single genomic match (unitags). These unitags are evenly distributed throughout the genome, with the number of tags from each chromosome correlating to the gene number (R2 = 0.970) and chromosome length (R2 = 0.966) (Supplementary Figure S2). Inspection of the 2880 tags without a match in the genome revealed that most result from improper trimming of the tags, likely as a result of MmeI cutting at positions other than the typical 18/20 bases downstream of the recognition site, or as a result of improper ligation of the MmeI generated two base 3′ overhangs. Most additional unmatched tags resulted from sequence errors, while strain sequence discrepancies, errors in the S288C sequence and possible post-transcription RNA editing (25) potentially account for the remaining non-matching tags. In some cases, these tag sequences could be edited and used in the analysis, but to exclude user bias they were not included in the results reported here.
For each unitag and corresponding mapping position on the chromosome, the associated gene with the same orientation was identified using the annotation from SGD (20). Of the 9738 unitags, we identified 8194 unitags locating in the region from 500 bp upstream of the ATG start codon to 100 bp downstream of the stop codon of annotated ORFs. As shown in Table 1, of the 8194 gene-associated unitags, >60% map to the putative 5′-untranslated region (5′-UTR), with 89% of multiple occurrence gene-associated unitags mapping to the 5′-UTR. We also noticed that ∼40% of unitags were mapped to the coding region and the putative 3′-UTR. These tags likely result from either premature reverse transcriptase termination or degraded mRNA template. Only 3.7% of unitags in the coding region and 4.3% in the 3′-UTR region occurred more than once, much lower than the 21.1% of unitags with occurrences in the putative 5′-UTR, suggesting that most of the coding region and 3′-UTR tags are spurious.
Table 1.
Gene-associated unitag positions relative to annotated S.cerevisiae genes
| Tag occurence | Putative 5′-UTR (−500, −1)a | Coding region | Putative 3′-UTR (+1, +100)a | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Unitags | Occurence | Unitags | Occurence | Unitags | Occurence | Unitags | Occurence | |
| =1 | 3895 (48%) | 3895 (33%) | 2754 (34%) | 2754 (23%) | 380 (4.6%) | 380 (3.2%) | 7029 (86%) | 7029 (59%) |
| >1 | 1041b (12%) | 4461 (38%) | 107b (1.3%) | 299 (2.5%) | 17b (2.1%) | 35 (0.3%) | 1165 (14%) | 4795 (41%) |
| Total | 4936c (60%) | 8356d (71%) | 2861 (35%) | 3053 (26%) | 397 (4.8%) | 415 (3.5%) | 8194 | 11 824 |
The number of 5′ SAGE unitags and corresponding number of total occurrence (Occurence) were categorized into three groups based on mapping positions and two groups based on the tag occurrence threshold. For each number, the percentage of the total unitag or occurrence (both in bold) is shown in the parenthesis.
aThe (−500, −1) is the position relative to the ATG start codon. The (+1, +100) refers to position relative to the STOP codon.
bThe numbers of multiple occurrence unitags. In the 5′-UTR category, ∼1041/4936 = 21.1% of unitags are multiple occurrence, while only 107/2861 = 3.7% and 17/397 = 4.3% of unitags mapping to coding region and 3′-UTR, respectively, are multiple occurrence.
cThe 4936 putative 5′-UTR mapped unitags come from 2231 genes, with a total of 660 genes represent at least one multiple occurrence unitag and 1571 genes represented by one or more single occurrence unitags.
dTotal tag occurrence of all 5′-UTR mapped unitags. We estimate the percentage of tags representing actual TSS as bounded by 8365/11 824 = 70.7% and (4461 + 1904)/11 824 = 53.8%, where 1904 is an estimate of the number of real single occurrence tag in the putative 5′-UTR.
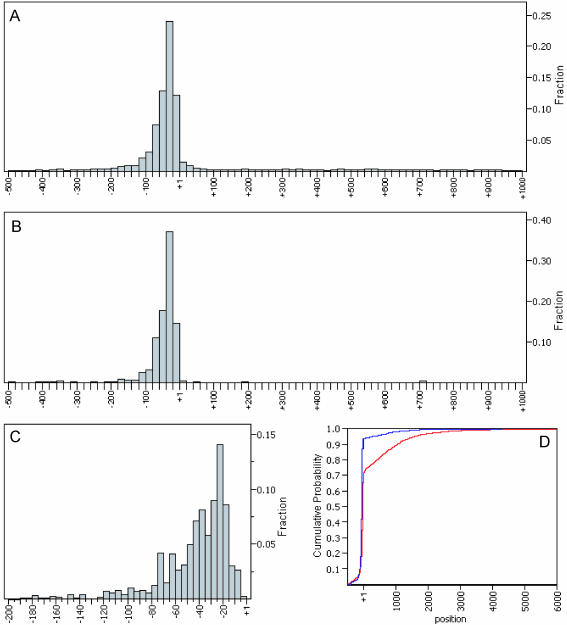
Figure 2 and Table 1 show the distribution of unitags weighted by occurrence from −500 to +1000 relative to the start codon. Both the histogram and the cumulative distribution function (CDF) plot indicate that >90% of multiple occurrence unitags and 70% of unitags are within the 500 bp putative 5′-UTR region. Furthermore, most transcripts start within the region 15–75 bp upstream of the start codon. From the data listed in Table 1 and Figure 2, we estimate that, among 11 824 occurrences of unitags, ∼54–70% appear to represent actual TSS, with the remainder spurious.
Figure 2.
5′ SAGE tag distribution around the ORF start codon. (A) Distribution of all 8194 unitags from 500 bp upstream of ATG to 1000 bp downstream of ATG. (B) Distribution of all 1165 multiple occurrence unitags from 500 bp upstream of ATG to 1000 bp downstream of ATG. (C) Zoom-in view of tag distribution shown in (B) within 200 bp putative 5′-UTR region. (D) Cumulative distribution function (CDF) plots of all unitags (red) and multiple occurrence unitags (blue).
Of the 4936 unitags from the 5′-UTR region, 2231 S.cerevisiae genes were identified. This is ∼40% of the protein-coding genes of S.cerevisiae. Among these 2231 genes, 660 genes were identified by at least one multiple occurrence unitag.
Correlation between tag occurrence, ORF length and gene expression level
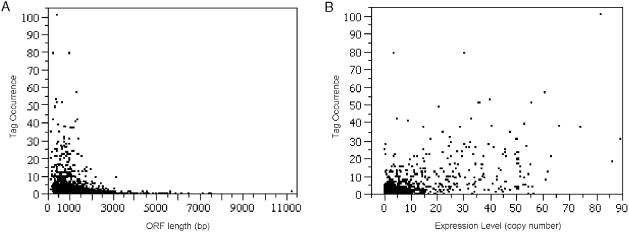
In conventional SAGE analysis, the tag occurrence is approximately proportional to the steady state level of transcripts in the cell. In 5′ SAGE, tag abundance appears to correlate not only to the level of gene expression but also to gene length (Figure 3). For this 5′ SAGE dataset, tag occurrence decreases with increasing ORF length, while tag occurrence increases with mRNA copy number.
Figure 3.
Correlation between tag occurrence and ORF length/gene expression level. Each 5′-UTR unitag occurrence was plotted with corresponding gene (A) ORF length, and (B) expression level (mRNA copies/cell). The expression level data is acquired from website http://web.wi.mit.edu/young/expression/ based on microarray data (24) and the ORF length was calculated based on SGD annotation (20). A linear regression model was applied (R2 = 0.3726, P < 2.2 × 10−16). Negative (−0.6320, P = 1.67 × 10−6) and positive (0.3804, P < 2 × 10−16) correlation coefficients were observed from the model for ORF length (bp) and expression level (mRNA copies/cell), respectively.
5′ SAGE tag coverage of highly expressed genes
As indicated above, the number of 5′ SAGE tags identified increased linearly with gene expression. To determine whether 5′ SAGE was selectively missing any protein-coding genes, we examined the 100 most highly expressed genes in the S.cerevisiae transcriptome (24) and found that 98 were identified by 5′ SAGE and that 88 were identified by multiple occurrences unitags in the 5′-UTR region. We also surveyed all 288 genes reported as having >10 mRNA copies per cell. Among these 288 genes, 269 genes have at least one 5′ SAGE tag in the 5′-UTR, and 211 have multiple occurrence tags (data not shown). These results suggest that 5′ SAGE does not show significant bias other than the bias against long transcripts and weakly expressed genes.
Validation of 5′ SAGE with TSS results from the published literature
As there is no database or reference work summarizing TSS in S.cerevisiae, we searched the primary literature for those genes where we have 5′ SAGE data. Forty-eight S.cerevisiae genes for which the TSS has been mapped and published are listed in Table 2. Because this data is generally found only in the text of the papers and not in the abstract, it is likely that some published results were overlooked. Most of these 48 gene TSS were mapped by primer extension (26) or S1 nuclease protection (27), the most commonly used conventional TSS mapping methods. Comparing the published TSS information for these genes with our data, we conclude that the 5′ SAGE determined TSS reported here are mostly consistent with the published results, either matching exactly or being within 1–5 bp (Table 2).
A total of 1128 tags identify multiple locations in the yeast genome. While it is impossible to identify the precise origin of these tags, three distinct multiple occurrence tags, with sequences ‘5′-ATAGAAGTCATCGAAA-3′’ (6 occurrences) and ‘5′-ATCGAAATAGATATTA(A)-3′’ (24 occurrences), mapped to the −68 and −59 of the multi-copy CUP1 locus, respectively, and are generally consistent with the previously published CUP1 TSS at positions −65 and −56 (28). Multiple location tags for the abundantly transcribed S.cerevisiae retrotransposons (29) were also found. Twenty-five distinct multiple occurrence tags were mapped to the upstream region of known TyA (Gag) and TyB (Gag-Pol) ORF of Ty1-4 as well as solo LTR elements. Three of these tags, ‘5′-AGGAGAACTTCTAG(TA)-3′’, with 19 total occurrences, were mapped to the TSS of Ty1, consistent with the previously TSS of ∼50 bp upstream of TyA/B protein start codon (30). Many of the remaining multiple location tags represent high occurrence sequences, such as ‘TAC(T)13’ and sequences from ribosomal RNA.
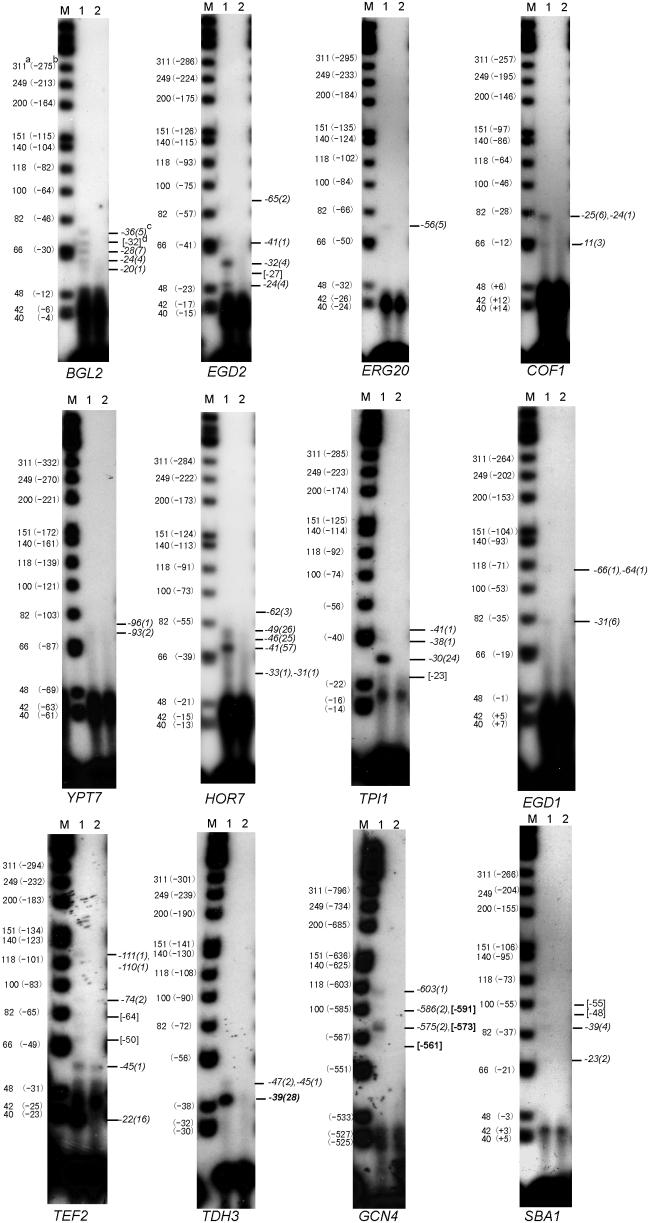
In order to further validate our 5′ SAGE results, we carried out TSS mapping by primer extension for 12 S.cerevisiae genes (Figure 4). Each of these 12 genes gave multiple TSS by 5′ SAGE, most of them having more than one multiple occurrence tag. Among these 12 genes, the TSS of TPI1, TEF2, TDH3 and GCN4 were previously published. The TSS data are included in the list of previously published TSS for S.cerevisiae for which we obtained TSS results in this study (Table 2). Our 5′ SAGE results and the primer extension results in Figure 4 generally agree with the previously published TSS results shown in Table 2, though there are some discrepancies. For example, the TSS of TEF2 reported by Nagashima et al. (31) is mapped to −23, yet our 5′ SAGE and primer extension results support an actual TSS of −22. For TPI1, the TSS has been mapped to −20 by Alber et al. (32,33), yet appears based on 5′ SAGE and primer extension to actually be at −30.
Figure 4.
Primer extension verification. Primer extension was used to map the TSS of 12 S.cerevisiae genes. For each gene, a gene-specific 32P-end-labeled primer was used to reverse transcribe to the 5′ end of the respective mRNA. Fragment sizes were analyzed by denaturing PAGE and autoradiograph. Lane M: Φ174 Hinf I DNA markers; lane 1: primer extension reaction; lane 2: reaction without RNA (negative control). (a) the marker actual size (nt); (b): the corresponding position (bp) to the ATG start codon; (c) the assigned 5′ SAGE tag position (bp) with occurrence in parenthesis, some are assigned to a single band because of the gel resolution; (d) The number in the bracket means the position (bp) estimation of apparent band without 5′ SAGE data.
The primer extension results shown in Figure 4 generally validate the 5′ SAGE results, with 35 out of 42 primer extension bands corresponding to 5′ SAGE predicted TSS, and with 7 out of 42 bands not represented in the 5′ SAGE data, and 4 out of 27 sites predicted by multiple occurrence 5′ SAGE tags not detected by these primer extension results. For these genes, all high abundance tags (>10 occurrences) corresponded to readily detected bands.
In addition to protein-coding genes, some non-coding RNAs, such as snoRNA (34) and the repression RNA SRG1 that regulates the expression of SER3 (35), are transcribed by RNA polymerase II. Similar to mRNA, these non-coding RNAs have 3′ poly(A) tails and 5′ caps (36), and thus can be detected by 5′ SAGE. We successfully mapped TSS unitags to nine non-coding RNAs (Supplementary Table S2), including SRG1. In this study, we identified tags representing both TSS of SER3 and SRG1, consistent with the published results.
Observations base on the TSS data set
The TSS information obtained from this study provides an opportunity to examine several aspects of the S.cerevisiae transcriptome as described below.
Distance between the TSS and TATA elements
Instead of transcription initiating 25–30 bp downstream of the TATA element as in higher eukaryotes (13), in S.cerevisiae the TSS have been previously reported to be 45–120 bp downstream of a TATA element (14). The 5′ SAGE determined TSS confirms this observation, with the TATA to TSS distance ranging from 50 to 125 nt for 80% of the 224 genes for which there is both 5′ SAGE TSS data and for which the TATA box was previously reported (19) (Supplementary Figure S3).
Identification of a TSS consensus sequence
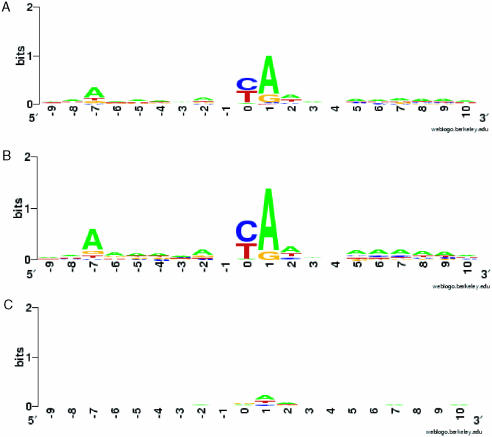
In S.cerevisiae sequence specificity in TSS selection has been reported (37), with the consensus reported as PyA(A/T)Pu (14,37), where A is the TSS. From the TSS information collected in this study, we selected 4937 unitags unambiguously mapping in the −500 to 0 bp region upstream of each ORF. The sequence of ±10 bp flanking each TSS was extracted from the S.cerevisiae genomic sequence and analyzed using WebLOGO (http://weblogo.berkeley.edu/) (Figure 5) (38,39). The consensus pattern found is A(Arich)5NPyA(A/T)NN(Arich)6, where A indicates the first transcribed nucleotide. This consensus pattern is unchanged when the analysis is carried out on the full set of genes, the set of TATA-containing genes, or the set of TATA-less genes, as classified by Basehoar et al. (19). This consensus is partially consistent with the previously reported consensus, the primary change being that an A is preferable at the –8 position. In addition, relatively A-rich regions were observed at +5 to +10 and −6 to −2 regions relative to the TSS. No significant pattern was identified around the putative TSS from the tags identified within or downstream of the coding region, further supporting the notion that these tags are spurious. The frequency of the consensus pattern in the 5′-UTR, coding, and 3′-UTRs of all S.cerevisiae protein-coding genes for 100 bp windows is shown in Supplementary Figure S4. The consensus pattern is overrepresented in the −100 to ATG region with 2.54 occurrences/kb, compared with 0.78 on the minus strand for this same region, which ranks highest among the regions surveyed.
Figure 5.
The consensus sequence of the TSS The sequence of ±10 bp flanking each TSS was extracted from the S.cerevisiae genomic sequence and analyzed using WebLOGO (http://weblogo.berkeley.edu/) (38,39). Sequence LOGO of TSS flanking sequences derived from (A) all 4936 unitags mapping to the putative 5′-UTR region, (B) 1041 multiple occurrence unitags mapping to the putative 5′-UTR and (C) 3258 unitags mapping to the coding region and putative 3′-UTR (negative control).
New gene prediction
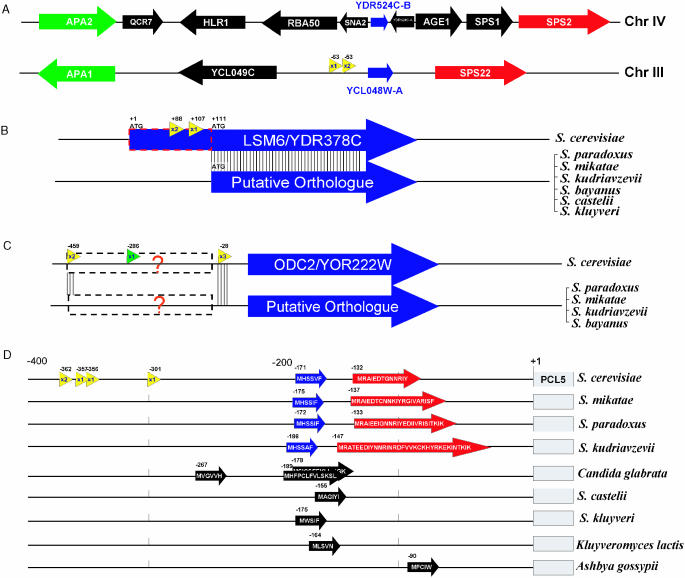
Conventional SAGE has identified previously unidentified genes in S.cerevisiae (40). Similarly, 5′ SAGE is uncovering yet more previously unidentified S.cerevisiae genes. Among 9738 unitags, 684 do not correlate with any known genes by criteria used in this study. Some of these ‘orphan’ unitags probably result from spurious transcripts, but some may correspond to transcripts of unknown protein or RNA coding genes. To identify potential previously unrecognized protein-coding genes, we looked for ORFs ranging in size from 30 to 100 codons in the sequences downstream of each of these 684 orphan unitags. A total of 1184 putative ORFs in this size range were identified. Comparative methods were used to identify evolutionarily conserved ORFs from these candidates. For example, the unitag 5′-AACGGCTAAAACAATT-3′ was mapped to the intergenic region between gene YCL049C and YCL048W of chromosome III. A putative ORF of 79 codons locates 53 bp downstream of 5′ end of the tag, coordinating position 41 488–41 727 on chromosome III. This new ORF has also been predicted by Cliften et al. (3) through multiple species comparison. Comparison of this gene with other hemiascomycete fungi genomic sequences indicates that this ORF is found only among the Saccharomyces sensu stricto. We also found this new ORF has an annotated paralog, YDR524C-B, on chromosome IV, synteny between these two regions being identified by comparison with the gene order in A.gossypii (Figure 6A). This supports this new gene prediction and suggests that this pair of genes arose as a result of the whole genome duplication in the S.cerevisiae lineage (1,41).
Figure 6.
New features predicted in S.cerevisiae insights from the TSS information from 5′ SAGE data combined with comparative genomics methods have versatile usages include: (A) New gene discovery: synteny view of S.cerevisiae chromosomal IV and III regions, which are believed to be duplicated regions resulting from the whole genome duplication. Each orthologous gene pair is shown in the same color. Two 5′ SAGE unitags with total three occurrences (yellow arrow) revealed a new gene, YCL048W-A, which is homologous to YDR524C-B. (B) Determine the real ATG start codon: Two unitags with one having multiple occurrences are mapped to the coding region of LSM6, while no tag is associated to its 5′-UTR. Protein sequence alignment to orthologs from other Saccharomyces species further supports the proposed LSM6 translation start position. (C) Search of putative regulatory RNA element similar to SRG1–SER3. Two multiple occurrence tags upstream of ODC2 coding region are shown (yellow arrow). The phylogenic comparison showed homology around these two TSS position among multiple species. There is also a conventional SAGE tag (green arrow) that maps to this region with position −286. (D) Example of uORF containing gene. Four unitags with one having multiple occurrences were mapped to 300+ bp upstream of PCL5 coding region. Two small uORFs (blue and red) are found and conserved among all four sensu stricto species in terms of position, length and sequences. Five other hemiascomycete species also contain similar putative uORF(s) in that region. In C.glabrata, three uORFs are present, with two overlapping in different reading frames.
Correction of gene annotation
One of the major caveats of genomic sequence based gene annotation is that it is difficult to determine the ATG start codon when there are multiple in-frame ATGs close to the 5′ end. By default, in S.cerevisiae ORFs were annotated starting from the most 5′ ATG. In some cases, this is not the actual start codon (3,42). Using TSS data obtained in this study combined with multi-species comparative analysis, we found 14 cases where it is likely that the incorrect ATG start codon is currently annotated. For each of these 14 genes (Table 3), no UTR TSS tags are present, while multiple occurrences TSS tags are found within the coding region as currently annotated. Alignment of the protein sequence with other recently sequenced fungal species (3,42) shows low sequence conservation in the putative 5′ domain and inconsistent occurrence of start codons analogous to the currently annotated S.cerevisiae start codon. These two lines of evidence suggest that a currently internal ATG codon is a more likely start codon. An example, LSM6/YDR378C, is shown in Figure 6B. However, it is also possible that some of these genes may be similar to HTS1 (43) where multiple transcription starts give rise to proteins with alternative N-terminal ends.
Table 3.
Annotation correction of ORF translational start position
| ORF | Gene | Unitag position (occurrence) | Chr | New ATG position | Predicteda |
|---|---|---|---|---|---|
| YDL208W | NHP2 | 6(1) 28(2) 48(1) | 4 | +51 | Yes |
| YDR378C | LSM6 | 88(2) 107(1) | 4 | +111 | No |
| YER030W | 0(3) 8(1) 12(1) 20(2) 404(1) | 5 | +21 | Yes | |
| YER050C | RSM18 | 133(2) 297(1) 571(1) | 5 | +192 | No |
| YGR088W | CTT1 | 8(2) 377(1) | 7 | +33 | No |
| YHR163W | SOL3 | 91(2) 848(1) | 8 | +93 | Yes |
| YIL043C | CBR1 | 82(1) 94(1) 102(3) | 9 | +114 | Yes |
| YIL053W | RHR2 | 35(5) 36(1) 349(1) 526(1) 852(1) | 9 | +63 | No |
| YIL076W | SEC28 | 177(4) 556(1) | 9 | +189 | No |
| YJL046W | 99(2) | 10 | +126 | No | |
| YKR042W | UTH1 | 48(3) 57(2) 413(1) 859(1) | 11 | +255 | Yes |
| YOR091W | 96(1) 106(2) | 15 | +168 | No | |
| YOR147W | MDM32 | 56(3) | 15 | +93 | No |
| YPR169W | JIP5 | 32(1) 50(1) 55(2) | 16 | +66 | Yes |
Proposed translational start codon changes for 14 genes based on 5′ SAGE TSS data and multiple orthologous sequences alignment. 5′ SAGE TSS position are shown relative to the currently annotated ATG, with unitag occurrence in parenthesis and multiple occurrence unitags are shown in bold. The proposed new downstream ATG start codon position relative to the original start position is listed.
a‘Predicted’ means the annotation change had been previously suggested by multiple Saccharomyces species comparison (42).
Finding a putative non-coding RNA gene
Following the SRG1–SER3 paradigm, we have identified an additional case of a gene with widely separated multiple occurrence tags in the 5′-UTR. This gene, ODC2/YOR222W, with multiple occurrence tags located at positions −459 and −28 is shown in Figure 6C. Alignment of orthologous sequence from related fungal species shows sequences around both TSS are conserved. A conventional SAGE tag 5′-CATGAAATAGTGGT-3′ uniquely mapping to the position −286 (44) further suggests that a small RNA coding transcript is transcribed from the 5′-UTR region of ODC2. Examination of this region shows no evidence of a conserved protein sequence, suggesting that this transcript is not protein coding.
uORF detection
Some S.cerevisiae genes, such as GCN4 and CPA1, have regulatory uORFs in their 5′ region (45). A major obstacle in identifying uORF-containing genes is the lack of TSS information for most S.cerevisiae genes. Examination of the 660 genes having multiple occurrence tags in their putative 5′-UTR region identifies a total of 24 genes that appear to contain at least one uORF in their 5′-UTR leader sequence (Supplementary Table S3). Comparison of these 5′-UTR regions with the corresponding regions of orthologous genes of several related hemiascomycete species (1,3,42,46) revealed that in several cases the uORFs are conserved, with PCL5 having the most conserved uORF pattern. Based on the 5′ SAGE tags, the length of PCL5 mRNA 5′-UTR ranges from 301 to 362 bp with this region containing two uORFs. The uORF1, located at position −171, appears to be present in Saccharomyces paradoxus, Saccharomyces mikatae and Saccharomyces kudriavzevii, and also in several more distantly related species (Figure 6D).
DISCUSSION
We have shown that 5′ SAGE is able to generate accurate TSS data for S.cerevisiae. This method combines the use of TypeIIS restriction enzymes and the ditag strategy from SAGE with the template switching TSS identification of SMART™ RACE (47,48), to determine the TSS. We have used a TS approach to capture the TSS instead of the cap-capture approach, which has been used successfully with human and mouse mRNA (8–10), showing that there are at least two workable approaches to 5′ SAGE. The accuracy of this method derives from the ditag strategy in 5′ SAGE that, as in SAGE, is essential to insure that each tag originates from a different mRNA and is not an artifact of PCR amplification. This confidence that multiple occurrence tags are independent is essential as we depend on multiplicity of occurrences to validate the individual unitag sites.
Carrying out 5′ SAGE on S.cerevisiae we have found that the TSS identified by 5′ SAGE agrees well with previously reported data, with significant disagreement for only 7 out of 48 genes. The seven genes (PIM1, HEM1, URA1, URA4, HSC82, ADH2 and TPI1) for which there is significant disagreement as to the location of the TSS need to be more closely investigated to identify the source of the discrepancy. For TPI1, primer extension results support the −30 position of the main TSS detected by 5′ SAGE, suggesting that the previous report (32) is erroneous. In some cases, the 5′ SAGE TSS data differed from published primer extension data by 1–5 bases. This 1–5 base discrepancy may result from the difficulties associated with obtaining single base pair resolution with either primer extension or nuclease protection assays. An example of this is TEF2, where our 5′ SAGE and primer extension results identify the 5′-UTR as being 1 nt shorter than the previously published (31). In some cases, including TDH3, GLN1, IMD2, RPS17A, GCN4, TEF2, ARO4 and HEM3, a small number of tags are upstream of the majority of the 5′ SAGE tags and the previously published TSS. It is unclear whether these tags are artifacts, represent rare transcripts from an upstream TSS, or represent regulatory RNAs similar to SRG1. For TEF2, TDH3 and GCN4, we have verified by primer extension the occurrence of these upstream TSS.
Our estimation of the accuracy of this method is also supported by the consistency of the TSS identified by 5′ SAGE with the previously reported TSS consensus sequence. Based on the frequency of tags falling outside of the 5′-UTR and the frequency of potentially spurious single occurrence tags within the 5′-UTR, we estimate that 54–70% of all 5′ SAGE TSS tags represent actual TSS. While 30–46% of 5′ SAGE tags do not represent true start sites and likely result from premature termination of reverse transcription or degraded mRNA, these false tags cause little problem as our basis for identifying TSS relies on multiple tag occurrence. Determination of the TSS for a specific gene by 5′ SAGE thus requires sequencing of the 5′ SAGE library to sufficient depth so that multiple TSS are represented for each gene. Approximately 10% of the tags had multiple positions in the genome, consistent with the fraction of repetitive sequences in this genome, and their transcription level.
The refined consensus sequence we have identified for S.cerevisiae differs significantly from that of human. The larger TSS consensus sequence in S.cerevisiae combined with the variable distance of TSS from the TATA element (49) has been previously suggested to indicate that the mechanism of transcription start in S.cerevisiae is significantly different than in human (50). It has been suggested that instead of assembly of the RNA polymerase II complex at the TATA element coupled to transcription initiation at an adjacent site (51), RNA polymerase II in S.cerevisiae may utilize a scanning model to identify the TSS (49). Supporting the idea that RNA polymerase may scan from the TATA to the TSS in S.cerevisiae is the observation from Giardina and Lis (52) that for the GAL1 and GAL10 promoters, the region of transcription-associated promoter melting is located adjacent to the TATA element as has been shown for mammalian TATA elements. In Schizosaccharomyces pombe, the TSS has been reported to be 25–40 bp from the TATA element (53), suggesting that having the transcription start adjacent to the site of polymerase assembly may be the more typical eukaryotic mode of transcription initiation, and that S.cerevisiae has an unusual mode of transcription initiation. The TSS consensus sequence found in this study is highly A-rich. As the S.cerevisiae genome is overall >60% AT, and the region of the TSS is >37% A, interpretation of the role of the A-rich regions in this consensus sequence is unclear, whether the poly(A) stretches surrounding the TSS are important in transcription initiation, or whether they are an artifact of some other aspect of genome structure. While some mutagenesis work on the TSS has been carried out in S.cerevisiae (54,55), further mutagenesis work in S.cerevisiae and TSS identification in one of the closely related but GC-rich hemiascomycetes, such as A.gossypii (1) will be necessary to refine our understanding of this consensus sequence. The identity of the TSS consensus sequence from TATA-containing genes and TATA-less genes suggests that the mechanism by which the TSS is selected in S.cerevisiae is independent of the presence of a TATA element. This is in contrast to transcription initiation in human, where, based on a small number of genes, it has been shown that for genes lacking a TATA element a 19 bp window including the TSS, the Inr element, is necessary and sufficient for transcription initiation (16).
We have identified multiple tag TSS for 660 protein-coding genes in this study, representing ∼11% of the ∼5800 protein-coding genes in S.cerevisiae (56). Consistent with the previously published results, 5′ SAGE data indicate that the majority of S.cerevisiae genes have multiple TSS. We have refined the TSS consensus sequence, refined the TSS to ATG and TSS to TATA average distance measurements, identified the location of a potential regulatory RNA upstream of ODC2 and identified 24 genes with potential uORFs in their 5′-UTR. We have also identified a previously overlooked protein-coding gene and have provided evidence, suggesting the starting methionine for 14 S.cerevisiae genes differs from that currently annotated. Thus, 5′ SAGE, in identifying the TSS of both protein-coding genes and RNA polymerase II transcribed RNA coding genes, provides data useful to understanding gene regulation as well as providing data refining the annotation of S.cerevisiae.
A variation on 5′ SAGE we are currently using involves replacing the oligo(dT) primer used in the reverse transcription step with a pool of 480 oligonucleotides selected from the minus strand at approximately +100 relative to the starting coding. These primers are selected to be unique in the genome, have similar melting temperatures and GC content. Initial experiments have revealed that it is also necessary to select the pool of primers to minimize primer–primer complementarity as the MMLV reverse transcriptase is able to extend complementary primers to create false tags. Using pools of primers, a method we refer to as 5′ SAGE II, we are currently generating and evaluating libraries. Preliminary results include the identification of the TSS of PFY1, the gene coding profiling, at −41, consistent with the previously published value of −41 as major TSS (57). Further refinements of these methods will allow the identification of the TSS of the set of transcribed S.cerevisiae genes, not just the most highly expressed genes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online. Raw data for this project is available at http://data.cgt.duke.edu/5sage.php.
Supplementary Material
Acknowledgments
The authors thank Andria Allen for help sequencing the 5′ SAGE library. The authors thank Phillipe Luedi, Shihua Lu, Rui Yi for valuable technical help and advice and Joe Heitman for providing yeast strains. The authors are grateful to Bryan Cullen, Joe Heitman and Douglas Marchuk for generously providing laboratory facilities and comments on the whole project. The authors also thank Mark DeLong for his careful reading and revising of the manuscript. The authors also want to thank Mike Cherry and the staff at SGD for useful discussion of this work, and for planning to add TSS data to SGD. Funding to pay the Open Access publication charges for this article was provided by discretionary funds of the corresponding author.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dietrich F.S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pohlmann R., Luedi P., Choi S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 2.Kellis M., Birren B.W., Lander E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 3.Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 4.Sherman D., Durrens P., Beyne E., Nikolski M., Souciet J.L. Genolevures: comparative genomics and molecular evolution of hemiascomycetous yeasts. Nucleic Acids Res. 2004;32:D315–D318. doi: 10.1093/nar/gkh091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z., Dietrich F.S. Verification of a new gene on Saccharomyces cerevisiae chromosome III. Yeast. 2003;20:731–738. doi: 10.1002/yea.996. [DOI] [PubMed] [Google Scholar]
- 6.Brachat S., Dietrich F.S., Voegeli S., Zhang Z., Stuart L., Lerch A., Gates K., Gaffney T., Philippsen P. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 2003;4:R45. doi: 10.1186/gb-2003-4-7-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannenhalli S., Levy S. Promoter prediction in the human genome. Bioinformatics. 2001;17(Suppl. 1):S90–S96. doi: 10.1093/bioinformatics/17.suppl_1.s90. [DOI] [PubMed] [Google Scholar]
- 8.Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei C.L., Ng P., Chiu K.P., Wong C.H., Ang C.C., Lipovich L., Liu E.T., Ruan Y. 5′ Long serial analysis of gene expression (LongSAGE) and 3′ LongSAGE for transcriptome characterization and genome annotation. Proc. Natl Acad. Sci. USA. 2004;101:11701–11706. doi: 10.1073/pnas.0403514101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimoto S., Suzuki Y., Kasai Y., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K. 5′-end SAGE for the analysis of transcriptional start sites. Nat. Biotechnol. 2004;22:1146–1149. doi: 10.1038/nbt998. [DOI] [PubMed] [Google Scholar]
- 11.Hurowitz E.H., Brown P.O. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 2003;5:R2. doi: 10.1186/gb-2003-5-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pesole G., Liuni S., Grillo G., Licciulli F., Mignone F., Gissi C., Saccone C. UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res. 2002;30:335–340. doi: 10.1093/nar/30.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buratowski S., Hahn S., Guarente L., Sharp P.A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 14.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smale S.T., Baltimore D. The ‘initiator’ as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 16.Weis L., Reinberg D. Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes. FASEB J. 1992;6:3300–3309. doi: 10.1096/fasebj.6.14.1426767. [DOI] [PubMed] [Google Scholar]
- 17.Weis L., Reinberg D. Accurate positioning of RNA polymerase II on a natural TATA-less promoter is independent of TATA-binding-protein-associated factors and initiator-binding proteins. Mol. Cell. Biol. 1997;17:2973–2984. doi: 10.1128/mcb.17.6.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 19.Basehoar A.D., Zanton S.J., Pugh B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 20.Cherry J.M., Ball C., Weng S., Juvik G., Schmidt R., Adler C., Dunn B., Dwight S., Riles L., Mortimer R.K., et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- 21.Burke D., Dawson D., Stearns T. Methods in Yeast Genetics; A Cold Spring Harbor Laboratory Course manual. Cold Spring Harbor, NY: CSHL Press; 2000. [Google Scholar]
- 22.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 23.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 24.Holstege F.C., Jennings E.G., Wyrick J.J., Lee T.I., Hengartner C.J., Green M.R., Golub T.R., Lander E.S., Young R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 25.Dance G.S., Beemiller P., Yang Y., Mater D.V., Mian I.S., Smith H.C. Identification of the yeast cytidine deaminase CDD1 as an orphan C → U RNA editase. Nucleic Acids Res. 2001;29:1772–1780. doi: 10.1093/nar/29.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickens M.P., Buell G.N., Schimke R.T. Synthesis of double-stranded DNA complementary to lysozyme, ovomucoid, and ovalbumin mRNAs. Optimization for full length second strand synthesis by Escherichia coli DNA polymerase I. J Biol. Chem. 1978;253:2483–2495. [PubMed] [Google Scholar]
- 27.Berk A.J., Sharp P.A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977;12:721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- 28.Karin M., Najarian R., Haslinger A., Valenzuela P., Welch J., Fogel S. Primary structure and transcription of an amplified genetic locus: the CUP1 locus of yeast. Proc. Natl Acad. Sci. USA. 1984;81:337–341. doi: 10.1073/pnas.81.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao X.B., Clare J.J., Farabaugh P.J. The upstream activation site of a Ty2 element of yeast is necessary but not sufficient to promote maximal transcription of the element. Proc. Natl Acad. Sci. USA. 1987;84:8520–8524. doi: 10.1073/pnas.84.23.8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fulton A.M., Rathjen P.D., Kingsman S.M., Kingsman A.J. Upstream and downstream transcriptional control signals in the yeast retrotransposon, TY. Nucleic Acids Res. 1988;16:5439–5458. doi: 10.1093/nar/16.12.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagashima K., Kasai M., Nagata S., Kaziro Y. Structure of the two genes coding for polypeptide chain elongation factor 1 alpha (EF-1 alpha) from Saccharomyces cerevisiae. Gene. 1986;45:265–273. doi: 10.1016/0378-1119(86)90024-7. [DOI] [PubMed] [Google Scholar]
- 32.Alber T., Kawasaki G. Nucleotide sequence of the triose phosphate isomerase gene of Saccharomyces cerevisiae. J. Mol. Appl. Genet. 1982;1:419–434. [PubMed] [Google Scholar]
- 33.Strathern J.N., Jones E.W., Broach J.R. Cold Spring Harbor Monograph Series; 11B. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. The molecular biology of the yeast Sacchromyces: metabolism and gene expression. pp. x, 680 p. [Google Scholar]
- 34.Morlando M., Greco P., Dichtl B., Fatica A., Keller W., Bozzoni I. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol. 2002;22:1379–1389. doi: 10.1128/mcb.22.5.1379-1389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martens J.A., Laprade L., Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 36.Morey C., Avner P. Employment opportunities for non-coding RNAs. FEBS Lett. 2004;567:27–34. doi: 10.1016/j.febslet.2004.03.117. [DOI] [PubMed] [Google Scholar]
- 37.Hahn S., Hoar E.T., Guarente L. Each of three ‘TATA elements’ specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1985;82:8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oshiro G., Wodicka L.M., Washburn M.P., Yates J.R., III, Lockhart D.J., Winzeler E.A. Parallel identification of new genes in Saccharomyces cerevisiae. Genome Res. 2002;12:1210–1220. doi: 10.1101/gr.226802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolfe K.H., Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 42.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 43.Natsoulis G., Hilger F., Fink G.R. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of Saccaromyces cerevisiae. Cell. 1986;46:235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- 44.Velculescu V.E., Zhang L., Zhou W., Vogelstein J., Basrai M.A., Bassett D.E., Jr, Hieter P., Vogelstein B., Kinzler K.W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 45.Vilela C., McCarthy J.E. Regulation of fungal gene expression via short open reading frames in the mRNA 5′ untranslated region. Mol. Microbiol. 2003;49:859–867. doi: 10.1046/j.1365-2958.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 46.Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., De Montigny J., Marck C., Neuveglise C., Talla E., et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 47.Chenchik A., Zhu Y.Y., Diatchenko L., Li R., Jill J., Siebert P.D. RT–PCR Methods for Gene Cloning and Analysis. MA, USA: Eaton, Natick; 1998. [Google Scholar]
- 48.Zhu Y.Y., Machleder E.M., Chenchik A., Li R., Siebert P.D. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 49.Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987;49:295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- 50.Russell P.R. Evolutionary divergence of the mRNA transcription initiation mechanism in yeast. Nature. 1983;301:167–169. doi: 10.1038/301167a0. [DOI] [PubMed] [Google Scholar]
- 51.Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980;209:1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- 52.Giardina C., Lis J.T. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- 53.Choi W.S., Yan M., Nusinow D., Gralla J.D. In vitro transcription and start site selection in Schizosaccharomyces pombe. J. Mol. Biol. 2002;319:1005–1013. doi: 10.1016/S0022-2836(02)00329-7. [DOI] [PubMed] [Google Scholar]
- 54.Healy A.M., Helser T.L., Zitomer R.S. Sequences required for transcriptional initiation of the Saccharomyces cerevisiae CYC7 genes. Mol. Cell. Biol. 1987;7:3785–3791. doi: 10.1128/mcb.7.10.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Healy A.M., Zitomer R.S. A sequence that directs transcriptional initiation in yeast. Curr. Genet. 1990;18:105–109. doi: 10.1007/BF00312597. [DOI] [PubMed] [Google Scholar]
- 56.Wood V., Rutherford K.M., Ivens A., Rajandream M.A., Barrell B. A Re-annotation of the Saccharomyces cerevisiae Genome. Comp. Funct. Genomics. 2001;2:143–154. doi: 10.1002/cfg.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Magdolen V., Oechsner U., Muller G., Bandlow W. The intron-containing gene for yeast profilin (PFY) encodes a vital function. Mol. Cell. Biol. 1988;8:5108–5115. doi: 10.1128/mcb.8.12.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagawa F., Fink G.R. The relationship between the ‘TATA’ sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1985;82:8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van den Heuvel J.J., Bergkamp R.J., Planta R.J., Raue H.A. Effect of deletions in the 5′-noncoding region on the translational efficiency of phosphoglycerate kinase mRNA in yeast. Gene. 1989;79:83–95. doi: 10.1016/0378-1119(89)90094-2. [DOI] [PubMed] [Google Scholar]
- 60.Wright R.M., Dircks L.K., Poyton R.O. Characterization of COX9, the nuclear gene encoding the yeast mitochondrial protein cytochrome c oxidase subunit VIIa. Subunit VIIa lacks a leader peptide and is an essential component of the holoenzyme. J. Biol. Chem. 1986;261:17183–17191. [PubMed] [Google Scholar]
- 61.Newton C.H., Shimmin L.C., Yee J., Dennis P.P. A family of genes encode the multiple forms of the Saccharomyces cerevisiae ribosomal proteins equivalent to the Escherichia coli L12 protein and a single form of the L10-equivalent ribosomal protein. J. Bacteriol. 1990;172:579–588. doi: 10.1128/jb.172.2.579-588.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paravicini G., Braus G., Hutter R. Structure of the ARO3 gene of Saccharomyces cerevisiae. Mol. Gen. Genet. 1988;214:165–169. doi: 10.1007/BF00340197. [DOI] [PubMed] [Google Scholar]
- 63.Dabeva M.D., Warner J.R. The yeast ribosomal protein L32 and its gene. J. Biol. Chem. 1987;262:16055–16059. [PubMed] [Google Scholar]
- 64.Escobar-Henriques M., Daignan-Fornier B., Collart M.A. The critical cis-acting element required for IMD2 feedback regulation by GDP is a TATA box located 202 nucleotides upstream of the transcription start site. Mol. Cell. Biol. 2003;23:6267–6278. doi: 10.1128/MCB.23.17.6267-6278.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huet J., Cottrelle P., Cool M., Vignais M.L., Thiele D., Marck C., Buhler J.M., Sentenac A., Fromageot P. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J. 1985;4:3539–3547. doi: 10.1002/j.1460-2075.1985.tb04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ogawa N., Saitoh H., Miura K., Magbanua J.P., Bun-ya M., Harashima S., Oshima Y. Structure and distribution of specific cis-elements for transcriptional regulation of PHO84 in Saccharomyces cerevisiae. Mol. Gen. Genet. 1995;249:406–416. doi: 10.1007/BF00287102. [DOI] [PubMed] [Google Scholar]
- 67.Cumsky M.G., Trueblood C.E., Ko C., Poyton R.O. Structural analysis of two genes encoding divergent forms of yeast cytochrome c oxidase subunit V. Mol. Cell. Biol. 1987;7:3511–3519. doi: 10.1128/mcb.7.10.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crabeel M., de Rijcke M., Seneca S., Heimberg H., Pfeiffer I., Matisova A. Further definition of the sequence and position requirements of the arginine control element that mediates repression and induction by arginine in Saccharomyces cerevisiae. Yeast. 1995;11:1367–1380. doi: 10.1002/yea.320111405. [DOI] [PubMed] [Google Scholar]
- 69.Faitar S.L., Brodie S.A., Ponticelli A.S. Promoter-specific shifts in transcription initiation conferred by yeast TFIIB mutations are determined by the sequence in the immediate vicinity of the start sites. Mol. Cell. Biol. 2001;21:4427–4440. doi: 10.1128/MCB.21.14.4427-4440.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Russell D.W., Smith M., Williamson V.M., Young E.T. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J. Biol. Chem. 1983;258:2674–2682. [PubMed] [Google Scholar]
- 71.Schneider J.C., Guarente L. Regulation of the yeast CYT1 gene encoding cytochrome c1 by HAP1 and HAP2/3/4. Mol. Cell. Biol. 1991;11:4934–4942. doi: 10.1128/mcb.11.10.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Minehart P.L., Magasanik B. Sequence of the GLN1 gene of Saccharomyces cerevisiae: role of the upstream region in regulation of glutamine synthetase expression. J. Bacteriol. 1992;174:1828–1836. doi: 10.1128/jb.174.6.1828-1836.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Freeman K.B., Karns L.R., Lutz K.A., Smith M.M. Histone H3 transcription in Saccharomyces cerevisiae is controlled by multiple cell cycle activation sites and a constitutive negative regulatory element. Mol. Cell. Biol. 1992;12:5455–5463. doi: 10.1128/mcb.12.12.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kunzler M., Paravicini G., Egli C.M., Irniger S., Braus G.H. Cloning, primary structure and regulation of the ARO4 gene, encoding the tyrosine-inhibited 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Saccharomyces cerevisiae. Gene. 1992;113:67–74. doi: 10.1016/0378-1119(92)90670-k. [DOI] [PubMed] [Google Scholar]
- 75.Zagorec M., Buhler J.M., Treich I., Keng T., Guarente L., Labbe-Bois R. Isolation, sequence, and regulation by oxygen of the yeast HEM13 gene coding for coproporphyrinogen oxidase. J. Biol. Chem. 1988;263:9718–9724. [PubMed] [Google Scholar]
- 76.Schneider J.C., Guarente L. The untranslated leader of nuclear COX4 gene of Saccharomyces cerevisiae contains an intron. Nucleic Acids Res. 1987;15:3515–3529. doi: 10.1093/nar/15.8.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pavlovic B., Horz W. The chromatin structure at the promoter of a glyceraldehyde phosphate dehydrogenase gene from Saccharomyces cerevisiae reflects its functional state. Mol. Cell. Biol. 1988;8:5513–5520. doi: 10.1128/mcb.8.12.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brindle P.K., Holland J.P., Willett C.E., Innis M.A., Holland M.J. Multiple factors bind the upstream activation sites of the yeast enolase genes ENO1 and ENO2: ABFI protein, like repressor activator protein RAP1, binds cis-acting sequences which modulate repression or activation of transcription. Mol. Cell. Biol. 1990;10:4872–4885. doi: 10.1128/mcb.10.9.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beacham I.R., Schweitzer B.W., Warrick H.M., Carbon J. The nucleotide sequence of the yeast ARG4 gene. Gene. 1984;29:271–279. doi: 10.1016/0378-1119(84)90056-8. [DOI] [PubMed] [Google Scholar]
- 80.Repetto B., Tzagoloff A. Structure and regulation of KGD1, the structural gene for yeast alpha-ketoglutarate dehydrogenase. Mol. Cell. Biol. 1989;9:2695–2705. doi: 10.1128/mcb.9.6.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sarokin L., Carlson M. Comparison of two yeast invertase genes: conservation of the upstream regulatory region. Nucleic Acids Res. 1985;13:6089–6103. doi: 10.1093/nar/13.17.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dohrmann P.R., Voth W.P., Stillman D.J. Role of negative regulation in promoter specificity of the homologous transcriptional activators Ace2p and Swi5p. Mol. Cell. Biol. 1996;16:1746–1758. doi: 10.1128/mcb.16.4.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nouraini S., Hu J., McBroom L.D., Friesen J.D. Mutations in an Abf1p binding site in the promoter of yeast RPO26 shift the transcription start sites and reduce the level of RPO26 mRNA. Yeast. 1996;12:1339–1350. doi: 10.1002/(SICI)1097-0061(199610)12:13%3C1339::AID-YEA31%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 84.Hahn S., Pinkham J., Wei R., Miller R., Guarente L. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol. Cell. Biol. 1988;8:655–663. doi: 10.1128/mcb.8.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Urban-Grimal D., Volland C., Garnier T., Dehoux P., Labbe-Bois R. The nucleotide sequence of the HEM1 gene and evidence for a precursor form of the mitochondrial 5-aminolevulinate synthase in Saccharomyces cerevisiae. Eur. J. Biochem. 1986;156:511–519. doi: 10.1111/j.1432-1033.1986.tb09610.x. [DOI] [PubMed] [Google Scholar]
- 86.Losson R., Fuchs R.P., Lacroute F. Yeast promoters URA1 and URA3. Examples of positive control. J. Mol. Biol. 1985;185:65–81. doi: 10.1016/0022-2836(85)90183-4. [DOI] [PubMed] [Google Scholar]
- 87.Kim G.J., Park J.H., Lee D.C., Ro H.S., Kim H.S. Primary structure, sequence analysis, and expression of the thermostable d-hydantoinase from Bacillus stearothermophilus SD1. Mol. Gen. Genet. 1997;255:152–156. doi: 10.1007/pl00008610. [DOI] [PubMed] [Google Scholar]
- 88.Erkine A.M., Adams C.C., Gao M., Gross D.S. Multiple protein–DNA interactions over the yeast HSC82 heat shock gene promoter. Nucleic Acids Res. 1995;23:1822–1829. doi: 10.1093/nar/23.10.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.