Abstract
The RNA-dependent RNA polymerase (RdRp) from hepatitis C virus (HCV), nonstructural protein 5B (NS5B), has recently been shown to direct de novo initiation using a number of complex RNA templates. In this study, we analyzed the features in simple RNA templates that are required to direct de novo initiation of RNA synthesis by HCV NS5B. NS5B was found to protect RNA fragments of 8 to 10 nucleotides (nt) from RNase digestion. However, NS5B could not direct RNA synthesis unless the template contained a stable secondary structure and a single-stranded sequence that contained at least one 3′ cytidylate. The structure of a 25-nt template, named SLD3, was determined by nuclear magnetic resonance spectroscopy to contain an 8-bp stem and a 6-nt single-stranded sequence. Systematic analysis of changes in SLD3 revealed which features in the stem, loop, and 3′ single-stranded sequence were required for efficient RNA synthesis. Also, chimeric molecules composed of DNA and RNA demonstrated that a DNA molecule containing a 3′-terminal ribocytidylate was able to direct RNA synthesis as efficiently as a sequence composed entirely of RNA. These results define the template sequence and structure sufficient to direct the de novo initiation of RNA synthesis by HCV RdRp.
Hepatitis C virus (HCV), a plus-strand RNA virus, is estimated to infect up to 3% of the world's population (44), causing liver cirrhosis and hepatocellular carcinoma (14). Following entry into the infected cell, the viral RNA directs the translation of a polyprotein that is proteolytically processed to produce 10 individual structural and nonstructural proteins (15, 32). Nonstructural protein 5B (NS5B) is at the C terminus of the polyprotein. NS5B is an RNA-dependent RNA polymerase (RdRp). Based on the paradigms of other RNA virus replication strategies (8), NS5B, along with viral and cellular proteins, forms a replicase that replicates the HCV genome. At present, functional HCV replicase has not been demonstrated in vitro. Therefore, studies of HCV RNA synthesis have focused on recombinant NS5B.
Recombinant HCV NS5B can catalyze a number of reactions. In the presence of a primer-template duplex, NS5B catalyzes template-dependent but relatively nonspecific RNA synthesis (5, 23–25, 45, 46). In addition, NS5B has recently been reported to direct de novo (oligonucleotide primer-independent) synthesis (26, 30, 47), a mechanism used for the replication of many plus-strand RNA viruses (8). De novo initiation of RNA synthesis may be especially relevant for HCV since, to our knowledge, it does not contain a VPg-like protein that could mediate protein-primed RNA synthesis, and there is no evidence for a cap-snatching mechanism (32). De novo RNA synthesis directed by HCV NS5B prefers a cytidylate template and the substrate nucleotide GTP (26, 42), although ATP can also be used as an initiation nucleotide (29, 42, 47). In general, RNA polymerases have a higher Km for the initiation nucleotide than for the same nucleotide during elongating RNA synthesis (for examples, see references 18, 26, 31, and 42).
The features of the template that direct RdRp binding and the initiation of HCV RNA synthesis remain poorly characterized. Several templates tested were unable to efficiently direct de novo RNA synthesis (30; D. Barket and B. Heinz, unpublished results; C. C. Kao, unpublished results). These results indicate that recombinant NS5B has some specific template requirements for de novo initiation, even in the absence of the other replicase components. The goal of this work was to determine the template requirements for efficient RNA synthesis. For the sake of simplicity, this work addresses only the role of cytidylate(s) as the template initiation nucleotide. A 25-nucleotide (nt) RNA, termed SLD3, was found to be capable of supporting efficient RNA synthesis. The secondary structure of SLD3 in solution was solved by one- and two-dimensional nuclear magnetic resonance (NMR) spectroscopy, and the features of SLD3 were systematically analyzed for the ability to direct RNA synthesis.
MATERIALS AND METHODS
RNA synthesis and purification.
Transcription reactions were carried out under the conditions described by Milligan et al. (27). Briefly, the DNA strands were purified via denaturing polyacrylamide gel electrophoresis and then adjusted to 8 μM. One microliter of each DNA was used in a 20-μl transcription reaction mixture containing final concentrations of 40 mM Tris (pH 8.1), 1 mM spermidine, 0.01% Triton X-100, 80 mg of polyethylene glycol 8000, and 4 mM each nucleoside triphosphate. The T7 RNA polymerase used was purified by the protocol of Grodberg and Dunn (12). RNAs of the correct length were purified by preparative denaturing gel electrophoresis and excised from the gel after UV shadowing. The gel slice was crushed and ground to small pieces, and the RNA was eluted from the polyacrylamide with 0.4 M ammonium acetate. Following precipitation with ethanol, the RNA concentration was determined by spectrophotometry and checked by toluidine blue staining on an analytical gel. Transcripts of SLD3 used for NMR spectroscopy were from a 40-ml transcription reaction. Chemically synthesized RNAs were purchased from Dharmacon (Boulder, Colo.), deprotected according to the supplier's instructions, and purified by denaturing gel electrophoresis as described above.
RdRp activity assay and product analysis.
Full-length recombinant HCV NS5B of genotype 1b was prepared from Escherichia coli as described previously (17, 42). The standard assay, described by Adkins et al. (1), consisted of a 40-μl reaction mixture containing 1 pmol of template (unless stated otherwise), 70 nmol of NS5B, 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12.5 mM dithiothreitol, 0.5% (vol/vol) Triton X-100, 1 mM MnCl2, 200 μM each ATP and UTP, 500 μM GTP, and 250 nM [α-32P]CTP (Amersham Inc.). Manganese was used to increase the level of RNA synthesis but was not required for RNA synthesis (10, 19) and does not alter the choice for the initiation site in the RNAs that we used (C. C. Kao, unpublished results). Reaction mixtures were incubated at 25°C for 60 min, extracted with phenol-chloroform, and ethanol precipitated in the presence of 5 μg of glycogen and 0.4 M ammonium acetate. Products were separated by electrophoresis on 10 to 20% denaturing (8 M urea) polyacrylamide gels. Gels were wrapped in plastic and exposed to film at −60°C. Product bands were quantified using a PhosphorImager (Molecular Dynamics). The results presented have been reproduced in at least three independent assays, all of which varied by less than 20%.
NMR spectroscopy.
RNA SLD3 used for NMR spectroscopy was dialyzed against NMR buffer (10 mM sodium phosphate, 0.1 mM sodium EDTA, and 100 mM sodium chloride in 90% H2O–10% D2O [pH 6.4]) for 48 h, with two buffer changes. The final NMR sample of 0.5 ml was 1.2 mM RNA.
All NMR experiments were recorded on a Bruker Advance 600-MHz spectrometer equipped with a triple-resonance, triple-axis gradient probe. Proton chemical shifts were compared to the residual water resonance (4.70 ppm at 21°C). NMR data were processed using the FELIX 98 program (Molecular Simulations, Inc.). Solvent suppression was achieved using the combination of a water flip-back selective pulse with a WATERGATE sequence (22). Radiation damping was reduced with a gradient pulse during the first time domain (39). The excitation maximum was set to the middle of the imino proton shift range (13.0 ppm). Nuclear Overhauser effect spectroscopy (NOESY) spectra in H2O were collected with a 2.0-s relaxation delay, a 13.0-kHz spectral window, and a data size of 4,096 × 512 at 21°C.
RNase protection assay.
RNA SLC1, used to obtain a preliminary assessment of the number of nucleotides protected by NS5B, was radiolabeled during transcription by T7 RNA polymerase. One picomole of RNA was then incubated with 1 to 20 ng of highly purified NS5B at 15°C for 15 min prior to the addition of 1 μl of 0.2-μg/μl RNase A for 5 min. Following extraction with a 1:1 mixture of phenol and chloroform, the RNA was precipitated with 8 volumes of ethanol, dried, and electrophoresed on a denaturing 20% polyacrylamide gel.
RESULTS
HCV NS5B can interact with RNAs that are not templates for RNA synthesis.
We sought to identify a template that can efficiently direct de novo RNA synthesis by HCV NS5B. However, our previous experience was that the 3′-terminal 25 or 60 nts of HCV genomic RNA cannot direct RNA synthesis (Barket and Heinz, unpublished; Kao, unpublished). A 35-nt stem-loop that contains the core promoter for brome mosaic virus (BMV) minus-strand RNA synthesis was also unable to direct RNA synthesis by the HCV RdRp (20). SLC1 has two 3′ cytidylates that can potentially serve as initiation nucleotides (Fig. 1A). The failure to direct synthesis may be due to an inability to bind NS5B and/or to direct the initiation of RNA synthesis. To distinguish these two possibilities, we attempted to determine whether NS5B is able to interact with radiolabeled SLC1 by protecting it from RNase A digestion in an assay similar to the one used for the poliovirus polymerase 3D (4). In the absence of NS5B, SLC1 was rapidly degraded by RNase A to oligonucleotides of 5 nts or shorter. However, in the presence of even 1 ng of NS5B, two prominent bands of ∼8 to 10 nts were observed (Fig. 1B, lanes 4 and 5). These fragments became more abundant with increasing amounts of NS5B, while the shorter oligonucleotides became less abundant (Fig. 1B, lanes 6 to 13). While these results do not identify the sequences recognized by the HCV NS5B, they demonstrate that NS5B can interact with an RNA that cannot direct RNA synthesis. Some features absent in SLC1 must be necessary to direct RNA synthesis by NS5B.
FIG. 1.
HCV NS5B can protect RNA from digestion by RNase A. (A) Sequence and secondary structure of SLC1 (20). (B) Autoradiogram of a 20% denaturing gel containing products protected from RNase A digestion by NS5B. SLC1, a 35-nt RNA (derived from nts 2039 to 2069 of plus-strand BMV RNA3), was labeled during transcription with 32P-CMP. The RNA products of the BMV replicase from proscript −20/13 are of 13 and 14 nts (34), and their positions are indicated to the left of the autoradiogram. RNase A (1 μl of a 0.2-μg/μl solution) and HCV NS5B additions are indicated above the autoradiogram.
Templates that can direct RNA synthesis by NS5B.
To elucidate the features necessary to direct RNA synthesis, several RNAs were tested for their ability to generate radiolabeled products. The RNAs selected had been previously shown to direct de novo RNA synthesis by the bovine viral diarrhea virus (BVDV) RdRp and the replicases of BMV and cucumber mosaic virus (19, 20, 34, 37, 38). All the RNAs possess a cytidylate at or near the 3′ terminus that could act as a potential initiation site for RNA synthesis (Fig. 2A). RNAs BV-21 and BV-22 were derived from the minus-strand 3′ terminus of BVDV RNA and were able to direct de novo initiation by the BVDV RdRp (Fig. 2A) (19). With HCV NS5B, neither BV-21 nor BV-22 could direct RNA synthesis (Fig. 2B, lanes 4 and 5). Similar results were obtained with the 3′-terminal 25 and 60 nts of HCV genomic RNA and with RNA −20/13, which efficiently directed the proper initiation of subgenomic RNA synthesis by the BMV RdRp (Fig. 2, lanes 2, 3, and 6). In contrast, RNAs B2(−)26G, C2(−)29G, SLC+8, and SLdel+8, derived from BMV and cucumber mosaic virus, were competent for RNA synthesis by HCV NS5B (Fig. 2B, lanes 7 to 10). Product synthesis required the presence of all 4 nts and was insensitive to the presence of rifampin and actinomycin D, both inhibitors of DNA-dependent RNA polymerases (Kao, unpublished), indicating that the products observed were generated by template-directed polymerization by NS5B.
FIG. 2.
RNAs tested for the ability to direct synthesis by NS5B. (A) Sequences and putative structures of several RNAs that were tested for the ability to direct RNA synthesis by HCV NS5B. The lengths of each RNA in nucleotides (nt) are shown in parentheses. The secondary structure of B2(−)26G was determined by NMR spectroscopy as described by Sivakumaran et al. (37). Other secondary structures were predicted by mfold. C2(−)29G and B2(−)26G are minus-strand RNAs that direct the initiation of genomic RNA2 synthesis by the BMV and cucumber mosaic virus replicases (37). SLC+8 and SLdel+8 are genomic plus-strand RNAs that can direct BMV minus-strand initiation (20). Cytidylates at or near the 3′ end that could potentially act as the initiation sites are in bold. (B) Autoradiograms of denaturing gels containing the products of synthesis by NS5B. Reactions in lanes 1 to 10 were from a 10% polyacrylamide gel, while reactions in lanes 11 to 13 were electrophoresed in a 12.5% polyacrylamide gel. Positions of monomeric RdRp products are indicated by white asterisks to the left of the bands. Φ above lane 1 denotes a reaction performed without any exogenous template. The names of the templates used in each reaction are shown at the top of each autoradiogram. The size of one of the products that comigrated with the 46-nt product made by the BMV replicase is labeled between the two autoradiograms. RNA synthesized from SLD3 comigrated with a preparation of SLD3 RNA that was end labeled using polynucleotide kinase. Also, an RNA ladder containing bands that are multiples of 12 nts and products of the BMV replicase were used as molecular size markers to determine the lengths of the HCV RdRp products (Kao, unpublished). Reactions in lanes 11 to 13 all contained RNA SLC+8 as the template but contained different polymerases. Φ, B, and H, reactions without any polymerase, with the BMV replicase, and with HCV NS5B, respectively.
HCV NS5B synthesized a complex array of products from these functional templates (Fig. 2B). This result is consistent with previous observations that the recombinant RdRps of BVDV and poliovirus can undergo template switch events that result in products that are multimeric relative to the length of the template (3, 19). A major product was approximately the length expected from de novo initiation at one of the 3′-most cytidylates and correct termination at or near the 5′ end of the template RNA (henceforth called the monomer). The BMV replicase, which tends to initiate from the penultimate cytidylate and terminate at the end of the template, was used to generate a 46-nt product that could be compared with the products of NS5B. The BMV replicase product is similar in length to the monomer-sized products of NS5B (Fig. 2B, lanes 12 and 13). However, while the product of the BMV replicase is relatively discrete, those produced by NS5B migrated as a series of bands differing by a few nucleotides, as well as an array of products of larger sizes. This ladder of products may be due to initiation from either of the two 3′ cytidylates and/or the addition of one to three nontemplate nucleotides to the nascent RNA. BVDV NS5B can initiate from either the 3′-terminal or the penultimate cytidylate (19). The expectation of nontemplate nucleotide addition is reasonable, since it is a common property of all well-characterized polymerases, including those of vaccinia virus, poliovirus, BMV, and BVDV (7, 28, 34, 48).
The templates that directed significant amounts of RNA synthesis contained at least one predicted stem of various lengths and a 3′ single-stranded region that might serve as the initiation cytidylate (Fig. 2A). Also, the templates that directed RNA synthesis, B2(−)26G, SLC+8, C2(−)29G, and SLdel+8, had 3′ single-stranded sequences of 3 to 21 nts, suggesting that a 3′ single-stranded tail is necessary for efficient RNA synthesis (Fig. 2B, lanes 7 to 10). RNAs BV-21, BV-22, and −20/13, predicted by the mfold program (16) to lack stable secondary structures, were unable to direct RNA synthesis, as was SLC1, known by NMR analysis to form a stem-loop structure with a side bulge (20; Kao, unpublished).
A short template competent for RNA synthesis by NS5B.
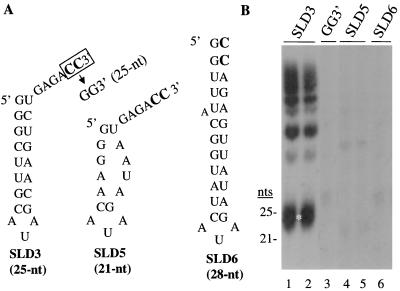
To identify short RNAs that can efficiently direct RNA synthesis by HCV NS5B, we made increasingly truncated versions of SLdel+8. A 25-nt RNA, termed SLD3, which lacks much of the single-stranded sequence in SLdel+8, directed RNA synthesis at 110% (±11%) relative to SLdel+8, after adjustment for radiolabeled CMP incorporation. Like other functional templates, SLD3 directed the synthesis of several bands (Fig. 3, lanes 1 and 2), with three monomeric products of 24 to 26 nts. The 24-nt RNAs were likely due to initiation from the penultimate cytidylate, with termination occurring at the end of the template. Some of the 25-nt products might have been due to initiation from the 3′-most cytidylate and precise termination at the end of the template, while others might have initiated at the penultimate cytidylate and contained one nontemplate nucleotide. The 26-nt RNA possessed at least one nontemplate nucleotide. The putative monomeric and multimeric products all initiated from the two 3′ cytidylates because RNA GG3′, in which the two cytidylates at the 3′ end of SLD3 are replaced with guanylates, was unable to direct synthesis (Fig. 3, lane 3). Deletion of the two cytidylates at the 3′ terminus also abolished RNA synthesis (Kao, unpublished). Removing 2 bp from the stem resulted in less than 20% of the synthesis seen with SLD3 (Kao, unpublished). Nucleotide substitutions that should result in more severe stem disruptions resulted in lower levels of synthesis (Fig. 3, lanes 4 and 5). Finally, RNA SLD6, possessing a stem and an AUA triloop but lacking a 3′ single-stranded tail, resulted in no synthesis (Fig. 3, lane 6). These findings confirm that the stem-loop and the single-stranded sequence in SLD3 are needed for efficient RNA synthesis.
FIG. 3.
SLD3 (25 nts) can direct RNA synthesis. (A) Sequences and putative structures of three RNAs tested to determine the minimal sequence necessary for efficient RNA synthesis. The potential initiation sites are in bold. An RNA named GG3′ (sequence not shown) contains a replacement of the 3′-terminal two cytidylates of SLD3 with two guanylates. (B) Autoradiogram of a 15% denaturing gel containing the products from the four RNAs. An RNA ladder with bands that are multiples of 12 nts was used to estimate the size of the 25-nt product indicated to the left of the autoradiogram. The white asterisk shows the position of monomeric RdRp product.
De novo initiation of RNA synthesis from SLD3.
The sizes of the products directed by SLD3 and the absence of products from RNA GG3′ suggest that initiation of RNA synthesis from SLD3 takes place by a de novo mechanism. In an attempt to confirm this possibility, we tested several RNAs whose 3′ ends cannot participate in phosphoryl transfer. BVDV NS5B has been shown to direct RNA synthesis from a template containing a 3′ ribose dideoxy analog (19). When the ribose at the 3′-most cytidylate of SLD3 was substituted with a C2′,3′-dideoxy or a 3′-amino moiety, the amount of product observed was decreased at least 50-fold in comparison to synthesis from SLD3 (Fig. 4A, lane 4) (Kao, unpublished). These results suggest that the ribose 3′-OH of the 3′-terminal cytidylate may be required to interact with NS5B during RNA synthesis, perhaps through the formation of an H bond. Next, we tested SLD3 containing a 3′-terminal puromycin in RNA, SLD3-Pmn. The nucleotide analog puromycin lacks 3′-OH and cannot participate in phosphoryl transfer. Instead, puromycin contains a 3′ amide bond that could potentially participate in H-bond formation. SLD3-Pmn directed efficient synthesis of the monomeric products, with the caveat that the products were decreased in length by 1 nt, likely due to the preferential use of the penultimate cytidylate in SLD3-Pmn rather than the 3′-terminal cytidylate (Fig. 4A, lane 3). Interestingly, SLD3-Pmn showed substantially decreased ability to direct the synthesis of dimeric and trimeric RNA products in comparison to SLD3 (compare Fig. 4A, lanes 2 and 3).
FIG. 4.
De novo initiation of RNA synthesis from SLD3. (A) Autoradiogram of a 20% denaturing gel containing NS5B products directed by variants of SLD3. RNA GG3′ has a replacement of the initiation cytidylates with guanylates. WT contains normal SLD3. C3′-Pmn contains a puromycin at the 3′ end of SLD3. C3′-NH2 is modified with an amino moiety. Lane 5 was the result of synthesis by BVDV NS5B from a 12-nt template that generated products of the lengths (in nucleotides) indicated to the right of the autoradiogram. (B) RNA synthesis from SLD3 requires high levels of GTP or a GTP analog. MantGTP {2′- (or 3′)-o[N-methyl(antraniloyl)GTP]} was from Molecular Probes (Eugene, Oreg.). The autoradiogram contains a 15% denaturing gel. All of the reactions were performed with limiting GTP (2 μM) that was not amended (reactions labeled with Φ) or that was amended with 100 mM GTP or GTP analogs shown above the autoradiogram.
A confirmation of de novo initiation was undertaken next. Initiation of viral RNA synthesis in vitro involves the RdRp, the initiation GTP (for HCV NS5B), a second nucleoside triphosphate (the i+1 nucleotide), and the template RNA. The apparent Km of the initiation nucleotide for RNA synthesis is severalfold higher than that of the i+1 nucleotide (18, 26). For the BMV replicase, the initiation GTP can be replaced with guanosine nucleotide analogs that can initiate synthesis but cannot function during elongating synthesis (18). To determine whether this is the case for HCV NS5B, reactions were performed with GTP limited to 2 μM, a concentration that severely reduces RNA synthesis, presumably because it is too low for initiation (Fig. 4B, lanes 1 and 2). The addition of GTP to 100 μM in these reactions restored RNA synthesis (Fig. 4B, lane 8). Various analogs, such as GMP, GDP, the dinucleotide GpG, and MantGTP, were found to partially restore RNA synthesis, while ADP did not (Fig. 4B, lanes 3 to 7). These results, along with the sizes of the monomeric products from SLD3 and the ability of SLD3-Pmn to initiate RNA synthesis, indicate that HCV NS5B initiated RNA synthesis by a de novo mechanism.
NMR analysis of SLD3 secondary structure.
The most stable secondary structure predicted for SLD3 by the mfold program (16) is shown in Fig. 5A. By virtue of its small size (25 nt), SLD3 is amenable to studies with proton NMR analysis. We sought to confirm or refute the predicted structure of SLD3 using NMR spectroscopy.
FIG. 5.
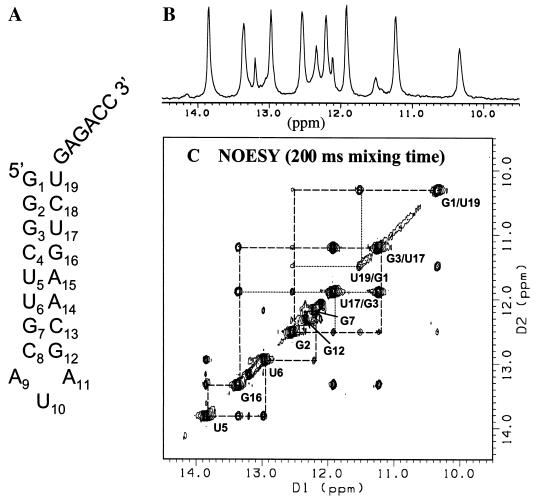
NMR analysis of the secondary structure of SLD3. (A) Secondary structure of SLD3 predicted by the mfold computer program. (B) Results of a one-dimensional analysis of SLD3 showing the spectrum of the imino protons between 14.5 and 9.5 ppm. (C) Results of two-dimensional NOESY performed with a 200-ms mixing time. The cross peaks that emanate from the signals present on the diagonal allow assignment of the connectivity of the imino protons participating in base pairing. The nucleotide assignments are superimposed on the spectrum. Axes labeled D1 and D2 correspond to proton dimensions 1 and 2, respectively.
SLD3 is predicted to have an 8-bp stem that contains 11 imino protons in one-dimensional NMR analysis, including the uridylate in the trinucleotide loop (Fig. 5A). The imino protons that are involved in base pairing or the formation of a stable conformation are usually protected from fast proton exchange with the water solvent, resulting in sharp imino proton peaks. We observed 12 sharp NMR peaks, indicating that the major conformation is quite stable.
Two-dimensional NMR spectra of an RNA molecule can be used to determine the secondary structure of the molecule and to help assign the protected imino protons that are located within a 5-Å space (43). Where 2 nts interact, cross peaks between imino peaks can be observed by NOESY (43). Furthermore, the NOESY cross-peak pattern allows us to distinguish between GC and AU base pairs using the characteristic cross peak between the uracil imino and the H2 amino proton of the AU pair versus the guanine imino and the cytosine amino protons of the GC pair. The expected GU wobble base pairs can be discerned by their characteristic strong imino cross peaks. The sequential connectivity of imino protons was made through the stem region (Fig. 5C). The key to this connectivity is the cross peaks between the G2 imino proton and imino protons of the adjacent two GU wobble base pairs. As expected from the predicted secondary structure, the G2 imino proton shows four cross peaks. Thus, the NMR results are in good agreement with the computer-predicted base-pairing pattern of SLD3.
Of all observable imino protons (Fig. 5B), only two (at 12.1 and 13.2 ppm) were left unassigned. It is known from the analysis of a similar triloop that the U10 in the triloop is partially stacked with the nucleotides in the stem and that additional interactions could account for unassigned peaks (20). At the present time, there is no NMR evidence that the 6 nts at the 3′ end of SLD3 are hydrogen bonded. Since no complementary sequence exists for this sequence, it seems likely that it is single stranded.
Features within SLD3 required for efficient RNA synthesis.
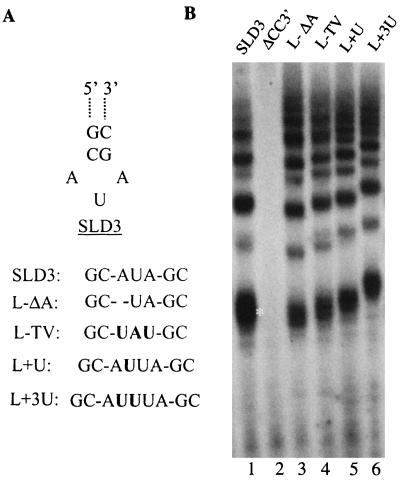
Next, we systematically examined the loop, stem, and single-stranded portions of SLD3 to see how they contribute to efficient RNA synthesis. The 3-nt loop of SLD3 was examined first with a series of changes (Fig. 6A). When an adenylate was removed, to result in RNA L-ΔA, synthesis was reduced to 50% that obtained with SLD3 (Fig. 6A and 6B, lane 3). Similar effects were observed when the loop nucleotides were changed to their Watson-Crick transversions (Fig. 6B, lane 4). Also, adding either 1 nt or 3 nts to the loop did not significantly decrease the amount of RNA synthesis, although the product sizes increased correspondingly, as expected (Fig. 6B, lanes 5 and 6). Thus, the sequence and length of the loop seem not to be major factors either in recognition by NS5B or in directing RNA synthesis.
FIG. 6.
Requirements in the loop of SLD3 for RNA synthesis. (A) Three-nucleotide loop and two closing base pairs of SLD3. Other parts of SLD3 are not shown. Nucleotides removed from the prototype SLD3 are indicated by dashes. Nucleotide additions and substitutions are indicated in bold. (B) Autoradiogram of a 15% denaturing gel containing NS5B products directed by the RNAs listed in panel A. ΔCC3′ is an RNA lacking the 3′-terminal two cytidylates of SLD3. The white asterisk shows the position of the monomeric (25-nt) RdRp product.
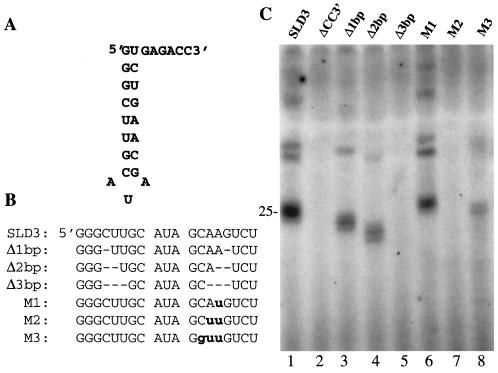
The SLD3 stem of 8 bp was subjected to a series of deletions and nucleotide substitutions. The removal of 1, 2, and 3 bp had an increasingly severe effect on RNA synthesis (Fig. 7B and 7C, lanes 3 to 5). To confirm and extend this observation, 1-, 2-, or 3-nt substitutions that should increasingly destabilize the stem were made and tested. The presence of a UU bp in the middle of the stem caused a slight reduction in RNA synthesis, to 60% (Fig. 7C, lane 6). However, two and three consecutive UU base pairs resulted in low levels of synthesis (Fig. 7C, lane 7 and 8). The last RNA may form an alternative structure that is recognized inefficiently by NS5B.
FIG. 7.
Requirements in the stem of SLD3 for RNA synthesis. (A) Sequence of the stem in SLD3. (B) Names of and changes in the sequences for SLD3. Deletions are indicated by a dash. Nucleotide substitutions are indicated by bold lowercase letters. The substitutions are Watson-Crick transversions that should disrupt or alter the stability of the stem in SLD3. (C) Autoradiogram of the products from SLD3 and the various stem mutants shown in panel B. ΔCC3′ denotes a reaction performed with an RNA identical to SLD3 except that the 3′-terminal two initiation cytidylates have been deleted.
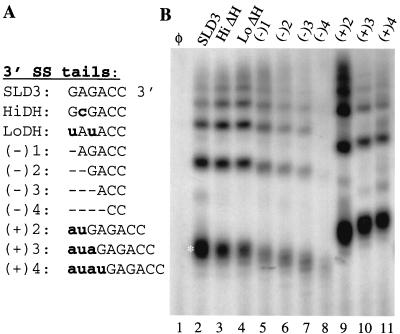
Several versions of the 3′ single-stranded tail of SLD3 were analyzed for their effects on RNA synthesis. The 6-nt tail of the prototypical SLD3 has the sequence 5′-GAGACC-3′. In the members of the alphalike virus superfamily, the sequence near the initiation site may regulate the level of RNA accumulation (38). Therefore, a change of the sequence to 5′-GCGACC-3′ was tested and found to result in 45% the synthesis seen with SLD3, after normalizing for 32P-CMP incorporation (Fig. 8B, lane 3). A change of the 3′ 6-nt sequence to 5′-UAUACC-3′ resulted in 70% the synthesis seen with SLD3 (Fig. 8B, lane 4). Therefore, the sequence of the single-stranded tail has only minor effects on RNA synthesis. Deletions of 1 nt to 4 nts in the single-stranded tail were found to increasingly reduce both the amount and the lengths of the RNA products (Fig. 8B, lanes 5 to 8). In contrast, an increase in the 3′ tail to 8 nts resulted in a 1.6-fold increase in synthesis, while further increases to 9 and 10 nts reduced synthesis to 91 and 70% that seen with SLD3, respectively (Fig. 8B, lanes 9 to 11). An 8-nt non-base-paired 3′ sequence is optimal.
FIG. 8.
Requirements in the 3′ single-stranded (SS) tail of SLD3 for RNA synthesis. (A) Sequences of the 3′ tail in SLD3 and in the various mutants. The sequences of the stem 5′ of the tail are not changed from those in SLD3 and are not shown. Deletions are denoted by dashes, and nucleotide substitutions and insertions are denoted by bold lowercase letters. (B) Autoradiogram of the products synthesized from SLD3 and the various mutants shown in panel A. Φ denotes a reaction performed in the absence of an exogenous template. The white asterisk shows the position of the monomeric (25-nt) RdRp product.
SLD3 chimeric for deoxy- and ribonucleotides.
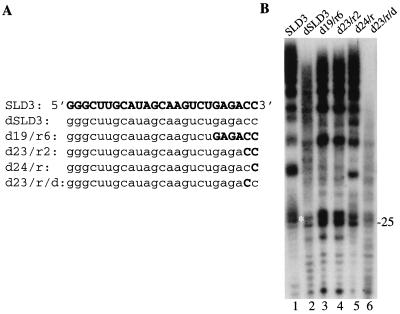
The inability of SLD3 containing a 3′ dideoxyribose to direct RNA synthesis indicates that HCV NS5B may interact with the 3′-terminal nucleotide through the ribose 2′-hydroxyl and/or 3′-hydroxyl. Several RdRps can direct synthesis from single-stranded DNAs, often at a reduced level (33, 35). To determine whether the riboses affect RNA synthesis, a version of SLD3 containing all deoxyribonucleotides, named dSLD3, was made and tested (Fig. 9). In comparison to SLD3, dSLD3 directed the synthesis of approximately 35% of the products. In addition, a higher abundance of products shorter than monomeric lengths was observed (Fig. 9B, compare lanes 1 and 2).
FIG. 9.
Ribonucleotides in SLD3 that contribute to efficient RNA synthesis. (A) RNA nucleotides are shown in bold uppercase letters, while DNA nucleotides are shown in lowercase letters. (B) Autoradiogram of the products synthesized by NS5B with the various RNA, DNA, and chimeric templates shown in panel A. The 25-nt product is indicated on the right. An RNA ladder with bands that are multiples of 12 nts (not shown) was used to determine the lengths of the products. Axes labeled D1 and D2 correspond to proton dimensions 1 and 2, respectively.
To elucidate the ribonucleotide(s) in SLD3 that may contribute to more efficient RNA synthesis, chimeras with ribo- and deoxyribonucleotides were made. We reasoned that more stringent requirements would be found for the sequence which participates in initiation than for that which participates in elongation. Therefore, the chimeras were designed to contain deoxyribonucleotides in their 5′ portion and ribonucleotides in their 3′ portion, near the initiation cytidylates. These RNAs are named 5′ to 3′ according to the number of deoxy- and ribonucleotides. RNA d19/r6 was found to direct RNA synthesis by NS5B at least as well as SLD3 (Fig. 9B, lane 3). A chimera that contained the two 3′ cytidylates composed of ribonucleotides gave high levels of RNA synthesis, indicating that the two riboses on the initiation cytidylates are primarily responsible for the different levels of synthesis observed from a ribo- versus a deoxyribose version of SLD3 (Fig. 9B, lane 4). To distinguish whether one or both of the two 3′ cytidylates are primarily responsible for efficient synthesis, d24/r and d23/r/d, containing, respectively, a 3′-terminal ribocytidylate and a penultimate ribocytidylate, were tested. Chimera d24/r gave levels of synthesis significantly higher than d23/r/d (Fig. 9B, lanes 5 and 6). The results indicate that the 2′-hydroxyl moiety at the 3′-most nucleotide of SLD3 contributes significantly to efficient RNA synthesis by NS5B. This result is consistent with our previous observation that an RNA with a 3′ dideoxyribose is a poor template.
DISCUSSION
Initiation of RNA synthesis by HCV NS5B is just beginning to be elucidated. The majority of studies to date have focused on polymerization from either oligonucleotide primers or templates that loop back on themselves (5, 23–25). De novo initiation is less thoroughly characterized, in part because a variety of templates, including the 3′-terminal 25 or 60 nts of the HCV RNA, were unable to direct RNA synthesis in vitro. In this work, we have determined that HCV NS5B can efficiently initiate RNA synthesis by a de novo mechanism. The RNA that efficiently directed de novo initiation possessed a stable stem-loop and a 6- to 8-nt single-stranded sequence containing a 3′ initiation nucleotide.
De novo initiation.
A de novo mechanism of initiation of RNA synthesis is appealing for HCV-infected cells, since it obviates the need for an additional enzyme either to generate the primer or to specifically cleave the linkage between the template and the newly synthesized RNA. We have several lines of evidence that SLD3 and other functional RNAs initiate synthesis by a de novo mechanism. First, a template blocked at the 3′ terminus with puromycin can nonetheless initiate RNA synthesis. This finding is consistent with the results of Luo et al. (26), who reported that an RNA with 3′-deoxyadenosine (cordycepin, which possesses a 2′-hydroxyl) retained the ability to direct RNA synthesis. Second, initiation requires the terminal cytidylates; substitutions with guanylates or deletion of the cytidylates abolishes RNA synthesis. Third, initiation can take place with several GTP analogs that cannot be hydrolyzed during phosphoryl transfer; hence, they must serve as the priming nucleotide by providing a 3′-hydroxyl for the formation of the first phosphodiester bond. Fourth, the higher concentration of GTP needed for initiation than for elongation is a feature consistent with de novo initiation. De novo initiation of RNA synthesis in vitro has now been demonstrated by a number of groups studying the HCV (26, 29, 42, 47) and BVDV (19) RdRps.
The initiation template nucleotide.
We have found that the initiation cytidylates provide several features that contribute to the efficiency of RNA synthesis. Changes to uridylates, guanylates, deoxycytidylates, or 2′-3′-deoxycytidylates all reduced RNA synthesis. These results indicate that the cytidine base may be specifically required to base pair with a GTP, while the ribose 2′-OH and 3′-OH may interact with NS5B. Our observations of the requirements of the initiation site are different from some previous observations. Zhong et al. (47) observed that initiation can take place from a purine nucleotide in the template, while others, including us, have observed that HCV NS5B has a specificity for initiation pyrimidines (26, 29, 42). In contrast to our observations, Zhong et al. (47) and Kao et al. (19) demonstrated that short templates containing a 2′-3′-dideoxynucleotide could direct de novo initiation by HCV NS5B and BVDV NS5B, respectively. The amount of de novo initiation products obtained from the dideoxy template by Zhong et al. (47) seems to be quite low in comparison to those of other RdRp products in their reactions, suggesting that synthesis was inefficient without a 3′-hydroxyl moiety. With regard to the difference between the results reported here and those obtained with BVDV RdRp (19), we believe that the two polymerases have different initiation requirements. Kim et al. (21) have confirmed the results of Kao et al. (19) that the template 3′-hydroxyl is not required for de novo initiation by BVDV RdRp. Another possible cause of the observed difference is that the recombinant NS5Bs used are slightly different, despite being from the same HCV strain (1b). Our results were generated with full-length NS5B, while others, including those of Kao et al., were obtained with proteins that lacked approximately 20 residues of the C-terminal tail (19, 26). The C-terminal tail of NS5B is present in the active site of the crystal structure and has been hypothesized to play a role in template discrimination (2). More analysis is needed to determine if the C termini of the BVDV NS5B and HCV NS5B play any role in template discrimination.
Template requirements.
An RNA composed of a stem-loop and a single-stranded sequence is commonly used to initiate viral RNA synthesis. Oh et al. (29), have independently found that a stem-loop and a non-base-paired region within the 98 nts present at the 3′ end of HCV RNA can direct RNA synthesis by NS5B. However, several of the sequences that directed RNA synthesis in our study are unrelated to HCV, demonstrating that HCV NS5B does not have exclusive recognition of sequences within the HCV 3′ region. Motifs similar to those within SLD3 are used to initiate RNA synthesis by the replicases of several plant-infecting RNA viruses, including the turnip yellow mosaic virus (9, 36), turnip crinkle virus (40, 41), and BMV (20). Perhaps a similar mode of recognition is to be expected, since RNA polymerases share similar structures and functions (6, 11, 13). Work with enriched multisubunit plant viral replicases, however, cannot yet distinguish which replicase subunit(s) contacts the template RNA, other than the initiation site, which must be recognized by RdRp proper. The observation that recombinant HCV NS5B has specific requirements for some structured RNA suggests that HCV RdRp could have additional roles in template discrimination.
Finally, the length of the 25-nt SLD3 and its ability to direct de novo initiation will be amenable to high-resolution analysis of the initiation of HCV RNA synthesis in vitro and to further probing of the NS5B-RNA interaction.
ACKNOWLEDGMENTS
We thank K. Kirkegaard, J. Colacino, K. Sivakumaran, R. Tayon, X.-L. Sun, and L. Kao for helpful discussions of this work and for critiques of the manuscript.
C.C.K. acknowledges a Linda and Jack Gill fellowship.
REFERENCES
- 1.Adkins S, Stawicki S S, Faurote G, Siegel R W, Kao C. Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA. 1998;4:455–470. [PMC free article] [PubMed] [Google Scholar]
- 2.Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct Fold Descr. 1999;7:1417–1426. doi: 10.1016/s0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 3.Arnold J J, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J Biol Chem. 1999;274:2706–2716. doi: 10.1074/jbc.274.5.2706. [DOI] [PubMed] [Google Scholar]
- 4.Beckman M T L, Kirkegaard K. Site size of cooperative single-stranded RNA binding by poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1998;273:6724–6730. doi: 10.1074/jbc.273.12.6724. [DOI] [PubMed] [Google Scholar]
- 5.Behrens S E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale R L, Mathieu M, De Francesco R. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown M, Dorson J W, Bollum F J. Terminal riboadenylate transferase: a poly(A) polymerase in purified vaccinia virus. J Virol. 1973;12:203–208. doi: 10.1128/jvi.12.2.203-208.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck K W. Comparison of the replication of positive-strand RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deiman B A L M, Konen A K, Verlaan P W G, Pleij C W A. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J Virol. 1998;72:3965–3972. doi: 10.1128/jvi.72.5.3965-3972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari E, Wright-Minogue J, Fang J W S, Baroudy B M, Lau J Y N, Hong Z. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J Virol. 1999;73:1649–1654. doi: 10.1128/jvi.73.2.1649-1654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu J-H, Gnatt A, Bushnell D A, Jensen G J, Thompson N E, Burgess R R, David P R, Kornberg R D. Yeast RNA polymerase II at 5 A resolution. Cell. 1999;98:799–810. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- 12.Grodberg J, Dunn J J. Purification of T7 RNA polymerase. J Bacteriol. 1988;170:1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen J L, Long A M, Schultz S C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 14.Hoofnagle J H. Hepatitis C: the clinical spectrum of disease. Hepatology. 1997;26:155–205. doi: 10.1002/hep.510260703. [DOI] [PubMed] [Google Scholar]
- 15.Houghton M. Hepatitis C viruses. In: Fields B, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippencott-Raven Press; 1996. pp. 1035–1058. [Google Scholar]
- 16.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson R, Sun X L, Hockman M A, Villarreal E C, Wakulchik M, Wang Q M. Specificity and mechanism analysis of hepatitis C virus RNA-dependent RNA polymerase. Arch Biochem Biophys. 2000;377:129–134. doi: 10.1006/abbi.2000.1749. [DOI] [PubMed] [Google Scholar]
- 18.Kao C C, Sun J H. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J Virol. 1996;70:6826–6830. doi: 10.1128/jvi.70.10.6826-6830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao C C, DelVecchio A M, Zhong W. De novo initiation of RNA synthesis by a recombinant Flaviviridae RNA-dependent RNA polymerase. Virology. 1999;253:1–7. doi: 10.1006/viro.1998.9517. [DOI] [PubMed] [Google Scholar]
- 20.Kim C H, Kao C, Tinoco I. RNA motifs that determine specificity between a viral RNA replicase and its promoter. Nat Struct Biol. 2000;7:415–423. doi: 10.1038/75202. [DOI] [PubMed] [Google Scholar]
- 21.Kim M-J, Zhong W, Hong Z, Kao C C. Template nucleotide moieties required for de novo initiation of RNA synthesis by a recombinant viral RNA-dependent RNA polymerase. J Virol. 2000;74:10312–10322. doi: 10.1128/jvi.74.22.10312-10322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lippens C, Dhalluin C, Wieruszeski J-M. Use of a water flip-back pulse in the homonuclear NOESY experiment. J Biomol NMR. 1995;5:327–331. doi: 10.1007/BF00211762. [DOI] [PubMed] [Google Scholar]
- 23.Lohmann V, Körner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohmann V, Roos A, Körner F, Koch J O, Bartenschlager R. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology. 1998;249:108–118. doi: 10.1006/viro.1998.9311. [DOI] [PubMed] [Google Scholar]
- 25.Lohmann V, Overton H, Bartenschlager R. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by NS5B. J Biol Chem. 1999;274:10807–10815. doi: 10.1074/jbc.274.16.10807. [DOI] [PubMed] [Google Scholar]
- 26.Luo G, Hamatake R K, Mathis D M, Racela J, Rigat K L, Lemm J, Colonno R J. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol. 2000;74:851–863. doi: 10.1128/jvi.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neufeld K L, Galarza J M, Richards O C, Summers D F, Ehrenfeld E. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol. J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh J-W, Sheu G-T, Lai M M C. Template requirements and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J Biol Chem. 2000;275:17710–17717. doi: 10.1074/jbc.M908781199. [DOI] [PubMed] [Google Scholar]
- 30.Oh J-W, Ito T, Lai M M C. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J Virol. 1999;73:7694–7702. doi: 10.1128/jvi.73.9.7694-7702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddy P S, Chatterji D. Evidence for a pyrimidine nucleotide-specific initiation site (the i site) on Escherichia coli RNA polymerase. Eur J Biochem. 1994;225:737–745. doi: 10.1111/j.1432-1033.1994.00737.x. [DOI] [PubMed] [Google Scholar]
- 32.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippencott-Raven Press; 1996. pp. 931–959. [Google Scholar]
- 33.Schiebel W, Haas B, Marinkovic S, Klanner A, Sanger H L. RNA-dependent RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J Biol Chem. 1993;263:11858–11867. [PubMed] [Google Scholar]
- 34.Siegel R W, Adkins S, Kao C. Sequence-specific recognition of a subgenomic promoter by a viral RNA polymerase. Proc Natl Acad Sci USA. 1997;94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegel R W, Bellon L, Beigelman L, Kao C C. Analysis of template requirements revealed that viral RNA-dependent RNA polymerase could bridge the transition from the RNA to the DNA world. J Virol. 1999;73:6424–6429. doi: 10.1128/jvi.73.8.6424-6429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh R N, Dreher T W. Specific site selection in RNA resulting from a combination of nonspecific secondary structure and CCR-boxes: initiation of minus-strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA. 1998;4:1083–1095. doi: 10.1017/s1355838298980694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sivakumaran, K., Y. Bao, M. Roossinck, and C. Kao. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicases of Brome mosaic virus and Cucumber mosaic virus. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 38.Sivakumaran K, Kim C H, Tayon R, Kao C. RNA sequence and structural determinants for the recognition and efficiency of RNA synthesis by an RNA replicase. J Mol Biol. 1999;294:667–682. doi: 10.1006/jmbi.1999.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sklenar V. Suppression of radiation damping in multidimensional NMR experiments using magnetic field gradients. J Magn Reson Ser A. 1993;102:241–245. [Google Scholar]
- 40.Song C, Simon A E. Requirement of a 3′ stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J Mol Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- 41.Stupina V, Simon A E. Analysis in vivo of turnip crinkle satellite RNA C variants with mutations in the 3′ terminal minus-strand promoter. Virology. 1997;238:470–477. doi: 10.1006/viro.1997.8850. [DOI] [PubMed] [Google Scholar]
- 42.Sun X L, Johnson R B, Hockman M A, Wang Q M. De novo initiation catalyzed by HCV RNA-dependent RNA polymerase. Biochem Biophys Res Commun. 2000;268:798–803. doi: 10.1006/bbrc.2000.2120. [DOI] [PubMed] [Google Scholar]
- 43.Varani G, Tinoco I., Jr Carbon assignments and heteronuclear coupling constants for an RNA oligonucleotide from natural abundance 13C-1H correlated experiments. J Am Chem Soc. 1991;113:9349–9354. [Google Scholar]
- 44.World Health Organization. Hepatitis C. Wkly Epidemiol Rec. 1997;72:65–69. [Google Scholar]
- 45.Yamashita T, Kaneko S, Shirota Y, Qin W, Nomura T, Kobayashi K, Murakami S. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J Biol Chem. 1998;273:15479–15486. doi: 10.1074/jbc.273.25.15479. [DOI] [PubMed] [Google Scholar]
- 46.Yuan Z H, Kumar U, Thomas H C, Wen Y W, Monjardino J. Expression, purification, and partial characterization of the HCV RNA polymerase. Biochem Biophys Res Commun. 1997;232:231–235. doi: 10.1006/bbrc.1997.6249. [DOI] [PubMed] [Google Scholar]
- 47.Zhong W, Uss A, Ferrari E, Lau J Y N, Hong Z. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol. 2000;74:2017–2022. doi: 10.1128/jvi.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]